Summary
Rodents are characterized by continuously renewing incisors whose growth is fueled by epithelial and mesenchymal stem cells housed in the proximal compartments of the tooth. The epithelial stem cells reside in structures known as the labial (toward the lip) and lingual (toward the tongue) cervical loops (laCL and liCL, respectively). An important feature of the rodent incisor is that enamel, the outer, highly mineralized layer, is asymmetrically distributed, as it is normally generated by the laCL but not the liCL. Here, we show that epithelial-specific deletion of the transcription factor Islet1 (Isl1) is sufficient to drive formation of ectopic enamel by the liCL stem cells, and also that it leads to production of altered enamel on the labial surface. Molecular analyses of developing and adult incisors revealed that epithelial deletion of Isl1 affected multiple, major pathways: Bmp (bone morphogenetic protein), Hh (hedgehog), Fgf (fibroblast growth factor), and Notch signaling were up-regulated and associated with liCL-generated ectopic enamel; on the labial side, up-regulation of Bmp and Fgf signaling, and Shh down-regulation, were associated with premature enamel formation. Transcriptome profiling studies identified a suite of differentially regulated genes in developing Isl1 mutant incisors. Our studies demonstrate that ISL1 plays a central role in proper patterning of stem cell-derived enamel in the incisor and indicate that this factor is an important upstream regulator of signaling pathways during tooth development and renewal.
Keywords: tooth development, mouse incisor, ectopic enamel, Isl1, amelogenesis
Introduction
Enamel, the outer covering of teeth is generated from ectoderm-derived epithelial cells, and it is the hardest physiological tissue in vertebrates. The mouse incisor provides a valuable model to study the molecular and cellular mechanisms of enamel formation, or amelogenesis. In the mouse incisor, enamel is normally deposited in an asymmetric fashion exclusively on the labial (toward the lip) surface. This asymmetry is important, because unlike the mouse molar, the incisor renews throughout life in a process that is fueled by mesenchymal and epithelial stem cells.(1) Thus, asymmetric enamel distribution favors abrasion of the softer, enamel-free, lingual (toward the tongue) surface, which maintains proper incisor length and sharpness in light of the continuous growth.
The dental epithelial stem cells (DESCs) responsible for mouse incisor enamel renewal are located in niches called the labial and lingual cervical loops (laCL and liCL, respectively) at the proximal end of the incisor (Fig. 1A,A′).(1-8) The laCL comprises several different cell types including the stellate reticulum (SR), the outer enamel epithelium (OEE), the inner enamel epithelium (IEE), and the transit-amplifying (TA) region, the last of which eventually differentiate into enamel-producing ameloblasts (Fig. 1A″).(9) In contrast, the rodent incisor liCL does not normally generate ameloblasts and enamel. Although several studies have focused on the laCL, very little is known about the liCL.
Figure 1. Expression of Isl1 during development of the mouse incisor.
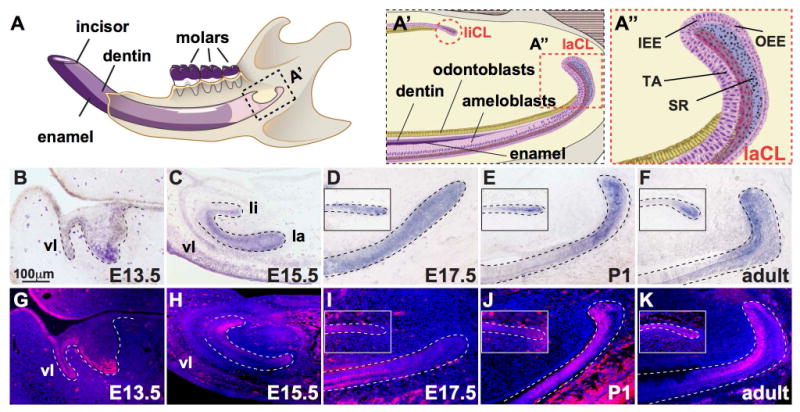
(A) Illustration of the mouse hemi-mandible showing the incisor and molars, as well as the mineralized dentin and enamel comprising the incisor. (A′) The proximal region of the incisor denoting the labial and lingual cervical loop (laCL and liCL, respectively, highlighted by dashed, red lines). (A″) Magnified view of the laCL showing the inner enamel epithelium (IEE), outer enamel epithelium (OEE), stellate reticulum (SR), and transit-amplifying cells (T-A). (B-F) In situ hybridization staining for Isl1 at embryonic day 13.5 (E13.5), E15.5, E17.5, post-natal day 1 (P1), and 6-week old (adult) showed Isl1 expression throughout mouse incisor development including the vestibular lamina (vl), and the lingual (li) and labial (la) aspects. In adults, Isl1 expression was predominant in the laCL and liCL. (G-K) Immunofluorescence staining for ISL1 during mouse incisor development showed similar expression profiles to Isl1 expression (B-F) with the exception of E15.5, where protein expression was increased on the lingual side.
The regulation of ectoderm-derived epithelium during amelogenesis and/or tooth development involves members of the major signaling pathways, including Bmp, Eda, Fgf, Notch, Shh, and Wnt family members.(1,2,10-17) To date, three distinct mouse models have shown that effects on Bmp or Fgf signaling can lead to the generation of ectopic enamel or ameloblasts by the liCL.(15,18,19)
Islet1 (Isl1) encodes a LIM-homeodomain (LIM-HD) transcription factor,(20) and mice lacking Isl1 die at embryonic day (E) 9.5.(21) Isl1 is involved in many different pathways and processes, including the control of motor neuron and interneuron specification(21) as well as pituitary,(22) pancreas,(23) heart,(24) and hindlimb development.(25) Isl1 also marks cardiovascular progenitor cells that give rise to cardiomyocyte, smooth muscle, and endothelial cell lineages.(26-30) Recently, Isl1 was shown to be highly expressed in dental epithelial tumors called ameloblastomas,(31) and was identified in a genome-wide association study (GWAS) to potentially play a role in caries development.(32)
Little is known about the function of Isl1 in tooth development. Isl1 is expressed in the oral and dental epithelium of the mouse incisor at E10, but its expression is limited to the epithelium in the enamel-free cusp region during molar development.(33) Early embryonic lethality after global deletion of Isl1 has precluded the study of Isl1 during tooth development in vivo, but in vitro studies have provided initial insights into the role of ISL1 in tooth patterning, including identification of a positive feedback loop between BMP4 and ISL1.(33) Interestingly, although SHH regulates Isl1 in neural tissues,(34) in vitro experiments suggested that SHH does not regulate Isl1 in oral epithelia.(33) Recently, a genome-wide gene expression analysis was performed on incisor, canine, and molar germs in 11-week old human fetuses, and Isl1 was observed to be expressed in incisor and canine, but not molar, germs.(35)
Here, we generated a conditional null mouse in which Isl1 was inactivated in epithelial tissue utilizing the Krt14Cre driver. Krt14Cre;Isl1fl/fl mice appeared normal and healthy, with the exception of incisor enamel defects, including hypo-mineralized enamel on the labial surface and ectopic enamel on the lingual surface, where enamel is not normally formed. Interestingly, molar teeth were not affected. Multiple signaling pathways, including Bmp, Fgf, Hh, and Notch, were perturbed in the laCL and liCL of control and mutant mice. Thus, Isl1 appears to play a critical, central role in incisor enamel formation.
Materials and Methods
Animals
All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) at UCSF and the mice were handled in accordance with the principles and procedure of the Guide for the Care and Use of Laboratory Animals under the approved protocol AN084146-02F. Mice were maintained in a temperature-controlled facility with access to mouse chow and water ad libitum. Mice carrying the Krt14Cre [Tg(KRT14-cre)1Amc](36) and Isl1fl [Isl1tm2Gan](37) alleles were mated to generate conditional, epithelial-specific, Isl1-inactivated mice, namely Krt14Cre/+;Isl1fl/fl and Isl1fl/fl mice (referred to as mutants and controls, respectively). To generate age-specific embryos, adult mice were mated overnight and females were checked for a vaginal plug in the morning. The presence of a vaginal plug was designated as embryonic day (E) 0.5. At least three 6-week old mice were examined at each time point for all experiments unless otherwise specified. Up to 5 mice of the same sex were housed together until time of sacrifice and no adverse events were reported. Both male and female mice were analyzed.
Histology, in situ hybridization, and immunohistochemistry
Mice were euthanized following standard IACUC protocols. Specimens at E13.5, E15.5, E17.5, post-natal day 1 (P1), and 6-week old were collected and fixed overnight in 4% paraformaldehyde at 4°C for 24-48 h, demineralized in 0.5M EDTA for 3-14 days if required, dehydrated, embedded in paraffin wax, and serially sectioned at 7 μm. Histological sections were stained with haematoxylin and eosin (H&E). For in situ hybridization analyses, sections were hybridized to DIG-labeled RNA probes for detection of RNA transcripts. Sections were treated with 10 μg/mL of proteinase K and acetylated prior to hybridization with probe. DIG-labeled RNA probes were synthesized from plasmids containing full-length cDNA or fragments of Isl1, Shh, Fst, Etv5, Fgf9, Fgf10, Spry2, and Bmp4. Immunohistochemistry was performed according to standard protocols. Antigen retrieval was performed by boiling the slides in Trilogy (Cell Marque) for 15 min and cooled at room temperature for 20 min after removing paraffin and rehydration. Primary antibodies used were as follows: anti-ISL1 (1:200; ab20670; Abcam), anti-AMEL (amelogenin; 1:200; Abcam), anti-AMBN (ameloblastin; 1:200; Abcam), anti-CLDN1 (Claudin1; 1:200; Abcam), anti-NICD (Cleaved Notch1 (Val1744); 1:200; Cell Signaling Technology). Goat anti-rabbit or mouse AlexaFluor 555 secondary antibodies were used (1:500, Invitrogen).
Detection of proliferating cells
Proliferating cells were identified by injection of 1 mg BrdU for 90 min followed by staining with a rat monoclonal anti-BrdU antibody (1:1000; Abcam). Slides were treated with 0.2N HCl prior to applying antibody, and BrdU-positive cells were visualized using a goat anti-rat AlexaFluor 555 secondary antibody (1:500, Invitrogen).
Microscopy
Fluorescent and bright field images were taken using a DM5000B microscope with a DFC500 camera (Leica). For confocal images, an SP5 Upright Confocal microscope (Leica) was used.
Micro-computed tomography (MicroCT)
MicroCT was performed on a MicroXCT-200 (Xradia, Pleasanton, CA, USA) through the MicroCT Imaging Facility at UCSF. Each specimen was scanned at 75 KVp and 6W at 4× magnification. Specimens were also imaged at the Small ANimal Tomographic Analysis (SANTA) facility located at the Seattle Children's Research Institute using a Skyscan 1076 micro-Computed Tomograph. Scans were done at an isotropic resolution of 17.21 μm using the following settings: 55kV, 179μA, 0.5mm Aluminum filter, 460ms exposure, rotation step of 0.7°, 180° scan, and 3 frame averaging. All data were reconstructed using Nrecon (v1.6.9.4) with the same greyscale threshold. Reconstructions were all converted to 3D volumes using Drishti v2.4 (www.sf.anu.edu.au).
RNA isolation, qPCR, and RNA-Seq
Total RNA was isolated using the RNeasy kit (Qiagen). DNA was removed in-column with RNase-free DNAse (Qiagen). All qPCR reactions were performed using the GoTaq qPCR Master Mix (Promega) in a Mastercycler Realplex (Eppendorf). PrimeTime Primers (IDT) were utilized for qPCR and primer sequences are available upon request. qPCR conditions were as follows: 95°C, 2 minutes; 40 cycles at 95°C,15 seconds; 58°C,15 seconds; 68°C, 20 seconds; followed by a melting curve gradient. Expression levels of the genes of interest were normalized to levels of Rpl19.
For RNA-Seq experiments, Krt14Cre/+;Isl1fl/fl mice were mated with RFP mice to generate Isl1fl/fl;RFP (control) or Krt14Cre/+;Isl1fl/fl;RFP (mutant) mice. Developing incisors from E15.5 embryos were dissected and total RNA isolated as described above. RNA-Seq experiments were performed at the SABRE Functional Genomics Core at UCSF (www.arrays.ucsf.edu.). Briefly, total RNA quality was assessed by spectrophotometry (NanoDrop, Thermo Fisher Scientific Inc.) and Agilent 2100 Bioanalyzer (Agilent Technologies). RNA sequencing libraries were generated using the TruSeq stranded mRNA sample prep kits with multiplexing primers, according to the manufacturer's protocol (Illumina), and library concentrations were measured using KAPA Library Quantification Kits (Kapa Biosystems, Inc.). Equal amounts of indexed libraries were pooled and sequenced on the Illumina HiSeq 2500 (Illumina). Data analysis involved de-multiplexing the results, trimming adapter sequences from the reads, and aligning unique reads to the mouse genome (mm10). Sequence alignment and splice junction estimation were performed using software programs Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)(38) and TopHat (http://tophat.cbcb.umd.edu/),(39) respectively. For differential expression testing, the genomic alignments were restricted to those mapping to an annotated transcriptome provided by Ensembl.(40) This subset of mappings was aggregated on a per-gene basis as raw input for the program DESeq.(41)
Gene ontology analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7; available at https://david-d.ncifcrf.gov).(42,43) Enriched GO terms for Biological Processes were detected and clustered using the default parameters for Functional Annotation Clustering. Annotation Clusters displaying an Enrichment Score of 1.3 and above were listed (Fig. S6). GO analysis was focused on genes differentially expressed by 2-fold or higher (up or down-regulation) with a false discovery rate less than 0.01. 22 out of 139 down regulated entries were not referenced in the DAVID platform, while 12 out of 131 up regulated entries were not referenced.
Scanning Electron Microscopy (SEM)
Mouse hemi-mandibles were dissected free of soft and connective tissue, fixed in 4% PFA in PBS overnight, then dehydrated in a graded ethanol series and dried in a vacuum desiccator. Hemi-mandibles were then embedded in epoxy resin (resin 105 and hardener 205 at a ratio of 5:1 w/w, WestSystem, Bay City, MI, USA), ground to the desired thickness on a plate grinder (EXAKT 400CS, Norderstedt, Germany) using 800 grit silicon carbide paper and polished with 2000 and 4000 grit silicon carbide paper (Hermes Abrasives, Mississauga, ON, Canada). The exposed tissue was etched with 10% phosphoric acid for 30 seconds, rinsed with water and dried in a vacuum desiccator. Samples were mounted on SEM stubs with carbon tape, surfaces coated with 7 nm gold using a sputter coating machine (Desk II, Denton Vacuum, Moorestown, NJ, USA), and imaged in a Philips SEM instrument (XL30 ESEM, Philips, Andover, MA, USA) operating at a beam energy of 20 keV in secondary electron or backscatter mode. Images were processed using Adobe Photoshop CS5.1 to adjust upper and lower limits of input levels in grayscale mode, and to apply auto balance and auto contrast settings.
Statistical analysis
All experiments were performed independently at least three times (i.e., N=3) in triplicates, and when applicable, presented as an average ± standard error of the mean. Student t-test was used to determine p-values and P<0.05 was deemed to be significant.
Results
Isl1 is expressed in the dental epithelium during development of the mouse mandibular incisor
We first analyzed the expression of Isl1 during mouse tooth development by in situ hybridization and immunofluorescence staining (Fig. 1B-K). Isl1 mRNA and protein were detected predominantly in the developing mandibular incisor (Fig. 1B-E,G-J) with little or no expression in molars (Fig. S1). At E13.5, Isl1 was expressed in the dental epithelium, vestibular lamina (vl), and the labial aspect of the incisor tooth bud (Fig. 1B,G). At E15.5, Isl1 expression was diminished in the dental epithelium and vl, but was observed on both the lingual (li) and labial (la) sides of the developing incisor (Fig. 1C,H). Interestingly, ISL1 protein appeared increased on the lingual aspect (Fig. 1H). At E17.5 and P1, Isl1 expression was restricted to the proximal regions of the incisor, namely the liCL and laCL (Fig. 1D,E,I,J). Lastly, in adult (i.e., 6 weeks old), mandibular incisors Isl1 expression was predominantly observed in the IEE, TA, and pre-ameloblast region of the laCL, whereas in the liCL, expression was limited to the most proximal region (Fig. 1F,K). We observed similar expression patterns in adult maxillary incisors (data not shown).
Conditional inactivation of epithelial Isl1 leads to incisor defects
Global inactivation of Isl1 causes early embryonic lethality.(21) Because Isl1 is predominantly expressed in dental epithelia (Fig. 1B-K), we generated mice carrying an epithelial-specific deletion of Isl1. Adult Krt14Cre;Isl1fl/fl mutant mice were viable and appeared healthy overall, but all mutant mice possessed white incisors with blunted tips, as compared to the yellow, sharp incisors in controls (Fig. 2A,E,M). We also observed a diastema or space, between the maxillary incisors in 30-50% of the mutants (Fig. 2B,C,F,G,M), and the bony socket housing the maxillary incisors appeared to be enlarged in mutants compared to controls (Fig. 2C,G).
Figure 2. Conditional inactivation of Isl1 in mouse epithelia leads to mice with enamel defects.
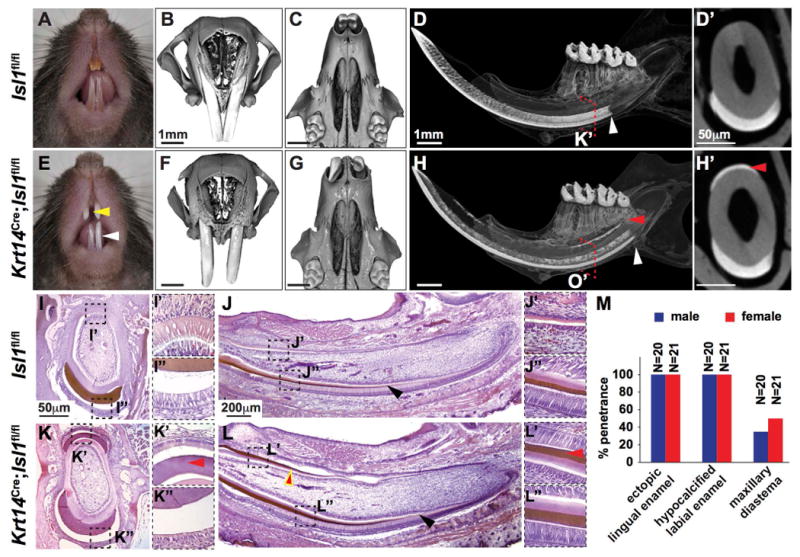
(A-H′) Comparison of control (Isl1fl/fl) and conditionally inactivated (Krt14Cre;Isl1fl/fl) adult mouse incisors. Control mice exhibited shiny, yellow shading of maxillary and mandibular incisors (A; white arrowheads), whereas mutant mice showed chalky, white enamel (E; white arrowheads). MicroCT analyses of maxillae in frontal (B,F) and ventral (C,G) view showed a diastema between the incisors in mutant (F,G) but not control (B,C) mice with enlarged tooth sockets in mutant mice (G; red arrowhead). MicroCT analyses of the hemi-mandible in sagittal view (D,H) and the incisor in cross-section (D′,H′) demonstrated the presence of ectopic enamel on the lingual surface (yellow arrowheads) where enamel is normally absent (D,D′). Furthermore, enamel appeared to be generated prematurely in mutant mice compared to controls (D,H; white arrowheads). (I-L″) Hematoxylin-eosin staining of P0 hemi-mandible in cross-section (I-I″,K-K″) and sagittal view (J-J″,L-L″) showed ectopic enamel matrix on the lingual side of mutant (K-K″, L,L′; red arrowheads) but not control (I-I″,J,J′) incisors. Enamel and dentin matrix appeared normal in mutants (L,L″) compared to controls (J,J″), however, enamel was generated prematurely in mutants (L; black arrowhead) compared to controls (J; black arrowhead). (M) There was 100% penetrance of the ectopic, lingual enamel and chalky, white (or hypocalcified labial) enamel phenotypes. The maxillary diastema was observed in less than half of the mutants.
We next examined the mineralized tissues in the mutants by microCT analysis of the mandibular incisors and found two distinct phenotypes. First, ectopic enamel was present on the lingual incisor surface in mutants both in sagittal (Fig. 2D,H) and cross-section (Fig. 2D′,H′) with 100% penetrance (Fig. 2M). Second, enamel mineralization occurred prematurely or closer to the laCL in mutants (Fig. 2D,H). The microCT results were confirmed histologically in P1 mice (Fig. 2I-L″). Incisor enamel matrix was observed on the lingual surface of the mutant but not control mandibular incisor in cross-sectional (Fig. 2I-I″,K-K″) and sagittal view (Fig. 2J-J″,L-L″). Moreover, premature enamel mineralization as indicated by the presence of enamel matrix was also confirmed histologically (Fig. 2J,L). In contrast to the incisors, we did not detect any differences in mutant molars using microCT (Fig. S2A,B) or SEM (Fig. S2C,D) analyses.
Deletion of Isl1 leads to altered labial enamel and enamel-like mineralized tissue on the lingual surface
SEM analyses of control and mutant hemi-mandibles in sagittal view revealed defects in labial enamel (Fig 3A-D‴). In control incisors, the enamel rods extend from the DEJ to near the surface (Fig. 3B). In mutants, enamel rods appeared normal near the dentino-enamel junction (DEJ) (Fig. 3D,D′) but became disorganized and lost their distinctive enamel rod pattern approximately halfway between the DEJ and surface (Fig. 3D-D‴). In addition, the lingual surface in mutants had enamel-like mineralization, with moderately organized enamel-like rods extending from the DEJ to the surface (Fig. 3E).
Figure 3. Conditional epithelial inactivation of mouse Isl1 leads to defects in enamel mineralization.
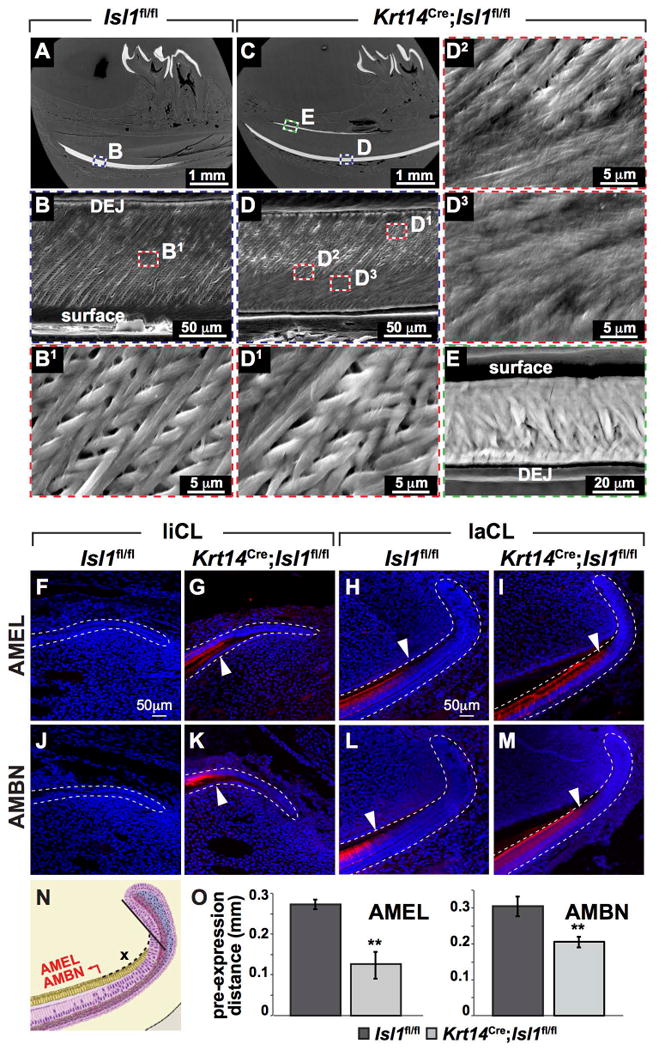
(A-E) Scanning electron microscopy (SEM) of the hemi-mandible in sagittal view of control (A-B1) and mutant (C-E) mice. Control enamel (B,B1) showed parallel enamel rods that ran continuously from the dentin-enamel junction (DEJ) to the surface with a 45° distal orientation. In mutants, the inner enamel (i.e., enamel near the DEJ) showed a similar pattern compared to controls (B1,D1), however, this pattern was lost approximately halfway between the DEJ and the surface (D2,D3). Ectopic lingual enamel showed enamel-like mineralization (E). (F-O) Amelogenin (AMEL) and ameloblastin (AMBN) showed ectopic and premature expression in mutant liCL and laCL respectively. AMEL and AMBN are normally not expressed in the liCL (F,J) but was expressed in mutant liCL (G,K). AMEL and AMBN were expressed in the laCL of control and mutant incisors, however, they were expressed prematurely (H,I,L,M; white arrowheads). (N,O) The premature expression of AMEL and AMBN was quantified and confirmed. **, P<0.01.
Interestingly, we could not detect differences in incisor labial enamel density using microCT (Fig. S2E,F) and SEM backscatter (Fig. S2G-J) analyses. The density of the ectopic lingual incisor enamel was also similar to labial enamel (Fig. S2E). In mutant labial enamel, the outer layer or approximately 1/3 of the enamel on the surface or at the side facing the embedding resin, appeared more uniform and highly mineralized than wild type specimens (Fig. S2G,H). This increased uniformity of mutant enamel was reflected in the narrower peak width of the enamel signal (gray scale value of ∼200) in the corresponding histograms (Fig. S2I,J), whereas the number of pixels of the mutant enamel peak was ∼8000) compared to the control enamel peak of ∼4000 was indicative of more densely mineralized enamel (Fig. S2I,J). However, mutant enamel showed a higher but narrower peak compared to the lower but broader peak in controls suggesting overall enamel densities may be similar. Moreover, we did not detect any differences in labial enamel volume or thickness when the mandibular incisor was analyzed between the distal incisor tip and 1st molar distal root to control for premature enamel mineralization in mutants (data not shown). Additional experiments are required to further analyze control and mutant incisor enamel but it was interesting that “white” enamel did not indicate hypo-mineralized enamel.
Abnormalities in enamel mineralization and ameloblast differentiation were also evidenced by immunofluorescence staining for two ameloblast markers, amelogenin (AMEL) and ameloblastin (AMBN) (Fig. 3F-M). AMEL and AMBN were detected in the mutant liCL (Fig. 3G,K), reflecting the ectopic enamel formation, in contrast to the control (Fig. 3F,J). In the laCL, both AMEL and AMBN were present in controls and mutants, but expression of both markers occurred earlier in mutants (Fig. 3H,I,L,M,N,O). Additional enamel proteins including enamelin (ENAM), kallikrein-4 (KLK4), and amelotin (AMTN) were assayed and shown to be expressed in the lingual aspect of the mutant incisor further supporting the generation of ectopic enamel or enamel-like tissue (Fig. S3A,B,E,F,I,J). Moreover, ENAM and KLK4 appeared to be prematurely expressed (Fig. S3C,D,G,H,M) similar to AMEL and AMBN. Interestingly, AMTN did not show premature, mutant expression (Fig. S3K-M).
The ectopic and premature expression of enamel proteins (e.g., AMEL, AMBN, AMTN, ENAM, and KLK4) in adult liCL and laCL suggested a change in the number of proliferative cells (Fig. 4). Indeed, we observed an increased number of proliferating cells in the mutant liCL, although there was no change in the size of the zone of BrdU+ cells (Fig. 4A-D). Conversely, we observed a decrease in the number of proliferative cells in the laCL of mutants, as well as a shortened zone of BrdU+ cells (Fig. 4E-H).
Figure 4. Conditional epithelial inactivation of Isl1 leads to effects on proliferation in the liCL and laCL of mandibular mouse incisors.
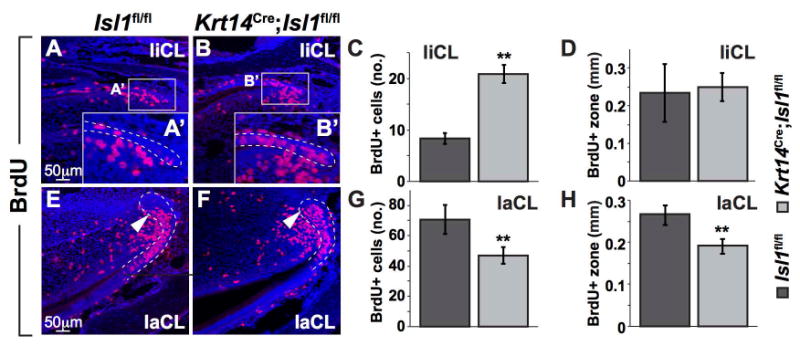
(A-H) BrdU was injected for 90 min to label proliferating cells in the liCL and laCL (denoted by white, dashed lines). Magnified view of the liCL (A′,B′) showed an increase in the number of proliferating cells in mutants (B′) compared to controls (A′). The number of BrdU+ cells was quantified and confirmed to be increased in mutants (C) however, the length of the BrdU+ zone was unchanged. Conversely, in the laCL, the number of BrdU+ cells was quantified and shown to be decreased in mutants compared to controls (E,F,G). Furthermore, the length of the BrdU+ zone was significantly shortened in mutants (H). **, P<0.01.
Epithelial-specific deletion of Isl1 affects multiple signaling pathways
Shh is an important regulator of the ability of dental epithelial stem cells to generate ameloblast progenitors in the laCL.(3) In adult incisors, we found differences in Shh expression in both the liCL (Fig. 5A-C) and laCL (Fig. 5J-L) using both in situ hybridization and qPCR. Interestingly, Shh expression was increased in the liCL, whereas it was decreased in the laCL, demonstrating that ISL1 functions in a context dependent fashion. Moreover, in the mutant liCL, Etv5, a readout of Fgf signaling,(44,45) and Fst, an antagonist of Bmp signaling(46,47) were up- and down-regulated, respectively (Fig. 5D-I). On the labial side, Etv5 expression showed no difference between control and mutant laCL, whereas Fst expression was decreased in mutant laCL (Fig. 5M-R).
Figure 5. Differential expression of genes from various signaling pathways in control and mutant mandibular incisors.
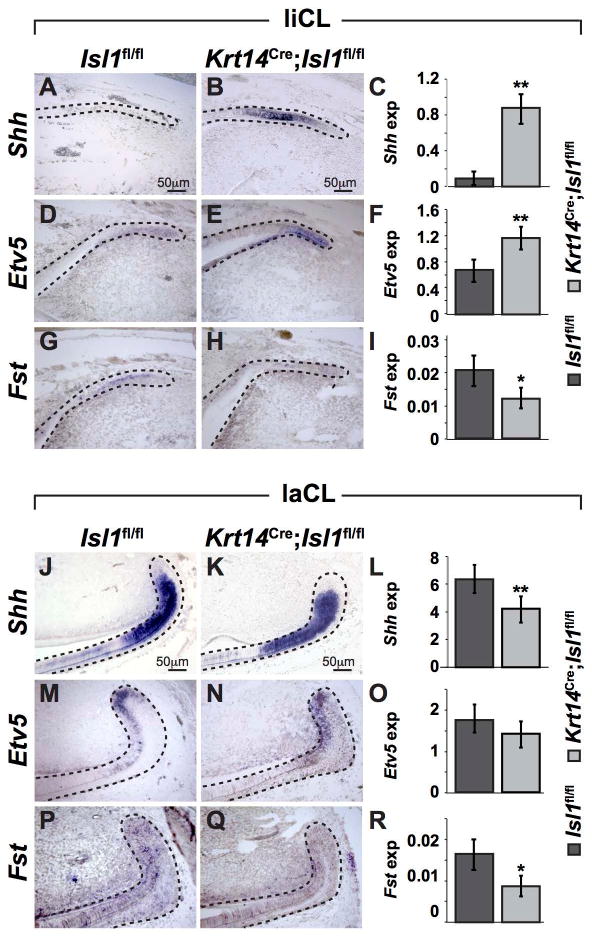
(A-I) Genes from various signaling pathways were analyzed by in situ hybridization and qPCR in the liCL. Expression of Shh and Etv5 was increased in mutant liCL (B,E) compared to controls (A,D), whereas Fst was decreased in mutant liCL (H) compared to controls (G). The in situ hybridization results were confirmed by qPCR analyses (C,F,I). (J-R) Genes from various signaling pathways were analyzed by in situ hybridization and qPCR in the laCL. Expression of Shh and Fst expression was decreased in mutants (J-L, P-R), and Etv5 showed no difference in expression between control and mutants (M-O). *, P<0.05; **, P<0.01.
We further analyzed genes in the Fgf and Bmp signaling pathways using in situ hybridization (Fig. S4). Sprouty2 (Spry2), a gene encoding an intracellular antagonist of Fgf signaling, appeared to be down-regulated in the mutant liCL (Fig. S4A,B), consistent with the up-regulation of Etv5 we observed in the mutant liCL (Fig. 5D-F). In the mutant laCL, there appeared to be no or little difference in Spry2 expression compared to controls (Fig. S4C,D). No obvious differences in Fgf9 and Fgf10 expression were observed in control and mutant liCL and laCL (Fig. S1E-L). Finally, Bmp4 expression appeared to be increased in the mutant liCL and laCL (Fig. S4M-P), which correlated with a decrease in Fst expression (Fig. 5P-R).
Immunofluorescence staining showed changes in expression of several key proteins between control and mutant liCL and laCL (Fig. 6). pMEK was up-regulated in mutant liCL and laCL compared to controls (Fig. 6A-D). Moreover, pMEK localization was altered in mutant laCL, with increased staining in the TA and pre-ameloblast regions (white arrowhead) and decreased staining in the OEE (yellow arrowhead; Fig. 6C,D). pSMAD1/5/8 was also up-regulated in mutant liCL and laCL (Fig. 6E-H). NICD or activated/cleaved NOTCH1 was up-regulated in mutant liCL, but no difference was noted in the laCL (Fig. 6I-L). Claudin1 (CLDN1), a tight junction component of epithelial polarized cells(48) that is potentially involved in mouse incisor(49) and human tooth development,(50) appeared to be increased in mutant liCL, but no difference was noted in control and mutant laCL (Fig. 6M-P).
Figure 6. Differential expression of proteins from various signaling pathways in control and mutant mandibular incisors.
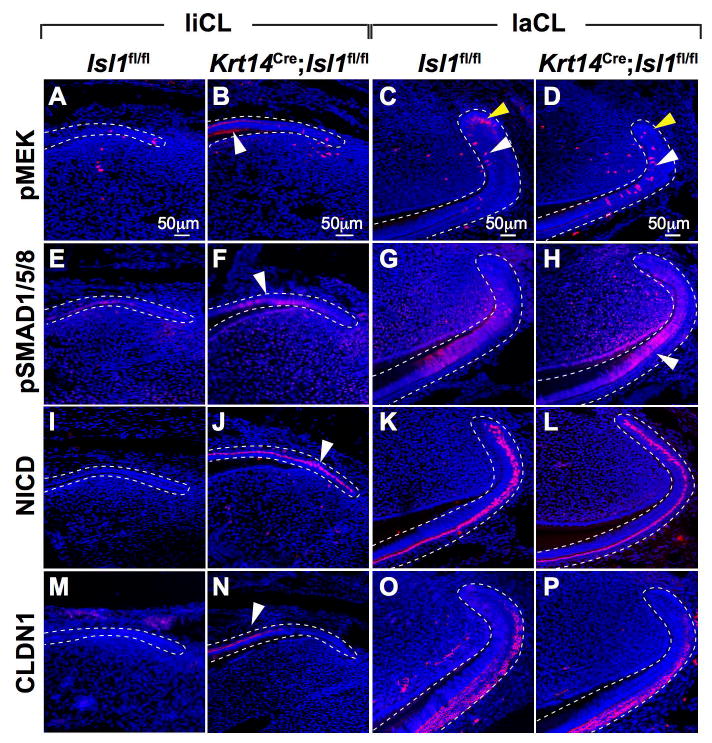
(A-P) Immunofluorescence staining showed differential expression of numerous proteins in the liCL and laCL regions. pMEK (A,B), pSMAD (E,F), NICD (I,J), and CLDN1 (M,N) were all upregulated in mutant liCL (white arrowheads). In the laCL, only pMEK (C,D) and pSMAD (G,H) appeared to be upregulated in mutants; NICD (K,L) and CLDN1 (O,P) remained unchanged.
Because NICD1, CLDN1, and Shh demonstrated differential expression in adult control and mutant incisors, we further assayed their expression during development (Fig. 7). Expression of NICD showed no differences between control and mutant incisors at E14.5, 15.5, and E17.5 (Fig. 7A-C,E-G). At P1, an important difference was observed, as NICD expression was maintained in the mutant but not control liCL (Fig. 7D,H) and this NICD expression persisted in adult mutant liCL (Fig. 6I,J). Expression of CLDN1 protein was similar in developing incisors at E14.5 and E15.5 in controls and mutants, but at E17.5 maintenance of intense CLDN1 expression was observed in mutant liCL (Fig. 7I-N), consistent with the strong CLDN1 expression in adult mutant liCL (Fig. 6M,N). Lastly, Shh expression was assayed during incisor development (Fig. 7O-T). Differences in expression patterns were first evident at E15.5, with Shh expression noted on the lingual side of the developing mutant incisor (Fig. 7P,S). This expression pattern was maintained in mutant incisors at E17.5 (Fig. 7Q,T), at P1 (data not shown), and in adults (Fig. 5A-C,J-L).
Figure 7. Expression of NICD, CLDN1 and Shh in the CLs during development.
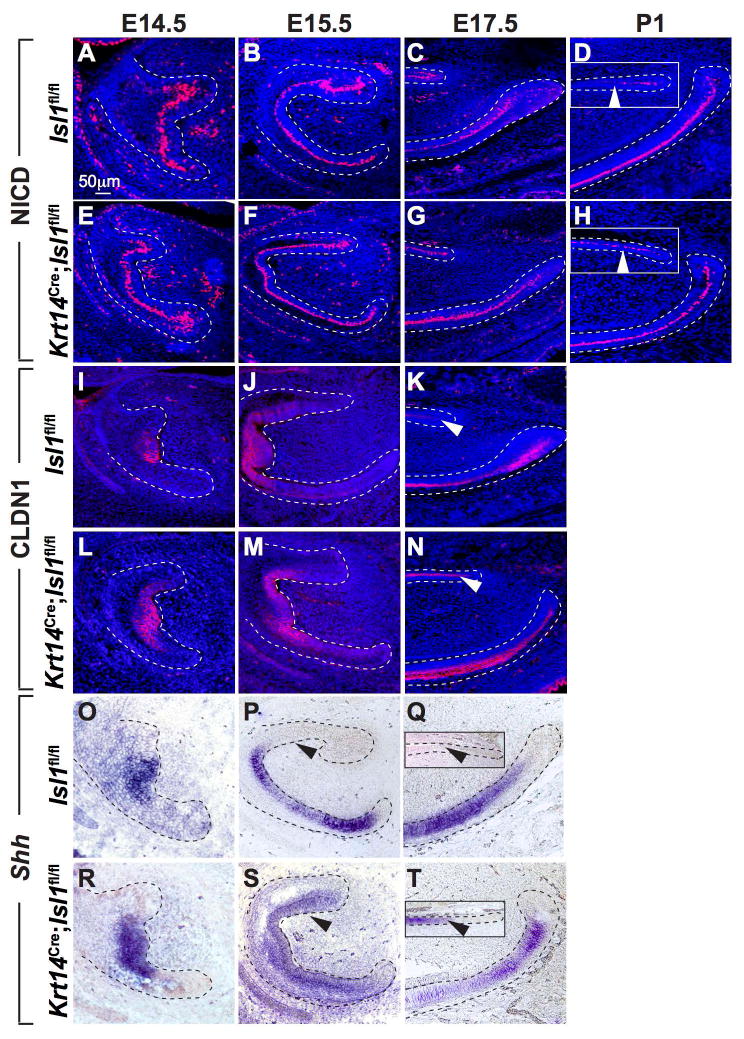
(A-H) Immunofluorescence staining for NICD showed similar patterns of expression in control and mutant mandibular incisors until E17.5 (C,G). At P1, NICD staining decreased in control liCL but remained in the mutant liCL (D,H insets; white arrowheads). There appeared to be little difference in NICD expression between control and mutant laCL. (I-N) Immunofluorescence staining for CLDN1 showed similar patterns of expression in the enamel knot area until E15.5 (I,J,L,M). However, similar to NICD staining at P1 (D,H insets), CLDN1 staining remained high in the mutant liCL but decreased in control liCL (K,N; white arrowheads). Again, there appeared to be little difference in CLDN1 expression in control and mutant laCL. (O-T) In situ hybridization analyses showed similar expression patterns in control and mutant mandibular incisors at E14.5 (O,R). At E15.5, Shh expression was evidenced on the lingual aspect of the mutant incisors but not in controls (P,S). At E17.5, Shh expression was present in mutant but not control liCL (Q,T insets; black arrowheads). At E17.5, consistent with observations in adult laCL (Fig. 5 J-L), there appeared to be decreased expression in mutant laCL compared to controls (Q,T).
RNA-Seq analysis points to numerous differentially expressed genes in control and mutant E15.5 mandibular incisors
In light of the myriad effects of Isl1 deletion on signaling pathways, we next took an unbiased approach to determine the effects of Isl1 deletion on gene expression. We analyzed the transcriptome of cells from developing incisors at E15.5 because this was the earliest stage at which we noted a difference in gene expression between control and mutant mice (Fig. 7O-T). RNA-Sequencing (Seq) analysis revealed 131 genes that were differentially expressed by 2-fold or higher (i.e., log2 fold change (FC) relative to the global average for differentially expressed genes) with a false discovery rate less than 0.01 (i.e., FDR < 0.01) (Fig. 8A, S5). The control and mutant groups clustered convincingly in our principle component analysis (PCA; Fig. S6A). From a list of all differentially expressed genes with an FDR < 0.01, we focused on 10 genes for confirmation based on previous known associations with ISL1, involvement in craniofacial development, and/or known expression in developing teeth at approximately E15.5 (Fig. 8B). Using qPCR, we confirmed that 6 of these 10 genes were indeed significantly differentially expressed. Otx1 and Tekt2 play a role in inner ear development;(51,52) Scg2 (also known as secretoneurin) interacts with STAT3, a known ISL1-interaction partner;(53,54) Ptprv is associated with bone and energy metabolism,(55) as well as P53-induced cell cycle exit;(56) Pdzk1ip1 (or Pdzk1-interacting protein 1) also interacts with Twist1, an important regulator of the Bmp signaling pathways;(57) and Stfa1 was observed to be overexpressed in a mouse psoriasis model.(58) Moreover, Otx1, Ptprv, and Scg2 are all expressed in E14.5 incisor tooth germs (genepaint.org). The remaining 4 genes, Kif27a, Frem3, Shisa7, and Hmgn2, showed similar trends to the RNA-Seq results but did not reach statistical significance (Fig. 8B,C).
Figure 8. Identification of differentially expressed genes between control and mutant mandibular incisors at E15.5 using RNA-Seq.
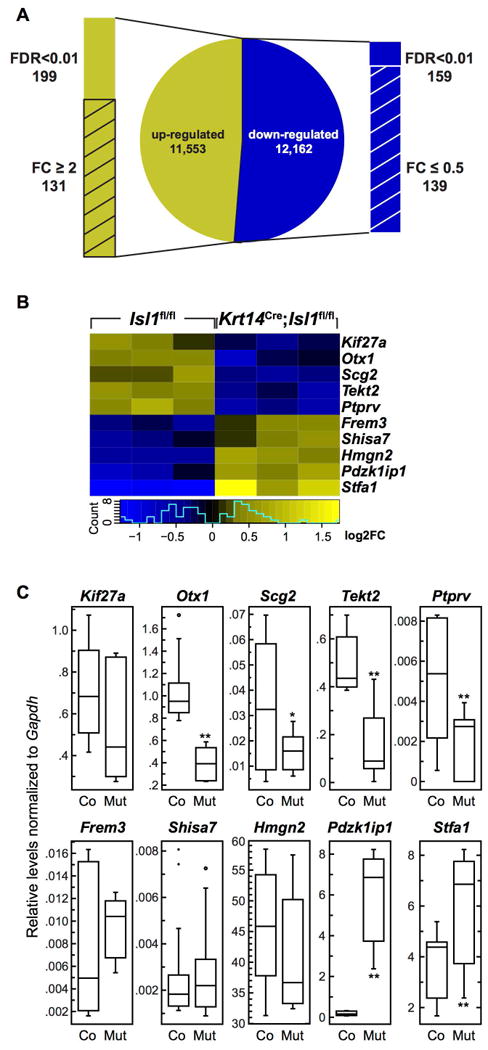
(A) RNA-Seq analysis revealed 357 differentially regulated genes with an FDR (false discovery rate) < 0.01. From this list, 131 genes were up-regulated 2-fold or higher (i.e., log2 FC (fold change)) and 139 genes were down-regulated 0.5-fold or lower. (B) Genes identified using RNA-Seq were confirmed to be differentially expressed between developing control (Co) and mutant (Mut) incisors using qPCR. Kif27a and Stfa1 expression levels were significantly higher in mutants compared to controls, whereas Otx2, Ptprv, Scg2, and Tekt2 were significantly lower in mutants. *, P<0.05; **, P<0.01).
Gene ontology analysis was conducted on genes differentially expressed by 2-fold or higher (up or down-regulation) with a false discovery rate < 0.01. This represents 139 down-regulated and 131 up-regulated genes. Gene ontology for our dataset consisted of 14 annotation clusters for the down-regulated entries (with 2 clusters displaying an enrichment score higher than 1.3), and 45 annotation clusters for the up regulated entries (with 12 clusters displaying an enrichment score higher than 1.3). The GO terms demonstrated that a diverse set of processes was impacted in the mutant (Fig. S6).
Discussion
Amelogenesis is a complex process that involves multiple signaling pathways. In the mouse incisor, enamel is generated on the labial surface (or crown-analog) by DESCs housed in the laCL. After proliferation in the TA region, the DESC progeny differentiate into enamel matrix-secreting ameloblasts. However, relatively little is known about the liCL, which normally does not make enamel on the lingual incisor surface (or the root-analog). Epithelial-specific deletion of Isl1 in Krt14Cre;Isl1fl/fl mutants resulted in incisors with altered enamel on the labial side and ectopic enamel-like tissue on the lingual side. Analyses of mouse models allow us to further dissect the cellular and molecular mechanisms of amelogenesis, which is potentially relevant to human tooth development.
Ectopic lingual enamel or ameloblasts has only been observed in three other genetically modified mice, all of which pointed to a central role for FGF and BMP signaling in the maintenance of asymmetric incisor enamel (i.e., enamel present only on the labial surface). First, alterations in Bmp signaling via inactivation of the extracellular antagonist follistatin (Fst) led to the presence of ectopic, lingual ameloblast-like cells.(15) Fst is normally expressed in the liCL (Fig. 5G), and its inactivation led to the expression of ectopic mesenchymal FGF3.(15) Because Fst-null mice exhibited perinatal lethality, the adult phenotype in these mice could not be studied. Second, the combined inactivation of the intracellular FGF antagonists Spry2 and Spry4 (Spry2+/-;Spry4-/- mice) resulted in ectopic lingual enamel production.(18) Ectopic mesenchymal Fgf3 and Fgf10 expression was detected in these mice near the liCL, and this was correlated with the differentiation of liCL-generated ameloblasts.(18) Third, deletion of the transcriptional repressor, Ctip2/Bcl11b, resulted in the inversion of Fgf3 and Fgf10 expression patterns.(19) This observation was correlated with a decrease in the size of the laCL along with abnormal ameloblasts, whereas the liCL was expanded in association with the generation of ameloblast-like cells.(19) However, similar to Fst-null mice, Bcl11b-null mice were perinatal lethal, which hindered comprehensive study of the cellular and molecular mechanisms of mutant incisors. Thus, our findings are distinct from the 3 prior mutants presenting with ectopic lingual enamel in that ISL1 is the first transcription factor to be identified whose conditional inactivation led to ectopic lingual enamel in viable and healthy mice.
Epithelial Isl1 inactivation resulted in alterations of all the major pathways that we tested, including the Bmp, Hh, Fgf, and Notch signaling pathways. The changes in the Bmp and Fgf signaling pathways were consistent with previous reports.(15,18,19) In the mutant liCL, Fgf signaling was increased, as evidenced by the up-regulation of Etv5 (Fig.5D-F) and pMEK (6A,B) and down-regulation of the intracellular Fgf antagonist Spry2 (Fig.S4A,B). Bmp signaling was also hyperactivated in mutant liCL, as demonstrated by the up-regulation of Bmp4 (Fig. S4M,N) and pSMAD1/5/8 (Fig. 6E,F), and the down-regulation of the Bmp antagonist Fst (Fig. 5G-I). NICD or NOTCH1 intracellular domain was expressed in mutant adult liCL (Fig. 6I,J), which appeared to be due to the retention of NICD expression in the liCL (Fig. 7A-H). The differences in NICD expression suggest a disruption in signaling between ameloblasts and the underlying SI.(59,60) Shh expression was also maintained in the mutant liCL (Fig. 5A-C). Differences in Shh expression were first detected at E15.5 (Fig. 7O-T), and SHH has been shown to regulate DESCs during differentiation of ameloblast progenitors in the laCL.(3) In summary, the epithelial deletion of Isl1 led to increased signaling through at least 4 major pathways, including Bmp, Fgf, Hh, and Notch. Moreover, we identified 217 genes that were differentially expressed greater than 2-fold in conditional Isl1 mutants. To date, we have confirmed 6 differentially expressed genes (i.e., Otx1, Scg2, Tekt2, Ptprv, Pdzk1ip1, and Stfa1) that are regulated by ISL1 (Fig. 8C). We will further analyze our RNA-Seq data and IPA-generated results (Fig. S6B,C) to discover additional information regarding the downstream ISL1 targets. Together, these findings point to ISL1 as a central factor in incisor amelogenesis.
Beyond the 4 major signaling pathways affected, we also observed that CLDN1 was present in mutant adult liCL, where it is not normally expressed (Fig. 6M,N). Differences in expression were first observed at E17.5, when CLDN1 localization was sustained in the mutant liCL (Fig. 7I-N). CLDN1 is a component of tight junctions essential for proper cell-cell adhesion and is expressed along with other claudins during mouse incisor and molar development.(61,62) Recently, it was shown that a deficiency in CLDN16 led to defects in the tight junctions of secretory ameloblasts that led to amelogenesis imperfecta.(63) SEM analysis of Isl1 mutant incisors revealed that labial inter-rods were disrupted near the tooth surface in the outer enamel but not near the DEJ in the inner (Fig. 3A-E), suggestive of defects in secretory ameloblasts.(64) Although differences in CLDN1 expression were not apparent on the labial side of Isl1 mutant incisors (Fig. 6O,P), it would be interesting to determine whether similar, altered inter-rod patterns are present in Cldn16-deficient mice.(63)
ISL1 functions in a context-dependent manner and is important in the determination of the crown vs. the root. The labial side of the mouse incisor is often considered to be the tooth crown analog, whereas the lingual side is the root analog. Enamel on the labial surface was prematurely mineralized in mutant incisors (Fig. 2D,H) likely due to premature differentiation of ameloblasts (Figs. 3F-O and S3A-H,M) and decreased proliferation of TA cells (Fig. 4E-H). SEM analysis revealed that labial enamel was altered in the mutant incisor (Fig. 3A-D3) but interestingly, we did not detect any density differences with additional microCT and SEM backscatter analyses (Fig. S2E-J). On the lingual side, we noted ectopic enamel (Fig. 2D,D′,H,H′) due to the presence of ectopic ameloblasts indicated by the expression of enamel proteins such as AMEL, AMBN, AMTN, ENAM, and KLK4 (Figs. 3F-O, S3), and the density of the ectopic lingual enamel was similar to that of labial enamel (Fig. S2E). Moreover, there was an increase in proliferative cells in the liCL (Fig. 4A-D), and differential expression of numerous genes between the liCL and laCL in control and mutant mice was observed. For example, Shh and Etv5 expression was up-regulated in mutant liCL but down-regulated in mutant laCL (Fig. 5). The seemingly opposite effects observed with epithelial Isl1 inactivation in the liCL and laCL, specifically in proliferation and gene expression, strongly suggest context-dependent regulation of ISL1 that will be fertile ground for future studies. It will be interesting to determine whether there are changes in molar root development in Isl1 mutant mice, as well as in 3 other mouse models that generate ectopic enamel and/or ameloblast-like cells from the liCL.(15,18,19)
The specific effect of epithelial Isl1 inactivation on continuously renewing mouse incisors but not molars (Figs. 2D,H, S2A-D). We did not detect any Isl1 expression during molar development (E14.5 to 6-week old). A previous report indicated slight expression of Isl1 during molar development at E11.5 and P4.(33) These prior experiments were whole-mount in situ hybridization that showed very limited Isl1 expression profiles on the lingual side of the developing molar at E11.5 and in the enamel-free cusp region at P4. Although it is possible that we did not analyze these specific sections in our study, it is difficult to confirm the prior report since no negative control sections were presented.(33) Regardless of the limited Isl1 expression in molars, we conclude that molar development was not affected by Isl1 inactivation (Fig. S2A-D). Our results strongly support a potential role for Isl1 in the development of anterior teeth (e.g., incisors and canines), as well as in the regulation of DESCs. This hypothesis is further supported by the specific expression of Isl1 in anterior teeth but not posterior teeth (e.g., molars) in humans.(35) Isl1 also appears to be a critical factor for maintaining labio-lingual asymmetry, as deletion of this gene leads the laCL and liCL regions to become more similar to one another.
Our RNA-Seq experiment comparing control and mutant E15.5 incisors produced an extensive list of differentially regulated genes to focus on in future studies (Fig. 8, S6). GO analysis revealed association of these genes to biological functions or components and highlighted annotation clusters showing the potential importance of protein translation, trafficking, localization, and cell signaling (Fig. S6). The relatively large presence of clusters encompassing immune response terms was not surprising, given that the dental mesenchyme at this stage houses numerous immune cells (65).
Our data suggest that there are least 3 distinct molecular mechanisms for the generation of enamel: molar ameloblasts do not require Isl1 to develop properly, laCL-generated ameloblasts do, and the absence of Isl1 leads to liCL-generated ectopic ameloblasts and enamel. Together, our findings demonstrate that ISL1 is a central factor in patterning of proper incisor amelogenesis and that it is an upstream regulator of multiple signaling pathways and genes.
Supplementary Material
Acknowledgments
MicroCT imaging work was performed in part by Sabra Djomehri in the UCSF Division of Biomaterials and Bioengineering Micro-CT Imaging Facility, supported by the Dept. of Health and Human Services/NIH S10 Shared Instrumentation Grant (S10RR026645) and the Departments of Preventive and Restorative Dental Sciences and Orofacial Sciences, School of Dentistry, UCSF. We would also like to thank the Laurel Foundation Endowment for Craniofacial Research at the University of Washington. The authors are funded by the National Institutes of Health (R35-DE026602 and R01-DE021420 to O.D.K., and R00-DE022059 to A.H.J.). A.N. was supported by the Université Paris Descartes – Sorbonne Paris Cité, Fondation Bettencourt-Schueller, Institut Servier, Fondation des Gueules-Cassées, Fondation Philippe, and Assistance Publique-Hôpitaux de Paris.
Footnotes
Disclosure – all authors have nothing to disclose
Authors' roles: Study design – AHJ, ODK; Animal treatment – AN, BM, TW, KBJ, AHJ; Histology, in situ hybridization, and immunostaining – AN, BZ, BM, MTS, TW, AHJ; microCT – TCC; SEM – BG, AHJ; qPCR data collection and analyses – BM, MP, AHJ; RNA-Seq analysis – PM, AHJ; Data interpretation – AN, BZ, AHJ, ODK; Manuscript preparation – AN, BZ, AHJ, ODK; Approval of final versions of manuscript - All; AHJ and ODK are responsible for the integrity of data analysis.
References
- 1.Jheon AH, Seidel K, Biehs B, Klein OD. From molecules to mastication: the development and evolution of teeth. Wiley Interdiscip Rev Dev Biol Mar-Apr. 2013;2(2):165–82. doi: 10.1002/wdev.63. Epub 2013/09/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999 Oct 4;147(1):105–20. doi: 10.1083/jcb.147.1.105. Epub 1999/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010 Nov;137(22):3753–61. doi: 10.1242/dev.056358. Epub 2010/10/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li CY, Cha W, Luder HU, Charles RP, McMahon M, Mitsiadis TA, et al. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev Biol. 2012 Jun 15;366(2):357–66. doi: 10.1016/j.ydbio.2012.03.012. Epub 2012/04/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. 2012 Aug 14;23(2):317–28. doi: 10.1016/j.devcel.2012.05.012. Epub 2012/07/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biehs B, Hu JK, Strauli NB, Sangiorgi E, Jung H, Heber RP, et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol. 2013 Jul;15(7):846–52. doi: 10.1038/ncb2766. Epub 2013/06/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith CE, Warshawsky H. Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat Rec. 1975 Dec;183(4):523–61. doi: 10.1002/ar.1091830405. [DOI] [PubMed] [Google Scholar]
- 8.Smith CE, Warshawsky H. Histological and three dimensional organization of the odontogenic organ in the lower incisor of 100 gram rats. Am J Anat. 1975 Apr;142(4):403–29. doi: 10.1002/aja.1001420402. [DOI] [PubMed] [Google Scholar]
- 9.Warshawsky H, Smith CE. Morphological classification of rat incisor ameloblasts. Anat Rec. 1974 Aug;179(4):423–46. doi: 10.1002/ar.1091790403. Epub 1974/08/01. [DOI] [PubMed] [Google Scholar]
- 10.Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998 Nov;125(21):4325–33. doi: 10.1242/dev.125.21.4325. Epub 1998/10/01. [DOI] [PubMed] [Google Scholar]
- 11.Mitsiadis TA, Hirsinger E, Lendahl U, Goridis C. Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol. 1998 Dec 15;204(2):420–31. doi: 10.1006/dbio.1998.9092. Epub 1999/01/12. [DOI] [PubMed] [Google Scholar]
- 12.Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, et al. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002 Mar;129(6):1533–41. doi: 10.1242/dev.129.6.1533. Epub 2002/03/07. [DOI] [PubMed] [Google Scholar]
- 13.Millar SE, Koyama E, Reddy ST, Andl T, Gaddapara T, Piddington R, et al. Over- and ectopic expression of Wnt3 causes progressive loss of ameloblasts in postnatal mouse incisor teeth. Connect Tissue Res. 2003;44(Suppl 1):124–9. Epub 2003/09/04. [PubMed] [Google Scholar]
- 14.Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004 Nov;7(5):719–30. doi: 10.1016/j.devcel.2004.09.012. Epub 2004/11/05. [DOI] [PubMed] [Google Scholar]
- 15.Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, et al. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007 Jun;5(6):e159. doi: 10.1371/journal.pbio.0050159. Epub 2007/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010 Nov-Dec;80(4-5):241–8. doi: 10.1016/j.diff.2010.06.004. Epub 2010/08/10. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Dangaria S, Andl T, Zhang Y, Wright AC, Damek-Poprawa M, et al. beta-Catenin initiates tooth neogenesis in adult rodent incisors. J Dent Res. 2010 Sep;89(9):909–14. doi: 10.1177/0022034510370090. Epub 2010/06/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008 Jan;135(2):377–85. doi: 10.1242/dev.015081. Epub 2007/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyrylkova K, Kyryachenko S, Biehs B, Klein O, Kioussi C, Leid M. BCL11B regulates epithelial proliferation and asymmetric development of the mouse mandibular incisor. PLoS One. 2012;7(5):e37670. doi: 10.1371/journal.pone.0037670. Epub 2012/05/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990 Apr 26;344(6269):879–82. doi: 10.1038/344879a0. Epub 1990/04/26. [DOI] [PubMed] [Google Scholar]
- 21.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996 Jan 26;84(2):309–20. doi: 10.1016/s0092-8674(00)80985-x. Epub 1996/01/26. [DOI] [PubMed] [Google Scholar]
- 22.Ericson J, Norlin S, Jessell TM, Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998 Mar;125(6):1005–15. doi: 10.1242/dev.125.6.1005. Epub 1998/05/09. [DOI] [PubMed] [Google Scholar]
- 23.Hunter CS, Dixit S, Cohen T, Ediger B, Wilcox C, Ferreira M, et al. Islet alpha-, beta-, and delta-cell development is controlled by the Ldb1 coregulator, acting primarily with the islet-1 transcription factor. Diabetes. 2013 Mar;62(3):875–86. doi: 10.2337/db12-0952. Epub 2012/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witzel HR, Jungblut B, Choe CP, Crump JG, Braun T, Dobreva G. The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev Cell. 2012 Jul 17;23(1):58–70. doi: 10.1016/j.devcel.2012.06.005. Epub 2012/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawakami Y, Marti M, Kawakami H, Itou J, Quach T, Johnson A, et al. Islet1-mediated activation of the beta-catenin pathway is necessary for hindlimb initiation in mice. Development. 2011 Oct;138(20):4465–73. doi: 10.1242/dev.065359. Epub 2011/09/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005 Feb 10;433(7026):647–53. doi: 10.1038/nature03215. Epub 2005/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006 Dec 15;127(6):1151–65. doi: 10.1016/j.cell.2006.10.029. Epub 2006/11/25. [DOI] [PubMed] [Google Scholar]
- 28.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009 Jul 2;460(7251):113–7. doi: 10.1038/nature08191. Epub 2009/07/03. [DOI] [PubMed] [Google Scholar]
- 29.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell Dec. 2003;5(6):877–89. doi: 10.1016/s1534-5807(03)00363-0. Epub 2003/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caputo L, Witzel HR, Kolovos P, Cheedipudi S, Looso M, Mylona A, et al. The Isl1/Ldb1 Complex Orchestrates Genome-wide Chromatin Organization to Instruct Differentiation of Multipotent Cardiac Progenitors. Cell Stem Cell. 2015 Sep 3;17(3):287–99. doi: 10.1016/j.stem.2015.08.007. Epub 2015/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heikinheimo K, Kurppa KJ, Laiho A, Peltonen S, Berdal A, Bouattour A, et al. Early dental epithelial transcription factors distinguish ameloblastoma from keratocystic odontogenic tumor. J Dent Res. 2015 Jan;94(1):101–11. doi: 10.1177/0022034514556815. Epub 2014/11/16. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Shaffer JR, Zeng Z, Begum F, Vieira AR, Noel J, et al. Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health. 2012;12:57. doi: 10.1186/1472-6831-12-57. Epub 2012/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsiadis TA, Angeli I, James C, Lendahl U, Sharpe PT. Role of Islet1 in the patterning of murine dentition. Development. 2003 Sep;130(18):4451–60. doi: 10.1242/dev.00631. Epub 2003/08/06. [DOI] [PubMed] [Google Scholar]
- 34.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993 Dec 31;75(7):1417–30. doi: 10.1016/0092-8674(93)90627-3. Epub 1993/12/31. [DOI] [PubMed] [Google Scholar]
- 35.Huang Z, Hu X, Lin C, Chen S, Huang F, Zhang Y. Genome-wide analysis of gene expression in human embryonic tooth germ. J Mol Histol. 2014 Dec;45(6):609–17. doi: 10.1007/s10735-014-9580-5. Epub 2014/08/06. [DOI] [PubMed] [Google Scholar]
- 36.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000 Nov;127(22):4775–85. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 37.Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008 Jun;135(11):1981–90. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012 Apr;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2014. Nucleic Acids Res. 2014 Jan;42(Database issue):D749–55. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 Jan;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001 Apr 3;11(7):503–7. doi: 10.1016/s0960-9822(01)00143-9. Epub 2001/06/20. [DOI] [PubMed] [Google Scholar]
- 45.Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006 Aug;11(2):181–90. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura T, Hasegawa Y, Sugino K, Kogawa K, Titani K, Sugino H. Follistatin inhibits activin-induced differentiation of rat follicular granulosa cells in vitro. Biochim Biophys Acta. 1992 Apr 30;1135(1):103–9. doi: 10.1016/0167-4889(92)90173-9. Epub 1992/04/30. [DOI] [PubMed] [Google Scholar]
- 47.Matzuk MM, Lu N, Vogel H, Sellheyer K, Roop DR, Bradley A. Multiple defects and perinatal death in mice deficient in follistatin. Nature. 1995 Mar 23;374(6520):360–3. doi: 10.1038/374360a0. Epub 1995/03/23. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Boulan E, Nelson WJ. Morphogenesis of the polarized epithelial cell phenotype. Science. 1989 Aug 18;245(4919):718–25. doi: 10.1126/science.2672330. Epub 1989/08/18. [DOI] [PubMed] [Google Scholar]
- 49.Inai T, Sengoku A, Hirose E, Iida H, Shibata Y. Differential expression of the tight junction proteins, claudin-1, claudin-4, occludin, ZO-1, and PAR3, in the ameloblasts of rat upper incisors. Anat Rec (Hoboken) 2008 May;291(5):577–85. doi: 10.1002/ar.20683. [DOI] [PubMed] [Google Scholar]
- 50.Bello IO, Soini Y, Slootweg PJ, Salo T. Claudins 1, 4, 5, 7 and occludin in ameloblastomas and developing human teeth. J Oral Pathol Med. 2007 Jan;36(1):48–54. doi: 10.1111/j.1600-0714.2006.00497.x. [DOI] [PubMed] [Google Scholar]
- 51.Morsli H, Tuorto F, Choo D, Postiglione MP, Simeone A, Wu DK. Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development. 1999 Jun;126(11):2335–43. doi: 10.1242/dev.126.11.2335. Epub 1999/05/05. [DOI] [PubMed] [Google Scholar]
- 52.Yoon H, Lee DJ, Kim MH, Bok J. Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol. 2011 Mar;44(1):69–78. doi: 10.5115/acb.2011.44.1.69. Epub 2011/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shyu WC, Lin SZ, Chiang MF, Chen DC, Su CY, Wang HJ, et al. Secretoneurin promotes neuroprotection and neuronal plasticity via the Jak2/Stat3 pathway in murine models of stroke. J Clin Invest. 2008 Jan;118(1):133–48. doi: 10.1172/JCI32723. Epub 2007/12/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao A, Novotny-Diermayr V, Bian W, Lin B, Lim CP, Jing N, et al. The LIM/homeodomain protein Islet1 recruits Janus tyrosine kinases and signal transducer and activator of transcription 3 and stimulates their activities. Mol Biol Cell. 2005 Apr;16(4):1569–83. doi: 10.1091/mbc.E04-08-0664. Epub 2005/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kassi E, Papavassiliou AG. A possible role of osteocalcin in the regulation of insulin secretion: human in vivo evidence? J Endocrinol. 2008 Nov;199(2):151–3. doi: 10.1677/JOE-08-0294. Epub 2008/07/23. [DOI] [PubMed] [Google Scholar]
- 56.Doumont G, Martoriati A, Marine JC. PTPRV is a key mediator of p53-induced cell cycle exit. Cell Cycle. 2005 Dec;4(12):1703–5. doi: 10.4161/cc.4.12.2207. Epub 2005/11/01. [DOI] [PubMed] [Google Scholar]
- 57.Hayashi M, Nimura K, Kashiwagi K, Harada T, Takaoka K, Kato H, et al. Comparative roles of Twist-1 and Id1 in transcriptional regulation by BMP signaling. J Cell Sci. 2007 Apr 15;120(Pt 8):1350–7. doi: 10.1242/jcs.000067. [DOI] [PubMed] [Google Scholar]
- 58.Lundberg KC, Fritz Y, Johnston A, Foster AM, Baliwag J, Gudjonsson JE, et al. Proteomics of skin proteins in psoriasis: from discovery and verification in a mouse model to confirmation in humans. Mol Cell Proteomics. 2015 Jan;14(1):109–19. doi: 10.1074/mcp.M114.042242. Epub 2014/10/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jheon AH, Prochazkova M, Meng B, Wen T, Lim YJ, Naveau A, et al. Inhibition of Notch Signaling During Mouse Incisor Renewal Leads to Enamel Defects. J Bone Miner Res. 2015 Jul 14; doi: 10.1002/jbmr.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, et al. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 2006 Feb 10;340(2):611–6. doi: 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 61.Ohazama A, Sharpe PT. Expression of claudins in murine tooth development. Dev Dyn. 2007 Jan;236(1):290–4. doi: 10.1002/dvdy.21001. [DOI] [PubMed] [Google Scholar]
- 62.Hoshino M, Hashimoto S, Muramatsu T, Matsuki M, Ogiuchi H, Shimono M. Claudin rather than occludin is essential for differentiation in rat incisor odontoblasts. Oral Dis Oct. 2008;14(7):606–12. doi: 10.1111/j.1601-0825.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- 63.Bardet C, Courson F, Wu Y, Khaddam M, Salmon B, Ribes S, et al. Claudin-16 Deficiency Impairs Tight Junction Function in Ameloblasts, Leading to Abnormal Enamel Formation. J Bone Miner Res. 2015 Oct 1; doi: 10.1002/jbmr.2726. [DOI] [PubMed] [Google Scholar]
- 64.Skobe Z. The secretory stage of amelogenesis in rat mandibular incisor teeth observed by scanning electron microscopy. Calcif Tissue Res. 1976 Oct 12;21(2):83–103. doi: 10.1007/BF02547385. [DOI] [PubMed] [Google Scholar]
- 65.Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, et al. Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. Elife. 2017 May 05;:6. doi: 10.7554/eLife.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


