Abstract
Aurora kinase A (AURKA) is essential for regulating mitosis and is frequently amplified in various cancer cell types. However, the effect of AURKA inhibition on the induction of apoptosis remains unclear. In the present study, it was reported that treatment with TCS7010, a specific inhibitor of AURKA, resulted in the accumulation of cells in the sub-G0/G1 phase of the cell cycle and increased the percentage of annexin V-binding cells. The cleavage of caspase-2, caspase-7, and poly(ADP-ribose)polymerase (PARP) significantly increased in a time-dependent manner following TCS7010 treatment. In addition, TCS7010 resulted in the production of reactive oxygen species (ROS) and stimulation of the unfolded protein response (UPR), leading to the upregulation of CCAAT/enhancer-binding protein-homologous protein (CHOP), and its downstream target BCL2 like 11 (BIM). Pretreatment with N-acetylcystein, a ROS scavenger, significantly abrogated TCS7010-induced accumulation of CHOP, BIM, cleaved caspase-7 and cleaved PARP. These results suggest that TCS7010 triggers apoptosis through the ROS-mediated UPR signaling pathway.
Keywords: aurora kinase A, TCS7010, unfolded protein response, reactive oxygen species, apoptosis
Introduction
Aurora kinases (AURKs) are members of the mitotic serine/threonine kinase family, which includes AURK A, B, and C. Among these kinases, AURKA is frequently amplified in various cancer cell types and can act as an oncogene (1). AURKA performs an essential role in centrosome maturation, chromosome alignment and proper formation of the mitotic spindle (2,3). Inhibition of AURKA has been demonstrated to arrest the cell cycle at the G2/M phase and suppress tumor growth in vivo (4–7). In prostate cancer cells, knockdown of AURKA expression by shRNA induces apoptosis through an upregulation of tumor protein p53 and its downstream target apoptosis regulator Bax (Bax) (8). However, the specific mechanism underlying the induction of apoptosis via AURKA inhibition remains unclear.
The endoplasmic reticulum (ER) is an organelle that has numerous cellular functions, including protein folding, post-translational modifications and trafficking. Cellular conditions associated with the accumulation of misfolded or unfolded proteins in the lumen of the ER induce a series of ER stress responses, known as the unfolded protein response (UPR) (9). Thus, this allows for restoration of protein folding homeostasis by inhibiting further protein translation, degrading misfolded proteins and increasing the synthesis of molecular chaperones to augment ER folding capacity (9). The UPR signaling pathway is mediated by ER transmembrane proteins, including double-stranded RNA-activated protein kinase-like ER kinase, inositol-requiring enzyme 1 (IRE1), and activating transcription factor 6 (9). However, if the ER stress is too severe and prolonged for the cell to overcome, the UPR signaling pathway can activate the caspase cascade and thus promote the cell death program through apoptosis (10).
The aim of the present study was to investigate the effects of TCS7010, a specific inhibitor of AURKA, on the induction of UPR-mediated apoptosis. The results demonstrated that TCS7010 produces reactive oxygen species (ROS) and triggers apoptosis through ROS-dependent UPR-mediated pathway.
Materials and methods
Cell culture and chemicals
The human colon carcinoma HCT116 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Cellgro; Corning Incorporated, Corning, NY, USA) at 37°C in a humidified incubator with an atmosphere containing 5% CO2. The cells were seeded onto 60-mm culture dishes at a density of 4×105 cells/dish and were further cultured until the cells reached up to a subconfluent monolayer (70–80% confluency). The cells were treated with the stated concentrations of TSC7010 (Tocris Bioscience, Bristol, UK).
Immunoblot analysis
Cells were harvested and lysed for 30 min at 4°C in a buffer consisting of 20 mM HEPES (pH 7.2), 1% Triton X-100, 10% glycerol, 150 mM NaCl, 10 µg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride. Protein concentrations were measured with a Pierce™ Bicinchoninic Protein Assay Kit (#23225; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The protein extracts (20 µg each) were separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. Following protein transfer, membranes were washed in Tris Buffered Saline with Tween 20 (TBST; Cell Signaling Technology, Inc., Danvers, MA, USA) 3 times for 5 min each at room temperature, followed by blocking in 5% non-fat milk for 30 min at room temperature. The blots were incubated with primary antibodies for 2 h at room temperature. Antibodies against phospho-Aurora A(Thr288)/B(Thr232)/C(Thr198) (#2914; 1:1,000 dilution), Cleaved caspase-7 (#9491; 1:1,000 dilution), IRE1 (#3294; 1:2,000 dilution), CHOP (#2895; 1:1,000 dilution), Bim (#2819; 1:1,000 dilution), Caspase-2 (#611022; 1:1,000 dilution) were purchased from Cell Signaling Technology, Inc. Antibodies against GAPDH (#sc-32233; 1:5,000 dilution), PARP (#sc-7150; 1:4,000 dilution) were obtained from Santa Cruz Biotechnology, Inc., (Dallas, TX, USA) and antibody against Caspase-2 (#611022; 1:1,000 dilution) was from BD Biosciences (San Jose, CA, USA). Following washing 3 times for 5 min each with TBST at room temperature, horseradish peroxidase-conjugated secondary antibody was incubated for 1 h at room temperature. Secondary antibody to anti-mouse IgG (Goat; #BR170-6516; 1:2,000 dilution) and anti-rabbit IgG (Goat; #BR170-6515; 1:2,000 dilution) were purchased from Bio-Rad Laboratories, Inc., (Hercules, CA, USA). Following incubation with secondary antibodies, membranes were washed 5 times with TBST for 5 min each at room temperature and the immunoreactivity of protein bands was visualized using an enhanced chemiluminescence detection system (GE Healthcare Life Sciences, Chalfont, UK). Each blot is representative of ≥3 separate experiments.
Cell viability assay
Cell viability was determined using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Rockville, MD, USA), according to the manufacturer's protocol. Briefly, exponentially growing cells were exposed to 5 or 10 µM TCS7010 for 24 h at 37°C followed by the addition of the CCK-8 solution for an additional 1 h at 37°C. The absorbance was measured at 450 nm using an Emax Endpoint ELISA microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA). Data were presented as the mean ± standard deviation of 2 independent experiments performed in triplicate.
Cell cycle analysis
Cellular DNA content was analyzed using flow cytometry, as previously described (11). Briefly, HCT116 cells were collected following 18 and 24 h of exposure to 5 µM TCS7010, fixed in 70% ethanol, and washed twice with phosphate-buffered saline. Subsequently, cells were stained with a 50 µg/ml propidium iodide (PI) solution containing 0.1% Triton X-100, 0.1 mM ethylenediaminetetraacetic acid and 50 µg/ml RNase A. Fluorescence was measured and analyzed using a NucleoCounter NC-3000 instrument (ChemoMetec, Allerød, Denmark). Data are representative of ≥3 separate experiments.
Quantification of apoptotic cells using annexin V staining
HCT116 cells (1×106 cells/sample) were treated with 5 µM TCS7010 for 24 h at 37°C. The cells were fixed with 4% paraformaldehyde, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated annexin V, according to the manufacturer's protocol (ChemoMetec). PI was used as a counterstain for dead cells. The fluorescence intensities of FITC-annexin V-positive cells (green) and PI-positive cells (red) were analyzed with a NucleoCounter NC-3000 instrument as described previously (12). Data are representative of ≥3 separate experiments.
Detection of intracellular ROS levels
Cells were trypsinized and incubated with 10 µM H2-2′, 7′-dichlorofluorescin diacetate (DCF-DA) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 60 min, followed by incubation with 5 µM TCS7010 for 3, 6, or 12 h. The fluorescence intensity was analyzed using a FACSCalibur flow cytometer (BD Biosciences), as described previously (11). CellQuest Pro software version 5.2 (BD Biosciences) was used for acquisition and analysis of the data. Data are representative of ≥3 separate experiments.
Analysis of X-box binding protein 1 (XBP-1) splicing
Total RNA was isolated using the phenol/guanidine-based Isol-RNA Lysis reagent (5 PRIME GmbH, Hamburg, Germany), and the synthesis of cDNA was performed using an iScript cDNA Synthesis kit (Bio-Rad, Inc.). Reverse transcription-polymerase chain reaction (RT-PCR) was performed using XBP-1-specific primers (forward, 5′-AGAGTAGCAGCTCAGACTGC-3′ and reverse, 5′-CATTAATGGCTTCCAGCTTG-3′) that correspond to nt 277–817, encompassing the IRE-1 cleavage site, as previously described (12). PCR products were further digested for 2 h at 37°C using PstI, whose recognition site is located at nt 556 within the 26-nt intron (nt 531–556) that is lost following IRE-1-mediated splicing (13). The PCR product of unspliced XBP-1 (uXBP1) mRNA is expected to be cleaved into two fragments of 279 bp and 261 bp following PstI digestion, while that of spliced XBP-1 (sXBP1) mRNA is resistant to PstI digestion, producing a 540-bp band (11). Data are representative of ≥3 separate experiments.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance followed by Sidak's multiple comparisons test using GraphPad Prism software (version 7.0; GraphPad Software Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results and discussion
Effect of TCS7010 on the phosphorylation of AURKA in HCT116 cells
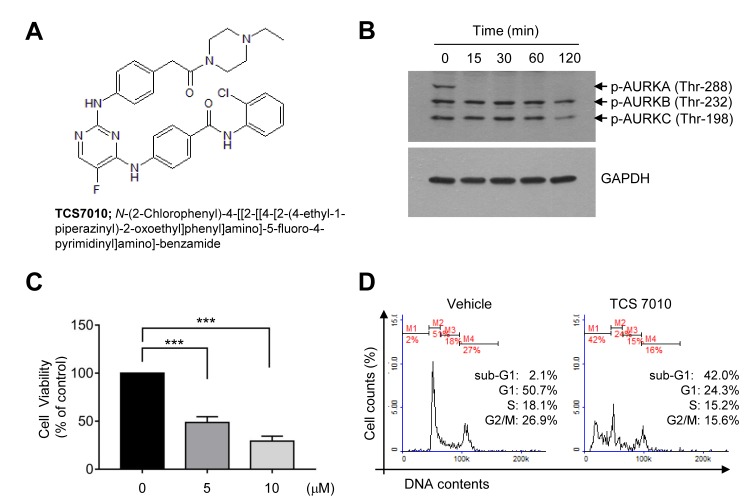
AURKA is overexpressed in a broad range of tumor cells (14). Immunoblot analysis was performed to evaluate whether TCS7010 inhibits AURKA in HCT116 colon cancer cells. The phosphorylation of all AURK subtypes (A, B and C) was highly elevated at the basal status (Fig. 1B). It was demonstrated that TCS7010 selectively inhibited AURKA phosphorylation on threonine-288 within 15 min of treatment (Fig. 1B).
Figure 1.
Effect of TCS7010 on the induction of apoptosis. (A) Chemical structure of TCS7010. (B) HCT116 cells were treated with 5 µM TCS7010 for different periods of time (0–120 min). Whole cell lysates were analyzed using immunoblotting. GAPDH was used as an internal control. (C) Cell viability assay in which HCT116 cells were treated with 5 µM TCS7010 for 24 h. Cell viability was determined using a Cell Counting Kit-8 assay. Data are presented as the mean ± standard deviation. ***P<0.001 (n=3), one-way analysis of variance followed by Sidak's multiple comparisons test. (D) Cell cycle analysis in which HCT116 cells (1×105 cells/sample) were treated with vehicle (DMSO) or 5 µM TCS7010, fixed with ethanol and stained with propidium iodide. The cellular DNA content was determined using flow cytometry. AURK, aurora kinase.
Effect of TCS7010 on the cell viability and cell cycle progression of HCT116 cells
To assess the cytotoxic effect of TCS7010, exponentially growing HCT116 cells were exposed to different TCS7010 concentrations for 24 h, and cell viability was measured using CCK-8. Treatment with TCS7010 significantly reduced HCT116 cell viability in a dose-dependent manner (Fig. 1C). The effective dose causing 50% of maximal cytotoxicity (ED50) was ~5 µM. To investigate the mechanism underlying TCS7010-induced cytotoxicity, the effect of TCS7010 on cell cycle progression was evaluated. HCT116 cells were treated with 5 µM TCS7010 for 24 h and cell cycle progression was monitored using flow cytometry. The number of G1 phase cells decreased (50.7 to 24.3%), with a concomitant increase in the number of sub-G1 phase cells (2.10 to 42.0%), which is indicative of a hypodiploid DNA cell population undergoing apoptosis (Fig. 1D). These results suggest that TCS7010 has a cytotoxic effect against HCT116 colon cancer cells, possibly via the induction of apoptosis.
Effect of TCS7010 on the induction of apoptosis
To determine whether TCS7010 triggers apoptosis, annexin V staining was performed. In the early stages of apoptosis, phosphatidylserine (PS) in the inner side of the plasma membrane translocates to the outer surface. As annexin V preferentially binds to PS (15), detection of PS using annexin V is widely used to identify apoptotic cells in response to cytotoxic agents. PI, which cannot enter live cells, was used to counterstain dead cells. HCT116 cells were treated with 5 µM TCS7010 for 24 h and then incubated with FITC-conjugated annexin V and PI. In Fig. 2A, the lower left quadrant of each cytogram (PI and FITC double-negative) represents viable cells, the lower right quadrant (PI negative/FITC positive) represents early apoptotic cells, and the upper right quadrant (PI and FITC double-positive) represents late apoptotic and dead cells. The PI-negative/FITC-positive population was increased in the cells treated with TCS7010 compared with the untreated control group (16 vs. 48%) following, suggesting that TCS7010 triggers apoptosis in HCT116 cells.
Figure 2.
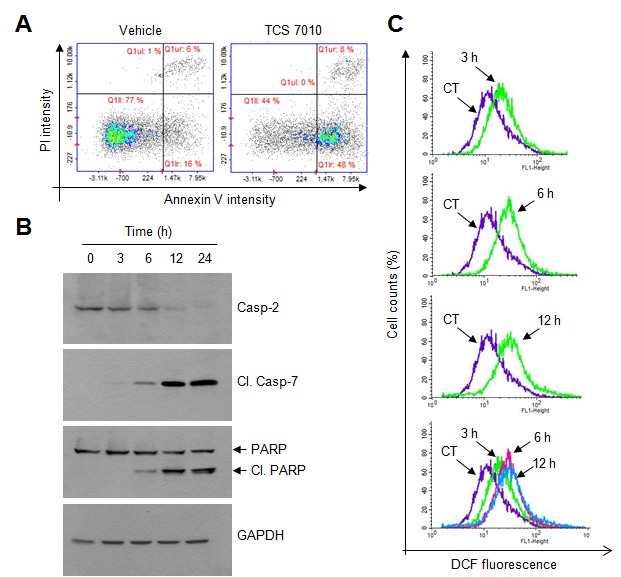
Effect of TCS7010 on the activation of caspases and produces reactive oxygen species. (A) Annexin V staining assay in which HCT116 cells were treated with vehicle (DMSO) or 5 µM TCS7010 for 24 h and then stained with FITC-annexin V and PI. Cells were harvested, washed and analyzed for fluorescence intensity. Scatter plots demonstrating the intensity of FITC-annexin V vs. that of PI. x-axis, Annexin V intensity; y-axis, PI intensity. (B) HCT116 cells were treated with 5 µM TCS7010 for various time periods (0–24 h). Whole cell lysates were prepared and analyzed using immunoblotting. GAPDH was used as an internal control. (C) HCT116 cells were incubated with 10 µM DCF-DA for 1 h, followed by the addition of 5 µM TCS7010 for 3, 6, and 12 h. CT, control treated with DCF-DA alone. Fluorescence was assessed using a FACSCalibur instrument. PI, propidium iodide; DCF-DA, H2-2′ 7′-dichlorofluorescin diacetate; casp, caspase; Cl., cleaved; PARP, poly(ADP-ribose) polymerase.
Caspases are a family of cysteine proteases that have an essential role in the progression of apoptosis (16). The effect of TCS7010 on the induction of caspase activation was determined in the present study. Upon TCS7010 treatment, the pro-caspase-2 level gradually decreased in a time-dependent manner and the amount of cleaved active caspase-7, and cleaved poly (ADP-ribose) polymerase (PARP), a well-known substrate for caspase-7, increased in a time-dependent manner (Fig. 2B). These data suggest that caspase activation is involved in TCS7010-induced apoptosis.
Effect of TCS7010 on the production of ROS
ROS are chemically reactive molecules containing oxygen, including superoxide anion radical, hydrogen peroxide and the highly reactive hydroxyl radical, all of which are formed as a byproduct of normal cellular metabolism. Abnormally excessive ROS levels may result in cellular damage, known as oxidative stress. Thus, under normal physiological conditions, various antioxidant enzymes, including catalase and superoxide dismutase maintain the proper cellular redox states. However, there is growing evidence for the beneficial effects of ROS on chemotherapy (17,18). As numerous cancer cells are more sensitive to ROS compared with normal cells, ROS production could be used for the selective targeting of cancer cells (12,19–21).
As ROS activates the caspase signaling pathway (22), ROS production levels were analyzed using a fluorescent DCF-DA probe. Upon TCS7010 treatment, ROS accumulation was detectable at 3 h and maintained up to 12 h following TCS7010 treatment (Fig. 2C). These data suggested that ROS may act as an upstream signal in TCS7010-induced apoptosis.
Effect of TCS7010 on the activation of the UPR signaling pathway
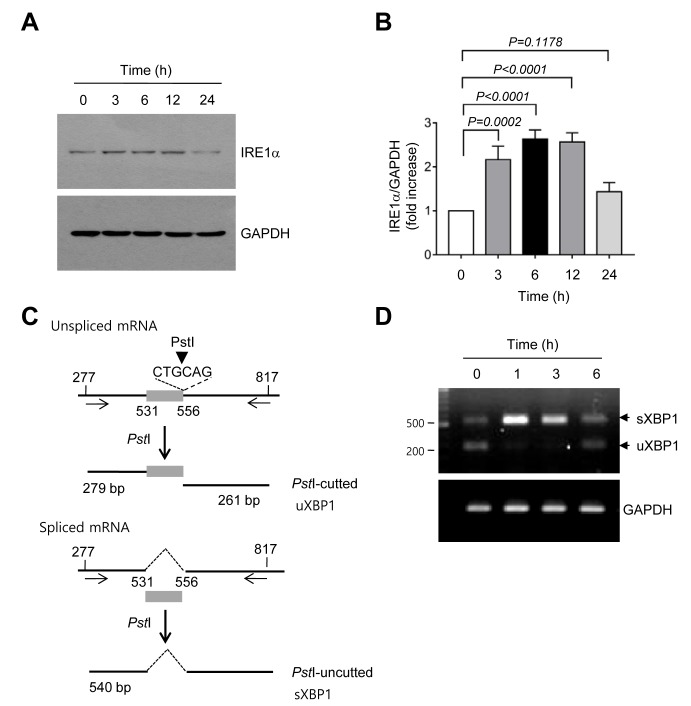
ROS can induce ER stress (23), and ER stress can trigger apoptosis through the UPR signaling pathway (24,25). Thus, whether TCS7010 activates the UPR signaling pathway was determined in the present study. A typical feature of the ER stress response is the accumulation of UPR sensor proteins. It was demonstrated that the amount of IRE1α protein significantly increased within 3 h compared with no treatment and then returned to the basal level at 24 h following TCS7010 treatment (3 h, P=0.0002; Sidak's multiple comparisons test; n=3; Fig. 3A and B). Activated IRE1 functions as a site-specific endoribonuclease that catalyzes the excision of an unconventional 26-nt intron from uXBP1 to generate sXBP1 mRNA (26). To investigate whether TCS7010 induces XBP1 mRNA splicing, RT-PCR was performed using primers that amplify the region spanning the 26-nt intron, followed by digestion with PstI. As the PstI restriction site is within the intron site, the uXBP1 PCR product should be cleaved into two fragments (279 and 261 bp) by PstI digestion, whereas the sXBP1 PCR product lacking the PstI site should produce a 540-bp band (Fig. 3C). As illustrated in Fig. 3D, TCS7010 caused the accumulation of a shifted PCR band (540 bp) between 1 and 6 h, reflecting the splicing of XBP1 mRNA. These data suggest that TCS7010 activates the ER stress-induced UPR signaling pathway.
Figure 3.
Effect of TCS7010 on the activation of the unfolded protein response signaling pathway. (A) HCT116 cells were treated with 5 µM TCS7010 for various time periods (0–24 h). Whole-cell lysates were subjected to immunoblotting using an antibody directed against IRE1α. GAPDH was used as an internal control. (B) Immunoblot band intensities were analyzed using a quantitative scanning densitometer. Vehicle (DMSO)-treated band intensity was set to 1 and the relative values of fold-increase were plotted as the mean ± standard deviation (n=3). Statistical analysis was performed using analysis of variance followed by Sidak's multiple comparisons test. (C) Schematic presentation of XBP-1 splicing. Numbers denote nucleotide positions with the transcription start site set at 1. The box indicates the 26-nt intron site (nt 531–556). The arrows indicate the RT-PCR primer positions. The arrowhead indicates the position of the PstI restriction enzyme. (D) HCT116 cells were treated with 5 µM TCS7010 for various time periods (0–6 h). Subsequently total RNA was isolated, XBP-1 mRNA splicing was determined using RT-PCR. GAPDH was used as an internal control. RT-PCR, reverse transcription-polymerase chain reaction; XBP1, X-box binding protein 1; s, spliced; u, unspliced; IRE1α, inositol-requiring enzyme 1α.
Effect of TCS7010 on the accumulation of CHOP and BIM proteins
Accumulating evidence has demonstrated that CHOP performs an essential role in ER-stress-induced apoptosis via upregulation of the pro-apoptotic protein BIM (24,25,27,28). BIM is a Bcl-2 homology domain 3 (BH3)-containing pro-apoptotic Bcl-2 family member. In general, BIM cooperates with other BH3-only proteins, including truncated BH3 interacting domain death agonist and BAX, to trigger mitochondria-mediated apoptosis (29). To determine whether CHOP expression is upregulated by TCS7010, HCT116 cells were treated with TCS7010 for different periods of time. Immunoblot analysis demonstrated that CHOP protein expression was maximal at 6 h following TCS7010 treatment and were maintained up to 24 h (Fig. 4A). The amount of BIM protein, a CHOP target, peaked at 12 h and then decreased (Fig. 4A). Densitometric analysis revealed that the increases of CHOP and BIM levels were significant (all P<0.0001; Sidak's multiple comparisons test; n=3) upon TCS7010 treatment. These results suggest that the UPR signaling pathway may be involved in TCS7010-induced apoptosis.
Figure 4.
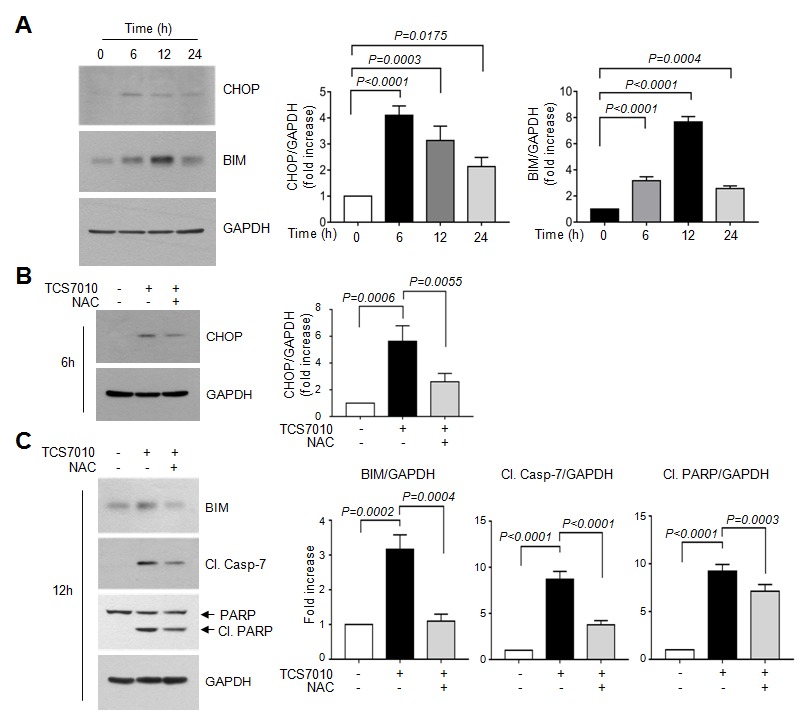
Effect of TCS7010 on the upregulation of CHOP and BIM proteins. (A) HCT116 cells were treated with 5 µM TCS7010 for various time periods (0–24 h). Whole-cell lysates were subjected to immunoblotting using antibodies directed against CHOP and BIM. GAPDH was used as an internal control. Immunoblot band intensities were analyzed using a quantitative scanning densitometer (right graph). Vehicle (DMSO)-treated band intensity was set to 1 and the relative values of fold-increase were plotted as the mean ± standard deviation (n=3). Statistical analysis was performed using analysis of variance followed by Sidak's multiple comparisons test. HCT116 cells were treated with 5 µM TCS7010 for (B) 6 h or (C) 12 h in the absence or presence of 2 mM NAC. Whole-cell lysates were subjected to immunoblotting. GAPDH was used as an internal control. Immunoblot band intensities were analyzed using a quantitative scanning densitometer (right graph). Vehicle (DMSO)-treated band intensity was set to 1 and the relative values of fold-increase were plotted as the mean ± SD (n=3). Statistical analysis was performed using analysis of variance followed by Sidak's multiple comparisons test. Cl., cleaved; Casp, caspase; NAC, N-acetylcystein.
Effect of N-acetylcystein (NAC) on TCS7010-induced CHOP accumulation
The association between ROS accumulation by TCS7010 and the UPR response was determined. Fig. 4B demonstrates that pretreatment of HCT116 cells with 2 mM NAC, a thiol-containing ROS scavenger, significantly abrogated TCS7010-induced CHOP accumulation compared with cells treated with TCS7010 alone (P=0.0055; Sidak's multiple comparisons test; n=3). Furthermore, pretreatment with NAC significantly reduced the TCS7010-induced accumulation of BIM and cleaved caspase-7 compared with cells treated with TCS7010 alone (P=0.0004 and P<0.0001, respectively; Sidak's multiple comparisons test; n=3; Fig. 4C). In addition, the TCS7010-induced cleavage of PARP was significantly reduced by pretreatment with NAC compared with cells treated with TCS7010 alone (P=0.0003; Sidak's multiple comparisons test; n=3). These results suggest that ROS production by TCS7010 is upstream of UPR-mediated apoptosis.
In conclusion, the results of the present study demonstrated that TCS7010 induces ROS-mediated apoptosis. Upregulation of BIM through the UPR signaling pathway may have an important role in TCS7010-induced apoptosis. Currently, the mechanism underlying AURKA regulation of the UPR signaling pathway remains unclear. Additional studies are required to investigate the role of AURKA in the control of ER stress responses.
Acknowledgements
The present study was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science and Future Planning (grant no. 2016R1A2B4008570). This paper was supported by the KU Research Professor Program of Konkuk University.
References
- 1.Mountzios G, Terpos E, Dimopoulos MA. Aurora kinases as targets for cancer therapy. Cancer Treat Rev. 2008;34:175–182. doi: 10.1016/j.ctrv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 3.Barr AR, Gergely F. Aurora-A: The maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 4.Qi W, Cooke LS, Liu X, Rimsza L, Roe DJ, Manziolli A, Persky DO, Miller TP, Mahadevan D. Aurora inhibitor MLN8237 in combination with docetaxel enhances apoptosis and anti-tumor activity in mantle cell lymphoma. Biochem Pharmacol. 2011;81:881–890. doi: 10.1016/j.bcp.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 6.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 7.Katayama H, Zhou H, Li Q, Tatsuka M, Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J Biol Chem. 2001;276:46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- 8.He W, Zhang MG, Wang XJ, Zhong S, Shao Y, Zhu Y, Shen ZJ. AURKA suppression induces DU145 apoptosis and sensitizes DU145 to docetaxel treatment. Am J Transl Res. 2013;5:359–367. [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 11.Lee da H, Jung Y Jung, Koh D, Lim Y, Lee YH, Shin SY. A synthetic chalcone, 2′-hydroxy-2,3,5′-trimethoxychalcone triggers unfolded protein response-mediated apoptosis in breast cancer cells. Cancer Lett. 2016;372:1–9. doi: 10.1016/j.canlet.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Shin SY, Lee JM, Lee MS, Koh D, Jung H, Lim Y, Lee YH. Targeting cancer cells via the reactive oxygen species-mediated unfolded protein response with a novel synthetic polyphenol conjugate. Clin Cancer Res. 2014;20:4302–4313. doi: 10.1158/1078-0432.CCR-14-0424. [DOI] [PubMed] [Google Scholar]
- 13.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y, Zhang Y, Lees E, Seghezzi W. AuroraA overexpression overrides the mitotic spindle checkpoint triggered by nocodazole, a microtubule destabilizer. Oncogene. 2003;22:8293–8301. doi: 10.1038/sj.onc.1206873. [DOI] [PubMed] [Google Scholar]
- 15.Andree HA, Reutelingsperger CP, Hauptmann R, Hemker HC, Hermens WT, Willems GM. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J Biol Chem. 1990;265:4923–4928. [PubMed] [Google Scholar]
- 16.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigas B, Sun Y. Induction of oxidative stress as a mechanism of action of chemopreventive agents against cancer. Br J Cancer. 2008;98:1157–1160. doi: 10.1038/sj.bjc.6604225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacker PT. Reactive oxygen species in cancer cells: Live by the sword, die by the sword. Cancer Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Adams DJ, Dai M, Pellegrino G, Wagner BK, Stern AM, Shamji AF, Schreiber SL. Synthesis, cellular evaluation, and mechanism of action of piperlongumine analogs; Proc Natl Acad Sci USA; 2012; pp. 15115–15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui X. Reactive oxygen species: The Achilles' heel of cancer cells? Antioxid Redox Signal. 2012;16:1212–1214. doi: 10.1089/ars.2012.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim BM, Rode AB, Han EJ, Hong IS, Hong SH. 5-Phenylselenyl- and 5-methylselenyl-methyl-2′-deoxyuridine induce oxidative stress, DNA damage, and caspase-2-dependent apoptosis in cancer cells. Apoptosis. 2012;17:200–216. doi: 10.1007/s10495-011-0665-2. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura M. The unfolded protein response triggered by environmental factors. Semin Immunopathol. 2013;35:259–275. doi: 10.1007/s00281-013-0371-y. [DOI] [PubMed] [Google Scholar]
- 24.Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 27.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Happo L, Strasser A, Cory S. BH3-only proteins in apoptosis at a glance. J Cell Sci. 2012;125:1081–1087. doi: 10.1242/jcs.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]