Abstract
Background
Arginase is an enzyme that converts L-arginine to urea and L-ornithine, an essential substrate for the polyamine pathway supporting Leishmania (Leishmania) amazonensis replication and its survival in the mammalian host. L-arginine is also the substrate of macrophage nitric oxide synthase 2 (NOS2) to produce nitric oxide (NO) that kills the parasite. This competition can define the fate of Leishmania infection.
Methodology/Principal findings
The transcriptomic profiling identified a family of oxidoreductases in L. (L.) amazonensis wild-type (La-WT) and L. (L.) amazonensis arginase knockout (La-arg-) promastigotes and axenic amastigotes. We highlighted the identification of an oxidoreductase that could act as nitric oxide synthase-like (NOS-like), due to the following evidences: conserved domain composition, the participation of NO production during the time course of promastigotes growth and during the axenic amastigotes differentiation, regulation dependence on arginase activity, as well as reduction of NO amount through the NOS activity inhibition. NO quantification was measured by DAF-FM labeling analysis in a flow cytometry.
Conclusions/Significance
We described an arginase-dependent NOS-like activity in L. (L.) amazonensis and its role in the parasite growth. The increased detection of NO production in the mid-stationary and late-stationary growth phases of La-WT promastigotes could suggest that this production is an important factor to metacyclogenesis triggering. On the other hand, La-arg- showed an earlier increase in NO production compared to La-WT, suggesting that NO production can be arginase-dependent. Interestingly, La-WT and La-arg- axenic amastigotes produced higher levels of NO than those observed in promastigotes. As a conclusion, our work suggested that NOS-like is expressed in Leishmania in the stationary growth phase promastigotes and amastigotes, and could be correlated to metacyclogenesis and amastigotes growth in a dependent way to the internal pool of L-arginine and arginase activity.
Introduction
Leishmaniasis is an important neglected tropical disease caused by the protozoa from Leishmania genus [1,2]. The infection caused by L. (L.) amazonensis can generate cutaneous and/or diffuse cutaneous manifestations [1,2]. This parasite has a dimorphic life cycle: the procyclic promastigote form can be found in the mid-gut of the phlebotomine sand fly; the metacyclic promastigote can be found in the insect foregut and stomodeal valve and corresponds to the infective form; and the amastigote form can be found into macrophages of the mammalian host [3,4]. Leishmania has a complex metabolic arsenal to enable its survival, replication and differentiation in different environments of both invertebrate and vertebrate hosts [3–5].
In L. (L.) amazonensis (La-WT), the L-arginine uptake is mediated by the amino acid permease 3 (AAP3) [6], then metabolized by arginase to produce urea and ornithine, being this last a substrate of polyamine pathway [7,8]. The availability of L-arginine and arginase activity is essential for the survival of Leishmania inside both invertebrate and mammalian hosts [9–12]. Additionally, L. (L.) amazonensis harbors NOS-like activity, producing NO [13–17]. L-arginine availability can interfere in the half-life of AAP3 transcripts, increasing the stability of the transporter and L-arginine uptake in L. (L.) amazonensis promastigotes [6]. The absence of arginase activity in L. (L.) amazonensis arginase knockout (La-arg-) leaded to an increase in L-arginine and citrulline levels and decrease in proline, ornithine and putrescine levels [18]. The availability of L-arginine and arginase influence the survival of Leishmania in axenic conditions and inside its mammalian host [9–12,19–22].
NOS is an oxidoreductase enzyme that catalyzes the conversion of L-arginine and molecular oxygen into L-citrulline and NO, using NADPH, heme, FAD, FMN and tetrahydro-L-biopterin (BH4) as cofactors, producing Nω-hydroxy-L-arginine (OH-arginine or NOHA), an intermediate product that is oxidized into citrulline with the release of NO [23–25]. In mammalian cells, the amplitude and concentration of NO are dependent on the NOS isoform activity and its cellular/tissue distribution [24–26]. Many NO functions have been described as directly dependent on its concentration in different tissues [26,27]. In mammals, NO regulates synapses (nNOS or NOS1), promotes the killing of pathogens (iNOS or NOS2) and modulates vascular smooth muscle contraction (eNOS or NOS3) [26], among other functions. In addition, other organisms such as plants can produce NO during germination and flowering [28].
NO production has also been described in Trypanosomatidae parasites. Paveto’s group (1995) purified a calcium-dependent NOS from Trypanosoma cruzi and demonstrated an L-arginine conversion to NO and L-citrulline [29], similar to neuronal-NOS (nNOS or NOS1) from mammalian cells [24]. Piacenza (2001) showed a NOS-dependent NO production and its role in suppressing T. cruzi apoptosis [30]. Pereira (2015) showed NOS activity, citrulline and NO production in T. cruzi and reduction in NO production during contact with the extracellular matrix of host cells [31]. Leishmania species also presents an enzyme that produces citrulline and NO [6,13,14,16–18,20]. The first demonstration of Leishmania-NOS-like protein was described by Basu (1997) for L. (L.) donovani [32]. Later, a NOS-like activity was also described for L. (L.) amazonensis, L. (V.) braziliensis and L. (L.) infantum chagasi [13–15,33]. A transcriptomic profiling of L. (L.) mexicana showed the presence of a NOS-like (LmxM.19.1450.1, EC 1.14.13.39), an enzyme with similar levels in promastigotes, and in both macrophage-derived and axenic amastigotes [34].
Based on RNA-seq data, here we identified a family of putative oxidoreductases in La-WT and La-arg- differentially expressed when we compared La-WT vs. La-arg- promastigotes and La-WT promastigotes vs. axenic amastigotes, as well a NOS-like gene expression, validated by qPCR. These findings indicated that NO production could be related to the parasite growth phase and to arginase activity. The quantification of NO production in the promastigote growth curve and in axenic amastigote forms of La-WT, La-arg- and the arginase addback (La-arg-/+ARG) revealed increased detection in the mid- to late-phase of the parasite growth. The absence of arginase activity induced early NO production compared with La-WT. The inhibition of NOS activity reduced NO amount and the frequency of metacyclic forms. Axenic amastigotes produced around 10-fold increased amount of NO compared with La-WT promastigotes, but the absence of arginase activity reduced NO production in both forms. Altogether, these data indicated a possible biological role for NO in parasite differentiation signaling.
Methods
Leishmania culture
La-WT (MHOM/BR/1973/M2269) promastigotes were maintained in culture at 25°C by inoculating 5x105 cells/mL in 10 mL of M199 medium, supplemented with 10% fetal bovine serum (FBS; Gibco, South America), 5 ppm hemin, 100 μM adenine, 100 U penicillin, 100 μg/mL streptomycin, 40 mM HEPES-NaOH and 12 mM NaHCO3, at pH 7.0, for a week-long culture at low passages (P1- 5). La-arg- promastigotes were maintained in the same M199 medium supplemented, as described above, with addition of 30 μg/mL hygromycin B, 30 μg/mL puromycin (Sigma, St. Louis, MO, USA) and 50 μM putrescine (Sigma, St. Louis, MO, USA) [20]. La-arg-/+ARG was maintained in the same medium of La-arg- with addition of 20 μg/mL phleomycin [20].
The growth curve was performed starting from an initial inoculum of 5x105 cells/mL from stationary growth phase, and then the number of parasites was determined using a Coulter Z1 particle counter (Beckman, Fullerton, CA, USA), on days 3, 5, 7 and 9.
Axenic amastigote differentiation
Mid-logarithmic growth phase promastigotes of La-WT, La-arg- or La-arg-/+ARG (5x106, each) were transferred to 10 mL of M199 medium supplemented as described above at pH 5.5 and maintained at 34°C. After 4 days of differentiation, amastigotes were flushed 5 times using a 22G syringe needle, washed twice with PBS (950 x g, 10 min, 4°C) and counted in Neubauer chamber [35,36].
NOS/NO inhibition assays
For analysis of differentiation of procyclic to metacyclic promastigote forms, promastigotes of La-WT and La-arg- at day 3 or 5 of culture were treated with 10 mM of ω-nitro-L-arginine methyl ester (L-NAME, NOS inhibitor) and/or 100 μM of 2-(4-Carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO, NO scavenger) for two days in M199 medium, as described above, at 25°C. Next, the frequency of metacyclic forms was analyzed by flow cytometry.
For analysis of NO production during amastigotes differentiation, mid-logarithmic growth phase promastigotes of La-WT and La-arg- were differentiated to amastigotes as described above, with 10 mM of L-NAME and/or 100 μM of cPTIO at days 0 or day 2 of culture and at day 4 of culture NO production was analyzed.
Total RNA isolation and library construction
Total RNA from La-WT and La-arg- promastigotes and axenic amastigotes was isolated using TRIzol reagent (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA samples were treated with DNase I (Thermo Scientific, Lithuania, EU), and the RNA concentration was determined by spectrophotometry (Nanodrop ND1000, Thermo Scientific, USA). In addition, the RNA integrity was evaluated using an Agilent 2100 Bioanalyzer and Pico Agilent RNA 6000 kit (Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer’s instructions. Library preparations from three independent biological replicates were performed using Strand-specific TrueSeq RNA-seq Library Prep (Illumina), according to the manufacturer’s instructions.
RNA-seq and data analysis
Paired-end reads (125 bp) were obtained using the Illumina HiSeq 2000 platform at the Norwegian Sequencing Centre at the University of Oslo. Trimmomatic was used to remove the Illumina adapter sequences [37]. The quality of the produced data was analyzed using FastQC by the Phred quality score [38]. Reads with Phred quality scores lower than 20 were discarded. The reads were aligned to the L. mexicana (MHOMGT2001U1103) genomic data obtained from TriTrypDB (www.tritryp.org) using TopHat [39,40]. Thereafter, read mapping was performed for transcript assembly using Cufflinks [41]. After assembly, the abundance of transcripts was calculated as the Fragments Per Kilobase of transcript per Million mapped reads (FPKM), which reflects the abundance of a transcript in the sample by normalization of the RNA length and the total read number [42]. Differentially expressed gene analysis was performed on four comparison pairs (La-WT promastigotes vs. La-arg- promastigotes; La-WT axenic amastigotes vs. La-arg- axenic amastigotes, La-WT promastigotes vs. La-WT axenic amastigotes; La-arg- promastigotes vs. La-arg- axenic amastigotes) [43].
RT-qPCR
Reverse transcription was performed using 2 μg of total RNA as a template, reverse transcriptase and random primers (Revertaid H minus Reverse Transcriptase kit, Thermo-Scientific, Canada), according to the manufacturer’s instructions. Equal amounts of cDNA were assessed in duplicates in a total volume of 25 μL containing LuminoCt SYBR Green qPCR ReadyMix (SigmaAldrich, St Louis,MO, USA) and the following primers (200 nM): NOS-L_F 5´- acagcgactccaaaccttcc—3´, NOS-L_R 5´- gggacctgtttcagaggacg -3´, GAPDH_F 5´-tcaaggtcggtatcaacggc-3´, GAPDH_R 5´-tgcaccgtgtcgtacttcat-3´, amastin-like (LmxM.33.0960)_F, 5´-ggagcgctacttcagctatgga-3´, amastin-like_(LmxM.33.0960)_R, 5´-cggatcatcaataagacgatgttg-3´, amastin-like (LmxM.08.0800)_F 5´-agttcctcgcgtttctctttgt-3´, amastin-like (LmxM.08.0800)_R 5´ggcactgtgtactggcaaacc-3´, amastin-like (LmxM.08.0760)_F 5´-cggctgccttttgctgtact-3´ and amastin-like (LmxM.08.0760)_R 5´-cagacaacgcaagctgtgaca-3´. The mixture was incubated at 94°C for 5 min, followed by 40 cycles at 94°C for 30 s and 60°C for 30 s. Reactions were carried out using an PikoReal 96 RealTime PCR System (Thermo Scientfic, Finland). A negative control without reverse transcriptase was included in RT-qPCR assays to certify no contamination with DNA in RNA samples. A non-template control was included in all qPCR plates. The Fold Change of the target genes (nosL and amastins-like) and reference gene (gapdh) were quantified in three to six biological replicate samples, using a relative quantification.
Quantification of NO production
Parasites were washed twice with PBS (1479 x g, 10 min, 4°C) and were incubated with 1 μM DAF-FM (4-amino-5methylamino-2’,7’-difluorofluorescein diacetate) (Life Technologies, Eugene, OR, USA) in PBS for 30 min at 25°C or 34°C for promastigotes and axenic amastigotes, respectively. Next, the cells were washed with PBS and were labeled with 1 ng/mL propidium iodide (PI) to analyze cell viability. Fluorescence acquisition was performed using a FACSCalibur (BD, Franklin Lakes, NJ, USA), and the collected data were analyzed using FlowJo Software (LLC, Ashland, OR, USA). The frequency of DAF-FM-positive cells and the mean fluorescence intensity (MFI) of DAF-FM-labeled cells (FL1 detector) were analyzed in viable cells (PI-; FL2 detector) from 20,000 events, based in forward scatter (FSC) and side scatter (SSC) features.
Statistical analysis
Statistical analysis was determined based on Student’s t-test, one-way ANOVA or Tukey’s comparison post-test, using the GraphPad Prism 6 Software (GraphPad Software, Inc., La Jolla, CA, USA) and considering p < 0.05 as significant.
Results
Oxidoreductases transcriptomic profiling of La-WT and La-arg- promastigotes and axenic amastigotes
The transcriptomic profiling data of La-WT and La-arg- promastigotes and axenic amastigotes are available on the NCBI BioProject under the accession number PRJNA380128 and Sequence Read Archive (SRA) under accession numbers SRX2661998 and SRX2661999 [43]. Based on this RNA-seq data, we identified oxidoreductases transcripts expression in the samples La-WT and La-arg- promastigotes and axenic amastigotes (Tables 1–4).
Table 1. Oxidoreductase-like transcripts expression of La-WT and La-arg- promastigotes.
| ID | EC number |
pro La-WT vs. pro La-arg- Product description |
Fold change |
|---|---|---|---|
| LmxM.03.0570 | 1.6.5.5 | quinone oxidoreductase, putative | 0.23 |
| LmxM.05.0980 | 1.6.5.3 | NADH-ubiquinone oxidoreductase, putative | 1.05 |
| LmxM.07.0600 | 1.5.5.1 | electron transfer oxidoreductase, putative | 0.88 |
| LmxM.12.1130 | 1.3.1.42 | NADH:flavin oxidoreductase/NADH oxidase, putative | 0.93 |
| LmxM.13.0720 | 1.6.5.5 | oxidoreductase-like protein | 0.75 |
| LmxM.17.0270 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 0.88 |
| LmxM.18.1480 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 1.08 |
| LmxM.20.0120 | no data | NADH: ubiquinone oxidoreductase, putative | 0.69 |
| LmxM.21.0420 | 1.18.1.6 | NADH: adrenodoxin oxidoreductase, putative | 0.60 |
| LmxM.23.0670 | no data | oxidoreductase-like protein | 1.09 |
| LmxM.23.0860 | 1.6.5.5 | quinone oxidoreductase, putative | 0.98 |
| LmxM.23.1590 | no data | oxidoreductase-like protein | 0.74 |
| LmxM.23.1600 | no data | oxidoreductase-like protein | 0.86 |
| LmxM.28.0940 | no data | oxidoreductase-like protein | 0.51 |
| LmxM.31.2210 | no data | NADH: adrenodoxin oxidoreductase, putative | 0.80 |
| LmxM.32.1770 | no data | oxidoreductase-like protein | 0.79 |
| LmxM.33.4320 | no data | NADH: adrenodoxin oxidoreductase, putative | 0.56 |
| LmxM.36.3230 | 1.1.99.1 | oxidoreductase-like protein | 0.59 |
| LmxM.36.4170 | 1.6.5.5 | oxidoreductase-like protein | 0.63 |
| LmxM.36.5660 | no data | FAD dependent oxidoreductase | 0.87 |
Comparison of the oxidoreductases-orthologs transcripts between pro La-WT vs. pro La-arg-, adjusted for p < 0.05. pro: promastigote, La-WT: L. (L.) amazonensis wild-type, La-arg-: L. (L.) amazonensis arginase knockout.
Table 4. Oxidoreductase-like transcripts expression of La-arg- promastigotes and axenic amastigotes.
| ID | EC number |
pro La-arg- vs. ama La-arg- Product description |
Fold change |
|---|---|---|---|
| LmxM.03.0570 | 1.6.5.5 | quinone oxidoreductase, putative | 2.27 |
| LmxM.05.0980 | 1.6.5.3 | NADH-ubiquinone oxidoreductase, putative | 0.73 |
| LmxM.07.0600 | 1.5.5.1 | electron transfer oxidoreductase, putative | 1.07 |
| LmxM.12.1130 | 1.3.1.42 | NADH:flavin oxidoreductase/NADH oxidase, putative | 1.66 |
| LmxM.13.0720 | 1.6.5.5 | oxidoreductase-like protein | 1.16 |
| LmxM.17.0270 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 0.97 |
| LmxM.18.1480 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 0.96 |
| LmxM.20.0120 | No data | NADH: ubiquinone oxidoreductase, putative | 1.09 |
| LmxM.21.0420 | 1.18.1.6 | NADH: adrenodoxin oxidoreductase, putative | 1.47 |
| LmxM.23.0670 | No data | oxidoreductase-like protein | 0.79 |
| LmxM.23.0860 | 1.6.5.5 | quinone oxidoreductase, putative | 0.86 |
| LmxM.23.1590 | No data | oxidoreductase-like protein | 1.59 |
| LmxM.23.1600 | No data | oxidoreductase-like protein | 0.68 |
| LmxM.28.0940 | No data | oxidoreductase-like protein | 2.64 |
| LmxM.31.2210 | No data | NADH: adrenodoxin oxidoreductase, putative | 1.37 |
| LmxM.32.1770 | No data | oxidoreductase-like protein | 2.27 |
| LmxM.33.4320 | No data | NADH: adrenodoxin oxidoreductase, putative | 1.27 |
| LmxM.36.3230 | 1.1.99.1 | oxidoreductase-like protein | 1.62 |
| LmxM.36.4170 | 1.6.5.5 | oxidoreductase-like protein | 1.52 |
| LmxM.36.5660 | No data | FAD dependent oxidoreductase | 2.14 |
Comparison of the oxidoreductases-orthologs transcripts between pro La-arg- vs. ama La-arg-, adjusted for p < 0.05. pro: promastigotes, ama: axenic amastigotes, La-WT: L. (L.) amazonensis wild-type, La-arg-: L. (L.) amazonensis arginase knockout.
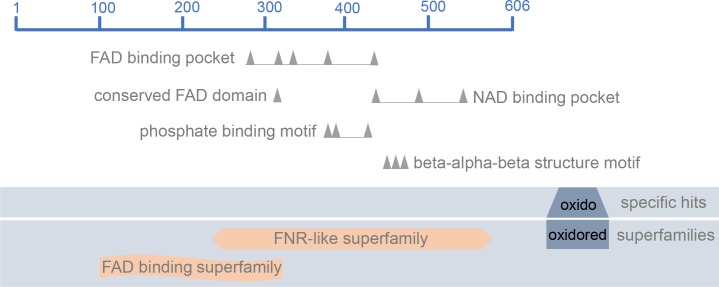
The oxidoreductase synthase-like (LmxM.19.1450/ EC 1.14.13.39) was identified and appeared up-regulated in La-arg- promastigotes when compared with La-WT promastigotes and in La-WT axenic amastigotes when compared with La-WT promastigotes, with fold-change 1.38 and 1.26, respectively. In silico search revealed the presence of the following oxidoreductase family domains: ferredoxin reductase (FNR)-like and flavin adenine dinucleotide (FAD) binding (Fig 1). In addition, previous data with metabolome fingerprints identified metabolic products such as NO and citrulline which could be produced by the enzyme (EC 1.14.13.39) [18].
Fig 1. NOS-like graphical summary of conserved domains.
Structure of the oxidoreductase synthase-like protein (LmxM.1950/EC 1.14.13.39) with the following conserved domains hits: ferredoxin reductase (FNR) binding domain, flavin adenine dinucleotide (FAD) binding pocket, conserved FAD binding motif, nicotinamide adenine dinucleotide (NAD) binding pocket, phosphate binding motif and beta-alpha-beta structure motif. The representation was based on in silico search of conserved domain of NCBI database.
In silico orthologs search revealed the presence of one gene copy per haploid genome in other Leishmania species: L. donovani, L. panamensis, L. braziliensis, L. major and L. infantum. The NOS-like alignment showed high identity among these species, confirming a biological role in the maintenance of parasite (S1 Fig). Additionally, a BLAST search for NOS-like (LmxM.19.1450) in the TriTryp database showed an identity of 99% with a contig of L. (L.) amazonensis MHOM/BR/71973/M2269 (KE391486.1). And a low identity was detected with Leptomonas seymouri, T. cruzi genome database (Dm28c), T. vivax (Y486), 74.2%, 21% and 16.2%, respectively (TriTryp database).
In addition, we analyzed the expression of nos-like by RT-qPCR in La-WT and La-arg- promastigotes and axenic amastigotes (Fig 2). As expected, an increase amount of nos-like was detected in La-arg- promastigotes when compared to La-WT.
Fig 2. Nos-like mRNA expression by RT-qPCR.
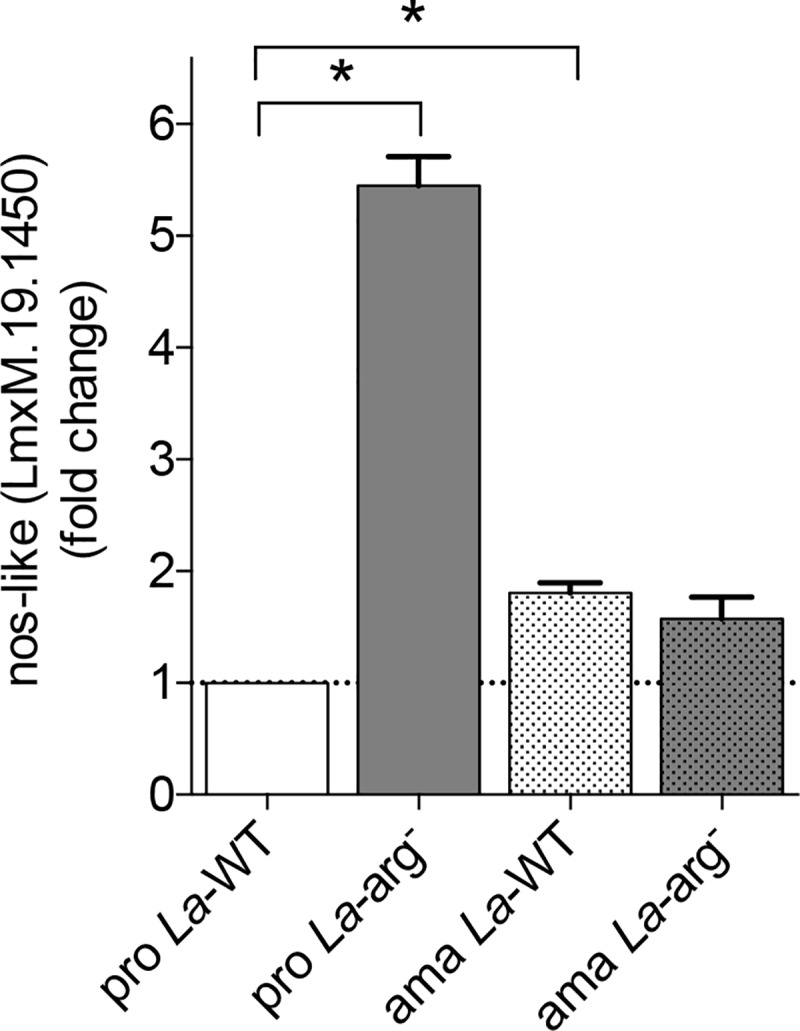
The nos-like mRNA levels were based on quantification of the target and were normalized by gapdh expression in promastigotes (pro) and amastigotes (ama) of La-WT and La-arg-. The values are the mean ± SEM of three independent biological replicates (n4-6). (*) p < 0.05.
Then, we analyzed the gene differential expression of other oxidoreductases identified through RNA-seq analysis. In Table 1, quinone oxidoreductase (LmxM.03.0570/EC 1.6.5.5) was down-regulated in La-arg- promastigotes compared to La-WT promastigotes. No differential expression gene was observed among the other oxidoreductases (Table 1).
In La-arg- axenic amastigotes compared to La-WT axenic amastigotes showed an up-regulation of FAD dependent oxidoreductase (LmxM.36.5660), and modest up-regulation of NADPH:ubiquinone oxidoreductase (Lmx>05.0980/EC 1.6.5.3), NADPH:ubiquinone oxidoreductases (LmxM.18.1480/EC 1.6.5.3, LmxM.20.0120/EC 1.6.5.3), NADH:adrenodoxin oxidoreductases (LmxM.21.0420/EC 1.18.1.6). No differential expression gene was observed among the other oxidoreductases (Table 2).
Table 2. Oxidoreductase-like transcripts expression of La-WT and La-arg- axenic amastigotes.
| ID | EC number |
ama La-WT vs. ama La-arg- Product description |
Fold change |
|---|---|---|---|
| LmxM.03.0570 | 1.6.5.5 | quinone oxidoreductase, putative | 1.08 |
| LmxM.05.0980 | 1.6.5.3 | NADH-ubiquinone oxidoreductase, putative | 1.22 |
| LmxM.07.0600 | 1.5.5.1 | electron transfer oxidoreductase, putative | 1.12 |
| LmxM.12.1130 | 1.3.1.42 | NADH:flavin oxidoreductase/NADH oxidase, putative | 1.18 |
| LmxM.13.0720 | 1.6.5.5 | oxidoreductase-like protein | 1.17 |
| LmxM.17.0270 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 1.00 |
| LmxM.18.1480 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 1.26 |
| LmxM.20.0120 | no data | NADH: ubiquinone oxidoreductase, putative | 1.43 |
| LmxM.21.0420 | 1.18.1.6 | NADH: adrenodoxin oxidoreductase, putative | 1.33 |
| LmxM.23.0670 | no data | oxidoreductase-like protein | 0.94 |
| LmxM.23.0860 | 1.6.5.5 | quinone oxidoreductase, putative | 1.07 |
| LmxM.23.1590 | no data | oxidoreductase-like protein | 1.06 |
| LmxM.23.1600 | no data | oxidoreductase-like protein | 0.59 |
| LmxM.28.0940 | no data | oxidoreductase-like protein | 1.22 |
| LmxM.31.2210 | no data | NADH: adrenodoxin oxidoreductase, putative | 0.67 |
| LmxM.32.1770 | no data | oxidoreductase-like protein | 1.01 |
| LmxM.33.4320 | no data | NADH: adrenodoxin oxidoreductase, putative | 0.88 |
| LmxM.36.3230 | 1.1.99.1 | oxidoreductase-like protein | 1.26 |
| LmxM.36.4170 | 1.6.5.5 | oxidoreductase-like protein | 0.60 |
| LmxM.36.5660 | no data | FAD dependent oxidoreductase | 1.81 |
Comparison of the oxidoreductases-orthologs transcripts between ama La-WT vs. pro La-arg-, adjusted for p < 0.05. ama: axenic amastigotes, La-WT: L. (L.) amazonensis wild-type, La-arg-: L. (L.) amazonensis arginase knockout.
In La-WT axenic amastigotes compared to promastigotes presented an up-regulation of NADH:flavin oxidoreductase/NADH oxidase (LmxM.12.1130/EC 1.3.1.42), NADH:adrenodoxin oxidoreductase (LmxM.31.2210) and oxidoreductases-like (LmxM.32.1770 and Lmx.36.4170/EC 1.6.5.5). No differential expression gene was observed among the other oxidoreductases (Table 3).
Table 3. Oxidoreductase-like transcripts expression of La-WT promastigotes and axenic amastigotes.
| ID | EC number |
pro La-WT vs. ama La-WT Product description |
Fold change |
|---|---|---|---|
| LmxM.03.0570 | 1.6.5.5 | quinone oxidoreductase, putative | 0.48 |
| LmxM.05.0980 | 1.6.5.3 | NADH-ubiquinone oxidoreductase, putative | 0.62 |
| LmxM.07.0600 | 1.5.5.1 | electron transfer oxidoreductase, putative | 0.84 |
| LmxM.12.1130 | 1.3.1.42 | NADH:flavin oxidoreductase/NADH oxidase, putative | 1.31 |
| LmxM.13.0720 | 1.6.5.5 | oxidoreductase-like protein | 0.74 |
| LmxM.17.0270 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 0.85 |
| LmxM.18.1480 | 1.6.5.3 | NADH: ubiquinone oxidoreductase, putative | 0.82 |
| LmxM.20.0120 | no data | NADH: ubiquinone oxidoreductase, putative | 0.53 |
| LmxM.21.0420 | 1.18.1.6 | NADH: adrenodoxin oxidoreductase, putative | 0.66 |
| LmxM.23.0670 | no data | oxidoreductase-like protein | 0.91 |
| LmxM.23.0860 | 1.6.5.5 | quinone oxidoreductase, putative | 0.78 |
| LmxM.23.1590 | no data | oxidoreductase-like protein | 1.11 |
| LmxM.23.1600 | no data | oxidoreductase-like protein | 0.99 |
| LmxM.28.0940 | no data | oxidoreductase-like protein | 1.11 |
| LmxM.31.2210 | no data | NADH: adrenodoxin oxidoreductase, putative | 1.64 |
| LmxM.32.1770 | no data | oxidoreductase-like protein | 1.79 |
| LmxM.33.4320 | no data | NADH: adrenodoxin oxidoreductase, putative | 0.81 |
| LmxM.36.3230 | 1.1.99.1 | oxidoreductase-like protein | 0.76 |
| LmxM.36.4170 | 1.6.5.5 | oxidoreductase-like protein | 1.60 |
| LmxM.36.5660 | no data | FAD dependent oxidoreductase | 1.03 |
Comparison of the oxidoreductases-orthologs transcripts between pro La-WT vs. ama La-WT, adjusted for p < 0.05. pro: promastigotes, ama: axenic amastigotes, La-WT: L. (L.) amazonensis wild-type.
Finally, in La-arg- axenic amastigotes compared to promastigotes identified an up-regulation of quinone oxidoreductases (LmxM.03.0570/EC 1.6.5.5), NADH:flavin oxidoreductase/NADH oxidase (LmxM.12.1130/EC 1.3.1.42) as well as oxidoreductases-like (LmxM.23.1590, LmxM.28.0949, LmxM.32.1770, LmxM.36.3230/EC 1.1.99.1, LmxM.36.4170/EC 1.6.5.5), NADH:adrenodoxin oxidoreductases (LmxM.21.0420/EC 1.18.1.6, Lmx.31.2210 and LmxM.33.4320) and FAD-dependent oxidoreductase (LmxM.36.5660) (Table 4).
The increase in NO production is arginase-dependent and indicative of metacyclogenesis in L. (L.) amazonensis promastigotes
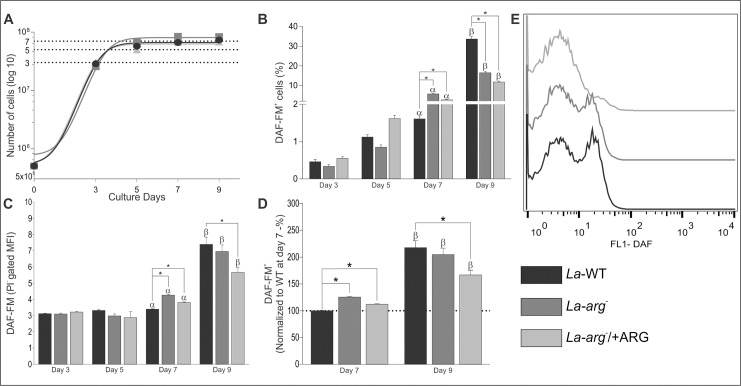
To correlate the expression of nos-like transcript and enzyme activity, we quantified NO amount in La-WT, La-arg-, and La-arg-/+ARG, along days 3, 5, 7 and 9 of a promastigote growth curve. We observed that all parasites lines reached the mid-logarithmic growth phase on day 3, the early-stationary growth phase occurred on day 5, the mid-stationary on day 7 and the late-stationary growth phase on day 9 (Fig 3A).
Fig 3. Growth curve and NO production in La-WT, La-arg- and La-arg-/+ARG on days 3, 5, 7 and 9.
L. (L.) amazonensis promastigotes wild-type (La-WT—black); arginase null knockout (La-arg-—dark gray) and the arginase addback (La-arg-/+ARG–light gray) growth curve and the NO production by DAF-FM labeling in flow cytometry. A total of 2x106 cells was labeled with 1 ng/mL propidium iodide (PI) and was analyzed using FACSCalibur (Becton Dickinson). The viable cells (PI-) were used for the analysis of the frequency of DAF-FM-labeled cells and the mean fluorescence (MFI). (A) Growth curves and the establishment of the logarithmic growth phase (day 3); early-stationary (day 5), mid-stationary (day 7) and late-stationary phase (day 9). (B) Frequency of DAF-FM+ cells. Each bar represents the mean of the percent ±SEM of DAF-FM+ cells (n = 3–9). (C) MFI of DAF-FM+ cells. Each bar represents the mean of the fluorescence intensity ±SEM of viable cells (n = 3–9). (D) Normalization of MFI values in relation to day 7 of La-WT (100%). (E) Histogram of MFI of the PI- gate at the 9th day of culture. These data were analyzed by Student’s t-test, and (*) p < 0.05, (α) p < 0.05 compared the day 7 with the days 3 and 5, (β) p < 0.05 compared the day 9 with the days 3, 5 and 7. The data are representative of 3 independent experiments.
We also analyzed the viability of cells by PI labeling using gating of unlabeled cells (PI-) (gate 1, S2 Fig); from this gating, we analyzed the DAF-FM+ cells (Fig 3B and gate 2, S2 Fig) and the mean of fluorescence intensity of DAF-FM using flow cytometry (Fig 3C and 3D, S2 Fig). Viable cells (PI- cells) represented approximately 99% of the total population (S2 Fig) for all tested promastigotes; only viable cells were used for DAF-FM analysis.
Then, we evaluated the NO production by DAF-FM labeling at every counting day in La-WT promastigotes in mid-logarithmic and early-stationary growth phase (days 3 and 5). We did not observe NO production in early growth phase of the parasite (Fig 3B). The detectable NO production was observed on day 7 at low levels, and high levels were detected on day 9 (Fig 3B), indicating that NO production can be related to the growth phase of the parasite.
The frequency of DAF-FM+ cells was nearly 1% on days 3, 5 and 7 in promastigotes La-WT and increased to 30% on day 9 in promastigotes late-stationary growth phase. In La-arg- promastigotes, NO-producing cells were induced at the mid-stationary growth phase (day 7), showing nearly 5-fold increase compared to La-WT (Fig 3B). Similarly, the mean DAF-FM per cell was 1.3-fold increase in La-arg- at the mid-stationary growth phase compared to La-WT (Fig 3C). In promastigotes La-WT at the late-stationary growth phase, the mean levels of DAF-FM per cell 2.5-fold increase compared to those on days 3, 5 or 7. The absence of arginase activity (La-arg-) or reduced arginase activity (La-arg-/+ARG) compared with La-WT [20] enabled an increase in DAF-FM+ cells in promastigotes at the mid-stationary growth phase, and the frequency was maintained in late-phase promastigotes, lower than La-WT, but the amount of NO per cell was increased in the absence of arginase activity (Fig 3B–3E). Altogether, these results suggested that NO production in L. (L.) amazonensis promastigotes is dependent on arginase activity and varies according to the growth phase of the parasite.
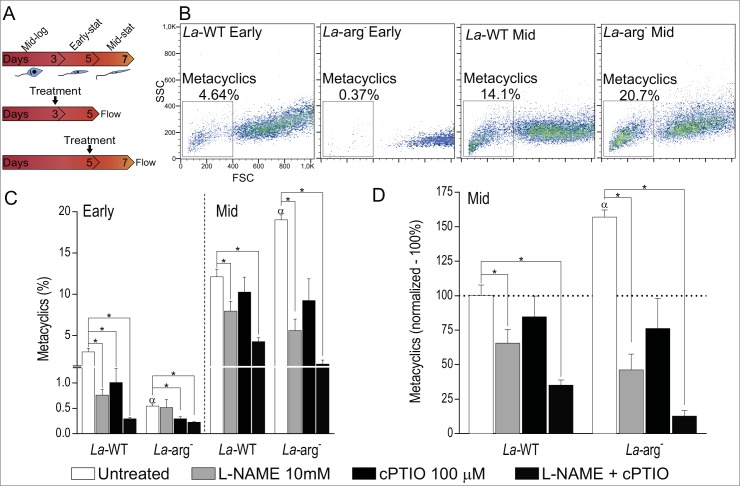
To analyze the function of NOS-like activity and NO production in metacyclogenesis, we used L-NAME (as NOS inhibitor), cPTIO (as NO scavenger) or L-NAME plus cPTIO treatments in mid-log (day 3) or early-stationary (day 5) growth phase of La-WT and La-arg- promastigotes (Fig 4A).
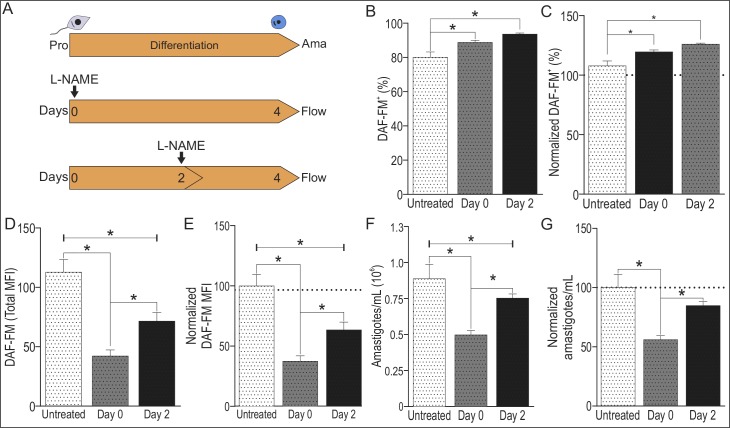
Fig 4. Inhibition of NOS activity or NO amount and metaclyclogenesis in La-WT and La-arg- promastigote.
(A) L. (L.) amazonensis wild-type (La-WT) and arginase null knockout (La-arg-) at logarithmic (log—day 3) and early-stat (early-stationary—day 5) phase promastigotes were untreated (white) or treated with 10 mM of L-NAME (light gray), 100 μM of cPTIO (dark gray) and L-NAME plus cPTIO (dark). After 2 days of culture metacyclic promastigotes forms were quantified by flow cytometry. (B) Dot plot of SSC and FSC features representative of metacyclics promastigote forms viable cells analyzed using FACSCalibur (Becton Dickinson). (C) Frequency of metacyclics promastigotes. Each bar represents the mean of the percent ±SEM of metaclyclic cells (n = 3–9). (D) Normalization of the percentage of metacyclic forms in relation to log phase of La-WT (100%). These data were analyzed by Student’s t-test, and (*) p < 0.05, (α) p < 0.05 compared the La-WT to La-arg-. The data are representative of 3 independent experiments (n = 6–9).
The frequency of metacyclic forms 4-fold from increased in early- to mid -stationary promastigote growth phase in La-WT of (untreated, Fig 4B and 4C). The inhibition of NOS activity or reduction of NO amount during the differentiation of mid-log to early-stationary growth phase reduced the frequency of metacyclic forms around 74% (4-fold) and 67%(3-fold), respectively. NOS activity and NO reduced in 90% (10-fold) the frequency of metacyclic forms (Fig 4C and 4D). However, treatments with L-NAME, cPTIO or L-NAME/cPTI during differentiation of early- to mid-stationary growth phase reduced this metacyclic frequency to nearly 34% (1.5-fold), 17% (1.2-fold) and 65% (2.8-fold), respectively (Fig 4C and 4D).
The absence of arginase activity reduced the frequency of metacyclic forms in early-stationary phase, but increased in mid-stationary. Despite that, we observed a decrease in frequency of metacyclic forms when NOS activity and NO was reduced in La-arg- (Fig 4C). The reduction on the frequency of metacyclics was pronounced during differentiation of early- to mid-stationary phase treated with L-NAME, cPTIO or/and L-NAME/cPTIO, to reduced 74%, 53% and 92%, respectively (Fig 4C and 4D). Thoroughly, these results correlated the NOS activity and NO levels to metacyclogenesis, as well as to dependence of arginase activity.
The increase in NO production in L. (L.) amazonensis axenic amastigotes
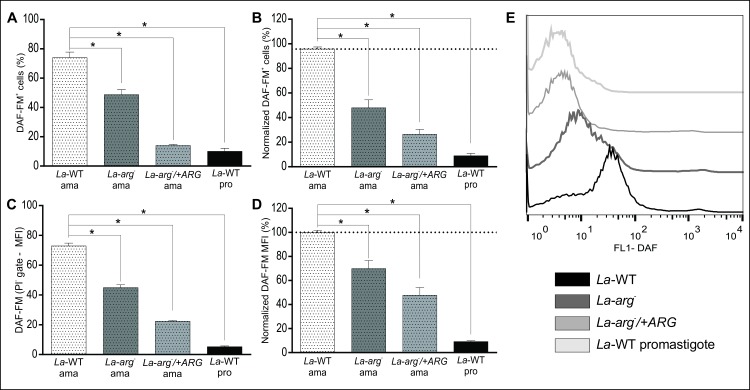
La-WT, La-arg- and La-arg-/+ARG promastigotes in the early-stationary growth phase were subjected to amastigote differentiation in M 199 medium, at 34°C and pH 5.5, for 4 days. The differentiation to amastigotes forms was confirmed by amastins gene expression (LmxM.08.00760, LmxM.08.0800 and LmxM.33.0960) by RT-qPCR. The amastins transcripts were increased in axenic amastigotes than stationary-phase promastigotes of La-WT or La-arg-, with exception of LmxM.33.0960 in La-arg- (S3 Fig). Next, the axenic amastigotes were labeled with DAF-FM and PI, as described for promastigotes. The viable cells (PI-) represented approximately 95% of the total population (S4 Fig). La-WT amastigotes represented the higher population labeled with DAF-FM, and approximately 75% of viable cells were DAF-FM+ (Fig 5A), a value higher than promastigotes. Compared to La-WT, NO-producing amastigotes were lower in La-arg- (50%) and La-arg-/+ARG (70%) (Fig 5A and 5B). The La-WT promastigotes in the late-stationary phase were used as the positive control of this experiment.
Fig 5. L. (L.) amazonensis axenic amastigotes produced NO in a higher amount than promastigotes.
La-WT, La-arg- and La-arg-/+ARG promastigotes at the early-stationary phase (day 5 – 5x106 cells/10 mL) were differentiated into amastigotes by changing the M199 medium to pH 5.5 and maintaining the culture at 34°C during 4 days. Next, the cells were labeled with 1 μM DAF-FM and 1 ng/mL PI and were analyzed by flow cytometry. La-WT promastigotes in the late-stationary phase were used as a positive control. The viable cells (PI-) were used to analyze the frequency of DAF-FM+ cells and MFI. (A) Frequency of DAF-FM+ cells. These are representative of 1 independent experiment (n = 3–4). (B) Normalization of the percentage of DAF-FM+ La-arg- and La-arg-/+ARG in relation to La-WT (100%). These are representative of 3 independent experiments (n = 9). (C) MFI of DAF-FM+ cells. These are representative of 1 independent experiment (n = 3–4). (D) Normalization of the MFI values of La-arg- and La-arg-/+ARG in relation to La-WT (100%). These data are representative of 3 independent experiments (n = 9). These data were analyzed were analyzed by one-way ANOVA and Tukey’s comparison post-test. Significance p-value is represented by *, p<0.05. (E) Histogram of MFI of DAF-FM.
Mean of fluorescence intensity (MFI) analysis of the tested amastigotes showed the same pattern regarding the percentage of cells producing NO. La-WT amastigotes was 2-fold and 3.5-fold increased amount of NO than La-arg- and La-arg-/+ARG, respectively (Fig 5C–5E). The NO production in La-WT amastigotes was 10-fold increased than La-WT promastigotes. These results showed a higher production of NO in axenic amastigotes of L. (L.) amazonensis and the absence or reduction of arginase activity led to a reduction of NO production.
The role of NOS activity and NO production during La-WT amastigotes differentiation was evaluated using a L-NAME (as NOS inhibitor) at day 0 or day 2 of culture of early-stationary promastigotes subjected to amastigotes differentiation for 4 days, at 34°C and pH 5.5 (Fig 5A). As observed previously, viable cells represented nearly 80% of total population of untreated or L-NAME treated cells. The frequency of DAF-FM+ cells presented a 10% slight increase after L-NAME treatment, (Fig 6B and 6C). However, the inhibition of NOS activity by L-NAME reduced the amount of NO per cell, in 58% and 29% in cells treated at day 0 or day 2, respectively (Fig 6C and 6D). The reduction in amastigotes numbers was pronounced after L-NAME treatment at day 0 compared to day 2 (Fig 6E and 6F), correlating reduction in NOS activity and NO production to number of amastigotes. These data revealed the NO production in the initial steps of amastigotes differentiation correlating with the replication of amastigotes forms.
Fig 6. Inhibition of NOS activity during La-WT and La-arg- amastigotes differentiation.
(A) L. (L.) amazonensis La-WT at early-stationary phase promastigotes (pro) were differentiated into amastigotes (ama), by changing the M199 medium to pH 5.5 and maintaining the culture at 34°C during 4 days, untreated (white) or treated with 10 mM of L-NAME (light gray) at day 0 or day of culture. Next, a total of 2x106 cells was labeled with 1 μM DAF-FM and 1 ng/mL propidium iodide (PI) and analyzed using FACSCalibur (Becton Dickinson). B) Frequency of of DAF-FM+ cells. (C) Normalization of the percentage of DAF-FM+ cells in relation to La-WT-untreated (100%). (D) MFI of DAF-FM+ cells. (E) Normalization of the MFI of DAF-FM+ cells in relation La-WT-untreated (100%). (F) Number of amastigotes forms. (G) Normalization of number of amastigote forms in relation La-WT-untreated (100%). Each bar represents the mean of the values ±SEM (n = 3–9). These data were analyzed by Student’s t-test, and (*) p < 0.05. The data are representative of 3 independent experiments.
Discussion
The biological function of NO in trypanosomatids is still not completely understood. The main controversy is to understand why the parasite produces NO, the same molecule produced by the host macrophages to kill them [19,22,44]. The key to answer this question can be related to the NO concentration. NO can perform multiple actions depending on its concentration, production time and exposure time. Previous studies with T. cruzi demonstrated that low NO concentration (nM levels) resulted in the induction of cGMP production and post-translational modifications, interfering in several biological processes, such as PKC signaling, cytochrome C regulation, caspase-cascade inactivation, protein degradation and control of the redox environment [45–50]. On the other hand, at high concentrations (mM), NO can be toxic, passing across various chemical reactions [26,51].
Physiological processes mediated by NO are modulated due the characteristics of the free radical with a short half-life [52] and quick diffusion away from the site of its synthesis [27]. Other modulation factor of these processes includes the source of L-arginine, presence of molecular oxygen and cofactors, such as NADPH, FAD, heme, FMN and BH4, the presence an active NOS, and the intracellular or extracellular environment limits/boundaries for the NO action [26,53,54]. Additionally, NO can react with other free radical species, such as superoxide, thiol radicals or lipid peroxides, or metal-coupled proteins such as hemoglobin [51,53].
In this work, we focused on oxidoreductases expression. Oxidoreductase is a class of enzymes that catalyze biological oxidation/reduction reactions [55]. The transcriptomic profiling analysis identified several oxidoreductases: (1) NADH:quinone oxidoreductase, previous described with important role in yeasts and prokaryotes to maintain the [NADH]/[NAD+] balance and for the entrance point of electrons into the respiratory chains of complex I coupling to the pumping of protons across the inner mitochondrial membrane [56,57]; (2) NADPH:ubiquinone oxidoreductases, previously described as related to the transfer of electrons from NADH to ubiquinone with the translocation of protons across the membrane in the respiratory complex I [58]; (3) electron transfer oxidoreductase or electron-transfer flavoprotein:ubiquinone oxidoreductase, previously described as a component of the mitochondrial respiratory chain, which catalyzes the electron-transfer of flavoprotein to the ubiquinone pool, as the electron acceptor [59]; (4) NADH:adrenodoxin oxidoreductases, previously described to initiate electron transport for cytochrome P450 receiving electrons from NADPH to reduce O2 to a superoxide radical [60]; (5) NADH:flavin oxidoreductase/NADH oxidase, previously described catalyzing the reduction of free flavins by NADPH or NADH; such enzymes are present in all microorganisms [61,62]; and (6) FAD-dependent oxidoreductase previously described as one of the assembly factors in the respiratory chains of complex I [63].
We highlighted one of them, the oxidoreductase-like (LmxM.19.1450/ EC 1.14.13.39). This oxidoreductase-like appeared up-regulated in La-arg- promastigotes compared to La-WT promastigotes and in La-WT axenic amastigotes compared to La-WT promastigotes, indicating a modulation during promastigote to amastigote differentiation as well as a dependence on arginase activity. According to the conserved domains content and to the metabolome data pointing to the expression of an enzyme (EC 1.14.13.39), which could be involved in the production of NO and citrulline [18], we suggested this oxidoreductase as a NOS-like. In addition, the multiple alignment revealed this NOS-like conserved among Leishmania spp.
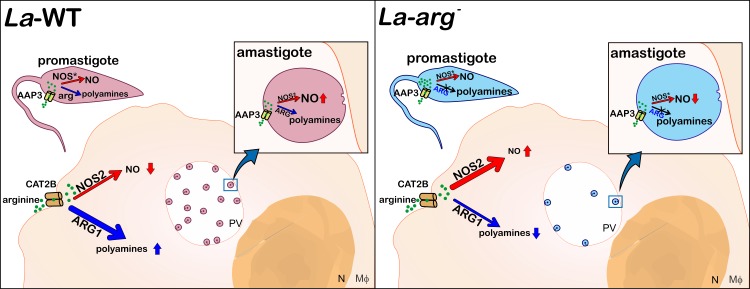
The fate of Leishmania infection in the mammalian host depends on dual role of macrophage L-arginine metabolism: the production of NO promoting the killing of the pathogen [19,22,44], or the production of ornithine, the polyamine precursor allowing the parasite replication [19,44,64]. Leishmania presents its own machinery to metabolize L-arginine: the AAP3 for the uptake and the arginase activity to metabolize the amino acid to polyamines production, which are essential for in vitro proliferation, as shown for L. (L.) amazonensis [6,8,20], L. donovani [65], L. (L.) mexicana [66] and L. (L.) major [66,67]. The lack of arginase activity reduces AAP3 and arginase levels and the infectivity of L. (L.) amazonensis [20,22], L. donovani [65], L. (L.) mexicana [66] and L. (L.) major [66,67]. In infected macrophages, L-arginine can be metabolized by both arginases, from the Leishmania (La-ARG) and from the macrophage (ARG1), supplying the polyamine pathway and enabling parasite survival [7,8,19,22,66]. Depending on substrate competition, L-arginine can be hydrolyzed by the macrophage NOS2 or by the parasite NOS-like [13–15,17,18]. Based on that, we proposed a schematic representation on the fate of the infection (Fig 7).
Fig 7. Schematic representation of L. (L.) amazonensis promastigote and amastigote NO production in the infection context.
In L. (L.) amazonensis, the AAP3 transporter takes up L-arginine, and this amino acid is metabolized by arginase into ornithine to produce polyamines or via NOS-like to produce NO. La-WT-infected macrophages increase the activity of the polyamine pathway, increasing the expression of the L-arginine transporter from the host (CAT2B) and parasite (AAP3), arginase1 (host) and parasite-arginase to metabolizing L-arginine to produce polyamines, consequently sustaining the basal level of NOS2 expression and NO production. Indeed, the La-WT amastigote differentiation induces NOS-like expression and NO production, higher levels than promastigotes, but lower than those produced by macrophages. The lack of arginase activity in promastigotes increases the L-arginine levels, even that the reduction in AAP3 expression, inducing the expression of NOS-like, NO and citrulline production. The La-arg—infected macrophages present lower levels of CAT2B and AAP3, which could reduce L-arginine uptake, and lower the levels of arginase 1 and polyamine production compared with La-WT infection allowing the increase in NOS2 expression and NO production, reducing infectivity. In addition, the La-arg- amastigote differentiation induces NOS-like expression but not AAP3 expression and L-arginine uptake sufficient to increase NO production compared with La-WT promastigotes. PV: parasitophorous vacuole.
The attenuation of infectivity in La-arg- [20,22], as well as in L. mexicana-arg- [68] was associated with NO overexpression by host macrophages but not in L. major-arg- [69]. The depletion of L-arginine in macrophages can inhibit both polyamines and NO production. In addition, the lower infectivity of La-arg- could correlate with lower levels of CAT2B [21] or AAP3 expression and amino acid uptake [70], reducing L-arginine availability for ARG1 in infected macrophages, reducing parasite survival. In this way, infectivity could be recovered by putrescine supplementation [21].
Our data showed NO production in L. (L.) amazonensis stationary phase promastigotes corroborating previous studies that detected NO in promastigotes of L. (L.) amazonensis M2269, L. (V.) braziliensis M2903, L. (L.) chagasi PP75, L. (L.) donovani AG83, L. (L.) donovani DD8, L. (L.) infantum IPT1, L. (L.) major 5ASKH, L. (L.) mexicana MIC and L. (V.) panamensis LS94 [17]. Indeed, the suggestion of NOS-like expression in Leishmania corroborates other studies that Leishmania harbors NOS-like activity to produce NO [13–15,17]. Genestra et al. (2006) purified a protein that exhibited NOS activity in L. (L.) amazonensis and showed similarities to T. cruzi NOS and neuronal NOS (nNOS). In addition, this group demonstrated NOS-like activity as Ca2+ and calmodulin dependent, corresponding to a constitutive form of protein [15]. Leon’s group also showed an increased amount of NO in L. (L.) amazonensis metacyclic form and in axenic amastigotes [15]. Our data correlated the NO production signaling to metacyclogenesis, since reduction in NOS activity and NO levels reduced the metacyclic differentiation, as well as reduced the amastigotes differentiation and growth.
The NO could confer parasite protection altering the steady state of promastigotes and amastigotes to resist to toxicity signals mediate by macrophage inflammatory. Corroborating with this idea, some studies show the influence of NO in protecting parasites from antimony-mediated oxidative stress promoting a higher H2O2 scavenging activity and increase in thiol levels [71]. Indeed, NO induction induces heat shock proteins expression [72] and could inhibit molecules involved in the apoptosis signal, such as cysteine proteinases [73], cis-aconitase and glyceraldehydes-3-phosphate dehydrogenase [74,75]. Although, NO-resistant parasites are able to differentiate from promastigotes to amastigotes and from amastigotes to promastigotes, but the differentiation of NO-resistant promastigotes to amastigotes occurs in lower levels than non-resistant parasites, probably by increased levels of 6-phosphogluconate dehydrogenase involved in NADPH and ribose-5-phosphate generation resulting in rapid consumption of glucose that accelerates the entry in stationary phase of growth [74].
Meanwhile, the high levels of NO detected in late-stationary growth phase of L. (L.) amazonensis promastigotes support the link between NO production and metacyclogenesis, similar to the previous observation that correlated a high amount of NO production in culture with a high number of metacyclic forms as well as with the infectivity [13]. In addition, L-arginine starvation of promastigotes L. (L.) amazonensis at the logarithmic growth phase reduced ornithine and putrescine levels [18]. By contrast, the absence of arginase activity increased the L-arginine availability and the citrulline levels, but decreased the levels of ornithine and putrescine in promastigotes [6,18,20]. As a consequence, it promoted NO production in the earlier steps of the stationary phase growth phase of promastigote differentiation (this communication). This evidence points to the importance of the competition for the same substrate, L-arginine, by arginase and NOS-like during metacyclogenesis and amastigotes differentiation, influencing the NO production.
Supporting information
Multiple alignment of the amino acid sequence of the nitric oxide synthase-like NOS-like of. L. amazonensis (Lam), L. mexicana (Lmx), L. donovani (Ldo), L. panamensis (Lpa), L. braziliensis (Lbr), L. major (Lmj) and L. infantum. The identical amino acids are highlighted in gray. The alignment was performed based on NCBI multiple alignment tool.
(TIF)
The DAF-FM labeling of La-WT, La-arg- and La-arg-/+ARG promastigotes and analysis were performed as follows: (1) DAF-FM labeling followed by PI (propidium iodide) staining; (2) The data were acquired using a FACSCalibur (BD) flow cytometer followed by analysis of the resulting pattern of labeling: PI-DAF-, PI-DAF+, PI+DAF- and PI+DAF+. Using FlowJo software (as described on methods section), the cells were gated based on the Forward Scatter (FSC) and the PI label was used to gate viable cells (PI-); (3) Using viable cells, DAF+ populations were selected for frequency analysis, and (4) the population producing NO was gated based on the histogram of the Mean of Fluorescence Intensity (MFI).
(TIF)
(A) The amastins LmxM.08.00760 (A), LmxM.0800800 (B) and LmxM.33.0960 (C) mRNA levels were based on quantification of the target and were normalized by gapdh expression in promastigotes (pro) and amastigotes (ama) of La-WT and La-arg-. The values are the mean ± SEM of three independent biological replicates (n = 4–6). (*) p < 0.05
(TIF)
DAF-FM labeling of La-WT, La-arg- and La-arg-/+ARG amastigotes and analysis were performed as follows: (1) DAF-FM labelling followed by PI (propidium iodide) staining; (2) The data were acquired on a FACSCalibur (BD) flow cytometer with analysis of the resulting pattern of labelling: PI-DAF-, PI-DAF+, PI+DAF- and PI+DAF+. Using FloJo software (as described on methods section), the cells were gated based on the Forward Scatter (FSC) and PI labeling was used to gate viable cells (PI-); (3) Using viable cells, DAF+ populations were selected for frequency analysis, and (4) the population producing NO was gated based on the histogram of the Mean Fluorescence Intensity (MFI).
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, www.cnpq.br: 307587/2014-2.) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, www.fapesp.br: 2014/50717-1, 2016/19815-2). SMA received a fellowship from FAPESP (2015/25942-4) and CNPq (103563/2013-0), and LMFW received a research fellowship from CNPq (307587/2014-2). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bogdan C (2008) Mechanisms and consequences of persistence of intracellular pathogens: leishmaniasis as an example. Cell Microbiol 10: 1221–1234. doi: 10.1111/j.1462-5822.2008.01146.x [DOI] [PubMed] [Google Scholar]
- 2.Liese J, Schleicher U, Bogdan C (2008) The innate immune response against Leishmania parasites. Immunobiology 213: 377–387. doi: 10.1016/j.imbio.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 3.Sacks D, Kamhawi S (2001) Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu Rev Microbiol 55: 453–483. doi: 10.1146/annurev.micro.55.1.453 [DOI] [PubMed] [Google Scholar]
- 4.Teixeira DE, Benchimol M, Rodrigues JC, Crepaldi PH, Pimenta PF, de Souza W (2013) The cell biology of Leishmania: how to teach using animations. PLoS Pathog 9: e1003594 doi: 10.1371/journal.ppat.1003594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Séguin O, Descoteaux A (2016) Leishmania, the phagosome, and host responses: The journey of a parasite. Cell Immunol 309: 1–6. doi: 10.1016/j.cellimm.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 6.Castilho-Martins EA, Laranjeira da Silva MF, dos Santos MG, Muxel SM, Floeter-Winter LM (2011) Axenic Leishmania amazonensis promastigotes sense both the external and internal arginine pool distinctly regulating the two transporter-coding genes. PLoS One 6: e27818 doi: 10.1371/journal.pone.0027818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Silva MF, Zampieri RA, Muxel SM, Beverley SM, Floeter-Winter LM (2012) Leishmania amazonensis arginase compartmentalization in the glycosome is important for parasite infectivity. PLoS One 7: e34022 doi: 10.1371/journal.pone.0034022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silva ER, Castilho TM, Pioker FC, Tomich de Paula Silva CH, Floeter-Winter LM (2002) Genomic organisation and transcription characterisation of the gene encoding Leishmania (Leishmania) amazonensis arginase and its protein structure prediction. Int J Parasitol 32: 727–737. [DOI] [PubMed] [Google Scholar]
- 9.Green SJ, Crawford RM, Hockmeyer JT, Meltzer MS, Nacy CA (1990) Leishmania major amastigotes initiate the L-arginine-dependent killing mechanism in IFN-gamma-stimulated macrophages by induction of tumor necrosis factor-alpha. J Immunol 145: 4290–4297. [PubMed] [Google Scholar]
- 10.Green SJ, Meltzer MS, Hibbs JB Jr., Nacy CA (1990) Activated macrophages destroy intracellular Leishmania major amastigotes by an L-arginine-dependent killing mechanism. J Immunol 144: 278–283. [PubMed] [Google Scholar]
- 11.Liew FY, Li Y, Moss D, Parkinson C, Rogers MV, Moncada S (1991) Resistance to Leishmania major infection correlates with the induction of nitric oxide synthase in murine macrophages. Eur J Immunol 21: 3009–3014. doi: 10.1002/eji.1830211216 [DOI] [PubMed] [Google Scholar]
- 12.Goldman-Pinkovich A, Balno C, Strasser R, Zeituni-Molad M, Bendelak K, Rentsch D, et al. (2016) An Arginine Deprivation Response Pathway Is Induced in Leishmania during Macrophage Invasion. PLoS Pathog 12: e1005494 doi: 10.1371/journal.ppat.1005494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genestra M, de Souza WJ, Cysne-Finkelstein L, Leon LL (2003) Comparative analysis of the nitric oxide production by Leishmania sp. Med Microbiol Immunol 192: 217–223. doi: 10.1007/s00430-003-0176-z [DOI] [PubMed] [Google Scholar]
- 14.Genestra M, Guedes-Silva D, Souza WJ, Cysne-Finkelstein L, Soares-Bezerra RJ, Monteiro FP, et al. (2006) Nitric oxide synthase (NOS) characterization in Leishmania amazonensis axenic amastigotes. Arch Med Res 37: 328–333. doi: 10.1016/j.arcmed.2005.07.011 [DOI] [PubMed] [Google Scholar]
- 15.Genestra M, Souza WJ, Guedes-Silva D, Machado GM, Cysne-Finkelstein L, Bezerra RJ, et al. (2006) Nitric oxide biosynthesis by Leishmania amazonensis promastigotes containing a high percentage of metacyclic forms. Arch Microbiol 185: 348–354. doi: 10.1007/s00203-006-0105-9 [DOI] [PubMed] [Google Scholar]
- 16.Sarkar A, Mandal G, Singh N, Sundar S, Chatterjee M (2009) Flow cytometric determination of intracellular non-protein thiols in Leishmania promastigotes using 5-chloromethyl fluorescein diacetate. Exp Parasitol 122: 299–305. doi: 10.1016/j.exppara.2009.04.012 [DOI] [PubMed] [Google Scholar]
- 17.Sarkar A, Saha P, Mandal G, Mukhopadhyay D, Roy S, Singh SK, et al. (2011) Monitoring of intracellular nitric oxide in leishmaniasis: its applicability in patients with visceral leishmaniasis. Cytometry A 79: 35–45. doi: 10.1002/cyto.a.21001 [DOI] [PubMed] [Google Scholar]
- 18.Castilho-Martins EA, Canuto GA, Muxel SM, da Silva MF, Floeter-Winter LM, Del Aguila C, et al. (2015) Capillary electrophoresis reveals polyamine metabolism modulation in Leishmania (Leishmania) amazonensis wild type and arginase knockout mutants under arginine starvation. Electrophoresis. [DOI] [PubMed] [Google Scholar]
- 19.Wanasen N, Soong L (2008) L-arginine metabolism and its impact on host immunity against Leishmania infection. Immunol Res 41: 15–25. doi: 10.1007/s12026-007-8012-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laranjeira-Silva MF, Zampieri RA, Muxel SM, Beverley SM, Floeter-Winter LM (2012) Leishmania amazonensis arginase compartmentalization in the glycosome is important for parasite infectivity. PLoS One 7: e34022 doi: 10.1371/journal.pone.0034022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laranjeira-Silva MF, Zampieri RA, Muxel SM, Floeter-Winter LM, Markus RP (2015) Melatonin attenuates Leishmania (L.) amazonensis infection by modulating arginine metabolism. J Pineal Res. [DOI] [PubMed] [Google Scholar]
- 22.Muxel SM, Laranjeira-Silva MF, Zampieri RA, Floeter-Winter LM (2017) Leishmania (Leishmania) amazonensis induces macrophage miR-294 and miR-721 expression and modulates infection by targeting NOS2 and L-arginine metabolism. Sci Rep 7: 44141 doi: 10.1038/srep44141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alderton WK, Cooper CE, Knowles RG (2001) Nitric oxide synthases: structure, function and inhibition. Biochem J 357: 593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacMicking J, Xie QW, Nathan C (1997) Nitric oxide and macrophage function. Annu Rev Immunol 15: 323–350. doi: 10.1146/annurev.immunol.15.1.323 [DOI] [PubMed] [Google Scholar]
- 25.Stuehr DJ (1999) Mammalian nitric oxide synthases. Biochim Biophys Acta 1411: 217–230. [DOI] [PubMed] [Google Scholar]
- 26.Pacher P, Beckman JS, Liaudet L (2007) Nitric oxide and peroxynitrite in health and disease. Physiol Rev 87: 315–424. doi: 10.1152/physrev.00029.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wink DA, Hines HB, Cheng RY, Switzer CH, Flores-Santana W, Vitek MP, et al. (2011) Nitric oxide and redox mechanisms in the immune response. J Leukoc Biol 89: 873–891. doi: 10.1189/jlb.1010550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planchet E, Kaiser WM (2006) Nitric oxide production in plants: facts and fictions. Plant Signal Behav 1: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paveto C, Pereira C, Espinosa J, Montagna AE, Farber M, Esteva M, et al. (1995) The nitric oxide transduction pathway in Trypanosoma cruzi. J Biol Chem 270: 16576–16579. [DOI] [PubMed] [Google Scholar]
- 30.Piacenza L, Peluffo G, Radi R (2001) L-arginine-dependent suppression of apoptosis in Trypanosoma cruzi: contribution of the nitric oxide and polyamine pathways. Proc Natl Acad Sci U S A 98: 7301–7306. doi: 10.1073/pnas.121520398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira M, Soares C, Canuto GA, Tavares MF, Colli W, Alves MJ (2015) Down regulation of NO signaling in Trypanosoma cruzi upon parasite-extracellular matrix interaction: changes in protein modification by nitrosylation and nitration. PLoS Negl Trop Dis 9: e0003683 doi: 10.1371/journal.pntd.0003683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basu NK, Kole L, Ghosh A, Das PK (1997) Isolation of a nitric oxide synthase from the protozoan parasite, Leishmania donovani. FEMS Microbiol Lett 156: 43–47. [DOI] [PubMed] [Google Scholar]
- 33.Temporal RM, Cysne-Finkelstein L, Echevarria A, Silva-Goncalves AJ, Leon LL, Genestra MS (2005) Amidine derivatives and Leishmania amazonensis: an evaluation of the effect of nitric oxide (NO) production on the parasite-macrophage interaction. J Enzyme Inhib Med Chem 20: 13–18. doi: 10.1080/14756360400015207 [DOI] [PubMed] [Google Scholar]
- 34.Fiebig M, Kelly S, Gluenz E (2015) Comparative Life Cycle Transcriptomics Revises Leishmania mexicana Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates. PLoS Pathog 11: e1005186 doi: 10.1371/journal.ppat.1005186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodgkinson VH, Soong L, Duboise SM, McMahon-Pratt D (1996) Leishmania amazonensis: cultivation and characterization of axenic amastigote-like organisms. Exp Parasitol 83: 94–105. doi: 10.1006/expr.1996.0053 [DOI] [PubMed] [Google Scholar]
- 36.Cysne-Finkelstein L, Temporal RM, Alves FA, Leon LL (1998) Leishmania amazonensis: long-term cultivation of axenic amastigotes is associated to metacyclogenesis of promastigotes. Exp Parasitol 89: 58–62. doi: 10.1006/expr.1998.4276 [DOI] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11.10.11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 43.Aoki JI, Muxel SM, Zampieri RA, Laranjeira da Silva MF, Muller KE, Nerland A, et al. (2017) RNA-seq transcriptional profiling of Leishmania amazonensis reveals an arginase-dependent gene expression regulation. PLoS Negl Trop Dis 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laranjeira-Silva MF, Floeter-Winter LM (2014) Arginase in Leishmania In: Andre LS Santos MHB, d'Avila-Levy Claudia M, Kneipp Lucimar F, Sodre Catia L, editor. Proteins and Proteomics of Leishmania and Trypanossoma. Springer: Springer. pp. 103–118. [Google Scholar]
- 45.Abello N, Kerstjens HA, Postma DS, Bischoff R (2009) Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J Proteome Res 8: 3222–3238. doi: 10.1021/pr900039c [DOI] [PubMed] [Google Scholar]
- 46.Hess DT, Stamler JS (2012) Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem 287: 4411–4418. doi: 10.1074/jbc.R111.285742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagawa H, Komai N, Takusagawa M, Miura Y, Toda T, Miyata N, et al. (2007) Nitration of specific tyrosine residues of cytochrome C is associated with caspase-cascade inactivation. Biol Pharm Bull 30: 15–20. [DOI] [PubMed] [Google Scholar]
- 48.Souza JM, Choi I, Chen Q, Weisse M, Daikhin E, Yudkoff M, et al. (2000) Proteolytic degradation of tyrosine nitrated proteins. Arch Biochem Biophys 380: 360–366. doi: 10.1006/abbi.2000.1940 [DOI] [PubMed] [Google Scholar]
- 49.Balafanova Z, Bolli R, Zhang J, Zheng Y, Pass JM, Bhatnagar A, et al. (2002) Nitric oxide (NO) induces nitration of protein kinase Cepsilon (PKCepsilon), facilitating PKCepsilon translocation via enhanced PKCepsilon -RACK2 interactions: a novel mechanism of no-triggered activation of PKCepsilon. J Biol Chem 277: 15021–15027. doi: 10.1074/jbc.M112451200 [DOI] [PubMed] [Google Scholar]
- 50.Ji Y, Neverova I, Van Eyk JE, Bennett BM (2006) Nitration of tyrosine 92 mediates the activation of rat microsomal glutathione s-transferase by peroxynitrite. J Biol Chem 281: 1986–1991. doi: 10.1074/jbc.M509480200 [DOI] [PubMed] [Google Scholar]
- 51.Wink DA, Mitchell JB (1998) Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med 25: 434–456. [DOI] [PubMed] [Google Scholar]
- 52.Lowenstein CJ, Dinerman JL, Snyder SH (1994) Nitric oxide: a physiologic messenger. Ann Intern Med 120: 227–237. [DOI] [PubMed] [Google Scholar]
- 53.Hall CN, Garthwaite J (2009) What is the real physiological NO concentration in vivo? Nitric Oxide 21: 92–103. doi: 10.1016/j.niox.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lancaster JR Jr. (1997) A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide 1: 18–30. doi: 10.1006/niox.1996.0112 [DOI] [PubMed] [Google Scholar]
- 55.May SW (1999) Applications of oxidoreductases. Curr Opin Biotechnol 10: 370–375. doi: 10.1016/S0958-1669(99)80067-6 [DOI] [PubMed] [Google Scholar]
- 56.Melo AM, Bandeiras TM, Teixeira M (2004) New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol Mol Biol Rev 68: 603–616. doi: 10.1128/MMBR.68.4.603-616.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brandt U (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem 75: 69–92. doi: 10.1146/annurev.biochem.75.103004.142539 [DOI] [PubMed] [Google Scholar]
- 58.Morina K, Schulte M, Hubrich F, Dorner K, Steimle S, Stolpe S, et al. (2011) Engineering the respiratory complex I to energy-converting NADPH:ubiquinone oxidoreductase. J Biol Chem 286: 34627–34634. doi: 10.1074/jbc.M111.274571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watmough NJ, Frerman FE (2010) The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim Biophys Acta 1797: 1910–1916. doi: 10.1016/j.bbabio.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 60.Hanukoglu I, Rapoport R, Weiner L, Sklan D (1993) Electron leakage from the mitochondrial NADPH-adrenodoxin reductase-adrenodoxin-P450scc (cholesterol side chain cleavage) system. Arch Biochem Biophys 305: 489–498. doi: 10.1006/abbi.1993.1452 [DOI] [PubMed] [Google Scholar]
- 61.Fieschi F, Niviere V, Frier C, Decout JL, Fontecave M (1995) The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli. J Biol Chem 270: 30392–30400. [DOI] [PubMed] [Google Scholar]
- 62.Fontecave M, Coves J, Pierre JL (1994) Ferric reductases or flavin reductases? Biometals 7: 3–8. [DOI] [PubMed] [Google Scholar]
- 63.Lemire BD (2015) Evolution of FOXRED1, an FAD-dependent oxidoreductase necessary for NADH:ubiquinone oxidoreductase (Complex I) assembly. Biochim Biophys Acta 1847: 451–457. doi: 10.1016/j.bbabio.2015.01.014 [DOI] [PubMed] [Google Scholar]
- 64.Darlyuk I, Goldman A, Roberts SC, Ullman B, Rentsch D, Zilberstein D (2009) Arginine homeostasis and transport in the human pathogen Leishmania donovani. J Biol Chem 284: 19800–19807. doi: 10.1074/jbc.M901066200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boitz JM, Gilroy CA, Olenyik TD, Paradis D, Perdeh J, Dearman K, et al. (2017) Arginase Is Essential for Survival of Leishmania donovani Promastigotes but Not Intracellular Amastigotes. Infect Immun 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roberts SC, Tancer MJ, Polinsky MR, Gibson KM, Heby O, Ullman B (2004) Arginase plays a pivotal role in polyamine precursor metabolism in Leishmania. Characterization of gene deletion mutants. J Biol Chem 279: 23668–23678. doi: 10.1074/jbc.M402042200 [DOI] [PubMed] [Google Scholar]
- 67.Reguera RM, Balana-Fouce R, Showalter M, Hickerson S, Beverley SM (2009) Leishmania major lacking arginase (ARG) are auxotrophic for polyamines but retain infectivity to susceptible BALB/c mice. Mol Biochem Parasitol 165: 48–56. doi: 10.1016/j.molbiopara.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaur U, Roberts SC, Dalvi RP, Corraliza I, Ullman B, Wilson ME (2007) An effect of parasite-encoded arginase on the outcome of murine cutaneous leishmaniasis. J Immunol 179: 8446–8453. [DOI] [PubMed] [Google Scholar]
- 69.Muleme HM, Reguera RM, Berard A, Azinwi R, Jia P, Okwor IB, et al. (2009) Infection with arginase-deficient Leishmania major reveals a parasite number-dependent and cytokine-independent regulation of host cellular arginase activity and disease pathogenesis. J Immunol 183: 8068–8076. doi: 10.4049/jimmunol.0803979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aoki JI, Muxel SM, Zampieri RA, Acuna SM, Fernandes JC, Vanderlinde RH, et al. (2017) L-arginine availability and arginase activity: characterization of amino acid permease 3 in Leishmania amazonensis. PLoS Negl Trop Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mandal G, Wyllie S, Singh N, Sundar S, Fairlamb AH, Chatterjee M (2007) Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology 134: 1679–1687. doi: 10.1017/S0031182007003150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adhuna A, Saltora P, Bhatnagar R (2000) Nitric oxide induced expression of stress proteins in virulent and avirulent promastigotes of Leishmania donovani. Immunol Lett 71: 171–176. [DOI] [PubMed] [Google Scholar]
- 73.Salvati L, Mattu M, Colasanti M, Scalone A, Venturini G, Gradoni L, et al. (2001) NO donors inhibit Leishmania infantum cysteine proteinase activity. Biochim Biophys Acta 1545: 357–366. [DOI] [PubMed] [Google Scholar]
- 74.Holzmuller P, Hide M, Sereno D, Lemesre JL (2006) Leishmania infantum amastigotes resistant to nitric oxide cytotoxicity: Impact on in vitro parasite developmental cycle and metabolic enzyme activities. Infect Genet Evol 6: 187–197. doi: 10.1016/j.meegid.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 75.Mauel J, Ransijn A (1997) Leishmania spp.: mechanisms of toxicity of nitrogen oxidation products. Exp Parasitol 87: 98–111. doi: 10.1006/expr.1997.4205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignment of the amino acid sequence of the nitric oxide synthase-like NOS-like of. L. amazonensis (Lam), L. mexicana (Lmx), L. donovani (Ldo), L. panamensis (Lpa), L. braziliensis (Lbr), L. major (Lmj) and L. infantum. The identical amino acids are highlighted in gray. The alignment was performed based on NCBI multiple alignment tool.
(TIF)
The DAF-FM labeling of La-WT, La-arg- and La-arg-/+ARG promastigotes and analysis were performed as follows: (1) DAF-FM labeling followed by PI (propidium iodide) staining; (2) The data were acquired using a FACSCalibur (BD) flow cytometer followed by analysis of the resulting pattern of labeling: PI-DAF-, PI-DAF+, PI+DAF- and PI+DAF+. Using FlowJo software (as described on methods section), the cells were gated based on the Forward Scatter (FSC) and the PI label was used to gate viable cells (PI-); (3) Using viable cells, DAF+ populations were selected for frequency analysis, and (4) the population producing NO was gated based on the histogram of the Mean of Fluorescence Intensity (MFI).
(TIF)
(A) The amastins LmxM.08.00760 (A), LmxM.0800800 (B) and LmxM.33.0960 (C) mRNA levels were based on quantification of the target and were normalized by gapdh expression in promastigotes (pro) and amastigotes (ama) of La-WT and La-arg-. The values are the mean ± SEM of three independent biological replicates (n = 4–6). (*) p < 0.05
(TIF)
DAF-FM labeling of La-WT, La-arg- and La-arg-/+ARG amastigotes and analysis were performed as follows: (1) DAF-FM labelling followed by PI (propidium iodide) staining; (2) The data were acquired on a FACSCalibur (BD) flow cytometer with analysis of the resulting pattern of labelling: PI-DAF-, PI-DAF+, PI+DAF- and PI+DAF+. Using FloJo software (as described on methods section), the cells were gated based on the Forward Scatter (FSC) and PI labeling was used to gate viable cells (PI-); (3) Using viable cells, DAF+ populations were selected for frequency analysis, and (4) the population producing NO was gated based on the histogram of the Mean Fluorescence Intensity (MFI).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.