Abstract
CC chemokine receptor 9 (CCR9) serves a role in the drug resistance and metastasis of tumors. In the present study, a peptide specifically bound to CCR9 was obtained and the effect on tumor cells was observed. A Ph.D.-12 phage display peptide library was used to screen for peptides binding specifically to the second extracellular loop of CCR9. The ratios of the input and output of phage clones increased gradually following three rounds of biopanning. A total of 8 positive phage clones were identified from DNA analysis. A phage clone, C-4, was identified which exhibited higher affinity and specificity for the second extracellular loop of CCR9 in vitro compared with other clones. A peptide (P1; VHWDFRQWWQPS) was identified which may inhibit the corresponding phage, C-4, binding to the second extracellular loop of CCR9. Furthermore, P1 was able to bind specifically with MOLT4 cells which exhibit marked expression of CCR9. In addition, P1 promoted the apoptosis of MOLT4 cells induced by doxorubicin, and inhibited the migration of MOLT4 cells in the presence of chemokine (C-C motif) ligand 25. It was suggested that decreased activity in the phosphorylation of protein kinase B in MOLT4 cells may be responsible for the inhibition. In conclusion, the peptide P1 derived from a screened phage is able to specifically bind to CCR9 and inhibit the activity of CCR9. It has potential use as an antagonist in the treatment of CCR9-overexpressed carcinoma.
Keywords: CC chemokine receptor 9, peptide, tumor, antagonist, apoptosis, migration
Introduction
CCR9 is a member of the family of G-protein-coupled receptors (GPCRs) and serves a role in T-cell differentiation and tissue-specific homing once bound to its cognate ligand chemokine (C-C motif) ligand 25 (CCL25) (1). CCR9 has also been implicated in numerous types of cancer including breast, lung, pancreas, prostate and colon cancer, in addition to adult T-cell leukemia; it is also reported that CCR9 mediates organ-selective metastasis and drug resistance (2–7). The functions of CCR9 in metastasis and drug resistance depend on the activation of the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway (8). Furthermore, the differences in the cell-surface profile between cancer cells and their normal counterparts may be used as a molecular signature for targeted therapy and drug delivery. Thus, CCR9 is a promising candidate for a target molecule based on its rare cell-surface expression in normal tissues and increased cell-surface expression in malignancies.
An immunotoxin CCL25-PE38 has previously been developed that is able to specifically kill high CCR9-expressing MOLT4 cells (9). However, several issues remain unresolved, including uptake and expression of CCL25-PE38. Peptides exhibit a short plasma half-life, increased tissue penetration ability, decreased immunogenicity, and are easily synthesized in comparison with macromolecular proteins (10).
Phage display is a molecular technology that permits the presentation of a number of peptides with high specificity and affinity for a target on the surface of a bacteriophage. For instance, the biopanning of phage display libraries on cellular receptors has been successful in selecting peptides that exhibit high affinity and specificity for the receptors (11–13). Therefore, peptides screened from phage display may be useful for the diagnosis and targeted therapy of tumors.
In the present study, the second extracellular loop of CCR9 was used to screen a 12-mer phage display library and a novel peptide (P1; VHWDFRQWWQPS) that was capable of specifically binding to CCR9 was identified. Furthermore, this peptide promoted the apoptosis of MOLT4 cells induced by doxorubicin (DOX) and inhibited the migration of MOLT4 cells in the presence of CCL25. The inhibitory effects may be associated with inhibition of the activation of Akt in MOLT4 cells. In conclusion, the novel peptide identified in the present study may inhibit the activity of CCR9 and is potentially useful in the research of novel treatments for CCR9-overexpressed carcinoma.
Materials and methods
Affinity screening procedure
The Ph.D.-12 phage display library (version 1.1; New England BioLabs, Ipswich, MA, USA) was used as the biopanning tool. The panning procedure was performed according to the manufacturer's protocol. Briefly, the second extracellular loop of CCR9 (AAADQWKFQTFMCKVVNSMYK; SBS Genetech Co., Ltd., Beijing, China) was used to coat microtiter plates at 10 µg/ml. Phages that specifically bound to the target were identified through three rounds of in vitro selection. In each round, the bound phages were rescued and amplified in ER2738 bacterial cells from the phage display library (New England BioLabs, Inc., Ipswich, MA, USA) for the subsequent panning, whereas the unbound phages were removed by washing with Tris-buffered saline with Tween-20 (TBST). The washing conditions for the second and third rounds were the same as those for the first round, with the exception that the Tween-20 concentration was increased from 0.1 to 0.5%, respectively. Following the elution step in the third round, the single clones of phage particles were harvested. The phages were subjected to DNA extraction, sequencing and an ELISA.
DNA sequencing of the selected phages
Following the third round of panning, 16 phage clones were randomly selected, amplified and purified. The clones were designated C-1 to C-16. Single-stranded DNA was extracted with M13 DNA Kit (Beijing BLKW Biotechnology Co., Ltd., Beijing, China) according to the manufacturer's protocol. DNA sequencing was performed by Sangon Biotech Co., Ltd. (Shanghai, China) using the −96gIII primer (5′-CCCTCATAGTTAGCGTAACG-3′). Sequence analysis was performed and phages with the same sequence were classified.
Phage capture ELISA
A 10-µg/ml sample of the second extracellular loop of CCR9 in 0.1 M NaHCO3, pH 8.6, was coated on ELISA plates. The ELISA plates were incubated overnight at 4°C in an air-tight humidified box. The excess solution was removed and the plates were blocked with a blocking buffer (0.1 M NaHCO3 (pH 8.6), 5 mg/ml BSA) for 1 h at 4°C. Subsequently, the plates were washed and phages including control phage, C-4, C-6, C-9 and C-10 [at 1011 plaque-forming units (pfu)] were added to the target-coated wells. The plates were incubated at room temperature for 1 h with agitation. Finally, a horseradish peroxidase-conjugated anti-M13 monoclonal antibody at 5 µg/ml (cat. no. 111973-MM05; Sino Biological, Inc., Beijing, China) was used to detect the bound phages. Data are reported from three independent experiments.
Peptide synthesis and labeling
The candidate peptide P1 (VHWDFRQWWQPS) and unrelated peptide (control P; WIRPPSGPMYSF) were synthesized by SBS Genetech, Co., Ltd. and Sangon Biotech Co., Ltd., respectively, using standard solid-phase chemistry. Fluorescein isothiocyanate (FITC) was conjugated to the N-terminus of P1 (FITC-P1) or the unrelated peptide (FITC-control P). The products were purified to a minimum purity of 95% using high-performance liquid chromatography and isolated by lyophilization. The sequence and structure of each peptide were characterized by mass spectrometry.
Competitive inhibition assay
To investigate the inhibition effect of P1 on C-4, a competitive inhibition ELISA was conducted. Briefly, wells were incubated with 10 µg/ml of the second extracellular loop of CCR9. P1 at various concentrations (0, 0.01, 0.1, 0.5, 1.0, 2.0 and 10 nmol/ml) was added to the cells. Subsequently, 1011 pfu C-4 was added to the wells prior to incubation at 37°C for 1 h. The subsequent procedure was similar to that of the ELISA performed for the aforementioned phage capture. The wells were washed with 1× PBS (pH 7.4) 3 times following incubation. Finally, a horseradish peroxidase-conjugated anti-M13 monoclonal antibody at 10 µg/ml (cat. no. 111973-MM05; Sino Biological, Inc.) was used to detect the bound phages. The rate of inhibition was calculated using the following formula: Rate of inhibition=[optical density (OD) of control P-OD of P1]/OD of control P × 100%. An unrelated peptide was used as the control P. Data are reported from three independent experiments.
Confocal microscopy analysis
MOLT4 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were maintained in RPMI-1640 medium (GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (GE Healthcare Life Sciences), 100 U/ml penicillin and 100 mg/ml streptomycin. The cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. The MOLT4 cells were washed three times with PBS. One group was washed three times with PBS and blocked with 2% bovine serum albumin (BSA; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C for 30 min and FITC-P1 (0.1 nmol/ml) was incubated with the cells for 1 h at 4°C. Meanwhile, another group was incubated with anti-CCR9 monoclonal antibody at 0.1 nmol/ml (anti-CCR9 mAb; cat. no. 112509; R&D Systems, Inc., Minneapolis, MN, USA) for 30 min at 37°C, blocked with 2% BSA and subsequently incubated with FITC-P1 at 4°C as above. Following three washes, the cells were placed on poly-L-lysine-coated slides and observed using a laser-scanning confocal microscope with the magnification, ×200 (LSM710; Zeiss GmbH, Jena, Germany). FITC-control P was used as the control.
Apoptosis assay
MOLT4 cells (4×105/well) were transferred to 24-well plates with each well containing 500 µl RPMI-1640 medium without FBS. Each well was treated with a distinct concentration of P1 (0, 0.1 and 1.0 nmol/ml) for 12 h, and then treated with DOX (1 µg/ml) for 12 h. The control groups were treated with various concentrations of P1 (0, 0.1 and 1.0 nmol/ml) for 24 h. The cells were washed twice, and subsequently detected using an Annexin V-FITC and propidium iodide (PI) double staining kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 15 min at 25°C in the dark, according to the manufacturer's protocol. Subsequently, the processed cells were rinsed with PBS and subjected to fluorescence-activated cell sorting (FACS) analysis. RBL-2H3 cells (ATCC, Manassas, VA, USA), were incubated with Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc.) at 4×105 cells/well. The subsequent procedure was the same as that for the MOLT4 cells. Results were presented as the percentage of Annexin V-FITC-positive cells, Annexin V-FITC- and PI-double-positive cells. Data are reported from three independent experiments.
Chemotaxis assay
The chemotaxis assay was performed in 24-well Transwell chambers (Corning Life Sciences, Cambridge, MA, USA) as previously described (14). MOLT4 cells (0.5×104 cells/well) were treated with P1 (0, 0.1 and 1.0 nmol/ml) and control P (0, 0.1 and 1.0 nmol/ml) for 1 h, prior to being added to the upper chamber. CCL25 (0.1 nmol/ml) was placed in the lower chamber (500 µl). Following incubation at 37°C in 5% CO2 for 2 h, cells were collected from the lower wells. The number of cells was counted using light microscopy. Spontaneous baseline migration (BM) was used as a control. The experiments were performed in triplicate.
Western blotting
To investigate the expression levels of Akt and phospho-Akt (p-Akt) in MOLT4 cells in the presence of P1, MOLT4 cells incubated in 24-well plates were serum-starved for 12 h and then treated with P1 at various concentrations (0, 0.1 and 1.0 nmol/ml) for 24 h. The protein extraction procedure was performed as described previously (15). The nitrocellulose membranes were incubated overnight at 4°C with anti-p-Akt (Ser473) at 1:100 (cat. no. 9271; Cell Signaling Technology, Danvers, MA, USA) or anti-Akt monoclonal antibody at 1:100 (cat. no. 9272, Cell Signaling Technology) and incubated with horseradish peroxidase-conjugated secondary antibody at 1:2,000 (cat. no. D110058-0100; Sangon Biotech Co., Ltd.). β-actin was used as the control. Finally, the membrane-bound proteins were detected using enhanced chemiluminescence substrate (Pierce ECL; cat. no. 32209, Thermo Fisher Scientific, Inc.). The blots were incubated with the ECL working solution for 1 min at room temperature, and then removed and placed in a film cassette with the protein side facing up in the dark for X-ray film exposure. The film was developed using the developing solution (cat. no., AR0132; BOSTER Biological Technology Co., Ltd, Wuhan, China) for 2 min and fixative solution (cat. no. AR0132; BOSTER Biological Technology Co., Ltd, Wuhan, China) for 1 min at room temperature following exposure of 60 sec. The experiments were performed in triplicate.
Statistical analysis
The data are expressed as the mean ± standard deviation or a percentage. Data analysis was performed using GraphPad Prism (Version 5; GraphPad Software, Inc., La Jolla, CA, USA). One-way analysis of variance and Student's t-test were used to perform statistical comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Screening of the Ph.D.-12 phage display library against CCR9
To obtain a specific peptide binding to the CCR9, a Ph.D.-12 phage display library was screened for binding to the second extracellular loop of CCR9. Following three rounds of biopanning, the titers of phages increased ~79-fold compared with the titer in the first round. The output/input ratio of phages following each round of panning was used to determine the enrichment efficiency, which increased from 5.2×10−6 to 2.4×10−4 (Table I). This increase indicated that phages capable of specifically binding to the second extracellular loop of CCR9 were significantly enriched following the biopanning.
Table I.
Enrichment of phages screened from the Ph.D.-12 library.
| Titer, pfu | ||||
|---|---|---|---|---|
| Round | Second extracellular loop of CCR9, µg | Input | Output | Recovery |
| First | 10 | 1.0×1011 | 5.2×105 | 5.2×10−6 |
| Second | 5 | 1.0×1011 | 3.6×106 | 3.3×10−5 |
| Third | 2 | 1.2×109 | 4.9×105 | 4.1×10−4 |
The output/input ratio was used to indicate the recovery of phages. CCR9, CC chemokine receptor 9; pfu, plaque-forming units.
DNA sequencing of the selected phage clones
The results demonstrated that 8 phage clones lacked an exogenous sequence and the remaining 8 clones were confirmed to be positive using DNA sequencing. A total of 8 phage clones and corresponding peptide sequences were obtained (Table II). Sequence alignment analysis did not identify homology among the peptide sequences.
Table II.
DNA sequences of the selected phage clones.
| Phage clone | DNA sequence | Peptide sequence | Frequency |
|---|---|---|---|
| C-1/−2/−4/13/−14 | GTGCATTGGGATTTTCGG | VHWDFRQWWQPS | 5 |
| CAGTGGTGGCAGCCTTCT | |||
| C-6 | GGGGATGGTAATTCGGTG | GDGNSVLKPGNW | 1 |
| CTGAAGCCGGGGAATTGG | |||
| C-9 | CATCTTAGTCTTCCGCTTTG | HLSLPLWKWEKS | 1 |
| GAAGTGGGAGAAGTCG | |||
| C-10 | GTGTTTGCTTATCATCTT | VFAYHLRIPSGD | 1 |
| AGGATTCCTTCTGGGGAT |
A total of 8 clones were identified to be endowed with four types of exogenous sequences following the third round of panning. The number of positive clones is presented as the frequency.
Phage-binding assay
An ELISA was performed to determine the affinity and specificity of positive phage clones. The results revealed that the phage C-4 appeared to exhibit increased affinity for the target molecule in comparison with the other phages (Fig. 1). The affinity of C-4 increased >8-fold compared with the control phage, a statistically significant increase (P<0.01). It also identified a significant difference between group C-4 and group C-6 (P<0.05), C-9 (P<0.05) and C-10 (P<0.05). Therefore, the displaying peptide (VHWDFRQWWQPS) derived from C-4 was designated P1 and was investigated further.
Figure 1.
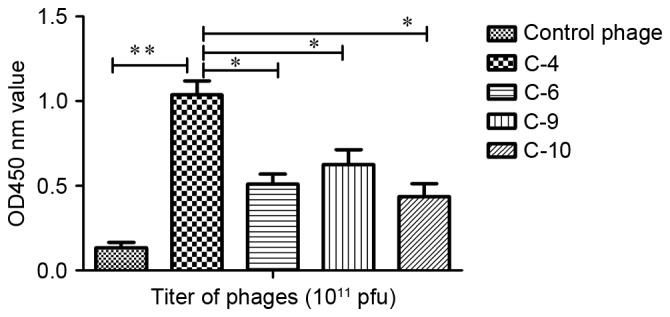
Identification of specific binding of phage clones to the second extracellular loop of CC chemokine receptor 9. Affinity was detected using ELISA. The affinity of C-4 increased >8-fold compared with the control phage, and was increased compared with the other phage clones (*P<0.05; **P<0.01). OD, optical density; pfu, plaque-forming units.
Competitive inhibition assay
To investigate whether the synthetic peptide P1 was able to inhibit the corresponding phage binding to the target molecule, competitive inhibition ELISA was performed. As presented in Fig. 2, C-4 binding to the target decreased following an increase in peptide P1. The peptide P1 appeared to inhibit C-4 binding to the second extracellular loop of CCR9 in a dose-dependent manner. When the concentration of P1 reached 1.0 nmol/ml, C-4 binding to the second extracellular loop of CCR9 was significantly inhibited (inhibition of 50.33%).
Figure 2.
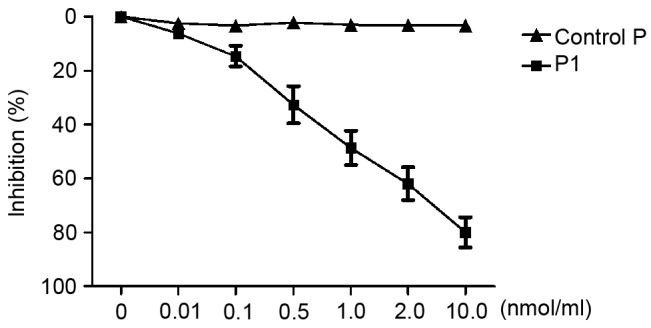
Inhibition of C-4 binding by P1. The binding of the positive phage C-4 to the target decreased following an increase in P1. When the concentration of P1 reached 1.0 nmol/ml, inhibition was >50.0%. An unrelated peptide was used as control P. Data are reported from three independent experiments.
Identification of P1 binding specifically to CCR9 high-expressing cells
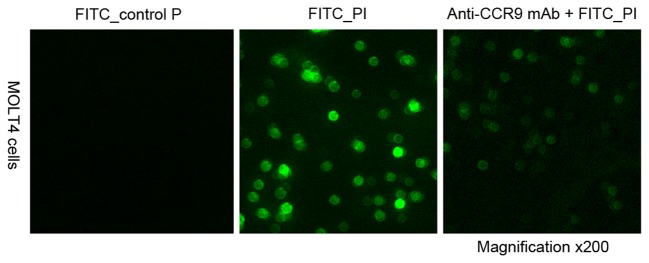
To investigate the peptide specifically binding to CCR9, confocal microscopy analysis was performed to evaluate the binding specificity of the peptide P1. It was revealed that FITC-P1 bound directly to MOLT4 cells, and the anti-CCR9 mAb was able to completely block the binding of P1. When a control peptide was incubated with MOLT4 cells, no cell binding was observed (Fig. 3). This suggests that P1 is able to specifically bind to CCR9 and is stable under physiological conditions.
Figure 3.
P1 binds specifically on cells which highly express CC chemokine receptor 9. FITC-control P (left panel) did not bind to MOLT4 cells; FITC-P1 (middle panel) did bind to MOLT4 cells; anti-CCR9 mAb (right panel) inhibited FITC-P1 binding to MOLT4 cells. Cells were observed using a laser-scanning confocal microscope; magnification, ×200. CCR9, CC chemokine receptor 9; mAb, monoclonal antibody; FITC, fluorescein isothiocyanate; PI, propidium iodide.
P1 increases apoptosis of MOLT4 cells induced by DOX
To investigate the effect of P1 on cell apoptosis, the number of apoptotic cells was measured using FACS following P1 and DOX treatments of MOLT4 cells and RBL-2H3 cells. As presented in Table III, the number of apoptotic MOLT4 cells induced by DOX increased to 64.7% following pre-treatment with 0.1 nmol/ml P1 (P<0.05); apoptotic MOLT4 cells further increased to 79.5% following pre-treatment with 1.0 nmol/ml P1 (P<0.01). On the other hand, the number of apoptotic RBL-2H3 cells induced by DOX revealed no significant difference between cells treated with P1 and those receiving no treatment. These results suggest that P1 may elicit its effect on MOLT4 cells by promoting an apoptotic pathway.
Table III.
Effect of P1 on apoptosis induced by DOX.
| A, MOLT4 cells | ||||
|---|---|---|---|---|
| P1, nmol/ml | ||||
| Treatment | 0 | 0.1 | 1.0 | |
| Control | 5.3±0.6 | 4.8±0.5 | 6.1±1.0 | |
| DOX | 52.4±2.5 | 64.7±3.1a | 79.5±4.2b | |
| B, RBL-2H3 cells | ||||
| P1, nmol/ml | ||||
| Treatment | 0 | 0.1 | 1.0 | |
| Control | 4.3±0.5 | 6.6±0.8 | 5.9±0.4 | |
| DOX | 50.1±3.0 | 52.3±2.6 | 51.5±2.9 | |
The effect of P1 on cell apoptosis induced by DOX was determined using fluorescence-activated cell sorting. Values are percentages. (A) For MOLT4 cells, P1 significantly increased number of apoptotic cells induced by DOX
P<0.05 (0.1 nmol/ml P1 and DOX group vs. only DOX group)
P<0.01 (1.0 nmol/ml P1 and DOX group vs. only DOX group). (B) For RBL-2H3 cells, no significant difference between groups treated with P1 and DOX compared with only DOX was identified. The data are presented as the mean ± standard deviation. The experiments were carried out three times. DOX, doxorubicin.
P1 inhibits migration of MOLT4 cells
To evaluate the effects of P1 on migration of MOLT4 cells, a Boyden chamber migration assay was performed. As presented in Fig. 4, P1 significantly inhibited the migration of CCR9-expressing MOLT4 cells induced by CCL25 in a dose-dependent manner. No statistical comparison between the different doses was performed. In contrast, the control P did not affect the migration of MOLT4 cells induced by CCL25. A statistically significant difference between P1 and group control P was identified (P<0.05), revealing that P1 may antagonize the effect of CCL25.
Figure 4.
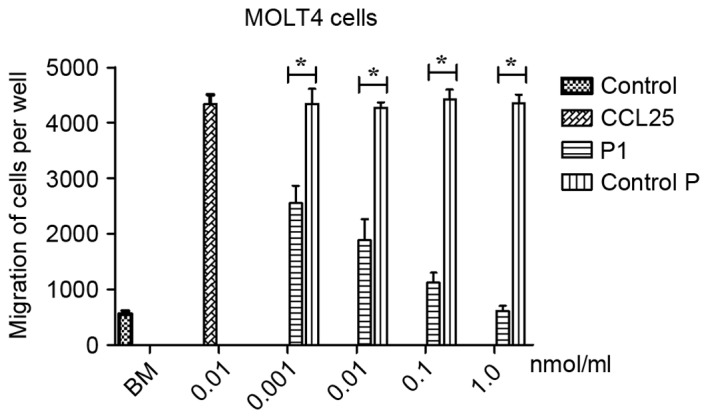
Chemotaxis of MOLT4 cells in the presence of P1. P1 may inhibit the migration of MOLT4 cells induced by CCL25. In contrast, control P had no effect on the migration of MOLT4 cells induced by CCL25 (*P<0.05). Spontaneous BM was used as a control; CCL25 was used as a positive control. BM, baseline migration.
P1 inhibits the phosphorylation of Akt in MOLT4 cells
As presented in Fig. 5, P1 caused a detectable inhibitory effect on the expression of p-Akt in MOLT4 cells. The level of p-Akt was significantly different when compared with Akt as a control. Each dose of P1 was also significantly different from the subsequent dose, indicating that the effect of P1 is dose-dependent (P<0.05). This result implies that P1 may block the downstream CCR9 signaling pathway.
Figure 5.
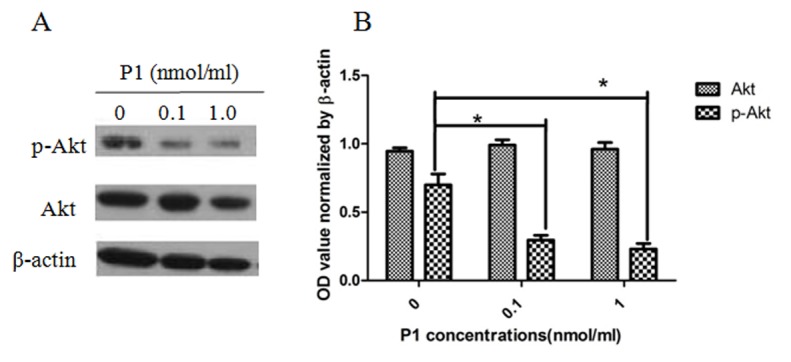
P1 inhibits the phosphorylation of Akt in MOLT4 cells. (A) The levels of Akt and p-Akt in MOLT4 cells were detected using western blot analysis following P1 treatment. (B) OD of Akt and p-Akt, normalized using β-actin. *P<0.05. Akt, protein kinase B; p-Akt, phospho-Akt; OD, optical density.
Discussion
Phage display has been proved to be an effective approach for screening the interaction of molecules, and to develop novel agents for the diagnosis and treatment of numerous types of cancer (16). In the present study, the second loop of CCR9 was used as the target molecule to screen the Ph.D.-12 phage display library for the first time, to the best of our knowledge. Following three rounds of biopanning, 8 positive phage clones expressing exogenous sequences were obtained. Of these positive clones, C-4 appeared five times and exhibited increased affinity compared with other phages detected by ELISA. Furthermore, the results revealed that P1 (VHWDFRQWWQPS) derived from C-4 exhibits specificity towards and affinity for the second extracellular loop of CCR9.
To investigate the ability of P1 binding to cells under physiological conditions, a fluorescently labeled peptide (FITC-P1) was synthesized and confocal microscopy analysis was performed. It was demonstrated that FITC-P1 may bind specifically to MOLT4 cells through CCR9. These results suggest that P1 may specifically bind to the second loop of CCR9.
CCR9 may serve a role in the drug resistance and metastasis of tumors. Furthermore, CCR9 may increase migration and invasion of tumor cells, and activate of PI3K/Akt signaling pathway in order to inhibit the apoptosis of tumor cells as induced by chemotherapeutic agents (5,8). Experiments were performed to determine the effect of P1 on CCR9-overexpressing cells and the phosphorylation of Akt. As expected, the results indicate that P1 may increase the apoptosis of MOLT4 cells as induced by chemotherapeutic agents. Furthermore, P1 may inhibit the migration of CCR9-expressing MOLT4 cells induced by CCL25. Furthermore, it was revealed that P1 may significantly decrease phosphorylation of Akt. The results of the present study identified that P1 may have an effect on CCR9-overexpressing cells, increasing their susceptibility to chemotherapy and decreasing chemotaxis.
In conclusion, a novel peptide, P1 (VHWDFRQWWQPS), was developed which may inhibit the activity of CCR9. P1 may possess potential as a novel treatment for types of cancer which overexpress CCR9. However, further investigation into the use of P1 as an inhibitor for carcinoma, including the plasma half-life, tissue penetration ability, immunogenicity and in vivo antitumor effect is required.
Acknowledgements
This study was supported, in part, by the Natural Science Foundation of Fujian Province, China (grant no. 2013D012) and National Science Foundation of China (grant no. 81371902). The authors thank Mr. Fuyi Huang (Institute of Urban Environment, Chinese Academy of Sciences, Xiamen, China) for support with technology.
References
- 1.Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 2.Nagakubo D, Jin Z, Hieshima K, Nakayama T, Shirakawa AK, Tanaka Y, Hasegawa H, Hayashi T, Tsukasaki K, Yamada Y, et al. Expression of CCR9 in HTLV-1+T cells and ATL cells expressing tax. Int J Cancer. 2007;120:1591–1597. doi: 10.1002/ijc.22483. [DOI] [PubMed] [Google Scholar]
- 3.Heinrich EL, Arrington AK, Ko ME, Luu C, Lee W, Lu J, Kim J. Paracrine activation of chemokine receptor CCR9 enhances the invasiveness of pancreatic cancer cells. Cancer Microenviron. 2013;6:241–245. doi: 10.1007/s12307-013-0130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta P, Sharma PK, Mir H, Singh R, Singh N, Kloecker GH, Lillard JW, Jr, Singh S. CCR9/CCL25 expression in non-small cell lung cancer correlates with aggressive disease and mediates key steps of metastasis. Oncotarget. 2014;5:10170–10179. doi: 10.18632/oncotarget.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma PK, Singh R, Novakovic KR, Eaton JW, Grizzle WE, Singh S. CCR9 mediates PI3K/AKT-dependent antiapoptotic signals in prostate cancer cells and inhibition of CCR9-CCL25 interaction enhances the cytotoxic effects of etoposide. Int J Cancer. 2010;127:2020–2030. doi: 10.1002/ijc.25219. [DOI] [PubMed] [Google Scholar]
- 6.Singh S, Singh UP, Stiles JK, Grizzle WE, Lillard JW., Jr Expression and functional role of CCR9 in prostate cancer cell migration and invasion. Clin Cancer Res. 2004;10:8743–8750. doi: 10.1158/1078-0432.CCR-04-0266. [DOI] [PubMed] [Google Scholar]
- 7.Chen HJ, Edwards R, Tucci S, Bu P, Milsom J, Lee S, Edelmann W, Gumus ZH, Shen X, Lipkin S. Chemokine 25-induced signaling suppresses colon cancer invasion and metastasis. J Clin Invest. 2012;122:3184–3196. doi: 10.1172/JCI62110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu Z, Xiao R, Xiong J, Tembo KM, Deng X, Xiong M, Liu P, Wang M, Zhang Q. CCR9 in cancer: Oncogenic role and therapeutic targeting. J Hematol Oncol. 2016;9:10. doi: 10.1186/s13045-016-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, Zhang L, Wu R, Han R, Jia Y, Jiang Z, Cheng M, Gan J, Tao X, Zhang Q. Specific killing of CCR9 high-expressing acute T lymphocytic leukemia cells by CCL25 fused with PE38 toxin. Leuk Res. 2011;35:1254–1260. doi: 10.1016/j.leukres.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Brasnjevic I, Steinbusch HW, Schmitz C, Martinez-Martinez P. European NanoBioPharmaceutics research initiative: Delivery of peptide and protein drugs over the blood-brain barrier. Prog Neurobiol. 2009;87:212–251. doi: 10.1016/j.pneurobio.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Wang FY, Zhang TY, Luo JX, He GA, Gu QL, Xiao F. Selection of CC chemokine receptor 5-binding peptide from a phage display peptide library. Biosci Biotechnol Biochem. 2006;70:2035–2041. doi: 10.1271/bbb.50654. [DOI] [PubMed] [Google Scholar]
- 12.Skelton NJ, Chen YM, Dubree N, Quan C, Jackson DY, Cochran A, Zobel K, Deshayes K, Baca M, Pisabarro MT, et al. Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1. Biochemistry. 2001;40:8487–8498. doi: 10.1021/bi0103866. [DOI] [PubMed] [Google Scholar]
- 13.Tang B, Li Z, Huang D, Zheng L, Li Q. Screening of a specific peptide binding to VPAC1 receptor from a phage display peptide library. PLoS One. 2013;8:e54264. doi: 10.1371/journal.pone.0054264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houimel M, Mazzucchelli L. Identification of biologically active peptides that inhibit binding of human CXCL8 to its receptors from a random phage-epitope library. J Leukoc Biol. 2009;85:728–738. doi: 10.1189/jlb.0608380. [DOI] [PubMed] [Google Scholar]
- 15.Li B, Wang Z, Zhong Y, Lan J, Li X, Lin H. CCR9-CCL25 interaction suppresses apoptosis of lung cancer cells by activating the PI3K/Akt pathway. Med Oncol. 2015;32:66. doi: 10.1007/s12032-015-0531-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Yin G, Yan D, Wei Y, Ma C, Huang Z, Liao X, Yao Y, Chen X, Hao B. In vitro screening of ovarian tumor specific peptides from a phage display peptide library. Biotechnol Lett. 2011;33:1729–1735. doi: 10.1007/s10529-011-0634-4. [DOI] [PubMed] [Google Scholar]