Abstract
The idea of eliminating the use of fertilizers which are sometimes environmentally unsafe is slowly becoming a reality because of the emergence of microorganisms that can serve the same purpose or even do better. Depletion of soil nutrients through leaching into the waterways and causing contamination are some of the negative effects of these chemical fertilizers that prompted the need for suitable alternatives. This brings us to the idea of using microbes that can be developed for use as biological fertilizers (biofertilizers). They are environmentally friendly as they are natural living organisms. They increase crop yield and production and, in addition, in developing countries, they are less expensive compared to chemical fertilizers. These biofertilizers are typically called plant growth-promoting bacteria (PGPB). In addition to PGPB, some fungi have also been demonstrated to promote plant growth. Apart from improving crop yields, some biofertilizers also control various plant pathogens. The objective of worldwide sustainable agriculture is much more likely to be achieved through the widespread use of biofertilizers rather than chemically synthesized fertilizers. However, to realize this objective it is essential that the many mechanisms employed by PGPB first be thoroughly understood thereby allowing workers to fully harness the potentials of these microbes. The present state of our knowledge regarding the fundamental mechanisms employed by PGPB is discussed herein.
Keywords: Biocontrol, Biofertilizer, Bioremediation, Phytohormones, Siderophore, Sustainable agriculture
Introduction
Plant growth promoting rhizobacteria
Bacteria are the most ubiquitous organisms known and tend not to be evenly distributed. That is, the concentration of bacteria around the roots of plants is generally much higher than the concentration found in the bulk soil. This reflects the fact that the small molecules (e.g. sugars, organic acids and amino acids) exuded from plant roots in large amounts (i.e. 5–30% of all photosynthetically fixed carbon) are commonly used as food sources for bacteria (Babalola 2010; Carvalhais et al. 2015).
Many of the bacteria that are found around plant roots (the rhizosphere) have the ability to facilitate plant growth and consequently are called plant growth-promoting bacteria or PGPB. In addition to organisms that inhabit the rhizosphere, PGPB also include various strains of rhizobia that form nodules on the roots of specific plants (legumes) and endophytes that can exist within the interior tissues of a plant (Santoyo et al. 2016). Today, several PGPB have been commercialized as either biocontrol agents or biofertilizers (Calvo et al. 2014; Reed and Glick 2013). This review looks at the different known mechanisms used by PGPB and comments regarding the relative importance of some of these mechanisms.
Mechanisms of actions of PGPB
PGPB can promote plant growth by both direct and indirect mechanisms (Glick 1995). Direct mechanisms are defined as employing those bacterial traits that result in the direct promotion of plant growth. They include the production of auxin, ACC deaminase, cytokinin, gibberellin, nitrogen fixation, phosphorous solubilization, and sequestration of iron by bacterial siderophores. Indirect mechanisms refer to bacterial traits that inhibit the functioning of one or more plant pathogenic organisms both fungi and bacteria. Indirect mechanisms include ACC deaminase, antibiotics, cell wall degrading enzymes, competition, hydrogen cyanide, induced systemic resistance, quorum quenching and siderophores. In addition to the aforementioned methods used by PGPB to control phytopathogens, biocontrol of some bacterial phytopathogens may be obtained through the selective use of bacteriophages (Balogh et al. 2010; Frampton et al. 2012). Bacteriophages have been used to control phytopathogens such as Erwinia amylovora (Boulé et al. 2011), and Ralstonia solanacearum (Fujiwara et al. 2011). Commercial applications such as Agriphage which is used for the control of Pseudomonas syringae pv. tomato and Xanthomonas campestris pv. Vesicatoria (Iriarte et al. 2007).
Various PGPB often possess one or more of the above mentioned traits so that in addition to certain bacteria being more suitable in specific environments (e.g. high versus low temperatures or specific pH ranges), there are a wide range of PGPB, each differing in activities under different environmental and soil conditions. In fact, no single organism has the ability to make use of all the available mechanisms that could be used to promote plant growth (Saharan and Nehra 2011). In addition, several PGPB inoculants that enhance plant growth through at least one of these mechanisms have been commercialized (Reed and Glick 2013).
Direct mechanisms
Auxin
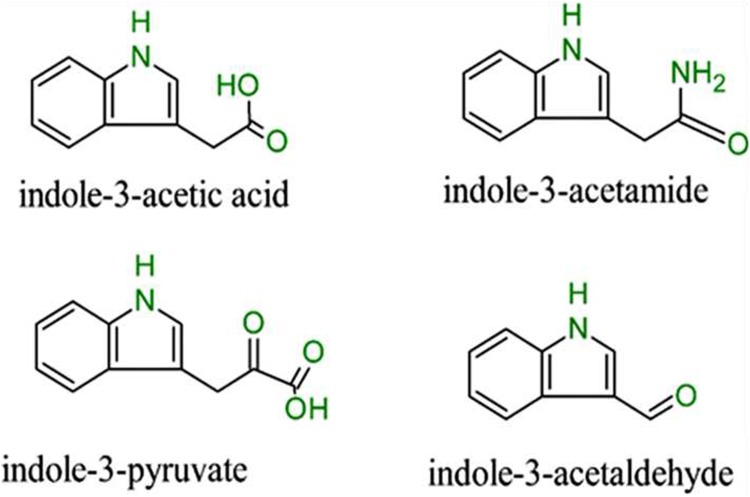
The most reported mechanism predominantly used to explain the positive PGPB effects on plant growth is their ability to produce auxin. Patten and Glick (1996) reported that about 80% of rhizosphere microbes could synthesize and release auxin as a secondary metabolite. Quite a number of known auxins occur naturally with indole-3-acetic acid (IAA) standing out as the most relevant (Spaepen et al. 2007b). In fact throughout the literature, auxin is often interchanged with IAA. Other similar compounds that have been reported to have auxin activity, some of which are actively involved in the anabolism of IAA are indole-3-acetamide, indole-3-pyruvate, and indole-3-acetaldehyde shown in Fig. 1.
Fig. 1.
Some known derivatives of IAA
Inactive forms of auxin include 4-chloroindole-3-acetic acid and other forms which can be conjugated with sugars, alcohols, amino acids, and glycoproteins (Korasick et al. 2013). Auxins function in geotropism and phototropism, vascular tissue differentiation, apical dominance, root initiation (lateral and adventitious), cell division, stem and root elongation (Grobelak et al. 2015). Rhizobacterial IAA changes plant auxin pools, ultimately increasing root length and surface area, and in the process increasing the level of root exudates available for uptake by plants as shown in the work of Ali et al. (2010).
IAA/Auxin synthesis occurs dominantly in the shoot apex, transported to the root apical meristem via the shoot vascular cambium and accrued in the quiescent center (QC), which is the columella initials and lateral root cap (Brunoud et al. 2012; Petrášek and Friml 2009). All known carrier proteins of auxin are PINs, ABCB and AUX1/LAX which are activated by the triggering of a signaling cascade involving the F-box protein Transport Inhibitor Response 1 (TIR1) auxin receptor (Petrášek and Friml 2009). During embryogenesis, there is the control of primary root development by the stem cell niche. Beyond embryogenesis, auxin-plethora complex stabilizes the positioning as well as maintenance of the niche. But plethora (PLT) which is the downstream transcription factor of auxin acts independently to the transcription factors shortroot (SHR) and scarecrow (SCR) (Aida et al. 2004; Grieneisen et al. 2007). When SHR moves to the nuclei of adjacent cells and activation of SCR is effected which allows the cell differentiation inhibition and maintenance of surrounding stem cells by down regulation of cytokinin sensing (Moubayidin et al. 2013). Alleviation of cytokinin sensing in the transit zone regulates PINs negatively leading to release of auxin again. The efflux of auxin causes differentiation to be initiated again in the cell (Ioio et al. 2008; Marhavý et al. 2011).
Different plants are sensitive to different levels of auxin; this includes different plant species and cultivars as well as plants of different ages (Cheng et al. 2013). In addition, the optimal level of auxin that is effective in promoting plant growth is approximately five orders of magnitude lower for roots when compared with the shoots (Glick 2012). Moreover, the concentration of plant-synthesized auxin determines its effect on the stimulation or inhibition of plant growth. Plant’s auxin concentrations may be either suboptimal or optimal so that the addition of bacterial auxin that can be taken up by the plant may change the hormone level in the plant to either optimal or supraoptimal (Glick 2012). Thus, bacterial IAA produced by a PGPB may either stimulate root development in cases where the plant’s concentration is suboptimal, or inhibit root development in cases where the auxin level is already optimal (Spaepen et al. 2007a).
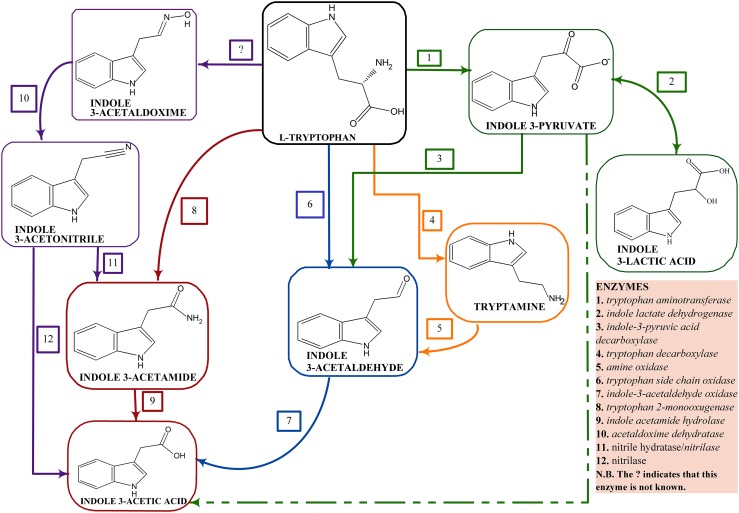
Most auxin/IAA is synthesized from the amino acid tryptophan present in plant root exudates at varying low concentrations based on the plant’s genotype. IAA appears to be synthesized by at least three different biosynthetic pathways with each pathway being named for a key intermediate within the pathway. These pathways include: the indole pyruvic acid (IPyA) pathway, the indole acetamide (IAM) pathway, the indole acetaldoxime (IAOx)/indoleacetonitrile (IAN) pathway (Duca et al. 2014), the indole acetaldehyde (IAH) pathway, and the tryptamine pathway (Fig. 2). It should be noted that various PGPB can have one, two or even three functional IAA biosynthesis pathways suggesting that the synthesis of IAA is clearly very important in the life and functioning of the bacterium. A schematic representation of bacterial IAA biosynthesis from tryptophan is shown in Fig. 2.
Fig. 2.
Schematic representation of the known pathways of IAA synthesis in bacteria. The dashed lines represent non-enzymatic reactions
The chromosome or the plasmid hots the auxin biosynthetic genes and this difference in hosts affects the IAA level with plasmid hosts found in many copies. A case study is that observed between P. savastanoi pv. savastanoi and P. syringae pv. Syringae where the genes are located on the plasmid and chromosomal DNA respectively (Aragón et al. 2014). The IAM pathway is majorly attributed to phytopathology, while the IPA pathway is connected to epiphytic and rhizosphere fitness. The ipdC gene which is one of the genes involved in IAA synthesis in PGPB are regulated by the amount of IAA produced which serve as a positive feed-back in up-regulating the expression of the gene (Spaepen and Vanderleyden 2011). This positive feed-back was reported in A. brasilience Sp245 as the first PGPB to show this observation (Broek et al. 2005). IAA is actively involved in its biosynthetic genes as it determines the activation, inactivation, over expression or under expression of these genes. Other regulatory mechanisms of IAA synthesis are reported in the work of Glick (2015b).
ACC deaminase
The presence of ethylene hormone in all higher plants, characterizes its importance in the modulation of normal cell development and plant growth as well as its significant role in helping plants to combat various levels of stress (Abeles et al. 1992). Nearly all plant tissues and stages of development are affected by ethylene. Ethylene synthesis in a particular plant is affected by the presence and concentration of other plant hormones, temperature, gravity, light, nutrition, and the presence of various degrees of biotic/abiotic stress which the plant may be subjected (Gamalero and Glick 2015). An increased concentration of ethylene in plants is a response to various stresses including the presence of metals, extreme temperatures, chemicals (both organic and inorganic), too much or too little water, ultraviolet light, insect and nematode damage, fungal and bacterial pathogens as well as mechanical wounding (Ali et al. 2014; Barnawal et al. 2012). Its production more than its threshold level by the action of ACC oxidase enzyme in plant tissues causes “stress ethylene” which affects the root and shoot development in plants. Colonization of “stress ethylene” plant rhizosphere by ACC deaminase producing PGPB help to alleviate this situation and restores normal plant development. The synthesis of “stress ethylene” includes the ethylene being synthesized in two peaks (Glick et al. 2007) with the first peak being a small fraction of the magnitude of the second peak. The first peak, which is small and difficult to measure, consumes the much of the existing 1-aminocyclopropane-1-carboxylate (ACC) in stressed plants and initiates the transcription of genes that encode plant defensive/protective proteins. The second, much larger, ethylene peak which occurs as a result of increased level of ACC in response to stress is detrimental to subsequent plant growth, initiating processes in the plant such as senescence, chlorosis and leaf abscission (Yim et al. 2013). The upregulated concentration of plant ethylene significantly worsen the effects of the original stress that triggered the ethylene response so that any treatment that down regulates the concentration of the second peak of stress ethylene should also be able to reduce/stop the plant damage resulting from the stress. In this regard, bacteria that express ACC deaminase, by lowering plant ACC levels (and subsequently plant ethylene levels) can decrease the detrimental effect on plants from different stresses (Glick 2014). The ACC is being converted by ACC deaminase in the PGPB to α-Ketobutyrate and ammonia (Fig. 3). Prior application of ACC deaminase-containing PGPB to plants typically lowers the concentration of ethylene synthesized by the plants in response to pathogen stress and thereby decreases the damage that the plant incurs from the pathogen (Glick 2012; Toklikishvili et al. 2010). To date, this approach has been shown to be quite effective with a variety of plants and with numerous different pathogens. However, these results have so far been limited to greenhouse and growth chamber experiments. Proteins encoding the gene that regulates ethylene biosynthesis are classified as the ETR proteins. The first member of these proteins was identified in mutant Arabidopsis in the works of Bleecker et al. (1988).
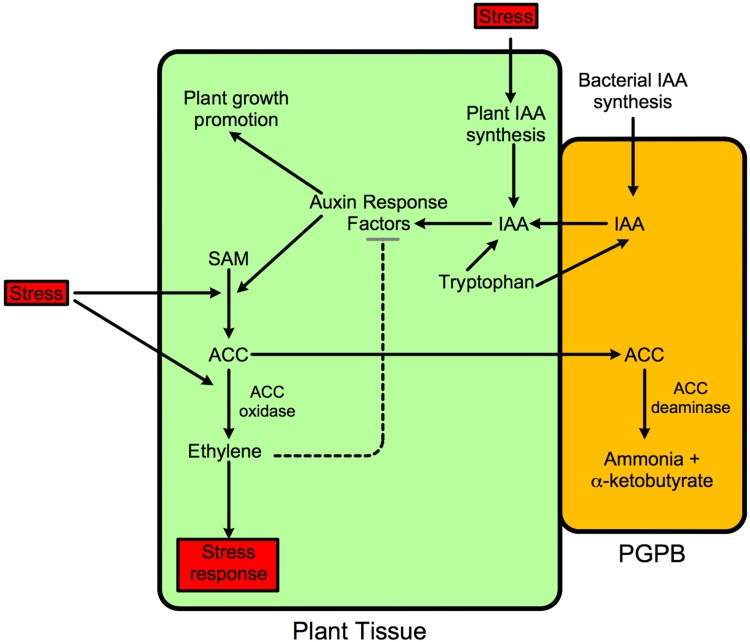
Fig. 3.
Schematic representation of the stimulation of plant growth by a PGPB containing ACC deaminase. Stress increases both IAA and ethylene synthesis within the plant ultimately decreasing plant growth. With PGPB strains that contain ACC deaminase, ethylene levels are decreased thereby relieving some of the growth inhibition that increased ethylene causes. This enables the bacterial IAA to continue to promote plant growth. Thus, PGPB that produce both IAA and ACC deaminase lower the extent that a wide range of environmental stresses inhibit plant growth. SAM S-adenosyl-methionine. These PGPB protect plants against the inhibitory effects of ethylene-producing stresses that include drought, flooding, temperature extremes, high salt, metal and organic contaminants, insect and nematode predation, and both fungal and bacterial phytopathogens
It was previously suggested that PGPB bound to plant roots (or found within plant tissues) can take up some of the tryptophan exuded by plants and convert the tryptophan to IAA, which is then secreted by the bacterium and taken up by the plant (Glick 2014). The increased amount of IAA can both facilitate plant growth and activate the transcription of the plant enzyme ACC synthase resulting in an increase in the level of ACC and hence the amount of ethylene within the plant. Thus, PGPB that synthesize IAA from plant tryptophan can both promote plant growth and inhibit plant growth (via the action of the ethylene that is eventually produced). Fortunately, PGPB that contain ACC deaminase decrease the level of (newly formed) ACC in the plant by the action of ACC deaminase enzyme, enabling bacterial IAA to promote plant growth without significantly inhibiting plant growth. In addition, by decreasing the level of ethylene in the plant, ethylene inhibition of auxin signal transduction is lowered allowing the bacterial auxin to further increase plant growth. Thus, by the down-regulation of plant ethylene levels, ACC deaminase facilitates the functioning of bacterial IAA. This model is depicted schematically in Fig. 3. The ACC is ultimately converted to ammonia and α-ketobutyrate.
Cytokinins
Cytokinins are widely distributed in algae, bacteria, and higher plants; however, relatively little information is available on the roles of bacteria-produced cytokinins. They are produced in the root tips and transported through the xylem to the shoot by translocation. Cytokinins control cell differentiation in plant meristematic tissues (De Rybel et al. 2016). Kinetin was the first cytokinin to be discovered, however, it is considered a “synthetic” cytokinin due to its source which is yeast, not plants (Miller et al. 1955). The widely known form in plants is zeatin, which was firstly isolated from corn (Zea mays) (Schäfer et al. 2015). In plants, cytokinins are primarily synthesized in roots although they are distributed throughout the plant. There are two groups of cytokinins based on their structure, the adenine type and the phenyl urea type. The adenine type includes kinetin and zeatin, while the phenyl urea type includes diphenyl urea and thidiazuron (Fig. 4). Cytokinins regulate apical dominance, cell division, root elongation, seed germination, xylem and chloroplast differentiation, flower and fruit development, nutritional signaling, leaf senescence, and plant-pathogen interactions (Sakakibara 2006).
Fig. 4.
a Adenine-derived cytokinins. b Diphenylurea-type cytokinins
Expression of cytokinin genes is relatively evident in several PGPB, and their addition to growing plants can largely alter the plant’s phytohormone composition. Cytokinin content and plant growth has been increased with the inoculation of lettuce with Bacillus subtilis (Arkhipova et al. 2005). Roles played by cytokinins in some experiments are based on the addition of purified hormones to individual plants. From these experiments, cytokinins have been shown to carry out senescence delay by accumulation of chlorophyll, cell tissue formation, root development, elongation and hair formation, initiation of stem, and expansion of leaves (Sakakibara 2006).
In one particular instance, a genetically engineered strain of Sinorhizobium meliloti which overproduces cytokinin was tested for the ability to protect alfalfa plants against the senescence resulting from drought stress (Xu et al. 2012). The transformed bacterium production of cytokinin was approximately five times more than the production by the wild type. Following a period of extreme drought stress, there was a tremendous increase in size of the alfalfa plants inoculated with the transformed strain compared to the plants inoculated with the non-transformed strain. This experiment indicates that rhizobial strains synthesizing higher than normal levels of cytokinin are able to improve the drought tolerance of alfalfa.
Mimicking the same actions in primary root development, auxin is being antagonized by cytokinin by preventing the development of lateral root system (Chang et al. 2013; Schaller et al. 2015). This is majorly shown in a study with Arabidopsis. While it is not decreased in the transition zone, the level of sensitivity to it seems reduced (Bielach et al. 2012; Marhavý et al. 2014) just as it was in the cytokinin inhibition in the quiescent center by SCR.
Gibberellin (GA)
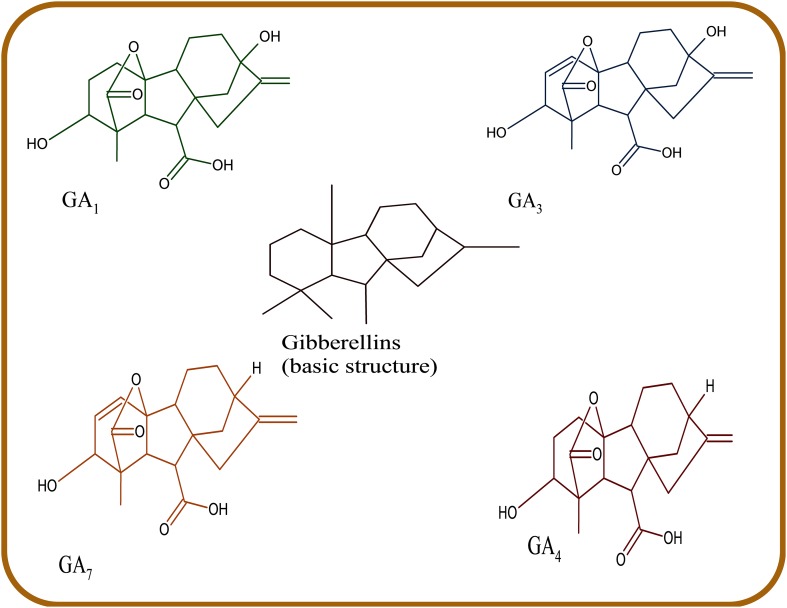
Gibberellins include a large group of tetracyclic diterpenoid carboxylic acids having either C20 or C19 carbon skeletons (Dodd et al. 2010; Hedden and Thomas 2012). 136 gibberellin structures have been identified and are represented as GA1–GA136 (Hedden and Thomas 2012). Only 4 GAs have been identified in bacteria; GA1,GA3, GA4, and GA20 (Gupta et al. 2016), with GA1 and GA4 being the most active (Nelson and Steber 2016). Gibberellins are known for growth stimulation and activation of important growth processes including stem elongation, seed germination, flowering, fruit setting (Zaidi et al. 2015), improve photosynthesis rate, and chlorophyll content (Khan et al. 2015; You et al. 2012). Along with other phytohormones, they are transducers of elicitor signals. However, despite the enormous abundance of different gibberellins, the biological activity and role of gibberellin molecules are largely unknown. GA1, GA3, GA4, and GA7 (Fig. 5) still remains the predominantly known bioactive forms.
Fig. 5.
The chemical structure of GAs
PGPB production of GAs has been observed in the following genera Achromobacter xylosoxidans, Gluconobacter diazotrophicus, Acinetobacter calcoaceticus, Rhizobia, Azotobacter spp., Bacillus spp., Herbaspirillum seropedicae, and Azospirillum spp. (Deka et al. 2015; Dodd et al. 2010). The biochemistry of gibberellins in bacteria is similar to that of plants with some small differences. The absence of GAs is readily observed as a reduction of the lateral root number and length (Dodd et al. 2010). As is the case for cytokinins, most of the currently attributed functional role of bacterially produced gibberellins in plant growth promotion is due to plant’s response to the exogenic addition of purified gibberellins to growing plants. Moreover, while the gibberellin biosynthetic pathways in plants and fungi are known, the same cannot be said about bacteria. Gibberellins can induce shoot growth and development and also inhibit root growth through the actions of the gibberellin signaling system, the, DELLA repressor which activates gibberellin-inducing genes (Martínez et al. 2016; Minguet et al. 2014; Nelson and Steber 2016; Wang et al. 2015).
Nitrogen fixation
Nitrogen is one of the important nutrients essential for the growth of all living organisms including plants and bacteria. The observation of nitrogen deficiency in soil has led to the use of large amounts of nitrogenous fertilizers in order to make up for the necessary plant requirements to achieve maximum plant yield in most soils (Zhang et al. 2015). Despite nitrogen’s abundance in the earth’s atmosphere, about 78%, this form of gaseous nitrogen [N2(g)] is not readily accessible to most organisms, i.e. it is not suitable for plant assimilation until it is first converted to ammonia (Baas et al. 2014). This conversion to ammonia requires a significant amount of energy because of the stability of the triple bond in N2(g). This energy may be provided at the expense of fossil fuels, through biological nitrogen fixation at the expense of ATP (Fig. 6), and through other mechanisms of nitrogen input in terrestrial systems. Broad range of nitrogen fixing bacteria have been identified including a number of organisms that fix nitrogen symbiotically with specific plants (mostly legumes). Examples of symbiotic nitrogen fixers are Rhizobium, Sinorhizobium, Azorhizobium, Allorhizobium, Mesorhizobium, Bradyrhizobium, Frankia, Azoarcus, Achromobacter, Burkholderia, and Herbaspirillum (Babalola 2010; Pérez-Montaño et al. 2014; Turan et al. 2016).
Fig. 6.
Biological nitrogen fixation driven by ATP hydrolysis
Rhizobia are gram-negative bacteria that form symbiotic relationships majorly with legumes; each rhizobial-plant interaction is specific. Aside legumes, some cereal crops have been reported to be colonized by rhizobia as some studies have been carried out to investigate nitrogen fixation in some cereal crops by rhizobial bacteria (Geddes et al. 2015; Oldroyd and Dixon 2014). Rhizobial bacteria colonize plant root cells and initiate a complex trend of developmental changes that lead root nodule formation (Gage 2004). In the root nodule, the bacteria exists as a bacteroid without a cell wall, fixing atmospheric nitrogen by means of the nitrogenase enzyme and producing ammonia. In this mutualistic relationship, the plant on its own part provides the bacterium with fixed carbon (organic acids) from photosynthesis which the bacterium requires for growth while as it continues to fix nitrogen in the nodules.
Various rhizobial strains have been used commercially to provide fixed nitrogen to crop plants for many years, and in numerous locales worldwide. On the other hand, while there are, numerous bacteria other than rhizobia that have the ability to fix atmospheric nitrogen, in practice most free-living bacteria fix only a limited amount of nitrogen and are therefore not employed commercially.
One unsuitable side reaction in the activities of the nitrogenase enzyme in nitrogen fixation is the reduction of H+ to H2 (hydrogen gas) because the hydrogen gas produced is lost to the atmosphere leading to a waste of the ATP expended in its production (Fig. 6). This side reaction significantly lowers the overall efficiency of the nitrogen-fixing process by approximately 30%. On the contrary, some strains of rhizobia have an enzyme called hydrogenase (Adams et al. 1981) that can take retrieve the lost H2 from the atmosphere and convert it back into H+ for the production of ATP which can be used for more nitrogen fixation. These strains help to conserve energy while fixing nitrogen at the same time.
Phosphate solubilization
Bacteria that solubilize phosphorus are referred to as phosphate solubilizing bacteria (Alori et al. 2017). They supply phosphate in a more acceptable way to the plants and are not deleterious to the environment. They convert insoluble organic and inorganic phosphate to a form which can be readily accessible to plants. Environmental conditions, plant and soil conditions, and bacterial strains all affect the actions of phosphate solubilizers (Gupta et al. 2015). According to Banerjee et al. (2005), the most powerful phosphate solubilizers are from the genera Bacillus, Rhizobium, and Pseudomonas, as well as non-symbiont nitrogen fixers such as Azotobacter and Azospirillum (Saharan and Nehra 2011).
The principal mechanism of inorganic phosphate solubilization is the use of mineral-dissolving compounds like hydroxyl ions, organic acids, protons, siderophores, and carbon dioxide (CO2) (Rodríguez and Fraga 1999). Organic acids produced together with their carboxyl and hydroxyl ions chelate cations or reduce the pH resulting in the release of phosphates (Khosro 2012). The organic acids which are either gluconic or keto gluconic acids are produced and excreted by phosphate solubilizing bacteria resulting in the acidification of the microbial cells and their surroundings, hence, phosphate ions are released. The lowered pH results from the production of proton/bicarbonate release and gaseous “O2/CO2” exchanges (Sharma et al. 2017). Thus, the pH of the rhizosphere and phosphorous availability are inversely connected.
The value of organic phosphorus in soil can be as high as 30–50%. The main sources of organic phosphorus in the soil are organic materials in the form of inositol hexaphosphate (phytate). Phytate (Fig. 7) is generally not biologically available to plants because plant roots produce very low amount of phytase enzyme which breaks down phytate. However, many PGPB can solubilize phytate. Other organic phosophorus compounds include phosphomonoesters, phosphodiesters, phospholipids, nucleic acids, and phosphotriesters (Rodríguez and Fraga 1999). Most of these organic compounds are high molecular-weight materials that must be broken down to lower molecular weight organic phosphate, before they can be assimilated by the cell (Peix et al. 2001). In fact, the term phosphorus mineralization refers to the solubilization of organic phosphorus and the degradation of the remaining portions of the molecule which is triggered by the unavailability of sufficient phosphate in the soil.
Fig. 7.
The chemical structure of phytate
Siderophores
Siderophores are small peptide molecules that have side chains and functional groups to which ferric ions can bind (Goswami et al. 2016). They are iron chelators that serve as iron carriers and have a high affinity for some ligands. Quite a large number of them have been screened and used from microbes and they can also be species-specific (Sandy and Butler 2009). Siderophore-producing microbes can prevent or lessen pathogen proliferation by reducing the amount of iron that is available to a pathogen (Shen et al. 2013). PGPB that synthesize siderophores prevent the proliferation of phytopathogens by secreting siderophores with an extremely high affinity for iron. These siderophores bind tightly to most of the Fe3+ that is present in the rhizosphere of the host plant taking up the bound iron into either the PGPB or the host plant. This prevents any fungal and bacterial pathogens in the host plant rhizosphere, where the biocontrol PGPB is bound, from acquiring enough iron for their growth. Thus, the pathogens are unable to proliferate because of a lack of iron, causing them to lose the ability to act as pathogens. The effectiveness of this method of biocontrol is based on the fact that PGPB siderophores have a much higher affinity for iron (typically by many orders of magnitude) than fungal siderophores (Kloepper et al. 1980).
Activities of siderophores as iron chelaters have been shown in different studies such as siderophores from Chryseobacterium spp. C138 which were effective in the supply of iron in tomato plants when delivered to the roots (Radzki et al. 2013), another case is seen in the supplementation of Pseudomonas strains which showed significant increase in germination and plant growth (Sharma and Johri 2003), another effect was seen in the work of Sharma and Johri (2003).
Iron uptake
Iron has an important role to play in the plant photosynthetic system as it is an integral part of the light-absorbing chlorophyll and is also involved in a wide range of different biosynthetic mechanisms. However, the amount of soluble iron present is often not sufficient for maximal crop yields.
Despite the fact that iron is one of the most abundant elements on the earth’s surface, plants and many soil microbes cannot readily absorb enough iron for their growth because of the insolubility of the iron, ferric (Fe3+) hydroxides, which is only slightly soluble and cannot be readily conveyed into the cells (Ganz 2013; Saha et al. 2013). To solve this problem, some bacteria, fungi and plants secrete low molecular mass (~ 400–1000 Da), specialized low molecular weight iron-binding molecules called siderophores into the soil to scavenge iron (DalCorso et al. 2013; Saha et al. 2013). In particular, siderophores produced by PGPB bind to Fe3+ with an exceptionally high affinity (i.e., Kd = 10−20–10−50). Once bound, the now soluble iron-siderophore complex is taken up by specific receptors on the surfaces of bacteria or plants, internalized and then following either reduction to the ferrous state (Fe2+) or cleavage of the siderophore molecule, the iron is released from the siderophore (Saha et al. 2013). Typically, siderophores produced by PGPB have a much higher affinity for iron than siderophores produced by either plants or fungi so that siderophores from PGPB can sequester even minute amounts of iron (Saha et al. 2016).
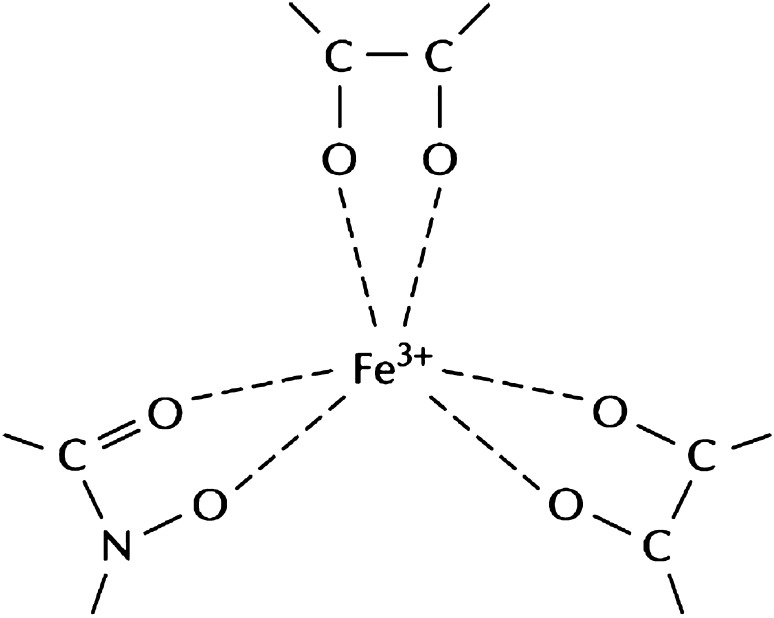
Siderophores are low molecular weight molecules with three iron-binding groups connected by a flexible backbone (Saha et al. 2013). Two oxygen atoms are connected to each functional group, or sometimes, nitrogen, that bind to iron. These functional groups are bidentate (Fig. 8), and trivalent ferric iron can successfully take up three of these groups thereby forming a six-coordinate complex (Glick 2015c). The functional groups on microbial siderophores are majorly hydroxamates or catecholates; or other functional groups such as carboxylate, citrate or ethylenediamine moieties (Laschat et al. 2017). These functional groups can be present in a combined form on a single siderophore molecule. Hydroxamate-type siderophores are common with fungi, while catecholates, which bind iron more tightly than hydroxamates, are common in bacterial siderophores (Glick 2015c; Saha et al. 2016). Linear hydroxy- and amino-substituted iminocarboxylic acids e.g. mugineic acid and avenic acid are plant siderophores, they tend to bind iron more efficiently than bacterial siderophores (Glick 2015c). Other negatively charged molecules have lower affinity for iron than bacterial siderophores. In addition, some other trivalent and divalent metal ions also bind bacterial siderophores, though with a much lower affinity.
Fig. 8.
Schematic representation of three bidentate groups of a siderophore molecule binding to iron
Indirect mechanisms
On average, various plant diseases reduce plant yields by around 10%/year in more developed countries and by about 20%/year in less developed countries of the world (http://www.fao.org/home/en/). In an effort to decrease the widespread use of chemicals as a means of preventing phytopathogen damage to plants, scientists have been developing the use of certain environmentally friendly PGPB as biocontrol agents (Glick and Bashan 1997; Lucy et al. 2004) with many of these organisms already available commercially.
Antibiotics
The major mechanism that used by PGPB to counter deleterious effects of phytopathogens is the synthesis of one or more antibiotics (Couillerot et al. 2009; Haas and Keel 2003; Raaijmakers and Mazzola 2012). However, an antibiotic that is recognised to control a pathogen thereby preventing plant damage from that pathogen might not be as such effective against another pathogen on the same plant, and the antibiotic-synthesizing PGPB may show varying differences in its actions at different field conditions. Moreover, the activity of a biocontrol bacterium can be altered by the method of cultivation and formulation of the biocontrol PGPB in the laboratory prior to use and its mode of application (Glick 2015c).
In general, antibiotics role for disease suppression by biocontrol PGPB comes from two types of experiments (de Jesus Sousa and Olivares 2016). On the one hand, non-antibiotic synthesizing mutant bacteria were concomitantly found to have lost all, or a large part, of the ability to prevent plants damage caused by the target phytopathogen(s) (Heimpel and Mills 2017). On the other hand, in those instances where it has been possible to isolate and purify specific antibiotics from biocontrol PGPB, it was subsequently shown that the purified antibiotics were inhibitory to the same spectrum of phytopathogens as the biocontrol PGPB strain itself (Glick 2015c).
Many antibiotics have been derived from bacteria of the genera Bacillus and Pseudomonas. They produce a variety of metabolites which serve as antifungal, antibacterial, antihelminthic, antiviral, antimicrobial, phytotoxic, antioxidant, cytotoxic, and antitumor agents. For Bacillus, they are either derived from the ribosome or the non-ribosomal peptide and/or polyketide synthetases (NRPSs/PKS). Examples include Tas A, sublancin, subtilosin, bacilysin, chlorotetain, subtilin, bacillaene, surfactin, iturin, and fengycin while from Pseudomonas we have Ecomycins, 2,4-Diacetyl Phloroglucinol (DAPG), Pseudomonic acid, Phenazine-1-carboxylic acid (PCA), Pyoluteorin, Pyrrolnitrin, OomycinA, Cepaciamide A, Viscosinamide, Butyrolactones, Zwittermycin A, Aerugine, Azomycin, Rhamnolipids, Cepafungins, Kanosamine, and Karalicin (Goswami et al. 2016). Antibiotic gene clusters have been identified in Bacillus subtilis 168 and Bacillus amyloquefaciens FZB42; they are srf, bmy, fen, nrs, dhb, bac, mln, bae, dfn, and act by coordinating the biosynthesis of peptides and polyketides by the NRPSs and PKS enzymes (Chang et al. 2007).
Cell wall degrading enzymes
Numerous plants respond to infection by fungal phytopathogens by activating the plant-encoded synthesis of a number of fungal cell wall degrading enzymes. The enzymes include chitinase which degrades chitin (Husson et al. 2017), it is a residue of β-(1, 4)-N-acetyl glucosamine polymer and an integral part of the cell wall of many phytopathogenic fungi; β-1,3-glucanase, another cell wall carbohydrate (Vaddepalli et al. 2017); protease, which can degrade cell wall proteins; and lipase, which can degrade some of the cell wall-associated lipid, all of which can to some extent individually lyse fungal cells (Friedrich et al. 2012; Gortari and Hours 2008). In addition to the plant-encoded cell wall degrading enzymes, some biocontrol PGPB synthesize a similar panel of cell wall degrading enzymes (Chernin et al. 1995). Proof of the efficacy of these enzymes typically comes from laboratory experiments in which PGPB strains that have been genetically transformed with genes encoding these cell wall degrading enzymes are shown to become more effective biocontrol agents (Koby et al. 1994). Chitinase genes can be overexpressed and strains co transformed with the insertion of acetamidase gene amds as was seen in different studies where the pyruvate constitutive promoter successfully improved chitinase activity in modified strains (Kowsari et al. 2016; Limon et al. 1999). Chitinases, peroxidases and β-1,3-glucanases are part of the PR proteins and their activation can actually induce ISR in plants (Yedidia et al. 1999). Bacillus sp. JS was shown to cause an upregulation of the PR-2 and PR-3 genes which encode β-1,3-glucanase and chitinase respectively (Kim et al. 2015).
Competition
In addition to mechanisms where a biocontrol PGPB produces substances that are inhibitory to phytopathogens per se, it is possible for some biocontrol PGPB to outcompete the phytopathogens, either for nutrients or for binding sites on the plant root (Barahona et al. 2011; Innerebner et al. 2011). Such competition can act to limit the binding of the phytopathogen to the plant thereby making it difficult for it to proliferate. However, since it is not always possible to create mutants of PGPB that are either more or less competitive for binding to the plant surface, there are a relatively limited number of unequivocal demonstrations of the ability of biocontrol PGPB to outcompete phytopathogens and thereby prevent their functioning. In fact, it is generally thought that PGPB competitiveness works together with other biocontrol mechanisms to thwart the functioning of phytopathogens. Example of competition for nutrients in biocontrol of Pythium aphanidermatum damping-off was studied in the work of Elad and Chet (1987). Another example was seen in the work of Porcel et al. (2014) showing the competitiveness of Bacillus megaterium in the enhancement of tomato plant growth.
Hydrogen cyanide
A number of biocontrol PGPB have the ability to synthesize hydrogen cyanide (HCN). If the HCN produced by these bacteria were the only biocontrol mechanism being used in most instances, the low level of HCN would not be particularly effective at preventing the proliferation of most fungal phytopathogens. However, it is often the case that biocontrol PGPB that can produce HCN also synthesize some antibiotics or cell wall degrading enzymes (Ramette et al. 2006). Moreover, it has been observed that the low level of HCN synthesized by the bacterium improves the effectiveness of antifungal directed against fungal pathogens thereby ensuring that the fungi do not develop resistance to the particular antifungal in question. Thus, HCN synthesized by PGPB appears to act synergistically with other methods of biocontrol employed by the same bacterium.
HCN toxicity is effected in its ability to inhibit cytochrome c oxidase as well as other important metallo-enzymes (Nandi et al. 2017). Many bacterial genera such as Rhizobium, Pseudomonas, Alcaligenes, Bacillus, and Aeromonas have shown to be HCN producers (Ahmad et al. 2008; Das et al. 2017; Zachow et al. 2017). Suppression of tomato root knot disease caused by Meloidogyne javanica have been attributed to the effect of HCN (Siddiqui et al. 2006) as well as the control of Odontotermes obesus, a crop pest in India (Kumar et al. 2015).
Induced systemic resistance (ISR)
Induced systemic resistance (ISR) is a process in which non-pathogenic microbes, including a number of PGPB, alleviate the deleterious effects of plant pathogens by activating a resistance mechanism in the plants (Van Loon et al. 1998). ISR has been studied in many rhizobacteria-inoculated plants (Halfeld-Vieira et al. 2006) and, as initially demonstrated by Van Peer and Schippers (1992), protected plants against growth inhibition, using Pseudomonas fluorescens strain WCS417r against the fungal pathogen Fusarium oxysporum f. sp. dianthi. Pre-treatment of plants with an appropriate PGPB can prime the plant to react faster and more strongly to a subsequent pathogen attack by inducing plant defense mechanisms. ISR does not target specific pathogens but rather primes the plant against a range of different pathogens, and it is not only expressed at the site of induction only.
Plants develop resistance in response to pathogen infection, insect attack, microbes’ colonization or after treatment with chemicals but this induced state is expressed by activating the “dormant” defense mechanisms which become expressed in response to external contacts from pathogens, insects etc. ISR confers a high level of protection which is controlled by a network of coordinated signaling pathways which are dominated and majorly regulated by plant hormones sharing signaling components (Pieterse et al. 2012, 2014; Walters et al. 2013).
Several different PGPB have been reported to produce salicylic acid (SA) that can act as a signal molecule to turn on a mechanism similar to ISR in plants that is called “systemic acquired resistance” or SAR (Chen et al. 1999). However, SAR is usually induced by phytopathogens per se, and salicylic acid is generally not considered to play a role in PGPB-induced plant resistance to phytopathogens (Zhang et al. 2002). Thus, while the PGPB that produce SA may activate plant phytopathogen-protective mechanisms, this trait is considered to be extremely rare in PGPB compared to ISR. Furthermore, SAR is coordinated by the activation of pathogenesis related (PR) genes which encode PR proteins. One of the best characterized PR gene is PR-1 which is mostly used as a biomarker for SAR (van Loon et al. 2006).
The protein involved majorly in the regulation of ISR and SAR is the redox-regulated protein nonexpressor of PR genes1 (NPR1). It is synthesized in the cytoplasm as an oligomer through intermolecular disulfide bonds and since its discovery in 1994, its function in transcriptional regulation has been well documented. Its importance has been shown in the jasmonic acid/ethylene dependent ISR activated by P. fluorescens WCS417r (Pieterse et al. 1998) as well as many other PGPBs (Abo-Elyousr et al. 2009; Lucas et al. 2014; Weller et al. 2012; Yi et al. 2013). The NPR-1 protein which is encoded by the npr-1 gene activates SAR establishment by activating PRs genes after receiving signal from the accumulation of SA (Pieterse et al. 1998).
PGPB-mediated ISR and SA-dependent SAR are coordinated by different signaling pathways which was supported by the observations that both PGPB-mediated ISR and pathogen-induced SAR are effective control mechanisms, but their degree of effectiveness are slightly different (Ton et al. 2002).
Quorum quenching
In the environment, bacterial cells use the mechanism of quorum sensing to detect the presence of similar (as well as different) types of bacteria. With growing bacterial cells, once they have attained a certain critical cell density, the bacteria “sense” the cell density (through the production of chemical signals) and start to alter their metabolism by turning on different sets of genes (Cornforth et al. 2014), so that similar bacteria that are proximal to one another may begin acting in a coordinate manner.
In most systems, bacteria synthesize low molecular weight chemicals called autoinducers that are typically secreted outside of the bacterial cells. When the bacterial cells population increases, the extracellular level of the autoinducers also increases until it exceeds some threshold level, binds to bacterial cellular receptors and triggers a signal transduction cascade thereby causing population-wide changes in bacterial gene expression with the actions of a unified group of cells, e.g. at a certain cell density a bacterial plant pathogen may begin to become more virulent (Huang et al. 2016). Disrupting this quorum sensing (Chan et al. 2011; Pei and Lamas-Samanamud 2014) (i.e. signaling amongst pathogens) can thwart the pathogen from becoming increasingly virulent and prevent it from inhibiting plant growth. There are a number of biological means of quenching the phenomenon of quorum sensing. One way is to utilize a PGPB that produces an enzyme called a lactonase that degrades the pathogen-produced autoinducer and pretreat plant seedlings (when they are most sensitive to many pathogens) with this PGPB, especially in situations where a particular bacterial pathogen is known to be particularly problematic (Glick 2015a). While this is a clever strategy that has been successful in lab, it has not yet been successfully tested in field.
Bacteriophages
Some bacterial phytopathogens may be lysed by specific bacteriophages, or bacterial viruses (Frampton et al. 2012). For this approach to work, the target bacterial phytopathogen must be unequivocally identified down to the strain level. Subsequently, it is possible to isolate and thoroughly characterize several different bacteriophages that can lyse only the target pathogen without affecting any other bacterial strains. To kill the target bacterial phytopathogen, the bacteriophages are sprayed onto an infected plant as a mixture of two or three different bacteriophage strains all directed against the targeted bacterial pathogen (Glick 2015a). The use of a mixture of bacteriophage strains decreases the possibility that bacteriophage-resistant mutants of the target pathogen will develop. This is because of the different binding sites of the bacteriophages on the surface of the bacterial pathogen.
Since most bacteriophages are quite sensitive to UV light (Buttimer et al. 2017), they are typically sprayed onto plants at dusk when the UV light intensity is low. Notwithstanding this precaution, some bacteriophages need to be applied weekly, or even daily, to be effective. At the present time, a few bacteriophage-based biocontrol agents have been licensed for use, e.g. for the bacterial pathogen Xanthomonas campestris pv. vesicatoria which causes bacterial spot of tomatoes and peppers, Pseudomonas syringae pv. actinidae which causes canker disease in kiwi fruit, and Pseudomonas syringae pv. tomato, which causes bacterial speck on tomatoes (Peitl et al. 2017; Yu et al. 2016).
The earliest reports of phages as biocontrol agents are presented in the studies of Coons (1925), Kotila and Coons (1925), and Moore (1926). Depending on their life cycle, they can be lytic phages which destroys host bacterial cells or lysogenic phages which incorporate their genome into that of the host and replicate without affecting the host bacterial cell. The treatment of bacterial infections with phages is known as phage therapy (Álvarez and Biosca 2017). It is a promising method for combating plant wilt diseases as it has been successfully used in treating plant phytopathogenic diseases (Bae et al. 2012; Balogh et al. 2010; Frampton et al. 2014; Fujiwara et al. 2011). More discussions on bacteriophages as biocontrol agents have been elaborately reviewed in the work of Buttimer et al. (2017), and others such as Abedon et al. (2017); Doss et al. (2017); Jones et al. (2007); O’Brien (2017) etc.
Conclusions and future prospects
The world that can support only a limited number of people. Unless new sustainable agricultural approaches and technologies are soon developed, food availability in the next 50 years approximately might be a great challenge for the growing population. To address this problem, one of the approaches that might be undertaken is the more widespread use of PGPB, initially in addition to, and possibly eventually instead of, the current use of agricultural chemicals.
The last 30–40 years have seen researchers developed an exhaustive, precise understanding of how PGPB facilitate plants growth so that the more widespread application of these organisms has now become feasible. For example, in the studies of Ali et al. (2014), Álvarez and Biosca, (2017) among others show the successful applications of PGPB in plant growth promotion. The latter using bacteriophages in biocontrol while the former shows the activities of PGPB in reduction of stress. However, in order to make this approach a worldwide reality, a number of steps must be undertaken. (1) New and improved techniques for the large-scale growth, storage, shipping, formulation and application of plant growth-promoting bacteria need to be developed. (2) Reasonable, safe, efficacious and consistent regulations for the use PGPB need to be developed in all countries of the world so that the technology may readily be transferred from one country to another. Also, unnecessary regulatory hurdles need to be kept to a minimum. (3) Broadly-based campaigns of public education regarding the nature of PGPB need to be initiated so that the public comes to understand that these bacteria are not sources of disease but are natural products playing a positive role. (4) Following additional fundamental work to better understand PGPB and their biochemistry, genetics and physiology, scientists, laymen and regulators need to accept that “optimal” PGPB strains may require some genetic manipulation and that the use of such genetically manipulated strains will not present any new hazards or risks to humans or the environment. (5) It is likely that different crops and varying situations will necessitate the use of PGPB that are either rhizospheric or endophytic. It will be necessary to delineate those situations where either rhizospheric or endophytic PGPB strains are most appropriate so that the most effective combination of plant and PGPB can always be applied. (6) Given that the growth of more than 90% of crop plants is enhanced by the plants’ interaction with mychorrhizae, it is necessary to develop a much better understanding of how PGPB and mychorrhizae interact in a way that optimally promotes plant growth. (7) As much as possible, this technology should be kept in the public domain so that a few large companies do not end up owning all of the key technology.
While there is still a lot more basic and applied work to be done, application of PGPB are already a success, on a relatively small scale, in several countries. If scientists, and the agencies that fund their work, direct their efforts toward addressing the above-mentioned and related issues, there is every reason to expect that agricultural practice worldwide can become both sustainable and highly efficacious. We are on the verge of a major paradigm shift in agriculture, a shift that should benefit both the developing and the developed world.
Acknowledgements
North-West University is gratefully acknowledged for school bursary to OOS. OOB would like to thank the National Research Foundation, South Africa for a grant (Ref: UID81192) that has supported research in her laboratory.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest whatsoever from the authors.
References
- Abedon ST, García P, Mullany P, Aminov R. Editorial: phage therapy: past, present and future. Front Microbiol. 2017;8:981. doi: 10.3389/fmicb.2017.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles F, Morgan P, Saltveit M., Jr . Ethylene in plant biology. 2. New York: Academic Press; 1992. [Google Scholar]
- Abo-Elyousr KAM, Hashem M, Ali EH. Integrated control of cotton root rot disease by mixing fungal biocontrol agents and resistance inducers. Crop Prot. 2009;28:295–301. doi: 10.1016/j.cropro.2008.11.004. [DOI] [Google Scholar]
- Adams M, Mortenson L, Chen J. Hydrogenase. Biochim Biophys Acta. 1981;594:105–176. doi: 10.1016/0304-4173(80)90007-5. [DOI] [PubMed] [Google Scholar]
- Ahmad F, Ahmad I, Khan MS. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res. 2008;163:173–181. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Aida M, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Ali B, Sabri AN, Hasnain S. Rhizobacterial potential to alter auxin content and growth of Vigna radiata (L.) World J Microbiol Biotechnol. 2010;26:1379–1384. doi: 10.1007/s11274-010-0310-1. [DOI] [Google Scholar]
- Ali S, Charles TC, Glick BR. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol Biochem. 2014;80:160–167. doi: 10.1016/j.plaphy.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Alori ET, Glick BR, Babalola OO. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol. 2017;8:971. doi: 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez B, Biosca EG. Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front Plant Sci. 2017;8:1218. doi: 10.3389/fpls.2017.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón IM, Pérez-Martínez I, Moreno-Pérez A, Cerezo M, Ramos C. New insights into the role of indole-3-acetic acid in the virulence of Pseudomonas savastanoi pv. savastanoi. FEMS Microbiol Lett. 2014;356:184–192. doi: 10.1111/1574-6968.12413. [DOI] [PubMed] [Google Scholar]
- Arkhipova T, Veselov S, Melentiev A, Martynenko E, Kudoyarova G. Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil. 2005;272:201–209. doi: 10.1007/s11104-004-5047-x. [DOI] [Google Scholar]
- Baas P, Mohan JE, Markewitz D, Knoepp JD. Assessing heterogeneity in soil nitrogen cycling: a plot-scale approach. Soil Sci Soc Am J. 2014;78:237–247. doi: 10.2136/sssaj2013.09.0380nafsc. [DOI] [Google Scholar]
- Babalola OO. Beneficial bacteria of agricultural importance. Biotechnol Lett. 2010;32:1559–1570. doi: 10.1007/s10529-010-0347-0. [DOI] [PubMed] [Google Scholar]
- Bae JY, Wu J, Lee HJ, Jo EJ, Murugaiyan S, Chung E, Lee S-W. Biocontrol potential of a lytic bacteriophage PE204 against bacterial wilt of tomato. J Microbiol Biotechnol. 2012;22:1613–1620. doi: 10.4014/jmb.1208.08072. [DOI] [PubMed] [Google Scholar]
- Balogh B, Jones JB, Iriarte F, Momol M. Phage therapy for plant disease control. Curr Pharm Biotechnol. 2010;11:48–57. doi: 10.2174/138920110790725302. [DOI] [PubMed] [Google Scholar]
- Banerjee MR, Yesmin L, Vessey JK, Rai M. Plant-growth-promoting rhizobacteria as biofertilizers and biopesticides. Handbook of microbial biofertilizers. New York: Food Products Press; 2005. pp. 137–181. [Google Scholar]
- Barahona E, Navazo A, Martínez-Granero F, Zea-Bonilla T, Pérez-Jiménez RM, Martín M, Rivilla R. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl Environ Microbiol. 2011;77:5412–5419. doi: 10.1128/AEM.00320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnawal D, Bharti N, Maji D, Chanotiya CS, Kalra A. 1-Aminocyclopropane-1-carboxylic acid (ACC) deaminase-containing rhizobacteria protect Ocimum sanctum plants during waterlogging stress via reduced ethylene generation. Plant Physiol Biochem. 2012;58:227–235. doi: 10.1016/j.plaphy.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Bielach A, et al. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell. 2012;24:3967–3981. doi: 10.1105/tpc.112.103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1090. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Boulé J, Sholberg P, Lehman S, O’gorman D, Svircev A. Isolation and characterization of eight bacteriophages infecting Erwinia amylovora and their potential as biological control agents in British Columbia, Canada. Can J Plant Pathol. 2011;33:308–317. doi: 10.1080/07060661.2011.588250. [DOI] [Google Scholar]
- Broek AV, Gysegom P, Ona O, Hendrickx N, Prinsen E, Van Impe J, Vanderleyden J. Transcriptional analysis of the Azospirillum brasilense indole-3-pyruvate decarboxylase gene and identification of a cis-acting sequence involved in auxin responsive expression. Mol Plant Microbe Interact. 2005;18:311–323. doi: 10.1094/MPMI-18-0311. [DOI] [PubMed] [Google Scholar]
- Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- Buttimer C, McAuliffe O, Ross RP, Hill C, O’Mahony J, Coffey A (2017) Bacteriophages and bacterial plant diseases. Front Microbiol 8 [DOI] [PMC free article] [PubMed]
- Calvo P, Nelson L, Kloepper JW. Agricultural uses of plant biostimulants. Plant Soil. 2014;383:3–41. doi: 10.1007/s11104-014-2131-8. [DOI] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Interact. 2015;28:1049–1058. doi: 10.1094/MPMI-01-15-0016-R. [DOI] [PubMed] [Google Scholar]
- Chan K-G, et al. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:51. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W-T, Chen Y-C, Jao C-L. Antifungal activity and enhancement of plant growth by Bacillus cereus grown on shellfish chitin wastes. Bioresour Technol. 2007;98:1224–1230. doi: 10.1016/j.biortech.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J Exp Bot. 2013;64:5021–5032. doi: 10.1093/jxb/ert291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Bélanger RR, Benhamou N, Paulitz TC. Role of salicylic acid in systemic resistance induced by Pseudomonas spp. against Pythium aphanidermatum in cucumber roots. Eur J Plant Pathol. 1999;105:477–486. doi: 10.1023/A:1008743502784. [DOI] [Google Scholar]
- Cheng X, Ruyter-Spira C, Bouwmeester H. The interaction between strigolactones and other plant hormones in the regulation of plant development. Front Plant Sci. 2013;4:199. doi: 10.3389/fpls.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernin L, Ismailov Z, Haran S, Chet I. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl Environ Microbiol. 1995;61:1720–1726. doi: 10.1128/aem.61.5.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coons GH. The transmissible lytic principle (bacteriophage) in relation to plant pathogens. Phytopathol. 1925;28:357–370. [Google Scholar]
- Cornforth DM, et al. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci USA. 2014;111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moënne-Loccoz Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol. 2009;48:505–512. doi: 10.1111/j.1472-765X.2009.02566.x. [DOI] [PubMed] [Google Scholar]
- DalCorso G, Manara A, Furini A. An overview of heavy metal challenge in plants: from roots to shoots. Metallomics. 2013;5:1117–1132. doi: 10.1039/c3mt00038a. [DOI] [PubMed] [Google Scholar]
- Das K, Prasanna R, Saxena AK. Rhizobia: a potential biocontrol agent for soilborne fungal pathogens. Folia Microbiol. 2017 doi: 10.1007/s12223-017-0513-z. [DOI] [PubMed] [Google Scholar]
- de Jesus Sousa JA, Olivares FL. Plant growth promotion by streptomycetes: ecophysiology, mechanisms and applications. Chem Biol Technol Agric. 2016;3:24. doi: 10.1186/s40538-016-0073-5. [DOI] [Google Scholar]
- De Rybel B, Mähönen AP, Helariutta Y, Weijers D. Plant vascular development: from early specification to differentiation. Nat Rev Mol Cell Biol. 2016;17:30. doi: 10.1038/nrm.2015.6. [DOI] [PubMed] [Google Scholar]
- Deka H, Deka S, Baruah C. Plant-growth-promoting rhizobacteria (pgpr) and medicinal plants. New York: Springer; 2015. Plant growth promoting rhizobacteria for value addition: mechanism of action; pp. 305–321. [Google Scholar]
- Dodd IC, Zinovkina NY, Safronova VI, Belimov AA. Rhizobacterial mediation of plant hormone status. Ann Appl Biol. 2010;157:361–379. doi: 10.1111/j.1744-7348.2010.00439.x. [DOI] [Google Scholar]
- Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses. 2017;9:50. doi: 10.3390/v9030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca D, Lorv J, Patten C, Rose D, Glick B. Microbial indole-3-acetic acid and plant growth. Anton Van Leeuwenhoek. 2014;106:85–125. doi: 10.1007/s10482-013-0095-y. [DOI] [PubMed] [Google Scholar]
- Elad Y, Chet I. Possible role of competition for nutrients in biocontrol of Pythium damping-off by bacteria. Phytopathol. 1987;77:190–195. doi: 10.1094/Phyto-77-190. [DOI] [Google Scholar]
- Frampton RA, Pitman AR, Fineran PC. Advances in bacteriophage-mediated control of plant pathogens. Int J Microbiol. 2012;2012:326452. doi: 10.1155/2012/326452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton RA, Taylor C, Moreno AVH, Visnovsky SB, Petty NK, Pitman AR, Fineran PC. Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae. Appl Environ Microbiol. 2014;80:2216–2228. doi: 10.1128/AEM.00062-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich N, Hagedorn M, Soldati-Favre D, Soldati T. Prison break: pathogens’ strategies to egress from host cells. Microbiol Mol Biol Rev. 2012;76:707–720. doi: 10.1128/MMBR.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara A, Fujisawa M, Hamasaki R, Kawasaki T, Fujie M, Yamada T. Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl Environ Microbiol. 2011;77:4155–4162. doi: 10.1128/AEM.02847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E, Glick BR. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015;169:13. doi: 10.1104/pp.15.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93:1721–1741. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- Geddes BA, Ryu M-H, Mus F, Costas AG, Peters JW, Voigt CA, Poole P. Use of plant colonizing bacteria as chassis for transfer of N 2-fixation to cereals. Curr Opin Biotechnol. 2015;32:216–222. doi: 10.1016/j.copbio.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Glick BR. The enhancement of plant growth by free-living bacteria. Can J Microbiol. 1995;41:109–117. doi: 10.1139/m95-015. [DOI] [Google Scholar]
- Glick BR (2012) Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012 [DOI] [PMC free article] [PubMed]
- Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Glick BR. Beneficial plant-bacterial interactions. New York: Springer; 2015. Biocontrol mechanisms; pp. 123–157. [Google Scholar]
- Glick BR. beneficial plant-bacterial interactions. Cham: Springer International Publishing; 2015. Modulating phytohormone levels; pp. 65–96. [Google Scholar]
- Glick BR. Beneficial plant-bacterial interactions. New York: Springer; 2015. Resource acquisition; pp. 29–63. [Google Scholar]
- Glick BR, Bashan Y. Genetic manipulation of plant growth-promoting bacteria to enhance biocontrol of phytopathogens. Biotechnol Adv. 1997;15:353–378. doi: 10.1016/S0734-9750(97)00004-9. [DOI] [PubMed] [Google Scholar]
- Glick BR, Cheng Z, Czarny J, Duan J. New perspectives and approaches in plant growth-promoting rhizobacteria research. New York: Springer; 2007. Promotion of plant growth by ACC deaminase-producing soil bacteria; pp. 329–339. [Google Scholar]
- Gortari MC, Hours RA. Fungal chitinases and their biological role in the antagonism onto nematode eggs: a review. Mycol Prog. 2008;7:221–238. doi: 10.1007/s11557-008-0571-3. [DOI] [Google Scholar]
- Goswami D, Thakker JN, Dhandhukia PC, Tejada Moral M. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2016;2:1127500. [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Grobelak A, Napora A, Kacprzak M. Using plant growth-promoting rhizobacteria (PGPR) to improve plant growth. Ecol Eng. 2015;84:22–28. doi: 10.1016/j.ecoleng.2015.07.019. [DOI] [Google Scholar]
- Gupta G, Parihar SS, Ahirwar NK, Snehi SK, Singh V. Plant growth promoting rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. J Microb Biochem Technol. 2015;7:096–102. [Google Scholar]
- Gupta S, Seth R, Sharma A. Plant growth-promoting rhizobacteria play a role as phytostimulators for sustainable agriculture. In: Choudhary DK, Varma A, Tuteja N, editors. Plant-microbe interaction: an approach to sustainable agriculture. Singapore: Springer; 2016. pp. 475–493. [Google Scholar]
- Haas D, Keel C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- Halfeld-Vieira BA, Vieira JR, Jr, Romeiro RS, Silva HSA, Mc BP. Induction of systemic resistance in tomato by autochthonus phylloplane resident Bacillus cereus. Pesq Agropec Bras. 2006;41:1247–1252. doi: 10.1590/S0100-204X2006000800006. [DOI] [Google Scholar]
- Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- Heimpel GE, Mills NJ. Biological control: ecology and applications. Cambridge: Cambridge University Press; 2017. [Google Scholar]
- Huang J, et al. Acyl-homoserine lactone-based quorum sensing and quorum quenching hold promise to determine the performance of biological wastewater treatments: an overview. Chemosphere. 2016;157:137–151. doi: 10.1016/j.chemosphere.2016.05.032. [DOI] [PubMed] [Google Scholar]
- Husson E, et al. The effect of room temperature ionic liquids on the selective biocatalytic hydrolysis of chitin via sequential or simultaneous strategies. Green Chem. 2017 [Google Scholar]
- Innerebner G, Knief C, Vorholt JA. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol. 2011;77:3202–3210. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioio RD, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Iriarte F, Balogh B, Momol M, Smith L, Wilson M, Jones J. Factors affecting survival of bacteriophage on tomato leaf surfaces. Appl Environ Microbiol. 2007;73:1704–1711. doi: 10.1128/AEM.02118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Jackson L, Balogh B, Obradovic A, Iriarte F, Momol M. Bacteriophages for plant disease control. Annu Rev Phytopathol. 2007;45:245–262. doi: 10.1146/annurev.phyto.45.062806.094411. [DOI] [PubMed] [Google Scholar]
- Khan AL, Waqas M, Hussain J, Al-Harrasi A, Hamayun M, Lee I-J. Phytohormones enabled endophytic fungal symbiosis improve aluminum phytoextraction in tolerant Solanum lycopersicum: an examples of Penicillium janthinellum LK5 and comparison with exogenous GA 3. J Hazard Mater. 2015;295:70–78. doi: 10.1016/j.jhazmat.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Khosro M. Phosphorus solubilizing bacteria: occurrence, mechanisms and their role in crop production. Resour Environ. 2012;2:80–85. [Google Scholar]
- Kim J-S, Lee J, Lee C-h, Woo SY, Kang H, Seo S-G, Kim S-H. Activation of pathogenesis-related genes by the hizobacterium, Bacillus sp. JS, which induces systemic resistance in tobacco plants. Plant Pathol J. 2015;31:195–201. doi: 10.5423/PPJ.NT.11.2014.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper JW, Leong J, Teintze M, Schroth MN. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature. 1980;286:885–886. doi: 10.1038/286885a0. [DOI] [Google Scholar]
- Koby S, Schickler H, Ilan C, Oppenheim AB. The chitinase encoding Tn7-based chiA gene endows Pseudomonas fluorescens with the capacity to control plant pathogens in soil. Gene. 1994;147:81–83. doi: 10.1016/0378-1119(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64:2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotila J, Coons G (1925) Investigations on the blackleg disease of potato. Michigan Agr Exper Sta Tech Bull 67
- Kowsari M, Zamani M, Motallebi M. Overexpression of chimeric chitinase 42 enhanced antifungal activity of Trichoderma harzianum against Fusarium graminearum. Mycol Iran. 2016;3:15–23. [Google Scholar]
- Kumar A, Bahadur I, Maurya B, Raghuwanshi R, Meena V, Singh D, Dixit J. Does a plant growth promoting rhizobacteria enhance agricultural sustainability. J Pure Appl Microbiol. 2015;9:715–724. [Google Scholar]
- Laschat S, Bilitewski U, Blodgett J, Duhme-Klair A-K, Dallavalle S, Routledge A, Schobert R (2017) Chemical and biological aspects of nutritional immunity-perspectives for new anti-infectives targeting iron uptake systems. Angew Chem Int Ed [DOI] [PubMed]
- Limon MC, Pintor-Toro JA, Benítez T. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathol. 1999;89:254–261. doi: 10.1094/PHYTO.1999.89.3.254. [DOI] [PubMed] [Google Scholar]
- Lucas JA, García-Cristobal J, Bonilla A, Ramos B, Gutierrez-Mañero J. Beneficial rhizobacteria from rice rhizosphere confers high protection against biotic and abiotic stress inducing systemic resistance in rice seedlings. Plant Physiol Biochem. 2014;82:44–53. doi: 10.1016/j.plaphy.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Lucy M, Reed E, Glick B. Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek. 2004;86:1–25. doi: 10.1023/B:ANTO.0000024903.10757.6e. [DOI] [PubMed] [Google Scholar]
- Marhavý P, et al. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell. 2011;21:796–804. doi: 10.1016/j.devcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Marhavý P, et al. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr Biol. 2014;24:1031–1037. doi: 10.1016/j.cub.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Martínez C, Espinosa-Ruiz A, Prat S. Gibberellins and plant vegetative growth. Annu Plant Rev. 2016;49:285–322. [Google Scholar]
- Miller CO, Skoog F, Okumura FS, Von Saltza MH, Strong F. Structure and synthesis of kinetin1. J Ame Chem Soc. 1955;77:2662–2663. doi: 10.1021/ja01614a108. [DOI] [Google Scholar]
- Minguet EG, Alabadí D, Blázquez MA. Gibberellin implication in plant growth and stress responses. In: Tran L-SP, Pal S, editors. Phytohormones: a window to metabolism, signaling and biotechnological applications. New York: Springer; 2014. pp. 119–161. [Google Scholar]
- Moore E. D’Herelle’s bacteriophage in relation to plant parasites. S Afr J Sci. 1926;23:306. [Google Scholar]
- Moubayidin L, et al. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell. 2013;26:405–415. doi: 10.1016/j.devcel.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi M, Selin C, Brawerman G, Fernando WGD, de Kievit T. Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol Control. 2017;108:47–54. doi: 10.1016/j.biocontrol.2017.02.008. [DOI] [Google Scholar]
- Nelson SK, Steber CM. Gibberellin hormone signal perception: down-regulating DELLA repressors of plant growth and development. Annu Plant Rev. 2016;49:153–188. [Google Scholar]
- O’Brien PA. Biological control of plant diseases. Aust Plant Pathol. 2017;44:1–12. doi: 10.1007/s13313-017-0466-3. [DOI] [Google Scholar]
- Oldroyd GE, Dixon R. Biotechnological solutions to the nitrogen problem. Curr Opin Biotechnol. 2014;26:19–24. doi: 10.1016/j.copbio.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- Pei R, Lamas-Samanamud GR. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl Environ Microbiol. 2014;80:5340–5348. doi: 10.1128/AEM.01434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitl DC, Araujo FA, Gonçalves RM, Santiago DC, Sumida CH, Balbi-Peña MI. Biological control of tomato bacterial spot by saprobe fungi from semi-arid areas of northeastern Brazil Semina. Ciências Agrárias. 2017;38:1251–1263. doi: 10.5433/1679-0359.2017v38n3p1251. [DOI] [Google Scholar]
- Peix A, Mateos P, Rodriguez-Barrueco C, Martinez-Molina E, Velazquez E. Growth promotion of common bean (Phaseolus vulgaris L.) by a strain of Burkholderia cepacia under growth chamber conditions. Soil Biol Biochem. 2001;33:1927–1935. doi: 10.1016/S0038-0717(01)00119-5. [DOI] [Google Scholar]
- Pérez-Montaño F, et al. Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol Res. 2014;169:325–336. doi: 10.1016/j.micres.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Petrášek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- Porcel R, Zamarreño ÁM, García-Mina JM, Aroca R. Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. BMC Plant Biol. 2014;14:36. doi: 10.1186/1471-2229-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, Mazzola M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol. 2012;50:403–424. doi: 10.1146/annurev-phyto-081211-172908. [DOI] [PubMed] [Google Scholar]
- Radzki W, Gutierrez Mañero FJ, Algar E, Lucas García JA, García-Villaraco A, Ramos Solano B. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek. 2013;104:321–330. doi: 10.1007/s10482-013-9954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Moënne-Loccoz Y, Défago G. Genetic diversity and biocontrol potential of fluorescent pseudomonads producing phloroglucinols and hydrogen cyanide from Swiss soils naturally suppressive or conducive to Thielaviopsis basicola-mediated black root rot of tobacco. FEMS Microbiol Ecol. 2006;55:369–381. doi: 10.1111/j.1574-6941.2005.00052.x. [DOI] [PubMed] [Google Scholar]
- Reed M, Glick BR. Applications of plant growth-promoting bacteria for plant and soil systems. Appl Microb Eng. 2013;CT:181–229. doi: 10.1201/b15250-8. [DOI] [Google Scholar]
- Rodríguez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999;17:319–339. doi: 10.1016/S0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol. 2013;53:303–317. doi: 10.1002/jobm.201100552. [DOI] [PubMed] [Google Scholar]
- Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res. 2016;23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- Saharan BS, Nehra V. Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res. 2011;21:1–30. [Google Scholar]
- Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G, Moreno-Hagelsieb G, del Carmen Orozco-Mosqueda M, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Schäfer M, Brütting C, Meza-Canales ID, Großkinsky DK, Vankova R, Baldwin IT, Meldau S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J Exp Bot. 2015;66:4873–4884. doi: 10.1093/jxb/erv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Johri BN. Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions. Microbiol Res. 2003;158:243–248. doi: 10.1078/0944-5013-00197. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kumar V, Tripathi RB. Isolation of phosphate solubilizing microorganism (PSMs) from soil. J Microbiol Biotechnol Res. 2017;1:90–95. [Google Scholar]
- Shen X, Hu H, Peng H, Wang W, Zhang X. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics. 2013;14:1471–2164. doi: 10.1186/1471-2164-14-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui IA, Shaukat SS, Sheikh IH, Khan A. Role of cyanide production by Pseudomonas fluorescens CHA0 in the suppression of root-knot nematode, Meloidogyne javanica in tomato. World J Microbiol Biotechnol. 2006;22:641–650. doi: 10.1007/s11274-005-9084-2. [DOI] [Google Scholar]
- Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol. 2011;3:a001438. doi: 10.1101/cshperspect.a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism plant signaling. FEMS Microbiol Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- Toklikishvili N, et al. Inhibitory effect of ACC deaminase-producing bacteria on crown gall formation in tomato plants infected by Agrobacterium tumefaciens or Agrobacterium vitis. Plant Pathol. 2010;59:1023–1030. doi: 10.1111/j.1365-3059.2010.02326.x. [DOI] [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, Pieterse CM. Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant Microbe Interact. 2002;15:27–34. doi: 10.1094/MPMI.2002.15.1.27. [DOI] [PubMed] [Google Scholar]
- Turan M, Kıtır N, Alkaya Ü, Günes A, Tüfenkçi Ş, Yıldırım E, Nikerel E. Making soil more accessible to plants: the case of plant growth promoting rhizobacteria. Plant growth. Rijeka: InTech; 2016. [Google Scholar]
- Vaddepalli P, Fulton L, Wieland J, Wassmer K, Schaeffer M, Ranf S, Schneitz K (2017) The cell wall-localized atypical β-1, 3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development 152231 [DOI] [PubMed]
- Van Peer R, Schippers B. Lipopolysaccharides of plant-growth promoting Pseudomonas sp. strain WCS417r induce resistance in carnation to Fusarium wilt. Neth J Plant Pathol. 1992;98:129–139. doi: 10.1007/BF01996325. [DOI] [Google Scholar]
- Van Loon L, Bakker P, Pieterse C. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 1998;36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Walters DR, Ratsep J, Havis ND. Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot. 2013;64:1263–1280. doi: 10.1093/jxb/ert026. [DOI] [PubMed] [Google Scholar]
- Wang G-L, Que F, Xu Z-S, Wang F, Xiong A-S. Exogenous gibberellin altered morphology, anatomic and transcriptional regulatory networks of hormones in carrot root and shoot. BMC plant biol. 2015;15:290. doi: 10.1186/s12870-015-0679-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller DM, Mavrodi DV, van Pelt JA, Pieterse CM, van Loon LC, Bakker PA. Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2, 4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology. 2012;102:403–412. doi: 10.1094/PHYTO-08-11-0222. [DOI] [PubMed] [Google Scholar]
- Xu J, Li X-L, Luo L. Effects of engineered Sinorhizobium meliloti on cytokinin synthesis and tolerance of alfalfa to extreme drought stress. Appl Environ Microbiol. 2012;78:8056–8061. doi: 10.1128/AEM.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia I, Benhamou N, Chet I. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl Environ Microbiol. 1999;65:1061–1070. doi: 10.1128/aem.65.3.1061-1070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H-S, Yang JW, Ryu C-M. ISR meets SAR outside: additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front Plant Sci. 2013;4:122. doi: 10.3389/fpls.2013.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim W, Seshadri S, Kim K, Lee G, Sa T. Ethylene emission and PR protein synthesis in ACC deaminase producing Methylobacterium spp. inoculated tomato plants (Lycopersicon esculentum Mill.) challenged with Ralstonia solanacearum under greenhouse conditions. Plant Physiol Biochem. 2013;67:95–104. doi: 10.1016/j.plaphy.2013.03.002. [DOI] [PubMed] [Google Scholar]
- You Y-H, et al. Fungal diversity and plant growth promotion of endophytic fungi from six halophytes in Suncheon Bay. J Microbiol Biotechnol. 2012;22:1549–1556. doi: 10.4014/jmb.1205.05010. [DOI] [PubMed] [Google Scholar]
- Yu J-G, Lim J-A, Song Y-R, Heu S, Kim GH, Koh YJ, Oh C-S. Isolation and characterization of bacteriophages against Pseudomonas syringae pv. actinidiae causing bacterial canker disease in kiwifruit. J Microbiol Biotechnol. 2016;26:385–393. doi: 10.4014/jmb.1509.09012. [DOI] [PubMed] [Google Scholar]
- Zachow C, Müller H, Monk J, Berg G. Complete genome sequence of Pseudomonas brassicacearum strain L13-6-12, a biological control agent from the rhizosphere of potato. Stand Genomic Sci. 2017;12:6. doi: 10.1186/s40793-016-0215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi A, Ahmad E, Khan MS, Saif S, Rizvi A. Role of plant growth promoting rhizobacteria in sustainable production of vegetables: current perspective. Sci Hortic. 2015;193:231–239. doi: 10.1016/j.scienta.2015.07.020. [DOI] [Google Scholar]
- Zhang S, Moyne A-L, Reddy M, Kloepper JW. The role of salicylic acid in induced systemic resistance elicited by plant growth-promoting rhizobacteria against blue mold of tobacco. Biol Control. 2002;25:288–296. doi: 10.1016/S1049-9644(02)00108-1. [DOI] [Google Scholar]
- Zhang S, Gao P, Tong Y, Norse D, Lu Y, Powlson D. Overcoming nitrogen fertilizer over-use through technical and advisory approaches: a case study from Shaanxi Province, Northwest China. Agric Ecosyst Environ. 2015;209:89–99. doi: 10.1016/j.agee.2015.03.002. [DOI] [Google Scholar]