Abstract
Binding of the retroviral structural protein Gag to the cellular plasma membrane is mediated by the protein’s matrix (MA) domain. Prominent among MA-PM interactions is electrostatic attraction between the positively charged MA domain and the negatively charged plasma membrane inner leaflet. Previously, we reported that membrane association of HIV-1 Gag, as well as purified Rous sarcoma virus (RSV) MA and Gag, depends strongly on the presence of acidic lipids and is enhanced by cholesterol (Chol). The mechanism underlying this enhancement was unclear. Here, using a broad set of in vitro and in silico techniques we addressed molecular mechanisms of association between RSV MA and model membranes, and investigated how Chol enhances this association. In neutron scattering experiments with liposomes in the presence or absence of Chol, MA preferentially interacted with preexisting POPS-rich clusters formed by nonideal lipid mixing, binding peripherally to the lipid headgroups with minimal perturbation to the bilayer structure. Molecular dynamics simulations showed a stronger MA-bilayer interaction in the presence of Chol, and a large Chol-driven increase in lipid packing and membrane surface charge density. Although in vitro MA-liposome association is influenced by disparate variables, including ionic strength and concentrations of Chol and charged lipids, continuum electrostatic theory revealed an underlying dependence on membrane surface potential. Together, these results conclusively show that Chol affects RSV MA-membrane association by making the electrostatic potential at the membrane surface more negative, while decreasing the penalty for lipid headgroup desolvation. The presented approach can be applied to other viral and nonviral proteins.
Introduction
For HIV and Rous sarcoma virus (RSV), formation of an infectious form of the virus requires interaction of the viral structural protein Gag with the acidic inner leaflet of the plasma membrane (PM). The resulting lateral Gag-Gag interactions lead to the formation of a Gag lattice and subsequently a budding virus particle. The N-terminal matrix (MA) domain of Gag mediates interaction with the PM by responding to multiple signals, including electrostatic and hydrophobic interactions, and in some cases, specific interaction with lipid headgroups (1). Cholesterol (Chol), an abundant PM lipid, is a critical component of the viral envelope (2). Compared with the PM, the viral envelope is enriched in sphingomyelin and anionic lipids, including phosphatidylserine (PS) and phosphatidyl(4,5)-inositol bisphosphate (PI(4,5)P2) (3, 4, 5). Moreover, viral budding sites colocalize with tetraspanin-rich domains (6). These data suggest that Gag selects or modulates compositionally distinct domains in the PM.
Neutron reflectometry (NR) has shown that for HIV and RSV Gag, the highly basic region of MA is oriented toward the membrane surface (7, 8), and that the Gag conformation changes upon addition of a nucleic acid (9). One drawback of NR is the requirement of a supported lipid bilayer that can influence lipid diffusion (10) and mixing behavior (11). Moreover, NR is primarily sensitive to the time-averaged matter distribution in the direction perpendicular to the bilayer plane, whereas in-plane structural information is much more difficult to access. However, a related technique, small-angle neutron scattering (SANS), can provide information about both in-plane and out-of-plane bilayer structure. Because neutrons are scattered differently by protium (1H) and hydrogen’s stable isotope deuterium (2H), mixtures of protiated and deuterated lipids generate a strong in-plane contrast upon clustering or phase separation, resulting in a distinct SANS signature (12), thus providing a unique tool to probe lipid lateral organization (13). SANS can also easily be applied to liposomes of the type used in biochemical or nuclear magnetic resonance (NMR) analyses of protein-membrane interaction (14, 15).
Complementing the time-averaged structural data from SANS, molecular dynamics (MD) simulations provide atomistic detail, allowing a focused examination of different protein binding modes and their dependence on membrane structure. For example, a coarse-grained MD study found that HIV MA can sequester multivalent but not monovalent acidic lipids upon anchoring in membranes without Chol (16). However, the cellular PM contains ∼40 mol % Chol (17), and liposome binding of both RSV and HIV Gag is stimulated by physiological Chol concentrations in model membranes (18). Both in vitro and in silico experiments have shown that the addition of Chol to fluid bilayers causes significant structural changes, including increased bilayer thickness (19, 20), increased headgroup spacing (21) and hydration (22), and reduced water penetration into the membrane hydrocarbon region (22, 23).
Here, using SANS, MD simulations, and liposome binding assays, we investigated the mechanism by which Chol influences RSV MA membrane binding, and whether protein binding changes the structure or lateral organization of lipids in the bilayer. Our results show that in bilayers with a fixed concentration of acidic lipids, Chol promotes binding of RSV MA by making the electrostatic potential above the membrane surface more negative, while at the same time decreasing the energetic penalty for lipid headgroup desolvation. We also find that MA selectively binds to preexisting patches of acidic lipids, but does not significantly affect their structure or composition. These findings reveal the intrinsic ability of lipid membranes to modulate the electrostatically driven binding of charged molecules, and have important implications for interpreting in vitro binding data.
Materials and Methods
Materials
Phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL) as dry powders and dissolved in HPLC-grade chloroform. Lipid stock concentrations were determined by inorganic phosphate assay (24) with an error <2%. Cholesterol powder was purchased from Nu-Chek Prep (Waterville, MN) and prepared as chloroform stock solutions at defined concentration using standard gravimetric procedures. Ultrapure H2O was obtained from a High-Q purification system (Wilmette, IL). D2O (99.96%), deuterated Tris buffer (Tris-D11, 98%), and deuterated glycerol (glycerol-D8, 99%) were purchased from Cambridge Isotopes (Andover, MA). Deuterium chloride (DCl) and Tris(2-carboxyethyl)phosphine (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO) and AMRESCO (Solon, OH), respectively. Buffer solutions were filtered through a prerinsed 0.2 μm filter before use.
Protein purification and liposome binding
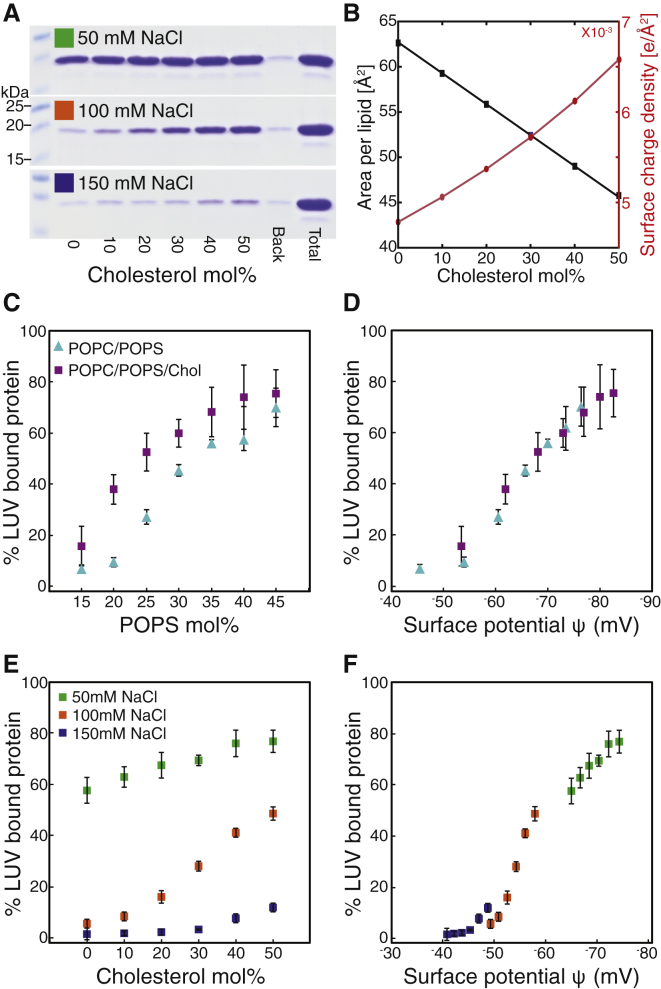
Protein was purified as previously described (8) and stored at –80°C until use. Preparation of 100 nm extruded liposomes and binding reactions were performed as previously described (25). Briefly, all binding reactions were performed with 15 μg (4.7 μM) protein and 50 μg (328−431 μM) lipid in 200 μL at 20 mM Tris-HCl pH 8, and adjusted with buffer to the stated NaCl concentration. Binding reactions were subjected to centrifugation at 90,000 RPM (relative centrifugal force = 351,955 × g) in a TLA-100 rotor (Beckman Coulter, Brea, CA) for 45 min at 4°C to pellet the liposome-bound protein. Pelleted protein was subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis analysis. Gels were stained with Coomassie Blue and then destained, and band intensity was determined by densitometry analysis using ImageQuant 5.2 (Molecular Dynamics, Sunnyvale, CA). Each binding reaction was repeated no fewer than four times. The binding data reported in Fig. 5 is the average, and error bars the SD, of measurements from these independent replicate samples.
Figure 5.
RSV MA membrane association is quantitatively explained by membrane surface potential. (A) Coomassie-stained gel of pelleted MA associated with LUVs with increasing Chol concentration (0–50 mol %, lanes 2–7). “Back” (lane 8) represents the amount of MA pelleted in the absence of LUVs and “Total” (lane 9) represents the total MA in each binding reaction. (B) Calculated membrane surface charge density (right axis, red) and average area per molecule (left axis, black) for POPC/POPS/Chol bilayers plotted versus Chol concentration. (C) Percent of LUV-bound MA plotted against POPS concentration for membranes without (light blue triangles) and with (purple squares) 36 mol % Chol. (D) Given here is binding data from (C) plotted versus calculated ψ. (E) Percent of MA bound to LUVs at 30 mol % POPS with increasing Chol and decreasing POPC concentration at different NaCl concentrations: 50 mM (green), 100 mM (orange), and 150 mM (blue). (F) Binding data from (E) is plotted versus calculated ψ. Legend in (A) corresponds to (A, E, and F); legend in (C) corresponds to (C and D). All data points are the average, and error bars are the SD, of no fewer than four independent replicate samples.
SANS sample preparation
Lipid mixtures were prepared by transferring lipid and cholesterol chloroform stocks to a glass vial with a glass syringe. Organic solvent was removed with a nitrogen stream and gentle heating, followed by vacuum drying for >12 h. Dry lipid films were hydrated to 40 mg/mL total lipid concentration with liposome storage buffer (D2O, 50 mM NaCl, 20 mM Tris-D11-DCl, 2 mM TCEP, pD 8), then vortexed vigorously to generate multilamellar vesicles. The multilamellar vesicle suspension was incubated for 1 h with intermittent vortexing, followed by 5 freeze/thaw cycles. Large unilamellar vesicles (LUVs) were prepared using a miniextruder (Avanti Polar Lipids) by passing the lipid suspension 31 times through a single 100-nm-pore-size polycarbonate filter. RSV-MA solution in protein storage buffer (D2O, 375 mM NaCl, 20 mM Tris-D11-DCl, 2 mM TCEP, 5 wt % glycerol-D8, pD 8) at 40 mg/mL was prepared from protein in H2O buffer by serial dilution and centrifugal filter concentration.
Samples for SANS measurements were prepared by combining and pipette-mixing 57 μL of the protein solution with 150 μL of the liposome solution and 390 μL pure D2O. By design, a small excess of osmolytes in the vesicle interior (liposome storage buffer) compared to the extravesicular buffer after mixing (D2O, 48 mM NaCl, 7 mM Tris-D11-DCl, 0.7 mM TCEP, 0.5 wt % glycerol-D8, pD 8) generated slightly hypotonic conditions that tended to swell the vesicles. This precaution was necessary to avoid distorted or partially collapsed vesicles, which result in complex scattering curves that cannot be analyzed with conventional spherical shell form factors. The final protein and lipid concentrations of SANS samples were 3.8 mg/mL (0.227 mM) and 10 mg/mL (13–16 mM, depending on the lipid composition), respectively, with all protein located in the extravesicular fluid. At these concentrations, enough protein was present to cover 70–80% of the vesicle surface area, assuming complete binding and average lipid and protein cross-sectional areas of 48–65 and 1375 Å2, respectively. The actual surface coverage determined by analysis of the scattering curves was 35–40% (Table S2), indicating that approximately half of the protein was bound to vesicles, with half remaining free in solution.
SANS data collection
SANS measurements were performed using the BL-6 extended Q-range small-angle neutron scattering (EQ-SANS) instrument of the Spallation Neutron Source at Oak Ridge National Laboratory, Oak Ridge, TN. Samples were loaded into 2 mm path-length quartz banjo cells (Hellma USA, Plainview, NY) and mounted in a temperature-controlled cell holder with ∼1°C accuracy. EQ-SANS data were taken at a 2.5 m sample-to-detector distance with a 2.5–6.0 Å wavelength band for a total scattering vector of 0.01 < q < 0.5 Å−1. Scattered neutrons were collected with a 2D (1 × 1 m) 3He position-sensitive detector (ORDELA, Oak Ridge, TN) with 256 × 192 pixels. The 2D data were reduced using the software package Mantid (26). During reduction, data were corrected for detector pixel sensitivity, dark current, and sample transmission, as well as background scattering from buffer. The 1D intensity I(q) (q = 4π sin(θ)/λ, where λ is the neutron wavelength and 2θ is the scattering angle relative to the incident beam), was obtained by radial averaging of the corrected 2D data.
SANS data analysis
SANS curves were fit with a laterally heterogeneous core-shell form factor (27) to account for coherent scattering contributions from both transverse (out-of-plane) and lateral (in-plane) neutron scattering length density (NSLD) variation within the vesicle (a detailed description of the model is provided in the Supporting Material). As shown schematically in Fig. S2 A, transverse NSLD variation arises primarily from the different atomic composition of the lipid headgroup and hydrocarbon layers, whereas lateral NSLD variation is primarily due to lipid clustering resulting from nonideal mixing or phase separation, and is pronounced when some lipid species are selectively deuterated. Both the transverse and lateral NSLD variation are further influenced by the presence of surface-bound protein, which has a different NSLD than lipid or water.
The heterogeneous core-shell form factor requires as input the transverse NSLD profiles of the domain and continuous phases, as well as the size, surface coverage, and spatial arrangement of the domains (Fig. S2 A). Our analysis allowed for the possibility of nonrandom lipid mixing, which we modeled as a PS-rich domain phase surrounded by a continuous phase depleted in PS. The compositions of the two phases were allowed to vary in the fit, but not independently; given a fixed overall vesicle composition, a domain composition, and phase area fractions, the composition of the continuous phase was constrained by material balance. Following Kučerka et al. (28), NSLD profiles for the domain and continuous phases were derived from underlying lipid volume probability distributions, modeled as the sum of separate distributions for the lipid headgroups and hydrocarbon chains. The total unit cell volume was calculated as a mole-fraction weighted sum of lipid component volumes obtained from the literature (Table S3) and constrained in the fit, leaving adjustable parameters for the area per lipid AL and headgroup thickness DH. Additional structural parameters, including the total bilayer thickness DB and the hydrocarbon thickness 2DC, were derived from relationships between the adjustable parameters and the lipid headgroup and hydrocarbon volumes as described in the Supporting Material. For MA-containing samples, a Gaussian volume probability distribution with adjustable parameters for position (zp) and width (σp) was added to the domain phase. NSLD profiles were then obtained as a sum of the separate headgroup, hydrocarbon, and protein volume probability distributions multiplied by their respective total scattering lengths. Lateral structure was modeled by assuming round domains with a 21 Å radius (corresponding to a cross-sectional area of 1375 Å2 for an MA monomer) that were randomly arranged on the vesicle surface. The in-plane contributions to the vesicle form factor were calculated to 200 expansion orders and included an adjustable parameter for the fraction of the vesicle surface area occupied by domains, ap.
For each nominal sample composition (i.e., POPC/POPS 70:30 mol % and POPC/POPS/Chol 34:30:36 mol %), separate SANS samples with different contrast were prepared using the four permutations of protiated and palmitoyl chain-perdeuterated variants of the two phospholipids (i.e., POPC or POPC-D31, mixed with POPS or POPS-D31; Fig. S2, C–F). Fitting was implemented in the softwares Mathematica 11.0 (Wolfram Research, Champaign, IL) and MATLAB R2013b (The MathWorks, Natick, MA). Each data set was fit independently, and the reported structural parameters represent the mean and SD from the four fits. A complete list of the structural parameters is found in Table S2.
High-resolution structure of MA
To provide the structural models required for the detailed interrogation of molecular interactions, we crystallized full-length RSV MA (155 amino acids) and determined an experimentally phased x-ray structure at 2.8 Å resolution. Residues 1−102 were well resolved but there was no interpretable electron density for the remainder of the molecule. We therefore characterized a truncated variant (MA2−102) with the disordered region removed. This variant crystallized in the same form as the full-length molecule (space group I4122), and also in an alternate form (space group I41). Structures were determined to 3.2 and 1.8 Å resolution, respectively. The three crystallographic models (Table S1) differ in detail from an earlier NMR solution structure for the first 88 amino acids (29). This likely reflects both the limited restraint set and the methodology used to derive the NMR model (30). However, consistent with NMR relaxation measurements (29), the basic surface loop (residues 14−22), which is critical for membrane association, is the most mobile element in the crystallographic models. For crystals in the space group I4122, the final helix of the N-terminal domain (helix 6, residues 89−102) interdigitates with the corresponding helix from a neighboring molecule, forming a symmetric dimer (Fig. S1 A). In contrast, for space group I41 crystals, the C-terminal sequence is disordered and neither helix 6 nor the dimer is observed. Because deletions in the C-terminal region of MA (amino acids 87−155) do not abrogate viral budding and infectivity (31), and because there is no evidence that MA dimerizes in solution (8) or when membrane associated (32), it is likely that the crystallographic dimer is biologically irrelevant. Because the monomer structure was not available when our MD simulations were carried out, one subunit of the dimer was adapted as the working model for the in silico experiments reported here (see below and Fig. S1).
MD simulations
All MD simulations were performed with the software NAMD (33) and the CHARMM36 force field for lipids and protein (34, 35). The simulations were run with a 2-fs time-step with all bonds to hydrogens constrained, in NPT ensemble with semiisotropic (for bilayer simulations) or isotropic (for water box simulation) pressure coupling at 1 bar and 293 K. Temperature and pressure were controlled with a Langevin thermostat and barostat, with a damping parameter set to 5 ps−1, a piston period of 200 fs, and a piston decay of 50 fs. The vdwForceSwitching option in NAMD was used for all membrane simulations (36). The van der Waals cutoff and switching distances were set to 12 and 10 Å, respectively. Particle mesh Ewald was used with a grid spacing of 1.0 Å.
To determine a starting conformation for MA in subsequent MA-membrane simulations, we first simulated one monomer from the crystal dimer structure of MA (Fig. S1 A) in an aqueous environment consisting of a water box with 13,470 water molecules and 20 mM NaCl. Following the hypothesis that the large angle between the C-terminal helix of MA (residues 91–102) and the rest of the protein body was dimer-specific and would not be a stable conformation of the MA monomer, we fixed the C-terminal helix in space and monitored the relaxation dynamics of the rest of the protein. The system was energy minimized for 5400 steps, then run for 500 ps with a 1-fs time step, followed by a production run of 110 ns. In the course of the simulation, the protein body moved closer to the C-terminal helix, adopting a stable compact conformation (Fig. S1, B and C). The protein coordinates from the last frame were used as the starting MA conformation for the MA-membrane simulations described below.
The two simulated bilayers (POPC/POPS 70:30 mol % and POPC/POPS/Chol 34:30:36 mol %) were taken from (37). After the production runs (191 and 270 ns, respectively), MA was placed on one or both sides of the bilayer and oriented with respect to the membrane surface in a way that optimized interactions between lysines and the surface as described for HIV MA (7), and for RSV MA (38). Multiple replica simulations of the bilayer-protein systems in 50 mM NaCl were run for ∼200 ns each and analyzed jointly as described in the Supporting Material.
Results and Discussion
MA binds to lipid headgroups without perturbing the overall bilayer structure
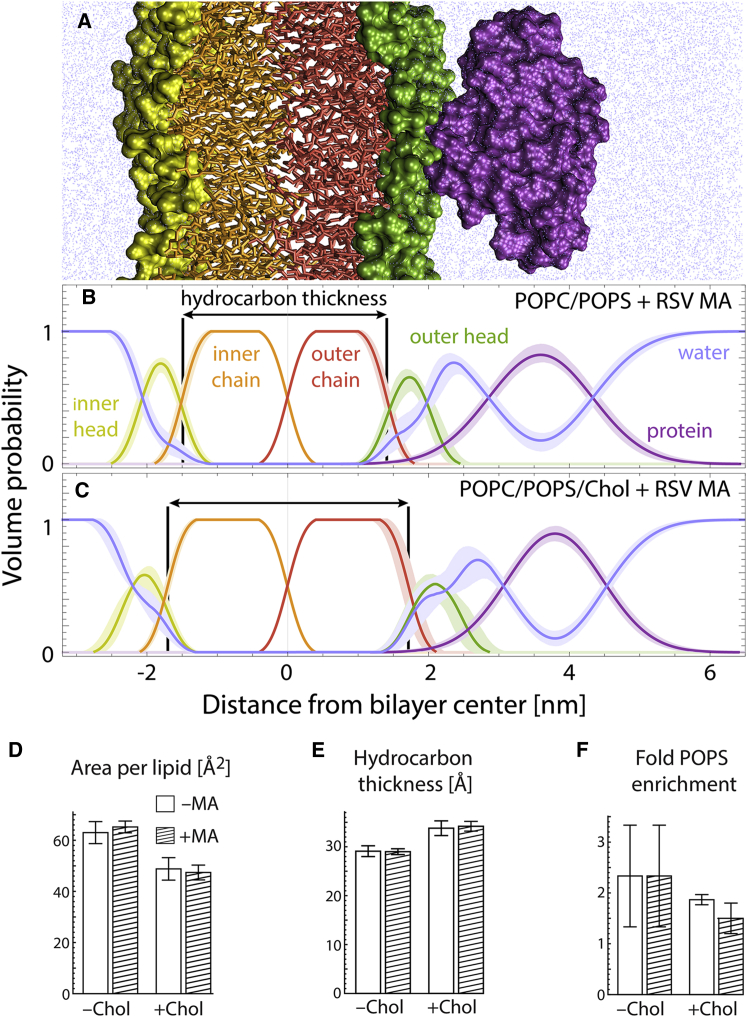
We used SANS to determine the bilayer structure before and after RSV MA binding to 100-nm-diameter unilamellar vesicles (i.e., LUVs, also referred to as “liposomes” throughout) at 20°C and 50 mM NaCl. Motivated by previous work showing that cholesterol enhances MA binding (18), we examined two compositions having a fixed amount of the negatively charged lipid POPS, namely POPC/POPS 70:30 mol % and POPC/POPS/Chol 34:30:36 mol %. The POPS concentration was based on estimates of the concentration of charged lipids in the PM inner leaflet (32 mol %; Table S4). These compositions allow for a direct comparison that isolates the effect of cholesterol, and are simple enough to be tractable in a SANS analysis. Scattering data (Fig. S2, C–F) were analyzed with a model describing the projected distribution of matter (i.e., lipid headgroups, lipid chains, protein, and water) along the bilayer normal, as a function of distance from the bilayer center. In particular, we used volume probability distributions to model the relative locations of the different components, which enabled the determination of bilayer structural parameters including area per lipid, total bilayer thickness, hydrocarbon thickness, and the position of the bound protein within the bilayer and the transverse water distribution (Supporting Material).
Fig. 1 shows the lipid and protein matter distribution for bilayers without (Fig. 1 B) and with Chol (Fig. 1 C) obtained from SANS analysis. The bulk of the protein density (purple curves) resides above the lipid headgroups (green curves), although a small degree of overlap suggests that some residues are able to intercalate between lipid headgroups. Importantly, practically no protein density overlaps with the lipid hydrocarbon region (red curves), an indication that MA does not penetrate the bilayer’s hydrophobic core, and binds in a manner consistent with electrostatic attraction to the negatively charged headgroup region. Structural parameters recovered from our analysis are in agreement with published values for POPC and POPS bilayers (39, 40). The replacement of 36 mol % of POPC with Chol (mimicking physiological Chol concentrations and charged-to-neutral headgroup ratios) reduced the area per lipid (defined here and throughout to include Chol) by ∼25%, from 63.0 to 48.8 Å2 (Fig. 1 D). Hydrocarbon chain thickness increased by nearly 5 Å, from 29.1 to 33.8 Å (Fig. 1 E), also consistent with published data (41). Neither the area per lipid nor the hydrocarbon thickness changed significantly in the presence of MA, suggesting that protein binding does not perturb the bilayer structure. This finding was supported by electron spin resonance (ESR) measurements showing that the lipid acyl chain order parameter did not change in the presence of MA (Fig. S3). A complete list of structural parameters obtained from the SANS analysis is found in Table S2.
Figure 1.
MA binds peripherally to lipid bilayers with minimal structural perturbation. (A) An MD simulation snapshot showing the different lipid and protein components modeled in the SANS analysis: inner leaflet headgroups (yellow) and chains (orange), outer leaflet chains (red) and headgroups (green), protein (purple), and water (blue). Below the cross-sectional view, the volume probability profiles obtained from experimental SANS data are displayed (thick lines surrounding the volume probability profiles reflect uncertainties): POPC/POPS 70:30 mol % with bound RSV MA (B); POPC/POPS/Chol 34:30:36 mol % with bound RSV MA (C). (D−F) Structural parameters obtained from SANS analysis of POPC/POPS bilayers without and with Chol, in the absence (white bars) or presence (hatched bars) of RSV MA: average area per molecule (D); lipid hydrocarbon thickness (E); and fold-enrichment of POPS over the average composition in PS-rich clusters (F). The reported structural parameters and errors represent the mean and SD from independent fits to four different neutron contrast data sets as described in the text and Supporting Material. A complete list of structural parameters is found in Table S2.
MA binds to POPS clusters
Neither POPC/POPS nor POPC/POPS/Chol bilayers are phase separated at 20°C (L. Goh, personal communication). Such mixtures may nevertheless be heterogeneous on small (<5 nm) length scales (42). To a first approximation, nonideality arises from the different nearest-neighbor interactions of unlike lipids (e.g., POPC and POPS). Interactions between unlike phospholipids can be unfavorable (43), resulting in clustering of like lipids to reduce unlike contacts. To account for the possibility of such nonrandom lipid mixing, we analyzed our scattering data using a form factor appropriate for laterally heterogeneous vesicles (27). We analyzed four combinations of protiated and palmitoyl chain-perdeuterated variants of POPC and POPS (Fig. S2 B) to take full advantage of neutron contrast for detecting possible lateral segregation of these lipids. The scattering model allowed for two distinct bilayer environments whose compositions were varied in the fit while maintaining overall matter balance (Fig. S2 A). In the analysis of data from MA-containing samples, the compositions of the protein-bound and protein-free portions of the bilayer were also allowed to vary. Consistent with previous reports (44), the SANS analysis indicated a patchy bilayer characterized by lipid clusters that are enriched approximately twofold in POPS (Fig. 1 F). In MA-containing samples, we found a similar amount of PS enrichment in the protein-bound bilayer composition (Fig. 1 F). The addition of Chol did not have a significant effect on the extent of nonideal mixing of POPC and POPS, either in the presence or absence of MA.
Although the lateral heterogeneity observed in our SANS analysis is statistically significant, the large uncertainties in PS enrichment (Fig. 1 F) do not allow us to draw any conclusions regarding the ability of MA to sequester charged lipids upon binding. To test this possibility, we applied a previously developed computational method based on continuum mean-field theory (45, 46) to quantify the extent of lipid redistribution in response to protein binding to an ideally mixed bilayer, and to calculate changes in the corresponding adsorption free energy upon the demixing process (Fig. S4; Supporting Material). Our calculations predict that MA cannot effectively sequester POPS as has been shown previously for other proteins (see (46) and references therein); minimal lipid segregation was achieved, which produced an insignificant change in the adsorption free energy (<1 kBT). Taken together with the SANS results, this calculation suggests that MA does not induce lipid redistribution. Rather, the protein takes advantage of the inherent patchiness of the nonideally mixed bilayers, binding to the membrane primarily at sites already enriched with anionic lipids.
Cholesterol enhances MA lysine-membrane contacts
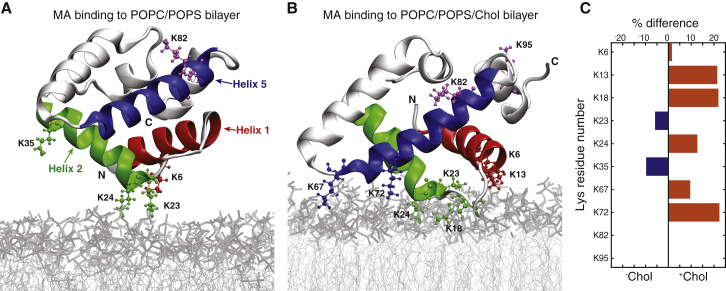
To investigate the atomic-level details of MA-membrane interactions, we performed MD simulations mimicking the SANS experimental conditions, i.e., monomeric MA in the presence of a lipid membrane composed of either POPC/POPS at 70:30 mol % (Fig. 2 A), or POPC/POPS/Chol at 34:30:36 mol % (Fig. 2 B) in 50 mM NaCl solution at 20°C. The MA structure used as a starting point in these simulations was obtained from the monomer of the dimer crystal after relaxation in water (see Materials and Methods), and is similar to the independently determined monomer crystal structure (the backbone RMSD between the two structures is 2.8 Å; Fig. S1 D).
Figure 2.
Cholesterol enhances MA-membrane contacts. Given here are snapshots of MA interacting with POPC/POPS (A) and POPC/POPS/Chol (B) bilayers after 100 ns of simulation (for corresponding movies, see Movie S1, A and B). The protein is displayed in silver with specific lysine residues labeled and shown in space-fill representation (“N” and “C” denote the protein termini). Lipid tails and headgroups are shown as light and dark gray lines, respectively, and water and ions are omitted for clarity. (C) Shown is the difference in the percent time that MA lysine residues spend in contact with the bilayer in the absence and presence of Chol, where contact is defined as a distance not greater than 3 Å between any residue atom and any lipid atom. K13, K18, and K72 spend a mean 22% more time in contact with the membrane when Chol is present, whereas K82 and K95 are never in contact with either bilayer.
The protein’s secondary structure remained stable during the simulations (Fig. S6 A), and the protein density did not penetrate further than the lipid headgroup region in either system (Fig. S5), consistent with the SANS results (Fig. 1, B and C). However, in the presence of Chol the helices’ tilts changed slightly, facilitating the exposure of K13, K18, K24, and K72 to the membrane (Fig. S6 C). Of the protein’s 10 lysine residues, two did not interact with either the POPC/POPS or POPC/POPS/Chol bilayer, whereas six exhibited a greater number of contacts with the +Chol membrane (Fig. 2 C). The slight conformational changes of the protein were accompanied by a moderate increase in the number of instantaneous lysine-POPS contacts (Fig. S6 B). For example, during the last 100 ns of the simulations when the bilayer contained Chol, up to seven lysine residues (as opposed to six in the POPC/POPS bilayer) simultaneously came into contact with PS, and the probability of a single PS lipid interacting with three lysine residues was 25 times greater than in the POPC/POPS bilayer. These results are consistent with a stronger electrostatic interaction between MA and the Chol-containing membrane.
The modes of RSV MA-membrane interaction observed in the simulations have been implicated in functional phenotypes in vivo (47). In the simulations, the basic residues in the first 35 amino acids of the protein were actively engaged in contacts with the bilayer. The exception was K35, which spent <20% of the time close to the membrane, consistent with the unaffected budding of virus particles in the K35Q mutant (47). In the simulations, neither K82, R61, or R85 (K82, K61, and K85 in the RSV strain studied in (47)) came in contact with the lipids, which agrees with the less severe reductions in virus particle release observed in the double mutants of K61Q/K82Q and K61Q/K85Q. Furthermore, in contrast to E25 and E70, the location of D52 on helix 4 keeps that residue away from the bilayer surface in the simulations, helping clarify the experimental observation of full versus partial rescue of a budding-defective mutant upon lysine substitution of the glutamic or aspartic acids, respectively (47).
Cholesterol increases the membrane surface charge density
Neither SANS nor MD simulations provided evidence for a direct interaction between MA and Chol, although SANS showed a reduction in area per lipid and an increase in hydrocarbon thickness for Chol-containing bilayers (Fig. 1, D and E). We therefore hypothesized that Chol indirectly influences MA binding by inducing changes in the bilayer structure that promote electrostatic interactions with the protein. To test this hypothesis, we first compared the structural properties of the two simulated bilayers in the absence of protein. It is important to note that because the ratio of negative charge to total lipid was fixed, the PC/PS ratio in the –Chol and +Chol bilayers was different (7:3 and 3.4:3, respectively). This choice was made to isolate the effects of cholesterol without changing the total negative charge.
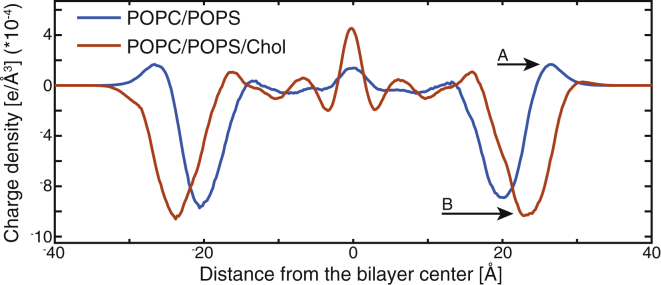
Incorporation of Chol increased the lipid acyl chain order parameter by >65% (Fig. S6 D), consistent with ESR measurements (18). This change was accompanied by a 28% decrease in the average area per lipid from 60.9 to 43.5 Å2, and a 6 Å increase in bilayer thickness from 40.5 to 46.9 Å, similar to SANS results (Fig. 1, D and E; Table S2). The reduction of bilayer area led to an increase in the PS surface density: for example, a 6.5 × 6.5-nm membrane patch would contain on average 42 or 60 charged headgroups in the absence or presence of 36 mol % Chol, respectively. The greater PS area density increased the bilayer’s negative surface charge density (i.e., charge per unit area), which together with the lower PC/PS headgroup ratio (see Fig. S7 for the individual contributions of PC and PS to the charge density) completely eliminated the small peak of positive charge density observed in the –Chol bilayer, thus making electrostatic interactions with the +Chol membrane more favorable (Figs. 3 and S7).
Figure 3.
Cholesterol influences the membrane surface charge density. Charge distribution along the bilayer normal is calculated from the two simulated bilayers in the absence of MA. Addition of Chol increases the negative charge density in the headgroup region (arrow B), eliminating the peak of positive density at the water-lipid interface present in the POPC/POPS bilayer (arrow A). See Fig. S7 for the individual contributions of the membrane components.
To relate the changes in surface charge density to MA binding, we calculated the bilayer electrostatic potential (defined here and henceforth as 3 Å above the surface) using nonlinear Poisson-Boltzmann theory and the Gouy-Chapman model of the electrical double layer (48). The potential is a function of salt concentration, temperature, and surface charge density, with the latter calculated as the ratio of POPS mole fraction to the average area per lipid and having units of e–/Å2 (the reference potential in the bulk water far from the surface is zero). This formulation assumes that the charges are uniformly distributed on a continuum flat surface and therefore reports an average surface potential. Calculations based on conditions used in the MD simulations yielded potentials of –67 and –76 mV for the respective −Chol and +Chol bilayers, suggesting that the preference of MA for Chol-containing membranes is due to a stronger electrostatic attraction, because MA bears a net +3 charge.
Nonideal mixing of POPC and POPS lipids has a small effect on surface potential
Bilayers for MD simulations were constructed from an initially random lateral arrangement of phospholipids and Chol by using CHARMM-GUI (49). In light of the SANS finding of POPS clusters (Fig. 1 F), and because the simulation length of ∼200 ns did not allow sufficient time for lipids to explore their conformational space through lateral diffusion, it is important to independently examine the effect of nonideal mixing on the bilayer’s electrostatic potential. To this end, we performed Monte Carlo lattice simulations of a binary mixture of neutral and charged lipids in a 70:30 ratio, to obtain their equilibrium lateral distribution (Supporting Material). In this simple model, the lipid distribution is a function of a single adjustable parameter, the excess mixing energy ΔEm of a PC/PS pair, which accounts for all nonideal interactions between these lipids (43). To examine different degrees of nonideal mixing we varied ΔEm from zero (i.e., random mixing) to +0.5 kBT, which is just below the threshold for complete phase separation (50).
Static snapshots of the equilibrium lipid distributions with an increasing degree of nonideal mixing are shown in Fig. S8 (see Supporting Material for details). Also shown (Fig. S8, lower panels) are corresponding maps of the electrostatic surface potential relative to that of a uniform bilayer with 30 mol % charged lipids (Supporting Material). Although some patchiness of the potential is observed with increasing nonideality, in most cases the local potential varied by <10% from the average value, indicating that for low to moderate degrees of nonideal mixing the average potential calculated with the uniform Gouy-Chapman model is a reasonable approximation for the potential above the bilayer surface. Taken together with the SANS results showing similar degrees of nonideal mixing of POPC and POPS in the presence and absence of Chol, as well as the calculated 13% decrease in the average potential in the +Chol membrane, these findings suggest that nonideal mixing cannot account for the Chol-enhanced MA/bilayer interaction observed both in vitro (18) and in silico (this work).
Cholesterol increases the MA/bilayer electrostatic interaction and decreases the penalty for headgroup desolvation
To examine the specific energetic contributions of the protein and bilayer in the MA-membrane interaction, we used the molecular mechanics-generalized Born surface area (MM-GBSA) approach to estimate the binding free energy ΔGbind of MA with the POPC/POPS and POPC/POPS/Chol bilayers, based on conformations sampled in the MD simulations (51, 52). In the MM-GBSA framework, ΔGbind is approximated as the sum of the interaction energy of the two binding partners (here, MA and bilayer) in vacuum , and the penalty for displacing water molecules upon protein binding. The latter is expressed as a difference in the energetic cost for desolvating the MA-bilayer complex and each partner separately, and is referred to as ΔΔGsolv.
Our analysis showed that in the presence of Chol, the binding free energy was >4 kcal/mol more favorable than in the absence of Chol (–5.8 ± 0.3 vs. –1.6 ± 0.2 kcal/mol, respectively). Decomposition of ΔGbind into electrostatic and desolvation contributions confirmed the much stronger electrostatic interaction of MA with the Chol-containing bilayer (–1531 vs. –1215 kcal/mol for +Chol and –Chol bilayers, respectively), while also revealing a similar trend in the polar desolvation penalties (1552 vs. 1242 kcal/mol), as shown in Table S5. When the two partners bind, the favorable electrostatic attraction between them is partially offset by the displacement of water molecules from the binding interface, which incurs an unfavorable desolvation penalty resulting from the high dielectric strength of water and charge screening by the salt.
Further insight was gained by decomposing into the separate contributions from each binding partner (52, 53). Whereas the balance between and ΔΔGsolv for MA was similar in the two MA-bilayer systems (Fig. 4 A, blue bars), the energetic cost for desolvating the lipids was lower for the +Chol bilayer, resulting in a more favorable MA-membrane interaction energy (Fig. 4 A, orange bars). The desolvation penalty is closely related to both the charge density and the solvent-accessible surface area (SASA) of the binding partners. Thus, the smaller desolvation penalty of the +Chol bilayer could be due to either a lower charge density, and/or smaller solvent-exposed molecular surface to desolvate in the presence of Chol. The area of the MA shadow (defined by the projection of the MA coordinates onto the bilayer plane) in the two bilayers was practically identical (Fig. S9 B), and the +Chol bilayer had on average one more POPS lipid than the –Chol bilayer (Fig. S9 E), thereby excluding the former possibility.
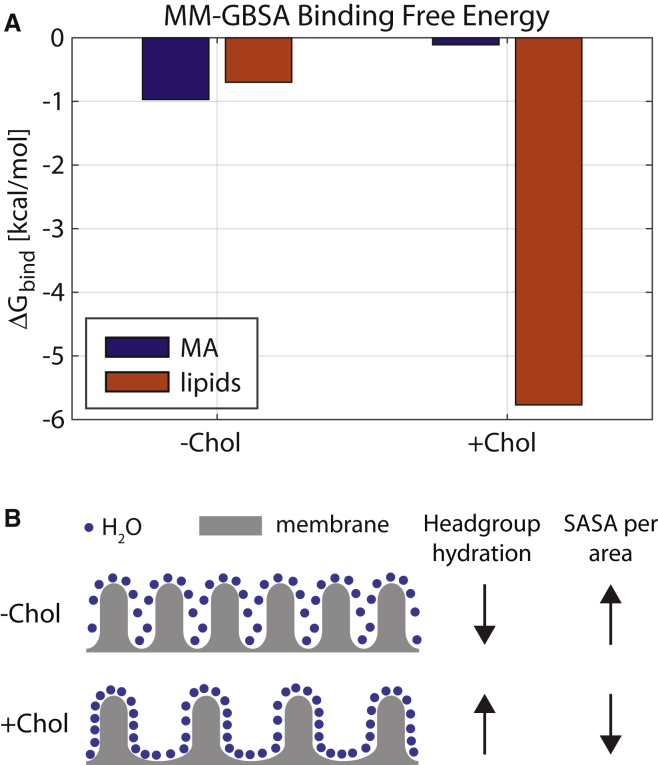
Figure 4.
Cholesterol decreases the energetic penalty for lipid desolvation. (A) Given here are contributions of MA and lipids to the total binding free energy ΔGbind of the MA-bilayer complex estimated using the MM-GBSA method. Each energetic contribution is the sum of the interaction energy in vacuum for the respective system and the solvation penalty calculated separately for MA and the lipids. The presence of Chol results in a 5 kcal/mol reduction in the solvation penalty of the lipids relative to , resulting in a more favorable bilayer contribution to ΔGbind compared to the PC/PS bilayer. For a detailed breakdown of the energy contributions and details of the underlying calculation, see Supporting Material. (B) A schematic illustration of the lipid headgroups (gray) on the bilayer surface in the +Chol and –Chol membranes, and the water molecules that solvate them (blue dots). Due to the area occupied by Chol itself, there are fewer lipid (PC or PS) headgroups in a bilayer patch of given size. Thus, even though Chol increases the number of bound waters per headgroup, it decreases the SASA per unit area of the bilayer (see text). Note that the spacing between the lipid headgroups in the +Chol schematic has been exaggerated to emphasize the lower packing density in this region.
To test for the latter, we compared the SASA of the top leaflet patch in the MA shadow in the two bilayers. As expected, the +Chol bilayer had a significantly smaller SASA than the –Chol bilayer (Fig. S9 A). At first glance, a smaller SASA in the presence of Chol suggests fewer waters per lipid. Interestingly, a count of the number of waters within 3 Å of a POPC or POPS lipid revealed that, consistent with experimental data (22), lipids were in fact better hydrated in the +Chol bilayer, having roughly 13.3 waters/headgroup compared to 12.3 in the Chol-free bilayer (Fig. S9 C). However, due to the surface area occupied by Chol itself, there were on average four fewer lipid headgroups under the protein in the presence of Chol (Fig. S9 D). This resulted in ∼30 fewer lipid-bound waters in the MA shadow in the +Chol bilayer compared to the –Chol bilayer (226 vs. 258), in agreement with the observed SASA difference. A count of the number of PC, PS, and Chol molecules in the MA shadow further confirmed that this difference was due to the replacement of POPC with Chol (Fig. S9 E). Thus, whereas Chol increases headgroup hydration, it decreases the overall bilayer SASA per unit of planar area in the bilayer, as illustrated schematically in Fig. 5 B, resulting in a smaller desolvation penalty of the POPC/POPS/Chol membrane upon protein binding. Therefore, our MM-GBSA analysis indicates that Chol both increases the electrostatic attraction between MA and the bilayer, and decreases the cost for headgroup desolvation when the protein binds. These two effects, in concert, result in the energetically more favorable interaction of MA with the +Chol membrane.
The membrane electrostatic potential is a key determinant of MA binding affinity
Although the effects of Chol on membrane thickness and lipid packing are well known (see (54)), the calculations of the previous sections demonstrate that bilayer structural perturbations induced by Chol constitute a general mechanism by which the molecule can indirectly mediate electrostatic interactions with proteins. To further explore retroviral MA-membrane association in the context of these interactions, we used liposome pelleting (25) to measure binding of RSV MA to LUVs with varying POPS and Chol concentrations, and at varying salt concentration. The data were analyzed in terms of calculated electrostatic potential above the membrane surface (Figs. 5 and S10).
Consistent with previous results for RSV and HIV MA and Gag (8, 25, 38, 55), increasing the POPS concentration from 15 to 45 mol % resulted in increased RSV MA binding to LUVs from ∼10 to 70% (Fig. 5 C, blue triangles). A significant enhancement in binding affinity was observed when 36 mol % POPC was replaced with Chol (Fig. 5 C, purple squares): for example, at 20 mol % POPS, Chol increased MA association with LUVs from 10 to 40%. We also directly tested the ability of Chol to enhance binding under a range of ionic strengths by incrementally replacing POPC with Chol at a fixed POPS concentration of 30 mol %. This replacement decreases the average area per molecule from 62 to 45 Å2, and increases the surface charge density from 4.8 × 10−3 to 6.6 × 10−3 e–/Å2 (Fig. 5 B). At 100 mM NaCl, we observed a significant increase in the amount of RSV MA associated with LUVs upon increasing Chol concentration (Fig. 5 E, red squares). Reducing the NaCl concentration to 50 mM resulted in increased binding at all Chol concentrations (Fig. 5 E, green squares), whereas increasing NaCl concentration to 150 mM practically eliminated binding (Fig. 5 E, blue squares).
Importantly, the binding trends for all bilayers with and without Chol, and at different ionic strength, collapse onto a single curve when plotted against the electrostatic potential above the membrane surface, ψ, calculated using the Gouy-Chapman model (Fig. 5, D–F). In all cases, MA shows little membrane affinity at ψ > –50 mV, whereas a dramatic increase in binding occurs as ψ drops to –70 mV. We also observed a similar trend when MA binding was measured as a function of varying NaCl concentration (Fig. S10). Taken together, these results reveal a sigmoidal dependence of binding on membrane surface potential, and demonstrate that the ability of Chol to enhance RSV MA membrane association can be explained by changes in bilayer structure that increase the surface charge density.
Implications for MA binding at the plasma membrane
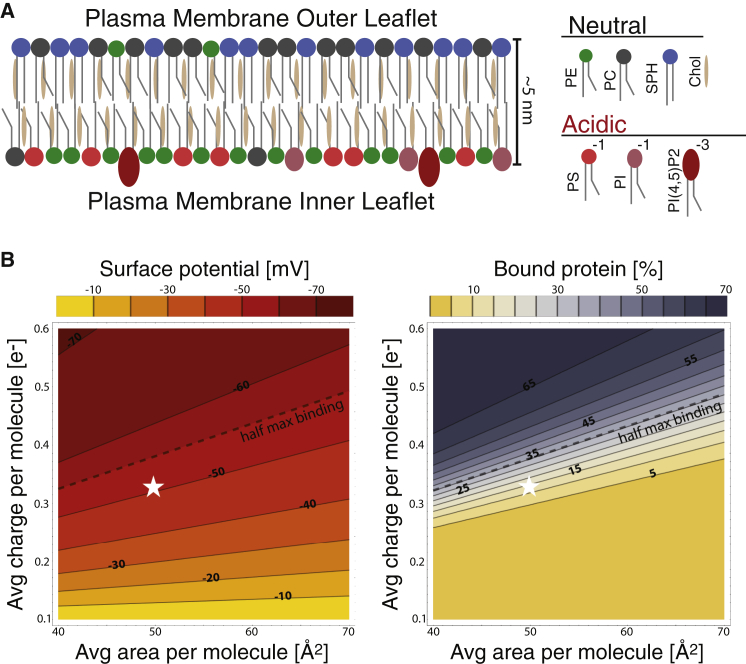
Fig. 6 A shows a model of a mammalian PM taken from published estimates of the outer and inner leaflet lipid composition (5, 17, 56). Using literature values for individual lipid areas, we calculated the average molecular area (48.9 Å2) and charge (0.32 e–) for the PM inner leaflet composition (Table S4), which results in an average surface charge density of 6.5×10−3 e–/Å2. It is instructive to consider this charge density in the context of our binding results. Fig. 6 B shows a contour plot of ψ as a function of average molecular area and charge at physiological ionic strength (150 mM NaCl) and temperature (37°C), calculated with the Gouy-Chapman model. Also shown is a contour plot of the percentage of membrane-bound MA protein under the same conditions, obtained by mapping the sigmoidal binding curve of Fig. 5 F to the calculated surface potential (Fig. S10; Supporting Material). Whereas ψ changes gradually as a function of surface charge density, the fraction of bound protein exhibits large changes over a relatively small range of ψ, from ∼–50 to –60 mV. The average PM composition is located approximately at the –50 mV contour, a point that corresponds to weak binding, but that is near the edge of the binding transition. Compositional perturbations such as nonideal mixing that increase the local concentration of charged lipid (corresponding to movement in the +y direction), or increase the local concentration of Chol (corresponding to movement in the –x direction), could therefore act as a binding switch. For example, a 10 Å2 reduction in the average lipid area—which corresponds to a 10−3 e–/Å2 increase in charge density, or roughly a 20% increase in the local PS density in a patch of average concentration of 30 mol %—could be sufficient to promote electrostatic binding. Thus, by tuning the local lipid composition in the inner leaflet, cells could create binding platforms enriched in monovalent charged lipids and Chol, which can attract proteins by means of electrostatic interactions.
Figure 6.
Cellular plasma membrane model. (A) Asymmetric distribution of PM lipids between the inner (Chol/PE/PC/PS/PI/PIP, 40:28:5:18:8:2 mol %) and outer (Chol/PE/PC/sphingomyelin, 40:7:27:26 mol %) leaflets (see Table S4). (B) Contour plots of electrostatic surface potential (left) and protein binding (right) as a function of average area and charge per molecule. Electrostatic potential was calculated from nonlinear Poisson-Boltzmann theory assuming physiological ionic strength (150 mM NaCl) and temperature (37°C). Protein binding was calculated from a sigmoidal function that maps the binding data displayed in Fig 5F to surface potential (Fig. S10; Supporting Material). The dashed line traces the corresponding contour of half-max binding, and the white star denotes the approximate molecular area (48.9 Å2) and charge (0.32 e–) of the PM’s inner leaflet estimated from the composition given above.
The mechanistic insights gained from this study also enable predictions for the effect on protein binding of Chol relative to other lipids. Our results indicate that Chol enhances protein binding through two synergistic effects: it both increases the charge density of the bilayer and decreases the energetic cost of displacing water from the lipid headgroups. As mentioned earlier, the penalty for headgroup desolvation is related to the SASA per unit of planar area. Thus, increasing the membrane’s charge density at fixed SASA (for example, by adding PIP2 lipids instead of Chol to a POPC/POPS membrane with a fixed PS mole fraction) should cause a large increase in protein binding. Indeed, this has been shown experimentally for a number of different proteins and model peptides (see Fig. 8 in (25)). On the other hand, decreasing the bilayer’s desolvation penalty (or SASA/unit area) at a fixed membrane charge density should result in a smaller increase in binding. One such example can be seen directly in the binding data in Fig. 5 C: namely, the –Chol bilayer with 25 mol % PS and the +Chol bilayer with 20 mol % PS have a similar charge density of ∼4 × 10−3 e–/Å2, yet the binding to the +Chol bilayer is stronger. This is likely due to Chol’s effects on the solvation properties of the membrane. PE lipids, which are abundant in the PM inner leaflet, have smaller headgroups (hence, smaller SASA) and smaller cross-sectional areas than corresponding PC or PS lipids (57). Due to their small SASA and ability to reduce the bilayer area, PE and Chol should have similar effects on binding. Indeed, although at 25 mol % PS, 27% and 53% MA is bound to POPC/POPS and POPC/POPS/Chol bilayers, respectively (Fig. 5 C), we find that 58% MA is bound to the corresponding POPE/POPS liposomes under the same conditions.
Conclusions
We have shown how the interactions crucial for Gag association with the PM can depend on the types of lipids present in the PM inner leaflet. Model membrane studies have previously shown that: 1) electrostatic interactions are critical for MA binding (1, 58), 2) cholesterol enhances MA and Gag binding (18, 59), and 3) cholesterol condenses the membrane area (60). Our work is the first to explain these observations by describing cholesterol's role using Gouy-Chapman electrostatics. For example, HIV virions are enriched in charged lipids compared to the cellular plasma membrane, and contain 30–40 mol % cholesterol (5), yet many studies of viral protein binding to model membranes do not include cholesterol. Our results suggest that cholesterol enhances MA binding by affecting both the electrostatic and solvation properties of the membrane. Thus, it is an essential component in model membranes, recreating a surface potential and binding conditions representative of the PM inner leaflet. Moreover, we found that the nonideal mixing behavior of neutral and charged lipids can promote the formation of anionic lipid clusters that act as protein binding sites. We conclude that membranes can mediate nonspecific electrostatic interactions with proteins even in the absence of multivalent acidic lipids, and that bilayer mixing behavior and structural properties should be considered when interpreting protein binding data.
Author Contributions
M.D., F.A.H., and R.A.D. designed the research, conducted experiments, analyzed data, cowrote the manuscript, and contributed equally to the work. R.L.K. conducted experiments, analyzed data, and cowrote the manuscript. G.K. and M.A.C. performed computational analysis and cowrote the manuscript. G.W.F. and V.M.V. designed the research and cowrote the manuscript. Y.W. conducted experiments. J.K. designed research.
Acknowledgments
We thank Jany Chan for construction of the expression plasmid and purification of the truncated RSV MA protein used for crystallography; Drew Marquardt and Chris Stanley for technical assistance with SANS measurements; Harel Weinstein for valuable discussions regarding the in silico analysis; Vincent Zoete for providing his implementation of the MM-GBSA free energy decomposition method, which is at the core of the modified version used here; Derek Shore for technical help with the MM-GBSA analysis; and the National Biomedical Center for Advanced ESR Technology at Cornell University and Siddarth Chandrasekaran for help with ESR data collection and analysis.
This work was supported by National Institutes of Health (NIH) grant R01GM107013 (to V.M.V.), National Institutes of Health (NIH) grant R01GM105684 (to G.W.F.), and National Institutes of Health (NIH) grant PO1DA012408 (for M.D., G.K., and M.A.C.). The work at Oak Ridge National Laboratory’s EQ-SANS instrument at the Spallation Neutron Source was sponsored by the Basic Energy Science (BES) Program, Office of Science, US Department of Energy (DOE) under contract no. DEAC05-00OR22725 with UT-Battelle. J.K. is supported through the Scientific User Facilities Division of the Department of Energy (DOE) Office of Science.
Editor: David Cafiso.
Footnotes
Milka Doktorova, Frederick A. Heberle, and Robert A. Dick contributed equally to this work.
Supporting Materials and Methods, thirteen figures, five tables, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30987-6.
Contributor Information
Frederick A. Heberle, Email: heberlefa@ornl.gov.
Robert A. Dick, Email: rad82@cornell.edu.
Supporting Material
Lipid acyl chains and headgroups are in light and dark gray, respectively. MA coloring is as described in the main text. All lysine residues are shown in ball-and-stick representation and colored according to the helix to which they belong, except for K18 (gold), which is positioned in a loop, and K95 (silver), which is positioned in helix 6. Water and ion atoms are omitted for viewing clarity.
Lipid acyl chains and headgroups are in light and dark gray, respectively. MA coloring is as described in the main text. All lysine residues are shown in ball-and-stick representation and colored according to the helix to which they belong, except for K18 (gold), which is positioned in a loop, and K95 (silver), which is positioned in helix 6. Water and ion atoms are omitted for viewing clarity.
References
- 1.Dick R.A., Vogt V.M. Membrane interaction of retroviral Gag proteins. Front. Microbiol. 2014;5:187. doi: 10.3389/fmicb.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono A., Freed E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brügger B., Glass B., Kräusslich H.G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan R., Uchil P.D., Wenk M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorizate M., Sachsenheimer T., Brügger B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell. Microbiol. 2013;15:292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- 6.Krementsov D.N., Rassam P., Thali M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanda H., Datta S.A., Curtis J.E. Electrostatic interactions and binding orientation of HIV-1 matrix studied by neutron reflectivity. Biophys. J. 2010;99:2516–2524. doi: 10.1016/j.bpj.2010.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick R.A., Datta S.A., Vogt V.M. Hydrodynamic and membrane binding properties of purified Rous sarcoma virus Gag protein. J. Virol. 2015;89:10371–10382. doi: 10.1128/JVI.01628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta S.A., Heinrich F., Nanda H. HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. J. Mol. Biol. 2011;406:205–214. doi: 10.1016/j.jmb.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machán R., Hof M. Recent developments in fluorescence correlation spectroscopy for diffusion measurements in planar lipid membranes. Int. J. Mol. Sci. 2010;11:427–457. doi: 10.3390/ijms11020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goksu E.I., Longo M.L. Ternary lipid bilayers containing cholesterol in a high curvature silica xerogel environment. Langmuir. 2010;26:8614–8624. doi: 10.1021/la9046885. [DOI] [PubMed] [Google Scholar]
- 12.Heberle F.A., Petruzielo R.S., Katsaras J. Bilayer thickness mismatch controls domain size in model membranes. J. Am. Chem. Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 13.Marquardt D., Heberle F.A., Katsaras J. On scattered waves and lipid domains: detecting membrane rafts with x-rays and neutrons. Soft Matter. 2015;11:9055–9072. doi: 10.1039/c5sm01807b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pencer J., Krueger S., Katsaras J. Method of separated form factors for polydisperse vesicles. J. Appl. Cryst. 2006;39:293–303. [Google Scholar]
- 15.Kucerka N., Nagle J.F., Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophys. J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlier L., Louet M., Floquet N. Coarse-grained simulations of the HIV-1 matrix protein anchoring: revisiting its assembly on membrane domains. Biophys. J. 2014;106:577–585. doi: 10.1016/j.bpj.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick R.A., Goh S.L., Vogt V.M. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. USA. 2012;109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nezil F.A., Bloom M. Combined influence of cholesterol and synthetic amphiphilic peptides upon bilayer thickness in model membranes. Biophys. J. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kucerka N., Perlmutter J.D., Sachs J.N. The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and x-ray scattering. Biophys. J. 2008;95:2792–2805. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh T.J., Magid A.D., Simon S.A. Cholesterol modifies the short-range repulsive interactions between phosphatidylcholine membranes. Biochemistry. 1989;28:17–25. doi: 10.1021/bi00427a004. [DOI] [PubMed] [Google Scholar]
- 22.Subczynski W.K., Wisniewska A., Kusumi A. Hydrophobic barriers of lipid bilayer membranes formed by reduction of water penetration by alkyl chain unsaturation and cholesterol. Biochemistry. 1994;33:7670–7681. doi: 10.1021/bi00190a022. [DOI] [PubMed] [Google Scholar]
- 23.Simon S.A., McIntosh T.J., Latorre R. Influence of cholesterol on water penetration into bilayers. Science. 1982;216:65–67. doi: 10.1126/science.7063872. [DOI] [PubMed] [Google Scholar]
- 24.Kingsley P.B., Feigenson G.W. Synthesis of a perdeuterated phospholipid—1,2-dimyristoyl-sn-glycero-3-phosphocholine-D72. Chem. Phys. Lipids. 1979;24:135–147. [Google Scholar]
- 25.Wen Y., Dick R.A., Vogt V.M. Effects of membrane charge and order on membrane binding of the retroviral structural protein Gag. J. Virol. 2016;90:9518–9532. doi: 10.1128/JVI.01102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold O., Bilheux J.C., Zikoysky J. Mantid—data analysis and visualization package for neutron scattering and mu SR experiments. Nucl. Instrum. Meth. A. 2014;764:156–166. [Google Scholar]
- 27.Heberle F.A., Anghel V.N.P., Katsaras J. Scattering from phase-separated vesicles. I. An analytical form factor for multiple static domains. J. Appl. Cryst. 2015;48:1391–1404. [Google Scholar]
- 28.Kučerka N., Nagle J.F., Balgavý P. Models to analyze small-angle neutron scattering from unilamellar lipid vesicles. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;69:051903. doi: 10.1103/PhysRevE.69.051903. [DOI] [PubMed] [Google Scholar]
- 29.McDonnell J.M., Fushman D., Cowburn D. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 1998;279:921–928. doi: 10.1006/jmbi.1998.1788. [DOI] [PubMed] [Google Scholar]
- 30.Andrec M., Snyder D.A., Levy R.M. A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins. 2007;69:449–465. doi: 10.1002/prot.21507. [DOI] [PubMed] [Google Scholar]
- 31.Nelle T.D., Wills J.W. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dick R.A., Barros M., Vogt V.M. Membrane binding of the Rous sarcoma virus Gag protein is cooperative and dependent on the spacer peptide assembly domain. J. Virol. 2015;90:2473–2485. doi: 10.1128/JVI.02733-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best R.B., Zhu X., MacKerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venable R.M., Brown F.L., Pastor R.W. Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem. Phys. Lipids. 2015;192:60–74. doi: 10.1016/j.chemphyslip.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doktorova M., Harries D., Khelashvili G. Determination of bending rigidity and tilt modulus of lipid membranes from real-space fluctuation analysis of molecular dynamics simulations. Phys. Chem. Chem. Phys. 2017;19:16806–16818. doi: 10.1039/c7cp01921a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalton A.K., Murray P.S., Vogt V.M. Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J. Virol. 2005;79:6227–6238. doi: 10.1128/JVI.79.10.6227-6238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kučerka N., Nieh M.P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 40.Pan J., Cheng X., Katsaras J. The molecular structure of a phosphatidylserine bilayer determined by scattering and molecular dynamics simulations. Soft Matter. 2014;10:3716–3725. doi: 10.1039/c4sm00066h. [DOI] [PubMed] [Google Scholar]
- 41.Kučerka N., Pencer J., Katsaras J. Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles. Eur. Phys. J. E Soft Matter. 2007;23:247–254. doi: 10.1140/epje/i2007-10202-8. [DOI] [PubMed] [Google Scholar]
- 42.Heberle F.A., Feigenson G.W. Phase separation in lipid membranes. Cold Spring Harb. Perspect. Biol. 2011;3:a004630. doi: 10.1101/cshperspect.a004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almeida P.F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Huang J., Swanson J.E., Feigenson G.W. Nonideal mixing of phosphatidylserine and phosphatidylcholine in the fluid lamellar phase. Biophys. J. 1993;64:413–425. doi: 10.1016/S0006-3495(93)81382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khelashvili G., Harries D. Modeling signaling processes across cellular membranes using a mesoscopic approach. Annu. Rep. Comput. Chem. 2010;6:236–261. [Google Scholar]
- 46.Khelashvili G., Weinstein H., Harries D. Protein diffusion on charged membranes: a dynamic mean-field model describes time evolution and lipid reorganization. Biophys. J. 2008;94:2580–2597. doi: 10.1529/biophysj.107.120667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callahan E.M., Wills J.W. Repositioning basic residues in the M domain of the Rous sarcoma virus gag protein. J. Virol. 2000;74:11222–11229. doi: 10.1128/jvi.74.23.11222-11229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andelman D. Introduction to electrostatics in soft and biological matter. In: Poon W.C.K., Andelman D., editors. Soft Condensed Matter Physics in Molecular and Cell Biology. Taylor & Francis; Abingdon, United Kingdom: 2006. pp. 97–122. [Google Scholar]
- 49.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 50.Huang J., Feigenson G.W. Monte Carlo simulation of lipid mixtures: finding phase separation. Biophys. J. 1993;65:1788–1794. doi: 10.1016/S0006-3495(93)81234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srinivasan J., Trevathan M.W., Case D.A. Application of a pairwise generalized Born model to proteins and nucleic acids: inclusion of salt effects. Theor. Chem. Acc. 1999;101:426–434. [Google Scholar]
- 52.Gohlke H., Kiel C., Case D.A. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 53.Zoete V., Meuwly M., Karplus M. Study of the insulin dimerization: binding free energy calculations and per-residue free energy decomposition. Proteins. 2005;61:79–93. doi: 10.1002/prot.20528. [DOI] [PubMed] [Google Scholar]
- 54.Pan J., Tristram-Nagle S., Nagle J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dalton A.K., Ako-Adjei D., Vogt V.M. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murate M., Kobayashi T. Revisiting transbilayer distribution of lipids in the plasma membrane. Chem. Phys. Lipids. 2016;194:58–71. doi: 10.1016/j.chemphyslip.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Kučerka N., van Oosten B., Katsaras J. Molecular structures of fluid phosphatidylethanolamine bilayers obtained from simulation-to-experiment comparisons and experimental scattering density profiles. J. Phys. Chem. B. 2015;119:1947–1956. doi: 10.1021/jp511159q. [DOI] [PubMed] [Google Scholar]
- 58.Murray P.S., Li Z., Murray D. Retroviral matrix domains share electrostatic homology: models for membrane binding function throughout the viral life cycle. Structure. 2005;13:1521–1531. doi: 10.1016/j.str.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 59.Barros M., Heinrich F., Lösche M. Membrane binding of HIV-1 matrix protein: dependence on bilayer composition and protein lipidation. J. Virol. 2016;90:4544–4555. doi: 10.1128/JVI.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heftberger P., Kollmitzer B., Pabst G. In situ determination of structure and fluctuations of coexisting fluid membrane domains. Biophys. J. 2015;108:854–862. doi: 10.1016/j.bpj.2014.11.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lipid acyl chains and headgroups are in light and dark gray, respectively. MA coloring is as described in the main text. All lysine residues are shown in ball-and-stick representation and colored according to the helix to which they belong, except for K18 (gold), which is positioned in a loop, and K95 (silver), which is positioned in helix 6. Water and ion atoms are omitted for viewing clarity.
Lipid acyl chains and headgroups are in light and dark gray, respectively. MA coloring is as described in the main text. All lysine residues are shown in ball-and-stick representation and colored according to the helix to which they belong, except for K18 (gold), which is positioned in a loop, and K95 (silver), which is positioned in helix 6. Water and ion atoms are omitted for viewing clarity.