Abstract
Background
The epidemiology of the pandemic A(H1N1) virus has been changing as population immunity continues to co-evolve with the virus. The impact of genetic changes in the virus on human’s susceptibility is an outstanding important question in vaccine design. In a community-based study, we aim to 1) determine the genetic characteristics of 2009–2015 pandemic H1N1 viruses, 2) assess antibody response following natural infections and 3) assess the correlation of A/California/07/09 antibody titers to protection in the 2013 and 2015 epidemics.
Methods
In a household transmission study, serum specimens from 253 individuals in Managua, Nicaragua were analyzed. Combined nose and throat swabs were collected to detect RT-PCR confirmed influenza infection and virus sequencing. Hemagglutination inhibition assays were performed and the protective titer for circulating H1N1pdm was determined.
Results
Clade 6b pandemic H1N1 viruses predominated in Nicaragua during the 2013 and 2015 seasons. Our household transmission study detected a household secondary attack rate of 17% in 2013 and 33% in 2015. Infected individuals, including vaccinees, showed an apparent antibody response to A/California/07/09. Baseline titers of A/California/07/09 antibodies were found to associate with protection in both seasons. A titer of ≥1:40 correlated to a 44% protection in children, a 29% protection in adults 15–49 years old and a 51% protection in adults 50–85 years old.
Conclusion
In 2013 and 2015, antibody titers to A/California/07/09 associated with an infection risk reduction amongst exposed household contacts. This is consistent with a detectable vaccine effectiveness reported in a number of studies.. Genetic changes in clade 6b viruses might have led to a reduced immunity in some whereas others might have been less affected. The use of human serologic data is important in virus characterization and if performed in a timely manner, could assist in vaccine strain selection.
Keywords: H1N1pdm virus, influenza, immune correlates, antibody titer, clade 6b
INTRODUCTION
Influenza is a major cause of respiratory infections with hospitalization and mortality rates typically highest amongst children and older adults [1–3]. Constant evolutionary change allows the virus to evade pre-existing immunity in the population resulting in reinfections with the same type/subtype of virus. Since its first emergence in 2009, pandemic A(H1N1) virus (H1N1pdm) has been undergoing constant genetic changes and it remains the predominate H1N1 virus worldwide [4, 5]. How genes changes the structure of virus epitopes and what its extrinsic impact is on human’s susceptibility are outstanding important questions in vaccine design [6]. Recent H1N1pdm viruses are most frequently clustered in genetic clade 6 and its contemporary subclades. Clade 6B viruses, characterized by a K163Q mutation, emerged in 2013 at the time when several regions reported more severe annual epidemics [7, 8]. It was uncertain whether genetic changes in H1Npdm increased population susceptibility, particularly amongst middle-age adults who were found more likely to show severe illness [9]. A study which sequentially infected ferrets revealed that pre-exposure to A/Chile/01/83 a seasonal H1N1 virus, but not several other seasonal H1N1 viruses or the Cal09 H1N1pdm virus, reduced cross-reactive antibody response to a reverse engineered 163Q virus in 38% of ferrets following a Cal09 re-challenge. This illustrates the unique advantage of using human serology data in antigenic characterization of H1N1pdm virus in addition to ferret serology – that infection history throughout individuals’ lifetime, an important base factor of how genetic changes in virus will translate to changes in pre-existing immunity of the human population, are well represented.
Human serologic data played a new key role in assessing vaccine virus candidates for the 2016–2017 season. This development creates a new paradigm in which immune response to a virus can be characterized among individuals of different ages, geographic locations and with different histories of influenza exposure. Retrospective studies of immune response to vaccine virus following natural infection have been used to evaluate vaccine match and help explain and validate early results of vaccine assessment. With regards to contemporary H1N1pdm, young and middle-aged adults were identified as a risk group that was disproportionately affected by the K163Q mutation, possibly owing to their exposure to specific seasonal H1N1 virus that was not shared with other birth cohorts [7, 10]. However, some individuals of this group remained capable of mounting strong 163Q virus antibody response in Cal09 vaccination studies [7]. Whereas influenza vaccine was found to be moderately efficacious against H1N1pdm in 2013–2014 in several regions [11–13], recent data found that there might be a lack of association between Cal09 antibodies and protection against clade 6B viruses [14]. Data related to this remains rare and using a community-based study design, we aim to further investigate the extent to which Cal09 antibodies are associated with protection against contemporary H1N1pdm in the general population.
In this report, we present genetic and serologic data collected in a household transmission study conducted in Managua, Nicaragua from 2013 through 2015. Our primary objectives are 1) to determine the genetic characteristics of influenza H1N1pdm viruses that circulated in each epidemic, 2) to assess antibody response to A/California/07/09 (Cal09) following natural infections and 3) to assess the correlation between hemagglutination inhibition antibody titers and protection against H1N1pdm infection in the general population.
METHODS
Study participants and procedures
Influenza index cases were recruited from a pediatric cohort study and the national influenza surveillance program at the Health Center Sócrates Flores Vivas (HCSFV), a government health center that provides free primary health care to the catchment area of District II in Managua [15–19]. Patients who 1) had a positive Influenza rapid test result; 2) experienced symptom onset of an acute respiratory infection within the previous 48 hours; 3) lived with at least one other household member; and 4) had no other household members experience any symptoms of acute respiratory infection within the previous two weeks were eligible to participate in the study with their household members.
All participants (index cases and household members) provided a nasal and oropharyngeal swab at enrollment. Daily symptom history and additional nasal and oropharyngeal swabs were collected from participants every 2–3 days over the 10–14 days following enrollment (for up to 5 visits) or when presenting to the HCSFV with respiratory symptoms or undifferentiated fever during the follow-up period (30–45 days). Symptoms monitored included fever or feverishness, cough, sore throat, runny nose, malaise, muscle or joint pain, difficulty breathing, respiratory sounds, nasal flaring, and chest indrawing. Paired venous blood specimens were collected at enrollment and at the end of the follow-up period from index cases and household contacts who were at least 6 months of age.
The study was approved by the Institutional Review Boards of the following institutions: 1) University of Michigan; 2) Centro Nacional de Diagnóstico y Referencia, Ministry of Health, Nicaragua; and 3) University of California, Berkeley. Written informed consent and parental permission from the parents or the legal guardians of children were obtained. Verbal assent was obtained from children 6 years of age or older.
Laboratory Methods
Combined nasal and oropharyngeal swabs were transported within 48 hours of collection to the National Virology Laboratory in a single tube of viral transport media maintained at 4°C. Samples were tested by real-time reverse-transcription polymerase chain reaction (rRT-PCR) using the Centers for Disease Control and Prevention validated protocols [20]. A subset of respiratory samples collected in this study was sent to J. Craig Venter Institute for sequencing. Additional viral sequences from another study performed out of the same health center in 2015 were also used to determine the circulating genotype. Blood specimens were sent to the National Virology Laboratory within 48 hours of collection. Haemagglutination inhibition assay (HAI) was performed on serum or plasma specimens to measure HAI antibody titers following the standard WHO protocol [21]. The WHO Influenza Reagent Kits were used to perform the assay, which included the influenza H1N1pdm09 control antigen A/California/07/2009 NYMC X-179A (Cal09), inactivated, Lot 1212H1AG. An initial sample dilution of 1:10 was used and serial two-fold dilutions were made until the endpoint titer was reached.
Statistical Methods
The primary outcome measure for influenza infection was rRT-PCR positivity. The reciprocal of HAI titers were log transformed to represent the primary outcome measure of reference virus antibodies. HAI titers of <1:10 were imputed as 1:5 in the analyses. HAI titers are log transformed so that the numerical values 0 to 8 represent titers 1:5 to 1:1280, with an increment of 1 per each two-fold increase in titer. Antibody response was defined as the number of two-fold rises in HAI antibody titer between enrollment (baseline) and end of the follow-up period. The weekly incidence of rRT-PCR-confirmed H1N1pdm infection in the Nicaraguan Pediatric Influenza Cohort Study [22] was used as a proxy representing the weekly activity level of H1N1pdm virus in the study population. A smoothing spline function [23] was used to generate a daily proxy which was then shifted upward by a negligibly small positive value to allow log incidence to be used in the analyses while preserving the shape of the epidemic curve. Since these households were recruited at different time points, the weekly H1N1pdm virus activity was used to adjust for time-varying risk of acquiring an infection from the community. The baseline HAI titer of household contacts was correlated with their risk of acquiring an infection using a structural equation regression model [24], which allows for both infection risk and antibody titer to vary with age. The number of study participants who were 50 years or older was small and there were no vaccinees in this age group. Therefore, stratified analyses for this group did not include age group or vaccination as predictor variables. Multiple sequence alignments were performed using ClustalX version 2.1 and AliView version 1.18.
RESULTS
Genetic characteristics of H1N1pdm in 2009–2013
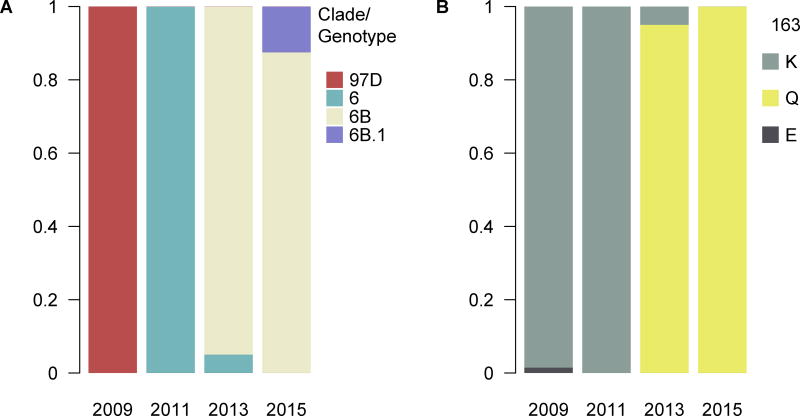
Clade 6 viruses are defined by a D97N mutation and Figure 1 shows that viruses circulated in 2009 in Managua remained to be the 97D genotype. Clade 6 viruses had represented all sequenced viruses isolated in 2011 and that dropped to only 5% in 2013. Almost all of the sequenced viruses isolated in 2013 and 2015 belonged to the 163Q genotype, also defined as clade 6B viruses, this change in the HA1 receptor-binding site likely contributes to antigenic change (see Appendix figures 1 and 2). Clade 6b.1 virus circulation was detected at the end of the 2015 season, however, a majority of viruses sequenced belonged to clade 6B.
Figure 1.
A) Genotype distribution of H1N1pdm in 2009–2015. B) Frequency distribution of amino acids at site 163 (H1 numbering) in 2009–2015.
The secondary attack rate of H1N1pdm
Between May and October 2013, 23 index patients who had a positive rRT-PCR result to H1N1pdm virus and 95 household contacts were followed up. Between September and December 2015, 58 index patients and 167 household contacts were followed up. The secondary attack rate amongst household contacts was 17% (95% C.I. 0.10, 0.26) in 2013 and 33% (95% C.I. 0.26, 0.41) in 2015 (Appendix Tables 1 and 2).
Pre-existing HAI antibodies and response to H1N1pdm infections
HAI antibody titers were measured at enrollment (baseline) and at the end of a 30–45 day follow-up period. Antibodies against Cal09 were measured in both study years. Infected contacts had a tendency to show lower baseline antibody titers compared to persons who remained healthy (Appendix Figures 3 and 4). In 2013, PCR-positive infected contacts showed a noticeable antibody response to Cal09, as illustrated by an increase in geometric mean titer (GMT) of 1:10 at baseline to 1:269 at follow-up. During the 2015 study, a baseline GMT of 1:7 increased to a post-infection GMT of 1:53 amongst PCR-positive infected contacts. Vaccinees were amongst those who showed a strong antibody response to infection.
HAI antibodies as a correlate of protection from secondary infection among household contacts
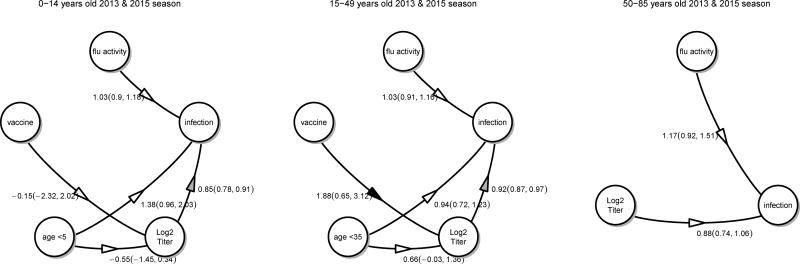
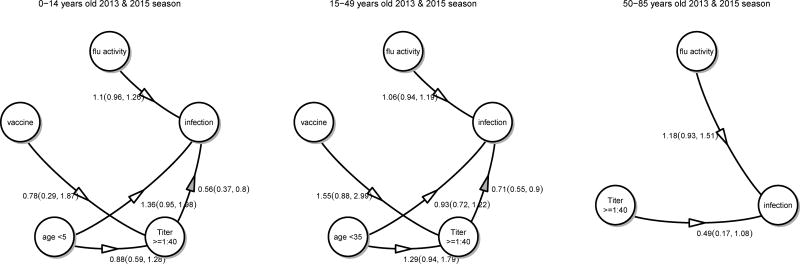
We estimated the protective efficacy of HAI antibodies among household contacts of influenza cases, adjusting for time-varying risk of community-acquired infection and age-specific risk difference in infection, and allowed antibody titers to vary with age. Cal09 antibody titer was associated with protection against PCR-positive infection in the 2013 and 2015 seasons. Figure 2 shows that each log2 increase in antibody titer was associated with 15% protection in children (adjusted OR 0.85; 95% C.I. 0.78, 0.91), 8% protection in adults 15–49 years old (adjusted OR 0.92; 95% C.I. 0.87, 0.97) and 12% protection in adults 50–85 years old (adjusted OR 0.88; 95% C.I. 0.74, 1.06). Alternative analyses using antibody titer of ≥ 1:40 as a predictor estimated a 44% protection in children (adjusted OR 0.56; 95%C.I. 0.37, 0.80), a 29% protection in adults 15–49 years old (adjusted OR 0.71; 95% C.I. 0.55, 0.90) and a 51% protection in adults 50–85 years old (adjusted OR 0.49; 95% C.I. 0.17, 1.08) (Figure 3). In the 2013 season, it was observed that adults who were vaccinated had on average a higher baseline titer compared with non-vaccinated adults, contributing to protection (Appendix Figures 5 and 6). Our analyses for the 2015 season also revealed that children under 5 years of age had lower baseline antibody titers compared to older children and young children were at an increased risk of infection (Appendix Figures 7 and 8).
Figure 2.
Association between log2 HAI titers, age, vaccination and community level of influenza activities and infection in the 2013 and 2015 household transmission study. Numbers represent odds ratios with the exception that the effects on log2 HAI titers were quantified using log2 increase in titers.
Figure 3.
Association between HAI titers (≥1:40), age, vaccination and community level of influenza activities and infection in the 2013 and 2015 household transmission study. Numbers represent odds ratios.
DISCUSSION
Through our community-based household transmission study, we detected a transition of H1N1pdm genotypes from D97N to clade 6 and 6B between 2009 and 2013. In 2013 when clade 6B viruses circulated, a high secondary attack rate was observed among household contacts, demonstrating high susceptibility to H1N1pdm despite several years of H1N1pdm circulation in Managua. We found Cal09 antibodies to associate with protection, a finding that is consistent with a detectable vaccine effectiveness reported by studies using a number of study designs [11–13, 25–27]. A robust Cal09 antibody response to H1N1pdm infection was detected in person of all ages in our study. Previous studies have found some individuals who were born between 1961 and 1983 showed reduced antibody response to K163Q virus [7, 10]. Data however also showed that H1N1pdm influenza vaccine in fact induced equally strong if not stronger antibody response to K163Q virus in many individuals in this group when compared with Cal09 [7]. Further investigation is required to identify factors that may favor a more broadly cross-reactive immune response, including more frequent updates of vaccine virus [28, 29] and using a vaccine that can induce more broadly protective antibodies [30–32]. Whereas other data have showed Cal09 antibodies had a null association with protection in the 2013–2014 season, it is possible that antibody specificity can vary between populations. One explanation was that an individual’s history of prior exposure to influenza can shape the landscape of antibody specificity [33–37]. Our study population could have been more frequently infected in the past compared to other populations due to a low vaccination rate. Geographical variation in the genotypes of H1N1pdm influenza virus might have also resulted in antigenic change that differed between populations. Findings from our study demonstrated the advantages of the newly adopted approach in using human serologic data and subgroup analyses in antigenic characterization [10]. Further studies are needed to confirm whether changing the vaccine strains more frequently will improve vaccine efficacy in highly vaccinated populations.
HAI titers were first proposed as an immune correlate of protection based on the animal studies data that showed passive transfer of HA-specific antibody conferred protection in naive animals. Human volunteer challenge studies have contributed substantively to establishing HAI titers as a quantitative surrogate measure of protection [38]. These studies may have limitations in identifying immune correlates of protection against antigenically drifted viruses. There are other mechanisms by which HA antibodies confer protection [30, 39]. These actions may be less readily detectable by HAI [40], particularly with antibodies that bind to the stalk region of HA [41]. The role of other classes of antibodies, for example anti-neuraminidase (NA) antibodies, should not be ignored. Major global efforts in standardizing serologic assessment have contributed to a high reproducibility of HAI assays results [42, 43]. However, these results need to be carefully interpreted in seasons when antigenic drift is substantial.
To conclude, we found Cal09 antibodies were associated with protection in the 2013 and 2015 seasons when clade 6B viruses predominated. Further study of different sources of serologic data will continue to improve antigenic characterization and assessment of the vaccine match and, if completed in a timely manner, could assist in vaccine strain selection.
Supplementary Material
Residue 163 visualized on a crystal structure of a 2009 H1N1 influenza virus hemagglutinin [44].
Residue 97 visualized on a crystal structure of a 2009 H1N1 influenza virus hemagglutinin [44].
Age-specific haemagglutination inhibition (HAI) titer of household contacts at baseline and on day 30–45 in the 2013 household transmission study. Individual HAI titers of infected household contacts are represented by red dots and those who were not infected are represented by blue dots. Antibody titers of vaccines were represented by green triangles.
Age-specific haemagglutination inhibition (HAI) titer of household contacts at baseline and on day 30–45 in the 2015 household transmission study. Individual HAI titers of infected household contacts are represented by red dots and those who were not infected are represented by blue dots. Antibody titers of vaccines were represented by green triangles.
Association between log2 HAI titers, age, vaccination and community level of influenza activities and infection in the 2013 household transmission study. Numbers represent odds ratios with the exception that the effects on log2 HAI titers were quantified using log2 increase in titers.
Association between HAI titers (≥1:40), age, vaccination and community level of influenza activities and infection in the 2013 household transmission study. Numbers represent odds ratios
Association between log2 HAI titers, age, vaccination and community level of influenza activities and infection in the 2015 household transmission study. Numbers represent odds ratios with the exception that the effects on log2 HAI titers were quantified using log2 increase in titers.
Association between HAI titers (≥1:40), age, vaccination and community level of influenza activities and infection in the 2015 household transmission study. Numbers represent odds ratios
Acknowledgments
FUNDING
This work was supported by the National institute of Allergy and Infectious Diseases, National Institutes of Health, under grant number U01AI088654 (to E.H. and A.G.) and contract number HHSN272201400006C, and was funded through a career development award from the John E. Fogarty International Center, National Institute of Health (K02TW009483 to A.G.), and a CEIRS training grant HHSN272201400008C (to S.N.).
We thank the families that participated in the study and our study staff at the Health Center Sócrates Flores Vivas and at the Centro Nacional de Diagnostico y Referencia of the Nicaraguan Ministry of Health.
Abbreviations
- Cal09
Influenza A/California/07/2009 virus
- C.I.
Confidence interval
- H1N1pdm
Pandemic influenza A(H1N1) virus
- HAI
Haemagglutination inhibition assay
- HCSFV
Health Center Sócrates Flores Vivas
- MN
Microneutralization assay
- NA
Neuraminidase
- NAI
Neuraminidase inhibition assay
- rRT-PCR
Real-time RT-PCR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
All authors declare no conflict of interests.
References
- 1.Chan PKS, Tam WWS, Lee TC, Hon KL, Lee N, Chan MCW, et al. Hospitalization Incidence, Mortality, and Seasonality of Common Respiratory Viruses Over a Period of 15 Years in a Developed Subtropical City. Medicine (Baltimore) 2015;94:e2024. doi: 10.1097/MD.0000000000002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matias G, Taylor RJ, Haguinet F, Schuck-Paim C, Lustig RL, Fleming DM. Modelling estimates of age-specific influenza-related hospitalisation and mortality in the United Kingdom. BMC Public Health. 2016;16:481. doi: 10.1186/s12889-016-3128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair H, Brooks WA, Katz M, Roca A, Berkley JA, Madhi SA, et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011;378:1917–30. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Weekly U.S. Influenza Surveillance Report. FluView. 2016 [Google Scholar]
- 5.Hong Kong Centre for Health Protection. Local Situation of Influenza Activity (as of Jul 27, 2016) Flu Express. 2016;13 [Google Scholar]
- 6.Harvey WT, Benton DJ, Gregory V, Hall JPJ, Daniels RS, Bedford T, et al. Identification of Low- and High-Impact Hemagglutinin Amino Acid Substitutions That Drive Antigenic Drift of Influenza A(H1N1) Viruses. PLoS Pathog. 2016;12:e1005526. doi: 10.1371/journal.ppat.1005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc Natl Acad Sci USA. 2014;111:15798–803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epperson S, Blanton L, Kniss K, Mustaquim D, Steffens C, Wallis T, et al. Influenza activity - United States, 2013-14 season and composition of the 2014-15 influenza vaccines. MMWR Morb Mortal Wkly Rep. 2014;63:483–90. [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen C, Moyes J, Tempia S, Groome M, Walaza S, Pretorius M, et al. Mortality amongst patients with influenza-associated severe acute respiratory illness, South Africa, 2009–2013. PLoS ONE. 2015;10:e0118884. doi: 10.1371/journal.pone.0118884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Recommended composition of influenza virus vaccines for use in the 2017 southern hemisphere influenza season. 2016. [Google Scholar]
- 11.Turner N, Pierse N, Huang QS, Radke S, Bissielo A, Thompson MG, et al. Interim estimates of the effectiveness of seasonal trivalent inactivated influenza vaccine in preventing influenza hospitalisations and primary care visits in Auckland, New Zealand, in 2014. Euro Surveill. 2014;19 [PMC free article] [PubMed] [Google Scholar]
- 12.team EC for DP and C (ECDC)-HCU-E editorial. [accessed December 23, 2016];Interim estimates of 2013/14 vaccine effectiveness against influenza A(H1N1)pdm09 from Canada’s sentinel surveillance network, January 2014. 2014 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20690.
- 13.Tewawong N, Prachayangprecha S, Vichiwattana P, Korkong S, Klinfueng S, Vongpunsawad S, et al. Assessing Antigenic Drift of Seasonal Influenza A(H3N2) and A(H1N1)pdm09 Viruses. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrie JG, Parkhouse K, Ohmit SE, Malosh RE, Monto AS, Hensley SE. Antibodies Against the Current Influenza A(H1N1) Vaccine Strain Do Not Protect Some Individuals From Infection With Contemporary Circulating Influenza A(H1N1) Virus Strains. J Infect Dis. 2016;214:1947–51. doi: 10.1093/infdis/jiw479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon A, Saborío S, Videa E, López R, Kuan G, Balmaseda A, et al. Clinical attack rate and presentation of pandemic H1N1 influenza versus seasonal influenza A and B in a pediatric cohort in Nicaragua. Clin Infect Dis. 2010;50:1462–7. doi: 10.1086/652647. [DOI] [PubMed] [Google Scholar]
- 16.Gresh L, Kuan G, Sanchez N, Azziz-Baumgartner E, Ojeda S, Melendez M, et al. Burden of Influenza and Influenza-associated Pneumonia in the First Year of Life in a Prospective Cohort Study in Managua, Nicaragua. Pediatr Infect Dis J. 2016;35:152–6. doi: 10.1097/INF.0000000000000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon A, Ortega O, Kuan G, Reingold A, Saborio S, Balmaseda A, et al. Prevalence and seasonality of influenza-like illness in children, Nicaragua, 2005–2007. Emerging Infect Dis. 2009;15:408–14. doi: 10.3201/eid1503.080238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon A, Kuan G, Aviles W, Sanchez N, Ojeda S, Lopez B, et al. The Nicaraguan pediatric influenza cohort study: design, methods, use of technology, and compliance. BMC Infect Dis. 2015;15:504. doi: 10.1186/s12879-015-1256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng S, Lopez R, Kuan G, Gresh L, Balmaseda A, Harris E, et al. The Timeline of Influenza Virus Shedding in Children and Adults in a Household Transmission Study of Influenza in Managua, Nicaragua. Pediatr Infect Dis J. 2016;35:583–6. doi: 10.1097/INF.0000000000001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 21.WHO Global Influenza. Manual for the laboratory diagnosis and virological surveillance of influenza. 2011. [Google Scholar]
- 22.Gordon A, Saborío S, Videa E, López R, Kuan G, Balmaseda A, et al. Clinical attack rate and presentation of pandemic H1N1 influenza versus seasonal influenza A and B in a pediatric cohort in Nicaragua. Clin Infect Dis. 2010;50:1462–7. doi: 10.1086/652647. [DOI] [PubMed] [Google Scholar]
- 23.Chambers JM, Hastie T. Statistical models in S. Pacific Grove, Calif: Wadsworth & Brooks/Cole Advanced Books & Software; 1992. [Google Scholar]
- 24.Hoyle RH. Handbook of structural equation modeling. New York: Guilford Press; 2012. [Google Scholar]
- 25.Chambers C, Skowronski DM, Sabaiduc S, Winter AL, Dickinson JA, De Serres G, et al. Interim estimates of 2015/16 vaccine effectiveness against influenza A(H1N1)pdm09, Canada, February 2016. Euro Surveill. 2016;21:30168. doi: 10.2807/1560-7917.ES.2016.21.11.30168. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan SG, Carville KS, Chilver M, Fielding JE, Grant KA, Kelly H, et al. Pooled influenza vaccine effectiveness estimates for Australia, 2012–2014. Epidemiol Infect. 2016;144:2317–28. doi: 10.1017/S0950268816000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapia MD, Sow SO, Tamboura B, Tégueté I, Pasetti MF, Kodio M, et al. Maternal immunisation with trivalent inactivated influenza vaccine for prevention of influenza in infants in Mali: a prospective, active-controlled, observer-blind, randomised phase 4 trial. Lancet Infect Dis. 2016;16:1026–35. doi: 10.1016/S1473-3099(16)30054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. PNAS. 1999;96:14001–6. doi: 10.1073/pnas.96.24.14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosterín Höpping A, McElhaney J, Fonville JM, Powers DC, Beyer WEP, Smith DJ. The confounded effects of age and exposure history in response to influenza vaccination. Vaccine. 2016;34:540–6. doi: 10.1016/j.vaccine.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe. 2016;19:800–13. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 2015;14:167–82. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 32.Nachbagauer R, Wohlbold TJ, Hirsh A, Hai R, Sjursen H, Palese P, et al. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. J Virol. 2014;88:13260–8. doi: 10.1128/JVI.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strengell M, Ikonen N, Ziegler T, Julkunen I. Minor changes in the hemagglutinin of influenza A(H1N1)2009 virus alter its antigenic properties. PLoS ONE. 2011;6:e25848. doi: 10.1371/journal.pone.0025848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med. 2013;5:187ra70. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koel BF, Mögling R, Chutinimitkul S, Fraaij PL, Burke DF, van der Vliet S, et al. Identification of amino acid substitutions supporting antigenic change of influenza A(H1N1)pdm09 viruses. J Virol. 2015;89:3763–75. doi: 10.1128/JVI.02962-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrie JG, Parkhouse K, Ohmit SE, Malosh RE, Monto AS, Hensley SE. Antibodies against the current influenza A H1N1 vaccine strain do not protect some individuals from infection with contemporary circulating H1N1 viral strains. J Infect Dis. 2016:jiw479. doi: 10.1093/infdis/jiw479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc Natl Acad Sci USA. 2014;111:15798–803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virelizier JL. Host defenses against influenza virus: the role of anti-hemagglutinin antibody. J Immunol. 1975;115:434–9. [PubMed] [Google Scholar]
- 39.Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, et al. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLOS Pathog. 2016;12:e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verschoor CP, Singh P, Russell ML, Bowdish DME, Brewer A, Cyr L, et al. Microneutralization assay titres correlate with protection against seasonal influenza H1N1 and H3N2 in children. PLoS ONE. 2015;10:e0131531. doi: 10.1371/journal.pone.0131531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Retamal M, Abed Y, Corbeil J, Boivin G. Epitope mapping of the 2009 pandemic and the A/Brisbane/59/2007 seasonal (H1N1) influenza virus haemagglutinins using mAbs and escape mutants. J Gen Virol. 2014;95:2377–89. doi: 10.1099/vir.0.067819-0. [DOI] [PubMed] [Google Scholar]
- 42.Zacour M, Ward BJ, Brewer A, Tang P, Boivin G, Li Y, et al. Standardization of Hemagglutination Inhibition Assay for Influenza Serology Allows for High Reproducibility between Laboratories. Clin Vaccine Immunol. 2016;23:236–42. doi: 10.1128/CVI.00613-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horby PW, Laurie KL, Cowling BJ, Engelhardt OG, Sturm-Ramirez K, Sanchez JL, et al. CONSISE statement on the reporting of Seroepidemiologic Studies for influenza (ROSES-I statement): an extension of the STROBE statement. Influenza Other Respir Viruses. 2017;11:2–14. doi: 10.1111/irv.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–60. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Residue 163 visualized on a crystal structure of a 2009 H1N1 influenza virus hemagglutinin [44].
Residue 97 visualized on a crystal structure of a 2009 H1N1 influenza virus hemagglutinin [44].
Age-specific haemagglutination inhibition (HAI) titer of household contacts at baseline and on day 30–45 in the 2013 household transmission study. Individual HAI titers of infected household contacts are represented by red dots and those who were not infected are represented by blue dots. Antibody titers of vaccines were represented by green triangles.
Age-specific haemagglutination inhibition (HAI) titer of household contacts at baseline and on day 30–45 in the 2015 household transmission study. Individual HAI titers of infected household contacts are represented by red dots and those who were not infected are represented by blue dots. Antibody titers of vaccines were represented by green triangles.
Association between log2 HAI titers, age, vaccination and community level of influenza activities and infection in the 2013 household transmission study. Numbers represent odds ratios with the exception that the effects on log2 HAI titers were quantified using log2 increase in titers.
Association between HAI titers (≥1:40), age, vaccination and community level of influenza activities and infection in the 2013 household transmission study. Numbers represent odds ratios
Association between log2 HAI titers, age, vaccination and community level of influenza activities and infection in the 2015 household transmission study. Numbers represent odds ratios with the exception that the effects on log2 HAI titers were quantified using log2 increase in titers.
Association between HAI titers (≥1:40), age, vaccination and community level of influenza activities and infection in the 2015 household transmission study. Numbers represent odds ratios