Introduction
Neurons are highly polarized and compartmentalized cells with processes that extend hundreds of microns away from the soma, forming up to thousands of synaptic connections with other neurons. Events that occur at synapses – such as synaptic activity, neurotrophic signaling, and injury – often result in changes in gene expression in the nucleus. Neurons employ several mechanisms to relay signals from synapses to the nucleus. Signals can be conveyed to the nucleus rapidly via electrochemical signaling (within milliseconds) or calcium waves (within seconds), or signals can be conveyed to the nucleus slowly through the physical translocation of signaling proteins from synapses to the nucleus (within minutes or hours). In addition to covering distinct domains of temporal coupling between stimulation and transcription, these various synapse-to-nucleus signaling pathways relay information from distinct subcellular domains, ranging from neuron-wide to synapse-specific. In this review, we highlight recent discoveries regarding the synapse-to-nucleus transport of individual signaling proteins. Rather than catalog all synaptonuclear signaling proteins, we discuss exemplar proteins that provide functional and mechanistic insights. We will focus primarily on synapse-to-nucleus communication during synaptic plasticity, but we will also include examples from neural development, disease, and injury.
Mechanisms of synapse-to-nucleus communication
Electrochemical signaling from the synapse to the nucleus occurs when glutamatergic synaptic stimulation generates an above-threshold excitatory postsynaptic potential (EPSP) to elicit action potentials. Back propagation of the action potential results in depolarization at the soma, opening L-type voltage-gated calcium channels. The resulting increase in intracellular calcium concentration acts on calcium-sensitive signaling proteins to regulate gene expression [1,2]. Electrochemical signals travel from synapses to the nucleus within milliseconds and this rapid mechanism of signaling provides a faithful indicator of firing frequency [3]. Since action potential firing depolarizes the entire neuron, electrochemical signaling cannot convey synapse-specific information about the site of stimulation, but rather serves as a neuron-wide means of signaling from cell-surface receptors to the genome.
A second mechanism of synaptonuclear communication involves the propagation of calcium waves through the endoplasmic reticulum (ER). Synaptic input can stimulate G protein-coupled receptors and subsequently create inositol triphosphate (IP3) molecules. IP3 activates its receptor on the ER (which extends from the nuclear membrane to the axon and distal dendrites) and causes calcium to be released from internal stores. The calcium wave is propagated to the soma by calcium-induced calcium release (CIRC), where calcium can act on second messengers and diffuse into the nucleus [1]. Calcium waves have been observed in slice preparations from hippocampus and cortex, but have not been examined in vivo [4]. The regenerative nature of ER calcium waves indicates that this signaling pathway would likely mediate axonal or dendritic branch-specific, rather than synapse-specific, signaling to the nucleus.
The transport of proteins from synapse-to-nucleus is a slower mechanism of signaling that is mediated by diffusion, active transport of proteins, and signaling endosomes. Protein transport occurs on the order of minutes to hours, allowing for gene expression to be impacted long after the stimulus has ended. Specific types of synaptic stimulation have been shown to trigger the translocation of signaling proteins and to regulate gene expression in the absence of long-range electrochemical or calcium signaling [5**,6]. However, in some cases, action potentials are sufficient to induce changes in gene expression even in the absence of synaptic connections [7]. Thus, both electrochemical and synaptonuclear protein signals are likely to be important for activity-dependent gene expression, with varying degrees of importance depending on the stimulation type and the time point examined. The existence of multiple synapse-to-nucleus signaling pathways allows the neuron to couple refined spatiotemporal integration of synaptic stimulation (ie integration of stimulation frequency and location) with the appropriate pattern of gene expression.
Signaling from the post-synaptic compartment to the nucleus
Dozens of proteins have been shown to undergo activity-dependent synapse-to-nucleus translocation, with important consequences for gene expression and synaptic plasticity (Figure 1). Here we review recent findings on a handful of synaptonuclear signaling proteins, selected because each illustrates a specific principle. We then briefly address the motor proteins and nuclear transporters that mediate their nuclear import.
Figure 1. Examples of activity-dependent signaling proteins that translocate from presynaptic and postsynaptic compartments to the nucleus.
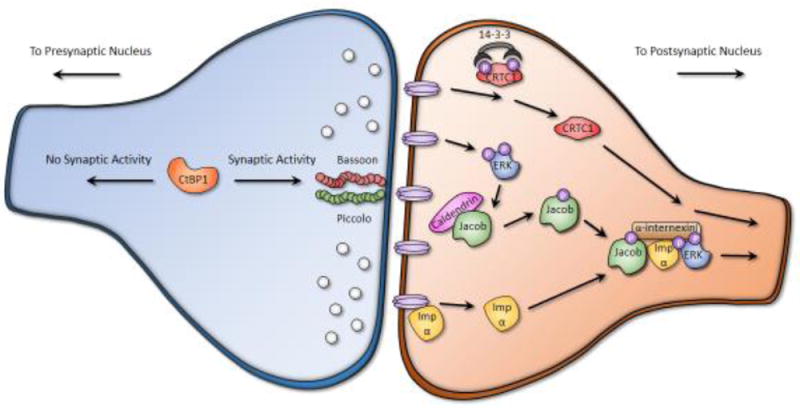
Presynaptic compartment to nucleus (left, in blue): In silenced neurons, CtBP1 translocates from the presynaptic terminal to the nucleus whereas, in stimulated neurons, CtBP1 is retained at the presynaptic compartment through an interaction with Bassoon and Piccolo [27**]. Postsynaptic compartment to nucleus (right, in orange): Upon NMDA receptor activation, CRTC1 is dephosphorylated by calcineurin (not shown) and released from 14-3-3 proteins, enabling the active transport of CRTC1 to the nucleus. Stimulation of synaptic NMDA receptors also activates ERK, which phosphorylates Jacob and releases it from caldendrin. Jacob is transported to the nucleus in a complex with ERK1/2, α-internexin, and importin α [9]. Notably, importin α associates with the cytoplasmic tail of the NMDA receptor and is released in an activity-dependent manner, enabling importin α to bind to and transport cargo following stimulation [35]. The presynaptic illustration is based on Kravchick and Jordan [46] and the postsynaptic illustration is based on Panayotis et al [12].
Jacob, a protein first characterized as a caldendrin-binding partner, undergoes synapse-to-nucleus translocation following either synaptic or extrasynaptic NMDA receptor activation [8]. Upon synaptic NMDA receptor activation, Jacob is phosphorylated by the MAP Kinase ERK1/2 at Ser180 [9]. Once in the nucleus, phosphorylated Jacob activates the transcription factor CREB and promotes plasticity-related gene expression. Interestingly, Jacob also translocates to the nucleus after activation of extrasynaptic NMDARs, albeit without phosphorylation at Ser180 [9]. Nuclear expression of non-phosphorylated Jacob promotes CREB shut-off and cell death. Jacob provides an example of how the phosphorylation state of a synaptonuclear signaling protein can have important functional consequences in the nucleus. To preserve the phosphorylation state of Jacob during the long-distance transport from synapse-to-nucleus, Jacob forms a stable complex with ERK1/2 and the intermediate filament α-internexin. This mechanism of intermediate filament binding is similar to how ERK phosphorylation is preserved during long-distance axon-to-nucleus signaling [10] (see section below). Knockout of Jacob in mice induces hippocampal dysplasia with impairments in hippocampal long-term potentiation (LTP) and hippocampal-dependent learning [11], consistent with a role for this synaptonuclear signal in neuronal development, long-lasting plasticity, and memory.
Phosphorylation of ERK can also serve as a signaling mechanism from postsynaptic sites to the nucleus. Zhai et al. recently demonstrated that activation of a few dendritic spines with glutamate uncaging produced phosphorylation of ERK in the nucleus within tens of minutes [5**]. In this experimental setup, local activation of dendritic spines did not produce any global electrochemical or calcium signaling; instead, the propagation of the signal to the nucleus was likely produced by local phosphorylation of ERK and subsequent translocation to the nucleus [5**,12]. Activation of between three and seven dendritic spines was sufficient to trigger ERK-dependent activation of the transcription factors CREB and Elk-1 in the nucleus. This study provides an example of how synaptonuclear signaling proteins can be sufficient to produce changes in gene expression, in the absence of electrochemical and calcium wave signaling mechanisms. The study also addressed the spatial pattern of synaptic stimulation necessary for signaling to the nucleus. The authors found that dispersed stimulation of synapses on multiple dendritic branches, rather than clustered stimulation of synapses on a single dendritic branch, was required to trigger nuclear ERK signaling.
CRTC1 is a transcriptional co-activator of CREB that undergoes activity-dependent synapse-to-nucleus translocation. CRTC1 is bound to 14-3-3 proteins at the synapse and, upon glutamate receptor activation, CRTC1 is dephosphorylated at specific residues, released from 14-3-3, and actively transported to the nucleus [13,14]. In the nucleus, CRTC1 binds to CREB and enhances the transcription of specific activity-dependent genes including Bdnf, Arc, and Fos [13,15]. Knockdown studies have demonstrated a role for CRTC1 in late-phase LTP and contextual fear conditioning, supporting the hypothesis that synaptonuclear protein signals are important for long-lasting plasticity [16,17]. Furthermore, CRTC1 exists in different phosphorylation states in response to different types of stimulations. This finding evokes the exciting possibility that stimuli are encoded in the phosphorylation state of CRTC1, coupling these stimuli to distinct patterns of gene expression in the nucleus [13].
Several novel synaptonuclear signaling proteins have been identified by characterizing the proteome of excitatory synapses and the interactome of NMDA receptors [18-21]. These systematic approaches allow for the identification of proteins with putative nuclear localization signals (NLS) within the synapse. Jordan et al. characterized the postsynaptic density of rat neurons using mass spectrometry, and identified 11 synaptic proteins containing a known NLS as well as additional synaptic proteins that were predicted to have nuclear expression [18]. This systematic approach led to the discovery of AIDA-1 as a synaptonuclear signaling protein that translocates to the nucleus after NMDAR activation to regulate nucleolar numbers and protein synthesis [19]. More recently, Dinamarca et al. characterized protein binding partners of the cytoplasmic tail of the GluN2A subunit of NMDARs, and identified the ring finger protein RNF10 as a novel binding partner [20]. The authors then demonstrated that RNF10 translocates to the nucleus during LTP to regulate gene expression. Similarly, PRR7 was previously identified in the postsynaptic density, and then recently shown to translocate to the nucleus following NMDAR activation, where it enhances c-Jun-dependent transcriptional activity during excitotoxicity [18,21]. These studies used proteomic profiling of synapses in combination with predicted nuclear localization data to contribute to the growing body of work identifying the population of proteins that undergo activity-dependent translocation.
Many of the aforementioned synaptonuclear signaling proteins undergo dynein-mediated active transport along microtubules to reach the nucleus [22]. Dendrites, unlike axons, have microtubules of mixed polarity and therefore both dyneins and kinesins could transport synaptonuclear signaling proteins to the nucleus. Only a few kinesins display dendritic localization, but recently Ghiretti et al. demonstrated a role for KIF21B in the retrograde trafficking of BDNF-bound Trk receptors in dendrites [23*]. The function of KIF21B was found to be activity-dependent, as neuronal stimulation enhanced the motility of KIF21B and reduced its microtubule remodeling ability. While this study focused on the role of KIF21B in mediating transport of signaling endosomes, similar activity-dependent regulation of motor proteins may exist for the retrograde transport of soluble proteins.
A critical aspect of synapse-to-nucleus signaling is the mechanism of import of signaling proteins into the nucleus. Small proteins can diffuse through the nuclear pore complex (NPC) passively, but proteins larger than 40-60 kDa require active transport by the family of importin (karyopherin) proteins [24]. In the classical nuclear import pathway, importin α binds to the NLS of a cargo protein and importin β binds to this complex to facilitate nuclear import. Previous work has shown that importins localize to synapses in silenced neurons, undergo translocation to the nucleus following neuronal stimulation, and that active nuclear import is required for long-lasting neuronal plasticity [25]. This evidence suggests that importins shuttle cargo from synapse-to-nucleus and that the transported cargo is crucial for long-term plasticity. Indeed, two of the aforementioned synaptonuclear signaling proteins, Jacob and RNF10, are cargoes of the importin α1 isoform at the synapse [8,20], while another synaptonuclear signaling protein, NF-κB, is a cargo of importin α2 [26]. These studies suggest that different importin isoforms have distinct cargo and relay different signaling proteins to the nucleus [24]. To support this claim and identify the potential cargoes of different importin isoforms, further work will be needed to systematically characterize the interactome of importin proteins at the synapse.
Signaling from the pre-synaptic compartment to the nucleus
Many proteins have been shown to undergo activity-dependent translocation from the post-synaptic compartment to the nucleus (see above), but less is known about activity-dependent signaling from the presynaptic compartment. Recently, Ivanova et al. demonstrated that the transcriptional co-repressor CtBP1 undergoes activity-dependent shuttling from the presynaptic compartment to the nucleus [27**]. In silenced neurons, CtBP1 translocates to the nucleus where it functions as a transcriptional co-repressor. During neuronal activity, however, CtBP1 is retained in the presynaptic compartment via an interaction with the scaffolding proteins Bassoon and Piccolo. This presynaptic localization of CtBP1 relieves the nucleus of co-repressor activity, allowing for upregulation of activity-dependent genes including Arc, Fos, Bdnf, Egr1, Egr4, and Junb. CtBP1 signaling provides a relatively slow mechanism of synapse-to-nucleus communication, as the translocation of CtBP1 takes several hours. This study contributes to previous work demonstrating that signaling proteins do undergo activity-dependent presynaptic synaptonuclear translocation to regulate gene expression. For example, in Aplysia neurons, MAPK translocates from the presynaptic terminal to the nucleus during 5-HT-induced long-term facilitation of sensory neurons, where it phosphorylates transcription factors important for long-lasting plasticity [6].
Axon-to-nucleus communication has been more widely studied in the context of neural development, disease, and injury. During development, retrograde transport of target-derived signals acts to promote neuron survival, axon outgrowth, and circuit formation [28]. Ji and Jaffrey recently demonstrated that two target-derived signals, BDNF and BMP4, can converge on the growth cone to induce local translation of the transcription factor SMAD. Subsequent retrograde transport of a SMAD-containing signaling endosome to the nucleus can modulate neuronal subtype specification in the developing trigeminal ganglia [29]. Synthesis of transcription factors within the synapse can also serve as a retrograde signal during neurodegeneration. Recently, Baleriola et al. used local application of Aβ1-42 as a model of Alzheimer’s disease and demonstrated that the transcription factor CREB2 (ATF4) is translated in axons in response to local application of Aβ1-42 [30*]. CREB2 then undergoes retrograde transport to the nucleus, where it promotes cell death by upregulating C/EBP homologous protein (CHOP) expression. The synaptonuclear signaling ability of CREB2 appears to be versatile, as Lai et al. previously demonstrated that CREB2 undergoes activity-dependent translocation from distal dendrites to the nucleus during long-term depression of rodent hippocampal neurons [31]. Axon-to-nucleus signaling is also involved in the response to nerve injury, as new gene transcription is required for regeneration. Axon-to-nucleus communication during nerve injury is reviewed extensively elsewhere and involves dynein-mediated transport of transcription factors including Stat3, kinases such as ERK, and other signaling molecules [10,32,33]. Parallels can be drawn with postsynaptic synaptonuclear signaling. In both pathways, importins play a critical role in transporting signaling molecules from distal processes to the nucleus and preservation of a distally generated signal (phosphorylated ERK) involves binding to an intermediate filament (vimentin) [10,34].
Cell type-specific and synapse type-specific signaling to the nucleus?
A majority of the research has focused on synaptonuclear signaling proteins at excitatory synapses [5**,19,26,35] and in excitatory neurons [13,36]. Recent work has shown that inhibitory interneurons employ a γCaMKI shuttle for translocation of Ca2+/CaM during activity-dependent surface-to-nucleus communication, which is in contrast to the γCaMKII shuttle employed by excitatory neurons [37,38**]. The authors note that parvalbumin-positive inhibitory interneurons fire more frequently and display larger calcium increases than excitatory neurons, which may necessitate this alternate signaling mechanism in inhibitory neurons. It is important to note that the translocation of γCaMKI/II occurs in response to calcium influx via the somatically-localized Cav1 channels, and therefore γCaMKI/II translocation is a result of electrochemical signaling rather than the local activation of signaling proteins at the synapse. It will be interesting to determine whether specific neuronal cell types also employ distinct sets of synaptonuclear signaling proteins, perhaps suited to their level of excitability and other properties. Of note, the synaptonuclear messengers Jacob and CRTC1 have been shown to translocate in excitatory but not inhibitory neurons [13,36], while CREB acts as an axon-to-nucleus messenger in developing DRG neurons but not in other neuronal cell types [39,40].
Studies of postsynaptic synaptonuclear signaling have focused predominantly on messengers that translocate from excitatory synapses to the nucleus. This focus is consistent with the fact that most studies of transcription-dependent LTP have focused on excitatory synapses [41,42]. However, long-term plasticity of inhibitory synapses has also been described, raising the question of whether and how the transport of proteins from inhibitory synapses to the nucleus contributes to persistent plasticity in these cell types [43]. Of great utility to these studies, Uezu et al. recently characterized the postsynaptic proteome of inhibitory synapses and identified many signaling proteins including kinases and phosphatases [44]. It will be interesting to determine whether these enzymes, in addition to modifying substrate proteins locally at the synapse, also undergo long-range signaling to the nucleus to impact gene expression.
The emerging appreciation that glia are critical elements of synapses, and play central roles during synaptic plasticity [45] suggests that it would also be of interest to explore long-distance signaling from distal glial processes to the nucleus during neuronal plasticity.
Conclusion
Protein transport is a mechanism of synapse-to-nucleus signaling that offers unique signaling capabilities. Protein transport is slower than electrochemical or calcium signaling, providing a longer time over which gene expression can be impacted. Certain synaptonuclear signals, such as Jacob and CRTC1, exist in multiple phosphorylation states, providing the capability for additional information about stimulus type (see [13]) or location (see [9]) to be encoded in the phosphorylation state of the signaling protein. A challenge for encoding information in the post-translation modifications (PTMs) of a protein is that the modification could be removed by enzymes during the long journey from the synapse to the nucleus. However, PTMs can be preserved when signaling proteins form stable complexes with other proteins during transport to the nucleus [9,10].
While the study of candidate synaptonuclear proteins has provided insight into the cell biological mechanisms of synapse-to-nucleus communication, further work is needed to globally identify the population of proteins that undergo synapse-to-nucleus translocation in response to different stimulations. Such an approach would provide systematic information about the types of signals that translocate to the nucleus; the identity and function of each synaptonuclear signaling protein would likely provide clues about the transcriptional programs that are initiated in response to synaptic stimuli. However, an important consideration when investigating novel synaptonuclear signaling proteins is whether the increased nuclear accumulation of the protein is due to translocation from stimulated synapses, rather than simply from translocation from the soma. A conclusive way to verify the synapse-to-nucleus translocation is to tag the protein of interest with a photoconvertible fluorophore, convert the protein at distal synaptic processes, and track the protein localization following synaptic stimulation [14].
Studies of synapse-to-nucleus signaling in neurons also provide a new, cell biological lens through which to understand how neurons integrate stimulation to encode information. Significant attention has been paid to the role of stimulation frequency in neural encoding of information. This provides a framework for studying how various forms of synapse-to-nucleus signaling couples temporal patterns of stimulation with specific patterns of gene expression. It may be that electrochemical signaling contributes to rapid changes in gene expression whereas slower protein translocation from synapse-to-nucleus contributes to more sustained changes. As opposed to the contribution of temporal patterns of neuronal firing, much less is known about how spatial patterns of stimulation contribute to neural coding. Identification and characterization of the protein signals that are relayed from specific stimulated synapses to the nucleus has the potential to provide fundamental insights into this open question.
Highlights.
Synapse to nucleus signaling couples synaptic stimulation with transcriptional regulation.
Stimulation of a few dendritic spines is sufficient to activate nuclear transcription factors.
The phosphorylation state of synaptonuclear signaling proteins can encode stimulus-specific information.
Activity regulates the nuclear import of both pre- and post-synaptic signaling molecules.
Mechanisms of excitation-transcription coupling are distinct in inhibitory and excitatory neurons.
Acknowledgments
We thank Sylvia Neumann, Jennifer Achiro, and Sarah Van Driesche for their comments on the manuscript, and members of the Martin lab for helpful discussion. Research on synapse to nucleus signaling in the Martin lab is funded by NIH R01MH077022 (to KCM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bading H. Nuclear calcium signalling in the regulation of brain function. Nat Rev Neurosci. 2013;14:593–608. doi: 10.1038/nrn3531. Internet. [DOI] [PubMed] [Google Scholar]
- 2.Greer PL, Greenberg ME. From Synapse to Nucleus: Calcium-Dependent Gene Transcription in the Control of Synapse Development and Function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Bengtson CP, Freitag HE, Weislogel JM, Bading H. Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys J. 2010;99:4066–4077. doi: 10.1016/j.bpj.2010.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross WN. Understanding calcium waves and sparks in central neurons. Nat Rev Neurosci. 2012;13:157–168. doi: 10.1038/nrn3168. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Zhai S, Ark ED, Parra-Bueno P, Yasuda R. Long-distance integration of nuclear ERK signaling triggered by activation of a few dendritic spines. Science. 2013;342:1107–11. doi: 10.1126/science.1245622. [Internet]. In this landmark study, the authors show that local activation of a few dendritic spines is sufficient to induce nuclear activation of ERK and downstream transcription factors, in the absence of electrochemical or calcium signaling. This study also demonstrates that stimulation of spines on multiple dendritic branches can induce nuclear changes more effectively than stimulating the same number of spines on a single dendrite, providing insight into how signals are integrated spatially during synapse-to-nucleus communication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin KC, Michael D, Rose JC, Barad M, Casadio A, Zhu H, Kandel ER. MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia. Neuron. 1997;18:899–912. doi: 10.1016/s0896-6273(00)80330-x. [DOI] [PubMed] [Google Scholar]
- 7.Eshete F, Fields RD. Spike frequency decoding and autonomous activation of Ca2+-calmodulin-dependent protein kinase II in dorsal root ganglion neurons. J Neurosci. 2001;21:6694–6705. doi: 10.1523/JNEUROSCI.21-17-06694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, König I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, et al. Caldendrin-Jacob: A protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6:0286–0306. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpova A, Mikhaylova M, Bera S, Bär J, Reddy PP, Behnisch T, Rankovic V, Spilker C, Bethge P, Sahin J, et al. Encoding and transducing the synaptic or extrasynaptic origin of NMDA receptor signals to the nucleus. Cell. 2013;152:1119–1133. doi: 10.1016/j.cell.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Perlson E, Michaelevski I, Kowalsman N, Ben-Yaakov K, Shaked M, Seger R, Eisenstein M, Fainzilber M. Vimentin Binding to Phosphorylated Erk Sterically Hinders Enzymatic Dephosphorylation of the Kinase. J Mol Biol. 2006;364:938–944. doi: 10.1016/j.jmb.2006.09.056. [DOI] [PubMed] [Google Scholar]
- 11.Spilker C, Nullmeier S, Grochowska KM, Schumacher A, Butnaru I, Macharadze T, Gomes GM, Yuanxiang PA, Bayraktar G, Rodenstein C, et al. A Jacob/Nsmf Gene Knockout Results in Hippocampal Dysplasia and Impaired BDNF Signaling in Dendritogenesis. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panayotis N, Karpova A, Kreutz MR, Fainzilber M. Macromolecular transport in synapse to nucleus communication. Trends Neurosci. 2015;38:108–116. doi: 10.1016/j.tins.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Ch’ng TH, Uzgil B, Lin P, Avliyakulov NK, O’Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ch’ng TH, DeSalvo M, Lin P, Vashisht A, Wohlschlegel JA, Martin KC. Cell biological mechanisms of activity-dependent synapse to nucleus translocation of CRTC1 in neurons. Front Mol Neurosci. 2015;8:48. doi: 10.3389/fnmol.2015.00048. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuchi M, Kirikoshi Y, Mori A, Eda R, Ihara D, Takasaki I, Tabuchi A, Tsuda M. Excitatory GABA induces BDNF transcription via CRTC1 and phosphorylated CREB-related pathways in immature cortical cells. J Neurochem. 2014;131:134–146. doi: 10.1111/jnc.12801. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One. 2006;1 doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nonaka M, Kim R, Fukushima H, Sasaki K, Suzuki K, Okamura M, Ishii Y, Kawashima T, Kamijo S, Takemoto-Kimura S, et al. Region-Specific Activation of CRTC1-CREB Signaling Mediates Long-Term Fear Memory. Neuron. 2014;84:92–106. doi: 10.1016/j.neuron.2014.08.049. [DOI] [PubMed] [Google Scholar]
- 18.Jordan Ba, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert Ta. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Jordan Ba, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- 20.Dinamarca MC, Guzzetti F, Karpova A, Lim D, Mitro N, Musardo S, Mellone M, Marcello E, Stanic J, Samaddar T, et al. Ring finger protein 10 is a novel synaptonuclear messenger encoding activation of NMDA receptors in hippocampus. Elife. 2016;5 doi: 10.7554/eLife.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kravchick DO, Karpova A, Hrdinka M, Lopez-rojas J, Iacobas S, Carbonell AU, Iacobas DA, Kreutz MR, Jordan BA. Synaptonuclear messenger PRR 7 inhibits c-Jun ubiquitination and regulates NMDA-mediated excitotoxicity. EMBO J. 2016 doi: 10.15252/embj.201593070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim AFY, Lim WL, Ch’ng TH. Activity-dependent synapse to nucleus signaling. Neurobiol Learn Mem. 2016 doi: 10.1016/j.nlm.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 23*.Ghiretti A, Thies E, Tokito M, Lin T, Ostap M, Kneussel M, Holzbaur E. Activity-Dependent Regulation of Distinct Transport and Cytoskeletal Remodeling Functions of the Dendritic Kinesin KIF21B. Neuron. 2016;92:857–872. doi: 10.1016/j.neuron.2016.10.003. This study provides a role for kinesin-4 KIF21B in activity-dependent trafficking in dendrites. The authors show that KIF21B is involved in retrograde transport of BDNF-bound TrkB receptors in dendrites and that neuronal activity enhances KIF21B’s motor activity while downregulating its cytoskeletal remodeling function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lever MB, Karpova A, Kreutz MR. An Importin Code in neuronal transport from synapse-to-nucleus? Front Mol Neurosci. 2015;8:33. doi: 10.3389/fnmol.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson KR, Otis KO, Chen DY, Zhao Y, O’Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity: A role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 26.Marcora E, Kennedy MB. The Huntington’s disease mutation impairs Huntingtin’s role in the transport of NF-κB from the synapse to the nucleus. Hum Mol Genet. 2010;19:4373–4384. doi: 10.1093/hmg/ddq358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Ivanova D, Dirks A, Montenegro-Venegas C, Schone C, Altrock WD, Marini C, Frischknecht R, Schanze D, Zenker M, Gundelfinger ED, et al. Synaptic activity controls localization and function of CtBP1 via binding to Bassoon and Piccolo. EMBO J. 2015;34:1056–1077. doi: 10.15252/embj.201488796. [Internet]. In this study, the authors demonstrate that the transcriptional co-repressor CtBP1 undergoes activity-dependent shuttling from the presynaptic compartment to the nucleus. In silenced neurons, CtBP1 translocates to the nucleus and impairs the expression of many activity-dependent genes. During synaptic activity, CtBP1 is retained in the presynaptic compartment via an interaction with scaffolding proteins Bassoon and Piccolo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrington AW, Ginty DD. Long-distance retrograde neurotrophic factor signalling in neurons. Nat Rev Neurosci. 2013;14:177–187. doi: 10.1038/nrn3253. Internet. [DOI] [PubMed] [Google Scholar]
- 29.Ji SJ, Jaffrey SR. Intra-axonal Translation of SMAD1/5/8 Mediates Retrograde Regulation of Trigeminal Ganglia Subtype Specification. Neuron. 2012;74:95–107. doi: 10.1016/j.neuron.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158:1159–1172. doi: 10.1016/j.cell.2014.07.001. The authors demonstrate a role for ATF4 (CREB2) as an axon-to-nucleus messenger during Alzheimer’s disease in basal forebrain cholinergic neurons that project to the dentate gyrus. Upon application of Aβ1-42 to axons, ATF4 is locally translated and undergoes retrograde transport to the nucleus, where it induces cell death via CHOP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai K-O, Zhao Y, Ch’ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci U S A. 2008;105:17175–80. doi: 10.1073/pnas.0803906105. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rishal I, Fainzilber M. Axon-soma communication in neuronal injury. Nat Rev Neurosci. 2014;15:32–42. doi: 10.1038/nrn3609. Internet. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31:1350–63. doi: 10.1038/emboj.2011.494. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry RBT, Doron-Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, Coppola G, McDonald MK, Gomes C, Geschwind DH, et al. Subcellular Knockout of Importin β1 Perturbs Axonal Retrograde Signaling. Neuron. 2012;75:294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffrey RA, Ch’ng TH, O’Dell TJ, Martin KC. Activity-dependent anchoring of importin alpha at the synapse involves regulated binding to the cytoplasmic tail of the NR1-1a subunit of the NMDA receptor. J Neurosci. 2009;29:15613–20. doi: 10.1523/JNEUROSCI.3314-09.2009. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikhaylova M, Karpova A, Bär J, Bethge P, YuanXiang P, Chen Y, Zuschratter W, Behnisch T, Kreutz MR. Cellular distribution of the NMDA-receptor activated synapto-nuclear messenger Jacob in the rat brain. Brain Struct Funct. 2014;219:843–860. doi: 10.1007/s00429-013-0539-1. [DOI] [PubMed] [Google Scholar]
- 37.Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, Zhang G, Neubert TA, Tsien RW. γcaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell. 2014;159:281–294. doi: 10.1016/j.cell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Cohen SM, Ma H, Kuchibhotla KV, Watson BO, Buzsáki G, Froemke RC, Tsien RW. Excitation-Transcription Coupling in Parvalbumin-Positive Interneurons Employs a Novel CaM Kinase-Dependent Pathway Distinct from Excitatory Neurons. Neuron. 2016;90:292–307. doi: 10.1016/j.neuron.2016.03.001. The authors demonstrate that, in inhibitory neurons, γCaMKI shuttles calmodulin from the cytoplasm to the nucleus in response to electrochemical signaling and that this γCaMKI shuttle is important for CREB phosphorylation. This mechanism is in contrast to the previously described γCaMKII shuttle found in excitatory neurons. The findings in this study suggest that different cell types employ different synaptonuclear signaling proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–59. doi: 10.1038/ncb1677. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tasdemir-Yilmaz OE, Segal RA. There and back again: Coordinated transcription, translation and transport in axonal survival and regeneration. Curr Opin Neurobiol. 2016;39:62–68. doi: 10.1016/j.conb.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Sci (New York, NY) 1994;265:1104–1107. doi: 10.1126/science.8066450. Internet. [DOI] [PubMed] [Google Scholar]
- 42.Chotiner JK, Khorasani H, Nairn AC, O’Dell TJ, Watson JB. Adenylyl cyclase-dependent form of chemical long-term potentiation triggers translational regulation at the elongation step. Neuroscience. 2003;116:743–752. doi: 10.1016/s0306-4522(02)00797-2. [DOI] [PubMed] [Google Scholar]
- 43.Mele M, Leal G, Duarte CB. Role of GABAA R trafficking in the plasticity of inhibitory synapses. J Neurochem. 2016 doi: 10.1111/jnc.13742. Internet. [DOI] [PubMed] [Google Scholar]
- 44.Uezu A, Kanak D, Bradshaw T, Soderblom E, Catavero C, Burette A, Weinberg R, Soderling S. Identification of an elaborate complex mediating postsynaptic inhibition. Sci (New York, NY) 2016;353:1123–1129. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stogsdill JA, Eroglu C. The interplay between neurons and glia in synapse development and plasticity. Curr Opin Neurobiol. 2017;42:1–8. doi: 10.1016/j.conb.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kravchick DO, Jordan BA. Presynapses go nuclear! EMBO J. 2015;34:984–6. doi: 10.15252/embj.201591331. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]


