Abstract
It has been suggested that the tumor microenvironment plays an important role in tumor progression, acquisition of androgen independence, and distant metastasis in prostate cancer (PC). However, little is known about the transcriptional basis of cellular interactions in the human PC microenvironment. To clarify the mechanism of PC progression and metastasis, we investigated the interaction of PC, epithelial, and stromal cells using genome-wide gene expression profiling. We hypothesized that PC cells could induce stromal cells to differentiate into so-called cancer-associated fibroblasts (CAFs), which might contribute to cancer invasion and metastasis. Genes upregulated in normal human prostate stromal cells (PrSC) co-cultured with human PC cells (LNCaP) included the mevalonate pathway enzymes 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). Knockdown of endogenous HMGCS1 or HMGCR in PC cells by shRNA resulted in a significant reduction of PC cell viability. Importantly, exogenous overexpression of HMGCS1 or HMGCR in either PC cells or prostate stromal cells stimulated PC cell growth, suggesting a possible autocrine/paracrine mechanism of action. Immunohistochemical analysis confirmed that HMGCS1 and HMGCR were overexpressed in PC stroma, especially in early stage PC. These results provide clues to the molecular mechanisms underlying PC invasion and metastasis, and suggest that HMGCS1 and HMGCR in PC, as well as in PC stroma, might serve as molecular targets for the treatment of PC.
Keywords: HMGCS1, HMGCR, mevalonate pathway, prostate cancer, stroma
Introduction
The prognosis of prostate cancer (PC) depends greatly on tumor invasion and distant metastasis. In PC it has been suggested that the tumor microenvironment plays an important role in progression, acquisition of androgen independence, and distant metastasis (1–3). We previously conducted a genome-wide loss of heterozygosity/allelic imbalance (LOH/AI) scan of DNA from the epithelium and stroma of 116 PCs and identified a total of 43 LOH/AI hot/cold spots, of which 17 were associated with both the epithelium and stroma, 18 were unique to the epithelium, and eight were unique to the stroma (4). We also identified 15 LOH/AI markers that were correlated with Gleason scores in that study. However, our understanding of the role of the microenvironment in human PC remains limited.
Epithelial-to-mesenchymal transition (EMT) has gained considerable attention as a conceptual paradigm to explain invasive and metastatic behavior during cancer progression (5). It has been proposed that EMT is induced by cancer cells during metastatic dissemination from a primary organ to secondary sites, but precisely how EMT occurs during PC invasion and metastasis remains uncertain.
To clarify the mechanism of tumor progression and metastasis in PC, and the role of the tumor microenvironment, we investigated the molecular interaction of PC cells, prostatic epithelium, and prostatic stroma through genome-wide gene expression profiling. We hypothesized that PC cells might act on stromal cells to induce their differentiation into cancer-associated fibroblasts (CAFs), thus contributing to tumor invasion and metastasis. Likewise, we hypothesized that CAFs could act on surrounding normal epithelial cells to change their characteristics into PC cells.
Materials and methods
Cell lines
The human PC cell lines LNCaP, PC-3, and 22Rv1 were obtained from the American Type Culture Collection (Rockville, MD, USA). The human normal prostate epithelial cell line PrEC and the human normal prostate stromal cell line PrSC were purchased from Lonza Group Ltd. (Basel, Switzerland). Cells were cultured as monolayers in appropriate medium supplemented with 10% fetal bovine serum (FBS), and maintained at 37°C in an atmosphere of humidified air with 5% CO2.
Co-culture experiments
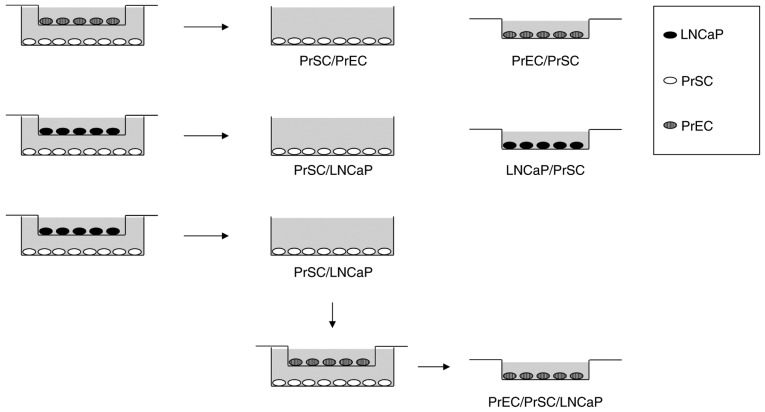
Co-culture experiments were performed as shown in Fig. 1. LNCaP, PrEC, and PrSC cell lines were cultured in 6-well plates or culture plate inserts (0.4 µm pore size; Corning Inc., Corning, NY, USA). On day 1, PrSC (2×105 cells/well) were plated onto 6-well plates using SCGM media (Lonza Group Ltd.) while PrEC (2×105 cells/well) and LNCaP (1×105 cells/well) were plated onto culture plate inserts using PrEGM (Lonza Group Ltd.) or DMEM plus 10% FBS media, respectively. On day two, culture media were replaced with keratinocyte-SFM (Thermo Fisher Scientific, Inc., Waltham, MA, USA) plus 2% FBS and the inserts containing PrEC or LNCaP were then transferred into 6-well plates containing PrSC cells to initiate the experiment. On day five, total RNA was isolated for microarray analysis. For PrEC/PrSC/LNCaP co-culture, the inserts containing PrEC were transferred into 6-well plates containing PrSC previously co-cultured with LNCaP (PrSC/LNCaP), and cultured for three days.
Figure 1.
Schematic illustration of co-culture experiments. PrSC/PrEC, PrSC co-cultured with PrEC; PrEC/PrSC, PrEC co-cultured with PrSC; PrSC/LNCaP, PrSC co-cultured with LNCaP; LNCaP/PrSC, LNCaP co-cultured with PrSC; and PrEC/PrSC/LNCaP, PrEC co-cultured with PrSC/LNCaP were used for subsequent microarray analysis. PrEC, normal human prostate epithelial cells; PrSC, normal human prostate stromal cells.
cDNA microarray analysis and data acquisition
Total RNA was extracted using the RNeasy Micro kit (Qiagen GmbH, Hilden, Germany) after co-culture experiments, according to the manufacturer's instructions. Briefly, total RNA was reverse-transcribed to cDNA with T7-Oligo(dT) primer. The cDNA synthesis product was used in an in vitro transcription (IVT) reaction involving T7 RNA polymerase. An unlabeled ribonucleotide mix was used in the first cycle of IVT amplification. Unlabeled cRNA was then reverse-transcribed in the first-strand cDNA synthesis step of the second cycle using random primers. Subsequently, the T7-Oligo(dT) promoter primer was used for the second-strand cDNA synthesis to generate a double-stranded cDNA template containing T7 promoter sequences. The resultant double-stranded cDNA was then amplified and labeled using a biotinylated nucleotide analog/ribonucleotide mix in the second IVT reaction. The labeled cRNA products were then fragmented, loaded onto the GeneChip® Human Genome U133 Plus 2.0 array (Affymetrix; Thermo Fisher Scientific, Inc.), and hybridized according to the manufacturer's instructions. GeneChip® array data were compared using the Kurabo custom analysis service (Kurabo Industries Ltd., Osaka, Japan; Kurabo Industries Ltd. is an authorized service provider for Affymetrix Japan K.K., Tokyo, Japan). Differences in gene expression were assessed using the following comparisons: PrSC/LNCaP vs. PrSC/PrEC, LNCaP/PrSC vs. LNCaP, and PrEC/PrSC/LNCaP vs. PrEC/PrSC (Fig. 1; PrSC/LNCaP, PrSC co-cultured with LNCaP; PrSC/PrEC, PrSC co-cultured with PrEC; LNCaP/PrSC, LNCaP co-cultured with PrSC; PrEC/PrSC/LNCaP, PrEC co-cultured with PrSC/LNCaP; PrEC/PrSC, PrEC co-cultured with PrSC). Raw intensity data from the GeneChip® array were analyzed using the GeneChip® Operating Software (Affymetrix; Thermo Fisher Scientific, Inc.) and hierarchical clustering analyses were conducted using Cluster and TreeView software (http://rana.lbl.gov/EisenSoftware.htm).
RNA isolation and semi-quantitative RT-PCR
Total RNA was isolated using the RNeasy Micro kit (Qiagen GmbH) from PC, PrEC, and PrSC cells according to the manufacturer's instructions and reverse-transcribed using Superscript II Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) with random primers, prior to performing PCR. PCR primers were: 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1; forward, 5′-CTCCCTGACGTGGAATGTCT-3′; reverse, 5′-GAACTGTCTGCCCAGGTGAT-3′), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR; forward, 5′-CTTGCCGAGCCTAATGAAAG-3′; reverse, 5′-TGACCCCCTGAGAAAGCTAA-3′), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; forward, 5′-CGGATTTGGTCGTATTGG-3′; reverse: 5′-TCCTGGAAGATGGTGATG-3′). PCR conditions were as follows: initial denaturation at 94°C for 9 min, followed by 28–30 cycles of denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, and elongation at 72°C for 60 sec on a C1000™ Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). Relative expression levels of mRNA were calculated in comparison to those of GAPDH.
Small hairpin RNA-expressing constructs and cell viability assay
We used SureSilencing short hairpin RNA (shRNA) plasmids (Qiagen GmbH) for examining RNA interference effects on the target genes. The target sequences of HMGCS1 and HMGCR were 5′-GAAGGAACGTGGTACTTAGTT-3′ (shHMG CS1) and 5′-CAAGGAGCATGCAAAGATAAT-3′ (shHMG CR), respectively. The scrambled sequence 5′-GGAATCTCATTCGATGCATAC-3′, which does not match any human, mouse, or rat gene, was used as a negative control (shNC vector). 22Rv1 cells that highly expressed both HMGCS1 and HMGCR were plated onto 10 cm dishes (2×106 cells/dish) and transfected with 6 µg of each shRNA plasmids (shHMG CS1, shHMG CR, or shNC) using FuGENE6 reagent (Promega Corporation, Madison, WI, USA) according to the supplier's protocol. Cells were selected in culture medium containing 1.0 mg/ml geneticin for 10 days, fixed with 100% methanol, and stained with 0.1% crystal violet to evaluate colony formation. Cell viability was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, with absorbance measured at 570 and 630 nm as a reference using a microplate reader (THERMOmax; Molecular Devices, LLC, Sunnyvale, CA, USA). Knockdown effects of these shRNA plasmids on endogenous HMGCS1 or HMGCR expression were validated 48 h following their transfection, by RT-PCR using the primers described above.
Autocrine/paracrine cell proliferation assay
Full-length human HMGCS1 and HMGCR cDNA (accession nos. NM002130 and NM000859) were amplified and cloned into the pcDNA3.1 (+) vector (Invitrogen; Thermo Fisher Scientific, Inc.). To examine the autocrine effect of HMGCS1 and HMGCR expression on PC cell growth, 22Rv1 cells were seeded into 6-well plates (5×105 cells/well) and transfected with pcDNA3.1 (+) empty vector (mock), pcDNA3.1 (+)-HMGCS1, or pcDNA3.1 (+)-HMGCR expression vectors at a final concentration of 0.6 µg/ml using FuGENE6 reagent (Promega), according to the manufacturer's instructions. The proliferation of 22Rv1 cells overexpressing HMGCS1 or HMGCR, or mock-transfected cells, was examined using a cell counter (TC10™ Automated Cell Counter; Bio-Rad Laboratories) during days 1–10. To examine the paracrine effect of HMGCS1 and HMGCR on PC cell growth, PrSC cells (1×105 cells/well) transfected with pcDNA3.1 (+) empty vector (mock), pcDNA3.1 (+)-HMGCS1, or pcDNA3.1 (+)-HMGCR expression vector at a final concentration of 0.6 µg/ml and selected with 1.0 mg/ml geneticin, were plated onto culture plate inserts, while 22Rv1 cells (1×105 cells/well) were grown on 6-well plates. The following day, culture media was replaced with keratinocyte-SFM (Thermo Fisher Scientific, Inc.) plus 2% FBS and the inserts containing transfected PrSC cells transferred into the 6-well plates containing the 22Rv1 cells to initiate co-culture. Growth of 22Rv1 cells was calculated during days 1–14 using a cell counter.
Tissue microarray samples and immunohistochemical study
To further investigate HMGCS1 and HMGCR expression in a larger number of tumor specimens, tissue microarray samples containing 80 cases of PC and 16 normal prostate tissue, in duplicate cores per case (PR1921; US Biomax, Inc., Rockville, MD, USA) were obtained. The deparaffinized tissue sections were heated in a microwave for 5 min for antigen retrieval. These sections were incubated with a 1:200 diluted solution of a rabbit anti-HMGCS1 polyclonal antibody (ab87246; Abcam, Cambridge, UK) or a 1:50 diluted solution of a mouse anti-HMGCR monoclonal antibody (C-1, sc-271595; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) overnight at 4°C and developed with peroxidase labeled-dextran polymer followed by diaminobenzidine (Dako EnVision+ System; Dako, Carpinteria, CA, USA). The sections were counterstained with hematoxylin. For negative controls, primary antibody was omitted. The stromal expression levels of HMGCS1 and HMGCR were determined by calculating average intensities of positive stromal cells, using a BZ-X Analyzer (Keyence Corporation, Osaka, Japan). Three random fields were analyzed with a magnification of 400x. Correlations between HMGCS1/HMGCR expression levels and clinicopathological variables (tissue type, Gleason grade, tumor stage, tumor classification, lymph node metastasis, distant metastasis, and prostate-specific antigen (PSA) expression levels) were evaluated using the Mann-Whitney U and Kruskal-Wallis tests. All statistical analyses were performed using the software JMP® (SAS Institute Inc., Cary, NC, USA). P<0.05 was considered to indicate a statistically significant differences.
Results
Candidate genes identified by cDNA microarray and cluster analysis
Before microarray analysis, we confirmed the overexpression of α-smooth muscle actin (ACTA2) in PrSC/LNCaP, which indicated the transition from normal stromal cells to CAFs (data not shown). We identified 10 genes that were upregulated in PrSC/LNCaP relative to PrSC/PrEC (signal log ratio >2.0), which included insulin induced gene 1 (INSIG1), HMGCS1, solute carrier family 14 member 1 (SLC14A1), and noggin (NOG; Table I). SLC14A1 has been reported to be regulated by androgens and potentially involved in prostate carcinogenesis (6), while NOG is involved in EMT (7). INSIG1 mediates feedback control of cholesterol synthesis by binding sterol regulatory element-binding protein (SREBP) cleavage-activating protein (SCAP) and HMGCR. HMGCR is the key enzyme of the mevalonate pathway and these facts prompted us to investigate the mevalonate pathway enzymes HMGCS1 and HMGCR together, although the signal log ratio of HMGCR was less than 2.0 (signal log ratio=1.4). We also identified 11 PC-specific genes that were upregulated in LNCaP/PrSC relative to LNCaP (signal log ratio >3.0), which included fatty acid synthase (FASN) and α-methylacyl-CoA racemase (AMACR; Table II). We additionally identified six genes that were upregulated in PrEC/PrSC/LNCaP relative to PrEC/PrSC (signal log ratio >4.0), which included the oncogene mitogen-activated protein kinase kinase kinase 8 (MAP3K8; Table III).
Table I.
Upregulated genes in PrSC/LNCaP compared with PrSC/PrEC.
| Gene symbol | Gene title | Function |
|---|---|---|
| SCD | Stearoyl-CoA desaturase (delta-9-desaturase) | Fatty acid synthesis |
| INSIG1 | Insulin induced gene 1 | Cholesterol synthesis, steroid metabolism |
| HMGCS1 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) | Cholesterol synthesis, lipid metabolism |
| HTR2B | 5-hydroxytryptamine (serotonin) receptor 2B | Serotonin receptor signaling pathway |
| OXTR | Oxytocin receptor | G-protein coupled receptor |
| PTHLH | Parathyroid hormone-like hormone | Inhibitor of osteoclastic bone resorption |
| IGFBP3 | Insulin-like growth factor binding protein 3 | Cell growth regulation |
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | Retinol metabolism |
| SLC14A1 | Solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | Urea transport |
| NOG | Noggin | BMP signaling pathway, EMT |
BMP, bone morphogenetic protein; EMT, epithelial-to-mesenchymal transition.
Table II.
Upregulated genes in LNCaP/PrSC compared with LNCaP.
| Gene symbol | Gene title | Function |
|---|---|---|
| EGR1 | Early growth response 1 | Transcriptional regulation, BMP signaling pathway |
| SOX9 | SRY (sex determining region Y)-box 9 | Skeletal development, PKB signaling cascade |
| PLA2G2A | Phospholipase A2, group IIA (platelets, synovial fluid) | phospholipid metabolism |
| STC1 | Stanniocalcin 1 | Renal phosphate reabsorption |
| DIO1 | Deiodinase, iodothyronine, type I | Hormone synthesis |
| FASN | Fatty acid synthase | Fatty acid synthesis |
| GINS2 | GINS complex subunit 2 (Psf2 homolog) | DNA replication |
| NFKBIZ | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | Regulation of NF-kappa-B |
| UHRF1 | Ubiquitin-like with PHD and ring finger domains 1 | DNA repair |
| AMACR | α-methylacyl-CoA racemase | Lipid metabolism |
| CLEC7A | C-type lectin domain family 7, member A | Innate immunity |
BMP, bone morphogenetic protein; PKB, protein kinase B.
Table III.
Upregulated genes in PrEC/PrSC/LNCaP compared with PrEC/PrSC.
| Gene symbol | Gene title | Function |
|---|---|---|
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | Oncogene |
| GABBR1 | γ-aminobutyric acid (GABA) B receptor, 1 | G-protein coupled receptor |
| TLR1 | Toll-like receptor 1 | Innate immunity |
| SOD2 | Superoxide dismutase 2, mitochondrial | Superoxide family member, mutated in cancer |
| HERC5 | Hect domain and RLD 5 | Interferon-induced E3 protein ligase |
| CMPK2 | Cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial | dUTP and dCTP synthesis in mitochondria |
Unsupervised hierarchical clustering using full gene expression profiles showed that the samples clustered into three distinct groups related to LNCaPs, PrSCs and PrECs, respectively (Fig. 2A). Interestingly, PrECs clustered close to LNCaPs when clustering analysis was restricted to the 767 genes most significantly upregulated or downregulated (i.e., with a log ratio of >2.0 or <-2.0) in PrEC/PrSC/LNCaP, that is PrEC co-cultured with CAFs (Fig. 2B).
Figure 2.
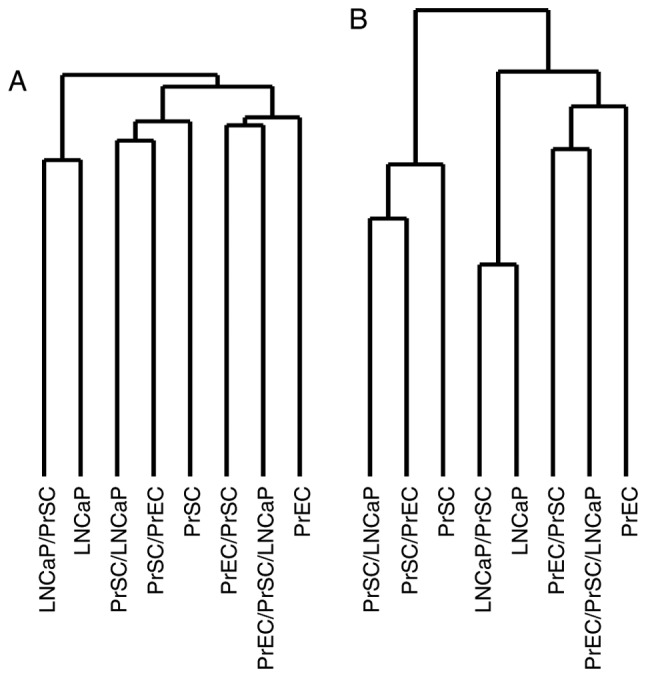
Unsupervised hierarchical clustering of cells based on gene expression profiles. (A) Analysis based on the full gene expression dataset revealed that samples clustered into three distinct groups (i.e., LNCaPs, PrSCs, and PrECs); (B) PrECs clustered close to LNCaPs when analysis was restricted to the 767 genes most significantly up- or downregulated in PrEC/PrSC/LNCaP. PrEC, normal human prostate epithelial cells; PrSC, normal human prostate stromal cells.
Knockdown of HMGCS1 or HMGCR expression by shRNA suppresses PC cell viability
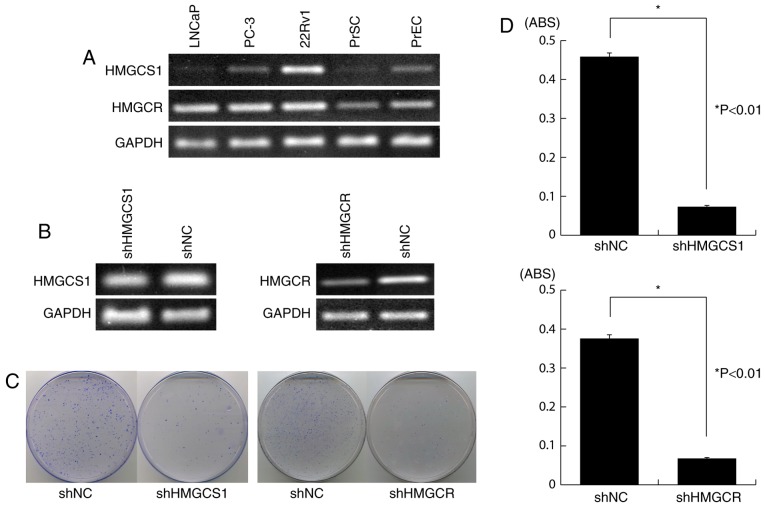
To investigate the biological role of HMGCS1 and HMGCR in PC cells, we knocked down their endogenous expression in 22Rv1 cells (Fig. 3A) using vector-based RNA interference technology. Transfection of the shRNA-expressing vectors shHMGCS1 or shHMGCR, clearly reduced endogenous expression of HMGCS1 and HMGCR, respectively (Fig. 3B), and resulted in significant growth suppression as measured by both colony formation assay and MTT assay (P<0.01; Fig. 3C and D, respectively). By contrast, transfection of the negative control vector (shNC) had little or no effect on HMGCS1 or HMGCR expression and did not affect the viability of 22Rv1 cells (Fig. 3B-D).
Figure 3.
Knockdown of HMGCS1 or HMGCR expression by shRNA attenuates PC cell viability. (A) Semi-quantitative RT-PCR showed that HMGCS1 and HMGCR were overexpressed in 22Rv1 cells, compared with normal prostate cells. GAPDH was used as a control for cDNA content; (B) Semi-quantitative RT-PCR analysis of the knockdown effect on endogenous HMGCS1 or HMGCR expression in 22Rv1 cells, following transfection of shRNA-expressing vectors (shHMGCS1 or shHMGCR) or the negative control vector, shNC; GAPDH was used as a quantitative control; (C) Colony formation assay showing decrease in the number of colonies in 22Rv1 cells transfected with shHMGCS1 or shHMGCR vector, compared to control (shNC); (D) MTT assay demonstrating significant suppression of PC cell viability following transfection of shHMGCS1 or shHMGCR vector (mean ± standard error). Experiments were carried out in triplicate, with statistical analysis performed by Student's t-test (*P<0.01). ABS, absorbance; PrEC, normal human prostate epithelial cells; PrSC, normal human prostate stromal cells; sh, short hairpin; NC, negative control; HMGCS1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; RT-PCR, reverse transcription-polymerase chain reaction; PC, prostate cancer.
HMGCS1 or HMGCR overexpression promotes PC cell proliferation through autocrine/paracrine regulation
To further investigate the potential oncogenic function of HMGCS1 and HMGCR, we examined the autocrine/paracrine effects of these proteins on PC cell growth. The cell proliferation assay revealed that 22Rv1 cells with exogenous overexpression of HMGCR grew more rapidly than 22Rv1 mock-transfected cells, indicating that HMGCR overexpression promotes PC cell proliferation in an autocrine fashion. HMGCS1 overexpression however, did not significantly stimulate PC cell growth (data not shown). We further examined the influence of HMGCS1 and HMGCR overexpression in PC stroma on PC cell growth using a co-culture experiment. Overexpression of HMGCS1 or HMGCR in PC stromal cells was found to induce a significantly higher growth rate of 22Rv1 cells than those that were mock transfected (P<0.01; Fig. 4), indicating a paracrine effect.
Figure 4.
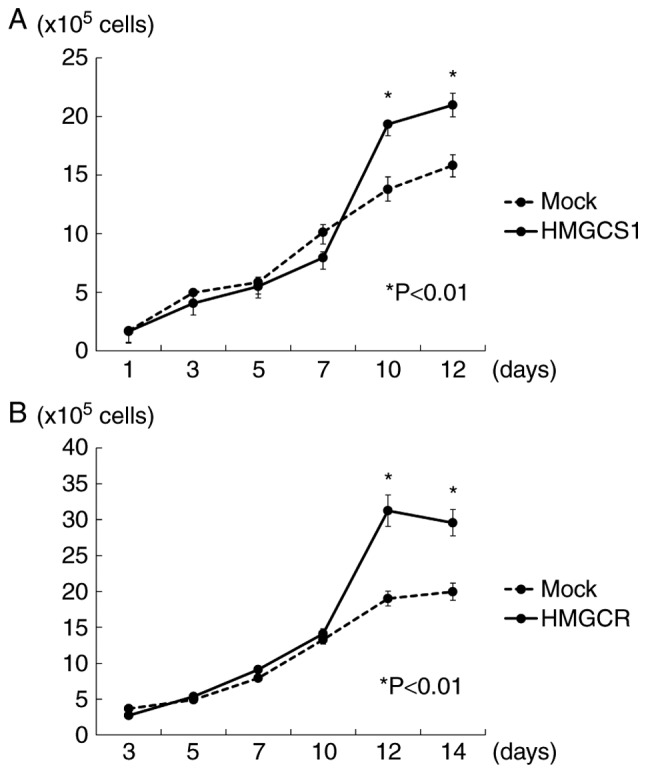
HMGCS1 and HMGCR promote prostate cancer cell proliferation through paracrine regulation. (A and B) In vitro growth curves (mean ± SE) revealed that 22Rv1 cells co-cultured with HMGCS1- or HMGCR-overexpressing PrSC cells (transfected with HMGCS1 or HMGCR expression vectors), grew more rapidly than those co-cultured with PrSC mock transfectants. Experiments were carried out in triplicate with statistical analysis performed by Student's t-test (*P<0.01). HMGCS1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase.
Immunohistochemical analysis of primary PC specimens
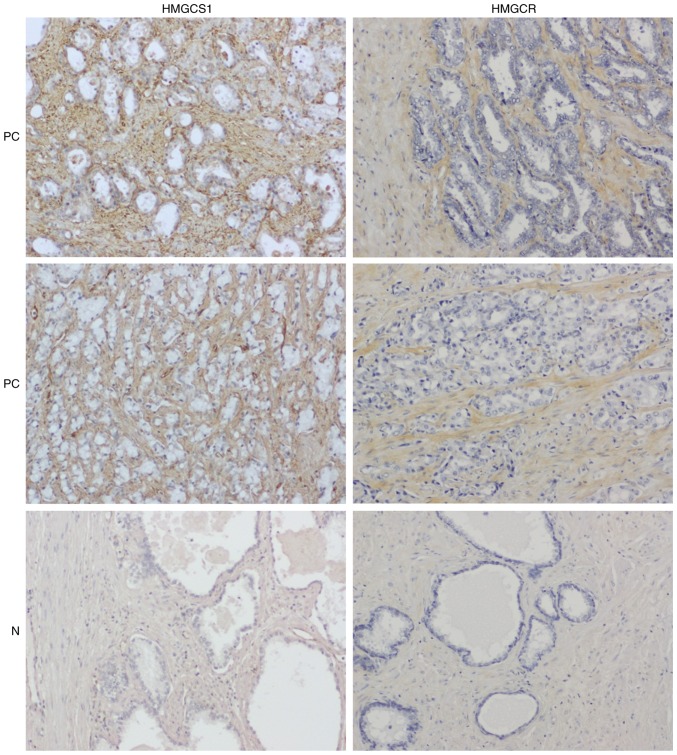
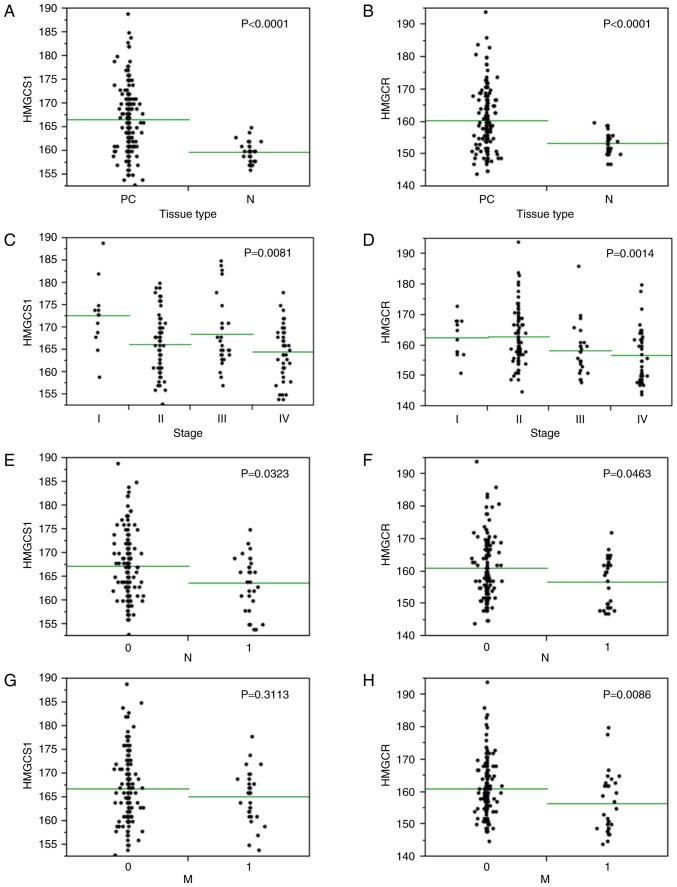
We subsequently performed immunohistochemical staining of 80 primary PC and 16 normal cases on tissue microarrays (a total of 192 cores) with anti-HMGCS1 or anti-HMGCR antibodies. Immunohistochemistry confirmed the overexpression of HMGCS1 and HMGCR in PC stroma compared with normal prostatic stroma (P<0.0001; Figs. 5, 6A and B). To investigate the clinicopathological significance of HMGCS1 and HMGCR stromal expression in PC tissues, we next analyzed the relationship between HMGCS1 or HMGCR stromal expression and the clinicopathological variables of PC specimens, which is summarized in Table IV. This analysis revealed a significant association between HMGCS1/HMGCR stromal expression and several clinicopathological factors. Most notably, we observed that HMGCS1 and HMGCR expression levels in PC stroma were inversely associated with tumor stage (P<0.01; Fig. 6C and D), with higher stage PCs associated with lower stromal expression levels of HMGCS1 and HMGCR. Likewise, HMGCS1 and HMGCR stromal expression was significantly lower in cases with lymph node metastasis (P<0.05; Fig. 6E and F). HMGCR stromal expression was also significantly lower in cases with distant metastasis (P<0.01; Fig. 6G), however HMGCS1 stromal expression levels had no correlation with distant metastatic status (P=0.3113; Fig. 6H). There was no relationship between HMGCS1 or HMGCR stromal expression and Gleason grade, tumor classification, or PSA expression levels (data not shown).
Figure 5.
Immunohistochemical stainings demonstrate overexpression of HMGCS1 and HMGCR in PC stroma. Representative stainings of PC and normal prostate specimens are shown (magnification, ×100). PC, prostate cancer; N, normal prostate; HMGCS1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase.
Figure 6.
Immunohistochemical analyses of PC specimens. (A) HMGCS1 and (B) HMGCR stromal expression was significantly upregulated in PC in comparison with normal prostate (P<0.0001); (C) HMGCS1 and (D) HMGCR expression in the PC stroma of higher stage cases was decreased compared with that of lower stage cases (P<0.01); (E) HMGCS1 and (F) HMGCR stromal expression was significantly lower in cases with lymph node metastasis (P<0.05). (G) There was no relationship between the stromal expression of HMGCS1 and distant metastatic status (P=0.3113), however the stromal expression of (H) HMGCR was significantly lower in cases with distant metastasis (P<0.01). In each panel, the x-axis shows clinicopathological variables and the y-axis indicates HMGCS1 or HMGCR stromal expression. Statistical analysis was performed using the Mann-Whitney U and Kruskal-Wallis tests. HMGCS1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; PC, prostate cancer; N, normal prostate.
Table IV.
Clinicopathological variables of PC cases.
| Number of cases (cores) | 80 (160) |
|---|---|
| Age, years [median (range)] | 69.5 (51–85) |
| Gleason grade, n, per core | |
| 3 | 16 |
| 4 | 74 |
| 5 | 60 |
| Tumor stage, n, per case | |
| I | 6 |
| II | 37 |
| III | 14 |
| IV | 22 |
| Tumor classification, n, per case | |
| T1 | 4 |
| T2 | 47 |
| T3 | 22 |
| T4 | 6 |
| Lymph node metastasis, n, per case | |
| Positive | 15 |
| Negative | 64 |
| Distant metastasis, n, per case | |
| Positive | 15 |
| Negative | 64 |
| PSA expression level (IHC), n, per core | |
| − | 52 |
| + | 28 |
| ++ | 45 |
| +++ | 32 |
Some data are missing in each clinicopathological variable. PC, prostate cancer; PSA, prostate-specific antigen; IHC, immunohistochemistry.
Discussion
Our data suggest that stromal cells may influence PC cell gene expression and contribute to their aggressiveness. In keeping with this concept, AMACR, which is widely used as a biomarker for PC, was included in the genes that were upregulated in PC cells co-cultured with prostate stromal cells. The cluster analyses support our hypothesis that CAFs might induce surrounding normal epithelial cells to change their characteristics towards PC cells. Interestingly, an oncogene, MAP3K8, was included among the genes that were upregulated in prostate epithelial cells when co-cultured with cancer-associated prostate stromal cells (i.e., prostate stromal cells previously co-cultured with PC cells). Among the genes identified in this study, we further investigated the mevalonate pathway genes, HMGCS1 and HMGCR. The mevalonate pathway is included in lipid metabolism and best known as the target of statins. There is a clear association between PCs and lipid metabolism or statins, which prompted us to investigate the mevalonate pathway genes, HMGCS1 and HMGCR. Notably, we investigated for the first time the roles of HMGCS1 in human PC cells and tissues. Tables I, II, and III are derived from normal prostate stromal cells, PC cells, and normal prostate epithelial cells, respectively, and these tables indicate that their gene expression could change by co-culture with the different type of cells. Because HMGCS1 and HMGCR were the upregulated genes in stroma, those were not among the genes in Tables II and III, which were derived from PC and epithelium.
HMGCS1 condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, which is the substrate for HMGCR. An association of HMGCS1 with PC has not been previously reported, although links with other cancers have been described (8–14). Lee et al suggested HMGCS1 as one of the candidates involved in tumor stem-like breast cancer cells (8), while Pandyra et al reported that dipyridamole acts as a potentiator of statin anticancer activity in multiple myeloma and acute myelogenous leukemia by attenuating the feedback response that upregulates HMGCS1 and HMGCR after statin treatment (10). In particular, these authors showed that direct targeting of multiple levels of the mevalonate pathway, including blockade the sterol-feedback loop initiated by statin treatment, is an effective and targetable anti-tumor strategy. HMGCS1 has also been linked to drug response or resistance in other studies (14,15). Such observations suggested the possibility that HMGCS1 and HMGCR may similarly represent molecular targets for the treatment of PC.
HMGCR is the rate-limiting enzyme for cholesterol synthesis and the pharmacological target for statins. The association of HMGCR with PC has been previously described in several studies (16–21). It has been reported that PC cells show high expression of HMGCR and that the inhibition of HMGCR with the use of statins lowers the viability of castration-resistant PC cells (17). Li et al identified two miRNAs, miR-185 and miR-342, that control lipogenesis and cholesterogenesis in PC cells by inhibiting SREBP-1 and SREBP-2 expression and downregulating their targeted genes, which includes HMGCR (18). miR-185 and 342 inhibit tumorigenicity, cell growth, migration, and invasion, and induce apoptosis through blockade of the SREBP metabolic pathway in PC cells, representing a novel targeting mechanism for PC therapy.
In the present study, overexpression of HMGCS1 or HMGCR in PC stroma promoted PC cell growth (Fig. 4). However, the stromal expression of HMGCS1 or HMGCR was associated with less aggressive PC (Fig. 6C-H). HMGCS1 and HMGCR might be needed for PC growth until PC progresses to aggressive disease, and then downregulated once PC invasion or metastasis has been developed. Several studies that support our findings have been reported on HMGCR (22–26), although there are no studies available on HMGCS1. Previous studies indicated that high HMGCR expression was associated with less aggressive tumor characteristics and HMGCR expression was a good prognostic marker in breast cancer (23–26). Associations of positive HMGCR expression with more favorable tumor characteristics and a prolonged survival were also shown in other type of cancer such as colorectal cancer (22). We analyzed and provided the stromal expression data of HMGCS1 and HMGCR using clinical PC specimens in this study, since PC specimens showed apparent stromal expression of HMGCS1 and HMGCR (Fig. 5). Unfortunately, there were no follow up data available for the tissue microarray, and we then need to find if HMGCS1 and HMGCR have an influence on survival or progression of PC patients in the future study.
Our findings suggest that overexpression of the mevalonate pathway genes, HMGCS1 and HMGCR is likely to be involved in PC cell growth through an autocrine/paracrine mechanism, supporting their potential of these proteins as molecular targets for PC therapy. Furthermore, immunohistochemical studies indicate that HMGCS1 and HMGCR stromal overexpression might be key factor in regulating the transition from organ-confined to metastatic disease in PC. We used 22Rv1 cell line for further experiments because only 22Rv1 overexpressed both HMHCS1 and HMGCR and should be relevant to these genes, while we confirmed the knockdown effect of shRNA on HMHCS1 and HMGCR by the mRNA expression results (not protein expression), as seen in the many studies published previously (27–32). The HMGCS1 and HMGCR mRNAs were overexpressed in 22Rv1 cells (Fig. 3A) and their knockdown effect by shRNA was validated on mRNA level (Fig. 3B). The use of a single cell line to verify the hypothesis and the absence of data regarding the knockdown effect on protein expression are two limitations of the present study. Further functional analysis of the role of HMGCS1 and HMGCR in regulating the interaction between PC and PC stroma are required, in order to fully elucidate their potential as molecular targets in the treatment of PC.
References
- 1.Chung LW, Baseman A, Assikis V, Zhau HE. Molecular insights into prostate cancer progression: The missing link of tumor microenvironment. J Urol. 2005;173:10–20. doi: 10.1097/01.ju.0000141582.15218.10. [DOI] [PubMed] [Google Scholar]
- 2.Sung SY, Hsieh CL, Law A, Zhau HE, Pathak S, Multani AS, Lim S, Coleman IM, Wu LC, Figg WD, et al. Coevolution of prostate cancer and bone stroma in three-dimensional coculture: Implications for cancer growth and metastasis. Cancer Res. 2008;68:9996–10003. doi: 10.1158/0008-5472.CAN-08-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. J Urol. 2001;166:2472–2483. doi: 10.1016/S0022-5347(05)65620-0. [DOI] [PubMed] [Google Scholar]
- 4.Ashida S, Orloff MS, Bebek G, Zhang L, Zheng P, Peehl DM, Eng C. Integrated analysis reveals critical genomic regions in prostate tumor microenvironment associated with clinicopathologic phenotypes. Clin Cancer Res. 2012;18:1578–1587. doi: 10.1158/1078-0432.CCR-11-2535. [DOI] [PubMed] [Google Scholar]
- 5.Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: Paradigm or puzzle? Nat Rev Urol. 2011;8:428–439. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaarala MH, Hirvikoski P, Kauppila S, Paavonen TK. Identification of androgen-regulated genes in human prostate. Mol Med Rep. 2012;6:466–472. doi: 10.3892/mmr.2012.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kestens C, Siersema PD, Offerhaus GJ, van Baal JW. BMP4 signaling is able to induce an epithelial-mesenchymal transition-like phenotype in barrett's esophagus and esophageal adenocarcinoma through induction of SNAIL2. PLoS One. 2016;11:e0155754. doi: 10.1371/journal.pone.0158755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee WJ, Kim SC, Yoon JH, Yoon SJ, Lim J, Kim YS, Kwon SW, Park JH. Meta-analysis of tumor stem-like breast cancer cells using gene set and network analysis. PLoS One. 2016;11:e0148818. doi: 10.1371/journal.pone.0148818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meerzaman DM, Yan C, Chen QR, Edmonson MN, Schaefer CF, Clifford RJ, Dunn BK, Dong L, Finney RP, Cultraro CM, et al. Genome-wide transcriptional sequencing identifies novel mutations in metabolic genes in human hepatocellular carcinoma. Cancer Genomics Proteomics. 2014;11:1–12. [PubMed] [Google Scholar]
- 10.Pandyra A, Mullen PJ, Kalkat M, Yu R, Pong JT, Li Z, Trudel S, Lang KS, Minden MD, Schimmer AD, Penn LZ. Immediate utility of two approved agents to target both the metabolic mevalonate pathway and its restorative feedback loop. Cancer Res. 2014;74:4772–4782. doi: 10.1158/0008-5472.CAN-14-0130. [DOI] [PubMed] [Google Scholar]
- 11.Pandyra AA, Mullen PJ, Goard CA, Ericson E, Sharma P, Kalkat M, Yu R, Pong JT, Brown KR, Hart T, et al. Genome-wide RNAi analysis reveals that simultaneous inhibition of specific mevalonate pathway genes potentiates tumor cell death. Oncotarget. 2015;6:26909–26921. doi: 10.18632/oncotarget.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Meer DL, Degenhardt T, Väisänen S, de Groot PJ, Heinäniemi M, de Vries SC, Müller M, Carlberg C, Kersten S. Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 2010;38:2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wali VB, Haskins JW, Gilmore-Hebert M, Platt JT, Liu Z, Stern DF. Convergent and divergent cellular responses by ErbB4 isoforms in mammary epithelial cells. Mol Cancer Res. 2014;12:1140–1155. doi: 10.1158/1541-7786.MCR-13-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao M, Li H, Bu X, Lei C, Fang Q, Hu Z. Quantitative proteomic analysis of cellular resistance to the nanoparticle abraxane. ACS Nano. 2015;9:10099–10112. doi: 10.1021/acsnano.5b03677. [DOI] [PubMed] [Google Scholar]
- 15.Rokosz LL, Boulton DA, Butkiewicz EA, Sanyal G, Cueto MA, Lachance PA, Hermes JD. Human cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase: Expression, purification, and characterization of recombinant wild-type and Cys129 mutant enzymes. Arch Biochem Biophys. 1994;312:1–13. doi: 10.1006/abbi.1994.1273. [DOI] [PubMed] [Google Scholar]
- 16.Bull CJ, Bonilla C, Holly JM, Perks CM, Davies N, Haycock P, Yu OH, Richards JB, Eeles R, Easton D, et al. Blood lipids and prostate cancer: A Mendelian randomization analysis. Cancer Med. 2016;5:1125–1136. doi: 10.1002/cam4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JH, Cox ME, Wasan KM. Effect of simvastatin on castration-resistant prostate cancer cells. Lipids Health Dis. 2014;13:56. doi: 10.1186/1476-511X-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Chen YT, Josson S, Mukhopadhyay NK, Kim J, Freeman MR, Huang WC. MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLoS One. 2013;8:e70987. doi: 10.1371/journal.pone.0070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menter DG, Ramsauer VP, Harirforoosh S, Chakraborty K, Yang P, Hsi L, Newman RA, Krishnan K. Differential effects of pravastatin and simvastatin on the growth of tumor cells from different organ sites. PLoS One. 2011;6:e28813. doi: 10.1371/journal.pone.0028813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murtola TJ, Syvälä H, Pennanen P, Bläuer M, Solakivi T, Ylikomi T, Tammela TL. The importance of LDL and cholesterol metabolism for prostate epithelial cell growth. PLoS One. 2012;7:e39445. doi: 10.1371/journal.pone.0039445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai M, Martinez-Arguelles DB, Aprikian AG, Magliocco AM, Papadopoulos V. De novo steroid biosynthesis in human prostate cell lines and biopsies. Prostate. 2016;76:575–587. doi: 10.1002/pros.23146. [DOI] [PubMed] [Google Scholar]
- 22.Bengtsson E, Nerjovaj P, Wangefjord S, Nodin B, Eberhard J, Uhlén M, Borgquist S, Jirström K. HMG-CoA reductase expression in primary colorectal cancer correlates with favourable clinicopathological characteristics and an improved clinical outcome. Diagn Pathol. 2014;9:78. doi: 10.1186/1746-1596-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgquist S, Djerbi S, Pontén F, Anagnostaki L, Goldman M, Gaber A, Manjer J, Landberg G, Jirström K. HMG-CoA reductase expression in breast cancer is associated with a less aggressive phenotype and influenced by anthropometric factors. Int J Cancer. 2008;123:1146–1153. doi: 10.1002/ijc.23597. [DOI] [PubMed] [Google Scholar]
- 24.Borgquist S, Jögi A, Pontén F, Rydén L, Brennan DJ, Jirström K. Prognostic impact of tumour-specific HMG-CoA reductase expression in primary breast cancer. Breast Cancer Res. 2008;10:R79. doi: 10.1186/bcr2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan DJ, Laursen H, O'Connor DP, Borgquist S, Uhlen M, Gallagher WM, Pontén F, Millikan RC, Rydén L, Jirström K. Tumor-specific HMG-CoA reductase expression in primary premenopausal breast cancer predicts response to tamoxifen. Breast Cancer Res. 2011;13:R12. doi: 10.1186/bcr2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustbée E, Tryggvadottir H, Markkula A, Simonsson M, Nodin B, Jirström K, Rose C, Ingvar C, Borgquist S, Jernström H. Tumor-specific expression of HMG-CoA reductase in a population-based cohort of breast cancer patients. BMC Clin Pathol. 2015;15:8. doi: 10.1186/s12907-015-0008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anazawa Y, Nakagawa H, Furihara M, Ashida S, Tamura K, Yoshioka H, Shuin T, Fujioka T, Katagiri T, Nakamura Y. PCOTH, a novel gene overexpressed in prostate cancers, promotes prostate cancer cell growth through phosphorylation of oncoprotein TAF-Ibeta/SET. Cancer Res. 2005;65:4578–4586. doi: 10.1158/0008-5472.CAN-04-4564. [DOI] [PubMed] [Google Scholar]
- 28.Anchi T, Tamura K, Furihata M, Satake H, Sakoda H, Kawada C, Kamei M, Shimamoto T, Fukuhara H, Fukata S, et al. SNRPE is involved in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression. Oncol Lett. 2012;3:264–268. doi: 10.3892/ol.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashida S, Furihata M, Katagiri T, Tamura K, Anazawa Y, Yoshioka H, Miki T, Fujioka T, Shuin T, Nakamura Y, Nakagawa H. Expression of novel molecules, MICAL2-PV (MICAL2 prostate cancer variants), increases with high Gleason score and prostate cancer progression. Clin Cancer Res. 2006;12:2767–2773. doi: 10.1158/1078-0432.CCR-05-1995. [DOI] [PubMed] [Google Scholar]
- 30.Satake H, Tamura K, Furihata M, Anchi T, Sakoda H, Kawada C, Iiyama T, Ashida S, Shuin T. The ubiquitin-like molecule interferon-stimulated gene 15 is overexpressed in human prostate cancer. Oncol Rep. 2010;23:11–16. [PubMed] [Google Scholar]
- 31.Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, Ashida S, Miki T, Fujioka T, Shuin T, et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–8140. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
- 32.Togashi A, Katagiri T, Ashida S, Fujioka T, Maruyama O, Wakumoto Y, Sakamoto Y, Fujime M, Kawachi Y, Shuin T, Nakamura Y. Hypoxia-inducible protein 2 (HIG2), a novel diagnostic marker for renal cell carcinoma and potential target for molecular therapy. Cancer Res. 2005;65:4817–4826. doi: 10.1158/0008-5472.CAN-05-0120. [DOI] [PubMed] [Google Scholar]