Abstract
Most terrestrial plants, including crops, engage in beneficial interactions with arbuscular mycorrhizal fungi. Vital to the association is mutual recognition involving the release of diffusible signals into the rhizosphere. Previously, we identified the maize no perception 1 (nope1) mutant to be defective in early signaling. Here, we report cloning of ZmNOPE1 on the basis of synteny with rice. NOPE1 encodes a functional homolog of the Candida albicans N-acetylglucosamine (GlcNAc) transporter NGT1, and represents the first plasma membrane GlcNAc transporter identified from plants. In C. albicans, exposure to GlcNAc activates cell signaling and virulence. Similarly, in Rhizophagus irregularis treatment with rice wild type but not nope1 root exudates induced transcriptome changes associated with signaling function, suggesting a requirement of NOPE1 function for presymbiotic fungal reprogramming.
Introduction
Arbuscular Mycorrhizal (AM) symbiosis is a mutually beneficial relationship between plants and fungi in which plant roots exchange photoassimilates for fungus-delivered soil minerals. The resulting interaction may profoundly influence plant performance, in both wild and cultivated systems. For the symbiosis to begin, plant roots and AM fungi (AMF) exchange signals via secretion of diffusible compounds1, including fungal chitin-based molecules (reviewed in2,3) detected by Lysine Motif (LysM) containing plasma membrane receptor-like kinases4,5. Central to the perception of AMF in rice is the α/β hydrolase DWARF14 LIKE (D14L) and the F-box protein DWARF3 (D3)6, although, in this instance, the signal molecules remain uncharacterized.
A number of plant-derived factors are known to stimulate morphological changes in AMF, promoting fungal-host encounters1, including flavonoids that enhance hyphal tip elongation7, 2-hydroxid fatty acids (2-OH-FA) that trigger hyphal branching8, and strigolactones (SL) that induce changes in fungal metabolism, coupled with profuse hyphal ramification9,10,11. Despite their pre-symbiotic effect on AMF growth, plant SL and flavonoid biosynthetic mutants are still partially or fully colonized12,13. Once the fungus has reached the plant’s surface, cutin monomers induce hyphopodium differentiation, the anchoring structure for entry of AMF into the root epidermal cell layer14. As the fungal genome lacks genes for the de novo biosynthesis of certain fatty acids15, cutin may have an additional nutritional role.
To better understand signalling during symbiotic establishment, we analyzed the maize no perception1 (nope1) mutant, which does not form arbuscular mycorrhizal symbioses16. We identified NOPE1 to encode an N-acetylglucosamine (GlcNAc) transporter, a function not described previously in plants, but characterized in fungi17. Notably, GlcNAc has been shown to stimulate the fungal pathogen Candida albicans to undergo morphological changes and increase expression of virulence genes that promote pathogenic interactions with the host18. Our analyses provide the first evidence that a previously unknown plant GlcNAc transporter plays a role in the initiation of root colonization by AMF.
Materials & Methods
Plant and fungal material
Oryza sativa ssp. japonica cv. Donjing, Zea mays inbred W22 and Arabidopsis thaliana ecotype Col-0 were used throughout the study. R. irregularis and G. rosea spores were either axenically produced5 or purchased (Premiertech, Rivière-du-Loup, Canada; Agronutrition, Toulouse, France). Piriformospora indica, Magnaporthe oryzae and Candida albicans strains were propagated as previously described1,4,7.
Identification of rice NOPE1
The maize nope1 locus was mapped to a ~10Mb interval on chromosome 10, defined by the markers UMC1336 (86.3Mb) and Phi071 (93.7Mb; Fig. S1A). The syntenic region on rice Chromosome 4S contains AM-inducible LOC_Os04g015208 (Fig. S1A). According to full cDNA and EST information LOC_Os04g01520 consists of two exons and one intron (Fig. S1B), producing an ORF of 1404 bp. The 5’ and 3’ RACE PCR analysis indicated a transcriptional start point at −70 bp and a 184 bp 3’UTR sequence. The gene product of LOC_Os04g01520 consists of 476 residues and has a predicted molecular weight of 50.34 kD.
Identification of OsNOPE1 mutant
Two rice lines, 4A-01057 and 3A-02512, were identified from public mutant collections19 with T-DNA insertions 158 bp downstream of ATG within the first exon, and 22 bp upstream of the 3’ intron splice-junction, respectively (Fig. S1B). RT-PCR based analysis of LOC_Os04g01520 mRNA levels detected no or wild type levels in 4A-01057 and 3A-02512, respectively (Fig. S1C). Amplicon sequencing from line 4A-01057 confirmed the predicted mutation and revealed the additional presence of ~800 bp of the backbone vector (pGA2517, Fig. S1B).
Genetic complementation of Osnope1 mutant
The genomic region of LOC_Os04g01520 including 1.5kb of promoter sequence were amplified and cloned into pGEM-T Easy (Promega, Dübendorf, Switzerland) to generate pRS909. The NOS terminator sequence was amplified with primers RS976 and RS977 (Supplemental Table S4) and inserted into the NdeI and SacI (Promega, Dübendorf, Switzerland) site of pRS909 to generate pRS934. Finally, pRS934 was digested with BglII and SacI (Promega, Dübendorf, Switzerland) and the resulting segment gel-isolated and cloned into binary vector pTF101.114 to generate pRS936. Stable rice transformation was performed as previously reported17.
Rice GlcNAc transport assays
Rice seedlings were grown in ½ MS medium (described in supplementary information) and transferred to equilibration solution, containing 10 mM MES-KOH at pH 6; 1 mM CaCl2 (ES) for 1h. Four seedlings were pooled per sample and 3 replicates per genotype were used. Seedlings were moved to incubation solution (IS), consisting of ES; 3kBq/ml [3H]GlcNAc (American Radiolabeled Chemicals, Saint Louis, MO). GlcNAc concentration was adjusted to 100 µM with cold GlcNAc (Sigma-Aldrich, Dorset, UK). For uptake experiments, roots were washed twice for 30 s with ice-cold washing solution (WS): ES; 100 µM GlcNAc (cold). For efflux experiments, roots were left for 2 h in IS, washed twice for 30 s with WS and transferred to a solution either lacking GlcNAc: ES, or supplemented with 50× GlcNAc: ES; 5 mM GlcNAc and incubated as indicated in results. To quantify [3H]GlcNAc content, roots were excised and placed in vials containing 3 ml of 0.1 N HCl for 1 h. Of the extracted fluid 1.6 ml was collected and [3H]GlcNAc was quantified by scintillation counting.
Genetic complementation of Candida albicans
Growth of C. albicans strains was examined by spotting a 10-fold cell dilution onto agar medium containing Yeast Nitrogen Base minimal medium and 50 mM of GlcNAc, glucose, or galactose (ThermoFisher Scientific, Grand Island, USA). Plates were incubated at 30°C for 2 days and then photographed. Induction of hyphal morphogenesis was examined by growing cells overnight at 37°C in minimal medium containing glucose, then resuspending them at 106 cells/ml in medium containing either 50 mM glucose or 50 mM GlcNAc and incubation for 2 h before documentation. The results were reproducible with different colonies and isolates obtained from two independent transformations.
Root exudate and GlcNAc treatment of Rhizophagus irregularis
Sampling of rice root exudates included five six-week old plants from sand-cultivated wild type and Osnope1 genotypes. The roots were well-washed, transferred to individual 1 l Erlenmeyer flasks containing ~750 ml of ½ HL (50 µM KH2PO4) solution and incubated with gentle agitation. After 3 days, exudates were harvested, sterilized using 0.2 µm filters (Sartorius Epsum, Surrey, UK) and immediately used. Groups of 80’000 spores of R. irregularis per replicate were germinated at 2% [CO2] and 30° C for 7 days in 8 ml of R. irregularis minimal medium5. Pre-germination solutions were replaced with 8 ml of wild type (Donjing) or Osnope1 rice exudates, or with 8 ml of 50 mM GlcNAc. Fungal material was collected at 0, 1 h, 24 h, and 7 d post treatment.
RNAseq sequencing and data analysis
Fungal transcriptome analysis was performed on three independently grown replicates. Please see supplementary information for detailed description of nucleic acid handling and library preparation. RNAseq sequencing involved the Illumina HiSeq2000 (Illumina Inc., San Diego, USA) using a 2 × 100 bp pair-end strategy with Illumina TruSeq SBS sequencing kits v3 (PN FC-401-3001, HiSeq2000). Sequencing was performed at the GeT (Genome & Transcriptome Core facilities, Toulouse, France, http://get.genotoul.fr/). RNAseq reads have been released at NCBI Gene Expression Omnibus (accession n° GSE65595).
Reads were mapped to the Gloin1 assembly21 to define transcript accumulation patterns by RNA-seq functionality of the CLC Genomic Workbench suite. Significantly differentially expressed genes were identified by calculating RPKM (Reads Per Kilobase of exon per Million fragments mapped) and proportion-based test statistics22 with a False Discovery Rate FDR23 correction for multiple testing (settings: minimum mapped read length fraction = 0.95; minimum similarity = 0.98). According to CLC recommendations, genes were significantly upregulated when meeting the requirements of "total difference reads mapped" >10, RPKM fold change >2 and FDR corrected p <0.05. Gene set enrichment involved the unconditional GOstats test of Falcon and Gentleman24 based on Gloin1 annotation available at http://genome.jgi.doe.gov/cgi-bin/ToGo?accession=all&species=Gloin1. The "p-value" corresponded to the tail probability of the hyper geometric distribution.
Data availability
R. irregularis RNAseq reads have been released at Gene Expression Omnibus (accession n° GSE65595). All other datasets (Supplemental Tables S1–S3) are included in this published article. C. albicans strains used in this study are available from J.B.K. (james.konopka@stonybrook.edu). All other data that support the findings of this study are available from the corresponding author upon request.
Results
Cloning of maize ZmNope1 on the basis of synteny with rice
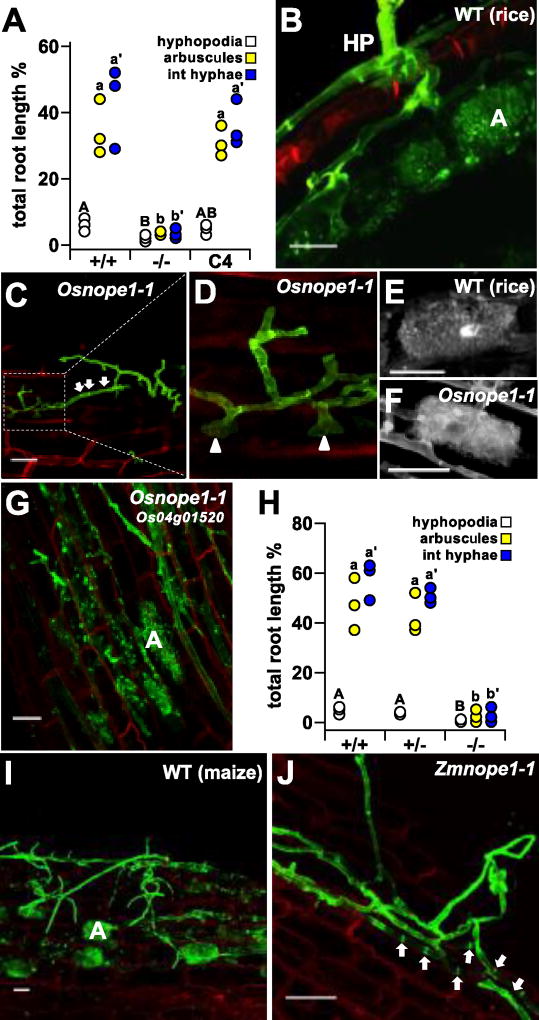
The maize Zmnope1 mutant is unable to establish AM symbioses16. Limited physical interaction of Zmnope1 with AMF suggested a failure in pre-symbiotic signal exchange. As genetic mapping had linked the Nope1 locus to the marker UMC1336 on chromosome 10, we searched for rice candidate genes exhibiting a transcriptional response to mycorrhizal root colonization located in the region syntenic to maize nope119, 20, identifying the gene LOC_Os04g0152019 (Fig. S1A). To investigate a potential role in AM symbiosis, rice plants segregating for a T-DNA insertion in LOC_Os04g01520 (4A-01057; Fig. S1B, C) were cocultivated with R. irregularis. A significant reduction in fungal root colonization was observed in plants homozygous for the insertion (p ≤ 0.05 for all structures tested; Fig. 1A). On the surface of mutant but not wild type roots, the fungus formed aberrant hyphopodia (Fig. 1B–C). Closer inspection of aberrant hyphopodia revealed multiple unsuccessful penetration attempts (Fig. 1D, arrowheads) and extensive hyphal septation, a sign of fungal stress (Fig. 1C, arrows). Infrequently, the fungus did succeed in invading the root cortex, and produced arbuscules that were of wild type morphology (Fig. 1E–F). To confirm that the phenotype was indeed linked to disruption of LOC_Os04g01520, we reintroduced a wild type copy of the gene under the native promoter, and observed a restoration of wild-type levels of AM fungal colonization (Fig. 1A, G; Fig. S2). In addition, transcript levels of previously described AM marker genes21 were reduced in Osnope1 homozygous plants, but restored to wild type levels in complemented lines (Fig. S4). The quantitative and qualitative phenotype of the rice insertion mutant was equivalent to that of the reported maize Zmnope1 mutant16 and, consequently, LOC_Os04g01520 was designated OsNOPE1, and the insertion 4A-01057, Osnope1-1. To investigate whether NOPE1 is required for susceptibility to different fungi known to invade rice roots21,22, wild type and Osnope1 mutants were inoculated with Piriformospora indica and Magnaporthe oryzae. Both fungi invaded mutant and wild type root tissue equally well (Fig. S3), suggesting that NOPE1 might be required specifically for interaction with AMF.
Figure 1. Mutation of the orthologous genes Os04g01520 and GRMZM2G176737 disrupted coloniztion by R. irregularis in rice and maize, respectively.
A, Percentage root length colonization in rice plants (n = 3) segregating Osnope1 (+/+, wild type, −/− homozygous Osnope1, C complemented line C4) at 6 weeks post infection (wpi). Means groups assigned for each fungal structure indicated by letters (adj. P < 0.05). B–G, WGA-staining of fungal structures and propidium-iodide counterstained plant cell walls of rice roots inoculated with R. irregularis at 6 wpi as examined by laser scanning confocal microscopy. B, Hyphopodium (HP) and arbuscule differentiation in a wild type (WT) root C, Misshapen and highly septate (arrows) hyphopodial hypha on the surface of the root of a Osnope1 homozygote. D, Detail of a hyphopodia on a Osnope1 homozygous plant, showing several aborted penetration attempts (arrowheads). Morphologically equivalent arbuscules formed in the roots of wild type (E) and Osnope1 homozygous (F) plants. G, Arbuscules formed in root cortical cells of the complemented line C4. H, Percentage R. irregularis root length colonization of maize plants segregating Zmnope1 (+/+, wild type, +/−, heterozygote, −/− homozygous mutant) at 6 wpi. Represented as A. I and J, WGA-staining of fungal structures and propidium-iodide counterstained plant cell walls of maize roots inoculated with R. irregularis at 6 wpi, as examined by laser scanning confocal microscopy. On wild type (I) roots the fungus develops normal hyphopodia and extensively colonizes the root forming frequent arbuscules. J, Misshapen hyphopodium on the roots of a nope1-1 homozygous mutant showing mutiple septa (arrows) and absence of root pentration. HP, hyphopodium; A, arbuscule. Scale bar = 50µm.
Mutation of maize ZmNope1 results in a phenotype equivalent to that of Osnope1-1
The maize nope1 mutant arose in a population with high levels of Mutator transposon activity16, and suppression of the phenotype of the original allele prevented further direct characterization of the maize nope1 mutation. Instead, we undertook a reverse genetics approach to verify the role of the maize homologue of OsNOPE1 in AM symbiosis. The maize genome contains a single gene, GRMZM2G176737, showing high similarity to OsNOPE1 (BLASTP; score=488, ID=83%, e-value=8.2×e−61), located on Chromosome 10 near to the mapped position of the nope1 mutation (Fig. S1A). A Dissociation (Ds) transposon linked to GRMZM2G176737 was identified and re-mobilized23,24, generating a novel insertion (Zmnope1-1) within the first exon, that resulted in an absence of transcript accumulation (Fig. S5A, B). Quantification of intraradical fungal structures in Zmnope1-1 homozygous plants revealed significantly lower fungal colonization (p < 0.05 for all structures tested) compared with wild type plants (Fig. 1H). Despite fungal proliferation on the root surface, hyphopodia were malformed and failed to penetrate (Fig. 1H–J). In addition, transcript levels of the maize homologues of the rice AM marker genes OsAM321 and OsPT1125, GRMZM2G135244 (ZmAM3) and GRMZM5G881088 (ZmPT6)26, were lower in inoculated roots homozygous for the transposon insertion as compared to hemizygous and wild-type siblings (Fig. S5C). The syntenic genetic location and equivalent loss-of-function phenotypes of rice and maize NOPE1 genes strongly suggests that they are orthologous, and that this maize gene was mutated in the previously reported nope1 mutant16.
NOPE1 belongs to the Major Facilitator Superfamily and is found in all land plants
Analysis of the predicted NOPE1 protein (http://phobius.binf.ku.dk/; http://smart.embl-heidelberg.de/) identified 12 transmembrane domains, no signal peptide and a domain of unknown function (DUF895) between amino acids 49 and 181 (Fig. S6). NOPE1 was identified as member of the Major Facilitator Superfamily (MFS, Pfam e-value 4.9e−13), suggesting a role in transport of small molecules across membranes. BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) indicated genes encoding NOPE1 to be present in the genomes of all land plants for which data was available, including non-mycorrhizal plant species (Fig. S6). The genome of Medicago truncatula contains two NOPE1 orthologs; Medtr3g093270 and Medtr3g093290; the gene products share 63% identity (79% similarity) and 64% identity (81% similarity) with OsNOPE1, respectively, providing an explanation for the absence of NOPE1-associated AM phenotypes from forward genetic screens in legumes. The genome of Arabidopsis thaliana, a non-host for AMF, also contains two NOPE1 orthologs (At1g18000 and At1g18010), expressed constitutively throughout the plant (https://bar.utoronto.ca/eplant/27). To determine the possible role of NOPE1 in Arabidopsis, RNAi-based silencing was used to down-regulate both genes simultaneously (Fig. S7), but we did not detect any phenotypic effect under standard conditions, with respect to germination, morphology, flowering time or seed set.
Rice NOPE1 mediates N-acetylglucosamine transport in Candida albicans
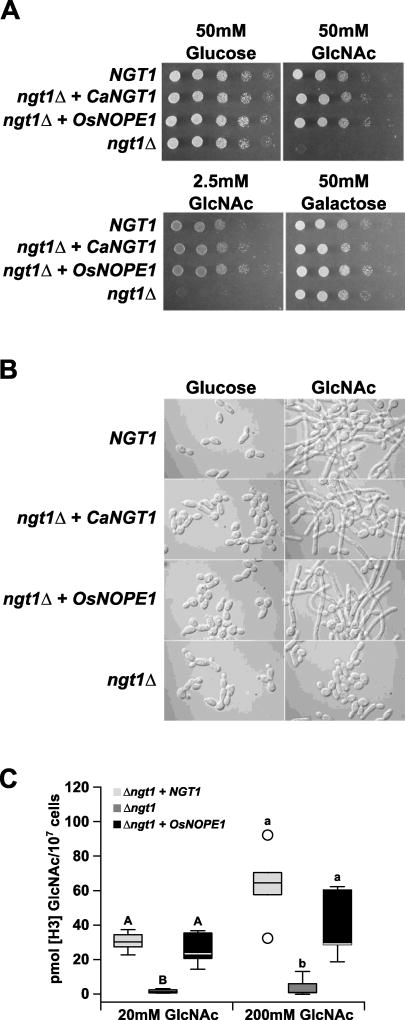
To investigate the mechanism by which NOPE1 influences AM symbioses, we sought to identify putative homologs with functional annotation. As no land plant orthologs had been characterized, we focused on the protein Ngt1 of the human pathogenic fungus Candida albicans. Ngt1 shows 41% identity at the protein level with OsNOPE1, and the two proteins are reciprocal best hits in a BLAST search. Ngt1 mediates N-acetylglucosamine (GlcNAc) transport across the plasma membrane of C. albicans, enabling growth when GlcNAc is the sole carbon source28. A key feature of C. albicans virulence is the ability to reversibly shift from isotropic budding to polarized filamentous growth in response to environmental signals29. Amongst the stimuli triggering this morphological switch is GlcNAc30. Deletion of the NGT1 gene impairs GlcNAc uptake, preventing cells from switching morphology and from proliferating on GlcNAc-containing medium17,31. To address whether OsNOPE1 is a functional GlcNAc transporter, a C. albicans codon-optimized version of the rice OsNOPE1, driven by the native fungal NGT1 promoter, was transformed into the C. albicans mutant ngt1Δ. Remarkably, introduction of rice OsNOPE1 restored growth and induction of filamentous hyphal differentiation on GlcNAc medium (Fig. 2A–C), indicating functional conservation of the protein across plant and fungal kingdoms.
Figure 2. Rice NOPE1 mediates GlcNAc transport in C. albicans.
A, Ten-fold cell dilution series of C. albicans strains spotted onto plates with indicated sugar. B, C. albicans strains grown overnight in glucose containing medium and resuspended in fresh medium containing either 50 mM glucose or 50 mM GlcNAc. C, [H3]GlcNAc uptake in C. albicans strains at 20 mM and 200 mM GlcNAc. GlcNAc, N-acetylglucosamine. Boxes show 1st quartile, median and 3rd quartile. Whiskers extend to the most extreme points within 1.5× box length; outlying values beyond this range are shown as unfilled circles. Means groups were calculated post hoc independently for the two GlcNAc treatments and are indicated by letters (p < 0.05). For description of strains see Supplemental Information Table S5.
To investigate substrate specificity of OsNOPE1, competition assays were performed in which an excess of cold hexoses was provided together with [3H]GlcNAc. Control studies showed that addition of a 2-fold excess of cold GlcNAc led to a partial decline in the uptake of radioactive GlcNAc but nearly complete inhibition at 20-fold excess cold GlcNAc (Fig. S8). OsNOPE1 showed strong specificity for transporting GlcNAc, since a 200-fold excess of glucosamine, dextrose, fructose or galactose did not significantly impact on the amount of [3H]GlcNAc transported into the cells. A 200-fold excess of N-acetylmannosamine partially competed with [3H]GlcNAc (p < 0.01 by non-parametric one-way ANOVA), suggesting that the N-acetyl moiety may be important for substrate specificity. Overall, these results demonstrated that OsNOPE1 exhibits a high specificity for transporting GlcNAc, similar to C. albicans Ngt1.
NOPE1 mediates GlcNAc transport in plants
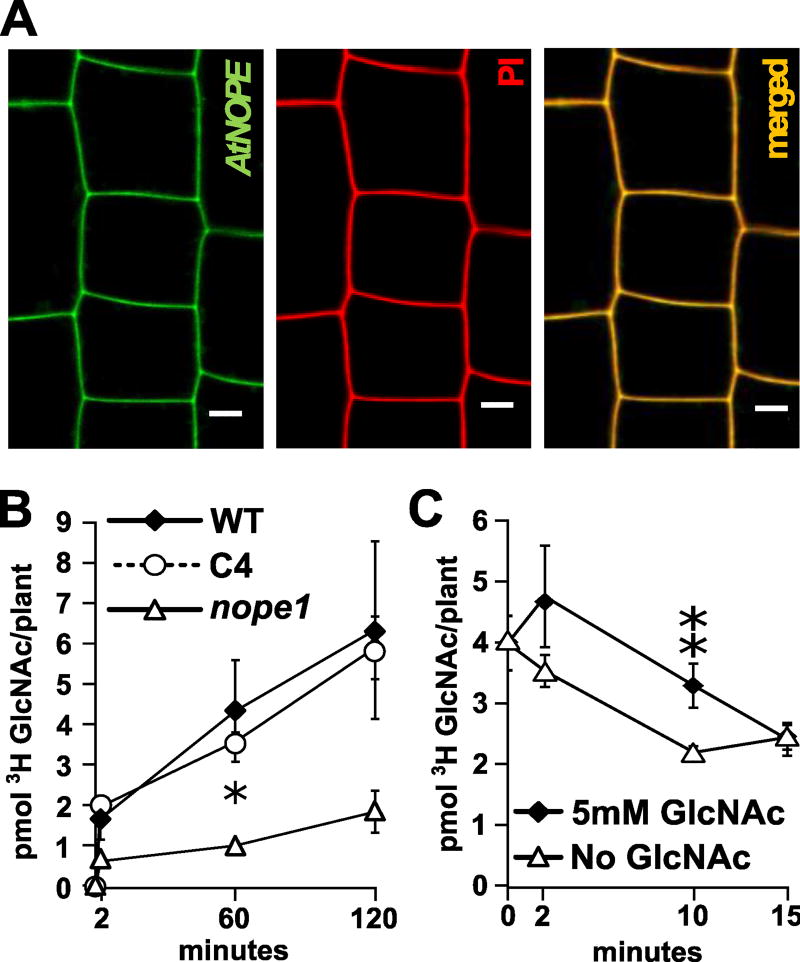
Heterologous expression in C. albicans suggested that NOPE1 may mediate GlcNAc transport across plasma membranes also in plants. We characterized subcellular localization of the Arabidopsis NOPE1 protein At1g1800 by stable, constitutive expression of an in-frame fusion to yellow fluorescent protein (YFP). Three independent transgenic lines showed a reproducible and clear signal consistent with plasma membrane localization (Figure 3A).
Figure 3. NOPE1 mediates GlcNAc transport in rice and Arabidopsis.
A, Laser scanning confocal microscopy of A. thaliana roots expressing Ubqprom::YPF::AtNOPE1. Three independent lines were analyzed to determine reproducible localization patterns, here shown for line At4731y-4 with the YFP-AtNOPE1 signal shown in green (left). Corresponding cells stained with Propidium Iodide (PI) shown in red (centre). Overlay (yellow, right). Scale bar: 5 µm. B, Time course of [3H]GlcNAc uptake in roots of Osnope1, wild type and genetically complemented mutant line C4. Means and SEs of three biologically independent experiments are shown (*P ≤ 0.05). Please note that the 0.5min value corresponds to unspecific adsorption of medium to the protoplasts. C, Time course of [3H]GlcNAc export activity of wild type rice roots at 0 and 5mM (50×) GlcNAc external concentration. Means and SEs of three biological replicates are shown. (**p ≤ 0.01).
To explore the capacity of OsNOPE1 to transport GlcNAc in planta, we measured [3H]GlcNAc root uptake in wild type, Osnope1 and complemented mutant rice seedlings. The rate of [3H]GlcNAc was significantly (p < 0.05) reduced in Osnope1 mutants compared with wild type or complemented plants (WT: 2.37 pmol h-1; C4: 2.00 pmol h-1; Osnope1: 0.624 pmol h-1; Fig. 3B). To test if NOPE1 supported transport of GlcNAc across cell membranes of non-mycorrhizal plants, we measured the [3H]GlcNAc uptake in protoplasts derived from the Arabidopsis At1g1800 overexpression line At4731y-4 and compared this uptake to that of the RNAi-silencing line AtMNC58. Uptake of [3H]GlcNAc was significantly (p < 0.01) higher in the overexpression line At4731y-4 compared to the RNAi-silenced line AtMNC58 (Fig. S10). Taken together, NOPE1 mediated GlcNAc uptake in whole roots and leaf protoplasts of mycorrhizal and non-mycorrhizal plant species, respectively.
We quantified GlcNAc efflux in the rice seedling system by first loading roots of wild type rice seedlings with 100 µM [3H]GlcNAc and then transferring them to a solution containing either unlabeled GlcNAc at 50× concentration or no GlcNAc. Monitoring the levels of root-retained [3H]GlcNAc revealed that in both cases GlcNAc was released from the roots, but that significantly less (p < 0.05) remained in the roots at shorter times when the external medium contained no GlcNAc (2.21 ± 0.10 [3H]GlcNAc/plant) as opposed to high GlcNAc medium (3.30 ± 0.36 [3H]GlcNAc/plant, Fig. 3C), indicating that substrate availability at the external side partially inhibited efflux.
Distinct transcriptional responses of R. irregularis to rice wild type and nope1 root exudates
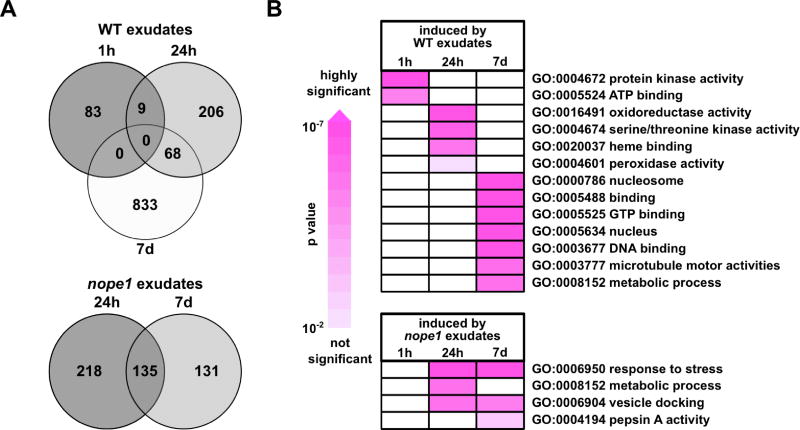
To characterize fungal responses to root-released compounds, RNAseq analysis was performed on pre-germinated R. irregularis spores exposed to either rice wild type or Osnope1 root exudates for 1h, 24h and 7 days. Interestingly, exposure to wild-type and Osnope1 exudates led to distinct expression profiles across the three time points. Treatment with wild-type exudates gradually enhanced the number of induced transcripts from 92 to 283 to 901 at 1 h, 24 h and 7 d (Fig. 4A). In contrast, treatment with Osnope1 root exudates did not result in a significant change in transcript accumulation patterns until 24 h, and then fewer genes were responsive (343 and 256 induced genes at 24 h and 7 d, respectively; Fig. 4A). In the wild type treatment, d.e. genes at 1 h were enriched for the gene ontology (GO) terms protein kinase and ATPase activity (Fig. 4B) suggesting an early induction of fungal signaling activities. At 24 h, both wild type and mutant exudates led to a significant change in the fungal transcriptome, however GO analysis suggested that while the fungus switched to an elevated oxidative status in response to wild-type exudates, mutant exudates induced stress responses. By 7 days, the stress response signature remained for the mutant exudate treatment, while d.e. genes with wild-type exudate treatment were enriched with GO terms corresponding to a higher energetic and metabolic status (Fig. 4B). Collectively, these data are consistent with an early and transient activation of fungal signaling, followed by the activation of genes involved in primary metabolism in wild type, but not Osnope1, root exudate treated fungus. We assayed also fungal growth responses, treating Gigaspora rosea with exudates from wild type and Osnope1, but no difference was observed in the number of hyphal apices and overall hyphal growth between treatments (Fig. S10), assigning the NOPE1 associated compound to a different functional class than the previously characterized SLs, 2-OH fatty acids or flavonoids, all of which trigger specific hyphal growth patterns.
Figure 4. R. irregularis transcriptional response to root exudates from rice wild type and Osnope1 mutant plants.
A, Venn diagrams indicating number of significantly induced fungal genes (P ≤ 0.05, one way ANOVA) in response to treatment with exudates from wild type relative to Osnope (top) and Osnope relative to wild type (bottom). B, Time-resolved Gene Ontology analysis for Biological Process terms (p ≤0.01) for fungal genes induced when treated with root exudates from wild type (top) or from Osnope1 (bottom). The colour code indicates the significance of gene enrichment (p-value).
RNAseq results were validated by qRT-PCR analysis (Fig. S11). The R. irregularis NGT1 homologue MIX9501_16_76,38 displayed a basal expression level but was not induced in response to treatment with either exudates or by GlcNAc treatments (Fig. S12). The specific fungal transcriptional response to rice wild type root exudates is consistent with the hypothesis that the NOPE1-mediated release of GlcNAc is required by R. irregularis for adequate reprogramming prior to host colonization. However, application of GlcNAc to R. irregularis-inoculated Osnope1 mutant plants for seven weeks at 1 mM, 10 mM or 100 mM GlcNAc did not complement the mutant phenotype. This may be due to application of GlcNAc outside the biologically active concentration, or that either the development of a GlcNAc gradient or efflux of a GlcNAc-conjugate might be necessary for stimulating the fungus. We examined whether the presence of total wild type exudates would restore AM colonization of the mutant when cocultivated within the same container, and, indeed, Osnope1 was fully colonized when grown together with wild type but not with mutant ‘donor’ plants (Fig. S13).
Discussion
NOPE1 is a plasma membrane GlcNAc transporter required for the initiation of AM symbiosis in both rice and maize. Current knowledge of plant-derived signals in AM symbiosis is largely limited to the stimulatory effects of SLs on fungal metabolism and development, although it has been anticipated that additionally secreted bioactive molecules are necessary for establishment of the symbiosis (34, for review see1). Wild-type, but not Osnope1, root exudates induced signaling-associated transcriptional responses in R. irregularis within the first hour of treatment, suggesting a priming of the fungus prior to establishment of AM symbiosis. The restoration of normal levels of colonization in Osnope1 by co-cultivation with wild-type plants further indicated bioactive molecules, essential for successful establishment AM symbiosis, to be absent from Osnope1 exudates. We hypothesize that a NOPE1 transported factor or factors act non-redundantly with other previously characterized plant-derived signals to prime the fungus, prior to establishment of AM symbiosis (see model Fig. S14).
NOPE1 mediates efficient GlcNAc import in C. albicans, rice and Arabidopsis. GlcNAc is commonly found in microbial environments, as a building block of fungal and bacterial cell walls, or as a signaling molecule, such as in rhizobial nod-factors, and GlcNAc transporter activities have been characterized in a number of microbial systems. NOPE1, however, represents, to our knowledge, the first plasma membrane GlcNAc transporter to be described in plants. Wild-type rice roots were shown to acquire and release GlcNAc, with uptake clearly dependent on NOPE1, although the role of NOPE1 in GlcNAc efflux was less clear. It has been shown previously that GlcNAc monomers are abundant in the leaves of Arabidopsis35. Although not directly quantified in roots, blocking the biosynthesis of the activated substrate for GlcNAc transfer, UDP-GlcNAc, in rice impaired cell expansion in the root elongation zone, leading to a short root phenotype, indicating an important role for GlcNAc metabolism in normal root development36.
Host-secreted GlcNAc is known to act as a potent signaling molecule for a number of microbial organisms, including the facultative human pathogenic fungus C. albicans18. Exposure to GlcNAc leads to the induction of invasive hyphal growth and the expression of virulence genes such as the adhesins that facilitate attachment to host cells (for review see28). Also, the thermally dimorphic pathogenic fungi Histoplasma capsulatum and Blastomyces dermatitidis respond to treatment with GlcNAc by a similar yeast-to-filament switch37. In these facultative human pathogens GlcNAc additionally functions as a source of sugar. The utilization of plant-derived GlcNAc as a substrate was recently reported for the plant-pathogenic bacteria Xanthomonas campestris pv. campestris while infecting leaves of Brassica oleracea38. The closely related vector-borne phytopathogenic bacterium Xylella fastidiosa enzymatically digests GlcNAc polymers (chitin) available in the foregut of the insect vector, also using the acquired GlcNAc as a nutrient source39. Remarkably, the effects of host-provided GlcNAc extend to the mutualistic association between bioluminescent squid and vibrio, where host-derived GlcNAc acts as a regulator of shifting the bacterial metabolism to provide optimal symbiont services to the host40. Determining the chemical identity of the bioactive molecule associated with NOPE1 remains an exciting future challenge, and whether NOPE1 contributes to microbe perception or influences root exudate composition requires further investigation.
Supplementary Material
Acknowledgments
We kindly thank Jacqueline Gheyselinck and Anne Bates for their technical assistance. We are grateful to John Arbuckle (DuPont/Pioneer) for helping with mapping the maize nope1 mutant and Sam Brockington for guidance with advanced BLAST searches. Research in the U.P. laboratories was supported by the Swiss National Science Foundation grants 3100A0-104132, PP00A-110874, PP00P3-130704 and by the Gatsby Charitable Foundation grant RG60824. S.N and J.B.K were supported by a grant from the National Institutes of Health (R01GM116048).
Footnotes
Author contribution: M.N., R.S., N.G., E.M., J.B.K., T.P.B. and U.P. designed the experiments. M.N., R.S., S.N., B.B., C.K., A.S., G.A., K.R.A., A.R., C.G., and C.R., performed the experiments. C.R. performed bioinformatics and statistical analyses of the RNAseq data. M.N., R.S., C.R., E.M., J.B.K. and U.P. wrote the manuscript.
The authors declare no conflict of interest.
References
- 1.Nadal M, Paszkowski U. Polyphony in the rhizosphere: presymbiotic communication in arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 2013;16:473–479. doi: 10.1016/j.pbi.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C, Oldroyd GE. Plant signalling in symbiosis and immunity. Nature. 2017;543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 3.Gutjahr C, Parniske M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annual review of cell and developmental biology. 2013;29:593–617. doi: 10.1146/annurev-cellbio-101512-122413. [DOI] [PubMed] [Google Scholar]
- 4.Miyata K, et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- 6.Gutjahr C, et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science. 2015;350:1521–1524. doi: 10.1126/science.aac9715. [DOI] [PubMed] [Google Scholar]
- 7.Bécard G, Douds DD, Pfeffer PE. Extensive In Vitro Hyphal Growth of Vesicular-Arbuscular Mycorrhizal Fungi in the Presence of CO(2) and Flavonols. Appl Environ Microbiol. 1992;58:821–825. doi: 10.1128/aem.58.3.821-825.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagahashi G, Douds DD., Jr The effects of hydroxy fatty acids on the hyphal branching of germinated spores of AM fungi. Fungal biology. 2011;115:351–358. doi: 10.1016/j.funbio.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 10.Besserer A, Becard G, Jauneau A, Roux C, Sejalon-Delmas N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 2008;148:402–413. doi: 10.1104/pp.108.121400. doi:pp.108.121400 [pii]10.1104/pp.108.121400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besserer A, et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006;4:e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. doi:nature07271 [pii] 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 13.Bécard G, Taylor L, Jr, D D, Pfeffer P, Doner L. Flavonoids are not necessary plant signals in arbuscular mycorrhizal symbiosis. Molecular Plant Microbe Interaction. 1995;8:252–258. [Google Scholar]
- 14.Wang E, et al. A Common Signaling Process that Promotes Mycorrhizal and Oomycete Colonization of Plants. Curr Biol. 2012;22:2242–2246. doi: 10.1016/j.cub.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 15.Wewer V, Brands M, Dormann P. Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus Rhizophagus irregularis during mycorrhization of Lotus japonicus. Plant J. 2014;79:398–412. doi: 10.1111/tpj.12566. [DOI] [PubMed] [Google Scholar]
- 16.Paszkowski U, Jakovleva L, Boller T. Maize mutants affected at distinct stages of the arbuscular mycorrhizal symbiosis. Plant J. 2006;47:165–173. doi: 10.1111/j.1365-313X.2006.02785.x. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Molecular biology of the cell. 2007;18:965–975. doi: 10.1091/mbc.E06-10-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naseem S, Konopka JB. N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens. PLoS pathogens. 2015;11:e1004947. doi: 10.1371/journal.ppat.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong DH, et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 20.Güimil S, et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci. 2005;102:8066–8070. doi: 10.1073/pnas.0502999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutjahr C, et al. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 2008;20:2989–3005. doi: 10.1105/tpc.108.062414. doi:tpc.108.062414 [pii] 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcel S, Sawers R, Oakeley E, Angliker H, Paszkowski U. Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell. 2010;22:3177–3187. doi: 10.1105/tpc.110.078048. doi:tpc.110.078048 [pii] 10.1105/tpc.110.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahern KR, et al. Regional mutagenesis using Dissociation in maize. Methods. 2009;49:248–254. doi: 10.1016/j.ymeth.2009.04.009. doi:S1046-2023(09)00092-9 [pii] 10.1016/j.ymeth.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Vollbrecht E, et al. Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell. 2010;22:1667–1685. doi: 10.1105/tpc.109.073452. doi:tpc.109.073452 [pii] 10.1105/tpc.109.073452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paszkowski U, Kroken S, Roux C, Briggs S. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci. 2002;99:13324–13329. doi: 10.1073/pnas.202474599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy R, et al. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.) Plant Biol. 2006;8:186–197. doi: 10.1055/s-2005-873052. [DOI] [PubMed] [Google Scholar]
- 27.Winter D, et al. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konopka JB. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo) 2012;2012 doi: 10.6064/2012/489208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteway M, Oberholzer U. Candida morphogenesis and host-pathogen interactions. Current opinion in microbiology. 2004;7:350–357. doi: 10.1016/j.mib.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature. 1974;250:344–346. doi: 10.1038/250344a0. [DOI] [PubMed] [Google Scholar]
- 31.Naseem S, Gunasekera A, Araya E, Konopka JB. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. The Journal of biological chemistry. 2011;286:28671–28680. doi: 10.1074/jbc.M111.249854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada K, et al. Monosaccharide absorption activity of Arabidopsis roots depends on expression profiles of transporter genes under high salinity conditions. The Journal of biological chemistry. 2011;286:43577–43586. doi: 10.1074/jbc.M111.269712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobae Y, et al. Up-regulation of genes involved in N-acetylglucosamine uptake and metabolism suggests a recycling mode of chitin in intraradical mycelium of arbuscular mycorrhizal fungi. Mycorrhiza. 2015;25:411–417. doi: 10.1007/s00572-014-0623-2. [DOI] [PubMed] [Google Scholar]
- 34.Gadkar V, et al. Root exudate of pmi tomato mutant M161 reduces AM fungal proliferation in vitro. FEMS microbiology letters. 2003;223:193–198. doi: 10.1016/S0378-1097(03)00357-4. [DOI] [PubMed] [Google Scholar]
- 35.Vanholme B, et al. Accumulation of N-acetylglucosamine oligomers in the plant cell wall affects plant architecture in a dose-dependent and conditional manner. Plant Physiol. 2014;165:290–308. doi: 10.1104/pp.113.233742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H, et al. A novel short-root gene encodes a glucosamine-6-phosphate acetyltransferase required for maintaining normal root cell shape in rice. Plant Physiol. 2005;138:232–242. doi: 10.1104/pp.104.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilmore SA, Naseem S, Konopka JB, Sil A. N-acetylglucosamine (GlcNAc) triggers a rapid, temperature-responsive morphogenetic program in thermally dimorphic fungi. PLoS genetics. 2013;9:e1003799. doi: 10.1371/journal.pgen.1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulanger A, et al. The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. MBio. 2014;5:e01527–01514. doi: 10.1128/mBio.01527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Killiny N, Prado SS, Almeida RP. Chitin utilization by the insect-transmitted bacterium Xylella fastidiosa. Appl Environ Microbiol. 2010;76:6134–6140. doi: 10.1128/AEM.01036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan M, Schwartzman JA, Dunn AK, Lu Z, Ruby EG. A Single Host-Derived Glycan Impacts Key Regulatory Nodes of Symbiont Metabolism in a Coevolved Mutualism. MBio. 2015;6:e00811. doi: 10.1128/mBio.00811-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
R. irregularis RNAseq reads have been released at Gene Expression Omnibus (accession n° GSE65595). All other datasets (Supplemental Tables S1–S3) are included in this published article. C. albicans strains used in this study are available from J.B.K. (james.konopka@stonybrook.edu). All other data that support the findings of this study are available from the corresponding author upon request.