Abstract
The aim of the present study was to investigate the effectiveness of the miR-17-92 cluster as a disease progression marker in prostate cancer (PCa). Reverse transcription-quantitative polymerase chain reaction analysis was used to detect the microRNA (miR)-17-92 cluster expression levels in tissues from patients with PCa or benign prostatic hyperplasia (BPH), in addition to in PCa and BPH cell lines. Spearman correlation was used for comparison and estimation of correlations between miRNA expression levels and clinicopathological characteristics such as the Gleason score and prostate-specific antigen (PSA). Receiver operating curve (ROC) analysis was performed for evaluation of specificity and sensitivity of miR-17-92 cluster expression levels for discriminating patients with PCa from patients with BPH. Kaplan-Meier analysis was plotted to investigate the predictive potential of miR-17-92 cluster for PCa biochemical recurrence. Expression of the majority of miRNAs in the miR-17-92 cluster was identified to be significantly increased in PCa tissues and cell lines. Bivariate correlation analysis indicated that the high expression of unregulated miRNAs was positively correlated with Gleason grade, but had no significant association with PSA. ROC curves demonstrated that high expression of miR-17-92 cluster predicted a higher diagnostic accuracy compared with PSA. Improved discriminating quotients were observed when combinations of unregulated miRNAs with PSA were used. Survival analysis confirmed a high combined miRNA score of miR-17-92 cluster was associated with shorter biochemical recurrence interval. miR-17-92 cluster could be a potential diagnostic and prognostic biomarker for PCa, and the combination of the miR-17-92 cluster and serum PSA may enhance the accuracy for diagnosis of PCa.
Keywords: microRNA-17-92 cluster, prostate cancer, diagnosis, biomarker
Introduction
Prostate cancer (PCa) is the second most frequently diagnosed type of cancer in men (1). Although a number of methods for cancer therapy have been developed, PCa remains the second leading cause of cancer-associated mortality among males in economically developed countries (2). In China, the incidence of PCa has increased over the past few years, and has become the sixth most common type of cancer and the ninth leading cause of cancer-associated mortality. Although multiple molecular and signaling pathways have characterized PCa proliferation, migration, and invasion, the mechanisms remain elusive (3). Therefore, an improved understanding of the biological mechanisms underlying PCa progression may contribute to improve the clinical management and therapy. Furthermore, it is essential to improve accuracy rate for tumor detection in males, as the low specificity of prostate-specific antigen (PSA) induces unnecessary biopsies and overtreatment (4). This could be achieved by identifying other more specific biomarkers that can be useful in the diagnosis of PCa.
microRNAs (miRNAs) are a type of endogenous, small noncoding RNA. Growing evidence has demonstrated that miRNAs may serve as potential tumor suppressors and oncogenes in PCa, regulating the expression of its specific target genes involved in cell development, and metastasis (5,6). The miR-17-92 cluster, also termed oncomiR-I, is composed of six members, including miR-17, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a (7). The miR-17-92 cluster has been validated to be upregulated in several types of malignant cancer (8). Hayashita et al (9) suggested that overexpression of the miR-17-92 cluster may perform an important role in the development of lung cancer, particularly in small-cell lung cancer. In Em-myc transgenic mice, B-cell lymphomas were accelerated when the miR-17-92 cluster was transduced into hematopoietic stem cells (10). Higher expression levels of various miR-17-92 cluster miRNAs were significantly associated with a lower overall survival rate in patients with osteosarcoma (11). In addition, a previous study indicated that the miR-17-92 cluster was downregulated following exposure to radiation in prostate cancer LNCaP cells (12). These findings demonstrate the oncogenic role of the miR-17-92 cluster. Therefore, the present study aimed to detect the differential expression of the miR-17-92 cluster between PCa and benign prostatic hyperplasia (BPH) tissue samples from patients, as well as between PCa, and BPH cell lines. In addition, the association between miR-17-92 cluster and PCa development was investigated.
Materials and methods
Patient samples
A total of 29 PCa tissues and 16 BPH tissues were obtained from male patients who underwent surgery at Beijing Chao-Yang Hospital, Capital Medical University (Beijing, China) from January 2014 to January 2015. The mean age of patients was 68.14 years (range, 55–76 years). The experiment was conducted in accordance with the Declaration of Helsinki (World Medical Association), and the study was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University. Written informed consent was obtained from each patient.
Cell cultures
The benign prostatic hyperplasia BPH1 cell line, and the LNCaP and PC3 PCa cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured routinely in RPMI-1640 supplemented with 5% fetal bovine serum, 100 U/ml penicillin and 50 µg/ml streptomycin (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cell lines were grown in a humidified incubator at 37°C with CO2.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Small RNA was extracted from tissues and cell lines grown to 80% confluence using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The primers for the miR-17-92 cluster (including miR-17-3p, cat. no., CD201-0017; miR-17-5p, cat. no., CD201-0016; miR-18a, cat. no., CD201-0018; miR-19a, cat. no., CD201-0021; miR-19b, cat. no., CD201-0278; and miR-92a, cat. no., CD201-0040), miRcute miRNA first-strand cDNA synthesis kit and the miRcute miRNA qPCR detection kit (SYBR-Green; both Tiangen Biotech Co., Ltd., Beijing, China) were used to detect the miR-17-92 cluster expression according to the manufacturer's protocol. The forward primer sequence for U6 was 5′-GCAAGGATGACACGCAAATTC-3′. A universal reverse primer was provided in the miRcute miRNA qPCR Detection kit. The PCR conditions included an initiation period at 94°C for 2 min, followed by a two-step PCR program consisting of 94°C for 20 sec and 60°C for 34 sec for 40 cycles. All samples were normalized against the internal control (U6 small nuclear RNA) and analyzed using the 2−ΔΔCq method (13).
Statistical analysis
All experiments were repeated three times, and all data were analyzed using SPSS software (version 19.0; IBM SPSS, Armonk, NY, USA). Quantified data are presented as the mean ± standard deviation. Comparisons between groups were performed using one-way analysis of variance followed by all pairwise multiple comparison procedures using Bonferroni correction and Student's t-tests or nonparametric tests. In addition, Spearman's correlation was used for comparison and estimation of correlations between miRNA expression levels, and clinicopathological characteristics, including the Gleason score (14) and PSA. Receiver operating curve (ROC) analysis was performed for evaluation of specificity and sensitivity of miR-17-92 cluster expression levels for distinguishing between patients with PC, and patients with BPH. All miRNAs significantly different between the PCa and BPH group were incorporated by logistic regression, and then multiplied by the relative quantities levels for each miRNA in each patient. Biochemical recurrence-free survival times of the patients with PCa were evaluated using the Kaplan-Meier method and log-rank tests according to the cutoff value of combined miRNA score: (−4.668+0.069) × (miR-17-3p+0.023) × (miR-17-5p+0.107) × (miR-18a+0.171) × (miR-19a+0.479) × (miR-19b-0.057) × (miR-92a). P<0.05 was considered to indicate a statistically significant difference.
Results
Differential expression of miR-17-92 cluster in PCa specimens compared with BPH
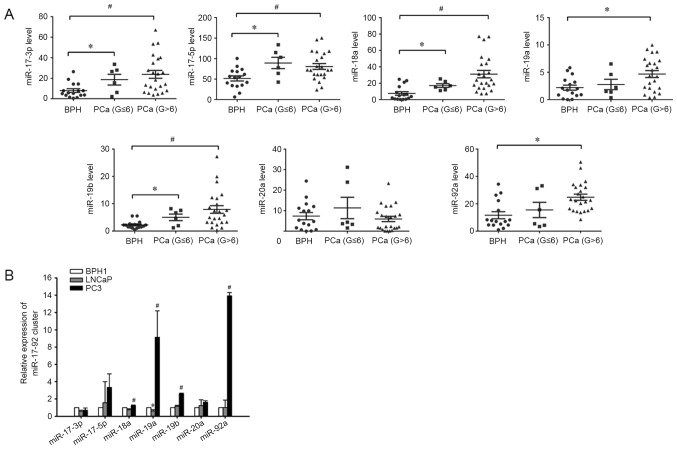
Total RNA was isolated from prostate tissues of 29 males with PCa and 16 with BPH. The clinicopathological characteristics of patients are presented in Table I. The RT-qPCR results revealed significantly increased expression levels of miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b and miR-92a in the cancer tissues of patients with PCa compared with patients with BPH, but no significant differences in the level of miR-20a were identified between the two groups (Fig. 1A). Notably, miR-19a and miR-92a were only significantly increased in the high-grade PCa group (Gleason >6; n=23), but similar levels were maintained in patients with low-grade PCa (Gleason ≤6; n=6) and BPH (n=16) (Fig. 1A).
Table I.
Clinicopathological data of patients with PCa and BPH.
| Value | ||
|---|---|---|
| Characteristic | PCa | BPH |
| Total, n | 29 | 16 |
| Mean age, years (range) | 68.14 (55–76) | 68.25 (49–77) |
| Mean PSA (range) | 56.004 (0.002–1000) | 5.080 (0.343–12.035) |
| Clinical stage, n | ||
| T1c | 2 | N/A |
| T2c | 11 | N/A |
| T3a | 6 | N/A |
| T3b | 8 | N/A |
| T4b | 2 | N/A |
| Gleason score, n | ||
| 3 + 3 | 6 | N/A |
| 3 + 4 | 6 | N/A |
| 4 + 3 | 8 | N/A |
| 4 + 4 | 6 | N/A |
| 4 + 5 | 2 | N/A |
| 5 + 4 | 1 | N/A |
PSA, prostate-specific antigen; PCa, prostate cancer; BPH, benign prostate hyperplasia.
Figure 1.
miR-17-92 cluster expression levels in patients with BPH, low-grade PCa, or high-grade PCa, and in BPH and PCa cell lines. (A) Expression of the miR-17-92 cluster was higher in PCa tissues compared with in BPH tissues. The expression of the miR-17-92 cluster in tissues was measured using reverse transcription-quantitative polymerase chain reaction. U6 served as an internal loading control. Each sample was detected in triplicate. *P<0.05 vs. BPH, #P<0.05 vs. BPH. (B) Expression of miR-18a, miR-19a, miR-19b and miR-92a was significantly higher in PC3 cells compared with in BPH1 and LNCaP cell lines, but the expression of miR-19a was significantly lower in LNCaP cells compared with BPH1 cells. *P<0.05 vs. BPH1, #P<0.05 vs. BPH1 and LNCaP. PCa, prostate cancer; miR, microRNA; BPH, benign prostatic hyperplasia; G, Gleason score.
miR-17-92 cluster expression in prostate cell lines
The miR-17-92 cluster expression pattern was determined in cultured prostate cell lines. Three different benign and PCa cell lines were used. All of the six miRNAs in miR-17-92 cluster were detectable in the human BPH1 cell line and PCa cell lines, LNCaP and PC3. Similarly, it was revealed that the expression level of miR-18a, miR-19a, miR-19b and miR-92a were significantly increased in the human PC3 cell line compared with that in the BPH1 and LNCaP cell lines (Fig. 1B).
Correlations between miR-17-92 cluster expression and clinicopathological features in PCa specimens
Among the 29 patients with PCa, the correlations of Gleason grade and PSA level with the progression risk of PCa were evaluated using bivariate correlation analysis. High expression of miR-17-3p, miR-18a, miR-19a, miR-19b and miR-92a was identified to be positively correlated with Gleason grade, but no correlation with PSA was observed (Table II).
Table II.
Correlation coefficients between miR-17-92 cluster expression and clinicopathological features in prostate cancer specimens.
| PSA | Gleason | |||
|---|---|---|---|---|
| Variable | R | P-value | R | P-value |
| miR-17a-3p | 0.018 | 0.928 | 0.602 | 0.001 |
| miR-17a-5p | 0.240 | 0.210 | −0.352 | 0.061 |
| miR-18a | −0.029 | 0.880 | 0.764 | <0.001 |
| miR-19a | 0.121 | 0.532 | 0.468 | 0.011 |
| miR-19b | −0.046 | 0.812 | 0.710 | <0.001 |
| miR-20a | −0.175 | 0.364 | 0.080 | 0.682 |
| miR-92a | −0.111 | 0.566 | 0.592 | 0.001 |
PSA, prostate-specific antigen; miR, microRNA.
ROC analysis of the miR-17-92 cluster expression in PCa specimens
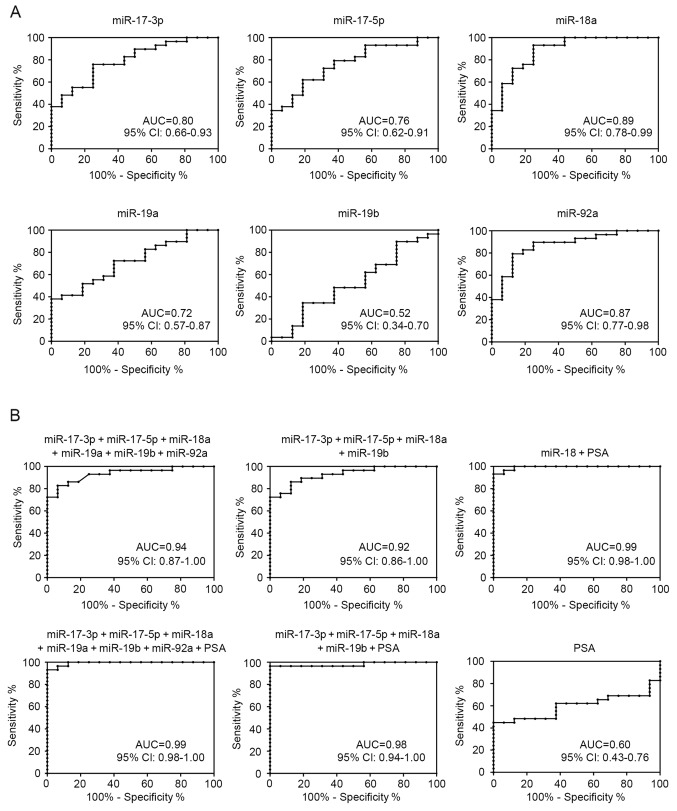
ROCs were constructed to explore the potential value of analyzed miR-17-92 cluster expression as diagnostic biomarkers for PCa (Fig. 2A). Notably, miR-18a demonstrated the most accurate discrimination [area under the ROC curve (AUC), 0.89; 95% confidence interval, 0.78–0.99; P<0.0001] between PCa and BPH. At the optimal cut-off value (9.54) of relative quantification, the sensitivity was 93.1% and specificity was 75.0%.
Figure 2.
ROC analysis using the miR-17-92 cluster to differentiate between PCa and BPH. (A) ROCs using individual differentially-expressed miRNAs to differentiate between PCa and BPH. (B) ROCs for PSA plus individual miRNAs or a combination of all differentially-expressed miRNAs with PSA to differentiate between PCa and BPH. ROC, receiver operating curve; miR/miRNA, mroRNA; PSA, prostate-specific antigen; PCa, prostate cancer; BPH, benign prostate hyperplasia; AUC, area under the curve; CI, confidence interval.
Combinations of miR-17-92 cluster and PSA improve diagnostic accuracy in ROC analysis
As presented in Fig. 2B, to evaluate the use of the miR-17-92 cluster as biomarkers of PCa diagnosis and progression, all the upregulated miRNAs were combined to distinguish between patients with PCa and BPH. In addition, four significantly increased miRNAs in the low-grade and high-grade PCa groups, including miR-17-3p, miR-17-5p, miR-18a, and miR-19b, were also grouped together to identify the presence of PCa. These multimarker ROC analyses demonstrated higher AUC, sensitivity and specificity compared with the use of each miRNA alone (Fig. 2A). Furthermore, certain combinations of these miRNAs and PSA improved the diagnostic accuracy in ROC analysis. Considering that the miR-18a alone demonstrated the most accurate discrimination between PCa and BPH, miR-18a was combined with PSA, and the combination significantly outperformed miR-18a use alone with an AUC of 0.99.
miR-17-92 cluster is a predictor of aggressive PCa
The median follow-up time was 15.6 months. A total of 9 patients exhibited biochemical PSA recurrence. For patients with a combined miRNA score above vs. below the cutoff value (high, >0.705; n=21 vs. low, ≤0.705; n=8), a high combined miRNA score of miR-17-92 cluster was identified to be associated with shorter biochemical recurrence interval (Fig. 3).
Figure 3.
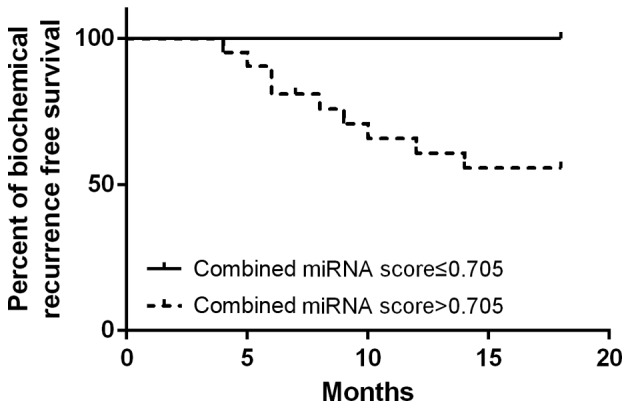
Survival analysis of the miR-17-92 cluster in predicting prostate cancer biochemical recurrence. miR, microRNA.
Discussion
Accumulating studies have demonstrated that aberrant expression levels of miRNAs are involved in the development of cancer. Aberrant expression of miRNA has been identified in various tumor types, and effect essential cellular processes involved in prostate tumorigenesis, including apoptosis-avoidance, cell proliferation and migration, and the androgen signaling pathway (15). Porkka et al (16) identified 51 differentially-expressed miRNAs between benign and carcinoma tumors with 37 downregulated, and 14 upregulated in carcinoma samples. Lichner et al (17) identified 25 differentially-expressed miRNAs between the high and low biochemical failure risk groups, and revealed that specific miRNAs could aid in predicting biochemical failure risk at the time of prostatectomy.
The polycistronic miR-17-92 cluster, located in the third intron of the chromosome 13 open reading frame 25 gene, was identified by Ota et al (18) in 2004. Expression of these miRNAs promoted the proliferation and suppressed apoptosis of cancer cells, and was identified to be dysregulated in several types of malignant tumor (19,20). In MYC transgenic mice, lymphoma development was accelerated following the transduction of the miR-17-92 cluster into hematopoietic cells (10). The miR-17-92 cluster was highly expressed in human hepatocellular carcinoma compared with the non-tumorous liver tissues (21). High miR-17-92 cluster expression exhibited poor prognostic implications in patients with diffuse large B-cell lymphoma (22). In addition, the miR-17-92 cluster was highly expressed in T precursor cells, and regulated lymphoproliferation in miR-17-92-transgenic models by targeting the phosphatase and tensin homolog (23). In addition, the miR-17-92 cluster was frequently and markedly overexpressed in lung cancer, particularly in those with small-cell lung cancer (9,24). The upregulation of the miR-17-92 cluster serves an important role in breast tumorigenesis and cell invasion, which contributes to the response of triple-negative breast to caloric restriction used in combination with radiation (25). A previous study reported that the miR-17-92 cluster was implicated in cervical cancer progression (26). Altogether, these findings demonstrated the potential of miR-17-92 cluster to disrupt the homeostasis of multiple organs and induce malignancy.
miRNAs in the miR-17-92 cluster have been demonstrated to function as oncogenes in PCa cells. Sylvestre et al (27) suggested that miRNAs from the miR-17-92 cluster serves as oncogenic miRNAs when overexpressed, by acting on essential regulators of the cell cycle and apoptosis. In addition, the miR-17-92 cluster can harbor androgen receptor binding sites, and their expression is sensitive to androgen stimulation in DUCaP and LNCaP cells (28). Members of the miRNA-17-92 cluster were also identified in PCa progression. Yang et al (29) confirmed that miR-17-5p and miR-17-3p could enhance PCa growth, and invasion by repressing the same target tissue inhibitor of metalloproteinase 3. Furthermore, serum levels of miR-19a and miR-19b were upregulated in patients with PCa (30). In contrast to the present results, the expression level of miR-20a was significantly higher in patients with a Gleason score of 7–10 compared with the patients with a Gleason score of 0–6, supporting the oncogenic role of miR-20a in PCa (31). This discrepancy may have been caused by differences in the source of miRNA obtained and the detection methods used.
Of note, miR-18a is a highly expressed miRNA in several types of cancer. The concentrations of miR-18a in plasma/serum of patients with cancer, including esophageal, pancreatic, hepatocellular and colorectal cancers, were all higher compared with that of healthy volunteers (32). Shen et al (33) revealed that the expression levels of miR-18a increased in lung cancer tissues, and Su et al (34) demonstrated that miR-18a could be a promising biomarker for the detection of gastric cancer, and its upregulation may be associated with prognosis of bladder cancer. Similar to these results, the present study also identified miR-18a to be upregulated in patients with PCa, and ROC analysis demonstrated that the increase of miR-18a in PCa tissues was the most accurate diagnostic biomarker and could distinguish patients with PCa from controls with high sensitivity and specificity.
Several studies have reported that numerous miRNAs may be independent biochemical recurrence prediction markers (35–38). The present study aimed to investigate the potential of miR-17-92 cluster derived from surgery at the initial visit in predicting the possibility of PCa progression or metastasis, and the results obtained demonstrated that the expression status of the miR-17-92 cluster was a good prognostic marker for time to progression to biochemical recurrence.
To the best of our knowledge, the present study was the first to demonstrate that the miR-17-92 cluster was upregulated in PCa tissue samples as compared with BPH control tissue, which suggests that these miRNAs are contributors to PCa oncogenesis. Furthermore, the high expression of the miR-17-92 cluster demonstrated increased sensitivity and specificity compared with PSA on diagnosis of PCa. Finally, the present findings suggested the high expression of the miR-17-92 cluster could be an independent prognostic marker for PCa progression. Therefore, miR-17-92 cluster is a potential diagnostic and prognostic biomarker for PCa, and possesses clinical utility to identify between patients with PCa and BPH. A combination of the miR-17-92 cluster and serum PSA can enhance the accuracy for diagnosis of PCa. However, the cellular mechanisms for the changes in miRNA levels in the prostate tissues with pathological progression are yet to be elucidated.
Acknowledgements
The present study was supported by the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (grant no. ZYLX201408), National Natural Science Foundation of China (grant no. 81200543), Beijing Natural Science Foundation (grant no. 7142057) and Beijing Municipal Health Bureau High-level Medical Professionals Promotion Project (grant no. 2013-3-016).
References
- 1.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 2.Saad F, Fizazi K. Androgen deprivation therapy and secondary hormone therapy in the management of hormone-sensitive and castration resistant prostate cancer. Urology. 2015;86:852–861. doi: 10.1016/j.urology.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 3.Ren SC, Chen R, Sun YH. Prostate cancer research in China. Asian J Androl. 2013;15:350–353. doi: 10.1038/aja.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanacore D, Boccellino M, Rossetti S, Cavaliere C, D'Aniello C, Di Franco R, Romano FJ, Montanari M, La Mantia E, Piscitelli R, et al. Micrornas in prostate cancer: An overview. Oncotarget. 2017 doi: 10.18632/oncotarget.16933. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuziwara CS, Kimura ET. Insights into regulation of the miR-17-92 cluster of miRNAs in cancer. Front Med (Lausanne) 2015;2:64. doi: 10.3389/fmed.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olive V, Li Q, He L. mir-17-92: A polycistronic oncomir with pleiotropic functions. Immunol Rev. 2013;253:158–166. doi: 10.1111/imr.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 10.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arabi L, Gsponer JR, Smida J, Nathrath M, Perrina V, Jundt G, Ruiz C, Quagliata L, Baumhoer D. Upregulation of the miR-17-92 cluster and its two paraloga in osteosarcoma-reasons and consequences. Genes Cancer. 2014;5:56–63. doi: 10.18632/genesandcancer.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.John-Aryankalayil M, Palayoor ST, Makinde AY, Cerna D, Simone CB, II, Falduto MT, Magnuson SR, Coleman CN. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178:105–117. doi: 10.1667/RR2703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Rees MA, Resnick MI, Oesterling JE. Use of prostate-specific antigen, Gleason score and digital rectal examination in staging patients with newly diagnosed prostate cancer. Urol Clin North Am. 1997;24:379–388. doi: 10.1016/S0094-0143(05)70384-0. [DOI] [PubMed] [Google Scholar]
- 15.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T. MicroRNA in prostate, bladder and kidney cancer: A systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 16.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 17.Lichner Z, Fendler A, Saleh C, Nasser AN, Boles D, Al-Haddad S, Kupchak P, Dharsee M, Nuin PS, Evans KR, et al. MicroRNA signature helps distinguish early from late biochemical failure in prostate cancer. Clin Chem. 2013;59:1595–1603. doi: 10.1373/clinchem.2013.205450. [DOI] [PubMed] [Google Scholar]
- 18.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.CAN-03-3773. [DOI] [PubMed] [Google Scholar]
- 19.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Han C, Wu T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis. 2015;36:1213–1222. doi: 10.1093/carcin/bgv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Go H, Jang JY, Kim PJ, Kim YG, Nam SJ, Paik JH, Kim TM, Heo DS, Kim CW, Jeon YK. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6:15035–15049. doi: 10.18632/oncotarget.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saki N, Abroun S, Soleimani M, Hajizamani S, Shahjahani M, Kast RE, Mortazavi Y. Involvement of microRNA in T-cell differentiation and malignancy. Int J Hematol Oncol Stem Cell Res. 2015;9:33–49. [PMC free article] [PubMed] [Google Scholar]
- 24.Osada H, Takahashi T. let-7 and miR-17-92: Small-sized major players in lung cancer development. Cancer Sci. 2011;102:9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 25.Jin L, Lim M, Zhao S, Sano Y, Simone BA, Savage JE, Wickstrom E, Camphausen K, Pestell RG, Simone NL. The metastatic potential of triple-negative breast cancer is decreased via caloric restriction-mediated reduction of the miR-17~92 cluster. Breast Cancer Res Treat. 2014;146:41–50. doi: 10.1007/s10549-014-2978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servín-González LS, Granados-López AJ, López JA. Families of microRNAs expressed in clusters regulate cell signaling in cervical cancer. Int J Mol Sci. 2015;16:12773–12790. doi: 10.3390/ijms160612773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 28.Pasqualini L, Bu H, Puhr M, Narisu N, Rainer J, Schlick B, Schäfer G, Angelova M, Trajanoski Z, Börno ST, et al. miR-22 and miR-29a are members of the androgen receptor cistrome modulating LAMC1 and Mcl-1 in prostate cancer. Mol Endocrinol. 2015;29:1037–1054. doi: 10.1210/me.2014-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ, Yang BB. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41:9688–9704. doi: 10.1093/nar/gkt680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SY, Shiboski S, Belair CD, Cooperberg MR, Simko JP, Stoppler H, Cowan J, Carroll PR, Blelloch R. miR-19, miR-345, miR-519c-5p serum levels predict adverse pathology in prostate cancer patients eligible for active surveillance. PLoS One. 2014;9:e98597. doi: 10.1371/journal.pone.0098597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesta M, Klecka J, Kulda V, Topolcan O, Hora M, Eret V, Ludvikova M, Babjuk M, Novak K, Stolz J, Holubec L. Importance of miR-20a expression in prostate cancer tissue. Anticancer Res. 2010;30:3579–3583. [PubMed] [Google Scholar]
- 32.Komatsu S, Ichikawa D, Takeshita H, Morimura R, Hirajima S, Tsujiura M, Kawaguchi T, Miyamae M, Nagata H, Konishi H, et al. Circulating miR-18a: A sensitive cancer screening biomarker in human cancer. In Vivo. 2014;28:293–297. [PubMed] [Google Scholar]
- 33.Shen Z, Wu X, Wang Z, Li B, Zhu X. Effect of miR-18a overexpression on the radiosensitivity of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:643–648. [PMC free article] [PubMed] [Google Scholar]
- 34.Su ZX, Zhao J, Rong ZH, Wu YG, Geng WM, Qin CK. Diagnostic and prognostic value of circulating miR-18a in the plasma of patients with gastric cancer. Tumour Biol. 2014;35:12119–12125. doi: 10.1007/s13277-014-2516-6. [DOI] [PubMed] [Google Scholar]
- 35.Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, Hu MM, Shen ZJ. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol. 2012;187:1466–1472. doi: 10.1016/j.juro.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi N, Uemura H, Nagahama K, Okudela K, Furuya M, Ino Y, Ito Y, Hirano H, Inayama Y, Aoki I, et al. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3:1455–1471. doi: 10.18632/oncotarget.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barron N, Keenan J, Gammell P, Martinez VG, Freeman A, Masters JR, Clynes M. Biochemical relapse following radical prostatectomy and miR-200a levels in prostate cancer. Prostate. 2012;72:1193–1199. doi: 10.1002/pros.22469. [DOI] [PubMed] [Google Scholar]
- 38.Goto Y, Kojima S, Nishikawa R, Enokida H, Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T, Seki N. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget. 2014;5:7748–7759. doi: 10.18632/oncotarget.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]