Highlights
-
•
Developed a high-throughput microarray for anti-microbial susceptibility testing (AST).
-
•
Demonstrated that the feasibility of the AST against clinical isolates of MRSA.
-
•
Platform is a low sample volume, rapid, high-throughput alternative to traditional assays.
Keywords: Antimicrobial susceptibility testing, High-throughput, Microarray
Abstract
We describe the development of a novel, high-throughput, nano-scale microarray platform for antimicrobial susceptibility testing (AST). The platform allows to process 480 samples at 50 nL volume on a single chip, analyze by fluorescence read-out with an easy dunk-and-rinse step, and the ability to process multiple samples and chips simultaneously. We demonstrate the applicability of this chip for culturing community acquired methicillin resistant Staphylococcus aureus (CA-MRSA), and perform AST against clinical isolates of CA-MRSA. The chip platform holds promise for an impact in microbial biotechnology as an attractive high-throughput, lower sample volume and quicker alternative to conventional AST such as the traditional broth microdilution or the newer automated systems.
1. Introduction
Antimicrobial resistance remains a major threat to human health throughout the world, largely due to the indiscriminate use of antibiotics. The liberal use of antibiotics is driven by the need to quickly administer active antimicrobial therapy in the face of diagnostic uncertainty. AST is commonly performed using principles based on broth microdilution or disk diffusion assays, and takes 2-3 days, which includes the time for culturing the clinical isolates followed by antibiotic susceptibility testing by exposing the cultures to a panel of antibiotics over a range of concentrations [1]. Typically, clinical samples isolated from patients are cultured to increase the inoculum and isolate the pathogen; and then the isolates are exposed to a range of antibiotic concentrations either in 96-well plate suspension cultures or drug-filled cassettes (broth microdilution) or to a single concentration of the drug-impregnated disc placed on an agar plate containing the organism (disk diffusion). Susceptibility is then assessed by visual inspection either manually or by an automated instrument. Thus, it is conceivable that obviating relatively large culture volume or cell density may increase the throughput, and decrease the time required to obtain AST results. It is now well established that faster availability of AST data can enable the clinician to initiate or switch out of broad-spectrum treatment regimen to appropriate antimicrobial therapy sooner, and significantly reduce health care costs and morbidity [2]. While there has been some progress in the recent years for rapid AST [3], [4], [5], the currently available tests are either limited to a few select organisms such as PCR-based methods [6], [7], may not predict drug response reliably [8], [9], [10], limited in throughput or are cost-prohibitive such as MALDI-TOF MS or NMR [11] to most labs even in developed countries. To address some of the aforementioned issues, we have designed a novel microarray platform for high-throughput antimicrobial susceptibility testing (AST-Chip) and tested against select community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) isolates.
2. Materials and methods
2.1. Culture conditions
Frozen stocks of Staphylococcus aureus strain UAMS 1 and clinical isolates were subcultured onto tryptic-soy-agar plates (TSA) (BD Difco, MD, USA), and propagated the in 10 ml of tryptic soy-broth (TSB) (BD Difco, MD, USA) in an orbital shaker at 37 °C. In order to capture cells in log-phase, 100 μl from the overnight liquid culture were subcultured into 10 ml of TSB for 3 h.
2.2. Preparation of AST-Chip
The biofilm chip was prepared by modifying glass slides, and the growth conditions were optimized to obtain maximum biofilm yield of S. aureus using factorial design as described in [12], [13]. The optimized media is a concoction of 2X Yeast Peptone Dextrose (YPD), 3X Brain Heart Infusion (BHI) along with 10% human serum. Antimicrobial stock solutions of doxycycline, vancomycin hydrochloride (Sigma, MO, USA), and clindamycin hydrochloride (RPI Corp., IL, USA), were diluted in phosphate buffered saline to a maximum concentration of 100 μg/ml. Any subsequent dilutions needed for the antimicrobial susceptibility assays were made in PBS, mixed in a suspension of 0.5% alginate, and optimized media, and 50 nL of this mixture was spotted using a non-contact microarray spotter (Omnigrid Micro, Digilab Inc., Holliston, MA) on modified glass slides. An array of 40 rows and 12 columns was printed at room temperature with relative humidity of 100%. The AST-chip was stored at 4 °C for up to one week before use.
2.3. S. aureus nano-biofilms and viability assay
S. aureus cells were adjusted to a density of 1 × 107 cells/ml in modified growth medium, and printed on top of the drug spots. After printing, the slides were then placed in a humidified hybridization cassette (Arrayit, Sunnyvale, CA, USA) to prevent evaporation of spots and incubated at 37 °C. All microarrayer functions such as sample loading, priming, printing, and spatial distribution of the array were controlled by AxSys programming (Digilab, MA, USA). The viability of nano-biofilms on the microarray was determined by staining with FUN-1 fluorescent dye as described in [13], [14]. The fluorescence from the chips were measured using a microarray scanner (GenePix Personal 4100A; Axon Instruments, Union City, CA), and the images were analyzed with GenePix Pro V7 (Axon instruments, Union City, CA) to determine the antimicrobial susceptibility by normalizing the intensity between 0% and 100% for dead and live cells, respectively. The susceptibility values were obtained as average of 20–30 spots for each treatment from a single chip, and this experiment was performed in duplicate on two different days. The IC50 values were calculated using GraphPad Prism [15].
2.4. Microscopy
Fluorescence microscopy of S. aureus biofilms was performed after staining the slides with BacLight (Life Technologies) following manufacturer’s protocol, and visualized at 40X (DMI6000, Leica). Scanning electron microscopy was performed after fixing the slides and visualized using FEI Quanta 200 [12].
2.5. Antimicrobial susceptibility testing using Vitek-2 assays
Antimicrobial susceptibility testing was performed on a Vitek 2 compact instrument using AST-GP75 cards following the manufacturer’s instructions. Briefly 3.0 ml of 0.45% saline was dispensed into two 12 × 75 mm tubes. Three to four well-isolated Staphylococcus aureus colonies on 5% sheep blood agar were selected with a rayon swab and transferred the first tube of saline. The suspension was homogenized by vigorously twirling the swab and vortexing. The inoculum density was adjusted to a 0.5 to 0.63 McFarland standard using a DensiCHEK instrument. The initial suspension was subsequently diluted by transferring 280 microliters into the second tube of saline. The Vitek AST-GP75 card and the second saline inoculum tube were then placed into a cassette and loaded into the instrument for filling and incubation. Accession numbers and organism identification data (S. aureus) were entered into the instrument database. A purity check plate was streaked from the second saline tube to assure a single morphotype was tested. Quality control as recommended by the manufacturer with appropriate ATCC strains was performed each day of testing. Results were printed and reviewed.
3. Results and discussion
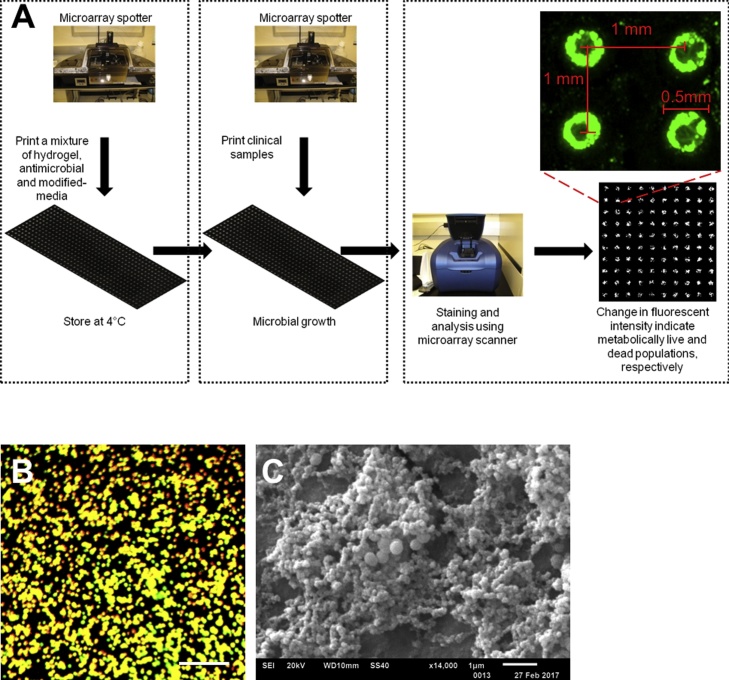
The AST-Chip is a chemically modified microscope glass slide with nano-scale islands of hydrogels, consisting of antimicrobial agents in modified-growth medium. The clinical isolates were printed on the islands, incubated with drugs, and the cell density in each spot was measured by a fluorescence assay to obtain AST Fig. 1. The key steps in chip fabrication are described in detail elsewhere [13]. Briefly, the fabrication consists of three steps: (i) modification of the surface of glass slides with poly(styrene-co-maleic anhydride) (PSMA); (ii) printing 50 nl mixture of poly-l-lysine and barium chloride creating the tie-layer using a robotic microarrayer (Microsys, Digilab); and (iii) printing a 50 nl mixture of 0.5% alginate solution, optimized-growth medium and antimicrobial agents at various concentrations on top of the tie-layer. This process produces hydrophilic, drug-loaded islands on a hydrophobic background of the modified glass slide. In its final format, the AST-Chip contains of 480 spots of hydrogels of 50 nL loaded with different drugs at various concentrations Fig. 1A.
Fig. 1.
High-throughput AST chip. (A) A schematic of high-throughput antimicrobial susceptibility testing using AST-Chip. Antimicrobial susceptibility testing using the chip can be conducted in 12 h; Microarray micrograph of S. aureus cultures formed on hydrogel spots of AST-Chip after 12–18 h. The spots are interspaced at 1 mm with a spot diameter of 0.5 mm; The nano-biofilms analyzed using FUN-1 is demonstrated in green color; (B) BacLight live/dead assay shows metabolically active cells in yellow-orange color, present as colonies. The scale bar is 100 μm; (C) SEM micrograph of S. aureus nano-biofilm colonies. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The growth medium in the mixture was optimized by factorial design to promote the growth of S. aureus [13]. We printed 50 nL spots of wild type (WT) Staphylococcus aureus (UAMS-1) in 0.5% alginate at 2500 cells/spot on top of alginate spots containing only optimized media Fig. 1B. We observed cell attachment to gel-spots in 30–45 min, following that the cells proliferated (1–8 h) and matured to a biofilm with the production of exopolymeric matrix material (8–16 h) [13]. After 12 h, the cell density at different spots was quantified using FUN-1, a vital cytoplasmic fluorescent dye compatible for analysis with a standard microarray scanner. A closer view of S. aureus biofilms is shown by fluorescence intensity distribution from FUN 1, which confirms the uniformity of cell viability among different spots (Fig. 1A). We also visualized the biofilm colonies closer by viability dye BacLight (Fig. 1B), and by scanning electron microscopy (SEM), both of which show the formation of S. aureus biofilm colonies (Fig. 1C).
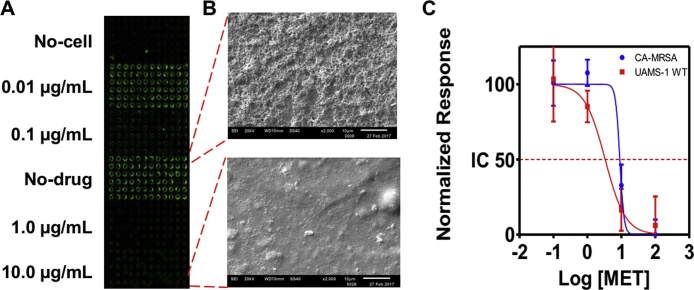
Having benchmarked the biofilm chip for the growth of S. aureus on top of hydrogel spots, we evaluated the antimicrobial-susceptibility of wild type (WT) Staphylococcus aureus (UAMS-1) and CA-MRSA (USA 300) strains against 0.1, 1.0, 10.0 and 100 μg/ml methicillin spotted on the AST-Chip. 50 nl of bacterial cell suspension in alginate was printed on top of the methicillin spots, and the chip was incubated for 12 h to allow for drug action, and stained with FUN1. The fluorescence intensities of control (no drug) and sodium hypochlorite-treated dead cells were arbitrarily set at 100% and 0% respectively. The inhibitory effects of the antibiotics were determined by the reduction in fluorescence intensity in comparison to the controls. As shown in Fig. 2A, the cell growth on the chip correlated with the drug concentration, and there was no cross-contamination across the spots despite their physical proximity and well-less culture. Consistent with the microarray scanner data, SEM micrographs confirmed that the untreated, control spots on the chip promoted S. aureus biofilm growth profusely, while methicillin completely inhibited biofilm growth (Fig. 2B). The susceptibility profile was deduced from the fluorescence intensities, and the data demonstrates that the AST-Chip accurately captures the decrease in susceptibility of CA-MRSA over WT strain (Fig. 2C).
Fig. 2.
Antimicrobial susceptibility profile of S. aureus. AST of methicillin against wild-type (UAMS-1WT) and community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA). (A) Microarray scanner image of AST-Chip with S. aureus cultures exposed to different concentrations of methicillin [MET]. The viability is indicated by the fluorescence intensity of the spots. (B) Scanning electron micrographs of S. aureus biofilms that are untreated (top) or treated with 10 μg/ml MET (bottom); (C) Dose-response profile of S. aureus cultures obtained using AST-Chip.
Next, we performed AST for CA-MRSA clinical isolates collected from patients with skin and soft tissue infections (SSTIs) in primary care practices affiliated with the South Texas Ambulatory Research Network (STARNet) Practice-Based Research Network (PBRN). CA-MRSA accounts for a vast majority of skin abscesses, has high rates of clinical failure and disease recurrence, puts close contacts and health care workers at risk of infection, is difficult to eradicate, and can cause fatal bacteremia. To demonstrate the applicability of the chip platform, we compared the susceptibility of five clinical isolates against three commonly used antibiotics, clindamycin (CLI), doxycycline (DOX), and vancomycin (VAN), with the clinically used Vitek-2 assay. Table 1 shows the results typically obtained from the Vitek-2 assay. The ‘R’ and ‘S’, show the concentrations at which the isolates are resistance or susceptible, respectively, and thus we find that A32 is susceptible to CLI at 4 μg/ml, and other isolates are susceptible to all three drugs.
Table 1.
Effect of clindamycin, doxycycline and vancomycin in inhibiting the growth of Staphylococcus aureus isolates on Vitek 2 AST-GP75 cards and AST-Chip. The letters “R” and “S” refer to the respective resistance and susceptibility to drugs at the specified concentration.
| Isolate | Age/Sex | Severity (Abcess) | Vitek |
AST- Chip Inhibition (%) |
||||
|---|---|---|---|---|---|---|---|---|
| CLI | DOX | VAN | CLI | DOX | VAN | |||
| A32 | 40/M | ≥ | >4 R | ≤ | <0.5 S | 13.2 | 31.23 | 50.0 |
| B2 | 52/F | Moderate-complicated (Yes) | ≤ | <0.5 S | <0.5 S | 64.8 | 30.1 | 51.1 |
| I27 | 37/M | Moderate-complicated (No) | ≤ | <0.5 S | <0.5 S | 27.8 | 36.6 | 50.7 |
| K2 | 53/F | Moderate-complicated (Yes) | ≤ | <0.5 S | <0.5 S | 46.5 | 37.7 | 52.5 |
| M2 | 38/M | Mild-Uncomplicated (Yes) | ≤ | <0.5 S | 1 S | 41.9 | 42.2 | 33.0 |
We tested the isolates, on the AST-chip, at their intermediate range of susceptibility of concentrations obtained from the Vitek-2 assay. The chip, consisting of 4 μg/ml of clindamycin (CLI), 4 μg/ml vancomycin (VAN), and 8 μg/ml of Doxycycline (DOX) with modified-media and alginate hydrogel was prepared, and the clinical isolates were tested. Table 1 shows the inhibition of bacterial growth in the presence of antibiotics using the biofilm chip, and it can be seen that the results are consistent with the Vitek-2 assay. The findings on the AST-Chip correlated well with the interpretive criteria of susceptible (S) and resistant (R), as determined by conventional AST from the Vitek 2 system. For instance, using Vitek 2, the isolate, A32, was classified as resistant to CLIN (MIC ≥ 4 μg/ml) and susceptible to VAN (MIC <0.5 μg/ml). The AST-Chip similarly demonstrated low percent inhibition (13%) to CLIN and high percent inhibition (50%) to VAN. In addition, the AST-Chip sensitively detected significant variation among the various clinical isolates, thus providing insights into the relative effectiveness of the drugs, which may not be available through conventional techniques. These results demonstrate concordance between conventional AST methods and the novel AST-Chip although future research will be needed to determine cut-off (‘breakpoints’) for this device. For instance, if we use an operational cut-off of <20% inhibition as ‘resistant’ and >20% as ‘susceptible’ strains for the AST-chip, as IC80 is often used to interpret near complete inhibition, we observe that the A32 strain is can be classified as resistant to CLI [16].
In summary, we have successfully developed a rapid, robust, high density AST platform for CA-MRSA. We expect that the reduction in volume and an increase in throughput can be advantageous from a practical standpoint in clinical microbiology labs because: (i) each AST-Chip consisting of 1200 assays require a total of only a few microliters, and hence the assay can be directly performed on the clinical samples without a need for the initial expansion of culture density with a potential savings of 24 h; (ii) a 1000-fold scale down in volume may conceivably decrease the time required to obtain susceptibility data to less than 12 h [17]. In its current format, a single AST-Chip replaces up to twelve 96-well plates and several agar plates, and has sensitivity comparable to the current standard assays used in the microbiology labs but requiring much smaller sample volume and potentially shorter assay duration. The technology is flexible enough to be adapted to other bacterial and fungal organisms, integration into other novel assay platforms such as paper-based microfluidics, and along with other recent advances, will herald a new era in phenotypic susceptibility testing [18].
Conflict of interest
The authors have no conflict of interest to declare.
Acknowledgements
This work was funded in part by grants from theAmerican Heart Association(13PRE17110093 to AS), the NIH (R01DE023510 to JLL-R; UL1TR001120 from NCATS to AKR and CRF), UTSA (TTM19-7296-01 to AS and AKR), and faculty start-up funds from SJSU. The content is solely the responsibility of the authors, and does not necessarily represent the official views of the funding agencies.
References
- 1.Jorgensen J.H., Ferraro M.J. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 1985;49(December 1 (11)) doi: 10.1086/647952. http://www.ncbi.nlm.nih.gov/pubmed/19857164 ([Internet]. Available from:) [DOI] [PubMed] [Google Scholar]
- 2.Qaseem A., Alguire P., Dallas P., Feinberg L.E., Fitzgerald F.T., Horwitch C. Appropriate use of screening and diagnostic tests to foster high-value, cost-conscious care. Ann. Intern. Med. [Internet]. 2012;156(Jan 17(2)):147–149. doi: 10.7326/0003-4819-156-2-201201170-00011. http://www.ncbi.nlm.nih.gov/pubmed/22250146 ([Internet]. Available from:) [DOI] [PubMed] [Google Scholar]
- 3.Wanger A., Mills K., Nelson P.W., Rex J.H. Comparison of etest and national committee for clinical laboratory standards broth macrodilution method for antifungal susceptibility testing: enhanced ability to detect amphotericin B-resistant Candida isolates. Antimicrob. Agents Chemother. 1995;(November 39(11)):2520–2522. doi: 10.1128/aac.39.11.2520. http://www.ncbi.nlm.nih.gov/pubmed/8585737 ([Internet]. Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ligozzi M., Bernini C., Bonora M.G., de Fatima M., Zuliani J., Fontana R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 2002;40(May 1(5)):1681–1686. doi: 10.1128/JCM.40.5.1681-1686.2002. http://jcm.asm.org/cgi/doi/10.1128/JCM.40.5.1681-1686.2002 [Internet](Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter S.S., Ferraro M.J. Manual of Clinical Microbiology. 10th edition. American Society of Microbiology; 2017. Susceptibility testing instrumentation and computerized expert systems for data analysis and interpretation; pp. 1144–1154.http://www.asmscience.org/content/book/10.1128/9781555816728.chap69 ([Internet] Available from) [Google Scholar]
- 6.Nightingale J. Clinical limitations of in vitro testing of microorganism susceptibility. Am. J. Hosp. Pharm. 1987;(January 44 (1)):131–137. http://www.ncbi.nlm.nih.gov/pubmed/3826085 ([Internet] Available from:) [PubMed] [Google Scholar]
- 7.Wolk D.M., Picton E., Johnson D., Davis T., Pancholi P., Ginocchio C.C. Multicenter evaluation of the cepheid xpert methicillin-resistant staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J. Clin. Microbiol. 2009;47(March 1 (3)):758–764. doi: 10.1128/JCM.01714-08. http://jcm.asm.org/cgi/doi/10.1128/JCM.01714-08 ([Internet] Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickert H., Machka K., Braveny I. The uses and limitations of disc diffusion in the antibiotic sensitivity testing of bacteria. Infection. 1981;(January 9 (1)):18–24. ([Internet] Available from: http://link.springer.com/10.1007/BF01640803) [Google Scholar]
- 9.Mak A., Miller M.A., Chong G., Monczak Y. Comparison of PCR and culture for screening of vancomycin-resistant enterococci: highly disparate results for vanA and vanB. J. Clin. Microbiol. 2009;47(December 1 (12)):4136–4137. doi: 10.1128/JCM.01547-09. http://jcm.asm.org/cgi/doi/10.1128/JCM.01547-09 ([Internet] Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.2013. Performance Standards for Antimicrobial Susceptibility Testing Twenty-third Informational Supplement. [Google Scholar]
- 11.Burckhardt I., Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011;(Sep 49 (9)):3321–3324. doi: 10.1128/JCM.00287-11. http://jcm.asm.org/lookup/doi/10.1128/JCM.00287-11 ([Internet] Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan A., Leung K.P., Lopez-Ribot J.L., Ramasubramanian A.K. High-throughput nano-biofilm microarray for antifungal drug discovery. MBio. 2013;4(4) doi: 10.1128/mBio.00331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan A., Torres N.S., Leung K.P., Lopez-Ribot J.L., Ramasubramanian A.K. nBiochip, a lab-on-a-chip platform of mono- and polymicrobial biofilms for high-throughput downstream applications. mSphere. 2017;2(3) doi: 10.1128/mSphere.00247-17. http://www.ncbi.nlm.nih.gov/pubmed/28680970 (Internet) (Available from): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jabra-Rizk M.A., Meiller T.F., James C.E., Shirtliff M.E. Effect of farnesol on staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 2006;50(April 1(4)):1463–1469. doi: 10.1128/AAC.50.4.1463-1469.2006. http://aac.asm.org/cgi/doi/10.1128/AAC.50.4.1463-1469.2006 [Internet] (Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan A., Leung K.P., Lopez-Ribot J.L., Ramasubramanian A.K. High-throughput nano-biofilm microarray for antifungal drug discovery. MBio. 2013;4(June 4(4)) doi: 10.1128/mBio.00331-13. http://mbio.asm.org/cgi/doi/10.1128/mBio.00331-13 [Internet] e00331-13-e00331-13 (Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huntjens D.R.H., Spalding D.J.M., Danhof M., Della Pasqua O.E. Correlation between in vitro and in vivo concentration-effect relationships of naproxen in rats and healthy volunteers. Br. J. Pharmacol. 2006;(June 148 (4)):396–404. doi: 10.1038/sj.bjp.0706737. http://www.ncbi.nlm.nih.gov/pubmed/16682968 [Internet] Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weibull E., Antypas H., Kjäll P., Brauner A., Andersson-Svahn H., Richter-Dahlfors A. Bacterial nanoscale cultures for phenotypic multiplexed antibiotic susceptibility testing. J. Clin. Microbiol. 2014;(September 52 (9)):3310–3317. doi: 10.1128/JCM.01161-14. http://www.ncbi.nlm.nih.gov/pubmed/24989602 [Internet] (Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurer F.P., Christner M., Hentschke M., Rohde H. Advances in rapid identification and susceptibility testing of bacteria in the clinical microbiology laboratory: implications for patient care and antimicrobial stewardship programs. Infect Dis Rep. 2017;9(March 30 (1)) doi: 10.4081/idr.2017.6839. http://www.pagepress.org/journals/index.php/idr/article/view/6839 [Internet] (Available from) [DOI] [PMC free article] [PubMed] [Google Scholar]