Abstract
This study was conducted to isolate and characterize Paenibacillus sp. MBT213 possessing β-glucosidase activity from raw milk, and examine the enzymatic capacity on the hydrolysis of a major ginsenoside (Rb1). Strain MBT213 was found to have a high hydrolytic ability on ginsenoside Rb1 by Esculin Iron Agar test. 16S rDNA analysis revealed that MBT213 was Paenibacillu sp. Crude enzyme of MBT213 strain exhibited high conversion capacity on ginsenoside Rb1 into ginsenoside Rd proven by TLC and HPLC analyses. The API ZYM kit confirmed that Paenibacillu sp. MBT213 exerted higher β-glucosidase and β-galactosidase activity than other strains. Optimum pH and temperature for crude enzyme were found at 7.0 and 35°C in hydrolysis of ginsenoside Rb1. After 10 d of optimal reaction conditions for the crude enzyme, ginsenoside Rb1 fully converted to ginsenoside Rd. Ginseng roots (20%) were fermented for 14 d, and analyzed by HPLC showed that amount of ginsenoside Rb1 significantly decreased, while that of ginsenoside Rd was significantly increased. The study confirmed that the β-glucosidase produced by Paenibacillus sp. MBT213 can hydrolyze the major ginsenoside Rb1 and convert to Rd during fermentation of the ginseng. The β-glucosidase activity of this novel Paenibacillus sp. MBT213 strain may be utilized in development of variety of health foods, dairy foods and pharmaceutical products.
Keywords: ginsenoside, β-glucosidase, β-galactosidase, Paenibacillus MBT213, raw milk
Introduction
Paenibacillus strains, a non-pathogenic and Gram-positive endospore-forming bacillus, are one of the most industrially significant facultative anaerobic bacteria. They are isolated from a variety of sources including soil, rhizosphere, water, diseased insect larvae and pasteurized food, cow manure, and raw and heat-treated milk (Daane et al., 2002; Scheldeman et al., 2004; Ziarno and Zareba, 2010). Paenibacillus strain can enter raw milk during milking, from the bedding materials, feed, or teat surfaces, during the transfer of milk to the tank car, and post-pasteurization recontamination in the dairy plant (Ziarno and Zareba, 2010). Paenibacillus strains display physiologically diverse characteristics, one distinctive characteri- stic of members of the genus is their ability to degrade a variety of macromolecules, amongst others proteins (Ash et al., 1993), polysaccharides (Kanzawa et al., 1995) and polyaromatic hydrocarbons (Daane et al., 2002). Some Paenibacillus strains also produce antibacterial (Martin et al., 2003; Piuri et al., 1998) or antifungal compounds (Beatty and Jensen, 2002; Chung et al., 2000).
The genus name of ginseng “Panax” is derived from the Greek pan (all) akos (cure), meaning “cure-all”. Although no single herb can be considered as a panacea, ginseng may come close to this effect. Ginseng is the most famous Asian medicinal plant, which has been used for therapeutic purpose for thousands of years (Li, 2004). Ginseng (the root of Panax ginseng C. A. Meyer, Araliaceae) has long been used as a medicinal purpose in Korea, China and Japan, since it exhibits anti-diabetic, anti-inflammatory (Shin et al., 2006), and anti-allergic activities (Attele et al., 1999). Active constituents found in most ginseng species include ginsenosides, polysaccharides, peptides, polyacetylenic alcohols, and fatty acids (Attele et al., 1999).
Ginsenoside Rd is the main hydrolyzed product of ginsenoside Rb1, Rb2, and Rc, which are contained more than 80% in ginseng roots (Son et al., 2008). Also ginsenoside Rd is the major precursor of ginsenoside Rg3, Rh2, F2 and Compound-K, which have high biological activities (Cheng et al., 2008). Among ginsenosides, minor ginsenoside Rd has been reportedly to have important biological functions including immunosuppressive (Wang et al., 2012), anti-inflammatory (Yang et al., 2012), immunological adjuvant (Han and Rhew, 2013), anticancer activity for gastric and breast cancer cell (Kim, 2013) and promotive effect on proliferation of neural stem cells (Palaniyandi et al., 2015).
Various transformation methods have been used to produce ginsenosides including mild acid hydrolysis (Han et al., 1982), enzymatic conversion (Ko et al., 2003), and microbial conversion (Bae et al., 2002). Of these methods, enzymatic conversion is the most desirable one for production of particular ginsenosides (Kim et al., 2005). β-glucosidase produced by various microorganisms such as Lactobacillus plantarum CRNB22 (Renchinkhand et al., 2015), Enterococcus faecalis CRNB-A3 (Renchinkhand et al., 2016) and Microbacterium sp. (Cheng et al., 2008) has been used for the production of biotransfomed ginsenosides.
We hypothesize that the β-glucosidase activity of the Paenibacillus sp. could be used to hydrolyze the ginsenoside Rb1. Therefore, objectives of this study were to: (1) isolate and characterize Paenibacillus sp. MBT213 possessing β-glucosidase activity from raw milk, and (2) examine the enzymatic capacity on the hydrolysis of a major ginsenoside (Rb1) in the Korean ginseng.
Materials and Methods
Ginsenoside standards and Ginseng
Ginsenoside standards Rb1, Rg3, Rd and Compound-K were kindly obtained from the Department of Pharmacology Science, College of Pharmacology, Chungnam National University (Korea) and Ginseng was purchased from Seungshin BST (Korea).
Isolation of β-glucosidase activity from fermented medicinal plants
Lactic acid producing spore forming strains were isolated from raw milk. Samples were serially diluted 10 times with 0.1 M phosphate buffered saline (0.85% NaCl, pH 7.2) and were plated on a BCP agar (Bromo Cresol Purple agar, Eiken Chemical Co. Ltd., Japan) at 35°C for 24 h. Strains produced lactic acid, and the color of the media was changed from purple to yellow. Morphologically different colonies with yellow color were selected, identified and individually plated, which were subsequently tested for their β-glucosidase production with esculin iron agar (Sigma-Aldrich, Chemie GmbH, Germany) at 35°C for 24 h. The esculin positive colonies were selected for dark brown zones after incubation for 24 h at 37 and 20°C (Kim et al. 2005).
Enzyme activity patterns
Enzyme activity patterns of the selected strains were examined using the API ZYM enzyme system (version 4.0 of API LAB plus, BioMérieux, France) according to the manufacturer’s instructions.
Conversion of ginsenoside Rb1 by selected strain MBT213
Esculin positive colonies were inoculated in Trypticase Soy (pH 7.0) (BBL, USA) and supernatants were collected by centrifugations at 7,000×g for 15 min at 4°C. Supernatants of selected cultures with β-glucosidase activity were tested for the hydrolysis of the major ginsenoside Rb1. The reaction mixture in the ratio 1:1 (v/v) of supernatant (pH 7.0) and each of 0.2 mg/mL of protopanaxadiol ginsenoside Rb1, Rb2 and protopanaxatriol ginsenoside Re (5 mM of sodium phosphate buffer, pH 7.0), respectively, were filtered through a 0.2 μm membrane filter (Toyo Roshi Kaisha Ltd, Japan). All reaction mixtures were incubated at 35°C for 7 d. The effect of pH on the conversion of ginsenoside by strain MBT213 was determined by examining the pH range of 4.0-9.0. These reaction mixtures were used to hydrolyze ginsenoside Rb1 at 30°C for 7 d. The effect of temperature on the conversion of ginsenoside by strain MBT213 was determined by hydrolyzing ginsenoside Rb1 in pH 7.0 at 30, 35 and 40°C for 7 d.
Thin layer chromatography (TLC) analysis
After incubation for 7 d, the reaction mixture was vigorously mixed with super butanol at the ratio 1:1 (v/v) and upper layer was collected for TLC analysis. Ginsenoside Rb1 hydrolysis was monitored with TLC performed on Silica gel 60 F254 plates (Merck, Germany). A solvent mixture of chloroform: methanol: water (65:35:10, v/v/v, lower phase) was used as the developing solvent. The resultant reaction spots were detected by spraying 10% sulfuric acid in methanol and heating under a lamp flame until the spots were clearly visible.
High performance liquid chromatography (HPLC) analysis of ginsenoside hydrolysates
Ginsenoside hydrolysates obtained from the β-glucosidase activity of strains MBT213 were analyzed using a reversed-phase HPLC system Shimadzu LC-6AD (Shimadzu Co., Ltd., Japan) and an ACE-5-C18 column (4.6×250 mm) equilibrated with solvent A (H2O). The ginsenoside hydrolysates were eluted with a linear gradient of solvent B (CH3CN) using an elution gradient system [0-35 min (20% B), 35-85 min (40% B), 85-105 min (50% B), 105-135 min (65% B), 135-145 min (85% B), 145-155 min (100% B), 155-160 min (100% B), 160-163 min (20% B), 163-165 min (20% B), respectively]. Runs were conducted at 40°C using the same HPLC system at a flow rate of 1.0 mL /min, and the absorbance of the elute samples was read at 203 nm using a UV detector 1260 Infinity (Agilent Technologies Inc., Japan). All samples were filtered through 0.5 µm Acrodisc Syringe Filters (Gelman Laboratory, USA).
16S rDNA sequencing of the strain MBT213
Selected strains were cultured in TS broth for 24 h, and 1 mL of culture was harvested via centrifugation. Genomic DNA was isolated using a Genomic DNA prep Kit (Solgent Co., Ltd., Korea). The primers used were 27F: 5'-AGAGTTTGATCACTGGC-TCAG-3' and 1492R: 5'-GGTTACCTTGTTACGACTT-3'. PCR was performed using the 2×PCR pre Mix (EF-taq) in a VeritiTM 96-Well Thermal Cycler (Applied Biosystems, USA). PCR conditions were as follows: an initial denaturation step for 3 min at 95°C, 30 cycles of denaturation at 94°C for 20 s, annealing at 56°C for 40 s, extension at 72°C for 40 s, and a final elongation step at 72°C for 3 min. PCR sequences were compared with those in the NCBI database using BLAST. Phylogenetic trees of selected bacteria, including MBT213, were drawn using Bioedit (Tom Hall Ibis Biosciences, An Abbott Co., USA) and the MEGA 5 program (Arizona State University, USA).
Fermentation of ginseng root by the strain MBT213
Ginseng roots were sliced using mixer for 5 min and sterilized at 75°C for 10 min. A 20% ginseng root solution was incubated with 3% strain MBT213 at 35°C for 14 d. After 3, 7 and 14 d, fermented ginseng root solutions were freeze-dried and resolved in 50% methanol for protein precipitation. Centrifugation was performed at 7000×g for 20 min and the supernatant was collected for freezing-drying. The supernatant was resolved in methanol (95%) and filtered using the 0.5 µm Acrodisc Syringe Filters (Gelman Laboratory, USA). Filtrates were used to TLC and HPLC analysis in order to determine conversion ginsenosides. Determination of viable cell counts was performed by plating in triplicate using a TS Agar (BBL, USA) and incubating at 37°C for 48 h after serial dilution with sterile 0.85% saline.
Statistical analysis
Statistical analyses were performed using the SPSS version 22 statistical package (SPSS Inc., USA). Analysis of variance and paired t-tests were applied to the data to determine significant difference, and a level of p<0.05 was considered statistically significant.
Results and Discussion
Isolation and screening of Paenibacillus strain exhibiting β-glucosidase activity
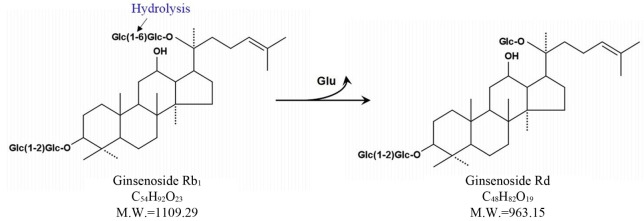
Esculin is chromogenic substrate for detection of β-glucosidase. Many researchers have used esculin as quick screening method for isolation of microorganisms exerting β-glucosidase activity (Gabriel et al., 2010). Using the esculin screening method in our study, a number of lactic acid producing bacteria were isolated from 14 types of Korean traditional fermented medicinal plants products. Among these bacterial strains, 37 selected strains were examined for β-glucosidase production capacities in the esculin agar for at 35°C for 24 h. It was found that MBT 213 was the most strong esculin positive strain. The results also showed that 18 strains, MBT213, 61L, 63L, 64L, 65L, 66L, 67L, 68L, 70L, 71L, 72bS, 72S, 73S, 73sS, 73bS, 103L, 122 and 123 exhibited the esculin positive reaction (Table 1). The 8 strains of these 18 esculin positive strains, the MBT213, 63L, 65L, 71L, 72S, 73S, 103L, 123 were selected for genomic DNA analysis based on cell morphology. The strain MBT213 had 4 types of enzymatic activity such as α-galactosidase, β-galactosidase, α-glucosidase and β-glucosidase in carbohydrate hydrolysis (Table 2). Among these enzymes, β-glucosidase can hydrolyze glycosidic linkage in ginsenosides. β-Glucosidase crude enzyme of strain MBT213 hydrolyzed glucose-glucose (6-1) linkages at C-20 in ginsenoside Rb1(molecular weight = 1109.29) and released one glucose molecule and ginsenoside Rd (molecular weight = 963.15) (Fig. 1). This would be the main conversion pathway of ginsenoside Rb1 to ginsenoside Rd by β-glucosidase enzyme, as shown in Fig. 1 (Suzuki et al. 2010). Among carbohydrate hydrolyzing enzymes, β-glucosidase can hydrolyze glucose-glucose (6-1) linkages (70%), glucose-arabinose (pyr) linkages (6-1) (20%), glucose-arabinose (fur) (6-1) linkages (20%) of ginsenoside Rb1 in ginseng, and β-galactosidase can hydrolyze glucose-glucose (2-1) linkages (100%), glucose-glucose (6-1) linkages (100%) of ginsenoside Rb1 in ginseng (Suzuki et al., 2010). This report supports the high hydrolyzing capacity of ginsenosides Rb1 by β-glucosidase in the strain MBT213 in our study. When the major ginsenoside Rb1, Rb2 and Re in the fermented ginseng samples were hydrolyzed by the strain MBT213, ginsenoside Rb1 was converted to Rd in minor quantity, while no conversions were occurred for gensenosides Rb2 and Re (Fig. 2).
Table 1. Isolation of strain with β-glucosidase activity from 14 samples at 35°C for 24 hr in esculin agar.
| Sample number | Colonies with yellow color in BCP agar | Esculin positive | Selected for DNA analysis |
|---|---|---|---|
| 1 | - | - | - |
| 2 | MBT213 | MBT213 | MBT213 |
| 3 | - | - | - |
| 4 | 41S, 42S, 43S, 44L, 45L, 46L, 47L | - | - |
| 5 | 51S | - | - |
| 6 | 61L, 63L, 64L, 65L, 66L, 67L, 68L, 70L | 61L, 63L, 64L, 65L, 66L, 67L, 68L, 70L | 63L, 65L |
| 7 | 71L, 72bS, 72S, 72’S, 73sS, 73bS | 71L, 72bS, 72S, 72’S, 73sS, 73bS | 71L, 72S, 73S |
| 8 | 84S, 85L | - | - |
| 9 | 96L, 97L | - | - |
| 10 | 101L, 102L, 103L, 104L | 103L | 103L |
| 11 | - | - | - |
| 12 | 122, 123, 124L, 125L | 122, 123 | 123 |
| 13 | - | - | - |
| 14 | 141, 142 | - | - |
| Total | 37 colonies | 18 colonies | 8 colonies |
Table 2. Enzymatic activities of strain MBT213 using API ZYM kit.
| Enzymes | Strain MBT213 |
| Control | 0* |
| Alkaline phophatase | 1 |
| Esterase (c4) | 3 |
| Esterase lipase (C8) | 1 |
| Lipase (c14) | 0 |
| Leucine arylamidase | 2 |
| Valine arylamidase | 2 |
| Cystine arylamidase | 0 |
| Trypsin | 0 |
| α-chymotrypsin | 0 |
| Acid phosphatase | 1 |
| Naphtol-AS-BO-phosphohydrolase | 2 |
| α-galactosidase | 1 |
| β-galactosidase | 3 |
| β-glucuronidase | 0 |
| α-glucosidase | 1 |
| β-glucosidase | 3 |
| N-acetyl-β-glucosamidase | 0 |
| α-mannosidase | 0 |
| α-fucosidase | 0 |
*Quantity of hydrolyzed substrate. 0: 0 nmol, 1: 5 nmol, 2: 10 nmol, 3: 20 nmol, 4: 30 nmol, 5: ≥40 nmol.
Fig. 1. Pathway on the conversion of ginsenoside Rb1 to Rd. (Suzuki et al., 2010).
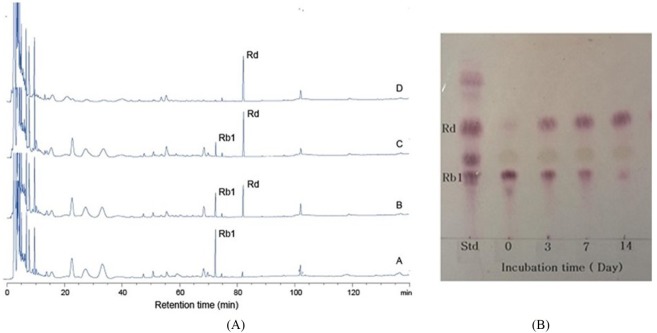
Fig. 2. TLC analysis of ginsenoside Rb1, Rb2 and Re by strain MBT213TLC was performed on Silica gel 60 F254 plates.
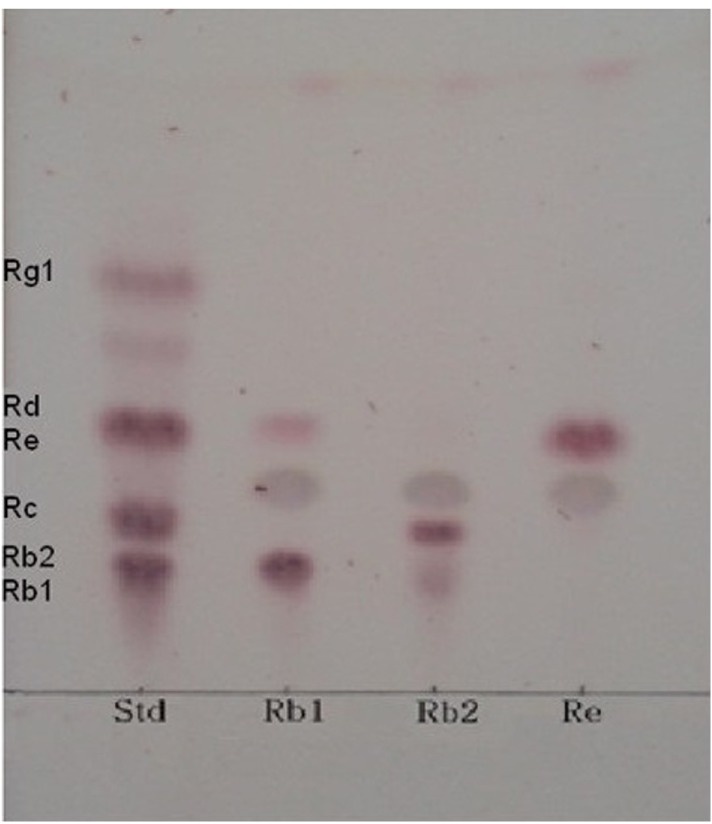
A solvent mixture of chloroform: methanol: water (65:35:10, v/v/v, lower phase) was used as the developing solvent.
Optimum pH and temperature for crude enzyme and conversion of Rb1
In conversion of ginsenoside Rb1, the optimum pH and temperature for the crude enzyme extracted from strain MBT213 were found to be pH 7.0 (Fig. 3A) and 35°C (Fig. 3B), respectively. Under these conditions, the β-glucosidase enzyme obtained from MBT213 converted ginsenoside Rb1 to ginsenoside Rd. After 14 d of hydrolysis, ginsenoside Rb1 was fully converted to ginsenoside Rd, and the chromatographic analyses of the conversion of ginsenoside Rb1 to ginsenoside Rd is as shown in HPLC (Fig. 4A) and TLC results (Fig. 4B). Under the above optimal conditions for 14 d, 3.794±0.061 mg/mL of ginsenoside Rb1 was fully hydrolyzed by crude enzyme of strain MBT213, and was released 0.110±0.01 mg/mL ginsenoside Rd (Table 2). On the other hand, in a study of isolation of Lactobacillus plantarum CRNB22 and its screening of LAB exhibiting β-glucosidase activity from Kimchi (Renchinkhand et al., 2015), observed that ginsenoside Rb1 was hydrolyzed under optimum condition (pH 6.0 or 7.0 at 35°C) for 7 d. In addition, under these optimal conditions, the crude enzyme obtained from CRNB22 converted the ginsenoside Rb1 to Rg3 (epimer S/R) and Rg5. After 7 d of hydrolysis, ginsenoside Rb1 was decreased from 1.019 mg to 0.773 mg, and then converted to 0.135 mg Rg3-S, 0.038 mg Rg3-R, and 0.072 mg Rg5. These reported data are in agreement with our observations in this study. Depending on types of enzymes and experimental conditions, different conversion compounds of ginsenoside Rb1 have been generated, such as Rd, Rg3, Compound-K and other compounds (Suzuki et al., 2010).
Fig. 3. The effect of pH and temperature on the conversion of ginsenoside by strain MBT213.
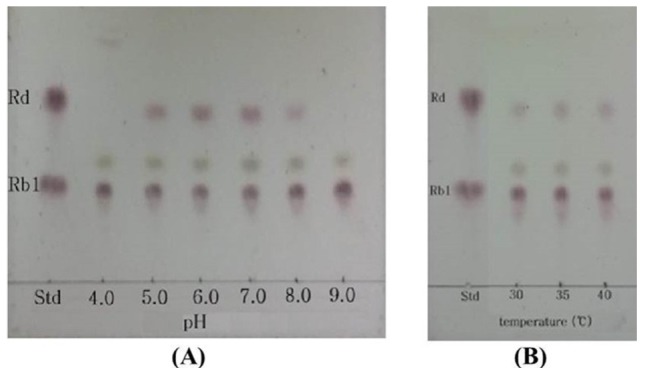
(A) pH, (B) Temperature. TLC was performed on Silica gel 60 F254 plates. A solvent mixture of chloroform: methanol: water (65:35:10, v/v/v, lower phase) was used as the developing solvent.
Fig. 4. Conversion of gincenoside Rb1 hydrolyzed by strain MBT213.
(A) HPLC analysis of ginsenoside Rb1 conversation at optimum temperature and optimum pH for 14 d hydrolysis by using ACE-5-C18 column (4.6×250 mm) equilibrated with solvent A (H2O). A-0 d, B-3 d, C-7 d, D-14 d; (B) TLC analysis of ginsenoside Rb1 at optimum temperature and pH for 14 d hydrolysis.
16S rDNA sequencing of the strain MBT213
Sequencing of 16S rDNA was performed to determine the correct species of the isolated strain MBT213. PCR produced many sequences, which corresponded to the full length 16S rDNA sequence of the strain MBT213. The sequence consisted of 1450 bp, which were compared to those in the NCBI database using BLAST and this strain is homology in 95% (1374 bp within 1450 bp) to Paenibacillus sp. JDR2 and is homology in 95% (1374 bp within 1450 bp) to Paenibacillus polyxa E681. As shown in Fig. 5, the sequence was identical to Paenibacillus according to the phylogenetic tree. The isolated MBT213 strain was thus named Paenibacillus sp. MBT213.
Fig. 5. Phylogenetic tree based on 16S rDNA sequences showing the position of strain MBT213, showing the phylogenetic relationships among strain MBT213 and related bacteria.
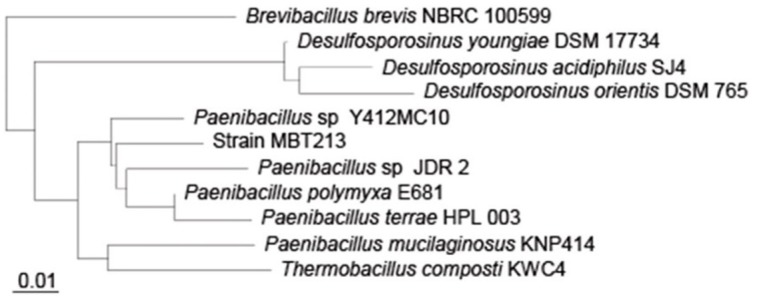
Scale length is 0.01.
Characteristics of fermented ginseng by Paenibacillus sp. MBT213
Table 3 shows the conversion of ginsenoside Rb1 to ginsenoside Rd by Paenibacillus sp. MBT213 in 20% fermented ginseng roots solution. In addition, it demonstrates the viable cell counts in the experimental fermented ginseng products. The initial cell count of Paenibacillus sp. MBT213 was 3.60±0.02 Log CFU/mL. Viable cell counts of Paenibacillus sp. MBT213 were increased to 8.07±0.021 Log CFU/mL for 7 d, and then after 14 d incubation, these cell counts were maintained at 7.16±0.016 Log CFU/mL level. Results of HPLC analysis of the fermented ginseng roots (Fig. 6) revealed that the peak of ginsenosides Rb1 was decreased and ginsenoside Rd was significantly increased after 14 d. In addition, after 14 d fermentation, the amount of ginsenoside Rb1 was decreased from 2.670±0.14 to 1.351±0.049 mg/mL during the fermentation, while amount of minor ginsenoside Rd was increased from 0 to 0.087±0.013 mg/mL (Table 3). The decrease in ginsenoside Rb1 and increase in Rd in the fermented of ginseng roots are apparently attributable to the conversion of ginsenoside Rb1 to Rd, which was fermentation time dependent. Park et al. (2016) reported that 6 types of lactic acid bacteria in fermented ginseng multiplied to approximately 1×109−1011 CFU/mL on first day of fermentation, and after 5 d of fermentation, viable cell counts of LAB were dropped to 4×107 CFU/mL. They also found the continuous decreases in the amounts of ginsenoside Rb1, Rb2, Rc in PPD type, while increases in ginsenoside Rd during fermentation by the LAB. Consequently, viable cell counts of fermented ginseng roots were increased, since ginseng roots contain various substrates, such as polysaccharides, peptides, vitamins, minerals as growth factors of the microorganisms. These findings in bacterial growth and ginsenoside changes in the fermented ginseng roots in their study are in agreement with the results that were observed in our study. This strain was deposited in the KACC database (http://www.kacc.rda.go.kr) under the registration number KACC-91831P and NCBI GenBank accession number is NCBINam.sqn A MF972642.
Table 3. Amounts of conversion of ginsenoside Rb1 to Rd and viable cell counts.
| Incubation time (d) | |||||
|---|---|---|---|---|---|
| 0 | 3 | 7 | 14 | ||
| A | Rb1 (mg/mL) | 3.794±0.061* | 1.599±0.031 | 0.868±0.032 | 0 |
| Rd (mg/mL) | 0 | 0.058±0.006 | 0.086±0.011 | 0.110±0.01 | |
| B | Rb1 (mg/mL) | 2.670±0.14 | 1.821±0.071 | 1.521±0.179 | 1.351±0.049 |
| Rd (mg/mL) | 0 | 0.017±0.005 | 0.069±0.011 | 0.087±0.013 | |
| C | Paenibacillus sp. MBT213 (Log CFU/mL) | 3.60±0.02 | 7.48±0.021 | 8.07±0.021 | 7.16±0.016 |
*Means±SD (n=3).
A) Amounts of conversion of ginsenoside Rb1 by supernatant of strain MBT213 during the 14 days; B) Amounts of conversion of ginsenoside Rb1 and Rd in ginseng root by strain MBT213 during the 14 days; C) Viable cell counts of strain MBT213 in 20% ginseng root.
Fig. 6. HPLC analysis.
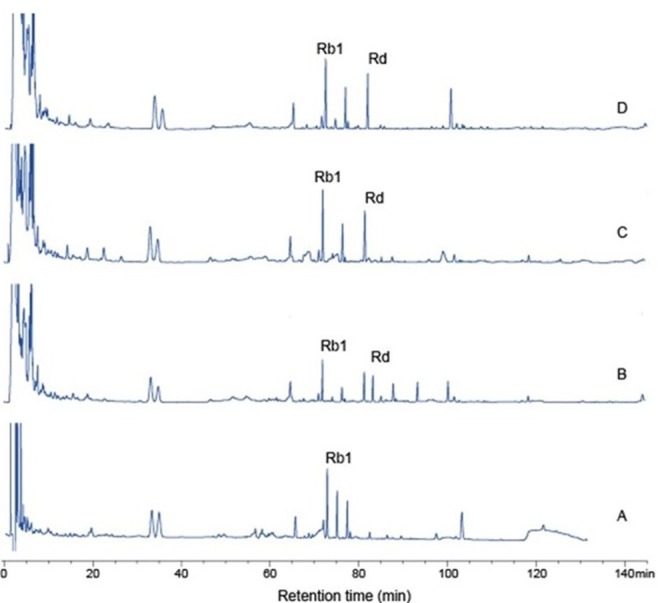
Ginsenosides conversion in 20% ginseng by strain Paenibacillus sp. MBT213 for 3 and 14 d; A-0 d, B-3 d, C-7 d, D-14 d.
Conclusion
The study has confirmed that the β-glucosidase produced by Paenibacillus sp. MBT213 can hydrolyze the major ginsenoside Rb1 and convert to Rd during fermentation of the ginseng. The β-glucosidase activity of this novel Paenibacillus sp. MBT213 strain may be utilized in development of variety of health foods and pharmaceutical products.
Acknowledgments
Gereltuya Renchinkhand and Soo Hyun Cho are equally qualified for senior authors. This study was supported by a grant (110060032SB020) from the Ministry for Food, Agriculture, Forestry and Fisheries of the Republic of Korea.
References
- Ash C., Farrow J. A. E., Wallbanks S., Collins M. D. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequence. Lett. Appl. Microbiol. 1991;13:2002–2006. [Google Scholar]
- Attele A. S., Wu J. A., Yuan C. S. Ginseng pharmacology: Multiple constituents and multiple actions. Biochem. Pharmacol. 1999;58:1685–1693. doi: 10.1016/S0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- Bae E. A., Han M. J., Cho M. K., Park S. Y., Kim D. H. Metabolism of 20(S) and 20(R) ginsenoside Rg3 by human intestinal bacteria and its relation to in vitro biological activities. Biol. Pharm. Bull. 2002;25:58–63. doi: 10.1248/bpb.25.58. [DOI] [PubMed] [Google Scholar]
- Beatty P. H., Jensen S. E. Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 2002;48:159–169. doi: 10.1139/w02-002. [DOI] [PubMed] [Google Scholar]
- Cheng L. Q., Na J. R., Bang M. H., Kim M. K., Yang D. C. Conversion of major ginsenoside Rb1 to 20(S)-ginsenoside Rg3 by Microbacterium sp. GS514. Phytochemistry. 2008;69:218–224. doi: 10.1016/j.phytochem.2007.06.035. [DOI] [PubMed] [Google Scholar]
- Chung Y. R., Kim C. H., Hwang I., Chun J. Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int. J. Syst. Evol. Microbiol. 2000;50:1495–1500. doi: 10.1099/00207713-50-4-1495. [DOI] [PubMed] [Google Scholar]
- Daane L. L., Harjono I., Barns S. M., Launen L. A., Palleroni N. J., Häggblom M. M. PAH-degradation by Paenibacillus spp. and description of Paenibacillus naphthalenovorans sp. nov., a naphthalene-degrading bacterium from the rhizosphere of salt marsh plants. Int. J. Syst. Evol. Microbiol. 2002;52:131–139. doi: 10.1099/00207713-52-1-131. [DOI] [PubMed] [Google Scholar]
- Gabriel P., Farina L., Barquet M., Boido E. A quick screening method to identify β-glucosidase activity innative wine yeast strains: application of Esculin Glycerol Agar (EGA) medium. World J. Microbiol. Biotechnol. 2010;27:47–55. [Google Scholar]
- Han B. H., Park M. H., Han Y. N., Woo L. K., Sankawa U., Yahara S., Tanaka O. Degradation of ginseng saponins under mild acidic conditions. Planta Med. 1982;44:146–149. doi: 10.1055/s-2007-971425. [DOI] [PubMed] [Google Scholar]
- Han Y. M., Rhew K. Y. Ginsenoside Rd induces protective anti-Candida albicans antibody through immunological adjuvant activity. Int. Immunopharmacol. 2013;17:651–657. doi: 10.1016/j.intimp.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Kanazawa Y., Harada A., Takeuchi M., Yokota A., Harada T. Bacillus curdlanolyticus sp. nov. and Bacillus kobensis sp. nov., which hydrolyze resistant curdlan. Int. J. Syst. Evol. Microbiol. 1995;45:515–521. doi: 10.1099/00207713-45-3-515. [DOI] [PubMed] [Google Scholar]
- Kim B. J. Involvement of melasatin type transient receptor potential 7 channels in ginsenoside Rd-induced apoptosis in gastric and breast cancer. J. Ginseng Res. 2013;37:201–209. doi: 10.5142/jgr.2013.37.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. K., Lee J. W., Lee K. Y., Yang D. C. Microbial conversion of major ginsenoside Rb(1) to pharmaceutically active minor ginsenoside Rd. J. Microbiol. 2005;43:456–462. [PubMed] [Google Scholar]
- Ko S. R., Choi K. J., Uchida K., Suzuki Y. Enzymatic preparation of ginsenosides Rg2, Rh1, and F1 from proto-panaxatriol-type ginseng saponin mixture. Planta Med. 2003;69:285–286. doi: 10.1055/s-2003-38476. [DOI] [PubMed] [Google Scholar]
- Martin N. I., Hu H., Moake M. M., Churey J. J., Whittal R., Worobo R. W., Vederas J. C. Isolation, structural characterization, and properties of mattacin (polymyxin M) a cyclic peptide antibiotic produced by Paenibacillus kobensis M. J. Biol. Chem. 2003;278:13124–13132. doi: 10.1074/jbc.M212364200. [DOI] [PubMed] [Google Scholar]
- Palaniyandi S. A., Damodharan K., Lee K. W., Yang S. H., Suh J. W. Enrichment of ginsenoside Rd in Panax ginseng extract with combination of enzyme treatment and high hydrostatic pressure. Biotechnol. Bioprocess Eng. 2015;20:608–613. doi: 10.1007/s12257-014-0857-z. [DOI] [Google Scholar]
- Park S. E., Na C. S., Yoo S. A., Seo S. H., Son H. S. Biotransformation of major ginsenosides in ginsenoside model culture by lactic acid bacteria. J. Ginseng Res. 2016 doi: 10.1016/j.jgr.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piuri M., Sanchez-Rivas C., Ruzal S. M. A novel antimicrobial activity of a Paenibacillus polymyxa strain isolated from regional fermented sausages. Lett. Appl. Microbiol. 1998;27:9–13. doi: 10.1046/j.1472-765X.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Renchinkhand G., Park Y. W., Cho S. H., Song G. Y., Bae H. C., Choi S. J., Nam M. S. Identification of β-glucosidase activity of Lactobacillus plantarum CRNB22 in Kimchi and its potential to ginsenoside Rb1 from Panax ginseng. J. Food Biochem. 2015;39:155–163. doi: 10.1111/jfbc.12116. [DOI] [Google Scholar]
- Renchinkhand G., Park Y. W., Song G. Y., Cho S. H., Choi S. J., Urgamal M., Bae H. C., Choi J-W., Nam M. S. Identification of β-glucosidase activity of Enterococcus faecalis CRNB-A3 in Airag and its potential to convert ginsenoside Rb1 from Panax ginseng. J. Food Biochem. 2016;40:120–129. doi: 10.1111/jfbc.12201. [DOI] [Google Scholar]
- Scheldeman P., Goossens K., Rodriguez-Diaz M., Pil A., Goris J., Herman L., Vos P. D., Logan N. A., Heyndrickx M. Paenibacillus lactis sp. Nov., isolated from raw and heat-treated milk. Int. J. Syst. Evol. Microbiol. 2004;54:885–891. doi: 10.1099/ijs.0.02822-0. [DOI] [PubMed] [Google Scholar]
- Shin Y. W., Bae E. A., Kim D. H. Inhibitory effect of ginsenoside Rg5 and its metabolite ginsenoside Rh3 in an oxazolone induced mouse chronic dermatitis model. Arch. Pharm. Res. 2006;29:685–690. doi: 10.1007/BF02968253. [DOI] [PubMed] [Google Scholar]
- Son J. W., Kim H. J., Oh D. K. Ginsenoside Rd production from the major ginsenoside Rb1 by β-glucosidase from Thermus caldophilus. Biotechnol. Lett. 2008;30:713–716. doi: 10.1007/s10529-007-9590-4. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Ko S. R., Choi K. J., Suzuki K., Kim Y. H., Cho B. G., Nho K. B., Jang D. S., Kim S. C., Kim C. M. Production of minor ginseng saponins and intestine bacterial ginseng saponin metabolites by enzymatic method; The 10th International Symposium on Ginseng; September 13-16; Seoul, Korea. 2010. p. 268. [Google Scholar]
- Wang L., Zhang Y., Chen J., Li S., Wang Y., Hu L., Wang L., Wu Y. Immunosuppressive effects of ginsenoside Rd on skin allograft rejection in rats. J. Surg. Res. 2012;176:267–274. doi: 10.1016/j.jss.2011.06.038. [DOI] [PubMed] [Google Scholar]
- Yang X. L., Guo T. K., Wang Y. H., Huang Y. H., Liu X., Wang X. X., Li W., Zhao X., Wang L. P., Yan S., Wu D., Wu Y. J. Ginsenoside Rd attenuates the inflammatory response via modulating p38 and JNK signaling pathways in rats with TNBS-induced relapsing colitis. Int. Immunopharm. 2012;12:408–414. doi: 10.1016/j.intimp.2011.12.014. [DOI] [PubMed] [Google Scholar]
- Ziarno M., Zareba D. Characteristics and occurrence of Paenibacillus bacteria in milk and milk products. Medycyna Weterynaryina. 2010;66:600–603. [Google Scholar]