Abstract
Fermented foods are known for their potential as main source of probiotics. The present study aimed at investigating the probiotic properties of bacteria isolated from fermented mango pickle. Non-hemolytic fermenting microbiota isolated from mango pickle was screened in vitro for their basic probiotic properties such as acid tolerance, bile salt, lysozyme and salt tolerance. They were also tested for their beneficial characters like cholesterol removal, bacterial adhesion to hydrocarbons, auto-aggregation, antimicrobial activity, β-galactosidase activity, exopolysaccharide production and adhesion to HT-29 cell line. Outputs of these parameters were subjected to principal component analysis (PCA) and these results were interpreted to select prospective bacterial isolates that can be used as potential probiotics. Out of eight isolates, PUFSTP35 (Bacillus licheniformis), PUFSTP38 (Bacillus amyloliquefaciens) and PUFSTP39 (Bacillus subtilis) showed similar trend to Weissella cibaria (MTCC 9814) that was used as a reference strain for profiling probiotic properties. B. licheniformis PUFSTP35 from fermented mango pickle appear to be the most potential candidate for use as a beneficial probiotic.
Keywords: Fermented food, Mango pickle, Bacillus sp., Probiotic properties, Health benefits
Introduction
Probiotics are ‘non-pathogenic live microorganisms’ that when administered in adequate amounts confer health benefit on the host (FAO/WHO 2006). Members of bifidobacteria and lactobacilli are most commonly used as probiotics (Sorokulova 2013). Probiotics may prevent the penetration of pathogens in human gut by increasing the production of mucin, reducing the gut permeability and releasing antimicrobial compounds or modulating the immune system. Probiotics are known to have additional beneficial effects like helping in the management of lactose intolerance, lowering serum cholesterol level, improving the uptake of nutrients and reducing the need for antibiotics (Guo et al. 2010). Routine use of probiotic is considered to be helpful to overcome various clinical conditions such as infantile diarrhoea, Helicobacter pylori infections, relapsing Clostridium difficile colitis, inflammatory bowel disease and uro-genital infections (Reid et al. 2003). Many Bacillus strains have been approved by regulatory agencies as probiotic strains for human consumption. From many centuries, alkaline-fermented foods are prepared by using Bacillus sp. and consumed (Wang and Fung 1996). In Russia and Ukraine, B. subtilis 3 and B. licheniformis 31 (Biosporin) are used as a drug for prophylaxis and gastro-intestinal infections (Gracheva et al. 1996). Bacillus spp. is the dominant microflora of Asian fermented foods as reported earlier from time to time (Meerak et al. 2007).
Early studies reported that fermented food kinema was rich in Bacillus spp. like B. circulans, Bacillus sphaericus, Bacillus thuringiensis, Bacillus subtilis and Bacillus licheniformis (Tamang 2003). Jeyaram et al. (2008) isolated B. cereus, B. licheniformis and B. subtilis from Indian soybean fermented food hawaijar.
Different types of fermented foods are produced and consumed throughout the world for several centuries owing to rich nutrients, quality proteins and amino acids, desirable taste and flavour. Fermented foods act as substrates for production of enzymes, proteins and lipids to enhance the flavor, texture, aroma (Steinkraus 2002). In Southern part of India, people consume different types of fermented foods such as curd, buttermilk, butter and fermented vegetables in the form of pickles that are made from mango, lemon, gooseberries, ginger, garlic, etc.
In the west coastal districts of India, a unique pickle based on wild tender mangoes is prepared by placing layers of tender mangoes in between layers of Salt. After a period of few months, pickle is prepared by mixing ground spice into the salt water and storing in a closed ceramic container for six to twelve months before consumption. During this period the product ferments with the rich salt tolerant bacterial species. Microbiology of the above product is uncharacterized. Current work has been planed with the hypothesis that product harbours rare salt tolerant microorganism which could be potentially probiotic. Microbiota from the mature pickle was isolated and characterized for their potential probiotic characteristic.
Materials and methods
Sample collection
Mango pickle (n = 15) samples were collected from Mangalore, Karnataka, India, located at Latitude 12.91 N and Longitude 74.85 E, and stored at room temperature at dark condition until processed.
Bacterial strains
Reference strain, Weissella cibaria (MTCC 9814) and the following test strains; Escherichia coli (MTCC 728), Staphylococcus epidermidis (MTCC 435), Enterococcus faecalis (MTCC 439), Enterobacter aerogenes (MTCC 2822), Yersinia enterocolitica (MTCC 861), Klebsiella pneumonia (MTCC 618), Proteus mirabilis (MTCC 425), Listeria monocytogenes (MTCC 657) and Pseudomonas aeruginosa (MTCC 424) were used for the study. All the above strains were procured from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology, Chandigarh, India.
Isolation of bacteria from fermented pickle
Bacterial strains were isolated by spread plating of appropriately diluted pickle samples on MRS (de Mann Rogosa Sharpe) agar (Himedia), and incubated at 30 °C for 48 h. Colonies were selected on the basis of different morphologies in MRS agar plate and sub-cultured until to get pure colony. All the pure colonies were stored in 4 °C as slant cultures and in − 80 °C as glycerol (70%) stock for further use.
Hemolytic activity
Hemolytic activity of the bacterial isolates was carried by streaking them on blood agar base supplemented with 5% sheep blood and incubated at 30 °C for 48 h. Only the non-hemolytic isolates were selected for further analysis of probiotic properties.
Probiotic characterization
Acid and bile tolerance
The experiment for tolerance of isolates (109 CFU/ml) to different pH conditions such as pH 2.0, 3.0 and 7.0 (control) was performed following the method described by Yu et al. (2013).
The bile tolerance of test isolates (109 CFU/ml) was determined using 0.5 and 1% of ox-bile (w/v) as stated by Vinderola and Reinheimer (2003). Viability was determined by spread-plate count using MRS agar.
Salt tolerance assay
The salt stress response of the selected isolates (109 CFU/ml) were screened in MRS broth containing different concentrations of NaCl ranging from 0 to 10% (w/v) as described by Adnan and Tan (2007).
Lysozyme tolerance
Selected isolates (109 CFU/ml) were screened for lysozyme (Himedia) tolerance as described by Zago et al. (2011).
Cholesterol removal
Cholesterol removal ability was determined using 0.3% ox-bile in MRS broth supplemented with filter sterilized cholesterol to a final concentration of 10 mg/ml (MRS-oxbile-cholesterol). Overnight grown cultures (109 CFU/ml) were inoculated in MRS-ox bile-cholesterol and un-inoculated broth served as control. The estimation of cholesterol was carried spectrophotometrically at 550 nm by o-phthalaldehyde method (de-Valdez and de-Taranto 2001).
where A0 is the control (MRS-ox bile-cholesterol without test isolate) and A1 is the test (MRS-ox bile-cholesterol with test isolates).
Antibiotic susceptibility test
The antibiotic susceptibility testing was performed using Mueller–Hinton agar overlaid with 0.1 ml (109 CFU/ml) of selected isolates against commercially available antibiotic discs viz., Amikacin (30 µg), Chloramphenicol (30 µg), Gentamycin (30 µg), Kanamycin (30 µg), Spectinomycin (100 µg), Bacitracin (10 µg) and Vancomycin (30 µg) (Himedia, India) and incubated at 37 °C for 24 h.
Antimicrobial activity
Antimicrobial activity of the isolates 0.1 ml (109 CFU/ml) of against selected food pathogens was determined by agar well diffusion method reported by Mishra and Prasad (2005).
β-Galactosidase activity
The β-Galactosidase activities of the selected isolates were determined as described by Angmo et al. (2016).
Screening for exopolysaccharide (EPS) production
Screening for EPS production was performed as stated by Angmo et al. (2016).
Bacterial adhesion to hydrocarbons (BATH assay)
BATH assay of the selected isolates (109 CFU/ml) were performed as per Saraniya and Jeevaratnam (2015) using Eon plate reader (Biotek, India).
Auto—aggregation
Overnight grown cultures were pelleted by centrifugation at 8000 g for 10 min at 4 °C, washed thrice and re-suspended in 100 mM (PBS) pH 7.4 and OD adjusted to 1.0 A560 using Eon plate reader (Biotek, India). Auto-aggregation was performed according to the method described by Osmanagaoglu et al. (2010). The percentage of auto-aggregation was determined by using the formula,
where ODI and ODF are initial and final absorbance, respectively.
Adhesion to human adenocarcinoma cell line HT-29
Adenocarcinoma cell line HT-29 was obtained from NCCS (National Centre for Cell Sciences), Pune, India and maintained in Dulbecco’s modified Eagle’s medium (DMEM, Himedia), with 10% fetal bovine serum (Himedia), 1X antibiotic–antimycotic solution (Himedia). Adhesion studies on HT-29 cells were performed as described by Arokiyaraj et al. (2014). After incubation, the bacterial suspensions were removed carefully; stained with Giemsa stain for 20 min at room temperature followed by methanol fixation, and visualized under inverted microscope. The cell suspension was then serially diluted, plated on MRS agar and incubated for 24 h at 37 °C.
where AO and A1 are initial and final colony counts, respectively.
Identification of selected isolates by 16S rDNA sequencing
The selected isolates were subjected to molecular characterization by sequencing of 16S rRNA using 27F (5′- AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) primers. The amplified product of 16S rRNA was sequenced and edited with Bio Edit 7.2.5 software and consensus sequences were compared with GenBank nucleotide database using Basic Local Alignment Search Tool (BLAST), (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Gene sequences of 16S rRNA were used to construct the phylogenetic tree for determination of nearest bacterial species by UPGMA method using MEGA 7.0.21 software (Kumar et al. 2016).
Statistical analysis
All data were analyzed using statistical software IBM SPSS Software version 20.0. (SPSS Inc., Chicago, IL, USA). Additionally, XLSTAT software 2016 (Addinsoft, New York, United States) was used to conduct principal component analysis (PCA) wherein the results of both qualitative and quantitative assays were changed into three coded values (0, 1 and 2) and used as input data.
Results
Characterization of probiotic properties
Hemolytic activity of isolates
A total of 106 isolates were isolated from fermented mango pickles. All the isolates were streaked on blood agar plates for hemolytic activity. Among 106 isolates screened, 8 isolates were non-hemolytic and were selected for further probiotic evaluation.
Acid and bile tolerance
Exposure of test isolates to simulated in vitro gastric juice of pH 2, revealed viability count ranging from 7.37 to 8.24 log CFU/ml with a significant (p ≤ 0.05) variability (Table 1). All test isolates retained similar level of viability after exposure to pH 3 for 3 h, and had higher tolerance than the reference strain, W. cibaria 9814 (SS1) at pH 2. Bile salt showed a significant influence on the viability of test isolate (Table 1). The survival rate of all isolates remained unaffected at 0.5% bile salt. The above isolates exhibited comparatively better tolerance than the reference strain at 1% bile salt (Table 1).
Table 1.
Effect of low pH, and bile salt on the viability of test isolates during 3 h incubation
| Isolates | pH Tolerance assay | Bile tolerance assay | ||||
|---|---|---|---|---|---|---|
| Survival rate (CFU/ml) | Survival rate (CFU/ml) | |||||
| pH 7 (control) | pH 3 | pH 2 | Control | 0.5% | 1% | |
| PUFSTP35 | 8.35 ± 0.01a | 8.05 ± 0.02b | 8.03 ± 0.03b | 8.61 ± 0.04a | 7.56 ± 0.01ab | 6.56 ± 0.04b |
| PUFSTP38 | 8.37 ± 0.02a | 8.26 ± 0.06b | 8.07 ± 0.03c | 8.40 ± 0.02a | 7.41 ± 0.02a | 6.31 ± 0.01b |
| PUFSTP39 | 8.39 ± 0.04a | 8.37 ± 0.02a | 7.98 ± 0.01b | 8.48 ± 0.06a | 7.38 ± 0.01b | 7.39 ± 0.03b |
| PUFSTP44 | 8.43 ± 0.01a | 8.39 ± 0.01a | 8.09 ± 0.03b | 8.40 ± 0.01a | 7.24 ± 0.02b | 7.24 ± 0.02b |
| PUFSTP70 | 8.46 ± 0.04a | 8.44 ± 0.08a | 8.12 ± 0.02b | 8.48 ± 0.04a | 7.43 ± 0.01b | 7.44 ± 0.01b |
| PUFSTP71 | 8.42 ± 0.01a | 8.36 ± 0.01b | 8.24 ± 0.03c | 8.60 ± 0.02a | 7.42 ± 0.02b | 7.34 ± 0.08b |
| PUFSTP74 | 8.39 ± 0.02a | 8.40 ± 0.02a | 8.13 ± 0.02b | 8.51 ± 0.01a | 7.50 ± 0.02a | 7.34 ± 0.01b |
| PUFSTP81 | 8.54 ± 0.06a | 8.29 ± 0.01b | 7.97 ± 0.03c | 8.67 ± 0.08a | 7.43 ± 0.01b | 7.36 ± 0.02c |
| SS1 | 8.63 ± 0.01a | 8.53 ± 0.06b | 7.37 ± 0.03c | 8.68 ± 0.01a | 7.49 ± 0.01b | 6.37 ± 0.01c |
The presented values are the means of three determinations (n = 3), with standard deviations for each column, different superscript letters differ significantly (p ≤ 0.05) by 2-sided Tukey’s HSD
The superscripts denote statistically significant values obtained by 2-sided Tukey analysis using SPSS program
Salt tolerance assay
Salt tolerance assay of all the tested cultures showed significant tolerance even at the highest tested concentration of 10% (Table 2). All the isolates showed significantly stronger tolerance when compared to the reference strain. One of the isolates (PUFSTP70) showed the highest salt tolerance.
Table 2.
Effect of salt and lysozyme on the viability of test isolates during 3 h incubation
| Isolates | Salt tolerance assay | lysozyme tolerance assay | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Survival rate (CFU/ml) | Survival rate (CFU/ml) | ||||||||||
| Control (0%) | 2% | 4% | 6% | 8% | 10% | Control | 50 µg/ml | 100 µg/ml | |||
| PUFSTP35 | 8.18 ± 0.03c | 8.37 ± 0.06b | 8.46 ± 0.02ab | 8.48 ± 0.03a | 8.25 ± 0.02c | 7.96 ± 0.04d | 8.12 ± 0.03a | 7.37 ± 0.04a | 6.12 ± 0.03b | ||
| PUFSTP38 | 8.39 ± 0.02a | 8.37 ± 0.09b | 8.26 ± 0.01b | 8.26 ± 0.01b | 8.42 ± 0.01a | 7.82 ± 0.01c | 8.30 ± 0.03a | 7.29 ± 0.03a | 6.30 ± 0.03a | ||
| PUFSTP39 | 8.06 ± 0.03b | 8.18 ± 0.11ab | 8.08 ± 0.04ab | 8.21 ± 0.04a | 8.18 ± 0.03ab | 7.70 ± 0.01c | 8.39 ± 0.01a | 7.45 ± 0.02b | 7.39 ± 0.02c | ||
| PUFSTP44 | 8.39 ± 0.01bc | 8.46 ± 0.02a | 8.38 ± 0.01c | 8.42 ± 0.06b | 8.05 ± 0.01d | 7.68 ± 0.03e | 7.87 ± 0.01a | 6.65 ± 0.56b | 6.64 ± 0.01c | ||
| PUFSTP70 | 8.61 ± 0.18b | 8.83 ± 0.03a | 8.64 ± 0.02ab | 8.48 ± 0.04bc | 8.35 ± 0.02c | 8.13 ± 0.02d | 8.51 ± 0.02a | 7.53 ± 0.04a | 7.51 ± 0.06a | ||
| PUFSTP71 | 8.37 ± 0.02ab | 8.56 ± 0.23a | 8.38 ± 0.01ab | 8.37 ± 0.03ab | 8.24 ± 0.00b | 7.77 ± 0.01c | 8.47 ± 0.04a | 8.55 ± 0.01a | 7.47 ± 0.02c | ||
| PUFSTP74 | 8.70 ± 0.03b | 8.76 ± 0.01a | 8.66 ± 0.02c | 8.45 ± 0.02d | 7.70 ± 0.00e | 7.68 ± 0.04e | 8.24 ± 0.05a | 7.57 ± 0.02a | 7.24 ± 0.05b | ||
| PUFSTP81 | 8.69 ± 0.01b | 8.79 ± 0.00a | 8.63 ± 0.04c | 8.49 ± 0.01d | 8.39 ± 0.01e | 7.83 ± 0.02f | 8.22 ± 0.02a | 7.22 ± 0.04b | 6.53 ± 0.06c | ||
| SS1 | 8.31 ± 0.01b | 8.46 ± 0.03a | 8.34 ± 0.02b | 7.36 ± 0.01c | 6.71 ± 0.01d | 6.74 ± 0.01d | 8.51 ± 0.03a | 7.57 ± 0.01b | 6.50 ± 0.02c | ||
The presented values are the means of three determinations (n = 3), with standard deviations for each column, different superscript letters differ significantly (p ≤ 0.05) by 2-sided Tukey’s HSD
The superscripts denote statistically significant values obtained by 2-sided Tukey analysis using SPSS program
Lysozyme tolerance
In lysozyme tolerance test, all the isolates showed strong resistance at 50 µg/ml of lysozyme (Table 2). Five of the eight isolates screened, namely, PUFSTP39, PUFSTP70, PUFSTP71, PUFSTP74 and PUFSTP81 consistently showed tolerance even at 100 µg/ml lysozyme, whereas isolates PUFSTP35, PUFSTP38, PUFSTP44 exhibited significantly lower tolerance to lysozyme. Reference strain W. cibaria SS1 showed reduction of 2.01 log CFU/ml viability in 100 µg/ml of lysozyme after 3 h of incubation.
Cholesterol-removal ability
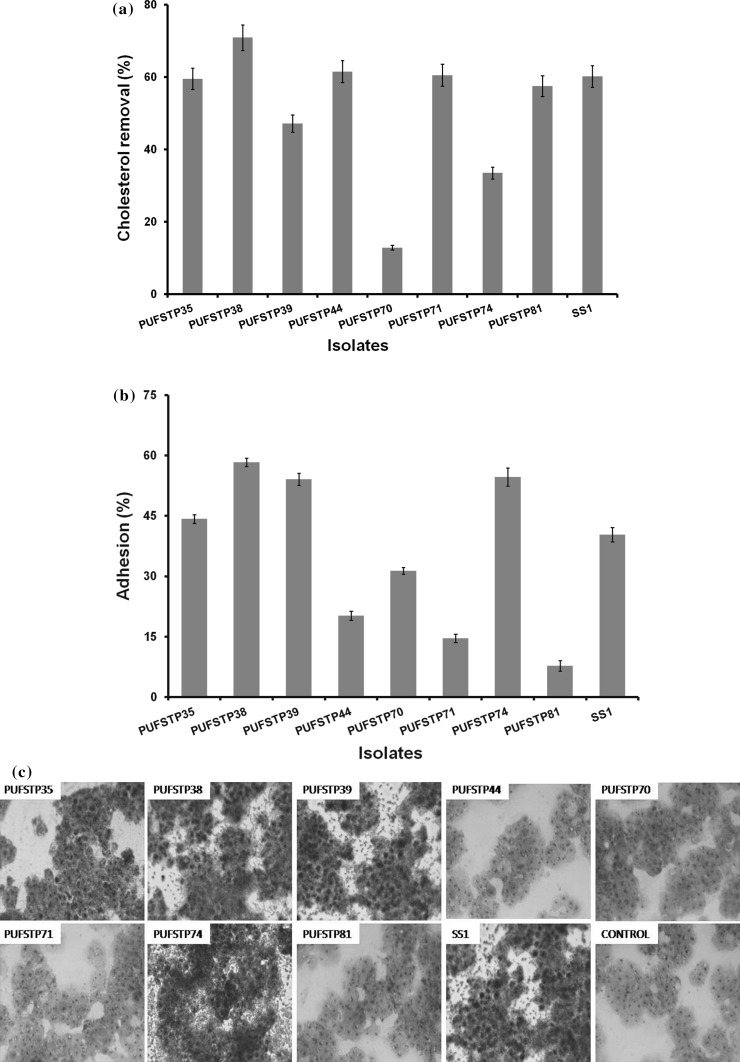
The cholesterol removal ability of the isolates ranged from 70.89 to 12.82% (Fig. 1a). Highest removal of cholesterol was observed with isolate PUFSTP38 (70.89%) and the lowest with PUFSTP70 (12.82%). Reference strain W. cibaria SS1 showed moderate levels 60.13% of cholesterol removal activity.
Fig. 1.
a Cholesterol removal by test isolates in de-Man Rogosa Sharpe broth at 37 °C and cholesterol removal was expressed in terms of percentage ± SE. b Adhesion of test isolates to human colon cell line HT-29 in de-Man Rogosa Sharpe agar at 37 °C and adhesion was expressed in terms of percentage ± SE. c Adhesion of selected bacterial strains on HT-29 cell line stained with Giemsa stain (40 ×)
Antibiotic resistance
All the isolates were susceptible to the tested antibiotics, namely Amikacin, Chloramphenicol, Gentamycin, Kanamycin and Spectinomycin. The isolates PUFSTP70, PUFSTP71, PUFSTP74 and PUFSTP81 were resistant to Vancomycin. All isolates except PUFSTP38 and PUFSTP44 were resistant to Bacitracin.
Antimicrobial activity
None of the selected isolates showed antimicrobial activity against any of the pathogens tested.
β–Galactosidase activity
Screening of eight isolates for probiotic potential revealed that the isolate PUFSTP81 only possessed the β–Galactosidase activity.
Exopolysaccharide production
Tested isolate PUFSTP35 was the only isolate that revealed exopolysaccharide production which was confirmed by ropy colony.
Bacterial adhesion to hydrocarbons (BATH assay)
The hydrophobicity of the isolates was evaluated by using xylene and n-hexadecane. Isolates exhibited hydrophobicity ranging from 1.25 to 57.33% and 7.07–57.51% towards n-hexadecane and xylene, respectively. Among the tested isolates, PUFSTP35 and PUFSTP74 showed higher maximum hydrophobicity compared to reference strain SS1 with hydrophobicity of 57.33 and 55.31% for n-hexadecane and xylene, respectively (Table 3).
Table 3.
Bacterial adhesion to hydrocarbons and auto-aggregation ability of test isolates
| Isolates | Bacterial adhesion to hydrocarbons assay | Auto-aggregation assay | ||
|---|---|---|---|---|
| n-hexadecane | Xylene | 3rd hour | 24th hour | |
| PUFSTP35 | 53.56 ± 2.32c | 57.51 ± 0.89b | 29.06 ± 0.48bc | 54.58 ± 0.57b |
| PUFSTP38 | 32.22 ± 2.62d | 47.46 ± 0.94a | 18.30 ± 0.54de | 48.57 ± 0.67c |
| PUFSTP39 | 25.14 ± 1.14e | 24.71 ± 1.94c | 22.61 ± 1.42 cd | 42.70 ± 0.76d |
| PUFSTP44 | 47.40 ± 2.53c | 54.71 ± 0.65b | 16.06 ± 1.70de | 37.52 ± 1.58e |
| PUFSTP70 | 3.81 ± 1.26e | 21.03 ± 1.23 cd | 34.28 ± 1.79ab | 43.56 ± 0.34d |
| PUFSTP71 | 5.54 ± 1.07e | 14.86 ± 1.64de | 12.60 ± 0.86e | 58.83 ± 0.91a |
| PUFSTP74 | 55.94 ± 2.16b | 54.98 ± 2.61b | 26.53 ± 1.19bc | 35.18 ± 0.44e |
| PUFSTP81 | 1.25 ± 1.00e | 7.07 ± 1.70e | 13.42 ± 0.02e | 34.49 ± 0.59e |
| SS1 | 57.33 ± 1.09a | 55.31 ± 1.60a | 36.98 ± 0.56a | 50.33 ± 0.49c |
The presented values are the means of three determinations (n = 3), with standard deviations for each column, different superscript letters differ significantly (p ≤ 0.05) by 2-sided Tukey’s HSD
The superscripts denote statistically significant values obtained by 2-sided Tukey analysis using SPSS program
Auto-aggregation
Auto-aggregation of the test isolates was highly variable (Table 3). Among the tested isolates, PUFSTP35, PUFSTP38 and PUFSTP71 showed comparatively higher aggregation (54.58, 48.57 and 58.83%, respectively) when compare to the other strains tested. Reference strain W. cibaria SS1 had auto-aggregation of 50.33%.
Adhesion to human adenocarcinoma cell line HT-29
The adhesion abilities of all the eight isolates are shown in Fig. 1b and c. The adhesion levels ranged from 8 to 58.33%, with the highest value for PUFSTP38 (58.33%) followed by PUFSTP74 (54.72%), PUFSTP39 (54.10%), PUFSTP35 (44.25%) and PUFSTP70 (31.37%). All the above strains showed stronger adhesion abilities when compare to reference strain.
Principal component analysis
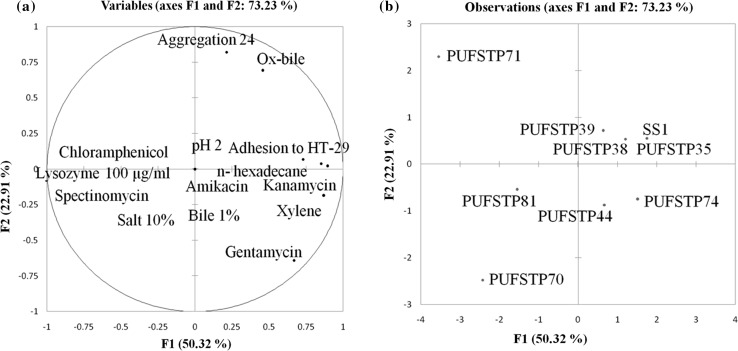
Principal component analysis (PCA) was performed to analyze the relationship between the probiotic properties. Four principal components (PC) of PCA revealed a total variation of 97.15%, while the first and second components of PCA were considered of 50.32 and 22.91%, respectively. In (Table 4), adhesion to HT-29, ox-bile, n-hexadecane, xylene, auto-aggregation, kanamycin, gentamycin was related to PC1 and PC2, variables analysed in the PCA are considered for the selection of most suitable isolates for further evaluation. The variables distributed on the PC1 and PC2 are represented in (Fig. 2a). Test isolates projected on the bi-dimensional space on the components of PCA are shown in (Fig. 2b). Test isolates PUFSTP35, PUFSTP38, PUFSTP39, PUFSTP44 and PUFSTP74 were the most potential candidates with the maximum probiotic characteristics. While isolate PUFSTP35 revealed higher resemblances with strain SS1 explaining the identical activity and probiotic characteristics of PUFSTP35 with respect to reference strain.
Table 4.
Correlations between variables and factors using PCA analysis
| Variables | PC 1 | PC 2 | PC 3 | PC 4 |
|---|---|---|---|---|
| Ph 2 | 0.000 | 0.000 | 0.000 | 0.000 |
| 1% bile | 0.000 | 0.000 | 0.000 | 0.000 |
| Salt 10% | 0.000 | 0.000 | 0.000 | 0.000 |
| Lysozyme 100 µg/ml | 0.000 | 0.000 | 0.000 | 0.000 |
| Adhesion to HT-29 | 0.730 | 0.068 | 0.592 | − 0.215 |
| Ox-bile | 0.458 | 0.692 | − 0.549 | − 0.031 |
| n-Hexadecane | 0.895 | 0.021 | − 0.081 | 0.407 |
| Xylene | 0.871 | − 0.187 | 0.206 | 0.364 |
| Aggregation24 | 0.213 | 0.820 | 0.462 | − 0.143 |
| Kanamycin | 0.852 | 0.038 | − 0.434 | − 0.288 |
| Spectinomycin | 0.000 | 0.000 | 0.000 | 0.000 |
| Gentamycin | 0.669 | − 0.641 | − 0.026 | − 0.350 |
| Amikacin | 0.000 | 0.000 | 0.000 | 0.000 |
| Chloramphenicol | 0.000 | 0.000 | 0.000 | 0.000 |
Significantly higher values are shown in bold
Fig. 2.
a Graphical representation of the variables formed by PC1 and PC2 analyzed by principal component analysis. b Prediction of the test isolates and reference strain Weisella cibaria (SS1) in the plane of PC1 and PC2
Species identification by 16S rDNA sequencing
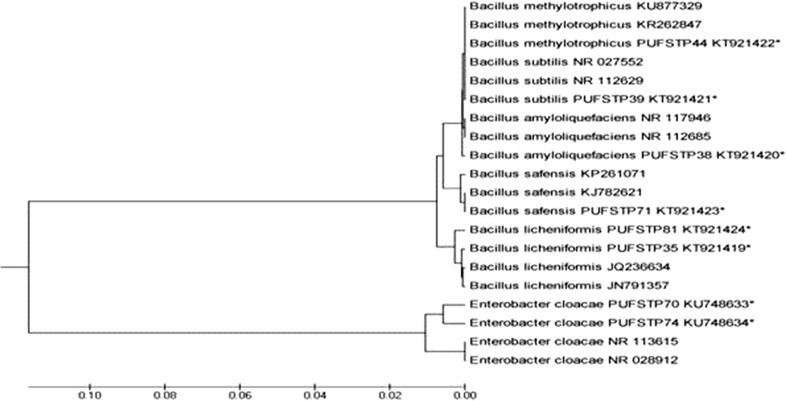
Test isolates were identified using amplification of conserved region of the 16S rRNA gene. BLAST analyses revealed that the identified probiotic isolates belong to the genera Bacillus and Enterobacter and were identified up to species level. Molecular phylogeny analysis and phylogenetic tree constructed using UPGMA method showed homology towards the similar species in public database (Fig. 3). The sequences of 16S rRNA genes were deposited to GenBank and accession numbers are given in Table 5.
Fig. 3.
UPGMA phylogenetic tree based on 16S rRNA sequences of selected isolates (*) with reference strains from NCBI database
Table 5.
Test isolates by 16S rRNA gene sequencing and their GenBank accession number
| Isolates | Species | Gen bank accession No. |
|---|---|---|
| PUFSTP35 | Bacillus licheniformis | KT921419 |
| PUFSTP38 | Bacillus amyloliquefaciens | KT921420 |
| PUFSTP39 | Bacillus subtilis | KT921421 |
| PUFSTP44 | Bacillus methylotrophicus | KT921422 |
| PUFSTP70 | Enterobacter cloacae | KU748633 |
| PUFSTP71 | Bacillus safensis | KT921423 |
| PUFSTP74 | Enterobacter cloacae | KU748634 |
| PUFSTP81 | Bacillus licheniformis | KT921424 |
Discussion
The present study was aimed at screening probiotic potential and functional properties of bacterial isolates from traditional fermented mango pickle. Basic characteristic of a potential probiotic strain includes tolerance towards highly acidic pH of stomach environment and its persistence in intestinal habitat. To achieve successive stable viability in Human intestinal tract, the probiotic strain should be capable of tolerating the extreme acidity, bile, salt and lysozyme. Tolerance of bacteria to acidic condition is important to overcome the stress in the gut environment and it also makes the strain capable in surviving in high acid-foods for longer time without any reduction in their viability of cell numbers (Wang et al. 2010). Our study revealed that eight selected isolates showed similar survival rate in both neutral and pH 3, which indicates their tolerance towards the acidic pH. Similar observations have recently been reported among the probiotic isolates from fermented mustard (Wang et al. 2014). Further, most of the selected isolates were even tolerant at pH 2 with marginal decrease in cell viability noticed in certain isolates. Our finding is in concurrence with the report of Guo et al. (2010). Wang et al. (2010) who reported that Bacillus species are capable of retaining their viability in stomach at pH as low as 2.0.
Most of the selected non hemolytic isolates were tolerant to lysozyme (100 µg/ml) under invitro conditions. The similar lysozyme concentration did not affect the endurance of L. plantarum isolates from fermented cheese in a simulated stomach passage (Zago et al. 2011). Salt tolerance can influence the physiology (water activity) and metabolism (enzyme activity) of bacterial strains, a probiotic isolate requires high osmotolerance to withstand excessive external osmotic pressure (Adnan and Tan 2007). Results from the present study on salt stress response were concurrent with Menconi et al. (2014). Nonexistence of transferable resistance against clinical antibiotics is one of the primary characteristic of probiotic (Georgieva et al. 2015). Earlier reports revealed probiotic bacterial isolates with β-galactosidase activity (Angmo et al. 2016). Beta-galactosidase activity alleviates lactose intolerance in the gut by breaking down lactose in the upper regions of small intestine (Vrese et al. 2001). Chabot et al. 2001 showed that EPS was involved in probiotic action, anti-tumour, anti-ulcer and immuno-modulatory activity and cholesterol lowering of isolate. Our study on antibiotic susceptibility, β-galactosidase activity and Exopolysaccharide production results revealed similar trend as reported by Angmo et al. (2016).
These isolates from mango pickle are shown to be potentially probiotic. However they must survive in the upper intestine that contains bile. Bile salts are secreted by liver, stored in gallbladder and transferred to intestinal tract through bile ducts and these salts are toxic for living cells, therefore tolerance to bile salt are considered as an important property needed for potential probiotic isolates to show their viability in the intestinal tract (Succi et al. 2005). Present study has revealed that all the selected isolates showed resistance to 0.5 and 1% of bile concentration. Similar tolerance towards bile has been reported for probiotics isolated from traditional fermented foods (Wang et al. 2014).
Many bacterial isolates have been identified to exhibit hypocholesterolemic activity through their effect on host by decreasing cholesterol levels in intestinal tract and preventing from coronary heart disease. Removal of cholesterol is carried through assimilation or de conjugation of bile acids promoting excretion or binding the cell surface of both dead and live cells of probiotic bacteria (Saraniya and Jeevaratnam 2015). In our study, considerable range of cholesterol removed in the medium was precipitated with inclusion of deconjugated bile acids (ox-bile). Pereira et al. (2003) reported earlier that removal of cholesterol by Bifidobacteria and lactic acid bacteria was higher in the presence of bile acids (ox-bile) in the media. Similar trend also was observed in our work.
Adherence to colon epithelial cells is one of the important properties and it’s the most desirable process of any probiotic to aggregate with the host cell for providing immunomodulatory and other health benefits. Cesena et al. (2001) reported that the isolates have the ability to aggregate and are generally in hydrophobic nature and showed viable in the intestinal tract. Many earlier reports advocated that BATH assay (Duary et al. 2011) and auto-aggregation ability (Jankovic et al. 2012) are related to the properties of cell adherence. In this study, test isolates showed varying in degrees of adherence in the BATH assay and isolates such as PUFSTP35, PUFSTP38, PUFSTP44 and PUFSTP74 demonstrated higher adhesion towards n-hexadecane and xylene. Duary et al. (2011) reported the adhesion property of faecal isolate Lactobacillus plantarum Lp91 showed high degree of hydrophobicity for toluene and n-hexadecane and it was followed by another isolate L. plantarum Lp9. In the present study, all the isolates exhibited auto-aggregation to various degrees. Potentiality of auto-aggregation was increased with time. Similar results for auto-aggregation were reported by Dias et al. (2013).
Adhesion to human intestinal tract is an important characteristic of probiotic bacteria. In-vitro and in vivo experimental models were reported to be used in investigating the adhesion properties of the proposed probiotic isolates (Tsai et al. 2004). Previous reports have shown that adhesion properties are strain and matrix dependent (Tallon et al. 2007). The result of our study was similar to the findings of Jensen et al. (2012) with similar adhesion properties.
Potential probiotic isolates were selected by using statistical application XLSTAT, where in multivariate data analysis of all the probiotic properties were performed. The most capable and potent probiotic isolates were namely PUFSTP35, PUFSTP38, PUFSTP39, PUFSTP44 and PUFSTP74 as they capable of survival at pH 2, bile 1%, lysozyme (100 µg/ml), 10% salt tolerance and cholesterol removal (ox-bile). Isolates PUFSTP35, PUFSTP38 and PUFSTP39 belonging to bacillus sp. showed similar trend with the reference strain (SS1) indicating their potential to be utilized commercially. Studies by Tamang et al. 2003 revealed the presence of bacillus sp. with probiotic potential in fermented kinema. Further it was suggested that Bacillus sp. including B. licheniformis as starter culture for ‘Hawaijar’, a traditional non-salted fermented soybean (Jeyaram et al. 2008).
Conclusion
The present investigation revealed that, PUFSTP35 (B. licheniformis), PUFSTP38 (B. amyloliquefaciens) and PUFSTP39 (B. subtilis) isolated from a traditional fermented mango brine pickle exhibited prominent probiotic properties in vitro. These isolates were shown to have most desirable probiotic properties; these isolates were originated from fermented products and considered safe for consumption, thus traditional foods can serve as potential source of probiotic bacteria. Our in vitro study has proved the potential of these isolates as probiotic candidates. Further confirmations of probiotic potential of the tested isolates using in vivo studies in future are essential.
Acknowledgements
Authors acknowledge Pondicherry University for all the facilities.
Compliance with ethical standards
Conflict of interest
Authors declare that they have no conflict of interest.
References
- Adnan AFM, Tan IK. Isolation of lactic acid bacteria from Malaysian foods and assessment of the isolates for industrial potential. Bioresour Technol. 2007;98:1380–1385. doi: 10.1016/j.biortech.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Angmo K, Kumari A, Savitri Bhalla TC. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT—Food Sci Technol. 2016;66:428–435. [Google Scholar]
- Arokiyaraj S, Islam VIH, Bharanidharan R, Raveendar S, Lee J, Kim DH, Oh YK, Kim E, Kim KH. Antibacterial, anti-inflammatory and probiotic potential of Enterococcus hirae isolated from the rumen of Bos primigenius. World J Microbiol Biotechnol. 2014;30:2111–2118. doi: 10.1007/s11274-014-1625-0. [DOI] [PubMed] [Google Scholar]
- Cesena C, Morelli L, Alander M, Siljander T, Tuomola E, Salminen S, Mattila-Sandholm T, Vilpponen-Salmela T, Wright VA. Lactobacillus crispatus and its non-aggregating mutant in human colonization trials. J Dairy Sci. 2001;84:1001–1010. doi: 10.3168/jds.S0022-0302(01)74559-6. [DOI] [PubMed] [Google Scholar]
- Chabot S, Yu H, Léséleuc LD, Cloutier D, Calsteren MV, Lessard M, Roy D, Lacroix M, Oth D (2001) Exopolysaccharides from Lactobacillus rhamnosus RQ-595 M stimulate TNF, IL-6 and IL-12 in human and mouse cultured immune-competent cells and IFN-g in mouse splenocytes. Le Lait, INRA Editions; pp. 683–697
- de-Valdez GF, de-Taranto MP. Probiotic properties of Lactobacilli: cholesterol reduction and bile salt hydrolase activity. In: Spencer JFT, de Spencer ALR, editors. Food microbiology protocols. New Jersey: Humana Press; 2001. pp. 173–182. [Google Scholar]
- Dias FS, Duarte WF, Schwan RF. Evaluation of adhesive properties of presumptive probiotic Lactobacillus plantarum strains. Biosci J. 2013;29:1678–1686. [Google Scholar]
- Duary RK, Rajput YS, Batish VK, Grover S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J Med Res. 2011;134:664–671. doi: 10.4103/0971-5916.90992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO/WHO . Probiotics in food, health and nutritional properties and guidelines for evaluation. FAO food and nutritional paper. No. 85. Rome: WHO/FAO; 2006. [Google Scholar]
- Georgieva R, Yocheva L, Tserovska L, Zhelezova G, Stefanova N, Atanasova A, Danguleva A, Ivanova G, Karapetkov N, Rumyan N, Karaivanova E. Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol Biotechnol Equip. 2015;29:84–91. doi: 10.1080/13102818.2014.987450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva NM, Gavrilov AF, Solov’eva AL, Smirnov VV, Sorokulova IB. The efficacy of the new bacterial preparation biosporin in treating acute intestinal infections. Zh Mikrobiol Epidemiol Immunobiol. 1996;1:75–77. [PubMed] [Google Scholar]
- Guo XH, Kim JM, Namb HM, Park SY, Kim JM. Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe. 2010;16:321–326. doi: 10.1016/j.anaerobe.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Jankovic T, Frece J, Abram M, Gobin I. Aggregation ability of potential probiotic Lactobacillus plantarum strains. IJSER. 2012;6:19–24. [Google Scholar]
- Jensen H, Grimmer S, Naterstad K, Axelsson L. In vitro testing of commercial and potential probiotic lactic acid bacteria. Int J Food Microbiol. 2012;153:216–222. doi: 10.1016/j.ijfoodmicro.2011.11.020. [DOI] [PubMed] [Google Scholar]
- Jeyaram K, Mohendro SW, Premarani T, Ranjita Devi A, Selina Chanu K, Talukdar NC, Rohinikumar SM. Molecular identification of dominant microflora associated with ‘Hawaijar’ a traditional fermented soybean (Glycine max (L.)) food of Manipur India. Int J Food Microbiol. 2008;122:259–268. doi: 10.1016/j.ijfoodmicro.2007.12.026. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerak J, Lida H, Watanabe Y, Miyashita M, Sato H, Nakagawa Y, Tahara Y. Phylogeny of poly-γ-glutamic acid-producing Bacillus strains isolated from fermented soybean foods manufactured in Asian countries. J Gen Appl Microbiol. 2007;53:315–323. doi: 10.2323/jgam.53.315. [DOI] [PubMed] [Google Scholar]
- Menconi A, Kallapura G, Latorre JD, Morgan MJ, Pumford NR, Hargis BM, Tellez G. Identification and characterization of lactic acid bacteria in a commercial probiotic culture. Biosci Microbiota Food Health. 2014;33:25–30. doi: 10.12938/bmfh.33.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V, Prasad DN. Application of in vitro methods for selection of Lactobacillus casei strains as potential probiotics. Int J Food Microbiol. 2005;103:109–115. doi: 10.1016/j.ijfoodmicro.2004.10.047. [DOI] [PubMed] [Google Scholar]
- Osmanagaoglu O, Kiran F, Ataoglu H. Evaluation of in vitro probiotic potential of Pediococcus pentosaceus OZF isolated from human breast milk. Probiotics Antimicrob Proteins. 2010;2:162–174. doi: 10.1007/s12602-010-9050-7. [DOI] [PubMed] [Google Scholar]
- Pereira DIA, McCartney AL, Gibson GR. An in vitro study of probiotic potential of a bile salt hydrolyzing Lactobacillus fermentum strain and determination of its cholesterol lowering properties. Appl Environ Microbiol. 2003;69:4743–4752. doi: 10.1128/AEM.69.8.4743-4752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Jass J, Sebulsky MT, McCormick JK. Potential use of probiotics in clinical practice. Clin Microbiol Rev. 2003;16:658–672. doi: 10.1128/CMR.16.4.658-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraniya A, Jeevaratnam K. In vitro probiotic evaluation of phytase producing Lactobacillus species isolated from Uttapam batter and their application in soy milk fermentation. J Food Sci Technol. 2015;52:5631–5640. doi: 10.1007/s13197-014-1686-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokulova I (2013) Modern status and perspectives of Bacillus bacteria as probiotics. J Prob Health, 1
- Steinkraus KH. Fermentations in world food processing. Compr Rev Food Sci Food Saf. 2002;1:23–32. doi: 10.1111/j.1541-4337.2002.tb00004.x. [DOI] [PubMed] [Google Scholar]
- Succi M, Tremonte P, Reale A, Sorrentino E, Grazia L, Pacifico S. Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol Lett. 2005;244:129–137. doi: 10.1016/j.femsle.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Tallon R, Arias S, Bressollier P, Urdaci MC. Strain- and matrix dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J Appl Microbiol. 2007;102:442–451. doi: 10.1111/j.1365-2672.2006.03086.x. [DOI] [PubMed] [Google Scholar]
- Tamang JP. Native microorganisms in fermentation of kinema. Indian J Microbiol. 2003;43:127–130. [Google Scholar]
- Tsai C, Huanga L, Lina C, Tsen H. Antagonistic activity against Helicobacter pylori infection in vitro by a strain of Enterococcus faecium TM39. Int J Food Microbiol. 2004;96:1–12. doi: 10.1016/j.ijfoodmicro.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Vinderola CG, Reinheimer JA. Lactic acid starter and probiotic bacteria: a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res Int. 2003;36:895–904. doi: 10.1016/S0963-9969(03)00098-X. [DOI] [Google Scholar]
- Vrese MD, Stegelmann A, Richter B, Fenselau S, Laue C, Schrezenmeir J. Probiotics-compensation for lactase insufficiency. Am J Clin Nutr. 2001;73:421–429. doi: 10.1093/ajcn/73.2.421s. [DOI] [PubMed] [Google Scholar]
- Wang J, Fung DYC. Alkaline-fermented foods: a review with emphasis on pidan fermentation. Crit Rev Microbiol. 1996;22:101–138. doi: 10.3109/10408419609106457. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Zhang L, Liu W, Zhang Y, Zhang X, Sun T. In vitro assessment of probiotic properties of Bacillus isolated from naturally fermented congee from Inner Mongolia of China. World J Microbiol Biotechnol. 2010;26:1369–1377. doi: 10.1007/s11274-010-0309-7. [DOI] [Google Scholar]
- Wang SC, Chang CK, Chan SC, Shieh JS, Chiu CK, Duh PD. Effects of lactic acid bacteria isolated from fermented mustard on lowering cholesterol. Asian Pac J Trop Biomed. 2014;4:523–528. doi: 10.12980/APJTB.4.201414B54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhang X, Li S, Li C, Li D, Yang Z. Evaluation of probiotic properties of Lactobacillus plantarum strains isolated from Chinese sauerkraut. World J Microbiol Biotechnol. 2013;29:489–498. doi: 10.1007/s11274-012-1202-3. [DOI] [PubMed] [Google Scholar]
- Zago M, Fornasari ME, Carminati D, Burns P, Suàrez V, Vinderola G, Reinheimer J, Giraffa G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011;28:1033–1040. doi: 10.1016/j.fm.2011.02.009. [DOI] [PubMed] [Google Scholar]