Abstract
Patients with chronic health conditions use online health communities to seek support and information to help manage their condition. For clinically related topics, patients can benefit from getting opinions from clinical experts, and many are concerned about misinformation and biased information being spread online. However, a large volume of community posts makes it challenging for moderators and clinical experts, if there are any, to provide necessary information. Automatically identifying forum posts that need validated clinical resources can help online health communities efficiently manage content exchange. This automation can also assist patients in need of clinical expertise by getting proper help. We present our results on testing text classification models that efficiently and accurately identify community posts containing clinical topics. We annotated 1,817 posts comprised of 4,966 sentences of an existing online diabetes community. We found that our classifier performed the best (F-measure: 0.83, Precision: 0.79, Recall:0.86) when using Naïve Bayes algorithm, unigrams, bigrams, trigrams, and MetaMap Symantic Types. Training took 5 seconds. The classification process took a fraction of 1 second. We applied our classifier to another online diabetes community, and the results were: F-measure: 0.63, Precision: 0.57, Recall: 0.71. Our results show our model is feasible to scale to other forums on identifying posts containing clinical topic with common errors properly addressed.
Keywords: Text mining, clinical topic, diabetes, online health communities, human-computer interaction, health information seeking, classification, patient
Graphical abstract
1. Introduction
Patients with chronic conditions visit online health communities to get help with managing their conditions [1]. In these communities, patients support one another through empathetic posts and consult on how to improve their daily health management strategies. At the same time, topics that can benefit from clinicians’ expertise frequently appear in these patient discussions [2, 3]. Messages containing such topics get buried in an overwhelming amount of posts, making it difficult for potential moderators to address them.
Moderators play an important role in online health communities. In addition to facilitating conversations, moderators add useful resources to posts containing clinically related questions [3]. Moderators also make sure information shared on their websites is not intended to be a substitute for professional medical advice by adding disclaimers or helping patients find relevant resources [3]. Patients self-moderate in online health communities where active, informal leaders exist [2, 4]. For newly developing communities, however, such moderation activities around clinically related topics can be hard to do due to overwhelming amount of posts [5]. Efficiently identifying the patients’ posts needing additional, validated clinical resource would improve the quality of information shared in online health communities.
Many online health communities do not have moderators who can redirect questions to those with relevant expertise. Especially for those information needing clinical expertise, the community can benefit from knowing when certain questions need specific expertise over another. An automated system could be added by the forum owners to identify clinically-related posts to act upon it. If the information can be verified against a known knowledge base, e.g. WebMD, the system could respond to the user’s post with either more information or additional verification that the advice is supported by the topic experts. If the information cannot be verified, or the concern can best be addressed by the user’s physician, then the system could notify the user of the need that the content can benefit from extra verification as current moderators do [2].
In this paper, we develop a classification method to efficiently identify clinically related posts in online health communities. We examine specifically whether the clinical post addresses a medical question, a symptom, or a treatment. Existing work begins to address this problem, but the performance of classifiers could be improved [6]. The classifier should also be able to scale to other communities. We used manually annotated data, feature design, feature selection methods, and comparisons across classifier algorithms to maximize the performance classifying clinically related posts in online diabetes communities. We also investigated the scalability of our classification model to other community context.
Our research questions include:
How can feature design and selection techniques improve performance?
Which classifier algorithm best perform in identifying topics from an online diabetes community?
How high is the performance on detecting clinically related sentences in online health community posts?
How much does our model built from one online health community generalize to other online health community contexts?
Below, we discuss related work, followed by the methods used to address these questions.
2. Related Work
Online health communities present significant benefits to patients receiving support toward managing chronic disease. Research has shown the effects of using online social networks for chronic disease management. Merolli et al. summarized and analyzed the health outcomes and effects reported in previous studies [7]. One important benefit patients receive is support, both informational and emotional. Vlahovic et al. analyzed the satisfaction of users with their received support based on the type of support they requested, and they found that users seeking informational support and receiving emotional support were less satisfied than users seeking emotional support and receiving informational support [8]. De Choudhury et al. surveyed users about their sharing and seeking health-related information on Twitter [9]. They found 20% of the participants sought health-related information from Twitter. In particular, over half of those seeking information from Twitter were about seeking treatment information. Bui et al. found the sentiments of posts in online social support networks evolved from negative to positive sentiment [10]. Hartzler et al. investigated connecting patients based on their shared interests [11, 12]. As such, existing work in online health social networks is focused on evaluating the efficacy of social support and devising ways to further augment support in online health communities. Further exploring work in improving qualities of sharing clinically related topics in online health communities can complement existing work around providing good quality social support to online health community members.
Huh et al. analyzed the roles of patients and moderators in online health communities [3]. They found that a majority of posts could benefit from clinical expertise, but there is not a sufficient number of clinical moderators to respond to all posts [13]. Even if moderators exist, sifting through a large number of community posts to identify posts needing clinical expertise can be overwhelming. To address this issue, a research team developed visualization tools to help moderators understand trends of aggregated online health community posts [14]. Furthermore, Huh et al. showed that moderators participate in online health communities to provide clinical expertise [3] and recommended patients to see a doctor [2]. A possible system to make these moderation activities more efficient is delivering moderators targeted posts needing their attention. To extract requirements for such system, Huh and Pratt interviewed clinicians while they read a subset of community threads to understand the challenges and necessary components of such a system [5]. The results indicated that clinicians identified clinically related keywords in posts as one of critical identifiers needing their attention and stated the importance of ”triaging” the posts based on the severity of the problem expressed by the patients in their posts.
Researchers have attempted to identify clinically related posts in social media settings. McRoy et al. developed a classifier for community-based question answering websites, where the classification scheme included: factual clinical questions, patient-specific questions, and non-clinical questions [15]. Researchers also examined ways to identify authors of online health community posts–whether they are health professionals, which could inform the authority of clinical advice shared [16, 17]. Abdaoui et al. used UMLS and other medical ontologies to determining whether the author of a post was a health professional or a lay man. Choumatare applied classification techniques to predict which patients had depression [18] to potentially provide help. Yang et al. used classification to detect posts that discuss adverse drug reactions [19]. Tuarob et al. classified whether or not each post from Twitter was health-related [20]. Akbari et al. proposed an algorithm to detect wellness events, which are activities performed related to diet, exercise, or health [21].
As such, researchers have actively begun to investigate ways to deliver high quality information to patients online, augment social support, and provide interventions based on their stories posted online. Our work builds on this line of work, contributing new and improved ways to efficiently identify when patients need clinical expertise.
3. Methods
3.1. Data Collection
Prior research has demonstrated that WebMD consists of active communities, where users discuss chronic health conditions [2, 3, 5, 6]. WebMD is a health information portal website which provides information and tools to users for managing their health [22]. One critical feature of WebMD includes Exchanges, which is online communities where users discuss anything about managing their medical conditions. Each community is dedicated to one specific health condition, e.g. Diabetes or Heart Disease. We focused on the diabetes community (WDC) because it had the most active participation regarding balance between informational and emotional posts shared [6].
From WDC, we collected all threads posted between July 2007 (the beginning of the community) and July 2014 (the last date of data collection). A thread is a series of posts, which begins with a thread initiating post, followed by replies from other users. Because patients often initiate discussions in thread initiating posts [3], we examined only the posts that initiate threads of conversation through replies and replies to replies. We extracted 9,576 thread initiating posts from the data we collected. We removed 538 duplicate posts. Each post contained one or more sentences. Figure 1 demonstrates that most posts have 10 or fewer sentences. One post can consist of sentences that include clinically relevant keywords and those that do not. To simplify the scope what is considered a clinically relevant post, we designated each sentence as a unit of analysis. Our process is shown in Figure 2.
Figure 1.
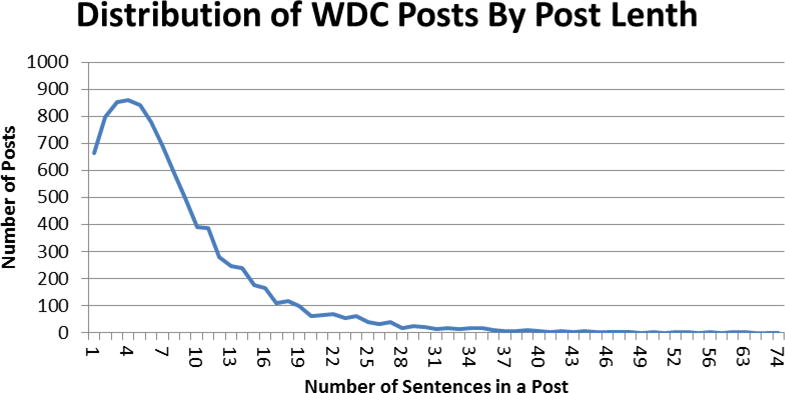
Frequency distribution of WebMD posts by number of sentences
Figure 2.
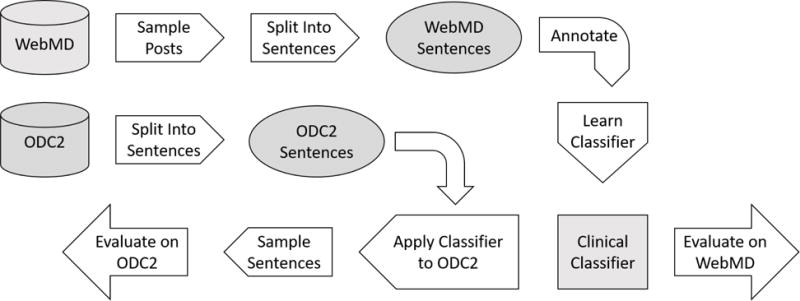
Workflow Process
We split all posts into sentences using Stanford’s Natural Language Toolkit (NLTK) sentence tokenizer [23]. NLTK split posts into sentences by splitting on periods. We used a regular expression to identify and merge incorrectly split sentences into a whole sentence. Additionally, some users used other symbols, e.g. commas, to separate sentences. To address this, we manually identified and split these sentences during annotation.
We also collected posts from another online diabetes community (ODC2). This data was provided by our collaborator, who agreed to share their deidentified forum posts with us for the purpose of research and improving their own community. Identical to WDC, the community post structure was thread-based: each thread began with a thread initiating post, followed by the replies. We received 23,473 thread initiating posts from ODC2. We split these post into sentences using the same method described for WDC, which generated 2,009,005 sentences in total. To test the performance of our models, we applied our best performing classifier to all sentences from this data set. We then randomly selected 250 clinical sentences and 250 non-clinical sentences, based on the classifier predicted label, for a human coder to annotate for gold standard. These sentences were presented to the annotator in a random order without the predicted labels to assess the efficacy of applying the classifier model built from WDC to another community.
3.2. Annotation for training data
We randomly sampled 20% of WDC posts (1,817 total posts) for manual annotation. Table 2 provides a breakdown of number of posts and the number of sentences in the sample. We then randomly selected 100 sentences from the total number of sentences in the sampled posts. Two coders iteratively worked through the posts to refine the definition of which sentences are clinically relevant or not. The two coders repeated coding independently and reviewed the disagreements together to improve the clarity of the codebook. The resulting codebook identified a sentence as clinical if it discussed one or more symptoms, treatments or medical conditions, or posed a health related question, such as ”how would jogging everyday affect my sugar levels?”. Otherwise the sentence was annotated as not clinical. One coder then finished the coding of the rest of the sentences from WDC. The same coder annotated the 500 sentences selected from ODC2. Examples of clinical and non-clinical sentences are in Table 1. There were 4,966 sentences in our sample, and the breakdown of clinical and non-clinical sentences is shown in Table 2.
Table 2.
This table shows the number of posts, sentences sampled, and sentences per class.
| Posts Sampled | 1,817 |
| Sentences in Sample | 4,966 |
| Clinical Sentences | 2,953 |
| Not Clinical Sentences | 2,013 |
Table 1.
Examples of Clinical and Non-Clinical Sentences
| Topic | Category | Text |
|---|---|---|
|
| ||
| Food | Not Clinical | “Did you know that January is National Oatmeal Month?!” |
| News | Not Clinical | “There’s another shooting in progress in Orlando!!!” |
| Users | Not Clinical | “where, s[sic] Rzbgy ??” |
| Symptom | Clinical | “I have noticed lately that my urine has had a strong odor every time I go to the bathroom.” |
| Monitor | Clinical | “Has any one had any problems with Reli On Glucose Meter from Wal-Mart?” |
| Drugs | Clinical | “Is there anyone currently using Humalog pens &/or Lantus pens?” |
3.3. Feature Selection
As a first step to construct our classification model, we selected the following features based on the lessons learned from previous literature and our own work:
Unigrams (U): unigrams have been widely used for information retrieval [24] and showed effectiveness in text classification [25, 26].
Bigrams and Trigrams (BT): bigrams and trigrams are commonly used for next word prediction [24] and have shown effectiveness in text categorization [27] and text classification [28, 29].
MetaMap Categories, Part of Speech, and Polarity (MCSP): Denecke and Nejdl proposed using the frequency of phrases tagged with one of three medical language semantic tagger (MetaMap) categories (Disorders, Procedures, and Chemicals & Drugs), frequency of terms used in one of three parts of speech (nouns, verbs, and adjectives), and the frequency of terms belonging to one of three polarities (positive, negative, and neutral) for retrieval of information type (informative or affective) [30].
MetaMap Semantic Types (MM): We explore broader medical concepts using the MetaMap [12].
Unigrams is commonly used for information retrieval for text data [24, 25, 26]. We used unigrams as our baseline feature set because of its simplicity and production of high performance [25, 26]. Researchers found adding bigrams and trigrams features improved performance [27, 28, 29]. Because our goal was to identify sentences discussing medical topics, we used the Unified Medical Language System (UMLS), which was developed by the National Library of Medicine (NLM). UMLS organizes medical concepts into categories, such as Disorders, Genes & Molecular Sequences, Physiology, and Procedures that include standardized biomedical vocabularies.
To find UMLS semantic types in our data set, we used MetaMap3, an application provided by the UMLS Terminology Services (UTS), an interactive system provided by UMLS, which searches a body of text and tags phrases for UMLS concepts. We count the frequency of each semantic type that is in our feature sets and added the frequency as a feature.
We compared two sets of semantic concepts from UMLS using MetaMap, which are listed in Table A.12 in the Appendix. For the first set of UMLS semantic concepts, we went through the list of UMLS concepts with our clinical collaborators based on its relevance to the codebook (e.g., treatments, medications, etc). This process is similar to how researchers select UMLS concepts [12]. We refer to this set of features as MM for the rest of the paper. The second set of concepts (Table A.12) were proposed by Denecke and Nejdl [30]. Because of the similarity in scope, we chose to add their choice of UMLS semantic concepts to our experiments. In their work, they added frequency of the categories as features rather than the frequency of each semantic type. Those categories, and their corresponding semantic types are listed in Table A.12. Additional features proposed by Denecke and Nejdl were the frequencies of words with positive sentiment, words with negative sentiment, objective words (neither positive nor negative sentiment), nouns, verbs, and adjectives. All of the features proposed by Denecke and Nejdl constitute the MetaMap Categories, Part of Speech, and Polarity (MCSP) feature set.
3.4. Feature Reduction
We applied several preprocessing techniques common in text classification [12]. We lowercased all characters. Prior to splitting the sentences into unigrams, we replaced non-alphanumeric characters with space. We did not want non-alphanumeric characters leading to unnecessary additional terms (e.g. “~19” is treated as “19”). To improve the classification results, we applied two common feature reduction techniques in NLP–stemming and stopword removal [24]. Stemming combines words that have the same morpheme, e.g. sugar and sugars share the morpheme sugar [24]. Porter Stemmer is the most widely used stemmer in information retrieval [24]. We applied the Snowball Stemmer [31], an improvement on the Porter Stemmer, to unigrams. We found that the Snowball Stemmer returned more human readable stems, which was important for understanding the terms that influenced performance. Stopwords are high frequency terms, such as pronouns and prepositions, that do not provide much semantic information. We removed stopwords from the unigram lists because they carry little semantic weight [24]. We removed singletons, features that only appear in only one sentence, because they would not be useful for predicting the label of other sentences.
We evaluated the following additional feature reduction techniques used in text classifications:
Number Replacement
Mutual Information
χ2 Statistic
Number Replacement (NR) is the process for replacing numbers with a common constant. Numbers are identified using a regular expression. We applied NR to all feature sets described in Section 3.3 to improve performance.
Mutual Information and χ2 Statistic worked differently than NR. These techniques generated a score to rank the existing features based on their likelihood of occurring in a clinical sentence [24]. Features with high scores were predictive of clinical sentences. We started with the set of features that have the highest performance, which was unigrams (U), bigrams and trigrams (BT), and MetaMap semantic types (MM), and tested whether Mutual Information or χ2 Statistic could improve performance. We selected the top k features, ranked by their scores. We tuned k by setting it to multiples of 1000, between 1000 and 20,000 features.
3.5. Classification algorithm
We explored the performance of classifiers on each classification task described in Section 3.2. Three classifiers had high performances on textual data, which is high-dimensional and sparse. They are: K-Nearest Neighbor (KNN) [32], Support Vector Machines (SVM) [33], and Naïve Bayes (NB). Support Vector Machine efficiently finds a hyperplane that separates the data by class, even in high dimensional space [34]. Because the hyperplane returned by SVM has the minimum error, it is less prone to overfitting than other classifiers [35]. Performance of SVM is dependent on the choice of kernel [34]. In our work, we found high performance using the simplest kernel, the linear kernel, so we did not explore other kernels.
When the data is arbitrarily shaped, K-Nearest Neighbor performs well [34]. Without knowing a priori whether our data is linearly separable, we tested both linear and nonlinear classifiers. Most often KNN is used when there are large number of features [35]. KNN has several disadvantages. Unlike other classifiers, classifying a new document is expensive because all of the training documents are stored in memory and are compared to the new document to find those most similar [34]. KNN is dependent on the choice of two parameters; the number of neighbors (k) and the similarity measure. We used cosine similarity, which is the widely accepted similarity measure for classification of text documents [24]. We explored different values for k and selected k that maximizes performance of our baseline, unigrams. We restricted k to odd values, to ensure there were no ties between the two classes, i.e. a sentence cannot have an equal number of non-clinical nearest neighbors as it has clinical neighbors.
Naïve Bayes classifier is a nonlinear classifier that performs well on natural language processing tasks like classification and word sense disambiguation [24]. By assuming terms are conditionally independent of each other given the label of the document, Naïve Bayes is not significantly influenced by irrelevant variables [34]. When terms are correlated, performance of Naïve Bayes degrades [34]. We observe this in our experiments, which we present in Section 4.
3.6. Performance metrics
We evaluated our classification model using four metrics to calculate performance of the classifiers: precision, recall, F-measure, and run time. For one of our classifiers, K-Nearest Neighbor, we must tune the parameter k. We use F-measure to determine which value of k has the best performance.
The training data used in this project was relatively small compared to the sentences on the Web. When we scale up to larger data, the run time will increase. In addition to the accuracy of the predictions, we also measured its runtime to test its future possibility in being trained and classifying in real-time. Support Vector Machines and Naïve Bayes spend more time on training but can classify new sentences quickly. K-Nearest Neighbor has no training time but spends a large amount of time making predictions [34]. We determined the run time of a classifier by first computing the total run time of 10-fold cross validation, then calculated the average run time per fold by dividing the total run time by the number of folds. These tests were run using Matlab R2015a and all of the classifier implementations are from the Statistics and Machine Learning Toolbox [36]. We ran the classification models an an ASUS Q550L laptop running Windows 10 with 8 GB of RAM installed.
To determine how generalizable the model is, we used 10-fold cross validation. To combine the results from the K folds, we took the harmonic mean [37] of the precision and recall across the 10 folds. To reduce the sampling bias, we then repeated 10-fold cross validation 10 times. Each time we split the data into 10 folds, we built 10 classifiers with each classifier missing 1 fold, i.e. 10% of the data. As a result, we built 100 classifiers total. We report the average performance of the 10 repeats of 10-fold cross validations.
To measure performance of our model on ODC2, we applied all of the trained models from our best feature design and best classifier to ODC2. We randomly selected 250 sentences where at least 50% of the models predicted the sentence was clinical, and 250 sentences with at least 50% of the models predicted the sentence was not clinical. These sentences were randomly ordered, then annotated without the annotator knowing the predictions of the models. Using the manual annotations, we computed the precision, recall, and f-measure of each model, and averaged them using the same method applied to WDC.
4. Results and Discussion
In this section, we analyze the performance of our feature designs, compare the performance of feature reduction techniques, and measure how the performance of our best performing model persist to a different data source.
4.1. Comparison of Feature Designs
In Section 3.3, we introduced four feature designs, and stated U was our baseline. The list of feature designs is shown in Table 3. All feature designs showed high performance, shown in Table 5 and Figures 3 and 4. The F-Measure was above 0.75 for all feature designs and all classifiers. For the majority of the feature designs, precision was above 0.7, meaning that of all the sentences predicted as clinically-related, our models were correct at least 70% of the time.
Table 3.
Feature design abbreviations
| Abbreviation | Feature Sets |
|---|---|
|
| |
| U | Unigrams only |
| U+BT | Unigrams, Bigrams, and Trigrams |
| U+MM | Unigrams and MetaMap Semantic Types |
| U+BT+MM | Unigrams, Bigrams, Trigrams, and MetaMap Semantic Types |
| U+MCSP | Unigram and MetaMap Categories, Parts of Speech, and Polarity |
| U+BT+MCSP | Unigrams, Bigrams, Trigrams and MetaMap Categories, Parts of Speech, and Polarity |
Table 5.
Performance of SVM on each feature design. SVM showed similar performance across all feature designs in terms of recall, precision, and F-measure.
| Feature Set | Precision | Recall | F-Measure |
|---|---|---|---|
|
| |||
| U | 0.794 | 0.782 | 0.788 |
| U+BT | 0.798 | 0.772 | 0.785 |
| U+MM | 0.798 | 0.781 | 0.789 |
| U+BT+MM | 0.799 | 0.777 | 0.788 |
| U+MCSP | 0.797 | 0.784 | 0.791 |
| U+BT+MCSP | 0.796 | 0.785 | 0.790 |
Figure 3.
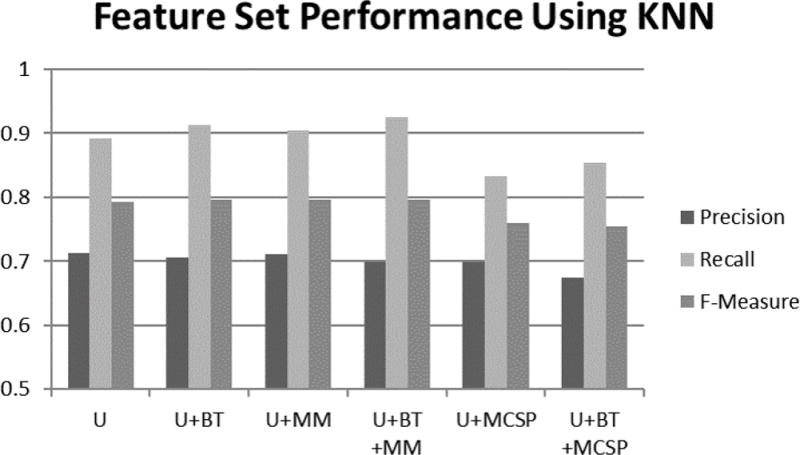
Performance of KNN for each feature design. KNN correctly predicted more clinical sentences as clinical, demonstrated by high recall. It predicted more non-clinical sentences as clinical which resulted in lower precision. Adding MM to the feature design improved recall and f-measure in the case of using KNN. The presence of MCSP features lowered precision, recall, and f-measure in the case of using KNN.
Figure 4.
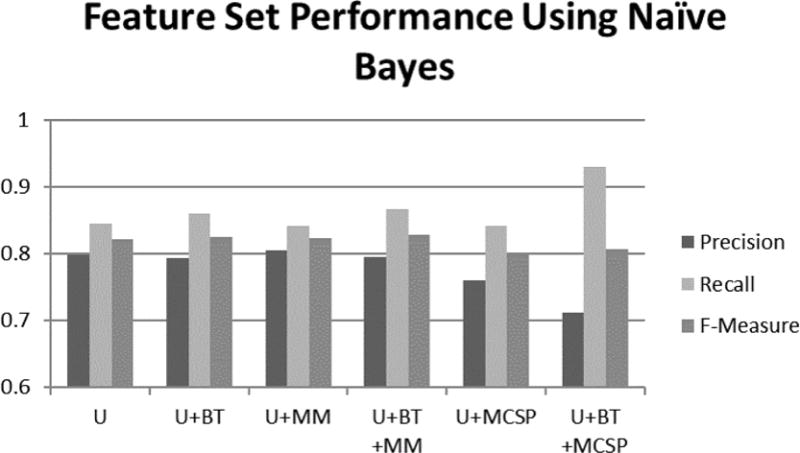
Performance of Naïve Bayes for each feature design. In the case of using Naïve Bayes classifier: BT and MM improved precision, recall, and f-measure; MCSP features lowered precision and improved recall; and the reduction in precision outweighed the improvement in recall, worsening f-measure.
Compared to our baseline, adding BT to the feature design improved results. This improvement was the highest for KNN, as shown in Figure 3. This improvement was consistent with prior work on text classification [28, 29]. For our first run of 10-fold cross validation using KNN, adding BT correctly predicted 69 additional clinical sentences at the cost of falsely labeling 38 non-clinical sentences as clinical. Overall precision increased while the F-Measure remained about the same.
Adding MM also improved performance, compared to our baseline, both before and after the addition of BT. This improvement was most visible for Naïve Bayes, as demonstrated in Figure 4. The confusion matrix using U, BT, and MM is in Table 4. By adding MM, 11 additional clinical sentences were correctly classified as clinical, and 10 additional nonclinical questions were correctly classified.
Table 4.
Confusion matrix for U, BT, and MM for Naïve Bayes
| Ground Truth | Predicted | |
|---|---|---|
| Clinical | Not Clinical | |
|
| ||
| Clinical | 2549 | 404 |
| Not Clinical | 642 | 1371 |
Using MCSP resulted in decreased performance, compared to our baseline, for Naïve Bayes and KNN. With MM, classifiers were presented with more fine grain types. Categories were too broad. For instance, the Chemicals & Drugs category contains several semantic types that relate to food. Thus, their presence did not indicate the sentence was clinically related.
All three classifiers showed a precision-recall trade off in performance. KNN and Naïve Bayes favored higher recall at the cost of lower precision. SVM had higher precision than recall.
We discovered two findings when we directly comparing the performance of the three classifiers, as shown in Figure 5. First, SVM had similar performances across all of the feature designs. Second, Naïve Bayes outperformed other classifiers on all feature designs. The best feature design is with U, BT, and MM.
Figure 5.
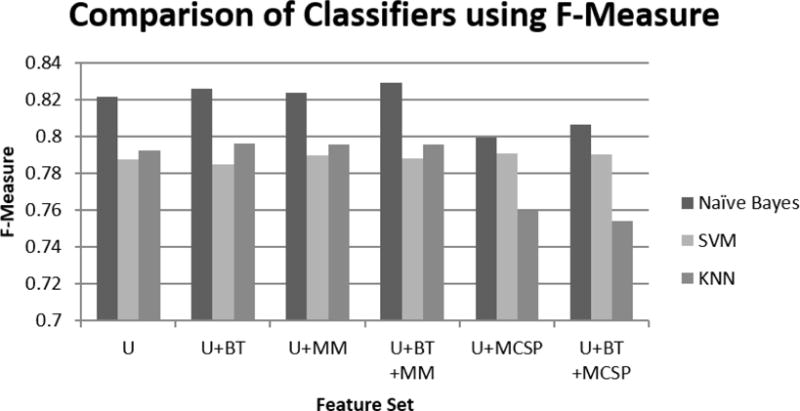
Comparison of each classifier, according to their F-Measure, for each feature design. Naïve Bayes had the best performance in comparison to SVM and KNN. KNN performed better than SVM on all feature designs except when adding MCSP.
4.2. Results on feature design and reduction
To further improve the performance, we applied three feature reduction techniques; Number Replacement, Mutual Information, and χ2 value. We found Number Replacement improved performance the most, shown in Tables 6, 7, and 8.
Table 6.
Comparison of performance of Support Vector Machine on whether or not to use Number Replacement. Using NR performed better than not using NR.
| Feature Design | Without NR | With NR |
|---|---|---|
|
| ||
| U | 0.788 | 0.790 |
| U+BT | 0.785 | 0.787 |
| U+MM | 0.789 | 0.791 |
| U+BT+MM | 0.788 | 0.792 |
| U+MCSP | 0.791 | 0.792 |
| U+BT+MCSP | 0.790 | 0.792 |
Table 7.
Using Number Replacement did not improve f-measure for KNN classifier, except for U+MCSP feature set. NR led to similar f-measure.
| Feature Design | Without NR | With NR |
|---|---|---|
|
| ||
| U | 0.792 | 0.791 |
| U+BT | 0.796 | 0.795 |
| U+MM | 0.796 | 0.794 |
| U+BT+MM | 0.796 | 0.795 |
| U+MCSP | 0.760 | 0.767 |
| U+BT+MCSP | 0.754 | 0.751 |
Table 8.
Number Replacement had no affect on Naive Bayes. For U+MCSP, F-measure improved marginally.
| Feature Design | Without NR | With NR |
|---|---|---|
|
| ||
| U | 0.821 | 0.822 |
| U+BT | 0.826 | 0.825 |
| U+MM | 0.824 | 0.823 |
| U+BT+MM | 0.829 | 0.828 |
| U+MCSP | 0.799 | 0.803 |
| U+BT+MCSP | 0.807 | 0.808 |
Number Replacement
Applying Number Replacement (NR) increased performance with U across all three classifiers, but had mixed effects on BT. The performance of KNN and Naïve Bayes, in Tables 7 and 8 respectively decreased. For SVM, NR improved performance across all feature designs, as shown in Table 6.
NR increasing the performance for U can attribute to the reduced number of features. When we applied NR to U, 296 number features were replaced by a single token. By replacing those features with a common token, we kept the relevant information without fitting the model to each unique number in the training design. We found 862 sentences out of 1082 containing at least one number were clinical sentences, showing that the knowledge of whether a sentence contains a number is a good predictor that the sentence is clinically related.
Applying NR of numbers to BT resulted in an increase in the number of features. NR produced 322 additional patterns. Phrases unique to only one sentence followed a pattern that existed in other sentences, so they included with NR. These additional patterns are the side effects of two choices, keeping stopwords in BT and replacing special characters with spaces. For example, a post described the nutrition information for two foods. One food had 0.552 fiber and the other had 1.25 fiber. The periods, as non-alphanumeric characters, were replaced with spaces rather than being removed because we found when splitting posts into sentences some users did not add spaces after commas or periods. If commas and periods were removed instead of replaced, two words would be combined into a single word instead of separated.
Mutual Information as feature reduction
We investigated selecting features based on ranking features by their mutual information. Features were selected from the set of features of our best performing feature design. We found that Naïve Bayes and KNN performed well, achieving 0.72 F-measure, with as few as 100 features. SVM performed worse than the other classifiers when using less than 2000 features. When using the top 2000 or more features, SVM has a similar performance as KNN. However, the performance of all three classifiers was worse than their performance when using all of the features.
χ2 Scores as feature reduction
Naïve Bayes consistently had a higher performance than KNN and SVM when at least 800 features were used ranked by the χ2 Scores, as shown in Figure 7. Below 800 features, the three classifiers had a similar performance. With at least the top 6000 features, Naïve Bayes had the same performance as the best feature design for KNN or SVM, at 0.8 F-Measure. For the top 700 to 1000 features, no significant information was added. For SVM and Naïve Bayes, there was not any performance improvement. For KNN, these features lead to clinical sentences be more similar to non-clinical sentences than to other clinical sentences, so the performance of KNN worsened. The performance of all three classifiers continued to improve from the top 1000 features as more features were added.
Figure 7.
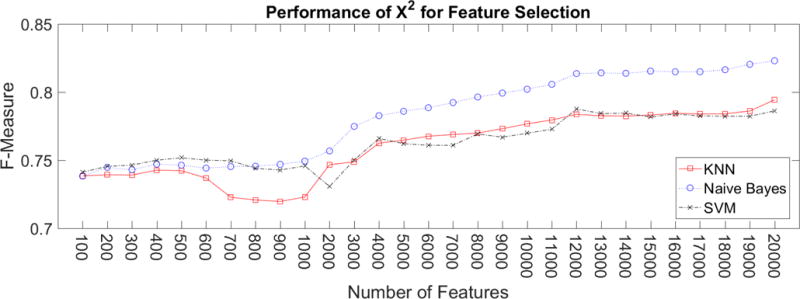
Performance using χ2 to select a reduced set of features. Naive Bayes consistently had higher f-measure than KNN and SVM.
Mutual Information and χ2 had a similar performance, which is evident in Figure 8. When using Naïve Bayes, Mutual Information produced a slightly higher performance. For KNN, χ2 improved performance when the number of features was between 4000 and 13,000.
Figure 8.
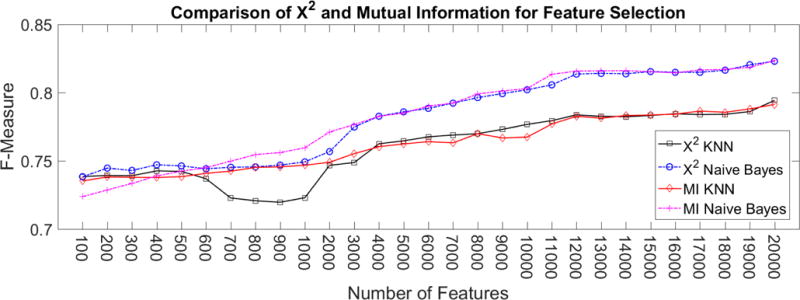
Comparison of performance between using Mutual Information and χ2 scoring techniques to select features. When the number of features is small, e.g. 1000, Mutual Information yields higher f-measure. As the number of features increase, χ2 and Mutual Information results in similar f-measure.
For both χ2 and Mutual Information, the performance was lower when using a subset of the features rather than all of the features. Although fewer features leads to worse performance, fewer features can reduce the run-time of training a classifier and prediction. Run-time is more important for some applications, e.g. classifying social media in real-time.
4.3. Run-time Performance
We investigated the speed of training and testing each classifier. We found Naïve Bayes was the fastest, across all feature designs. On feature designs that only included U, Naïve Bayes could train a classifier and make a prediction in under 1 second. When BT were included, it took approximately 5 seconds to train the classifier. KNN and SVM were significantly slower. SVM took 30 seconds to train and test a classifier using only U, and over 3 minutes when using U and BT. KNN was slightly faster than SVM, taking 18 seconds on U, and approximately 3 minutes on U and BT. Adding the MM or MCSP features did not change the run-time, while adding BT greatly increased the run-time. The run-times of each classifier are in Figure 9.
Figure 9.
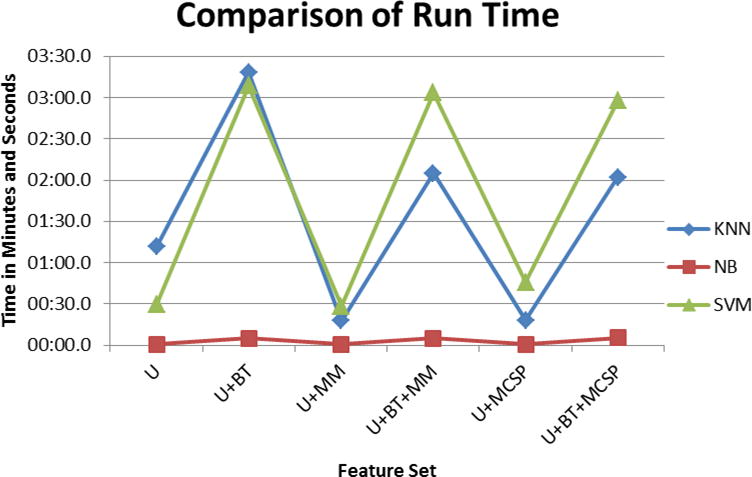
Comparison of the run-time of each classifier to train a model from the training set and apply it to a test set. Naive Bayes was the fastest, taking less than 15 seconds to learn a model and apply it to the test set. SVM was the slowest when MM or MCSP features were present. KNN was slowest on U and U+BT. More features lead to model learning and applying take more time.
4.4. Summary on classifiers
In this study, we compared the performance of three classifiers on a variety of feature designs. We evaluated the models using four measures, precision, recall, F-measure, and run-time. Naïve Bayes had the best performance, in F-measure and in run-time. Naïve Bayes can build an accurate model, with a minimum precision, recall, and F-measure of 0.7, 0.8, and 0.79 respectively, for every feature designs. Building and applying the model to new sentences takes seconds.
SVM had similar performance across all feature sets, with over 0.78 F-measure. It performed worse than Naïve Bayes and KNN on all feature designs except when MCSP features were included in the model. SVM was the only classifier that had similar F-measure when MCSP was present compared to when it was absent. SVM was the slowest classifier. When BT were included in the feature set, it took around 3 minutes to build an SVM classifier and apply it to the validation set.
KNN performed slightly better than SVM on all measures when MCSP was not part of the feature design. F-measure was over 0.78 when MCSP features were absent and over 0.74 when MCSP features were present. KNN performed marginally better than SVM, but significantly worse than Naïve Bayes (For feature design U+BT+MM, SVM, KNN, and Naïve Bayes had F-measures 0.788, 0.796, and 0.828, respectively). The run-time of KNN to predict sentences in the validation set was approximately two thirds of the run-time needed for SVM to train a model and make predictions. Although all three classifiers had high performance, Naïve Bayes was the best because it had the highest F-measure and lowest run-time.
4.5. Analysis of Top Features
In this research, we identified the most predictive features for identifying clinical sentences using three measures; χ2, Mutual Information, and Naïve Bayes, which identifies the posterior probability of a clinical sentence given the presence of a feature. The top 10 features are listed in Table 9. Two MM features were among the top 10 features in all three feature ranking schemes–Pharmacologic Substances and Disease or Syndrome. Based on our definition that clinical sentences discuss treatments or their shared disease, we expect these UMLS concepts to be prevalent in clinical sentences. Additionally, ”blood” and ”sugar” terms are ranked high. This result makes sense because managing one’s blood sugar is a key self-management activity in diabetes.
Table 9.
Top 10 features based on χ2 and Mutual Information ranking, and by Naïve Bayes posterior probability. Parentheses denote which feature design the feature belonged. “Sub” is an abbreviation for Substance. “Proc” is an abbreviation for Procedure. “Syn” is an abbreviation of Syndrome
| χ2 | Mutual Information | Naïve Bayes |
|---|---|---|
|
| ||
| weekend (U) | Pharmacologic Subs. (MM) | IsNumber (NR) |
| post (U) | Finding (MM) | Finding (MM) |
| blood (U) | Laboratory Proc. (MM) | Pharmacologic Subs. (MM) |
| diabet (U) | Disease or Syn. (MM) | diabet (U) |
| IsNumber (NR) | Therapeutic/Preventive Proc. (MM) | Disease or Syn. (MM) |
| sugar (U) | blood (U) | incid (U) |
| Pharmacologic Subs. (MM) | Diagnostic Proc. (MM) | therapi (U) |
| blood sugar (BT) | Sugar (U) | sugarmi (U) |
| Disease or Syn. (MM) | Weekend (U) | wheat (U) |
| webmd (U) | diabet(U) | blood (U) |
The different feature order of these algorithms gave additional insights as well. Mutual Information ranked more MM in the top 10 features. Both χ2 and Mutual Information ranked “weekend” as an indicator of clinical sentences, however Naïve Bayes indicated that sentences are more likely not clinical given the presence of the term weekend.
4.6. Applying models to another community
To measure how much our results apply to other communities than the community we used for building the model, we applied our best performing classifier to the sentences from ODC2. We applied Naïve Bayes using the feature design of U, BT, and MM with NR. We evaluated this classifier on its performance on randomly selected 500 sentences classified as either clinical or non-clinical. Our annotator found 201 of those sentences as clinical. The remainder were non-clinical. Our classifier results were biased towards non-clinical sentences. Precision, Recall, and F-Measure of the clinical class were 0.57, 0.71, and 0.63 respectively. The confusion matrix is in Table 11. For the application of suggesting posts to a moderator, such high recall score is beneficial because the moderator would be less likely to miss important clinical sentences.
Table 11.
Confusion for Naïve Bayes on another online diabetes community
| Ground Truth | Predicted | |
|---|---|---|
| Clinical | Not Clinical | |
|
| ||
| Clinical | 143 | 58 |
| Not Clinical | 107 | 192 |
We then performed the error analysis to understand the classifier errors. We randomly selected a set of 20 false positives, i.e. non-clinical sentences predicted as clinical, and 20 false negatives. We found two themes from analyzing the false negatives. First, 9 of the false negatives (45%) were about diet choices, including the example shown in Table 10. Because diabetes is a diet-related illness, it may be challenging for a classifier to identify which sentences about food are clinically relevant, e.g. diet management, and which are not, e.g. recipes. As the next step, we can build a consumer oriented food vocabularies to inform the classifier. Second, the error occurred in sentences that referenced healthcare in a broad way, e.g. providing general advice to see doctor when in doubt and questions during appointments. Although these sentences referred to healthcare, e.g. doctor, they did not discuss the clinical aspects of the illness directly. The context, e.g. predicted labels of sentences before and after the current sentence, could be used to improve the classifier performance.
Table 10.
Examples of a false positive and a false negative from ODC2
| Error Type | Sentence |
|---|---|
|
| |
| False Positive | waited two hrs |
| False Negative | so I’m looking for foods that can be served to all yet be tasty and diabetic friendly |
Analysis of the false positives, non-clinical sentences falsely predicted as clinical showed 3 themes in why the error occurred. First, 30% of the false positives were short sentences, having less than 5 words. These sentences contained several stopwords, which provide no information about which class this sentence belongs to. When the Naive Bayes classifier does not have sufficient information about the sentence, it will predict the sentence to belong to the larger class, i.e. the class that had more examples in the training set, which was the clinical class. Increasing the number of non-clinical sentences in the training set would lead to short sentences being predicted as non-clinical. Second, the number terms, e.g. two, or numerals, e.g. 2, occurred more frequently in clinical sentences than non-clinical sentences. 25% of the false positives had number terms or numerals, like the one shown in Table 10. Including more non-clinical sentences that contain numerals or number terms in the training set would reduce these false positives. Third, some sentences were ambiguous in whether they could be clinical or not, depending on the context. These sentences provided general advice which mentioned controlling their pain or being kind to one’s body. Such ambiguity has been a challenge for classifying when we need clinical experts’ help in patients’ online conversations [6]. The classifier identified symptom words, e.g. pain, as clinical, but the other terms did not influence enough to sway the classifier to predict the sentence as not clinical. This theme encompassed 20% of the false positives. To improve performance, having a stricter codebook and providing a classifier with more examples of borderline sentences, e.g. non-clinical sentences that mention body or pain, would help the classifier better differentiate between clinical and non-clinical sentences.
5. Discussion and Future Work
5.1. Strengths and Limitations
From applying our best performing model to WebMD and ODC2, we observed that this model has several strengths and limitations. Among its strengths, Naïve Bayes was fast to train and apply to unseen sentences, and its F-measure was high. The reason for high F-measure was due to high recall, i.e. our model was able to identify most of the clinically-related sentences. However, our approach had lower precision; 20% of sentences predicted to be clinically related were not clinically related. A second limitation is related to the MetaMap API. Feature designs which used MetaMap (MM and MCSP) were the slowest to generate. For smaller forums, MetaMap can be applied without significant delays in prediction time. For large forums which grow rapidly, e.g. Facebook or Twitter, it may not be feasible to apply MetaMap to all posts.
5.2. Productionization
Forum owners and administrators can benefit from integrating the algorithm proposed in this research. We can develop a dashboard for forum owners to help them to monitor which posts are most clinically-related, ranked by its predicted accuracy, so that the post can be redirected to users with relevant expertise or moderators if there are any. A post’s rank will be based on the probability output from the classifier. Posts with multiple clinically-related sentences or few sentences that have high probability of being clinically-related will receive higher rank.
From this dashboard, forum owners will be able to respond to the posts and provide feedback to the system about its prediction. The dashboard can use this feedback to improve the classifier, which will allow the classifier to learn over time.
6. Conclusion
In this research, we demonstrated that by using data mining, we are able to predict which sentences discuss clinical information with high F-measure. We found that the models learned from WDC have high recall when applied to other health-related Online Social Networks but low precision. Short sentences, discussions of diet, and discussions of general healthcare pose challenges to predicting whether sentences are clinical or not clinical. Adding more annotated sentences and adding context from earlier sentences may improve performance.
Our methods can also inform other applications, such as: detecting disease outbreaks. General online social networks, e.g. Twitter, have gained attention recently for detecting disease outbreaks [38, 39]. One challenge is identifying the relevant keywords. Using the method proposed in this research, researchers can identify clinically-related tweets and use those tweets to identify the keywords relevant to the specific disease.
In future work, we will apply our best classifier to a stream of online forum posts, to annotate the unlabeled posts. For this task, our classifier will need two performance strengths; make fast accurate predictions and the model can be stored in memory. SVM made fast predictions, but they were the least accurate of all 3 classifiers. KNN made accurate predictions, but they were slow. Naïve Bayes made fast and accurate predictions. Both SVM and Naïve Bayes have small models that only depend on the number of features, but KNN has a large model because it stores all of the training data. Naïve Bayes is the best classifier for this task, in terms of F-measure, run time, and model size.
We will also explore predicting which sentences can be addressed with an automatic response versus human’s responses, which would depend on the complexity of the clinical problem patients were attempting to address. A new kind of codebook should be developed for what needs human help and what is sufficient in using automated responses. In addition to the binary classification task we showed in this work, determining the severity and immediacy of the information presented by the patient would further sophisticate this line of research. We would prioritize posts by severity of the attention needed for the moderators, such as a triaging process [5]. Posts that suggest the patient will adopt harmful behavior should be ranked the highest and addressed by a moderator before they address posts with lower severity.
Our study developed, compared, and evaluated classification models that will help to identify when patients discuss clinically related topic for the moderators to efficiently respond. We also tested the feasibility of applying our model to other communities. We suggested solutions to improve performance for applying and modifying this model to classify clinical topics of other communities. Our work will contribute to improving the quality of information delivered to patients in today’s information environment bombarded with internet-based information.
Figure 6.
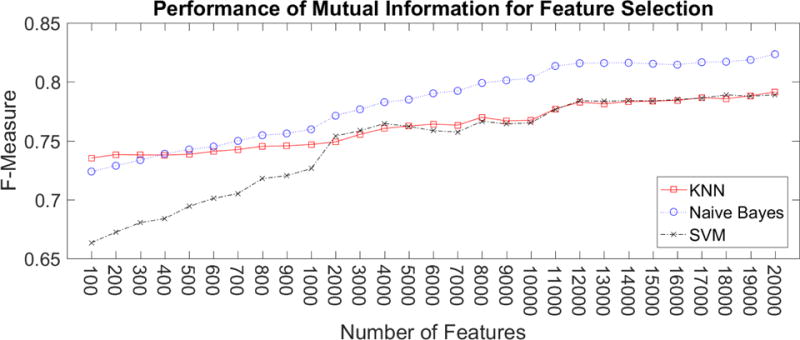
Performance using Mutual Information to select a reduced set of features. Naïve Bayes consistently had higher f-measure than KNN and SVM.
Highlights.
Automatically identify forum posts that need clinical resources
Analyze feature designs and feature selection techniques
Evaluate the performance of classifier algorithms
Analyze performance of features and classifier on another online health community
Acknowledgments
This work has been partially funded by NIH grant 1 K01 LM011980-01. We would like to thank Alliance Health and John Crowley for their collaboration and support.
Appendix A. MetaMap Types
Table A.12 presents the UMLS semantic types used in this paper. Each proposed type was added as a feature in the Metamap Semantic Types (MM) feature set. The frequency of MetaMap categories, measuring by the sum of the frequency of each of their semantic types were used in the MetaMap Categories feature set (MCSP), and were originally proposed by Denecke and Nejdl [30].
Table A.12.
UMLS Semantic Concepts selected as features. Italicized concepts appear in both sets of semantic types.
| Proposed MetaMap Types | Denecke and Nejdl MetaMap Types |
|---|---|
| Laboratory or Test Result; Mental Process; Genetic Function; Physiologic Function; Organ or Tissue Function; Molecular Function; Cell Function; Phenomenon or Process; Human-caused Phenomenon or Process; Environmental Effect of Humans; Natural Phenomenon or Process; Biologic Function; Organism Function; Pharmacologic Substance; Finding; Sign or Symptom; Cell or Molecular Disfunction; Disease or Syndrome; Mental or Behavioral Dysfunction; Neoplastic Process; Pathologic Function; Experimental Model of Disease; Diagnostic Procedure; Health Care Activity; Laboratory Procedure; Therapeutic or Preventive Procedure; Injury or Poison; |
Chemicals & Drugs: Amino Acid, Peptide, or Protein; Antibiotic; Biologically Active Substance; Biomedical or Dental Material; Carbohydrate; Chemical, Chemical Viewed Functionally; Chemical Viewed Structurally; Clinical Drug; Eicosanoid; Element, Ion, or Isotope; Enzyme; Hazardous or Poisonous Substance; Hormone; Immunologic Factor; Indicator, Reagent, or Diagnostic Aid; Inorganic Chemical; Lipid; Neuroreactive Substance or Biogenic Amine; Nucleic Acid, Nucleoside, or Nucleotide; Organic Chemical; Organophosphorus Compound; Pharmacologic Substance; Receptor; Steroid; Vitamin Disorders: Acquired Abnormality; Anatomical Abnormality; Cell or Molecular Dysfunction; Congenital Abnormality; Disease or Syndrome; Experimental Model of Disease; Finding; Injury or Poisoning; Mental or Behavioral Dysfunction; Neoplastic Process; Pathologic Function; Sign or Symptom Procedures: Diagnostic Procedure; Educational Activity; Health Care Activity; Laboratory Procedure; Molecular Biology Research Technique; Research Activity; Therapeutic or Preventive Procedure |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fox S, Purcell K. Chronic Disease and the Internet. 2010 http://www.pewinternet.org/2010/03/24/chronic-disease-and-the-internet/ accessed May 31, 2017.
- 2.Huh J. Clinical questions in online health communities: The case of ”see your doctor” threads; Proceedings of the 18th ACM Conference on Computer Supported Cooperative Work & Social Computing, CSCW ’15, 2015; pp. 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huh J, McDonald DW, Hartzler A, Pratt W. Patient moderator interaction in online health communities; AMIA Annual Symposium Proceedings, AMIA 2013; 2013. pp. 627–36. [PMC free article] [PubMed] [Google Scholar]
- 4.Kraut RE, Resnick P, Kiesler S, Burke M, Chen Y, Kittur N, Konstan J, Ren Y, Riedl J. Building Successful Online Communities: Evidence-Based Social Design. The MIT Press; 2012. [Google Scholar]
- 5.Huh J, Pratt W. Weaving clinical expertise in online health communities; Proceedings of the 32Nd Annual ACM Conference on Human Factors in Computing Systems, CHI ’14; New York, NY, USA: ACM; 2014. pp. 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huh J, Yetisgen-Yildiz M, Pratt W. Text classification for assisting moderators in online health communities. Journal of Biomedical Informatics. 67(6) doi: 10.1016/j.jbi.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merolli M, Gray K, Martn-Snchez F. Health outcomes and related effects of using social media in chronic disease management: A literature review and analysis of affordances. Journal of Biomedical Informatics. 46 doi: 10.1016/j.jbi.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Vlahovic TA, Wang YC, Kraut RE, Levine JM. Support matching and satisfaction in an online breast cancer support community; Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, CHI ’14; 2014. pp. 1625–1634. [Google Scholar]
- 9.De Choudhury M, Morris MR, White RW. Seeking and sharing health information online: Comparing search engines and social media; Proceedings of the 32Nd Annual ACM Conference on Human Factors in Computing Systems, CHI ’14; ACM; 2014. pp. 1365–1376. [DOI] [Google Scholar]
- 10.Bui N, Yen J, Honavar V. Social Computing, Behavioral-Cultural Modeling, and Prediction. Vol. 9021. Springer International Publishing; 2015. Temporal causality of social support in an online community for cancer survivors. [Google Scholar]
- 11.Hartzler AL, McDonald DW, Park A, Huh J, Weaver C, Pratt W. Evaluating health interest profiles extracted from patient-generated data; AMIA Annual Symposium Proceedings, AMIA 2014; 2014. p. 626635. [PMC free article] [PubMed] [Google Scholar]
- 12.Park A, Hartzler LA, Huh J, McDonald WD, Pratt W. Automatically detecting failures in natural language processing tools for online community text. J Med Internet Res. 17(8) doi: 10.2196/jmir.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huh J, Patel R, Pratt W. Tackling dilemmas in supporting ’the whole person’ in online patient communities; Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, CHI ’12; 2012. pp. 923–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon BC, Kim SH, Lee S, Choo J, Huh J, Yi JS. Visohc: Designing visual analytics for online health communities. IEEE Trans Vis Comput Graph. 22 doi: 10.1109/TVCG.2015.2467555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McRoy S, Jones S, Kurmally A. Toward automated classification of consumers cancer-related questions with a new taxonomy of expected answer types. Health Informatics Journal. 2015:1460458215571643. doi: 10.1177/1460458215571643. [DOI] [PubMed] [Google Scholar]
- 16.Abdaoui A, Azé J, Bringay S, Grabar N, Poncelet P. Predicting medical roles in online health fora; The 2nd International Conference on Statistical Language and Speech Processing (SLSP 2014); 2014. pp. 247–258. [Google Scholar]
- 17.Huh J, Yetisgen-Yildiz M, Hartzler A, McDonald DW, Park A, Pratt W. Text classification to weave medical advice with patient experiences; AMIA 2012, American Medical Informatics Association Annual Symposium; Chicago, Illinois, USA. November 3–7, 2012; 2012. [Google Scholar]
- 18.Chomutare T. Text classification to automatically identify online patients vulnerable to depression; Pervasive Computing Paradigms for Mental Health, 4th International Symposium, MindCare 2014; Tokyo, Japan. May 8–9, 2014; 2014. pp. 125–130. Revised Selected Papers. [Google Scholar]
- 19.Yang M, Kiang M, Shang W. Filtering big data from social media - building an early warning system for adverse drug reactions. J of Biomedical Informatics. 54(C) doi: 10.1016/j.jbi.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Tuarob S, Tucker CS, Salathe M, Ram N. Discovering health-related knowledge in social media using ensembles of heterogeneous features; Proceedings of the 22Nd ACM International Conference on Conference on Information & Knowledge Management, CIKM ’13; 2013. pp. 1685–1690. [Google Scholar]
- 21.Akbari M, Hu X, Liqiang N, Chua TS. From tweets to wellness: Wellness event detection from twitter streams; Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, AAAI’16; AAAI Press; 2016. pp. 87–93. [Google Scholar]
- 22.L. WebMD. About WebMD. 2005 http://www.webmd.com/about-webmd-policies/default.htm?ss=ftr. accessed January 22, 2016.
- 23.Loper E, Bird S. Nltk: The natural language toolkit; Proceedings of the ACL-02 Workshop on Effective Tools and Methodologies for Teaching Natural Language Processing and Computational Linguistics - Volume 1, ETMTNLP ’02; Stroudsburg, PA, USA. Association for Computational Linguistics; 2002. pp. 63–70. [Google Scholar]
- 24.Jurafsky D, Martin JH. Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linquistics, and Speech Recognition. 2nd. Pearson Education, Inc.; Upper Saddle River, New Jersey: 2009. [Google Scholar]
- 25.Cao Y, Liu F, Simpson P, Antieau L, Bennett A, Cimino JJ, Ely J, Yu H. Askhermes: An online question answering system for complex clinical questions. Journal of biomedical informatics. 44(2) doi: 10.1016/j.jbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tausczik YR, Pennebaker JW. The psychological meaning of words: Liwc and computerized text analysis methods. Journal of Language and Social Psychology. 2010;29(1):24–54. doi: 10.1177/0261927X09351676. [DOI] [Google Scholar]
- 27.Tan CM, Wang YF, Lee CD. The use of bigrams to enhance text categorization. Information Processing & Management. 2002;38(4):529–546. doi: http://dx.doi.org/10.1016/S0306-4573(01)00045-0. [Google Scholar]
- 28.Zhang D, Lee WS. Question classification using support vector machines, in. Proceedings of the 26th Annual International ACM SIGIR Conference on Research and Development in Informaion Retrieval, SIGIR ’03, ACM. 2003:26–32. doi: 10.1145/860435.860443. [DOI] [Google Scholar]
- 29.Saleh MR, Martn-Valdivia M, Montejo-Rez A, Urea-Lpez L. Experiments with {SVM} to classify opinions in different domains. Expert Systems with Applications. 2011;38(12):14799–14804. doi: http://dx.doi.org/10.1016/j.eswa.2011.05.070. [Google Scholar]
- 30.Denecke K, Nejdl W. How valuable is medical social media data? content analysis of the medical web. Inf Sci. 2009;179(12):1870–1880. [Google Scholar]
- 31.Porter M. The English (Porter2) stemming algorithm. 2006 http://snowball.tartarus.org/algorithms/english/stemmer.html. accessed January 22, 2016.
- 32.Cover T, Hart P. Nearest neighbor pattern classification. Information Theory, IEEE Transactions on. 13(1) [Google Scholar]
- 33.Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Proceedings of the Fifth Annual Workshop on Computational Learning Theory, COLT ’92. 1992:144–152. [Google Scholar]
- 34.Tan PN, Steinbach M, Kumar V. Introduction to Data Mining. First. Addison-Wesley Longman Publishing Co., Inc.; Boston, MA, USA: 2005. [Google Scholar]
- 35.Stark DG, Hart PE, Duda RO. Pattern Classification. Second. Wiley-Interscience; 2000. [Google Scholar]
- 36.MathWorks. Matlab Statistics and Machine Learning Toolbox (Version 2015b) [Computer software] 2015 http://www.mathworks.com/products/statistics/
- 37.Kenny JF, Keeping ES. Mathematics of statistics. Third. Van Nostrand company; New York, NY, USA: 1954. [Google Scholar]
- 38.Culotta A. Towards detecting influenza epidemics by analyzing twitter messages, in. Proceedings of the First Workshop on Social Media Analytics, SOMA ’10, ACM. 2010:115–122. doi: 10.1145/1964858.1964874. [DOI] [Google Scholar]
- 39.Tegtmeyer R, Potts L, Hart-Davidson W. Tracing and responding to foodborne illness. Proceedings of the 30th ACM International Conference on Design of Communication, SIGDOC ’12, ACM. 2012:369–370. doi: 10.1145/2379057.2379131. [DOI] [Google Scholar]


