Abstract
The long path from initial research on oligonucleotide therapies to approval of antisense products is not unfamiliar. This lag resembles those encountered with monoclonal antibodies, gene therapies, and many biological targets and is consistent with studies of innovation showing that technology maturation is a critical determinant of product success. We previously described an analytical model for the maturation of biomedical research, demonstrating that the efficiency of targeted and biological development is connected to metrics of technology growth. The present work applies this model to characterize the advance of oligonucleotide therapeutics. We show that recent oligonucleotide product approvals incorporate technologies and targets that are past the established point of technology growth, as do most of the oligonucleotide products currently in phase 3. Less mature oligonucleotide technologies, such as miRNAs and some novel gene targets, have not passed the established point and have not yielded products. This analysis shows that oligonucleotide product development has followed largely predictable patterns of innovation. While technology maturation alone does not ensure success, these data show that many oligonucleotide technologies are sufficiently mature to be considered part of the arsenal for therapeutic development. These results demonstrate the importance of technology assessment in strategic management of biomedical technologies.
Keywords: oligonucleotide therapeutics, antisense, ribozyme, small interfering RNA, RNAi, miRNA, FDA approval, technology forecasting, data analytics, technology management
Introduction
With the recent approval of the antisense therapeutic nusinersen for the treatment of spinal muscular atrophy, jointly developed by Biogen and Ionis Pharmaceuticals, oligonucleotide technologies may have finally yielded a clinically and commercially successful biopharmaceutical product. This long-anticipated success was the subject of a series of recent review articles that have chronicled the difficult, 30-year path that led to this important milestone.1, 2, 3, 4
The long path from salient scientific discoveries to successful products should not be unfamiliar to those who follow biotechnology. As previous reviews have noted, the >30-year interval between the emergence of oligonucleotide technologies in the 1980s and the present successes is similar to the decades-long lag between the first description of hybridoma technologies by Kohler and Milstein in the early 1970s and approval of Rituxan, the first successful commercial antibody product, in the late 1990s. Similar lags have been seen between discoveries of molecular targets and the first approval of products associated with those targets. For example, there was a >20-year lag between the first description of the tyrosine kinase activity of retroviral oncogenes in the 1970s and approval of the first tyrosine kinase inhibitor, and there was a similar lag between the discovery of tumor necrosis factor (TNF) in 1975 and approval of the first TNF inhibitors. These are not isolated examples. A recent analysis of 113 first-in-class products by Eder et al. showed that there has been, on average, a 22-year lag between publication of research describing a novel drug target, therapeutic concept, or chemotype and first approval of a therapeutic product associated with this research.5 These data are consistent with the average 17-year lag noted in a 2011 literature review from RAND Europe6 and the 18-year lag between basic research funding and new drug approvals incorporated in an economic model of drug discovery and development by the Congressional Budget Office.7
The recent series of expert reviews chronicled a long series of insights, false starts, successes, and failures that mark the path from the initial discoveries of nucleotide therapeutics to the approval of nusinersen.1, 2, 3, 4, 8 Analogous expert reviews have been written for other technologies that experienced decades-long lags between an enabling scientific insight or invention and approval of a first-in-class therapeutic based on that advance. For example, the ability to make monoclonal antibodies, first described by Kohler and Milstein, only generated therapeutic products when their methods for producing murine antibodies was supplanted by methods for producing chimeric, humanized, and, finally, fully human monoclonal antibodies,9 while the discovery of TNF only led to anti-TNF therapeutics after the failure of TNF as a cancer therapy and recognition of the role played by TNF in arthritis and other inflammatory disorders.10 While this type of analysis by experts in a field can faithfully relate the complex, and often convoluted, path of translational science, such post hoc reflections have not produced a generalized explanation for the characteristic innovation lag between scientific insights and inventions and first approval of successful therapeutic products based on this science.6 A 2011 RAND report highlighted the importance of such an understanding in concluding: “Despite their policy salience, little is known about time lags and how they should be managed. This lack of knowledge puts those responsible for enabling translational research at a disadvantage.6”
Explanations for this characteristic lag have come from research on the process of innovation itself, including the roles of organizational behavior, strategy, social networks, and technology management and the dynamics of scientific, intellectual, human, and economic capital in the innovation ecosystem. One aspect of this research has focused on the temporal relationship between scientific and technological progress and successful product development, demonstrating that technological maturation or “readiness” is a critical determinant in the ability to develop successful products. A 1999 General Accounting Office report on the management of technologies noted, for example: “.…no element is more important than having technology, advanced enough to meet requirements but also mature enough to be predictably managed, available at the start of the product development cycle. Maturing new technology before it is included on a product is perhaps the most important determinant of the success of the eventual product.11” As a result, strategic technology management often uses tools such as technology roadmapping12, technological forecasting,13, 14 or technology readiness assessment15, 16, 17 to assess the maturation of critical path technologies and their ability to satisfy the performance specifications for innovative products over time.
While most of this research involves computers and communication technologies, mechanical engineering, aerospace engineering, or defense systems, we have applied these theories18, 19, 20 to develop analogous models for biopharmaceutical development. Specifically, we have asked whether the characteristic lag between biomedical discoveries and successful biopharmaceutical development is analogous to the lag related to technological maturation or readiness observed in other technology sectors.21, 22, 23, 24, 25 Studies have shown that many different technologies mature through a characteristic “S curve,19, 26, 27” in which an initial insight or invention initiates a period of exponential, technological advance, which slows as the technology approaches its limits. Our initial studies suggested that there were qualitative parallels between patterns of innovation observed in other technology sectors and the accumulation of publications for monoclonal antibodies, nucleotide therapeutics, and gene therapies.21 We then described a bibliometric-based analytical model, the Technology Innovation Maturation Evaluation model (“TIME model”), which allows for a quantitative assessment of research maturation based on publication activity over time. Briefly, the model posits that peer-reviewed research papers embody a quantum of new knowledge related to a research area; some represent positive contributions, while others may represent insignificant, or even negative, contributions. Integrated over large numbers of published papers, the number of publications may be considered a metric for the advance of that technology. We have applied this model to more than 200 discrete drug targets along with technologies for monoclonal antibodies and gene therapies and have shown that the accumulation of publications for the large majority of technologies examined can be modeled as an exponentiated logistic function (“S curve”).22, 23, 24, 28 This curve is characterized by a point of initiation (“Ti”) representing the point of maximum acceleration into a period of exponential growth, which slows as the technology passes an established point (“Te”), defined as the point of maximum slowing, and approaches a limit (Figure 1). The analytically defined point of initiation and the established point provide objective, quantitative metrics for asking whether successful development of biopharmaceutical products is linked to maturation of associated technologies, as observed in other technology sectors.
Figure 1.
Schematic of TIME Model for Technology Growth
Technology growth is modeled as an S curve using an exponentiated logistic function (solid green line) fit to cumulative publications (N) in a PubMed search. The technology initiation point (Ti) is calculated as the point of maximum acceleration of publication activity (maximum d2N/dx2), representing the beginning of a phase of exponential growth. The established point (Te) is calculated as the point of maximum slowing of publication activity (minimum d2N/dx2), representing the end of the phase of exponential growth and slowing as the technology approaches its limits.
In studies of >400 new molecular entities (NMEs) using the TIME model, we have shown that few products discovered using targeted screening or biological products are approved before the associated technologies pass the established point,23, 24, 25 and that interval between the initiation point of a new research area and first approval of a drug based on this research is 36 years.24, 25 In contrast, there was no association between metrics of technology growth and approval of product discovered through phenotypic methods.24, 25 These observations are consistent with the expectation that targeted discovery is based on accumulated knowledge of potential drug targets and their relationship to disease processes, whereas phenotypic methods are not based on such knowledge.29
These studies suggest that the characteristic lag between the initiation of new areas of research and first approval of biopharmaceutical products based on this research is an intrinsic property of targeted and biologic strategies for drug discovery and development. In this report, we extend these studies to oligonucleotide therapeutics, asking whether quantitatively similar patterns are evident in this field. The results suggest that innovation of oligonucleotide therapeutics has followed largely predictable patterns, with successful products emerging only after research related to the specific oligonucleotide technology, component technologies, and the molecular targets past the established point. This analysis explores how the theoretical understanding of biomedical innovation can be used to inform more efficient product development and effective business strategy in the future.
Results and Discussion
Mechanism-Based Technologies for Oligonucleotide Therapeutics
There have been many outstanding reviews of oligonucleotide technologies, which address the historical development of the field, essential technologies, and the current state of clinical development.2, 30 We will not attempt to add to this literature.
Briefly, oligonucleotide therapeutics are products composed of single- or double-stranded nucleotides and have many different mechanisms of action. Antisense oligonucleotides are single-stranded nucleotides designed to bind complementary mRNA sequences and alter splicing (splice-switching oligonucleotides), suppress translation, or activate RNase H cleavage of the mRNA. Ribozymes have a complex nucleotide structure that cleaves mRNA through RNA-mediated catalysis.31 RNAi includes both small interfering RNAs (siRNAs) and microRNAs (miRNAs) and involves the use of double-stranded nucleotides, which are incorporated into an RNA-induced silencing complex (RISC) that guides the nucleotides to complementary mRNA sequences and alters post-transcriptional gene expression. siRNAs are ∼21–23 nt long and degrade mRNA through precise complementary binding and subsequent cleavage of the mRNA. miRNAs are larger oligonucleotides that form stem-loop structures with double-stranded segments, which are processed into smaller segments that imperfectly bind to a cRNA sequence and cause translational repression rather than cleavage of the mRNA.32 Aptamers are single-stranded oligonucleotides designed to target proteins rather than nucleic acids.33 CRISPR is also a nucleotide therapeutic, but it does not target post-transcriptional gene expression; instead, it uses nucleotide sequences as a guide for targeted gene editing. We have included it in our analysis because it provides a complementary example of an early-stage, emerging technology. For the purpose of this analysis, each of these approaches represent discrete, ordinal technologies that arose from a discrete scientific insight or invention with an identifiable, though sometimes overlapping, literature that defines the design parameters, performance, optimization, and potential applications of that mechanism.
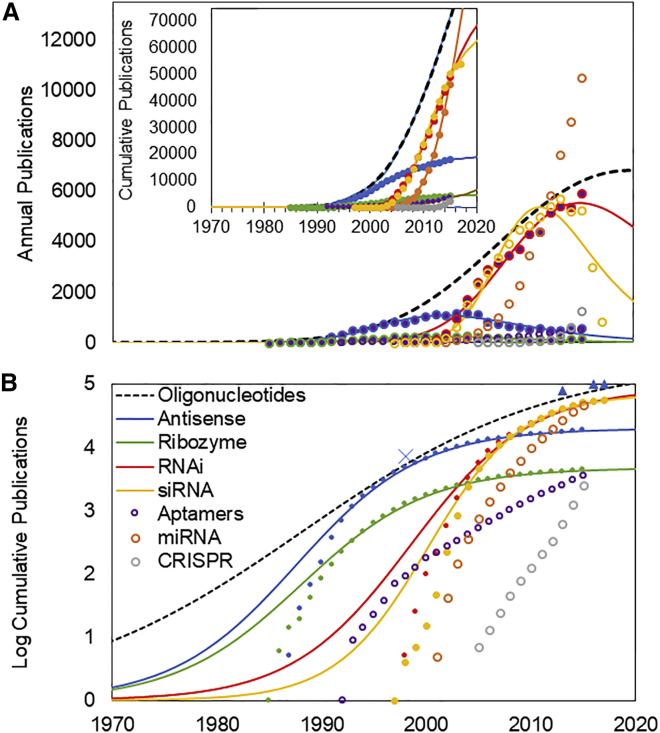
We used the TIME model to characterize the growth of each technology (Figure 2). The annual and cumulative numbers of publications are shown in Figure 2A. The exponentiated logistic function assumes a logistic, S-curve form on log scales, as shown in Figure 2B.
Figure 2.
TIME Models for Growth of Oligonucleotide Technologies
(A) Annual publications in PubMed related to oligonucleotide therapeutics (markers) and best fit TIME model (lines). The TIME model uses the best fit exponentiated logistic function based on the cumulative number of publications (inset graph). Data indicated with open markers could not be fit to this function. (B) The logistic (“S curve”) representation of the TIME model is shown on a log scale. Note that the apparent residuals are exaggerated at the low end of the log scale (see Materials and Methods). Dates of FDA approval of oligonucleotide therapeutics are indicated, including those for fomivirsen (1998, subsequently withdrawn from market), mipomersen (2013), nusinersen (2016), and eteplirsen (2016).
For each curve, we assessed the validity of the initiation point by examining the correspondence between the calculated initiation point, which introduces the phase of exponential growth, and seminal advances noted in expert reviews. Specifically, we found that many reviews of antisense technologies cite the 1978 work of Zamecnik et al., on the synthesis of a 13-mer antisense oligonucleotide34 as a seminal contribution. Similarly, seminal events in the initiation of ribozyme technologies include the Nobel-prize-winning work by Cech, published in 1982,35 and of Altman, published in 1983.36 Each of these dates are within close proximity of the calculated Ti of 1980 for these technologies. The discovery of RNAi originated in the mid-1990s with experimental work in petunias37 and C. elegans.38 While these dates are somewhat later than the calculated Ti of 1991, this is consistent with the expectation that prior work on oligonucleotide therapeutics made a contribution to the rate of growth of RNAi.
Figure 2B shows that three technologies—antisense, ribozymes, and RNAi—exhibit an “S-curve pattern” of growth. Both antisense and ribozymes passed the point of maximum slowing, the established point, in the mid-1990s; RNAi technology reached this point a decade later. Two technologies shown in Figure 2, microRNA and CRISPR, do not exhibit an exponential pattern of growth and could not be modeled with the TIME model. These more recent technologies exhibit an exponential, or near-exponential, growth pattern characteristic of technologies that have not yet reached the established point.
Component Technologies for Oligonucleotide Therapeutics
A feature of technology growth cycles is the premise that technological progress occurs as limits of precursor or invention-stage technologies are reached and new ordinal technologies emerge.39 Oligonucleotide therapeutics emerged from basic research on mRNA function and processing as well as the observation that exogenously administered nucleotides could interfere with cognate gene expression or translation in various in vitro and in vivo systems. McNamee and Ledley observed that the rapid growth of this field was enabled by the emergence of several enabling technologies, including automated oligonucleotide synthesis, transfection, and delivery. This sequential emergence of ordinal technologies can be approximated by a logistic S-curve; in this case, the TIME model. The growth cycles of oligonucleotide therapeutics are generally understood to be influenced by several challenges, including chemical synthesis, stability, and therapeutic delivery.2, 3, 30, 31, 40 Additionally, the understanding of the effect of synthetic oligonucleotides on human biology, such as its interaction with the innate immune system, has also been critical to the development of drugs that were safe for administration and also opened novel mechanisms of action for a new class of immune-active drugs.41
Much of the focus on research and development of oligonucleotide therapeutics has focused on novel chemistries that improve the pharmacodynamic properties of the oligonucleotide, efficient manufacturing and control, chemical stability, and delivery.2, 3, 30, 31, 40 These include phosphorothioate chemistry, locked and bridged nucleic acids, phosphorodiamidate morpholino chemistry, 2′-OME chemistry, and phosphotriester oligonucleotide chemistry. Many delivery technologies have been studied that were not addressed in this study.
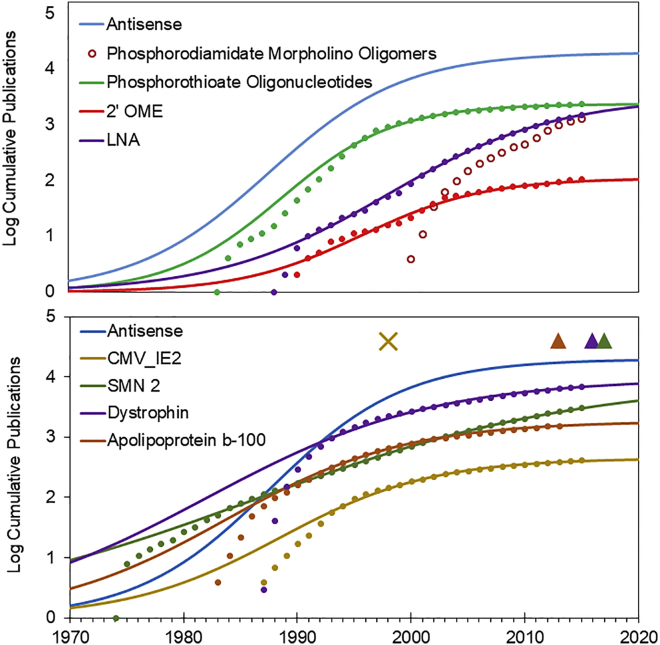
TIME analysis was done to examine the technology growth of several ordinal technologies that have been utilized in the current approved therapeutics, which include phosphorothioate chemistry, phosphorodiamidate morpholino chemistry, 2′-O-methoxyethyl (2′-MOE) chemical modifications, and locked nucleic acid (LNA) (Figure 3A). While the applicability of TIME models to growth of chemistry technologies has not been previously investigated, these few test cases do show the S-curve pattern. Many of these technology growth cycles were concurrent or occurred after antisense oligonucleotide, ribozyme, and RNAi growth cycles. As broader understanding of oligonucleotide therapeutic technology matured, the underlying chemical technologies also matured and represent a necessary step toward successful drug development for project-specific approaches.
Figure 3.
TIME Models for Growth of Component Technologies and Biological Targets
Top: cumulative publications (markers) and best fit exponentiated logistic functions (lines) are shown for chemistries incorporated in approved oligonucleotide therapeutics. Open markers indicate datasets that could not be fit with the TIME model. Bottom: cumulative publications (markers) and best fit exponentiated logistic functions (lines) are shown for the biological targets of approved oligonucleotides. Dates of FDA approval of oligonucleotide therapeutics are indicated in color corresponding to their biological target.
Molecular Targets for Oligonucleotide Therapeutics
In addition to the growth cycles of ordinal chemical and delivery technologies, oligonucleotide therapeutic development is also affected by project-specific biological targets. From Pharmaprojects searches, the biological targets for therapeutic candidates that have reached phase 3 or approval were identified, resulting in 19 unique targets. TIME model analysis shows that the biological targets that are reaching phase 3 are relatively mature (Table S1). The TIME models for biological targets of approved antisense oligonucleotide therapeutics show that matured biological targets are correlated with the successful launch of a product. Interestingly, the launch of fomivirsen was ahead of the established point of its biological target (CMV-IE2) and antisense oligonucleotides and was subsequently withdrawn from the market. The more recent approvals more closely align with the established points of their underlying technology growth cycles. (Figure 3B).
Approval and Development of Oligonucleotides in Context
Research on innovation in many different technology sectors has shown that there are characteristic patterns to the process by which scientific and technological advances are translated into novel products. One such pattern is the recurring relationship between the maturation of technologies and the ability to develop successful products based on those technologies. The present data suggest that similar patterns may be evident in the emergence of oligonucleotide therapeutics. There are several dimensions to this association.
The first dimension concerns the series of distinct oligonucleotide technologies. While there are many commonalities to antisense, ribozymes, RNAi, and miRNA, the ordinal succession of these technologies does not reflect sequential improvement on previous technologies the way that chimeric, humanized, and fully human monoclonal antibodies each improved on previous technologies.21 In this context, the emergence of nucleotide therapies more closely resembles the growth of gene therapy, which has involved a series of distinct technologies, including retrovirus-, adenovirus-, adeno-associated-virus-, and lentivirus-based vectors.22 While there is clearly more overlap between the ordinal oligonucleotide technologies than those for gene therapy, we have modeled the growth of each oligonucleotide technology separately in assessing the relationship between technology maturation and product approvals.
This analysis suggests that the emergence of antisense therapeutics exhibits the same pattern seen previously for monoclonal antibodies and drug targets. The first oligonucleotide therapeutic to be approved, Vitravene (fomivirsen) received Food and Drug Administration (FDA) approval in 1998, before antisense technologies reached the established point, and was subsequently withdrawn from the market in 2006. This experience parallels the launch of the first monoclonal antibody product, Orthoclone OKT3 (muromonab-CD3), a murine monoclonal antibody that was approved before monoclonal antibody technologies reached the established point and that was subsequently withdrawn from the market. Both experiences are consistent with observations in other technology sectors that immature technologies may be able to produce products; however, those products are less likely to be able to compete with products developed from mature technologies. In contrast, the recent approvals of mipomersen in 2013 and of nusinersen and eteplirsen in 2016 came well after the established point of antisense technologies.
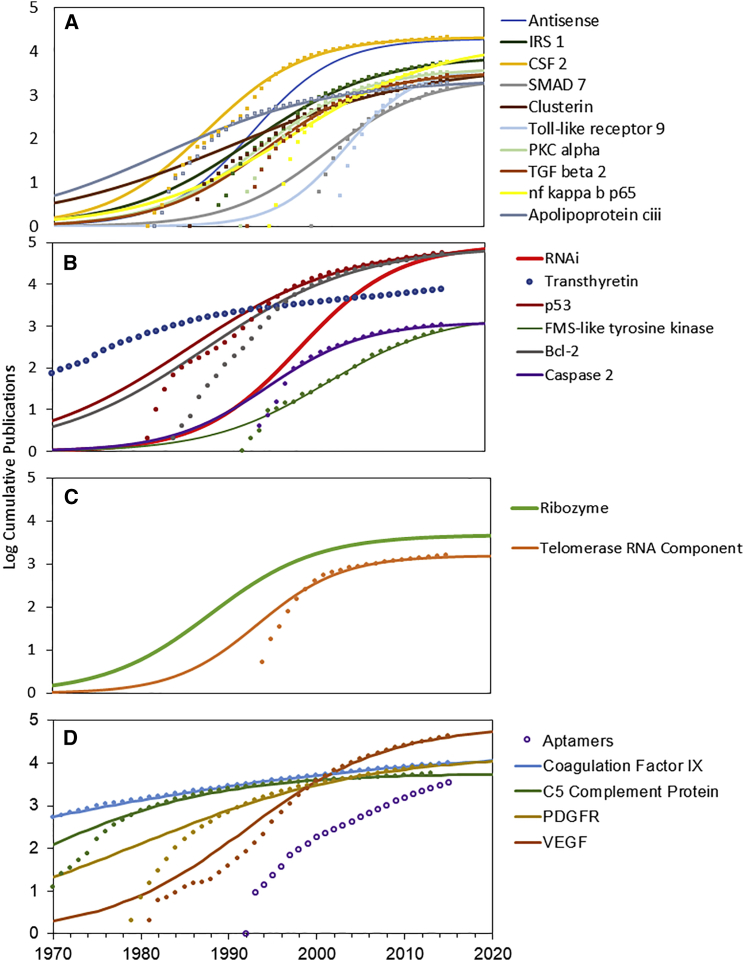
The second dimension concerns the drug target. A series of papers examining patterns of innovation for more than 400 NMEs has demonstrated a consistent relationship between metrics describing the growth of research on drug targets and first approval of targeted and biological NMEs.23, 24, 25 This analysis of RNA therapeutics shows that each of the approved oligonucleotides involves targets that had progressed past the established point when the products were developed (Figure 3). Furthermore, our data suggest that, independent of the maturation of oligonucleotide technologies, there are few, if any, targeted therapeutics approved until research on the target has matured. Finally, TIME model evaluation of biological targets in phase 3 development for antisense, RNAi, and ribozyme therapeutics, as determined from Pharmaprojects, shows few to no immature technologies making it to phase 3 (Figure 4).
Figure 4.
TIME Models for Growth of Technologies for Oligonucleotide Therapeutics in Phase 3 Clinical Trials and Their Biological Targets
Best fit exponentiated logistic functions are shown (lines). Open markers indicate datasets that could not be fit with this function. (A) Antisense products in phase 3. (B) RNAi products in phase 3. (C) Ribozyme products in phase 3. (D) Aptamer products in phase 3.
The third dimension involves the nucleotide chemistries that have proved essential to producing oligonucleotide therapeutics with acceptable pharmaceutical and pharmacodynamic properties. We have not previously applied our bibliometric-based TIME model to chemical technologies, nor do we think that bibliometric-based metrics are the best measure of such technologies, since chemical properties of oligonucleotides can be readily measured against parameters required by the product design. Nevertheless, Figure 3B shows that a series of chemical technologies that are incorporated in recently approved products are no longer in an exponential growth phase and have passed the point that would be considered established.
Taken together, this analysis suggests that the long-awaited success of oligonucleotide therapeutics exhibits a pattern that is quantitatively similar to the characteristic patterns observed in other technology sectors as well as previous studies of biopharmaceutical development using the TIME model.23, 24, 25 These results suggest that the lag in developing such products reflects, at least in part, an intrinsic property of technological innovation. In this context, it should be noted that the established point identified by the TIME model is the point of maximum slowing of publication activity, most of which reflects basic research, as opposed to research on specific processes or products. This slowing may reflect a point in the advance of this basic science when research achieves a more nuanced and detailed understanding of the system but produces fewer unpredictable results, leading to large numbers of publication, and may diminish grant support. If so, this point would also represent a stage when the science is sufficiently advanced to provide meaningful validation of molecular targets, selection of lead compounds, and achievable clinical indications, which are essential determinants of product success.
Technological maturation is not sufficient for successful product development. There are many reasons that products fail in clinical trials; not only inadequate efficacy relative to placebo or existing therapies, but also unexpected, sometimes idiosyncratic, side effects as well as “commercial failures” when sponsors choose to discontinue development for strategic reasons or concern about economic viability of the product.42 It may also be true that certain technologies, even when mature, prove not to have utility for drug discovery and development. Mipomersen, for example, has achieved limited sales despite FDA approval because small molecules continue to have significant advantage over oligonucleotides in terms of oral administration and cost. Certain molecular targets are traditionally considered “undruggable,” despite intensive research and development efforts,43 and technologies for murine monoclonal antibodies as well as gene therapy using murine retroviral vectors have been abandoned in favor of superior technologies. Ribozymes may prove to be an example of a technology that, even when fully mature, will not generate products that are competitive with other oligonucleotide technologies, biological products, or small molecules.
Implications for Strategic Management of Future Development
In other technology sectors, management tools such as technology roadmapping, technology forecasting, and technology readiness assessment enable managers and investors to consider the state of maturation of enabling technologies in product design and development. The previously referenced report from the Government Accountability Office (GAO) summarized the importance of these tools: “…resolving technology problems before product development begins results in 10 times the savings compared to correcting problems afterward. In this sense, technology maturity breeds product success.11”
The present results suggest that oligonucleotide technologies have now matured to a level equivalent to that of monoclonal antibodies when those technologies first achieved commercial success. Moreover, our analysis of oligonucleotide therapeutics currently in phase 3 clinical trials shows a robust development pipeline comprising products that incorporate oligonucleotide technologies and address targets that have passed the established point of technology growth. We did note that the targets for RNAi products in development are somewhat less mature than those being targeted by antisense technologies (Figure 4). The present results also suggest that newer oligonucleotide technologies, including miRNA, are significantly less mature than other technologies that have generated successful biopharmaceutical products (Figure 2B).
Our conclusion is not that innovators or investors can be sanguine about success of oligonucleotide technologies in the future; only that the maturation of critical path technologies should be a factor in the design of candidate therapeutics and in assessing the opportunity and risk of biopharmaceutical technologies, as it is in other technology sectors. The present results suggest that there are recurring patterns to the maturation of basic research on various oligonucleotide technologies and potential targets for these products. As such, these results are similar to those of others who have characterized the progression of research on RNAi from an initiating MeSH domain to the subsequent expansion of research to other research domains.44 We suggest that these observations might be extended with the application of machine learning and artificial intelligence to develop expert systems that are useful in assessing the maturation of technologies as part of the decision to proceed with development of products based on these technologies.
These results also point to the need for caution in designing initiatives aimed at accelerating translational science. These initiatives should accelerate the maturation of science to the point that it can sustain successful product development, and they do not prematurely advance candidate products into development that are associated with immature technologies. As noted in our analysis of gene therapy, the failure of investments and clinical investigations of immature technologies can create unwarranted pessimism concerning these technologies and stall successful development.22
Materials and Methods
Data Sources
Pharmaprojects was used to identify oligonucleotide therapeutics that are in development, have been approved, or have been recently discontinued that were designed to work by antisense, RNAi, or ribozyme. For each candidate product, data on the therapeutic target, stage of development, and the licensee and originator companies (current to May 2017) were retrieved. Component technologies that are incorporated into the design of these oligonucleotide therapeutics were ascertained from literature review. The list of products and associated technologies is shown in Table S1.
TIME Analysis
We identified publications in PubMed associated with oligonucleotide technologies and the mechanism of action of these therapeutics, as well as the biological targets and component technologies associated with approved products or candidate products in phase 3 trials. Boolean search terms and NCBI Query Translations were optimized to minimize incomplete ascertainment of relevant papers due to immature vocabularies as well as identification of research on unrelated topics for each technology. These search terms are shown in Table S1.
The TIME model fits an exponentiated logistic function to the cumulative number of publications over time, from the first year of continuous, annual publication through 2015, using methods described previously21, 24, 28 (Figure 1). The exponentiated logistic function has the form:
or
where N represents the number of publications, L represents the calculated upper limit of publications, r represents the growth rate, t represents time, and t0 represents the midpoint of exponential growth. The parameters were fit to time series publication data using a non-linear least-squares implementation of the Levenberg-Marquardt algorithm (http://lmfit.github.io/lmfit-py/). This asymmetric sigmoidal function, exhibits the common logistic sigmoid function over log scales. Calculations were performed in Python using scripts that are freely available at http://lmfit.github.io/lmfit-py/. Publication records for some technologies could not be fit to the exponentiated logistic function and no further analyses of these technologies was possible. These technologies are indicated by open circles of raw data points in the figures.
The initiation (Ti) and established (Te) points approximate the boundaries of a period of exponential growth. The point of initiation (Ti) is calculated as the point of maximum acceleration of publication accumulation. The established point (Te) is calculated as the point of maximum slowing of publication accumulation. These points have the form logN’’(t)max,min and are determined analytically by
SEs from the Levenberg-Marquardt analysis were used to educate a Monte-Carlo simulation of the fitted parameters. The average SE of this simulation was used as the metric for goodness of fit. For each technology, the analytically defined initiation point calculated from the best-fit TIME model was qualitatively validated by comparing this date with seminal scientific milestones identified in expert reviews. The error range for Ti and Te is estimated from the mean of the residuals.
Author Contributions
Conceptualization, J.M.B., L.M.N., and F.D.L.; Methodology, J.M.B., L.M.N., and F.D.L.; Verification, J.M.B., L.M.N., and F.D.L.; Formal Analysis, J.M.B., L.M.N., and F.D.L.; Investigation, J.M.B., L.M.N., and F.D.L.; Data Curation, J.M.B., L.M.N., and F.D.L.; Writing – Original Draft, J.M.B., L.M.N., and F.D.L.; Writing – Review & Editing, J.M.B., L.M.N., and F.D.L.; Visualization, J.M.B., L.M.N., and F.D.L.; Supervision, F.D.L.; Project Administration, F.D.L.; Funding Acquisition, F.D.L.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors acknowledge the analytical advice and assistance from Drs. Michael Walsh and Ekaterina Cleary and the generous insights provided by Dr. Thomas Cech, as well as the Executives in Residence at the Center for Integration of Science and Industry, Drs. Michael Boss and Nancy Hsiung. This work was supported by a grant from the National Biomedical Research Foundation.
Footnotes
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.10.017.
Supplemental Information
References
- 1.Editorial The commercial tipping point. Nat. Biotech. 2017;35:181. doi: 10.1038/nbt.3829. [DOI] [PubMed] [Google Scholar]
- 2.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 4.Garber K. Worth the RISC? Nat. Biotechnol. 2017;35:198–202. doi: 10.1038/nbt.3810. [DOI] [PubMed] [Google Scholar]
- 5.Eder J., Sedrani R., Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 2014;13:577–587. doi: 10.1038/nrd4336. [DOI] [PubMed] [Google Scholar]
- 6.Morris Z.S., Wooding S., Grant J. The answer is 17 years, what is the question: understanding time lags in translational research. J. R. Soc. Med. 2011;104:510–520. doi: 10.1258/jrsm.2011.110180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austin D.H. U.S. Congress and U.S. Congressional Budget Office; 2006. Research and Development in the Pharmaceutical Industry. [Google Scholar]
- 8.Singh N.N., Howell M.D., Androphy E.J., Singh R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017;24:520–526. doi: 10.1038/gt.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichert J.M. Monoclonal antibodies in the clinic. Nat. Biotechnol. 2001;19:819–822. doi: 10.1038/nbt0901-819. [DOI] [PubMed] [Google Scholar]
- 10.Palladino M.A., Bahjat F.R., Theodorakis E.A., Moldawer L.L. Anti-TNF-alpha therapies: the next generation. Nat. Rev. Drug Discov. 2003;2:736–746. doi: 10.1038/nrd1175. [DOI] [PubMed] [Google Scholar]
- 11.GAO (1999). Best practices: better management of technology development can improve weapon system outcomes. Report to the Chairman and Ranking Minority Member, Subcommittee on Readiness and Management Support, Committee on Armed Services, U.S. Senate. July 1999, GAO/NSIAD-99-162. http://www.gao.gov/assets/160/156673.pdf.
- 12.Carvalho M., Fleury A., Lopes A.P. An overview of the literature on technology roadmapping (TRM): Contributions and trends. Technol. Forecast. Soc. Change. 2013;80:1418–1437. [Google Scholar]
- 13.Meredith J.R., Mantel S.J., Jr. Appendix C: technological forecasting. In: Meredith J.R., Mantel S.J. Jr., Shafer S.M., editors. Project Management: A Managerial Approach. 9th ed. Wiley; 2014. http://www.wiley.com/college/meredith [Google Scholar]
- 14.National Research Council . National Academies Press; 2010. Persistent Forecasting of Disruptive Technologies—Report 2. [Google Scholar]
- 15.Oliveira M.G., Rozenfeld H. Integrating technology roadmapping and portfolio management at the front-end of new product development. Technol. Forecast. Soc. Change. 2010;77:1339–1354. [Google Scholar]
- 16.Ding M., Dong S., Eliashberg J., Gopalakrishnan A. Portfolio management in new drug development. In: Ding M., Eliashberg J., Stremersch S., editors. Innovation and Marketing in the Pharmaceutical Industry. Springer; 2014. pp. 83–118. [Google Scholar]
- 17.Cowlrick I., Hedner T., Wolf R., Olausson M., Klofsten M. Decision-making in the pharmaceutical industry: analysis of entrepreneurial risk and attitude using uncertain information. R&D Manag. 2011;41:321–336. [Google Scholar]
- 18.Christensen C.M., Raynor M.E. Harvard Business School Press; 2003. The Innovator’s Solution: Creating and Sustaining Successful Growth. [Google Scholar]
- 19.Christensen C.M. Harvard Business School Press; 2013. The Innovator’s Dilemma: When New Technologies Cause Great Firms to Fail. [Google Scholar]
- 20.Foster R.N. Summit Books; 1986. Innovation: The Attacker’s Advantage. [Google Scholar]
- 21.McNamee L.M., Ledley F.D. Patterns of technological innovation in biotech. Nat. Biotechnol. 2012;30:937–943. doi: 10.1038/nbt.2389. [DOI] [PubMed] [Google Scholar]
- 22.Ledley F.D., McNamee L.M., Uzdil V., Morgan I.W. Why commercialization of gene therapy stalled; examining the life cycles of gene therapy technologies. Gene Ther. 2014;21:188–194. doi: 10.1038/gt.2013.72. [DOI] [PubMed] [Google Scholar]
- 23.McNamee L.M., Ledley F.D. Modeling timelines for translational science in cancer; the impact of technological maturation. PLoS ONE. 2017;12:e0174538. doi: 10.1371/journal.pone.0174538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNamee L.M., Walsh M.J., Ledley F.D. Timelines of translational science: From technology initiation to FDA approval. PLoS ONE. 2017;12:e0177371. doi: 10.1371/journal.pone.0177371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beierlein J.M., McNamee L.M., Walsh M.J., Kaitin K.I., DiMasi J.A., Ledley F.D. Landscape of innovation for cardiovascular pharmaceuticals: from basic science to new molecular entities. Clin. Ther. 2017;39:1409–1425.e20. doi: 10.1016/j.clinthera.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Christensen C.M. Exploring the limits of the technology S-curve. Part II: architectural technologies. Prod. Oper. Manag. 1992;1:358–366. [Google Scholar]
- 27.Christensen C.M. Exploring the limits of the technology S-curve. Part I: component technologies. Prod. Oper. Manag. 1992;1:334–357. [Google Scholar]
- 28.Beierlein J.M., McNamee L.M., Walsh M.J., Ledley F.D. Patterns of innovation in Alzheimer’s disease drug development: a strategic assessment based on technological maturity. Clin. Ther. 2015;37 doi: 10.1016/j.clinthera.2015.07.003. 1643–51.e3. [DOI] [PubMed] [Google Scholar]
- 29.Swinney D.C. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther. 2013;93:299–301. doi: 10.1038/clpt.2012.236. [DOI] [PubMed] [Google Scholar]
- 30.Lundin K.E., Gissberg O., Smith C.I. Oligonucleotide therapies: the past and the present. Hum. Gene Ther. 2015;26:475–485. doi: 10.1089/hum.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnett J.C., Rossi J.J. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Fougerolles A., Vornlocher H.-P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephenson M.L., Zamecnik P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 36.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen R.A., Cluster P.D., English J., Que Q., Napoli C.A. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 38.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 39.Kurzweil R. Penguin; 2005. The Singularity Is Near: When Humans Transcend Biology. [Google Scholar]
- 40.Prakash T.P. An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem. Biodivers. 2011;8:1616–1641. doi: 10.1002/cbdv.201100081. [DOI] [PubMed] [Google Scholar]
- 41.Robbins M., Judge A., MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 42.Kaitin K.I. Deconstructing the drug development process: the new face of innovation. Clin. Pharmacol. Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makley L.N., Gestwicki J.E. Expanding the number of “druggable” targets: non-enzymes and protein-protein interactions. Chem. Biol. Drug Des. 2013;81:22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leydesdorff L., Rotolo D., de Nooy W. Innovation as a nonlinear process, the scientometric perspective, and the specification of an “innovation opportunities explorer.”. Technol. Anal. Strateg. Manage. 2013;25:641–653. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.