Abstract
Chromobacterium violaceum is an abundant component of the soil and water microbiota in tropical and subtropical regions around the world. For many years, it was mainly known as a producer of violacein and as a reporter for the discovery of quorum sensing molecules. However, C. violaceum has recently emerged as an important model of an environmental opportunistic pathogen. Its high virulence in human infections and a mouse infection model involves the possession of several predicted virulence traits, including two type III secretion systems (T3SSs). In this article, in addition to providing an update on the new clinical cases of human C. violaceum infections, we will focus on recent advances in understanding the molecular mechanisms regarding C. violaceum pathogenesis. It has been demonstrated that the C. violaceum Cpi-1 T3SS plays a pivotal role in interaction with host cells. It is required for the secretion of effector proteins and is the agonist recognized by the Nod-like receptor CARD domain-containing protein 4 (NLRC4) inflammasome from innate immune cells. Pyroptosis and its release of hepatocytes for killing by neutrophils are key events required for the clearance of C. violaceum. Given the prominent role of T3SSs in C. violaceum virulence, we examine their occurrence in the Chromobacterium genus, taking advantage of several draft genome sequences of Chromobacterium species that have recently become available. Our finding that the Cpi-1 T3SS is widespread among Chromobacterium species points toward the pathogenic potential of this genus for humans or to novel roles of the T3SS in the interaction of Chromobacterium species with other organisms.
Keywords: Chromobacterium violaceum, Chromobacterium species, genome sequencing, comparative genomics, pathogenicity island, type III secretion system
Introduction
For many years, studies on Chromobacterium violaceum have been focused on investigating small molecules of biotechnological interest derived from its secondary metabolism, while aspects related to the pathogenicity of C. violaceum have been neglected. In fact, there are many reviews describing the biotechnological and pharmacological importance of C. violaceum and its secondary metabolites, mainly the purple pigment violacein (Durán and Menck, 2001; Durán et al., 2007, 2016), but none have focused on the mechanisms of C. violaceum virulence. However, this situation has changed in recent years, with numerous works advancing toward revealing multiple facets of the biology of C. violaceum and its interaction with mammalian hosts (Miki et al., 2010, 2011; Maltez et al., 2015; Previato-Mello et al., 2017). In this work, we summarize recent advances in the knowledge of the pathogenesis of C. violaceum infections and update the scenario regarding clinical cases and deaths caused by C. violaceum. Moreover, we evaluate the presence and genomic organization of the genes encoding type III secretion systems (T3SSs) in members of the Chromobacterium genus.
Overview of the Chromobacterium Genus
Chromobacterium is a genus of soil- and freshwater-associated Gram-negative bacteria within the Neisseriaceae family of Betaproteobacteria. Despite its saprophytic, free-living lifestyle, the species type of the genus, C. violaceum, has been associated with infections in humans and other animals (Durán and Menck, 2001; Yang and Li, 2011). One particular characteristic of this genus is the production of violacein, a purple pigment for which the synthesis is regulated by quorum sensing (Mcclean et al., 1997). However, non-pigmented isolates have also been identified (Sivendra and Tan, 1977). Violacein is a pigment with high biotechnological interest due to its in vitro activity against bacteria, fungi, protozoa, viruses, and tumor cells (Durán et al., 2007, 2016). Environmental isolates of Chromobacterium have the potential to be used in other biotechnological applications, including biocontrol of plant diseases caused by insect pests (Chromobacterium sp. strain C-61) (Kim et al., 2014), prevention of disease transmission by Anopheles gambiae and Aedes aegypti mosquitoes (Chromobacterium sp. Csp_P) (Ramirez et al., 2014), hydrogen cyanide-mediated gold recovery from electronic waste (Tay et al., 2013), and production of the anti-tumoral depsipeptide FR901228 (C. violaceum) (Durán and Menck, 2001; VanderMolen et al., 2011).
Although C. violaceum has been recognized as the single species of the Chromobacterium genus for a long time, nine novel species have been proposed since 2007: C. subtsugae (Martin et al., 2007), C. aquaticum (Young et al., 2008), C. haemolyticum (Han et al., 2008), C. piscinae (Kämpfer et al., 2009), C. pseudoviolaceum (Kämpfer et al., 2009), C. vaccinii (Soby et al., 2013), C. amazonense (Menezes et al., 2015), C. alkanivorans (Bajaj et al., 2016), and C. rhizoryzae (Zhou et al., 2016). Additionally, the great genetic variability found in Chromobacterium isolates collected from distinct tropical regions (Hungria et al., 2005; Lima-Bittencourt et al., 2007) supports the trend to attempt to recognize novel species. Most of the Chromobacterium species were isolated from environmental samples (mainly from water, soil, and rhizosphere) and have not yet been associated with human infections (Table 1). Exceptions include C. violaceum, isolated from both environmental and clinical samples and associated with several cases of fatal infections (Yang and Li, 2011), and C. haemolyticum, isolated from a patient’s sputum culture (Han et al., 2008) and associated with a human case of bacteremia (Okada et al., 2013). With respect to violacein production, the non-purple species of this genus are C. aquaticum, C. haemolyticum, C. alkanivorans, and C. rhizoryzae (Table 1).
Table 1.
Summary of the Chromobacterium species and selected strains with genome sequences available.
| Species/strain with published genomea | Isolation source | Biological interaction | Colony color | Reference of genome sequencing |
|---|---|---|---|---|
| C. violaceum/strains ATCC 12472 and CV017 (derived from ATCC 31532) | Soil and water worldwide; human infections | Human and animal pathogen | Violet | Vasconcelos et al., 2003; Wang et al., 2016 |
| C. subtsugae/strains PRAA4-1; MWU12-2387; MWU3525; MWU2576; and MWU2920 | Forest soil and rhizosphere in United States | Toxic to insect larvae | Violet | Vöing et al., 2015b, 2017; Blackburn et al., 2016 |
| C. aquaticum/strain CC-SEYA-1 | Spring water in Taiwan | Non-described | Tan | Soby, 2017a |
| C. haemolyticum/strain T124 | Sputum from a patient in United States; bacteremia in Japan | Human pathogen | Gray | Miki and Okada, 2014 |
| C. piscinae/strain ND17 | Pond water in Malaysia | Non-described | Violet | Chan and Yunos, 2016 |
| C. pseudoviolaceum/strain LMG 3953T | Unclear | Non-described | Violet | Soby, 2017b |
| C. vaccinii/strains MWU205 and MWU328 | Soil and rhizosphere in United States | Toxic to insect larvae | Violet | Vöing et al., 2015a |
| C. amazonense | River water in Brazil | Non-described | Violet | – |
| C. alkanivorans | Contaminated soil in India | Degradation of halogenated alkanes | Tan | – |
| C. rhizoryzae | Rhizosphere in China | Inhibition of fungal pathogens | Tan | – |
| Chromobacterium sp./strain C-61 | Rhizosphere in Korea | Inhibition of fungal pathogens | Kim et al., 2011 | |
aAll 10 proposed Chromobacterium species are shown. In respect to genome sequence availability, we listed only the Chromobacterium strains whose genome sequence were published. A more comprehensive list of Chromobacterium genomes publicly available can be found in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genome/?term=chromobacterium).
Updates Regarding Clinical Reports of Chromobacterium violaceum Infections
Although rare, human infections with C. violaceum are associated with high mortality rates; bacteria spreading rapidly to several organs, especially the liver, lungs, and spleen; and life-threatening sepsis (Sneath et al., 1953; Durán and Menck, 2001). The main clinical manifestations are fever, abdominal pain, skin lesions, and formation of metastatic abscesses. The most common route of transmission involves the exposure of wounds and traumatic lesions to soil and water containing C. violaceum (Martinez et al., 2000; Durán and Menck, 2001; Baker et al., 2008; Ansari et al., 2015). Due to the rapid clinical course of the chromobacteriosis, one important predisposing risk factor in C. violaceum infections is inappropriate antimicrobial therapy. It has been reported that C. violaceum is resistant to several antibiotics, mainly to some beta-lactams, but it is sensitive to others, such as carbapenems and quinolones (Aldridge et al., 1988). Indeed, most of the treatments which were successful in controlling infections involved the use of the antibiotics ciprofloxacin and meropenem (Nanayakkara et al., 2008; Ke et al., 2012; Campbell et al., 2013; Pant et al., 2017).
The last comprehensive compilation of human cases of C. violaceum infection analyzed 106 patients with C. violaceum infections between 1952 and 2009 (Yang and Li, 2011). Here, our update on the published clinical reports of C. violaceum infections (searching the PubMed database from 2010 to 2017) reveals that infections due to C. violaceum are still rare and are associated with high mortality (Supplementary Table S1). Overall, there are less than 150 published clinical reports describing human C. violaceum infections. During the period of our analysis, 23 new cases of human infection were reported, with a mortality rate of 35% (eight fatal cases) (Supplementary Table S1). This is a smaller value than what was seen at the time of the last update (53%) (Yang and Li, 2011). The reduction in the rate of fatal cases could be attributed to an improvement in antibiotic administration or better diagnostics. The tendency for the distribution of cases to be worldwide was maintained in our compilation, despite the most fatal cases having been described in developing countries (Supplementary Table S1). Moreover, recent reports of C. violaceum infection found that, in addition to systemic infections, this bacterium causes urinary tract infections and pneumonia in hospital environments, which raises concerns about its potential as a nosocomial pathogen (Hagiya et al., 2014; Swain et al., 2014; Pant et al., 2015, 2017).
In addition to C. violaceum, another species of potential medical interest in the same genus is C. haemolyticum (Table 1). Although only a few cases of human C. haemolyticum infection have been reported so far, and none of them were fatal, this bacterium has shown remarkable hemolytic activity against human and sheep erythrocytes (Han et al., 2008; Okada et al., 2013). It has several uncharacterized potential virulence factors, as predicted from the draft genome sequence of one clinical isolate (Miki and Okada, 2014). Interestingly, C. haemolyticum isolates collected from a tropical freshwater lake exhibited strong beta-hemolytic activity and high resistance to beta-lactam antibiotics, as observed in clinical isolates (Lima-Bittencourt et al., 2011).
Molecular Pathogenesis of C. violaceum Infections
The sequencing of the complete genome of C. violaceum strain ATCC 12472 shed light on the virulence mechanisms of this bacterium by revealing the presence of many predicted virulence factors (Vasconcelos et al., 2003). The most remarkable of these predicted virulence factors was the type III secretion system (T3SS), which is a needle-like multiprotein complex that injects various bacterial effectors into host cells (Galán et al., 2014). Surprisingly, genomic data have revealed that C. violaceum has two T3SSs whose genes were clustered in Chromobacterium pathogenicity islands 1 and 2 (Cpi-1 and Cpi-2) (Vasconcelos et al., 2003; Alves de Brito et al., 2004). These islands were located next to each other on the chromosome, but while the genes from Cpi-2 were all grouped together, some genes from Cpi-1, encoding the needle complex, were located distantly from Cpi-1, in a cluster called the Cpi-1a (Betts et al., 2004). C. violaceum Cpi-1/1a and Cpi-2 resemble the well-characterized Salmonella pathogenicity islands Spi-1 and Spi-2 (Betts et al., 2004).
In 2010, it was demonstrated that deletion of genes from Cpi-1/1a, but not from Cpi-2, causes a profound reduction in C. violaceum virulence in a mouse model of infection, positioning Cpi-1/1a as the major determinant for C. violaceum pathogenicity (Miki et al., 2010). In addition, the capacity of C. violaceum to cause fulminant hepatitis in mice through the induction of cytotoxicity and cell death in hepatocytes was shown to be dependent on the Cpi-1/1a-encoded T3SS (Miki et al., 2010). Despite of the absence of a clear requirement for Cpi-2 for systemic infection by C. violaceum (Miki et al., 2010), more studies are necessary to understand the role of this T3SS in the interaction of C. violaceum with host cells. Cpi-2 could be involved in the survival of C. violaceum within macrophages, as described for the Salmonella Spi-2 system (Hensel, 2000).
Further investigations demonstrated that the repertoire of Cpi-1/1a-encoded T3SS effectors translocated into hepatocytes includes at least 16 effector proteins, but the role of the most of them have yet to be determined (Miki et al., 2010, 2011). The authors discovered that one of these effectors, called CopE, plays a key role in C. violaceum invasion of non-phagocytic epithelial cells and is required for C. violaceum virulence in mice (Miki et al., 2011). This study demonstrated that CopE acts as a guanine exchange factor (GEF) that activates Rac1 and Cdc42 in HeLa cells, resulting in the induction of actin rearrangement. Consequently, this promotes C. violaceum invasion of non-phagocytic cells (Miki et al., 2011). Interestingly, it has been recently discovered that C. violaceum escapes from the phagosome to the cytosol in epithelial cells by a mechanism involving CipC, a translocon apparatus protein of the Cpi-1 T3SS (Du et al., 2016).
The roles of transcriptional regulators in C. violaceum virulence have been investigated. For instance, a study based on expression and mutagenesis analyses of five putative regulators located within the Cpi-1 and Cpi-2 islands (CilA, CivF, ArmR, CsrB, and CsrC) revealed that CilA is the master transcriptional activator of most of the Cpi-1/1a genes (Miki et al., 2011). This is consistent with the previous finding that a cilA-mutant strain was fully attenuated for virulence in mice (Miki et al., 2010). The signals that turn on the expression of the CilA-regulated genes in C. violaceum, including the Cpi-1/1a genes, remain largely unknown. More recently, it has been reported that the MarR family transcriptional regulator OhrR is important for the virulence of C. violaceum in mice (Previato-Mello et al., 2017). In C. violaceum, OhrR is a sensor of organic hydroperoxides that regulates the expression of a few genes related to antioxidant defense, synthesis of cyclic di-GMP, and the production of virulence-related, secreted enzymes (da Silva Neto et al., 2012; Previato-Mello et al., 2017). Finally, the involvement of a quorum sensing system in the ability of C. violaceum to kill Caenorhabditis elegans has been determined (Swem et al., 2009), allowing this nematode to be used as an alternative model for identifying virulence genes of C. violaceum.
An elegant study demonstrated that the molecular detection of C. violaceum by human macrophages involves the recognition of the Cpi-1a T3SS needle protein CprI by the NAIP protein; human NAIP recognizes CprI and promotes Nod-like receptor CARD domain-containing protein 4 (NLRC4) inflammasome oligomerization, which is followed by caspase-1 activation and pyroptosis (Zhao et al., 2011). Subsequent investigations using a murine model revealed that C. violaceum infection is promptly controlled in healthy mice by the NLRC4 inflammasome via two pathways that release bacteria from intracellular niches: pyroptosis and Natural Killer (NK) cell cytotoxicity (Maltez et al., 2015). These mechanisms eject the intracellular bacteria from macrophages and hepatocytes and expose them to the action of neutrophils (Maltez et al., 2015). In fact, neutrophil deficiencies in NADPH oxidase, as seen in patients with chronic granulomatous disease (CGD), drastically increase susceptibility to C. violaceum infections (Segal et al., 2003; Yang and Li, 2011; Maltez et al., 2015). In addition, it was verified that knockout mice in these immune system pathways are extremely susceptible to infection by C. violaceum and Burkholderia thailandensis, two CGD-associated pathogens (Maltez et al., 2015). Therefore, a hypothesis was proposed, stating that inflammasomes evolved as a form of defense against infection due to environmental bacteria with virulence traits that did not evolve with vertebrate hosts (Maltez and Miao, 2016).
Prevalence of T3SSs in the Chromobacterium Genus
Bacterial T3SSs have been demonstrated to be key determinants of virulence for many Gram-negative plant and animal pathogens via delivery of effector proteins into the cytosol of host eukaryotic cells (Deng et al., 2017). Gene clusters encoding T3SSs are also found in genomes of non-pathogenic bacteria, and the roles of these T3SSs are not restricted to pathogenesis, but seem to include other processes during interactions involving bacteria and their hosts in diverse ecological contexts (Nazir et al., 2017). As mentioned above, earlier genome sequencing of the strain type C. violaceum ATCC 12472 revealed the presence of two T3SSs (Cpi-1/1a and Cpi-2), of which, Cpi-1/1a is absolutely required for virulence (Vasconcelos et al., 2003; Miki et al., 2010). Recently, several draft genome sequences of Chromobacterium species were published (Table 1) and many other have become publicly available (39 Chromobacterium genomes, as searched on 15 June, 2017 in the NCBI GenBank database), allowing for a detailed inspection of the occurrence and organization of T3SSs in members of the Chromobacterium genus and among strains of the same species.
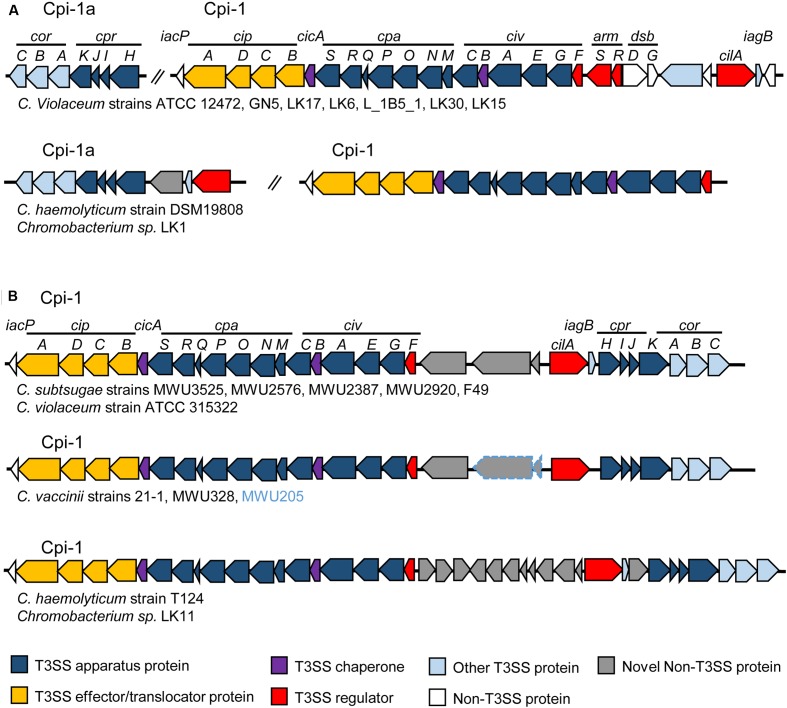
We performed such an analysis by searching for genes of the Cpi-1/1a- and Cpi-2-encoded T3SSs of C. violaceum ATCC 12472 in the draft genome sequences of 22 Chromobacterium species/strains (Supplementary Table S2), using tools available in the Integrated Microbial Genomes and Microbiome (IMG/M) system (Chen et al., 2016). Some interesting findings arose from this analysis (Supplementary Table S2 and Figure 1): (i) the widespread occurrence of the Cpi-1/1a T3SS in the Chromobacterium genus, since its absence was observed only in C. piscinae (Supplementary Table S2); (ii) the existence of two genomic organizations for the Cpi-1/1a genes, which were found either as two separated gene clusters (Cpi-1 and Cpi-1a, as seen in Figure 1A, for instance in C. violaceum strains), or as a single cluster of contiguous genes (Cpi-1, as seen in Figure 1B, for instance in C. subtsugae strains); and (iii) the narrow distribution of Cpi-2 in the Chromobacterium genus, since the occurrence of most Cpi-2 genes was restricted to C. piscinae and C. vaccinii (Supplementary Table S2). These data support the hypothesis that the presence of Cpi-1 is ancient in the C. violaceum genome (and perhaps in the Chromobacterium genus) and that Cpi-2 was acquired more recently (Betts et al., 2004).
FIGURE 1.
Genomic organization of Chromobacterium pathogenicity island 1 (Cpi-1) in members of the Chromobacterium genus. The T3SS genes are either split into two gene clusters (Cpi-1/1a) (A); or grouped together (Cpi-1) on the chromosome (B). Comparison was performed using T3SS gene clusters from C. violaceum ATCC 12472 (Cpi-1a, CV2417-CV2423 and Cpi-1, CV2615-CV2642) as previously annotated (Betts et al., 2004). Genes are colored according to functional category (genes in gray are that absent in C. violaceum ATCC 12472). For C. vaccinii, the two genes surrounded in blue are present only in MWU205 strain.
Concluding Remarks and Perspectives
In recent years, we have begun to learn how C. violaceum causes severe infection in mammalian hosts, despite its evolution as an environmental free-living bacterium. Evolutionarily, the most relevant event that makes it an opportunistic pathogen is very likely to be the acquisition and/or maintenance of Cpi-1, a pathogenicity island containing the T3SS, which is essential for C. violaceum virulence. This T3SS is important for the pathogenesis of C. violaceum because it causes damage to hepatocytes and promotes the invasion of non-phagocytic cells. Also, it is the signal that triggers activation of the NLRC4 inflammasome, resulting in an effective clearance of the infection by the innate immune system. The widespread occurrence of intact Cpi-1 in many Chromobacterium species (most of them isolated from environmental sources and yet not associated with human infection) raises questions about the potential of these species to be pathogenic for humans. An alternative hypothesis is that the Cpi-1 T3SS contributes to the interaction of Chromobacterium with other organisms. In fact, several Chromobacterium species have been isolated from the roots of plants and are able to suppress plant disease by killing insect larvae or antagonize fungal pathogens. Future studies involving the T3SSs and other virulence factors of C. violaceum will contribute to a better understanding of the pathogenesis of this rare but deadly human pathogen.
Author Contributions
JB and JdS conceived the idea, wrote the manuscript, and prepared the figures and tables. JB performed the update regarding clinical reports of infections. JdS performed the analysis of prevalence and organization of T3SS genes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the São Paulo Research Foundation (FAPESP; grant number 2012/20435-9) and Fundação de Apoio ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da FMRP-USP (FAEPA). JB was supported by a fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02213/full#supplementary-material
References
- Aldridge K. E., Valainis G. T., Sanders C. V. (1988). Comparison of the in vitro activity of ciprofloxacin and 24 other antimicrobial agents against clinical strains of Chromobacterium violaceum. Diagn. Microbiol. Infect. Dis. 10 31–39. [DOI] [PubMed] [Google Scholar]
- Alves de Brito C. F., Carvalho C. B., Santos F., Gazzinelli R. T., Oliveira S. C., Azevedo V., et al. (2004). Chromobacterium violaceum genome: molecular mechanisms associated with pathogenicity. Genet. Mol. Res. 3 k148–161. [PubMed] [Google Scholar]
- Ansari S., Paudel P., Gautam K., Shrestha S., Thapa S., Gautam R. (2015). Chromobacterium violaceum isolated from a wound sepsis: a case study from Nepal. Case Rep. Infect. Dis. 2015:181946. 10.1155/2015/181946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj A., Kumar A., Yadav S., Kaur G., Bala M., Singh N. K., et al. (2016). Isolation and characterization of a novel Gram-negative bacterium Chromobacterium alkanivorans sp. nov., strain IITR-71T degrading halogenated alkanes. Int. J. Syst. Evol. Microbiol. 66 5228–5235. 10.1099/ijsem.0.001500 [DOI] [PubMed] [Google Scholar]
- Baker S., Campbell J. I., Stabler R., Nguyen H. V. M., To D. S., Nguyen D. V., et al. (2008). Fatal wound infection caused by Chromobacterium violaceum in Ho Chi Minh City, Vietnam. J. Clin. Microbiol. 46 3853–3855. 10.1128/JCM.01068-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts H. J., Chaudhuri R. R., Pallen M. J. (2004). An analysis of type-III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 12 476–482. 10.1016/j.tim.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Blackburn M. B., Sparks M. E., Gundersen-Rindal D. E. (2016). The genome of the insecticidal Chromobacterium subtsugae PRAA4-1 and its comparison with that of Chromobacterium violaceum ATCC 12472. Genom. Data 10 1–3. 10.1016/j.gdata.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. I., Lan N. P. H., Qui P. T., Dung L. T., Farrar J. J., Baker S. (2013). A successful antimicrobial regime for Chromobacterium violaceum induced bacteremia. BMC Infect. Dis. 13:4. 10.1186/1471-2334-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. G., Yunos N. Y. (2016). Whole-genome sequencing analysis of Chromobacterium piscinae strain ND17, a quorum-sensing bacterium. Genome Announc. 4:e81–16. 10.1128/genomeA.00081-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. A., Markowitz V. M., Chu K., Palaniappan K., Szeto E., Pillay M., et al. (2016). IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45 507–516. 10.1093/nar/gkw929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Neto J. F., Negretto C. C., Netto L. E. (2012). Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a cys-based redox sensor regulated by thioredoxin. PLOS ONE 7:e47090. 10.1371/journal.pone.0047090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Marshall N. C., Rowland J. L., Mccoy J. M., Worrall L. J., Santos A. S., et al. (2017). Assemble, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15 323–337. 10.1038/nrmicro.2017.20 [DOI] [PubMed] [Google Scholar]
- Du J., Reeves A. Z., Klein J. A., Twedt D. J., Knodler L. A., Lesser C. F. (2016). The type III secretion system apparatus determines the intracellular niche of bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 113 4794–4799. 10.1073/pnas.1520699113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán N., Justo G. Z., Durán M., Brocchi M., Cordi L., Tasic L., et al. (2016). Advances in Chromobacterium violaceum and properties of violacein-its main secondary metabolite: a review. Biotechnol. Adv. 34 1030–1045. 10.1016/j.biotechadv.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Durán N., Justo G. Z., Ferreira C. V., Melo P. S., Cordi L., Martins D. (2007). Violacein: properties and biological activities. Biotechnol. Appl. Biochem. 48 127–133. 10.1042/BA20070115 [DOI] [PubMed] [Google Scholar]
- Durán N., Menck C. F. (2001). Chromobacterium violaceum: a review of pharmacological and industrial perspectives. Crit. Rev. Microbiol. 27 201–222. 10.1080/20014091096747 [DOI] [PubMed] [Google Scholar]
- Galán J. E., Lara-Tejero M., Marlovits T. C., Wagner S. (2014). Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68 415–438. 10.1146/annurev-micro-092412-155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiya H., Murase T., Suzuki M., Shibayama K., Kokumai Y., Watanabe N., et al. (2014). Chromobacterium violaceum nosocomial pneumonia in two Japanese patients at an intensive care unit. J. Infect. Chemother. 20 139–142. 10.1016/j.jiac.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Han X. Y., Han F. S., Segal J. (2008). Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. Int. J. Syst. Evol. Microbiol. 58 1398–1403. 10.1099/ijs.0.64681-0 [DOI] [PubMed] [Google Scholar]
- Hensel M. (2000). Salmonella pathogenicity island 2. Mol. Microbiol. 36 k1015–1023. 10.1046/j.1365-2958.2000.01935.x [DOI] [PubMed] [Google Scholar]
- Hungria M., Astolfi-Filho S., Chueire L. M., Nicolás M. F., Santos E. B., Bulbol M. R., et al. (2005). Genetic characterization of Chromobacterium isolates from black water environments in the Brazilian Amazon. Lett. Appl. Microbiol. 41 17–23. 10.1111/j.1472-765X.2005.01724.x [DOI] [PubMed] [Google Scholar]
- Kämpfer P., Busse H. J., Scholz H. C. (2009). Chromobacterium piscinae sp. nov. and Chromobacterium pseudoviolaceum sp. nov., from environmental samples. Int. J. Syst. Evol. Microbiol. 59 2486–2490. 10.1099/ijs.0.008888-0 [DOI] [PubMed] [Google Scholar]
- Ke L., An K. P., Heng S., Riley M., Sona S., Moore C. E., et al. (2012). Paediatric Chromobacterium violaceum in Cambodia: the first documented case. Trop. Doct. 42 178–179. 10.1258/td.2012.120054 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Choi H. S., Yang S. Y., Kim I. S., Yamaguchi T., Sohng J. K., et al. (2014). Both extracellular chitinase and a new cyclic lipopeptide, chromobactomycin, contribute to the biocontrol activity of Chromobacterium sp. C61. Mol. Plant Pathol. 15 122–132. 10.1111/mpp.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Park J. Y., Han S. H., Lee J. H., Rong X., McSpadden-Gardener B. B., et al. (2011). Draft genome sequence of the biocontrol bacterium Chromobacterium sp. strain C-61. J. Bacteriol. 193 6803–6804. 10.1128/JB.06191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Bittencourt C. I., Astolfi-Filho S., Chartone-Souza E., Santos F. R., Nascimento A. M. A. (2007). Analysis of Chromobacterium sp. natural isolates from different Brazilian ecosystems. BMC Microbiol. 7:58. 10.1186/1471-2180-7-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Bittencourt C. I., Costa P. S., Barbosa F. A., Chartone-Souza E., Nascimento A. M. A. (2011). Characterization of a Chromobacterium haemolyticum population from a natural tropical lake. Lett. Appl. Microbiol. 52 642–650. 10.1111/j.1472-765X.2011.03052.x [DOI] [PubMed] [Google Scholar]
- Maltez V. I., Miao E. A. (2016). Reassessing the evolutionary importance of inflammasomes. J. Immunol. 196 956–962. 10.4049/jimmunol.1502060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltez V. I., Tubbs A. L., Cook K. D., Aachoui Y., Falcone E. L., Holland S. M., et al. (2015). Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity 43 987–997. 10.1016/j.immuni.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P. A., Gundersen-Rindal D., Blackburn M., Buyer J. (2007). Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int. J. Syst. Evol. Microbiol. 57 993–999. 10.1099/ijs.0.64611-0 [DOI] [PubMed] [Google Scholar]
- Martinez R., Velludo M. A. S. L., Santos V. R., Dinamarco P. V. (2000). Chromobacterium violaceum infection in Brazil. A case report. Rev. Inst. Med. Trop. Sao Paulo 42 111–113. 10.1016/j.jcma.2011.08.013 [DOI] [PubMed] [Google Scholar]
- Mcclean K. H., Winson M. K., Fish L., Taylor A., Chhabra S. R., Camara M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology 143 3703–3711. 10.1099/00221287-143-12-3703 [DOI] [PubMed] [Google Scholar]
- Menezes C. B., Tonin M. F., Corrêa D. B., Parma M., de Melo I. S., Zucchi T. D., et al. (2015). Chromobacterium amazonense sp. nov. isolated from water samples from the Rio Negro, Amazon, Brazil. Antonie Van Leeuwenhoek 107 1057–1063. 10.1007/s10482-015-0397-3 [DOI] [PubMed] [Google Scholar]
- Miki T., Akiba K., Iguchi M., Danbara H., Okada N. (2011). The Chromobacterium violaceum type III effector CopE, a guanine nucleotide exchange factor for Rac1 and Cdc42, is involved in bacterial invasion of epithelial cells and pathogenesis. Mol. Microbiol. 80 1186–1203. 10.1111/j.1365-2958.2011.07637.x [DOI] [PubMed] [Google Scholar]
- Miki T., Iguchi M., Akiba K., Hosono M., Sobue T., Danbara H., et al. (2010). Chromobacterium pathogenicity island 1 type III secretion system is a major virulence determinant for Chromobacterium violaceum-induced cell death in hepatocytes. Mol. Microbiol. 77 855–872. 10.1111/j.1365-2958.2010.07248.x [DOI] [PubMed] [Google Scholar]
- Miki T., Okada N. (2014). Draft genome sequence of Chromobacterium haemolyticum causing human bacteremia infection in Japan. Genome Annouc. 2:e1047–14. 10.1128/genomeA.01047-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanayakkara G. M., Pethiyagoda P., Jayasinghe P. N., Premachandra U. (2008). Chromobacterium violaceum infection in a provincial hospital in Sri Lanka. Ceylon Med. J. 53 156–157. [PubMed] [Google Scholar]
- Nazir R., Mazurier S., Yang P., Lemanceau P., van Elsas J. D. (2017). The ecological role of type three secretion systems in the interaction of bacteria with fungi in soil and related habitats is diverse and context-dependent. Front. Microbiol. 8:38. 10.3389/fmicb.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Inokuchi R., Shinohara K., Matsumoto A., Ono Y., Narita M., et al. (2013). Chromobacterium haemolyticum-induced bacteremia in a healthy young man. BMC Infect. Dis. 13:406. 10.1186/1471-2334-13-406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant N. D., Acharya S. P., Bhandari R., Yadav U. N., Saru D. B., Sharma M. (2017). Bacteremia and urinary tract infection caused by Chromobacterium violaceum: case reports from a tertiary care hospital in Kathmandu, Nepal. Case Rep. Med. 2017:7929671. 10.1155/2017/7929671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant N. D., Sharma M., Khatiwada S. (2015). Asymptomatic bacteriuria caused by Chromobacterium violaceum in an immunocompetent adult. Case Rep. Med. 2015:652036. 10.1155/2015/652036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previato-Mello M., Meireles D. A., Netto L. E. S., da Silva Neto J. F. (2017). Global transcriptional response to organic hydroperoxide and the role of OhrR in the control of virulence traits in Chromobacterium violaceum. Infect. Immun. 85:e00017–17. 10.1128/IAI.00017-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J. L., Short S. M., Bahia A. C., Saraiva R. G., Dong Y., Kang S., et al. (2014). Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLOS Pathog. 10:e1004398. 10.1371/journal.ppat.1004398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal B. H., Ding L., Holland S. M. (2003). Phagocyte NADPH oxidase, but not inducible nitric oxide synthase, is essential for early control of Burkholderia cepacia and Chromobacterium violaceum infection in mice. Infect. Immun. 71 205–210. 10.1128/IAI.71.1.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivendra R., Tan S. H. (1977). Pathogenicity of nonpigmented cultures of Chromobacterium violaceum. J. Clin. Microbiol. 5 514–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath P. H., Whelan J. P., Bhagwan- Singh R., Edwards D. (1953). Fatal infection by Chromobacterium violaceum. Lancet 265 276–277. 10.1016/S0140-6736(53)91132-5 [DOI] [PubMed] [Google Scholar]
- Soby S. D. (2017a). Draft genome sequence of Chromobacterium aquaticum CC-SEYA-1, a nonpigmented member of the genus Chromobacterium. Genome Annouc. 5:e1661–16. 10.1128/genomeA.01661-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soby S. D. (2017b). Draft genome sequence of Chromobacterium pseudoviolaceum LMG 3853T, an enigmatic member of the genus Chromobacterium. Genome Announc. 5:e1632–16. 10.1128/genomeA.01632-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soby S. D., Gadagkar S. R., Contreras C., Caruso F. L. (2013). Chromobacterium vaccinii sp. nov., isolated from native and cultivated cranberry (Vaccinium macrocarpon Ait.) bogs and irrigation ponds. Int. J. Syst. Evol. Microbiol. 63 1840–1846. 10.1099/ijs.0.045161-0 [DOI] [PubMed] [Google Scholar]
- Swain B., Otta S., Sahu K. K., Panda K., Rout S. (2014). Urinary tract infection by Chromobacterium violaceum. J. Clin. Diagn. Res. 8 DD01–DD02. 10.7860/JCDR/2014/9230.4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem L. R., Swem D. L., O’Loughlin C. T., Gatmaitan R., Zhao B., Ulrich S. M., et al. (2009). A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35 143–153. 10.1016/j.molcel.2009.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S. B., Natarajan G., Rahim M. N. A., Tan H. T., Chung M. C. M., Ting Y. P., et al. (2013). Enhancing gold recovery from electronic waste via lixiviant metabolic engineering in Chromobacterium violaceum. Sci. Rep. 3:2236. 10.1038/srep02236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderMolen K. M., Mcculloch W., Pearce C. J., Oberlies N. H. (2011). Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J. Antibiot. 64 525–531. 10.1038/ja.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos A. T. R., Almeida D. F., Hungria M., Guimarães C. T., Antônio R. V., Almeida F. C., et al. (2003). The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. U.S.A. 100 11660–11665. 10.1073/pnas.1832124100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vöing K., Harrison A., Soby S. D. (2015a). Draft genome sequence of Chromobacterium vaccinii, a potential biocontrol agent against mosquito (Aedes aegypti) larvae. Genome Announc. 3:e477–15. 10.1128/genomeA.00477-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vöing K., Harrison A., Soby S. D. (2015b). Draft genome sequences of three Chromobacterium subtsugae isolates from wild and cultivated cranberry bogs in southeastern Massachusetts. Genome Announc. 3:e998–15. 10.1128/genomeA.00998-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vöing K., Harrison A., Soby S. D. (2017). Draft genome sequence of Chromobacterium subtsugae MWU12-2387 isolated from a wild cranberry bog in Truro, Massachusetts. Genome Announc. 5:e1633–16. 10.1128/genomeA.01633-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hinshaw K. C., Macdonald S. J., Chandler J. R. (2016). Draft genome sequence of Chromobacterium violaceum strain CV017. Genome Announc. 4:e80–16. 10.1128/genomeA.00080-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Li Y. H. (2011). Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J. Chin. Med. Assoc. 74 435–441. 10.1016/j.jcma.2011.08.013 [DOI] [PubMed] [Google Scholar]
- Young C. C., Arun A. B., Lai W. A., Chen W. M., Chou J. H., Shen F. T., et al. (2008). Chromobacterium aquaticum sp. nov., isolated from spring water samples. Int. J. Syst. Evol. Microbiol. 58 877–880. 10.1099/ijs.0.65573-0 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y. N., Lu Q., Xu H., et al. (2011). The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477 596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zhou S., Guo X., Wang H., Kong D., Wang Y., Zhu J., et al. (2016). Chromobacterium rhizoryzae sp. nov., isolated from rice roots. Int. J. Syst. Evol. Microbiol. 66 3890–3896. 10.1099/ijsem.0.001284 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.