Abstract
The transcriptional represser Mnt is a functional antagonist of the proto-oncoprotein Myc. Both Mnt and Myc utilise Max as an obligate partner for DNA binding, and Myc/Max and Mnt/Max complexes compete for occupancy at E-box DNA sequences in promoter regions. We have previously shown in transgenic mouse models that the phenotype and kinetics of onset of haemopoietic tumours varies with the level of Myc expression. We reasoned that a decrease in the level of Mnt would increase the functional level of Myc and accelerate Myc-driven tumorigenesis. We tested the impact of reduced Mnt in three models of myc transgenic mice and in p53+/− mice. To our surprise, mnt heterozygosity actually slowed Myc-driven tumorigenesis in vavP-MYC10 and Eμ-myc mice, suggesting that Mnt facilitates Myc-driven oncogenesis. To explore the underlying cause of the delay in tumour development, we enumerated Myc-driven cell populations in healthy young vavP-MYC10 and Eμ-myc mice, expecting that the reduced rate of leukaemogenesis in mnt heterozygous mice would be reflected in a reduced number of preleukaemic cells, due to increased apoptosis or reduced proliferation or both. However, no differences were apparent. Furthermore, when mnt+/+ and mnt+/− pre-B cells from healthy young Eμ-myc mice were compared in vitro, no differences were seen in their sensitivity to apoptosis or in cell size or cell cycling. Moreover, the frequencies of apoptotic, senescent and proliferating cells were comparable in vivo in mnt+/− and mnt+/+ Eμ-myc lymphomas. Thus, although mnt heterozygosity clearly slowed lymphomagenesis in vavP-MYC10 and Eμ-myc mice, the change(s) in cellular properties responsible for this effect remain to be identified.
c-Myc (hereafter Myc) is a basic helix–loop–helix–leucine zipper (bHLHZip) transcription factor that regulates a multitude of cellular processes, including cell growth, proliferation, metabolism, DNA damage response, malignant transformation and apoptosis.7, 10, 14, 16, 20, 48, 49 In normal cells, Myc expression is tightly regulated during the cell cycle but in many cancers control is abrogated, due to chromosomal translocation, mutation or amplification of the myc gene, or as a result of mutations affecting upstream regulatory pathways. Whether Myc targets specific groups of genes or serves as a global amplifier of the transcriptional programme inherent to individual cell types remains the subject of vigorous debate and experimentation.27, 29, 34 Irrespective, the level of Myc expression critically influences cellular outcome.33
Myc heterodimerises with a ubiquitous bHLHZip protein, Max, and binds the E-box CACGTG to activate gene transcription.5 Max also binds transcriptional repressors containing Myc-related bHLHZ domains, such as the Mxd proteins and Mnt.10, 22, 55 Since these repressors compete with Myc for available Max and also bind E boxes, they function as Myc antagonists. Hence their loss would be predicted to mimic Myc overexpression. Deletion of Mxd proteins in mice did not produce a phenotype resembling Myc overexpression, perhaps because expression of Mxd proteins is primarily confined to differentiated cells.22 In contrast, loss of Mnt, which is expressed in both proliferating and differentiating cells in all tissues tested,24 resulted in accelerated proliferation, susceptibility to transformation by oncogenic Ras and sensitivity to apoptosis, all characteristics of Myc overexpression.22, 25, 35 Surprisingly, these effects of Mnt knockdown were observed also in cells lacking Myc, leading to the suggestion that Myc functions as a Mnt antagonist, relieving Mnt-mediated repression, rather than by transcriptional activation as is widely believed.35 However, irrespective of whether Mnt controls Myc or Myc controls Mnt, the level of Mnt would be expected to influence the oncogenic potential of Myc.
Mnt can serve as a tumour suppressor, as tissue-specific deletion of Mnt in mice resulted in mammary adenocarcinomas and T-cell lymphomas.8, 25, 47 Conversely, Mnt overexpression mimicked Myc knockout, producing growth defects and embryonic lethality25 as well as reducing cell cycle entry and proliferation of MEFs in vitro.53 In further support of a tumour suppressor role, deletion of the region encompassing the MNT locus has been reported in human chronic lymphocytic leukaemia12 and in Sezary syndrome,52 a cutaneous T-cell lymphoma/leukaemia. A recent study has suggested that the dominant role of Mnt, at least in proliferating T cells, is suppression of Myc-driven apoptosis.30
Transgenic mice have been a powerful tool for analyzing the role of Myc in malignancy.32 Our Eμ-myc transgenic mice,1, 21 which model the 8;14 translocation found in Burkitt’s B lymphomas, have provided many insights into Myc-driven lymphomagenesis. The overexpression of Myc produces a polyclonal increase in pre-B cells in young mice, accompanied by reduced differentiation to mature B cells.28 However, Myc overexpression also enhances susceptibility to apoptosis,3, 17, 45 which restrains progression of preleukaemic cells to malignancy. Consequently, anti-apoptotic mutations synergise with myc to promote malignant transformation4, 13, 15, 40, 44, 51 and the pre-B and B lymphomas that eventually arise in Eμ-myc mice often harbour mutations that mutate p53 or its upstream regulators,15 as do Burkitt lymphomas.18, 19, 31, 54
More recently we developed several transgenic lines that express Myc via haemopoietic-specific regulatory elements from the vav gene.42, 43 The tumour phenotype of the vavP-MYC mice varied between lines, depending on the level of Myc achieved: high expression resulted in rapid-onset T-cell lymphomas whereas low expression resulted in late-onset myeloid tumours and intermediate levels produced both tumour types.
Reasoning that the level of Mnt would affect Myc activity, in this study we have investigated the impact of mnt heterozygosity on Myc-driven tumorigenesis, using both vavP-MYC and Eμ-myc mouse models. We also investigated whether mnt heterozygosity influenced tumorigenesis in p53+/− mice.
Results
Mnt heterozygosity confers reduced Mnt expression
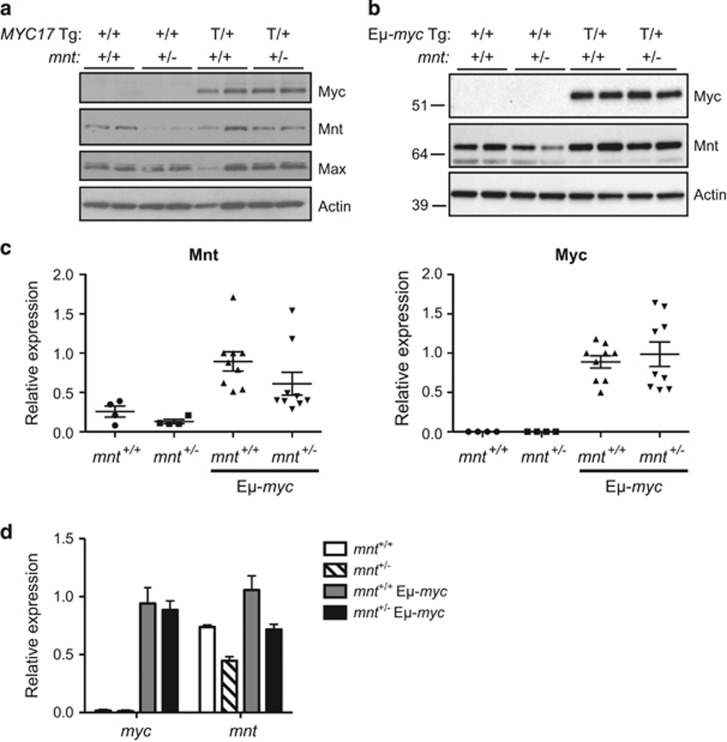
We first ascertained how mnt heterozygosity impacted expression of Mnt and Myc. Western blot analysis verified that the level of Mnt protein was lower in mnt+/− compared with mnt+/+ thymocytes, both on the WT and vavP-MYC17 transgenic background (Figure 1a) and in pre-B cells from WT and Eμ-myc mice (Figures 1b and c) and the level of transgenic Myc was unaltered. Quantitative PCR analysis confirmed that mnt RNA was ~2-fold lower in mnt+/− Eμ-myc versus mnt+/+ Eμ-myc pre-B cells (Figure 1d).
Figure 1.
Expression analysis of mnt+/+ and mnt+/− lymphoid cells from WT and myc transgenic mice. (a) Mnt and transgenic MYC protein expression in thymocytes from mnt+/+ and mnt+/− WT and vavP-MYC17 mice. Western blots of lysates of thymocytes from four-week-old mice of the indicated genotypes, probed with antibodies specific for Myc, Mnt, Max and β-actin (see Materials and Methods). Each lane represents an individual mouse. Comparable results were obtained for thymocytes from mnt+/+ and mnt+/− vavP-MYC10 mice. (b) Mnt and transgenic Myc protein expression in pre-B cells from mnt+/+ and mnt+/− Eμ-myc mice. Western blot analysis of pre-B cells (B220+sIg−) sorted by flow cytometry from bone marrow of 4-week-old mice of indicated genotypes. Each lane represents an individual mouse and MWs (kD) of markers run in parallel are indicated. Endogenous Myc was not detectable in WT control cells. (c) Quantification of Mnt and Myc protein expression in mnt+/+ and mnt+/− WT and Eμ-myc mice. The blot shown in (b) and two additional blots using independent samples were quantified using ChemiDoc and Image Lab software (Bio-Rad), with normalisation to actin expression. Expression is relative to the first mnt+/+ Eμ-myc sample on each blot. (d) Quantitative PCR analysis of myc and mnt RNA in pre-B cells isolated using magnetic beads coated with CD19 antibody from the bone marrow of 4-week-old mice of the indicated genotypes (n=2–15). Data are normalised to myc expression in mnt+/+ Eμ-myc mice
Decreased Mnt does not accelerate tumorigenesis in vavP-MYC transgenic mice
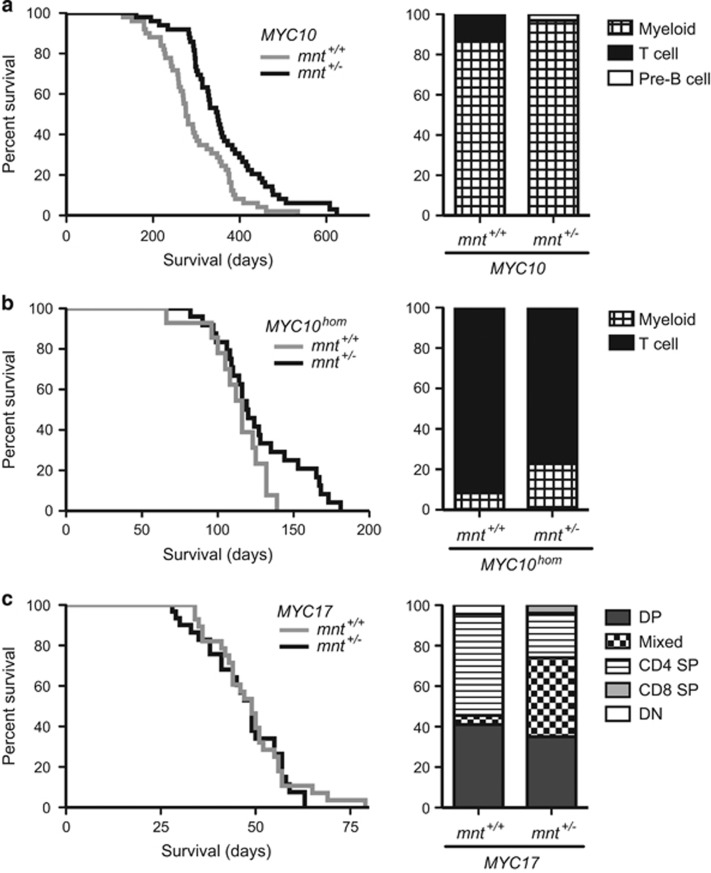
To study the impact of mnt heterozygosity on Myc-driven lymphomagenesis, we turned first to vavP-MYC10 mice because the level of MYC protein expressed in this line represents a critical functional threshold: When expression is increased ~2-fold (by generating mice homozygous for the transgenic locus; hereafter vavP-MYC10hom mice), the mice develop predominantly T lymphomas rather than the mixture of T lymphomas and myeloid tumours found in mice heterozygous for the transgene.43 We reasoned that lowering the level of Mnt in vavP-MYC10het mice would increase the level of MYC/Max heterodimers and drive the phenotype towards that seen in vavP-MYC10hom mice; viz more rapid tumour onset, and primarily T lymphomas rather than a mixture of T and myeloid tumours.43
Cohorts of mnt+/+vavP-MYC10 (n=54) and mnt+/−vavP-MYC10 (n=50) transgenic mice) were monitored for tumour onset. Surprisingly, the outcome was the opposite from that predicted: Mnt heterozygosity (black line) resulted in slower tumour-associated morbidity (P<0.001) (Figure 2a, left panel) and, with the exception of one pre-B lymphoma, all tumours were myeloid (Figure 2a, right panel).
Figure 2.
Impact of mnt heterozygosity on tumorigenesis in vavP-MYC mice. (a) Tumorigenesis in vavP-MYC10 mice. Left panel: Kaplan–Meier survival curve showing delayed morbidity in mnt+/− vavP-MYC10 mice (black line, n=50, median survival 349 days) versus mnt+/+vavP-MYC10 mice (grey line, n=54, median survival 276 days); P<0.001, Log-rank (Mantel–Cox) test. Right panel: Incidence of different tumour types, identified by immunostaining and flow cytometry (see Materials and Methods). For mnt+/+vavP-MYC10 mice, 13% of tumours were T lymphoid (CD4+CD8+) and 87% were myeloid (Mac1+); for mnt+/− vavP-MYC10 mice, 3% of tumours were pre-B cell (CD19+ B220+ IgM−IgD−) and 97% were myeloid (Mac1+). (b) Tumorigenesis in vavP-MYC10hom mice. Left panel: Kaplan–Meier survival curve for mnt+/+vavP-MYC10hommice (grey line, n=14, median 116 days) and mnt+/−vavP-MYC10hommice (black line n=25, median 120 days). Right panel: For mnt+/+vavP-MYC10hom mice, 92% of tumours were T lymphoid (CD4+CD8+) and 8% were myeloid (Mac1+); for mnt+/− vavP-MYC10hom mice, 77% of tumours were T lymphoid (CD4+CD8+) and 23% were myeloid. (c) Tumorigenesis in vavP-MYC17 mice. Left panel: Kaplan–Meier survival curve showing similar survival of mnt+/+vavP-MYC17 mice (grey line, n=33; median 7 weeks) and mnt+/− vavP-MYC17 mice (black line, n=34; median 7 weeks). Right panel: All tumours were T lymphoid. For mnt+/+ vavP-MYC17 mice, 41% were DP (CD4+ CD8+); 50% were CD4 SP (CD4+CD8−); 4.5% were DN (CD4−CD8−); 4.5% were mixed (DP and SP or DN). For mnt+/− vavP-MYC17 mice, 35% were DP; 22% were CD4 SP; 4% were CD8 SP (CD4−CD8+); 39% were mixed
In MYC10hom mice expressing higher MYC levels, mnt heterozygosity also retarded morbidity, although not reaching statistical significance (Figure 2b, left panel). Furthermore, the frequency of T lymphomas was reduced (77% in mnt+/−vavP-MYC10hom mice versus 92% in mnt+/+vavP-MYC10hom mice) and morbidity due to myeloid tumours increased (23% versus 8%) (Figure 2b, right panel). Taken together, these results suggested that Mnt heterozygosity reduces the risk of Myc-driven T lymphomagenesis in vavP-MYC10 transgenic mice.
Next, the impact of mnt heterozygosity was tested in vavP-MYC17 mice (Figure 2c). The level of vavP-MYC transgene expression is highest in this line and lymphomagenesis much faster.43 Therefore it was not particularly surprising to find that mnt heterozygosity had no impact on the kinetics of morbidity (Figure 2c, left panel). Interestingly, however, while all mice still succumbed to T lymphoma, there appeared to be a shift in tumour phenotype, a decrease in the frequency of CD4 SP (CD4+CD8−) T lymphomas (50% down to 22%) and an increase in those having a mixed (DP plus SP or DN) phenotype (4.5% up to 39%), suggestive of an impact on T lymphoid differentiation (Figure 2c, right panel).
Decreased Mnt retards tumorigenesis in Eμ-myc transgenic mice
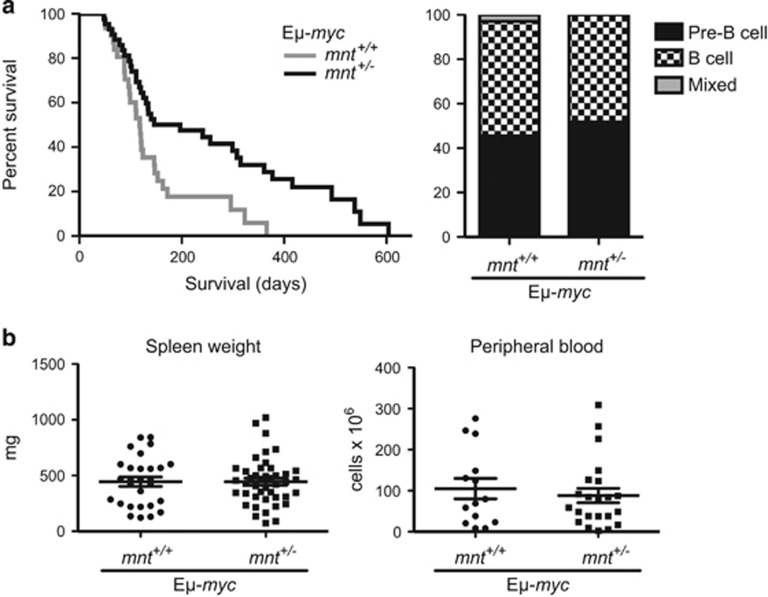
To ascertain whether mnt heterozygosity also impaired B lymphoid tumour development, we crossed mnt+/− mice with Eμ-myc mice, in which transgene expression is largely restricted to the B lymphoid lineage. Once again, mnt heterozygosity slowed rather than accelerated tumour development, the median onset being 197 days for mnt+/− Eμ-myc mice versus 118 days for mnt+/+ Eμ-myc littermates (P=0.004) (Figure 3a, left panel). There was no detectable change in tumour phenotype, both genotypes developing pre-B and B-cell lymphomas at about the same relative frequency (Figure 3a, right panel). Furthermore, there was no significant difference in the severity of disease, as reflected in the spleen weight or peripheral leucocyte count of moribund mice (Figure 3b).
Figure 3.
Impact of mnt heterozygosity on lymphomagenesis in Eμ-myc mice. (a) Left panel: Kaplan–Meier survival curve showing delayed morbidity from tumours in mnt+/− Eμ-myc mice (black line, n=43, median survival 28 weeks) compared with mnt+/+ Eμ-myc mice (grey line, n=31, median survival 17 weeks). P=0.0042. By 12 months, all mnt+/+ Eμ-myc mice had succumbed to tumours, whereas 29% of mnt+/− Eμ-myc mice were still alive. Right panel: mnt heterozygosity had no impact on lymphoma phenotype in Eμ-myc mice. Of 33 tumours from mnt+/+ Eμ-myc mice analysed, 45.5% were pre-B cell, 51.5% were B cell and 3% were mixed pre-B/B-cell tumours. Similarly, of 27 tumours from mnt+/− Eμ-myc mice, 52% were pre-B cell and 48% were B-cell tumours. (b) Tumour burden of moribund mice. Left panel: Spleen weight of moribund mnt+/+ Eμ-myc (n=28) and mnt+/− Eμ-myc mice (n=43) mice. Right panel: white blood cell count of moribund mnt+/+ Eμ-myc (n=14) and mnt+/− Eμ-myc (n=22) mice. Bars represent mean ±S.E.M. No significant differences were observed
Impact of Mnt heterozygosity in p53+/− mice
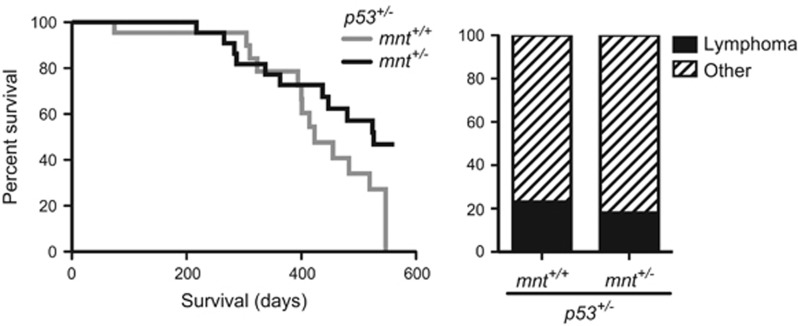
We also tested the impact of mnt heterozygosity in p53-deficient mice, in which Myc can have a major impact on tumorigenesis. Lymphomas develop rapidly and at high frequency in p53−/− mice but are rare in p53+/− mice, which develop a range of other cancers, albeit after a long latency.11, 26, 37 We had reasoned that if decreased Mnt increased the level of functional Myc in lymphoid cells of p53 heterozygotes, then the frequency of lymphomas would increase and the latency shorten, as it does if an Eμ-myc transgene is introduced into p53+/− mice.6, 23, 40 Once again, however, mnt heterozygosity tended to retard tumour development, although not reaching statistical significance (Figure 4, left panel). The frequency of lymphomas versus other tumour types was not altered (Figure 4, right panel).
Figure 4.
Impact of mnt heterozygosity in p53 heterozygous mice. Left panel: Kaplan–Meier survival curve for mnt+/+ p53+/− mice (grey line, n=24, median survival 60 weeks) and mnt +/− p53+/− mice (black line, n=25, median survival 75 weeks). Survival curves for p53+/− mnt+/+ and p53+/− mnt+/− are not significantly different. Right panel: Tumour types were unaffected by mnt heterozygosity. Lymphoma incidence was 23% in mnt+/+ p53+/− mice and 18% in mnt+/− p53+/− mice; other tumour-related deaths were predominantly due to osteosarcoma and sarcoma, as previously reported for p53+/− mice26
Does Mnt heterozygosity perturb preleukaemic cell populations in vivo?
In contrast to earlier studies,8, 25, 47 these results had provided no evidence for a tumour suppressor role for Mnt in any of the mouse models tested. Indeed, they suggested instead that Mnt facilitates Myc’s oncogenic role.
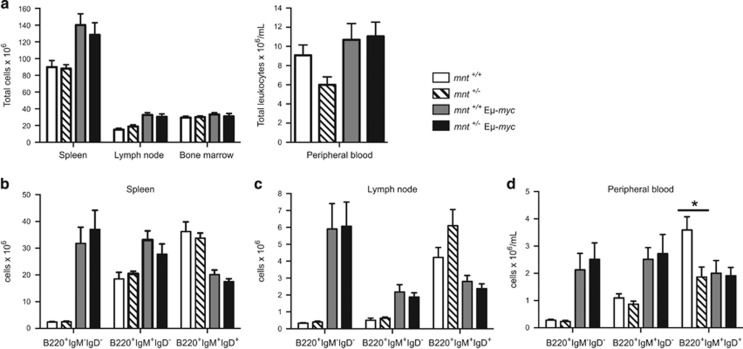
To gauge any impact of mnt heterozygosity on the premalignant Myc phenotype, we analysed haemopoietic tissues of healthy young mice, prior to tumour onset. Young Eμ-myc mice harbour excessive numbers of immature B lymphoid cells, accompanied by reduced numbers of mature B cells.28 No major differences were apparent between 6-week-old mnt+/− and mnt+/+ Eμ-myc mice (Figure 5 and Supplementary Table S1). In both genotypes, the bone marrow, spleen and lymph nodes contained abnormally high numbers of pre-B (B220+sIg−) and immature B (B220+sIgM+sIgD−) cells, and diminished numbers of mature B cells (B220+sIgM+sIgD+). As expected,28 homoeostasis had improved by 12 weeks (Supplementary Table S2) but there were no significant differences in B lymphoid populations in mnt+/− Eμ-myc and mnt+/+ Eμ-myc mice, in any tissue, at either time point.
Figure 5.
Impact of mnt heterozygosity on pre-tumour phenotype of Eμ-myc mice. Enumeration of cell populations in 6-week-old mnt+/+ (white; n=5–6), mnt+/− (diagonal stripe; n=6–7), mnt+/+ Eμ-myc (light grey; n=6–7) and mnt+/− Eμ-myc (black; n=3–5) mice. (a) Total leucocytes and (b–d) indicated B lymphoid populations in spleen, lymph nodes, bone marrow and peripheral blood. Bars represent mean±S.E.M.; see also Supplementary Table S1. Statistical significance is shown for only mnt+/+ vs mnt+/− and mnt+/+Eμ-myc vs mnt+/−Eμ-myc mice *P<0.05 as calculated by Student’s t-test
vavP-MYC10 mice were analysed at 9–10 weeks of age (Supplementary Table S3). Once again, no statistically significant differences were found between mnt+/+ and mnt+/− transgenic mice in the populations expressing the transgene, either lymphoid (B or T) or myeloid.
Due to the early onset of malignancy in the vavP-MYC17 line, these mice were analysed at 2 weeks. Interestingly, the transient B lymphoid hyperplasia found in these mice42 was significantly diminished in mnt+/− mice compared with mnt+/+vavP-MYC17 littermates, as was the (modest) elevation in peripheral T cells (Supplementary Table S4). Thus, reduced expression of Mnt correlated with reduced Myc-driven expansion of B- and T-cell numbers in healthy young vavP-MYC17 mice.
In summary, analyses of preleukaemic populations in the vavP-MYC10 and Eμ-myc mice yielded little insight into why mnt heterozygosity had retarded Myc-driven tumorigenesis in these mice. While changes were observed in young vavP-MYC17 mice, these changes did not impact on the kinetics of morbidity and the unfortunate loss of this line has prevented any further analysis.
Assessing impact of mnt heterozygosity on cell function
To look more closely at which Myc-induced cellular phenotypes might have been altered by reduction in Mnt, we decided to focus our attention on the Eμ-myc model. Characteristics previously reported for cells overexpressing Myc include increased cell size, increased proportion of cells in the S phase of the cell cycle and increased sensitivity to apoptosis,3, 17, 28, 41 many of which have also been reported for cells lacking Mnt.25, 30, 35
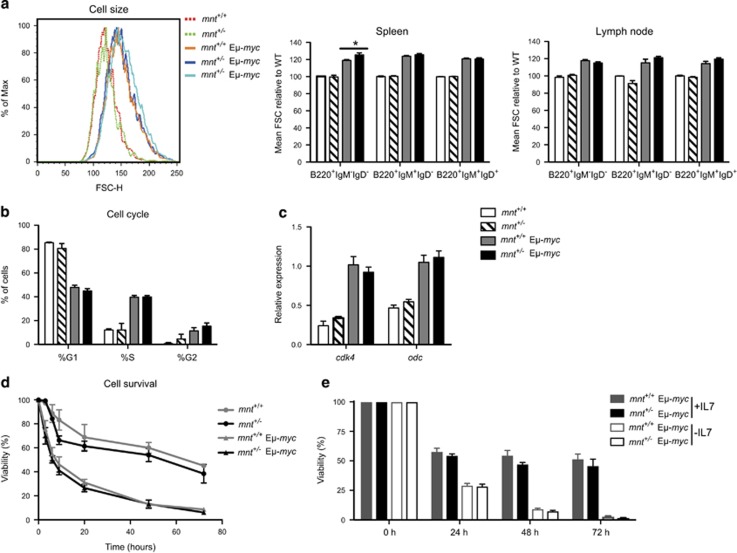
In order to avoid the complication of additional oncogenic changes, we analysed preleukaemic B lymphoid cell populations from healthy young (4- to 6-week-old) Eμ-myc mice. Cell size was assessed by determining mean forward light scatter by flow cytometry (Figure 6a). Representative size profiles are shown in the left panel and the adjacent panels summarise the mean forward light scatter of pre-B (B220+sIg−), immature B (B220+IgM+IgD−) and mature B (B220+IgM+IgD+) cells, expressed relative to that of non-trangenic cells analysed contemporaneously. As expected, 28 the mean size of each B lymphoid cell type was greater for myc transgenic than non-transgenic cells. Mnt heterozygosity did not affect the size profile of non-transgenic or transgenic cells of any major B lymphoid type from lymph nodes (right panel) or bone marrow or peripheral blood (not shown), with the exception of pre-B cells from the spleen of mnt+/− Eμ-myc mice, which were somewhat larger than those in mnt+/+ Eμ-myc mice (middle panel).
Figure 6.
Impact of mnt heterozygosity on pre-neoplastic Eμ-myc pre-B cells. (a) Mnt heterozygosity has no discernible impact on size of B lymphoid cells in 6-week-old mnt+/+ and mnt+/− WT and Eμ-myc mice. Cells were immunostained then gated for pre-B cells (B220+IgM−IgD−), immature B cells (B220+IgM+IgD−) and mature B cells (B220+IgM+IgD+) cells, and mean forward light scatter (FSC-H) determined. Values were normalised to WT cells. Left panel shows representative histogram of pre-B cells from a mnt+/+ (red dashed line), mnt+/− (green dashed line), Eμ-myc (orange solid line) and two mnt+/− Eμ-myc (blue solid lines) 6-week-old mice. Right panels show B lymphoid cell populations from spleen (centre) and lymph nodes (right) of 6-week-old WT (white; n=5–6), mnt+/− (diagonal stripe; n=6–7), Eμ-myc (grey; n=6–7) and mnt+/− Eμ-myc (black; n=3–5) mice. Bars represent mean±S.E.M. Statistical significance is shown for only WT vs mnt+/− and Eμ-myc vs mnt+/− Eμ-myc; *P<0.05 as calculated by Student’s t-test. (b and c) Mnt heterozygosity has no discernible impact on cycling of pre-B cells. (b) Proportion of pre-B cells in the indicated phases of the cell cycle, determined using Nicoletti stain, flow cytometry and Dean/Jett/Fox modelling using FlowJo software (see Materials and Methods). Pre-B cells were isolated from bone marrow of 4-week-old mice of the indicated genotypes using magnetic beads coated with CD19 antibody. (c) Quantification of cdk4 and odc transcripts in sorted pre-B cells by q-PCR analysis. Expression is indicated relative to mnt+/+ Eμ-myc cells. (d and e) Mnt heterozygosity has no discernible impact on apoptosis of pre-B cells in vitro. (d) Pre-malignant pre-B cells isolated from the bone marrow of 4-week-old mice using magnetic beads coated with CD19 antibody were cultured in conventional medium without additional cytokines and viability determined at the indicated time points by flow cytometry (see Materials and Methods). For each genotype, viability is expressed relative to that at t=0. Similar results were obtained for pre-B cells isolated using a MoFlow cell sorter (not shown). (e) Pre-B cells isolated from bone marrow of 4-week-old mice by flow cytometry were cultured in medium containing IL-7 as described previously.50 After 5 days, cells were washed three times to remove IL-7, and then replated with or without IL-7. Viability was determined at 0, 24, 48 and 72 h by flow cytometry. Results shown for n=6 mice of each genotype, analysed in two independent experiments; mean±S.E.M. For each genotype, viability is expressed relative to that at t=0
Cell cycle analysis of preleukaemic pre-B cells from bone marrow was performed by flow cytometry. As reported previously,28 the proportion of cells in the S and G2 phases was greater for cells from Eμ-myc than non-transgenic mice. However, mnt heterozygosity had no significant impact on the cycling of either transgenic or non-transgenic pre-B cells (Figure 6b). Moreover, it did not affect the level of transcripts from the cdk4 and odc genes, both of which are targeted by Myc during cell cycling (Figure 6c).2, 20, 25
Sorted pre-B cells were cultured in conventional medium for 3 days to ascertain their susceptibility to apoptosis (Figure 6d). As reported previously,45 the Eμ-myc cells died at a faster rate than their non-transgenic counterparts, but mnt heterozygosity had only marginal impact. Similarly, when IL-7 was removed from IL-7-supported cultures of pre-B cells, mnt heterozygosity had no significant impact (Figure 6e).
Impact of mnt heterozygosity on Eμ-myc lymphoma cells
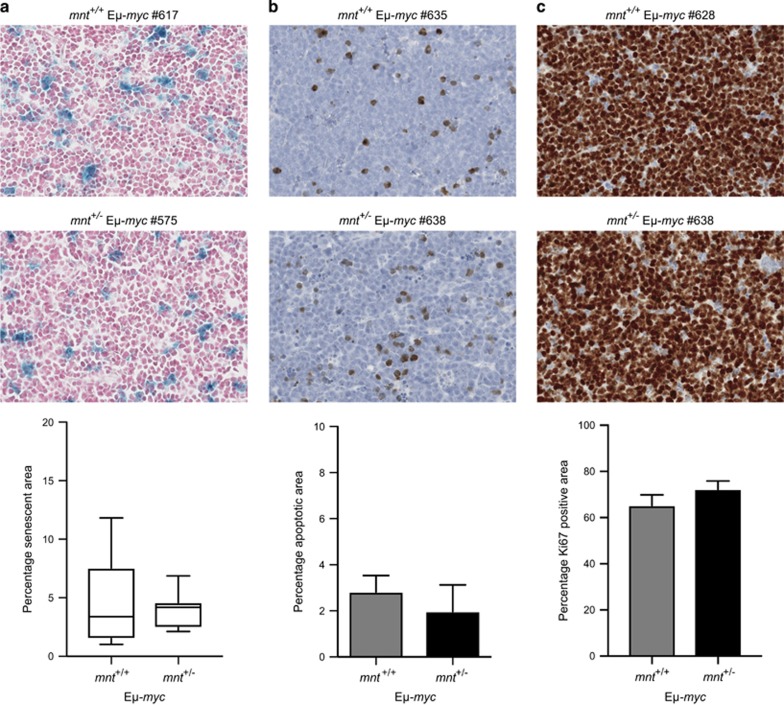
Having failed to observe any changes in the preleukaemic cell populations that might account for the slower onset of tumour-associated morbidity, we decided to assess Eμ-myc lymphomas to ascertain whether reduced Mnt had increased the probability of malignant Myc-driven cells undergoing cell death in vivo or reduced their proliferation rate. Tumour sections were stained for β-galactosidase activity; cleaved caspase-3 or Ki-67 and scanned digitally to quantify positive cells. No significant differences were detected between mnt+/+ and mnt+/− Eμ-myc lymphomas in the frequency of senescent (β-galactosidase-positive) or apoptotic (cleaved caspase-3-positive) cells (Figures 7a and b). Neither was there any difference in proliferation, as judged by quantitation of Ki-67 staining (Figure 7c).
Figure 7.
Impact of mnt heterozygosity on Eμ-myc lymphoma cells. (a) mnt heterozygosity has no impact on senescence. Sections of lymph node lymphomas of mnt+/+ Eμ-myc (n=12) and mnt+/− Eμ-myc (n=7) mice were stained for β-galactosidase activity, counterstained with nuclear fast red (see Materials and Methods) and scanned on an Aperio Digital Pathology Slide Scanner. β-Galactosidase-positive cells in extracted images were quantified using Fiji software. Data shown are mean±S.E.M. (b) mnt heterozygosity does not impact upon apoptosis. Sections of lymphomas in lymph node and/or spleen of mnt+/+ Eμ-myc (n=10) and mnt+/− Eμ-myc mice (n=5) were stained using an antibody specific for cleaved caspase-3 followed by a haematoxylin counterstain (see Materials and Methods). Cells positive for cleaved caspase-3 were quantified as in (a) above. Data shown are mean±S.E.M. (c) Proliferation in Eμ-myc lymphomas is not altered by mnt heterozygosity. Sections of lymphomas in lymph node and/or spleen of mnt+/+ Eμ-myc (n=11) and mnt+/− Eμ-myc mice (n=4) were stained for Ki-67 with haematoxylin counterstain. The Ki-67 positive area was quantified as in (a) above. Data shown are mean±S.E.M.
Discussion
The bHLHZip protein Myc activates transcription of a diverse array of genes, thereby coordinating cellular proliferation in response to cytokines, mitogens and nutrients. In normal cells, Myc is tightly regulated at transcriptional, post-transcriptional and post-translational levels, preventing inappropriate expansion of cell numbers and thereby reducing the risk of malignancy. As a further safeguard, Myc increases the propensity of cells to undergo apoptosis when cytokines and nutrients become limiting.3, 17, 41 Thus, somewhat paradoxically, Myc has properties of both a proto-oncoprotein and a tumour suppressor protein. The level of Myc expression can influence the balance between proliferation and apoptosis, because apoptosis was reported to be robust at high levels of Myc but minimal when Myc levels are low, perhaps reflecting differential affinities for target genes.33
The transcriptional repressor Mnt, also a bHLHZip protein, is a functional antagonist of Myc. Mnt competes with Myc for dimerisation with their mutual obligate partner Max and for binding at E-box sites in an overlapping set of target genes.22 Mnt overexpression can suppress Myc-dependent cell proliferation24 and tissue-specific loss of Mnt in mice resulted in mammary adenocarcinomas and T-cell lymphomas.8, 25, 47 Thus Mnt has been regarded as a tumour suppressor.
In this study, we set out to determine the impact of reduced Mnt levels on Myc-driven tumorigenesis. VavP-MYC10 mice were selected for the initial study because they are a useful barometer of MYC leukaemogenic function: Mice bearing one transgenic MYC allele develop both early onset T lymphomas and late myeloid tumours, and homozygosity greatly increases the frequency of T lymphomas and accelerates morbidity.43 We thought that reducing Mnt levels would be comparable to increasing Myc and hence expected to see accelerated tumorigenesis and increased T lymphoma frequency in mice heterozygous for vavP-MYC10 and heterozygous for mnt. In fact, the opposite occurred: mnt+/− vavP-MYC10 mice developed fewer T lymphomas and morbidity was slower (Figure 2a). These results suggested that mnt heterozygosity had reduced susceptibility to Myc-driven T lymphomagenesis. Tumorigenesis was not significantly retarded in mice homozygous for the vavP-MYC10 transgene or in another T lymphoma-susceptible line (vavP-MYC17), possibly because Mnt heterozygosity is insufficient to counter the higher levels of Myc in these strains.
When the experiment was repeated with Eμ-myc mice, in which transgene expression is largely restricted to the B lymphoid lineage, mnt heterozygosity again resulted in slower tumour onset. Thus, reducing Mnt by 50% can impede both B and T lymphomagenesis in two independent transgenic myc models.
Similar findings have been made recently using a different model: T-cell specific acute deletion of mnt and a ROSA-26 locus-driven mycT58A mutant transgene.30 While mycT58A mice succumbed to thymic lymphomas with a median survival of 220 days, mnt deletion resulted in a median survival of 355 days and none of the mnt-/− mycT58A mice developed thymic lymphoma. Link et al. also showed that Mnt-deficient thymocytes and concanavalin A-activated splenic T cells were more susceptible to apoptosis in vitro and concluded that the dominant physiological role of Mnt is suppression of apoptosis. The inference is that, by counteracting Myc’s capacity to promote apoptosis, Mnt facilitates Myc’s oncogenic drive.
We considered that diminished suppression of apoptosis was also a likely explanation for why mnt heterozygosity retards lymphomagenesis in our vavP-MYC10 and Eμ-myc mice. Nevertheless, we were unable to provide direct evidence for this. Increased susceptibility to apoptosis might have been expected to reduce the number of Myc-driven preleukaemic cells. However, mnt heterozygosity provoked no diminution of pre-B-cell numbers in healthy young Eμ-myc mice, nor of lymphoid (B or T) or myeloid populations in vavP-MYC10 mice (Supplementary Tables S1), although the transient pre-B-cell population in neonatal mnt+/− vavP-MYC17 mice was significantly reduced (Supplementary Table S4). Furthermore, when we cultured preleukaemic Eμ-myc pre-B cells, we found no evidence that mnt heterozygosity increased their susceptibility to apoptosis (Figures 6d and e).
Reasoning that mnt heterozygosity might be insufficient to enhance susceptibility to Myc-driven apoptosis in vitro but might have conferred increased susceptibility in vivo to particular physiological signals or to other oncogenic mutations, we quantitated cell death in sections of mnt+/+ versus mnt+/− Eμ-myc lymphomas. No significant differences were found in the frequency of either apoptotic or senescent cells (Figure 7). Furthermore, the reduced level of Mnt did not diminish the proliferation of the tumour cells, as judged by Ki-67 staining.
Although we have been unable to pinpoint the change(s) responsible for the slower onset of morbidity in our mnt+/− myc transgenic mice, our study provides evidence that Mnt facilitates Myc-driven tumorigenesis.55 Given the plethora of genes regulated by opposing Myc/Max and Mnt/Max heterodimers, functions other than enhanced apoptosis may contribute. We are currently undertaking conditional (lymphoid-specific) homozygous deletion of mnt in Eμ-myc and vavP-MYC10 mice to further explore role of Mnt in apoptosis and leukaemogenesis.
Materials and Methods
Mice
The vavP-MYC mice (which express human MYC cDNA42, 43), Eμ-myc mice (which express mouse myc cDNA21) and p53-deficient mice26 used in this study were all on a C57BL/6 background and bred at the Walter and Eliza Hall Institute (WEHI) in accordance with the WEHI animal ethics committee regulations. The mnt+/− mice, originally on a 129/S6 background,46 were backcrossed to C57BL/6 mice for several generations prior to being sent to WEHI, where backcrossing to C57BL/6 was continued. All mnt+/− mice used in this study had been backcrossed for >9 generations. The rapid T lymphoma onset in the vavP-MYC17 line made it difficult to maintain a breeding colony,43 so cryopreserved sperm from vavP-MYC17 mice was used to fertilise oocytes from super-ovulated mnt+/− females (67 offspring) and vavP-MYC17 mnt+/− sperm was then collected and used to fertilise oocytes from super-ovulated wild-type females (132 offspring). Moribund mice were autopsied and organs collected for histology and immunophenotyping.
Antibodies
Western blots, prepared by standard procedures, were probed using the following antibodies: c-Myc (N-262), Max (C-17) and Mnt (M-132) all from Santa Cruz Biotechnology (Dallas, TX, USA), or c-Myc (D84C12) from Cell Signaling Technology (Danvers, MA, USA) and Mnt (A303-626A) from Bethyl Laboratories (Montgomery, TX, USA) and β-actin (AC-74, Sigma-Aldrich, St. Louis, MO, USA). Blots in Figure 1b were imaged on a ChemiDoc Touch (Bio-Rad, Hercules, CA, USA) and analysed using Image Lab software (Bio-Rad). Monoclonal antibodies used for flow cytometry, produced and labelled with FITC, PE or APC in house, included the following: RB6-8C5, anti-Gr1; MI/70, anti-Mac1; YTA3.2.1, anti-CD4; 53.6.7.2, anti-CD8; Ter119, anti-erythroid marker; ID3, anti-CD19; RA3-6B2, anti-CD45R-B220; 5.1, anti-IgM; 11-26C, anti-IgD; 145-2C11, anti-CD3; E13.161.7, anti-Sca-1; 57, anti-CD43; T329.1, anti-Thy1; A20.1 anti-Ly5.1.
Cell phenotype analysis
An ADVIA haematology analyser (Bayer, Leverkusen, Germany) or flow cytometer (LSR1; Becton Dickinson, Franklin Lakes, NJ, USA) was used to analyse peripheral blood following treatment with a buffer containing 0.168 M ammonium chloride to deplete red blood cells. Single-cell suspensions were prepared from bone marrow (one femur), thymus, lymph nodes (axillary, brachial, inguinal) and spleen. Viable cells were counted using a haemocytometer and trypan blue exclusion or enumerated on a CASY Counter (Scharfe Roche, Penzberg, Germany). Cell immunophenotype was determined by staining with surface marker-specific antibodies followed by flow cytometry analysis. Stained single-cell suspensions were sorted using a MoFlo (Cytomation, Fort Collins, CO, USA) high speed sorter. Cell viability was determined by flow cytometry following staining with propidium iodide and annexin V-FITC. Statistical significance was determined using the Student’s t-test (two-tailed, assuming equal variance).
Cell cycle analysis
Pre-B cells were isolated from bone marrow using MACS columns and CD19 beads (Miltenyi, Bergisch Gladbach, Germany), resuspended in 0.1% sodium citrate, 0.1% Triton X and 50 μg/ml propidium iodide (Nicoletti stain)38 and incubated on ice in the dark for 1 h prior to FACS analysis. Cell cycle profiles were characterised using FlowJo software (Tree Star, Ashland, OR, USA).
Cell survival assays
Pre-B cells were isolated by magnetic purification using CD19 MicroBeads (Miltenyi Biotech, Bergisch Gladbach, Germany) and MS columns (Miltenyi Biotech) according to the manufacturer’s protocols. Cells were cultured at 0.2–0.5 × 106 cells/ml in high-glucose Dulbecco’s modified Eagle’s medium supplemented with 10% foetal calf serum (Sigma-Aldrich), 50 μM 2-mercaptoethanol (2-ME; Sigma-Aldrich, St Louis, MO, USA) and 100 μM asparagine (Sigma-Aldrich) without additional cytokines. Cell viability was determined by staining with FITC-conjugated annexin V and propidium iodide followed by flow cytometry. IL-7 cytokine withdrawal assay was performed as published.50
Histology
For cleaved caspase-3 or Ki-67 immunohistochemistry (IHC), spleens and lymph nodes from lymphoma-bearing mice were fixed in 10% formalin and then embedded in paraffin. Sections were stained for cleaved caspase-3 using the SignalStain Apoptosis (Cleaved Caspase-3) IHC Detection Kit (Cell Signaling Technology) according to the manufacturer’s protocol and counterstained with haematoxylin. Ki-67 IHC was performed using D3B5 Rabbit mAb (Cell Signaling Technology). To assess senescence, we followed the protocol of Post et al.36 Briefly, lymph nodes from lymphoma-bearing mice were fixed for 2 h in 4% paraformaldehyde, and then incubated for at least 6 h in 10% sucrose/Hanks’ Balanced Salt Solution (HBSS), followed by 15% sucrose/HBSS, then 20% sucrose/HBSS, before snap freezing in Tissue-Tek O.C.T. Compound (Sakura Finetek, Torrance, CA, USA). Cryosections were stained for SA-β-galactosidase as previously described,9 and counterstained with nuclear fast red. Complete slides were scanned on an Aperio Digital Pathology Slide Scanner and regions to be analysed were extracted using ImageScope software (Leica Biosystems, Wetzlar, Germany). Cleaved caspase-3 or β-galactosidase-positive cells were quantified using Fiji software (ImageJ, NIH, Bethesda, MD, USA).39
PCR analysis
Lymphocyte populations were purified as described above. Total RNA was isolated using RNeasy mini kit (Qiagen, Hilden, Germany). In total, 0.2 μg of RNA was reverse transcribed using the TaqmanRT system (Roche, Basel, Switzerland). Real-time PCR analysis was performed on cDNA using QuantiTect SYBR green PCR kit (Qiagen) on an ABI Prism 7900 (Applied Biosystems, Foster City, CA, USA). Relative fold expression was determined using the comparative threshold cycle method and β-actin. Primer sequences were as follows: c-myc sense caaatcctgtacctcgtccgattc, antisense cttcttgctcttcttcagagtcgc; mnt sense cagtccctgaagaggaagga, antisense ccggagcacacgatctatct; ornithine decarboxylase sense gaccttgtgaggagctgctgat, antisense tggcagtcaaactcgtccttag; cdk4 sense cagtcagtggtgccagagatggag, antisense cagcgagggtttctccaccaag; β-actin sense tattggcaacgagcggttc, antisense ccatacccaagaaggaaggct.
Statistical analysis
Statistical comparisons were made using a two-tailed Student’s t-test with Prism v7.0 software (GraphPad, San Diego, CA, USA). Data are shown as means±S.E.M. with P-values <0.05 considered statistically significant. Mouse survival analysis was carried out using GraphPad Prism (Version 7.0) and significance determined using log-rank (Mantel–Cox) test.
Acknowledgments
We thank our colleagues JM Adams, A Strasser, C Scott and P Bouillet for useful discussions; ML Bath for performing in vitro fertilisations; B Helbert, K Mackwell and C Young for genotyping; M Robati for technical assistance; K Hughes, C D’Alessandro and G Siciliano for mouse husbandry, WEHI cytometry and histology services; and L Whitehead for imaging analysis. This work was supported by postdoctoral fellowships from EMBO and the Human Frontier in Science Program (to KJC); a PhD fellowship from Leukaemia Foundation of Australia (to NSA); research grants from National Health and Medical Research Council (NHMRC) (program grant 461221) and National Cancer Institute (CA43540); and operational infrastructure grants through the Australian Government IRISS and the Victorian State Government OIS.
Author contributions
Designed research: KJC, CJV, NSA and SC. Performed research: KJC, CJV and NSA. Provided mnt+/− mice, advice: PJH. Analysed and interpreted experimental data and wrote/edited manuscript: KJC, CJV, NSA and SC.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JM Hardwick
The authors declare no conflict of interest.
Supplementary Material
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985; 318: 533–538. [DOI] [PubMed] [Google Scholar]
- Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci 1998; 3: D250–D268. [DOI] [PubMed] [Google Scholar]
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 1991; 6: 1915–1922. [PubMed] [Google Scholar]
- Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 1992; 359: 552–554. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 1991; 251: 1211–1217. [DOI] [PubMed] [Google Scholar]
- Cory S, Vaux DL, Strasser A, Harris AW, Adams JM. Insights from Bcl-2 and Myc: malignancy involves abrogation of apoptosis as well as sustained proliferation. Cancer Res 1999; 59: 1685s–1692s. [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 2009; 15: 6479–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezfouli S, Bakke A, Huang J, Wynshaw-Boris A, Hurlin PJ. Inflammatory disease and lymphomagenesis caused by deletion of the Myc antagonist Mnt in T cells. Mol Cell Biol 2006; 26: 2080–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolaiti D, McFerrin L, Carroll PA, Eisenman RN. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim Biophys Acta 2015; 1849: 484–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CAJ, Butel JS et al. Mice deficient for p53 are developmentally normal but are susceptible to spontaneous tumours. Nature 1992; 356: 215–221. [DOI] [PubMed] [Google Scholar]
- Edelmann J, Holzmann K, Miller F, Winkler D, Buhler A, Zenz T et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 2012; 120: 4783–4794. [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA 2004; 101: 6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes Dev 2008; 22: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by myc-induced suppression of Bcl-XL or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol 2001; 21: 5063–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G, Littlewood T. A matter of life and cell death. Science 1998; 281: 1317–1321. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 1992; 69: 119–128. [DOI] [PubMed] [Google Scholar]
- Farrell PJ, Allan GJ, Shanahan F, Vousden KH, Crook T. p53 is frequently mutated in Burkitt's lymphoma cell lines. EMBO J 1991; 10: 2879–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA 1991; 88: 5413–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000; 16: 653–699. [DOI] [PubMed] [Google Scholar]
- Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The Eμ-myc transgenic mouse: a model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med 1988; 167: 353–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker CW, Hurlin PJ. Of Myc and Mnt. J Cell Sci 2006; 119: 208–216. [DOI] [PubMed] [Google Scholar]
- Hsu B, Marin MC, El-Naggar AK, Stephens LC, Brisbay S, McDonnell TJ. Evidence that c-myc mediated apoptosis does not require wild-type p53 during lymphomagenesis. Oncogene 1995; 11: 175–179. [PubMed] [Google Scholar]
- Hurlin PJ, Queva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev 1997; 11: 44–58. [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Zhou ZQ, Toyo-oka K, Ota S, Walker WL, Hirotsune S et al. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J 2003; 22: 4584–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol 1994; 4: 1–7. [DOI] [PubMed] [Google Scholar]
- Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 2015; 15: 593–607. [DOI] [PubMed] [Google Scholar]
- Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in Eμ-myc transgenic mice. Cell 1986; 47: 11–18. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012; 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link JM, Ota S, Zhou ZQ, Daniel CJ, Sears RC, Hurlin PJ. A critical role for Mnt in Myc-driven T-cell proliferation and oncogenesis. Proc Natl Acad Sci USA 2012; 109: 19685–19690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 2012; 44: 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8: 976–990. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 2008; 14: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Maclean KH, Keller UB, Pendeville H, Baudino TA, Cleveland JL. Mnt loss triggers Myc transcription targets, proliferation, apoptosis, and transformation. Mol Cell Biol 2004; 24: 1560–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Terzian T, Smith C, Eischen CM, Lozano G. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene 2010; 29: 1260–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie CA, Harrison DJ, Peter A, Dobbie L, White S, Howie SE et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene 1994; 9: 603–609. [PubMed] [Google Scholar]
- Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc 2006; 1: 1458–1461. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 1999; 13: 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Glynn JM, Guilbert LJ, Cotter TG, Bissonnette RP, Green DR. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science 1992; 257: 212–214. [DOI] [PubMed] [Google Scholar]
- Smith DP, Bath ML, Harris AW, Cory S. T-cell lymphomas mask slower developing B-lymphoid and myeloid tumors in transgenic mice with broad hematopoietic expression of MYC. Oncogene 2005; 24: 3544–3553. [DOI] [PubMed] [Google Scholar]
- Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood 2006; 108: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 1990; 348: 331–333. [DOI] [PubMed] [Google Scholar]
- Strasser A, Elefanty AG, Harris AW, Cory S. Progenitor tumours from Em-bcl-2-myc transgenic mice have lymphomyeloid differentiation potential and reveal developmental differences in cell survival. EMBO J 1996; 15: 3823–3834. [PMC free article] [PubMed] [Google Scholar]
- Toyo-oka K, Hirotsune S, Gambello MJ, Zhou ZQ, Olson L, Rosenfeld MG et al. Loss of the Max-interacting protein Mnt in mice results in decreased viability, defective embryonic growth and craniofacial defects: relevance to Miller-Dieker syndrome. Hum Mol Genet 2004; 13: 1057–1067. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Bowen TJ, Hirotsune S, Li Z, Jain S, Ota S et al. Mnt-deficient mammary glands exhibit impaired involution and tumors with characteristics of myc overexpression. Cancer Res 2006; 66: 5565–5573. [DOI] [PubMed] [Google Scholar]
- Tu WB, Helander S, Pilstal R, Hickman KA, Lourenco C, Jurisica I et al. Myc and its interactors take shape. Biochim Biophys Acta 2015; 1849: 469–483. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10: 301–309. [DOI] [PubMed] [Google Scholar]
- Vandenberg CJ, Motoyama N, Cory S. FoxO3 suppresses Myc-driven lymphomagenesis. Cell Death Dis 2016; 6: e2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988; 335: 440–442. [DOI] [PubMed] [Google Scholar]
- Vermeer MH, van Doorn R, Dijkman R, Mao X, Whittaker S, van Voorst Vader PC et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res 2008; 68: 2689–2698. [DOI] [PubMed] [Google Scholar]
- Walker W, Zhou ZQ, Ota S, Wynshaw-Boris A, Hurlin PJ. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J Cell Biol 2005; 169: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiman KG, Magnusson KP, Ramqvist T, Klein G. Mutant p53 detected in a majority of Burkitt lymphoma cell lines by monoclonal antibody PAb240. Oncogene 1991; 6: 1633–1639. [PubMed] [Google Scholar]
- Yang G, Hurlin PJ. MNT and emerging concepts of MNT-MYC antagonism. Genes (Basel) 2017; 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.