Abstract
Biological public goods are broadly shared within an ecosystem and readily available. They appear to be widespread and may have played important roles in the history of life on Earth. Of particular importance to events in the early history of life are the roles of public goods in the merging of genomes, protein domains and even cells. We suggest that public goods facilitated the origin of the eukaryotic cell, a classic major evolutionary transition. The recognition of genomic public goods challenges advocates of a direct graph view of phylogeny, and those who deny that any useful phylogenetic signal persists in modern genomes. Ecological spillovers generate public goods that provide new ecological opportunities.
This article is part of the themed issue ‘Reconceptualizing the origins of life’.
Keywords: major evolutionary transitions, public goods, evolutionary theory
1. Introduction
Individuals of the bacterium Escherichia coli generally have 4000–5500 genes, but a recent study estimated that only 3188 gene families were ‘core’ (which they defined as being present in 95% of genomes) from a total of approximately 90 000 unique gene families found in E. coli [1]. The majority of the genes form a pool of accessory genes, which individuals may take up depending on environmental conditions. The fluidity of the E. coli genome, and similar examples in other taxa, have led to the concept of a pan-genome where genes may be shared not just in a market of one species but also across widely divergent clades [2]. This challenges the orthodoxy of using tree-like topologies in order to depict evolutionary relationships. We have argued for the inclusion of ‘goods-thinking’ within biology as a distinct conceptual viewpoint from the phylogenetic or tree-based thinking prevalent among evolutionary biologists, and the emphasis on trophic food webs among ecologists. Public goods are widespread in biology, including genes that are widely shared among ‘prokaryotes’, oxygen generated by oxygenic photosynthesis, transposable elements and the provision of ecological services via ecosystem engineering and niche construction [3–5]. In each case, the relevant aspect of a biological component is not information, vertical inheritance, nor trophic interactions. Rather these components are behaving as biological goods, analogous to economic goods. Here, we focus on major events in the early evolution of life and the importance of biological public goods through the merging of evolving objects, and through ecological facilitation (figure 1).
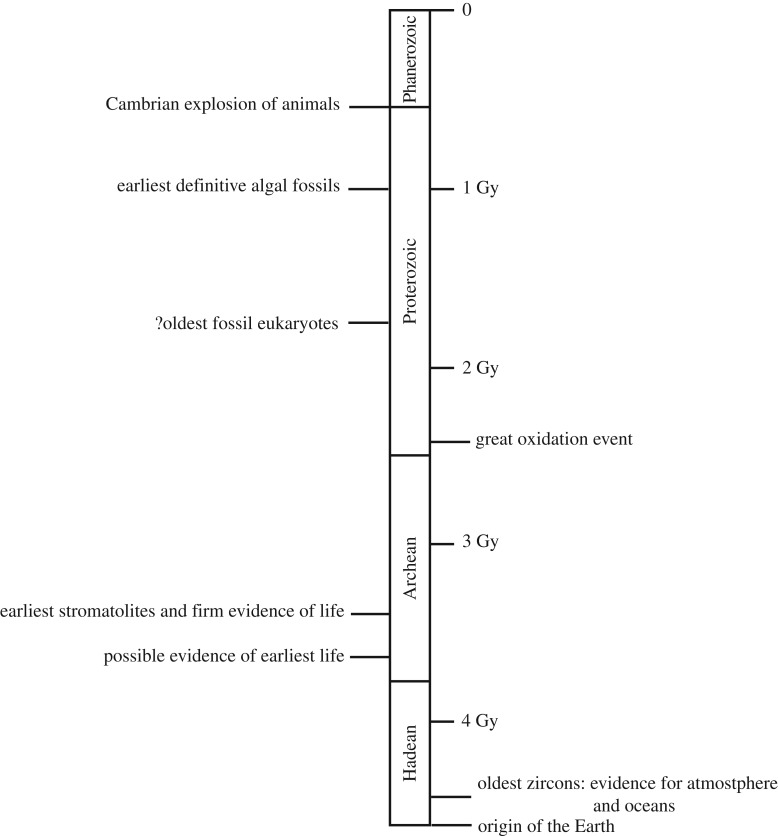
Figure 1.
Generalized timeline of major events in the early history of the Earth, focusing on those associated with the generation of public goods. Note that there is now little evidence for a ‘Late Heavy Bombardment’ around 4.0–3.8 Gy. Gy—billion years ago.
The importance of public goods in evolutionary innovation also highlights the influence of ecological processes on the evolution of life. Particularly in discussions of the origin and early history of life, the prevalence of phylogenetic methods, or ‘tree-thinking’, has led to a neglect of ecological processes. The ability to reconstruct vertical lines of descent has proved invaluable and has highlighted the frequency of horizontal gene transfer (HGT). However, many biological components may be shared widely within a broader clade, as genes, metabolic products or non-trophic means that provide scaffolds for physical or chemical elements of the environment.
2. Nature of public goods
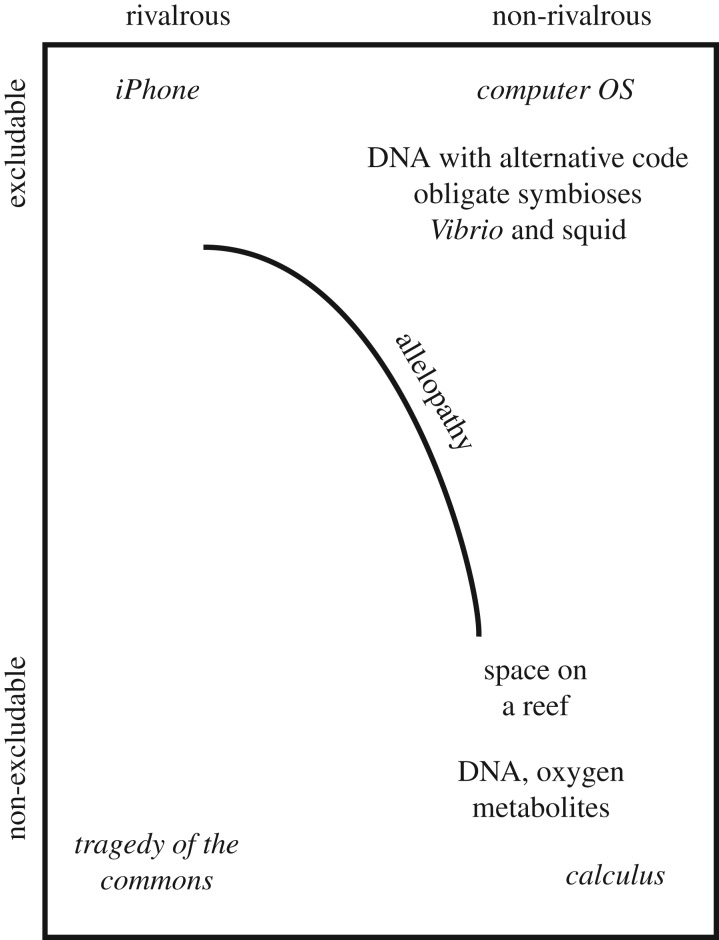
The classic definition of an economic good is an object or service that has a benefit and can be bought or sold. But economists have recognized that this definition is too narrow, as many important goods cannot be readily traded. Some goods anyone can consume without reducing their availability to others, a condition known as ‘non-rivalry’ and for which it is difficult to exclude others from using the good or ‘non-excludability’. Because they are readily available, these are known as public goods. The calculus (and knowledge in general) is a classic public good: there is no limit to the number of integrals that can be solved at one time, and one person's activity does not limit that of any one else. Language, the air traffic control system, oxygen in the air and national defence are all examples of public goods (the term ‘the public good’ is also used for shared societal benefits such as education, but this is not the same as the economic definition). A matrix of excludability versus rivalrousness defines four classes of economic goods (figure 2). Club goods are also of interest: they are non-rivalrous but excludable. Examples include an encoded GPS signal, tollways or academic societies. In each case, access to the good can be controlled and its benefits are available only to members of the club. Some economists have argued that public goods are the foundation of much economic growth [6].
Figure 2.
The relationship between different classes of economic goods (in italics) and biological goods based upon the relationship between rivalry and excludability. Private goods are both rivalrous and excludable; club goods are excludable but non-rivalrous; and public goods are non-rivalrous and non-excludable. In economics, patent law and similar mechanisms serve to convert public goods to private goods. In biology, mechanisms such as allelopathy enable organisms secreting particular chemicals to protect their space from others who might encroach upon it. Allelopathic interactions are common among benthic organisms on reefs, for example.
In this contribution, we will largely focus on public goods, though biological club goods are also of interest. These are products where organisms have evolved means to restrict the use of the good to a particular clade. An example is the usage of alternative genetic codes. We find that when clades such as the Mycoplasma/Spiroplasma group of bacteria have evolved an alternative genetic code (in this case, the reassignment of the UGA codon so that it encodes tryptophan), many protein-coding genes are only useful to members of this clade and horizontal transfer of these genes outside of this clade will not result in useful proteins. Protein-coding genes can be made excludable by altering the genetic code by which they are decoded.
Many biological products are analogous to economic goods, and we have found the concept of biological goods an important corrective to an overly tree-based view of evolutionary history. Public goods also expand the relationship between ecology and evolution, emphasizing the importance of feedback between ecological processes and evolutionary outcomes. The most obvious example of a public good is the oxygen in the atmosphere, as produced via oxygenic photosynthesis [7]. The origin of cyanobacteria, probably before 2.7 billion years ago (Ga), resulted in the progressive oxygenation of the atmosphere and eventually the oceans [8,9]. The establishment of a redox gradient within marine sediment or a soil, from aerobes through denitrifiers, manganese reducers to sulfate reducers and eventually methanogens, represents a series of coupled biogeochemical cycles where microbes of one class produce the environment necessary for microbes with another metabolism. In such cases, the public goods are metabolic by-products which change the surrounding environment (globally in the case of oxygenic photosynthesis), generating new opportunities for other clades [4]. A related class of public goods represent ecosystem-constructing activities that modify the environment for that population and for other species in the same environment. Often described as niche construction [5,10] or ecosystem engineering [11], examples include burrowing (bioturbation) of sediments, beaver dams and the construction of reefs. A final class of public goods involve the reusability of the macromolecules of inheritance. This includes the merging of evolving objects through a number of mechanisms, including HGT via plasmids or phage, naked DNA in soil or water, or viruses [12], as well as symbioses, and some of what have been described as major evolutionary transitions, such as the origin of eukaryotes. Unlike the first two classes, which generate new ecological and evolutionary opportunities, this last class generates new evolutionary variation [4]. The theory of public goods has recently been applied to cooperation in microbial systems [13,14] and social evolution [15,16], placing these observations of cooperation and social evolution of microbes on a firm theoretical footing.
3. Merging of evolving objects
Evolving objects come in a variety of forms and are organized into many units upon which natural selection can act. Each part of a gene—whether it is a promoter region, a coding region, a eukaryotic intron or an upstream or downstream enhancer of expression—can be made available to other genes and is thus acting as a public good. Its merging with another part of a gene can become permanent if this proves to be beneficial, or at least not detrimental to function. Genes, however, are not the only objects of selection. Selection can act on operons, gene clusters, entire genomes or small collections of genes organized into plasmids or phages or viruses [17]. Indeed, as we shall see, entire genomes can be available as public goods for integration with other entire genomes. When we look at the evolutionary history of all these evolving objects, we see that it is a history of continuous merging, balanced with deletions and splitting of evolving objects. The appropriate way to think about this kind of evolutionary history is not to focus solely on the use of phylogenetic trees, because they can only accurately represent vertical evolutionary histories. Instead, it is necessary to employ network models.
In 1997, Tatusov et al. [18] constructed a dataset of 17 967 proteins from seven genomes and used network structures in order to define gene families. That same year, Park et al. [19] used the idea of ‘intermediate’ sequences in order to identify distant homology. Later, ‘Rosetta stone’ sequences were used to describe a fusion or composite protein in one species where the protein is in two parts in other species [20]. The Rosetta Stone idea was based on a functional perspective of protein evolution. Rosetta Stone proteins would provide clues about the evolution of protein functions. Fusion of two protein domains into a single protein could reasonably be inferred to reflect complementary activities within a cell.
Enright et al. further took the idea of identifying fusion proteins as a way to enhance protein–protein interaction prediction [21]. Network structures have also been used to analyse community structure [22], detect remote homologues [23] or display relationships [24]. The global structure of genomic data is highly connected, though with very clear breaks—termed ‘genetic worlds’ [24]. Dagan & Martin [25] carried out a community analysis of genome networks of prokaryotes and detected massive levels of HGT that were not restricted to particular kinds of gene or genome. These complex evolutionary histories have been summarized as the process of ‘introgressive descent’ [17]. The tree-thinking perspective, while obviously useful for analysing evolutionary histories that are tree-like, are not applicable for situations where evolution has not been tree-like. When evolution has been net-like, then this history is appropriately analysed using net-like evolutionary models. Indeed, the recurrent, independent, homoplastic merging of protein domains into whole proteins has been observed and proteins that have independently arisen by this process are now named ‘epaktologs’ [26].
Sequence similarity networks (SSNs) have been used in order to reconstruct the formation of composite genes [27,28]. Several SSN motifs have been formally described and what these motifs might imply has been explored [17]. Dagan & Martin [29] have described networks as more complete and accurate pictures of genome evolution. Jachiet et al. [28] have formalized the problem of detecting composite genes in SSNs. These composite genes act as ‘glue’ that holds networks together and facilitates the growth of connected components. Most recently, SSNs have been used to show that standard approaches in evolutionary biology struggle to describe the evolution of composite genes and at times can mislead evolutionary interpretations [27].
Currently, the comparison of proteins that differ in their domain composition is treated in an ad hoc way, with extra or missing domains often pruned from analyses. However, going against this trend, Bjorklund et al. [30] explicitly mapped domain similarities onto phylogenetic trees to explain protein evolution. ‘Cleaning up’ of alignments is an excellent solution if the goal is to infer the phylogenetic history of parts of proteins, but it does remove true evolutionary events. The use of tree-thinking approaches to understand evolution preceded our understanding that composite gene formation was so frequent and that genes behaved as public goods.
The merging of two or more protein domains to form a new protein can be considered to be a merging of evolving objects ‘of the same level’. However, empirical evidence says that mergings do not always have to be of the same level [17]. For instance, the horizontal acquisition of genes by organisms is not a merging of objects of the same level. The whole organism is at one level and the transferred gene is at a different level. Similarly, plasmids being integrated into a host cell, or even genes being integrated into a plasmid are also examples of mergings of different levels. Inspired by the ideas of ‘egalitarian’ and ‘fraternal’ major evolutionary transitions, we suggest that we could also classify the mergings of evolutionary objects into ‘fraternal mergers’, where the merging objects are at the same level, and ‘egalitarian mergers’ to denote mergers where the partners are at different levels.
The most iconic and important fraternal merger is the formation of the first eukaryotic cells [31,32]. It has long been recognized that the mitochondrion and its relatives (mitosomes, hydrogenosomes, etc.) are descended from an alpha-proteobacterium that was involved in a merger with another cell [31–34]. Recent analysis of metagenomic data suggests that the other partner in this merger was an early member of the ASGARD group of archaebacteria [35,36]. The nascent eukaryotic cell seems to have been able to combine several features of both partners in this merger, in order to produce new versions of DNA replication, transcription, RNA processing, translation and metabolism [31]. This is a situation where a fraternal merger has facilitated a major evolutionary transition. In a similar way, the origin of the eukaryotic plastid (chloroplast and relatives such as the apicoplast in apicoplastids) involved the merger of a cyanobacterial cell with a eukaryotic cell. The selective pressure that made this merger successful was metabolic, with the chloroplast providing energy from sunlight to its host. Subsequent to the acquisition of the chloroplast by the host, thousands of genes were transferred from the plastid to the nucleus, a process known as endosymbiotic gene transfer [37].
Fraternal mergers frequently involve the merging of protein domains with protein domains. We also see fraternal merging in the context of whole-genome duplications, which are known to have happened to some yeast species [38] and at the origin of vertebrates [39]. In addition, polyploidy in plants is common and these can also be viewed as a fraternal merging of genomic information. Palaeoploidy refers to ancient whole-genome duplication events that have persisted in some form for many millions of years. In these cases, the merging involves the combination of two identical genomes into a single organism or cell [40]. This is facilitated because genomes can behave as public goods.
HGT is an example of an egalitarian merger and is better known in prokaryotes than eukaryotes, but is certainly not restricted to prokaryotes. Several confirmed horizontal transfers of prokaryote DNA into eukaryotes are known (e.g. [41,42]) and are usually associated with significant fitness effects. However, the rate of HGT in prokaryotes seems to be significantly higher than in eukaryotes [43,44] and there is no known mechanism that facilitates HGT in eukaryotes [45]. HGT involves two different events—the transfer event and the retention event. These will usually have quite different rates. When we think of molecular sequences acting as public goods, the likelihood of retention is not completely relevant. For instance, it is generally accepted that air is a public good. It satisfies the criteria for being a public good—it is not excludable and not rivalrous. However, methanogenic archaea grow only in anaerobic conditions and thus do not avail themselves of this public good. The lesson here is that any public good does not need to be beneficial for all organisms, in order to be considered a public good. Hanage [46] has conflated public goods with group selection and argued that all genes must be beneficial to all organisms; otherwise they cannot be categorized as public goods. This is a mistaken interpretation of public goods.
Finally, just as genes and plastids served as public goods later in the history of life, the earliest phases in the origin of life may have offered plentiful opportunities for public goods. Woese proposed that discrete evolutionary lineages developed from an earlier phase in which cells shared many components [47]. The machinery for translation may have been among the earliest to anneal into a discrete unit and thus became a public good broadly shared among different cells. Similarly, cell membranes may initially have been a public good. Many proposed environments for early life, such as hydrothermal vents, would have facilitated the sharing of different components among protocells. For example, in a discussion of the encapsulation of replicators within a mineral honeycomb, Branciamore et al. suggest that mild parasites may have acquired the ability to form membranes or replicases [48]. But these may have begun as shared resources. As Woese noted, the early history of life may have included more shared goods than generally appreciated.
4. Ecological facilitation as a public good
Although evolutionary biologists seldom appreciate it, the redox history of the Earth is a three-act play, and most of the major evolutionary events, from the origin of cells and various metabolic pathways through the origin of eukaryotes to the origin of multicellularity occurred on an Earth much different from today's Earth. The advent of oxygenic photosynthesis eventually led to a dramatic rise in atmospheric oxygen about 2.4 Ga, transforming the redox state of the Earth from essentially anoxic to one with a slightly (by modern standards) oxygenated atmosphere and variably anoxic to sulfur-rich (euxinic) ocean. This redox state persisted until 800–550 million years ago when a second oxygenation event resulted in the redox state we have today. Although the causes of this second episode remain controversial, biological contributions via bioturbation of sediment via early animal burrowing and carbon sequestration with the origin of a through-gut in bilaterian metazoans were an important contributor [49]. Thus, significant public goods were generated during each redox transition, opening new evolutionary opportunities for a variety of clades.
There are many other examples of organisms generating public goods through activities that modify their external environment. Niche construction and ecosystem engineering are both forms of ecological facilitation, where biological activities, particularly non-trophic activities, modify the surrounding environment sufficiently to impact the fitness and thus evolutionary trajectory of the niche-constructing taxa. If these activities persist over generations, they create an ecological inheritance, as in the case of the oxygenation events described above, which will often spill over and impact the macroevolutionary dynamics of other species [5].
5. Implications for understanding the early evolution of life
Although economics and evolutionary biology each trace their intellectual lineage to political economy (via Darwin and Malthus), ideas from economics have had relatively little impact on evolutionary theory. The theory of public goods provides an alternative and complementary view of evolution to the tree-based paradigm of phylogenetics, to the gene-centric view of many evolutionary biologists and emphasizes the impact that non-trophic interactions can have in ecology. We have discussed three ways in which the generation of public goods has influenced evolution: the generation of metabolic by-products, broad utilization of genomic components and modification of the external environment through ecological facilitations. The sharing of genetic, cellular or ecological resources across clades is consistent with the view that the early history of life more closely resembled a network than a phylogenetic tree. More importantly, however, many economists have argued that public goods have been critical to innovation and economic growth and we are intrigued by the possibility that the same may be true in biological evolution. By generating new types of genetic variation, facilitating the rapid spread of valuable genomic components, and by modifying the environment through both metabolic by-products such as oxygen and through other ecosystem engineering and niche-constructing activities, biological public goods have modified selection pressures. Early in the history of life, the shared resource of pan-genomes may have facilitated the more rapid formation of viable cells. Ecological public goods may also have played a role. Among the avenues to be explored are the role of biological activity in changing the chemical environment for the origin of life and biological scaffolds. Microbial biofilms involve ecological facilitation among many different microbes and probably arose early in the history of life. The public goods viewpoint may also allow the identification of particular genes or gene suites whose interactions may reduce their utility for phylogenetic reconstruction. One avenue for future exploration is the identification of domains, genes and other genomic components, which have been extensively used as public goods.
6. Conclusion
Tracing phylogenetic relationships among clades emphasizes the vertical transmission of genetic information. Recognition of the extent of HGT has led many authors to discuss the network-like nature of descent, particularly early in the history of life, and to question whether a meaningful phylogeny can be recovered. Our argument here is for the addition of an ecological view, in which the generation of metabolic by-products, ecological facilitation and ecosystem inheritance, and the merging of evolving objects reflect the construction and utilization of public goods. We have provided examples where public goods have served as a source of new variation and have generated ecologic and evolutionary opportunities. Focusing on the role of public goods throughout the history of life brings ecology and evolution closer together, and offers a fascinating new window through which to explore major events in the history of life. The focus on tree-like patterns resulted in a mistaken view of the eukaryote cell as a primary domain of life, whereas incorporating ideas about the sharing and merging of genetic components has revealed it to be a chimerical domain. Future work on reconstructing Earth's history should include a goods-thinking perspective.
Data accessibility
This article has no additional data.
Authors' contributions
This paper was conceived, written and edited jointly by the two authors.
Competing interests
We declare we have no competing interests.
Funding
D.H.E. acknowledges the support of NASA through the National Astrobiology Institute to the MIT node. J.McI. acknowledges the support of the John Templeton Foundation (JTF grant 60579) and the Biotechnology and Biological Sciences Research Council (grant number BB/N018044/1).
References
- 1.Land M, et al. 2015. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15, 141–161. (doi:10.1007/s10142-015-0433-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fullmer MS, Soucy SM, Gogarten JP. 2015. The pan-genome as a shared genomic resource: mutual cheating, cooperation and the black queen hypothesis. Front. Microbiol. 6, 728 (doi:10.3389/fmicb.2015.00728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McInerney JO, Pisani D, Bapteste E, O'Connell MJ. 2011. The public goods hypothesis for the evolution of life on Earth. Biol. Direct 6, 41 (doi:10.1186/1745-6150-6-41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erwin DH. 2015. A public goods approach to major evolutionary innovations. Geobiology 13, 308–315. (doi:10.1111/gbi.12137) [DOI] [PubMed] [Google Scholar]
- 5.Erwin DH. 2008. Macroevolution of ecosystem engineering, niche construction and diversity. Trends Ecol. Evol. 23, 304–310. (doi:10.1016/j.tree.2008.01.013) [DOI] [PubMed] [Google Scholar]
- 6.Romer PM. 1990. Endogenous technological change. J. Polit. Econ. 98, S71–S102. (doi:10.1086/261725) [Google Scholar]
- 7.Maynard Smith J, Szathmary E.. 1995. The major transitions in evolution. New York, NY: W. H. Freeman. [Google Scholar]
- 8.Lenton TM, Boyle RA, Poulton SW, Shields-Zhou G, Butterfield NJ. 2014. Co-evolution of eukaryotes and ocean oxygenation in the Neoproterozoic era. Nat. Geosci. 7, 257–265. (doi:10.1038/ngeo2108) [Google Scholar]
- 9.Lyons TW, Reinhard CT, Planavsky NJ. 2014. The rise of oxygen in Earth's early ocean and atmosphere. Nature 506, 307–315. (doi:10.1038/nature13068) [DOI] [PubMed] [Google Scholar]
- 10.Odling-Smee FJ, Laland KN, Feldman MW.. 2003. Niche construction: the neglected process in evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- 11.Jones CG, Lawton JH, Shachak M. 1997. Positive and negative effects of organisms as physical ecosystem engineers. Ecology 78, 1946–1957. (doi:10.1890/0012-9658(1997)078[1946:PANEOO]2.0.CO;2) [Google Scholar]
- 12.Koonin EV. 2016. Viruses and mobile elements as drivers of evolutionary transitions. Phil. Trans. R. Soc. B 371, 20150442 (doi:10.1098/rstb.2015.0442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachmann H, Fischlechner M, Rabbers I, Barfa N, Branco dos Santos F, Molenaar D, Teusink B. 2013. Availability of public goods shapes the evolution of competing metabolic strategies. Proc. Natl Acad. Sci. USA 110, 14 302–14 307. (doi:10.1073/pnas.1308523110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner GD, et al. 2014. Evolution of microbial markets. Proc. Natl Acad. Sci. USA 111, 1237–1244. (doi:10.1073/pnas.1315980111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin SA. 2014. Public goods in relation to competition, cooperation, and spite. Proc. Natl Acad. Sci. USA 111 (Suppl. 3), 10 838–10 845. (doi:10.1073/pnas.1400830111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobata S, Tsuji K. 2013. Public goods dilemma in asexual ant societies. Proc. Natl Acad. Sci. USA 110, 16 056–16 060. (doi:10.1073/pnas.1309010110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bapteste E, Lopez P, Bouchard F, Baquero F, McInerney JO, Burian RM. 2012. Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc. Natl Acad. Sci. USA 109, 18 266–18 272. (doi:10.1073/pnas.1206541109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278, 631–637. (doi:10.1126/science.278.5338.631) [DOI] [PubMed] [Google Scholar]
- 19.Park J, Teichmann SA, Hubbard T, Chothia C. 1997. Intermediate sequences increase the detection of homology between sequences. J. Mol. Biol. 273, 349–354. (doi:10.1006/jmbi.1997.1288) [DOI] [PubMed] [Google Scholar]
- 20.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. 1999. Detecting protein function and protein-protein interactions from genome sequences. Science 285, 751–753. (doi:10.1126/science.285.5428.751) [DOI] [PubMed] [Google Scholar]
- 21.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90. (doi:10.1038/47056) [DOI] [PubMed] [Google Scholar]
- 22.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584. (doi:10.1093/nar/30.7.1575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolten E, Schliep A, Schneckener S, Schomburg D, Schrader R. 2001. Clustering protein sequences—structure prediction by transitive homology. Bioinformatics 17, 935–941. (doi:10.1093/bioinformatics/17.10.935) [DOI] [PubMed] [Google Scholar]
- 24.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. 2010. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl Acad. Sci. USA 107, 127–132. (doi:0908978107[pii]10.1073/pnas.0908978107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dagan T, Martin W. 2007. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl Acad. Sci. USA 104, 870–875. (doi:0606318104[pii]10.1073/pnas.0606318104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy A, Banyai L, Patthy L. 2011. Reassessing domain architecture evolution of metazoan proteins: major impact of errors caused by confusing paralogs and epaktologs. Genes (Basel) 2, 516–561. (doi:10.3390/genes2030516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haggerty LS, et al. 2014. A pluralistic account of homology: adapting the models to the data. Mol. Biol. Evol. 31, 501–516. (doi:10.1093/molbev/mst228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jachiet PA, Pogorelcnik R, Berry A, Lopez P, Bapteste E. 2013. MosaicFinder: identification of fused gene families in sequence similarity networks. Bioinformatics 29, 837–844. (doi:10.1093/bioinformatics/btt049) [DOI] [PubMed] [Google Scholar]
- 29.Dagan T, Martin W. 2009. Getting a better picture of microbial evolution en route to a network of genomes. Phil. Trans. R. Soc. B 364, 2187–2196. (doi:10.1098/rstb.2009.0040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A. 2005. Domain rearrangements in protein evolution. J. Mol. Biol. 353, 911–923. (doi:10.1016/j.jmb.2005.08.067) [DOI] [PubMed] [Google Scholar]
- 31.McInerney J, Pisani D, O'Connell MJ.. 2015. The ring of life hypothesis for eukaryote origins is supported by multiple kinds of data. Phil. Trans. R. Soc. B 370, 20140323 (doi:10.1098/rstb.2014.0323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInerney JO, O'Connell MJ, Pisani D. 2014. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat. Rev. Microbiol. 12, 449–455. (doi:10.1038/nrmicro3271) [DOI] [PubMed] [Google Scholar]
- 33.Horner DS, Hirt RP, Kilvington S, Lloyd D, Embley TM. 1996. Molecular data suggest an early acquisition of the mitochondrion endosymbiont. Proc. R. Soc. Lond. B 263, 1053–1059. (doi:10.1098/rspb.1996.0155) [DOI] [PubMed] [Google Scholar]
- 34.Sagan L. 1967. On the origin of mitosing cells. J. Theor. Biol. 14, 255–274. (doi:10.1016/0022-5193(67)90079-3) [DOI] [PubMed] [Google Scholar]
- 35.McInerney JO, O'Connell MJ. 2017. Microbiology: mind the gaps in cellular evolution. Nature 541, 297–299. (doi:10.1038/nature21113) [DOI] [PubMed] [Google Scholar]
- 36.Zaremba-Niedzwiedzka K, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358. (doi:10.1038/nature21031) [DOI] [PubMed] [Google Scholar]
- 37.Martin W, et al. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA 99, 12 246–12 251. (doi:10.1073/pnas.182432999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfe KH. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2, 333–341. (doi:10.1038/35072009) [DOI] [PubMed] [Google Scholar]
- 39.Dehal P, Boore JL.. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314 (doi:10.1371/journal.pbio.0030314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson ES, Roth EM, Woods DD, Chow R, Gomes JO. 2004. Handoff strategies in settings with high consequences for failure: lessons for health care operations. Int. J. Qual. Health Care 16, 125–132. (doi:10.1093/intqhc/mzh026) [DOI] [PubMed] [Google Scholar]
- 41.Husnik F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578. (doi:10.1016/j.cell.2013.05.040) [DOI] [PubMed] [Google Scholar]
- 42.He D, Fu CJ, Baldauf SL. 2016. Multiple origins of eukaryotic cox15 suggest horizontal gene transfer from bacteria to jakobid mitochondrial DNA. Mol. Biol. Evol. 33, 122–133. (doi:10.1093/molbev/msv201) [DOI] [PubMed] [Google Scholar]
- 43.Ku C, et al. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524, 427–432. (doi:10.1038/nature14963) [DOI] [PubMed] [Google Scholar]
- 44.Chewapreecha C, et al. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46, 305–309. (doi:10.1038/ng.2895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. (doi:10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- 46.Hanage WP. 2016. Not so simple after all: bacteria, their population genetics, and recombination. Cold Spring Harb. Perspect. Biol. 8, a018069 (doi:10.1101/cshperspect.a018069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woese C. 1998. The universal ancestor. Proc. Natl Acad. Sci. USA 95, 6854–6859. (doi:10.1073/pnas.95.12.6854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Branciamore S, Gallori E, Szathmary E, Czaran T. 2009. The origin of life: chemical evolution of a metabolic system in a mineral honeycomb? J. Mol. Evol. 69, 458–469. (doi:10.1007/s00239-009-9278-6) [DOI] [PubMed] [Google Scholar]
- 49.Erwin DH, Valentine JW.. 2013. The Cambrian explosion: the construction of animal biodiversity. Greenwood, CO: Roberts & Co. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.