Abstract
Over the last several hundred years of scientific progress, we have arrived at a deep understanding of the non-living world. We have not yet achieved an analogous, deep understanding of the living world. The origins of life is our best chance at discovering scientific laws governing life, because it marks the point of departure from the predictable physical and chemical world to the novel, history-dependent living world. This theme issue aims to explore ways to build a deeper understanding of the nature of biology, by modelling the origins of life on a sufficiently abstract level, starting from prebiotic conditions on Earth and possibly on other planets and bridging quantitative frameworks approaching universal aspects of life. The aim of the editors is to stimulate new directions for solving the origins of life. The present introduction represents the point of view of the editors on some of the most promising future directions.
This article is part of the themed issue ‘Reconceptualizing the origins of life’.
Keywords: origins of life, astrobiology, statistical physics, prebiotic chemistry
1. Introduction
The origin of life is widely regarded as one of the most important open problems in science. It is also notorious for being one of the most difficult. It is now almost 100 years since scientific efforts to solve the problem began in earnest, with the work of Oparin [1] and Haldane [2]. Both proposed that organic material, subjected to suitable primitive environmental conditions, could begin to increase in complexity, eventually giving rise to living cells. It was not until about 30 years later in 1953 that the proof-of-concept for the ‘primordial soup’ hypothesis was demonstrated by Stanley Miller and Harold Urey with the abiotic synthesis of biological components from simple starting materials under what were thought to be prebiotic environmental conditions at the time [3]. Within days their experiment produced an organic goo that remarkably contained amino acids. This was a surprise and there was some optimism that, had the experiment been left running, living creatures would soon be crawling out of the laboratory. In the ensuing decades, researchers have been able to generate nearly all components of living cells under different plausible scenarios for prebiotic environments. But, these ‘bottom-up’ approaches have not yet generated anything nearly as complex as a living cell. At most we are lucky to generate short polypeptides or polynucleotides or simple vesicles—a far cry from the complexity of anything living.
A major challenge is that modern cells are highly evolved, being the product of over 3.5 billion years of evolutionary refinement. Starting from the ‘top-down’ and removing as much complexity as possible yields ‘minimal’ cells [4]. The simplest living example to date is a stripped down version of Mycoplasma mycoides, JCV-syn3.0, with 473 genes [5]. While simple by biological standards, this organism is extraordinarily complex by chemical ones. If instead of working to construct a minimal functional life form in the laboratory, we extrapolate backwards in time from extant life to its most primitive ancestors, we uncover the properties of a last universal common ancestor (LUCA): the most ancient life on Earth whose properties we can infer based on genomic analysis [6]. Like JCV-syn3.0, this ‘simple’ life is not so simple. We are still largely in the dark about the properties of LUCA (Where did it live? What metabolism did it have?). Whatever LUCA was, it was a highly evolved cellular life form, complete with genetic translation machinery.
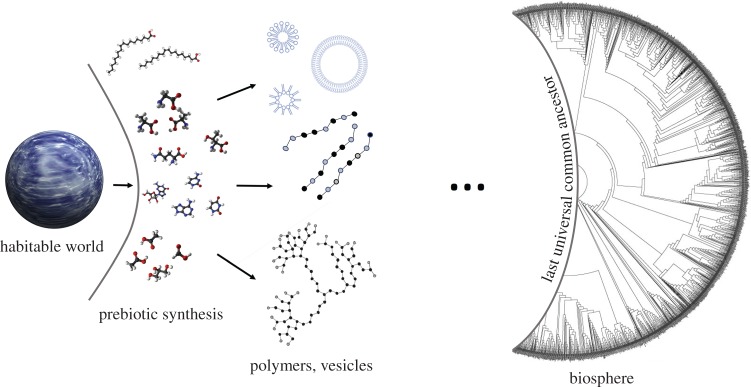
It is reasonable to expect that approaches working from the bottom-up and the top-down will eventually converge, with life emerging somewhere along the way. Yet, despite significant progress on both ends, a vast gulf persists separating our understanding of the geochemistry of early Earth, and the biomolecules it could produce, from what we know of the most ancient life (figure 1). LUCA represents the most ancient form of life for which we have direct evidence. A rough analogy can be drawn with the surface of last scattering in cosmology [7], which represents the point in cosmological history when the Universe became transparent to photons, such that information could propagate freely. We have only indirect evidence of the epochs preceding the decoupling of photons in the early Universe, just as we have only indirect evidence of the evolutionary epoch before the translation machinery solidified [8,9], enabling genetic information to propagate freely. In the case of cosmology, it is widely accepted that we must have an understanding of the physics governing earlier epochs to understand what happened. Likewise, to infer what happened before LUCA, we need to better understand the universal properties of life [10].
Figure 1.
Research on the origins of life approaches the problem from bottom-up, starting from geochemical synthesis of biomolecules, or top-down, uncovering the properties of ancient life: a vast gulf persists in connecting these two approaches, necessitating new conceptual frameworks for addressing life's origins. (Online version in colour.)
It is an open question whether we will arrive at a deep understanding of life in the same manner we have come to understand the non-living world, or whether our current approaches to physics are adequate to solve this problem, or if new physics is necessary [11]. One commonly held viewpoint is that life is so complex, we must first better understand it as it exists today, before gaining traction on its origins. However, a counterargument can be made that it is precisely in attempting to understand the transition from non-life to life where we have our best hope of understanding those properties of life that are its most fundamental. With this viewpoint in mind, a workshop held at the Carnegie Institution for Science in 2015 on ‘Re-conceptualizing the Origins of Life’ [12], from which the current theme issue of the Philosophical Transactions of the Royal Society A emerged. We here briefly summarize some of the most exciting ideas emerging in this new era of origins research, from the perspective of the editors. It is our view that more abstract, universal approaches to the origins of life will not only enable a re-conceptualization of the origin-of-life problem and set new directions for progress, but also provide new paths forward in making synthetic life in the laboratory and in our search for life on other worlds.
2. Revisiting chemistry, revisiting history
In the course of re-conceptualizing the origins of life (in general), it is notable that the question of life's origin(s) has been historically dominated by chemists, opening the question of whether re-conceptualizing the chemical approach is worth consideration. Historically, chemistry appeared to have been an obvious place to start, when one considered the origin of the molecular building blocks of life (e.g. amino acids, nucleobases, sugars, … etc.); and as a point of conjecture from which the next stage of self-assembly presumably occurred. This early chemistry is historically typified by the Miller–Urey (MU) experiments whereby amino acids were detected as products arising from electric discharge in environments with reduced gases (CH4, NH3 and H2) [3]. This was not the first foray into origins-of-life chemistry for chemists [13,14], but it is arguably one of the most historic and influential.
A recurrent theme in origins-of-life chemistry is that once a potential solution to a specific biosynthetic challenge has been identified, e.g. the MU experiment for amino acid synthesis, arguments are then promoted to support that specific chemistry/mechanism as being integral to life's origins. For example, if it is understood that amino acids in the MU reaction form through the Strecker synthesis, where HCN is the critical carrier of nitrogen, and that it has been shown that nucleobases can also form from HCN polymerization [15], then it might be conjectured that the presence of HCN was integral to the origins of life and environments that support an abundance of HCN were the environments where life probably emerged, i.e. from a synthesis mechanism arises a potential geophysical constraint.
From a biochemical perspective, invoking HCN as the primary nitrogen source for abiotic synthesis of critical nitrogenous biomolecules appears odd as all extant biochemistry relies on nitrogen in the form of ammonium cation (NH4+) as a basis for all nitrogen metabolism. Furthermore, HCN is unstable in water and will spontaneously decompose into formate and ammonium ion in relatively short times. So, even in an environment that forms HCN in the presence of water, aqueous ammonium would be expected to naturally accumulate and probably dominate the active nitrogen pool. The abiotic synthesis of amino acids through the reductive amination of keto acids is not more difficult than the Strecker synthesis and, therefore, there appears no actual imperative to require electric discharge and HCN chemistry to enable the presence of amino acids on a primitive Earth for utilization of an emergent living system. Either an MU atmosphere and electric discharge or an environment conducive to the formation of keto acids and reduced nitrogen (ammonia) would be equally favourable for the formation of amino acids.
Moving beyond the MU experiment, the next dominant chemical foray into origins-of-life chemistry were the key chemical steps towards the development of the ‘RNA world’, where self-replication of small RNA oligomeric strands works to mimic the self-replicating nature of life as we know it [16,17]. The elegant simplicity of the RNA world hypothesis is ultimately offset by the reality that a natural environment capable of synthesizing small strands of RNA would be expected to also produce a myriad of other molecular entities, i.e. the idea of a ‘naked’ RNA world, with no interference from other molecules, would be extremely unlikely [18]. This statement also ignores the critical problem of establishing homochirality—a virtually unaddressed issue.
The next revolution in prebiotic chemistry will have to embrace the essentially ‘messy’ nature of any special environment capable of the simultaneous and broad range of molecular synthesis probably required for the emergence of first life. The first essential step to developing molecular complexity is the capacity to make carbon–carbon bonds; starting from simple precursors like CO2, CO and H2, carbon bond formation can proceed readily through heterocatalytic carbonyl insertion reactions that can form both linear and branched products that are typically terminated by carboxylic groups [19]. However, the products of such reactions (beyond the carboxyl groups) are typically or dominantly fully saturated and, therefore, unreactive to additional functionalization. If, however, the environment also provides the capacity of partial oxidation of saturated products, activation of molecular products for further reaction would be possible. Such a situation was observed in the NiS-catalysed hydrothermal reaction of methacrylic acid + CO + H2O to form methylsuccinic acid (expected saturated product) and two thiolated products (unexpected partially oxidized products), where the partial oxidation was due to the corresponding reduction of NiS to Ni3S2 [20].
If, in a simple molecular system, one could simultaneously have conditions reducing enough to support catalysed carbonyl insertion reactions and also allow for partial oxidation of alkanes to alkenes, this would further enable hydration and dehydration reactions; amination and deamination reactions; oxidative decarboxylation; and Aldol and retro-Aldol reactions: all enabling considerable molecular complexity to develop, spontaneously. For example, if one considers a hypothetical chemical system starting with simple products such as butanoic acid and subsequent isobutanoic acid, both readily formed from catalysed carbonyl insertion, and hydrolysis starting with propene (e.g. Cody et al. [19]) and allow for each of the reactions listed above to operate on all products, one finds (on ‘paper’) that in excess of 100 different molecules could plausibly form. These would include many saturated and unsaturated polycarboxylic acids, amino acids, keto acids and alcohol acids.
To the best of our knowledge, the development of such molecular complexity has not been demonstrated experimentally for a system governed principally by these reactions. There is, however, evidence that such a scenario has occurred naturally, and more significantly a system appearing to require establishment of a dynamic (regenerative) organic reaction network. Recently, Cooper et al. [21] identified a number of keto acids, hydroxyl tricarboxylic acids, saturated tricarboxylic acids and a tetracarboxylic acid. In this group of molecules, pyruvic acid, citric acid and isocitric acid particularly stand out. Whereas pathways to such molecules are known, all three molecules are reactive and subject to decomposition under hydrothermal conditions. It is estimated that warm wet conditions persisted in the interiors of primitive planetismals for potentially upwards of millions to tens of millions of years—with heat generated from the radioactive decay of short-lived radionuclides. It, therefore, appears these compounds would have had to have been continuously regenerated up to the point that all reactions ceased to occur (upon freezing with reduction in the rate of radiogenic heating).
The next level of complexity (messiness) probably arose through the spontaneous synthesis of the ‘abiotic’ world's first ‘abio’-polymers. During mild heating, mixtures of polycarboxylic acids (e.g. di- and tricarboxylic acids) and polyols (e.g. glycerol) readily polymerize to form random hyperbranched polymers (HBPs) [22,23]. It is intriguing to consider that such primitive macromolecules may have served as the most primitive ‘proto-enzymes’ providing micro-environments that could enhance reactions, which are less probable in solution. Wet–dry cycling can occur in many environments driven by different mechanisms: in any natural environment such wet–dry cycling appears to be inevitable at some frequency. A chemical system capable of spontaneously synthesizing functionalized polycarboxylic acid molecules, polyols and perhaps amino-alcohols also has the capacity to form functionalized HBPs.
In re-conceptualizing the chemical origins of life, it appears necessary to embrace the seemingly inevitable ‘messy’ chemistry that naturally occurring dynamic organic reaction networks must exhibit. Frontier experiments establishing sustainable dynamic organic reaction networks provide the opportunity to gain understanding of chemical system evolution and may better provide more information about the core-level chemical constraints that must factor into the emergence of Earth's first living systems. The challenge is analytical (potentially hundreds of dynamic reactions operating in parallel) and it appears likely that standard molecule-collecting analytics (chromatography with detection, mass spectrometry or other) will not be sufficient to unravel the core science here. Looking forward, a linking of complex (messy) chemistry with a theoretical physics (prediction) approach, informed by statistical mechanics as discussed by Guttenburg et al. [24], may be the path forwards.
3. Quantifying life
The origin of life is hard because we do not understand what life is (or why life is). Schrödinger's seminal 1943 lectures titled What is life? [25] opened a challenge for physicists to explain the properties of living matter from what we know of physics. Now, over 70 years later we have not made much progress: hundreds of definitions for life exist (see figure 2 for common words in the definitions of life). The majority of these tend to be phenomenologically motivated rather than motivated by first principles. The state of the field is in such disarray that serious attempts have been made to propose new definitions based on consensus of older ones [26].
Figure 2.

Some of the most commonly used words in the definitions of life. Shown are words common to 123 definitions, as compiled by Trifonov [26]. (Online version in colour.)
Our uncertainties in the most fundamental properties of life have left the origins-of-life researchers with a conundrum–-without an understanding of what life is, how can we approach understanding its origins? Current hypotheses for the origins of life are motivated by the varied definitions for life (and tend to lie along disciplinary boundaries). One popular working definition for life is ‘a self-sustaining chemical system capable of Darwinian evolution’ (first emerging from the NASA Exobiology Discipline Working Group [27]), motivated by an assumption that it is genetic inheritance with vertical descent that is life's most basic property. This definition drives the engineering of simple, molecular Darwinian systems in the laboratory, which have been successful in both demonstrating that Darwinian evolution is possible in molecular systems [28] and providing new insights into the relationship between genotype and phenotype in cases where the mapping between the two can be directly studied [29]. However, so far laboratory examples have not been truly self-sustaining, nor have any such systems been produced under prebiotically plausible conditions. It is also debated whether Darwinian evolution emerged early [30] or late [8]. The capability to undergo Darwinian evolution also necessitates a population and is not a property of individuals [31], muddling our ability to interpret what it means for the capacity to undergo Darwinian evolution to be a key property of life (or at least changing how we define life). In the context of life's origins, the Darwinian definition provides few insights into the systems that could have preceded the first Darwinian replicator. Other definitions are fraught with similar challenges.
The challenge with current definitional frameworks for understanding life is they focus on life as a system or thing, rather than a process. This leads to a black-or-white distinction between life and non-life: something is alive or it is not. This provides little insight into the chemical networks of ‘almost life’ that span the stages of complexity between prebiotic molecules and fully functioning cellular life, such as the peptide networks discussed in [32]. We recognize abiotically produced molecules as not life, and LUCA as life, but have no capacity to characterize the things in between. Ideally, in assessing the validity of any model for the origins of life, empirical or computational, we should have metrics for identifying how ‘life-like’ a given system is. If we are to understand the processes that transform matter to life, we must start focusing on understanding life too as a process [33,34].
This necessitates a re-conceptualization of the origins of life, removing the imposed hard boundary between non-life and life, and recognizing there may exist physical processes that we do not yet understand which are most prominent in living systems but are not necessarily absent elsewhere. One candidate is the physics of information: it is often speculated that information may be a key factor in the origins of life [35,36]. Just as massive bodies represent ideal example systems to study gravity, life could represent the structures in physical reality where the effects of information are most prominent. However, we do not understand how information can structure matter (or precisely what ‘information’ is for that matter), yet it seems apparent this is critical to structuring living systems across the hierarchy of living processes from cells to cities. Flack [37] suggests that one way this could mechanistically operate is via coarse-graining, where components of a biological system tune behaviour in response to slowly changing estimates of aggregate properties, which capture coarse-grained regularities in the external world, or even in a system's own internal state.
Schrödinger himself conceded towards the end of his book that ‘living matter while not eluding the “laws of physics” as established up to date, is likely to involve “other laws of physics” hitherto unknown’ [25]. These could manifest as fundamental bounds on the architecture of life, for example in scaling laws that characterize the limits of efficiency for living entities [38]. If we can quantify life in terms of the informational architecture of physical systems, it could lead to new directions with both comparing different hypotheses for life's emergence on an equal footing, by contrasting the structure of information flows in chemical networks and by providing a metric for quantifying how ‘alive’ a physical system is (e.g. how much information it generates, processes, etc.). This can be approached extrinsically or intrinsically, as approached by Cronin and co-workers [39] and Marshall et al. [40], respectively.
Cronin and co-workers placed their emphasis on distinguishing physical objects that would require a ‘program’ to generate them [39]. A measure called ‘pathway complexity’ is proposed, counting the number of possible ways a given object could be assembled, which can provide quantitative metrics for biosignatures under the assumption that there is a critical threshold of pathway complexity above which an object could only be produced by life. It is, therefore, a candidate for quantifying life (or at least its artefacts). This criterion assumes that it is only living architectures that can produce the specified sequence of steps (or at least it is highly improbable for non-living systems to do so). For this to be the case assumes something about the living state: particular sequences of events can happen because of stored information, e.g. defining a ‘non-trivial' trajectory [12,41]. That is, it assumes there are some trajectories that (sequences of transformations) cannot happen without exchange of information (which must be stored to make those sequences of transformations repeatable).
Marshall et al. [40] instead focus on the intrinsic complexity of living processes. Building on earlier work by Kim et al. [42] (see also [43]) and leveraging the quantitative framework of integrated information theory [44], they demonstrate how causal analysis can reveal where the boundaries of a living system might lie, and importantly how, within those boundaries, the system generates its own future states. Their analysis is done on the Boolean model of the fission yeast cell cycle gene regulatory network [45]. Previously, a small set of nodes in this network had been identified as a ‘control kernel’ that could be externally manipulated to drive the network to biologically functional states. The analysis of Marshall et al. reveals how the network internally regulates these nodes, providing a window into how (at least a toy model) a biological system is structured such that it can control its own transformations. That is, how it can construct itself.
There is an intense area of research centred on understanding the relationship between thermodynamics and information [46], and it may be that this provides insights into the physics underlying the origins of life, and in particular why life has taken the trajectories through chemical space that it has to yield the biosphere we observe today.
4. Universal properties of life: synthesizing theory, data and simulation
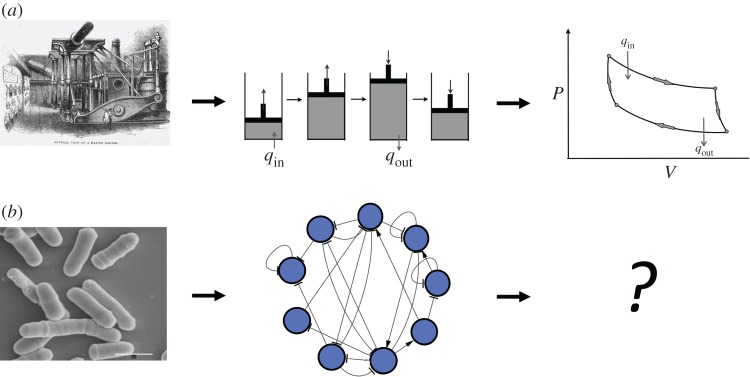
Understanding life and, in particular, the origins of life presents unique difficulties from a theoretical perspective. We would like to understand universal properties of life, but we have, in a concrete sense, only one exemplar, only one biosphere to study. Study of our single exemplar can, however, give us some insights into life's universal properties by taking a theoretical perspective that places life in the largest possible context, viewing it as a complex dynamical process, and trying to glean its universal properties from comparing data from our single exemplar of life with theoretical constructs (figure 3).
Figure 3.
To advance our understanding of the origins of life, we need to identify the appropriate abstractions for identifying universal principles. Thermodynamics was successful in describing the operation of heat engines because of an abstract understanding of thermodynamic cycles (a). In biology, it is unknown whether equivalently powerful abstractions will be identified (b) (industrial engine from [47], fission yeast cells from [48]). (Online version in colour.)
This can be accomplished through both theory and data. Goldenfeld et al. [49] present universal attributes of life by taking a physics-rooted approach seeking to describe early life. Within this framework, they focus on two phenomena that have empirical consequence: the origin of homochirality and the transition from a ‘collective networked phase’ of interacting chemical–biological entities to a Darwinian phase, where Darwinian selection is operative. They show how both can be in terms of a statistical mechanics framework, bringing quantitative tools from physics directly to bear on some of the most challenging open problems in origins research.
One of the main sources of data to drive theoretical perspectives is the genome. Kacar et al. [50] analyse contemporary genomes to infer the chemistry of early life, long since extinct, from phylogenetically embedded genomic structures. They ‘resurrect inferred ancestral protein sequences’ of early life, essentially inferring prehistoric phenotypic information from contemporary genomes. This research represents a fascinating introduction to a new set of tools to study evolution as a process sampling many possible biogeochemical states, all in the context of our single exemplar of life.
Both Goldenfeld et al. and Kacar et al. illustrate how theory and data analysis can be used to obtain universal properties of life even with only one exemplar of biology, but this limitation may be overcome in other ways, if we widen our attention from the study of the details of our one example to the study of imagined other examples. One source of stimulation for such imagination is observations of other planets, leading to the study of the origins of life in astrobiology. Another source of stimulation is computer simulation, which has led to a burgeoning new field of artificial life. Chris Langton, the founder of artificial life, articulated this expansion of our horizons thus [51]: ‘In addition to providing new ways to study the biological phenomena associated with life here on Earth, life-as-we-know-it, Artificial Life allows us to extend our studies to the larger domain of the “bio-logic” of possible life, life-as-it-could-be, whatever it might be made of and wherever it might be found in the universe’.
The astrobiological perspective considers life at a planetary level, explored by McInerney & Erwin [52] in terms of the roles of biological and ecological ‘public goods’. The concept of public goods was originally an economic concept as goods that have value but that are freely available to all, without exclusion. The concept extends quite naturally to the biological realm, where certain chemical substances (like oxygen in the atmosphere) are freely available. McInerney and Erwin argue that such public goods can be crucial in the process of niche formation, and even in evolutionary transitions such as the advent of the eukaryotic cell.
The perspective of artificial life offers a broad range of models seeking to capture universal properties of life. One particularly strong intellectual thread in the field begins with a purely computational abstract form of ‘chemical’ processes [53,54], and proceeds to purely digital ‘organisms’ interacting with each other in an abstract computational environment (Core War, Tierra). Avida (Adami) is currently the most developed digital life platform and has evolved to such an extent that evolution within its purely computational universe begins to have properties that parallel the properties of evolution in our single exemplar of the biosphere. Nitash et al. [55] use the Avida platform to explore the emergence of a digital LUCA.
Artificial chemistry [56] is a class of artificial life models that aims to abstract chemical processes, keeping enough complexity to obtain properties that have a hope of being universal enough to apply to real chemistry. The models have enormous range, from purely computational chemistry of interacting programs [53,54] to models that seek explicit approximation of real chemistry. Meringer & Cleaves [57] and Andersen et al. [58] are both on the latter side of the spectrum. Meringer and Cleaves use chemistry to motivate the construction of graph-based structure generation, aiming to understand the prebiotic formation of the chemical playing field, the universe of compounds used by biology, with an eye towards the potential design of novel (i.e. not currently found in biology) types of living systems. Andersen et al. form an abstraction of chemistry that consists of generative graph grammars, motivated by actual chemical interactions, to study the emergence of essential prebiotic chemical processes such as autocatalysis.
Finally, artificial life has led us to consider life in a broader context than purely biological. We are led to consider life not as a collection of biological objects, but as a living process. This informs new thinking on chemical origins of life, with regard to whether the process was inevitable or not [59] and what energy sources could have driven it [60]. The study of life as a living process also leads quite naturally to enlarging the process from chemically interacting primitive biological entities to socially interacting high-level biological entities (e.g. human beings). Inclusion of social interactions of entities then leads naturally to inclusion of non-biological, technological elements (such as the Internet, the World Wide Web and social media) into the living process. The result is to consider life as a hybrid technological–biological process, a perspective explored by Ikegami et al. [61].
5. Conclusion
It is an exciting time for research into life's origins. Progress is being made on many fronts including embracing the ‘messy chemistry’ inherent in prebiotic mixtures, and in adopting universal approaches to understanding life and quantifying its properties, as discussed throughout this theme issue. One motivation for the increased pace of research is the discoveries of planets orbiting other stars: astrobiology now has new, and plentiful, targets in our search for life on other worlds. Understanding the mechanisms governing how life arises could greatly inform our search for life on these worlds and those within our own Solar System. In turn, better understanding the diversity of planetary environments in which life could emerge and persist will place important constraints on our theories for its origins. Both the origins of life and the search for life on other worlds will benefit from re-conceptualizing the nature of life, leading to new approaches and new progress on long-standing questions. This theme issue introduces some work in this exciting area, but it will be up to an emerging community of scholars to develop these approaches into new frameworks, merging theory and experiment, to solve the problem of the origins of life and start the next chapter on one of the great open questions in science.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
Acknowledgements
The ‘Re-conceptualizing the Origins of Life’ meeting was sponsored by the Carnegie Institution for Science, the National Science Foundation, and the National Aeronautics and Space Administration. We thank the sponsors and all who attended and participated for the chance to engage in so many stimulating discussions that led to this theme issue being possible.
References
- 1.Oparin AI. 1953. The origin of life, viii p 2, l New York, NY: The Macmillan Company. [Google Scholar]
- 2.Haldane JBS. 1957. Genesis of life. In The Earth and its atmosphere (ed. DR Bates), pp. 287–301. New York, NY: Basic Books, Inc. [Google Scholar]
- 3.Miller SL. 1953. A production of amino acids under possible primitive earth conditions. Science 117, 528–529. ( 10.1126/science.117.3046.528) [DOI] [PubMed] [Google Scholar]
- 4.Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES. 2017. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439. ( 10.1038/nchem.2644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchison CA, III, et al. 2016. Design and synthesis of a minimal bacterial genome. Science 351, aad6253 ( 10.1126/science.aad6253) [DOI] [PubMed] [Google Scholar]
- 6.Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136. ( 10.1038/nrmicro751) [DOI] [PubMed] [Google Scholar]
- 7.Smoot GF, et al. 1991. Preliminary results from the COBE differential microwave radiometers–-large angular scale isotropy of the cosmic microwave background. Astrophys. J. 371, L1–L5. ( 10.1086/185988) [DOI] [Google Scholar]
- 8.Woese CR. 2002. On the evolution of cells. Proc. Natl Acad. Sci. USA 99, 8742–8747. ( 10.1073/pnas.132266999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi N, Hogeweg P, Kaneko K. 2017. Conceptualizing the origin of life in terms of evolution. Phil. Trans. R. Soc. A 375, 20160346 ( 10.1098/rsta.2016.0346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldenfeld N, Woese C. 2011. Life is physics: evolution as a collective phenomenon far from equilibrium. Annu. Rev. Condens. Matter Phys. 2, 375–399. ( 10.1146/annurev-conmatphys-062910-140509) [DOI] [Google Scholar]
- 11.Walker SI. 2017. Origins of life: a problem for physics, a key issues review. Rep. Prog. Phys. 80, 092601 ( 10.1088/1361-6633/aa7804) [DOI] [PubMed] [Google Scholar]
- 12.Cronin L, Walker SI. 2016. Origin of life. Beyond prebiotic chemistry. Science 352, 1174–1175. ( 10.1126/science.aaf6310) [DOI] [PubMed] [Google Scholar]
- 13.Brack A. 1998. The molecular origins of life: assembling pieces of the puzzle. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 14.Fox SW, Klaus O.1977. Molecular evolution and the origin of life. New York, NY: Marcel Dekker Inc.
- 15.Oro J, Kimball AP. 1961. Synthesis of purines under possible primitive Earth conditions. I. Adenine from hydrogen cyanide. Arch. Biochem. Biophys. 94, 217–227. ( 10.1016/0003-9861(61)90033-9) [DOI] [PubMed] [Google Scholar]
- 16.Gilbert W. 1986. Origin of life: the RNA world. Nature 319, 618 ( 10.1038/319618a0) [DOI] [Google Scholar]
- 17.Joyce GF. 1989. RNA evolution and the origins of life. Nature 338, 217–224. ( 10.1038/338217a0) [DOI] [PubMed] [Google Scholar]
- 18.Shapiro R. 1996. Prebiotic syntheses of the RNA bases: a critical analysis. Orig. Life Evol. Biosph. 26, 238–239. ( 10.1007/BF02459733) [DOI] [Google Scholar]
- 19.Cody GD, Boctor NZ, Brandes JA, Filley TR, Hazen RM, Yoder HS. 2004. Assaying the catalytic potential of transition metal sulfides for abiotic carbon fixation. Geochim. Cosmochim. Acta 68, 2185–2196. ( 10.1016/j.gca.2003.11.020) [DOI] [Google Scholar]
- 20.Cody GD, Boctor NZ, Hazen RM, Brandes JA, Morowitz HJ, Yoder HS. 2001. Geochemical roots of autotrophic carbon fixation: hydrothermal experiments in the system citric acid, H2O-(±FeS)−(±NiS). Geochim. Cosmochim. Acta 65, 3557–3576. ( 10.1016/S0016-7037(01)00674-3) [DOI] [Google Scholar]
- 21.Cooper G, Reed C, Nguyen D, Carter M, Wang Y. 2011. Detection and formation scenario of citric acid, pyruvic acid, and other possible metabolism precursors in carbonaceous meteorites. Proc. Natl Acad. Sci. USA 108, 14 015–14 020. ( 10.1073/pnas.1105715108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamajanov I, Callahan MP, Dworkin JP, Cody GD. 2015. Prebiotic alternatives to proteins: structure and function of hyperbranched polyesters. Orig. Life Evol. Biosph. 45, 123–137. ( 10.1007/s11084-015-9430-9) [DOI] [PubMed] [Google Scholar]
- 23.Mamajanov I, Cody GD. 2017. Protoenzymes: the case of hyperbranched polyesters. Phil. Trans. R. Soc. A 375, 20160357 ( 10.1098/rsta.2016.0357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttenberg N, Virgo N, Chandru K, Scharf C, Mamajanov I. 2017. Bulk measurements of messy chemistries are needed for a theory of the origins of life. Phil. Trans. R. Soc. A 375, 20160347 ( 10.1098/rsta.2016.0347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrödinger E. 1943. What is life? Cambridge, UK: Cambridge University Press. [Google Scholar]
- 26.Trifonov EN. 2011. Vocabulary of definitions of life suggests a definition. J. Biomol. Struct. Dyn. 29, 259–266. ( 10.1080/073911011010524992) [DOI] [PubMed] [Google Scholar]
- 27.Deamer DW, Fleischaker GR. 1994. Origins of life: the central concepts. Boston, MA: Jones & Bartlett Pub. [Google Scholar]
- 28.Lincoln TA, Joyce GF. 2009. Self-sustained replication of an RNA enzyme. Science 323, 1229–1232. ( 10.1126/science.1167856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadler PF, Stadler BMR. 2006. Genotype-phenotype maps. Biol. Theory 1, 268–279. ( 10.1162/biot.2006.1.3.268) [DOI] [Google Scholar]
- 30.Pross A. 2011. Toward a general theory of evolution: extending Darwinian theory to inanimate matter. J. Syst. Chem. 2, 1 ( 10.1186/1759-2208-2-1) [DOI] [Google Scholar]
- 31.Mix LJ. 2015. Defending definitions of life. Astrobiology 15, 15–19. ( 10.1089/ast.2014.1191) [DOI] [PubMed] [Google Scholar]
- 32.Taran O, Chen C, Omosun TO, Hsieh M-C, Rha A, Goodwin JT, Mehta AK, Grover MA, Lynn DG. 2017. Expanding the informational chemistries of life: peptide/RNA networks. Phil. Trans. R. Soc. A 375, 20160356 ( 10.1098/rsta.2016.0356) [DOI] [PubMed] [Google Scholar]
- 33.Dupré J. 2017. The metaphysics of evolution. Interface Focus 7, 20160148 ( 10.1098/rsfs.2016.0148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith E, Morowitz HJ. 2016. The origin and nature of life on Earth: the emergence of the fourth geosphere. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 35.Adami C. 2015. Information-theoretic considerations concerning the origin of life. Orig. Life Evol. Biosph. 45, 309–317. ( 10.1007/s11084-015-9439-0) [DOI] [PubMed] [Google Scholar]
- 36.Walker SI, Davies PCW. 2013. The algorithmic origins of life. J. R. Soc. Interface 10, 20120869 ( 10.1098/rsif.2012.0869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flack JC. 2017. Coarse-graining as a downward causation mechanism. Phil. Trans. R. Soc. A 375, 20160338 ( 10.1098/rsta.2016.0338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kempes CP, Wolpert D, Cohen Z, Pérez-Mercader J. 2017. The thermodynamic efficiency of computations made in cells across the range of life. Phil. Trans. R. Soc. A 375, 20160343 ( 10.1098/rsta.2016.0343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall SM, Murray ARG, Cronin L. 2017. A probabilistic framework for identifying biosignatures using Pathway Complexity. Phil. Trans. R. Soc. A 375, 20160342 ( 10.1098/rsta.2016.0342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall W, Kim H, Walker SI, Tononi G, Albantakis L. 2017. How causal analysis can reveal autonomy in models of biological systems. Phil. Trans. R. Soc. A 375, 20160358 ( 10.1098/rsta.2016.0358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams A, Zenil H, Davies PCW, Walker SI. 2017. Formal definitions of unbounded evolution and innovation reveal universal mechanisms for open-ended evolution in dynamical systems. Sci. Rep. 7, 997 ( 10.1038/s41598-017-00810-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, Davies P, Walker SI. 2015. New scaling relation for information transfer in biological networks. J. R. Soc. Interface 12, 20150944 ( 10.1098/rsif.2015.0944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker SI, Kim H, Davies PCW. 2016. The informational architecture of the cell. Phil. Trans. R. Soc. A 374, 20150057 ( 10.1098/rsta.2015.0057) [DOI] [PubMed] [Google Scholar]
- 44.Oizumi M, Albantakis L, Tononi G. 2014. From the phenomenology to the mechanisms of consciousness: integrated information theory 3.0. PLoS Comput. Biol. 10, e1003588 ( 10.1371/journal.pcbi.1003588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davidich MI, Bornholdt S. 2008. Boolean network model predicts cell cycle sequence of fission yeast. PLoS ONE 3, e1672 ( 10.1371/journal.pone.0001672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parrondo JMR, Horowitz JM, Sagawa T. 2015. Thermodynamics of information. Nat. Phys. 11, 131–139. ( 10.1038/nphys3230) [DOI] [Google Scholar]
- 47.Project Gutenberg. 2016. Harper’s New Monthly Magazine, Volume 2, No. 12, May, 1851. Project Gutenberg.
- 48.Morgan DO. 2007. The cell cycle: principles of control. Sunderland, MA: New Science Press. [Google Scholar]
- 49.Goldenfeld N, Biancalani T, Jafarpour F. 2017. Universal biology and the statistical mechanics of early life. Phil. Trans. R. Soc. A 375, 20160341 ( 10.1098/rsta.2016.0341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kacar B, Guy L, Smith E, Baross J. 2017. Resurrecting ancestral genes in bacteria to interpret ancient biosignatures. Phil. Trans. R. Soc. A 375, 20160352 ( 10.1098/rsta.2016.0352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langton CG, Taylor C, Farmer JD, Rasmussen S. 1992. Artificial life II. Reading, MA: Addison-Wesley.
- 52.McInerney J, Erwin DH. 2017. The role of public goods in planetary evolution. Phil. Trans. R. Soc. A 375, 20160359 ( 10.1098/rsta.2016.0359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCaskill JS. 1988. Polymer chemistry on tape: a computational model for emergent genetics. Göttingen, Germany: Max-Planck-Society. [Google Scholar]
- 54.Fontana W. 1992. Algorithmic chemistry. In Artificial life II (eds Langton CG, Taylor C, Farmer JD, Rasmussen S), pp. 159–210. Reading, MA: Addison-Wesley. [Google Scholar]
- 55.CG N, LaBar T, Hintze A, Adami C. 2017. Origin of life in a digital microcosm. Phil. Trans. R. Soc. A 375, 20160350 ( 10.1098/rsta.2016.0350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banzhaf W, Yamamoto L. 2015. Artificial chemistries. Cambridge, MA: MIT Press. [Google Scholar]
- 57.Meringer M, Cleaves HJ. 2017. Exploring astrobiology using in silico molecular structure generation. Phil. Trans. R. Soc. A 375, 20160344 ( 10.1098/rsta.2016.0344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersen JL, Flamm C, Merkle D, Stadler PF. 2017. An intermediate level of abstraction for computational systems chemistry. Phil. Trans. R. Soc. A 375, 20160354 ( 10.1098/rsta.2016.0354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hazen RM. 2017. Chance, necessity and the origins of life: a physical sciences perspective. Phil. Trans. R. Soc. A 375, 20160353 ( 10.1098/rsta.2016.0353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adam ZR, Zubarev D, Aono M, Cleaves HJ. 2017. Subsumed complexity: abiogenesis as a by-product of complex energy transduction. Phil. Trans. R. Soc. A 375, 20160348 ( 10.1098/rsta.2016.0348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikegami T, Mototake Y, Kobori S, Oka M, Hashimoto Y. 2017. Life as an emergent phenomenon: studies from a large-scale boid simulation and web data. Phil. Trans. R. Soc. A 375, 20160351 ( 10.1098/rsta.2016.0351) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.