Abstract
Breast cancer is the leading cause for cancer-associated mortality in women. Although great progress has been made in the earlier diagnosis and systemic therapy of patients with breast cancer in recent years, recurrence or distant metastasis continue to present major barriers to the successful treatment of breast cancer. Therefore, fully understanding the molecular mechanisms underlying the progression of breast cancer may be critical for the development of effective therapeutic strategies against breast cancer. The aim of the present study was to explore the expression, function and molecular mechanisms of microRNA-154 (miR-154) in human breast cancer. It was demonstrated that miR-154 was significantly downregulated in breast cancer tissue and cell lines. The restoration of miR-154 expression suppressed the proliferation, migration and invasion of breast cancer cells. ADAM metallopeptidase domain 9 (ADAM9) was identified as a novel direct target for miR-154 in breast cancer. It was demonstrated that miR-154 acted as a tumor suppressor in breast cancer by targeting ADAM9. The results of the present study suggest that the restoration of miR-154 expression may be an effective therapeutic strategy for the treatment of breast cancer in the future.
Keywords: microRNA-154, breast cancer, growth, metastasis, ADAM metallopeptidase domain 9
Introduction
In 2012, breast cancer was the most prevalent type of malignant tumor and the leading cause of cancer-associated mortality in females worldwide (1). It is estimated that there were 231,840 new cases of invasive tumors, 60,290 new cases of noninvasive, in situ tumors and 40,290 mortalities resulting from breast cancer in 2015 in the United States (2). The highest reported prevalence of breast cancer is in economically developed countries; however, there is an increasing incidence and mortality in developing countries (3). This phenomenon is predominantly due to the adoption of a western lifestyle, the lack of breast cancer awareness and the poor access to screening and healthcare services (4–7). At present, the primary methodology for breast cancer therapy is resective surgery followed by hormonal therapy, chemotherapy, radiotherapy and/or biological therapy (8,9). Although great progress has been made in the earlier diagnosis and systemic therapy of patients with breast cancer in recent years, recurrence or distant metastasis continue to present major barriers to the successful treatment of breast cancer (10,11). Therefore, fully understanding the molecular mechanisms underlying the progression of breast cancer may be critical for the development of effective therapeutic strategies against breast cancer.
MicroRNAs (miRNAs) are a large family of evolutionarily conserved, 20–24 nucleotide, non-coding and single-stranded RNA molecules which commonly occur in plant, animal and viral genomes (12). miRNAs post-transcriptionally regulate gene expression by base-pairing with complementary nucleotide sequences in the 3′-untranslated regions (3′UTRs) of specific target mRNAs, leading to the transcriptional repression or degradation of the target genes (13,14). miRNAs have attracted considerable attention due to their ability to regulate a large number of mRNAs and influence a wide range of cell functions, including cell growth, metabolism, development, migration, invasion and survival (15,16). The importance of miRNAs in tumor initiation and development has been recognized; the abnormal expression of miRNAs serves a significant role in human cancer, caused by a variety of mechanisms, including deletions, amplifications, epigenetic silencing or mutations in miRNA loci (17). Emerging evidence has demonstrated that miRNAs can function as oncogenes or tumor suppressor genes, depending on the roles of their target genes (18). The inactivation of oncogenic miRNAs (19,20) or restoration of tumor-suppressor miRNAs (21,22) may have potential in cancer treatment.
In the present study, it was demonstrated that miR-154 was downregulated in breast cancer; enforced miR-154 expression repressed breast cancer cell proliferation, migration and invasion. Additionally, ADAM metallopeptidase domain 9 (ADAM9) was identified as a novel direct target of miR-154 in breast cancer.
Materials and methods
Tissue samples
A total of 45 samples of human breast cancer and corresponding non-tumor breast tissue samples (age range, 37–71 years; mean age, 56 years) were obtained during surgery at Binzhou Medical University Hospital (Binzhou, China) between September 2011 and December 2014. None of the patients had received chemotherapy or radiotherapy prior to surgery. The study was approved by the Ethics Committee of Binzhou Medical University Hospital; all patients provided written, informed consent.
Cell lines, cell culture and vectors
Breast cancer cell lines, including MCF-7, SKBR3 and MDA-MB-231, were purchased from the American Type Culture Collection (Manassas, VA, USA). The MCF-10A normal mammary epithelial cell line and 293T cells were acquired from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (All Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) throughout the study.
An miR-154 mimic, a corresponding negative control (NC) and luciferase reporter vectors [PmirGLO-ADAM9-3′UTR wild-type (Wt) and mutant (Mut)] were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). miR-154 potential target genes were predicted using miRanda (www.microrna.org) and TargetScan (www.targetscan.org) software. The ADAM9 vector and a corresponding blank vector control were synthesized and purified by Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was prepared from tissues or cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. To quantify miR-154 expression, TaqMan MicroRNA Reverse Transcription Kit (cat. no., 4366597, Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to perform reverse transcription, followed by quantitative polymerase chain reaction with a TaqMan microRNA assay kit (cat. no., 4326614, Applied Biosystems; Thermo Fisher Scientific, Inc.) was applied according to the manufacturer's protocol. To quantify ADAM9 mRNA expression, cDNA was generated using the ReverTra Ace qPCR RT Kit (Toyobo Life Science, Osaka, Japan) and qPCR was performed using SYBR Green Real-time Master mix (Toyobo Life Science). This reaction includes 2 µl cDNA (100 ng), 2 µl forward primer, 2 µl reverse primer, 10 µl SYBR Green PCR Master Mix and 4 µl ddH2O. The thermocycling conditions for qPCR were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers were designed as follows: ADAM9 forward, 5′-TGTGGGAACAGTGTGTTCAAGGA-3′; ADAM9 reverse, 5′-CCAATTCATGAGCAACAATGGAAG-3′; GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′; and GADPH reverse 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The relative quantification of miRNA and mRNA were achieved by normalization to U6 and GADPH, respectively. Each sample was analysed in triplicate and repeated three times. The relative expression was calculated by the 2−∆∆Cq method (23).
Cell proliferation assay
MCF-7 or MDA-MB-231 cells were seeded in 6-well plates overnight and then transfected with miR-154 mimics or NC, and/or an ADAM9 or empty vector, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were trypsinized 24 h after transfection, counted and seeded in 96-well plates at a density of 3,000 cells/well. A cell proliferation assay was performed at 24, 48, 72 and 96 h after seeding. Briefly, 10 µl Cell Counting kit-8 (CCK8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) solution was added to each well. Following incubation at 37°C for a further 2 h, the absorbance at 450 nm was detected using an ELISA reader. Each assay was performed in quintuplicate and repeated three times.
Transwell migration and invasion assay
Transwell inserts with an 8 µm pore size from Corning Incorporated (Corning, NY, USA) were used to assess cell migration and invasion abilities. For transwell migration assays, MCF-7 or MDA-MB-231 cells were seeded in 6-well plates overnight and transfected with an miR-154 mimic or NC, and/or an ADAM9 or empty vector, using Lipofectamine 2000 according to the manufacturer's protocol. Following incubation at 37°C for 48 h, cells were trypsinized and counted. Then, 4×104 cells were resuspended in FBS-free DMEM and seeded in the top chambers. A total of 500 µl DMEM containing 20% FBS was added to the lower chamber. At 48 h, cells remaining on the upper membrane were removed carefully with a cotton swab. The migrated cells attached to the lower surface of the membrane were fixed with methanol at room temperature for 10 min, stained with 0.5% crystal violet at room temperature for 10 min and counted under an inverted microscope (Olympus Corporation, Tokyo, Japan) in five random fields. For the transwell invasion assay, the process was the same as the transwell migration assay, except that the transwell inserts were coated with pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA). All assays were repeated three times.
Protein extraction and western blot
MCF-7 or MDA-MB-231 cells were washed in PBS (Gibco; Thermo Fisher Scientific, Inc.), and lysed in 1X radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 48 h after transfection with an miR-154 mimic or NC, or an ADAM9 or empty vector, with Lipofectamine 2000, according to the manufacturer's protocol. The protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal quantities of protein (30 µg) were subjected to 10% SDS-PAGE, and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline with 0.1% Tween (TBST), followed by incubation with the primary antibodies overnight at 4°C, including a monoclonal mouse anti-human ADAM9 antibody (dilution, 1:1,000; cat. no., ab57934) and a monoclonal mouse anti-human GADPH antibody (dilution, 1:1,000; cat. no., ab9484; both Abcam, Cambridge, UK). Subsequent to washing with TBST, a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (dilution, 1:5,000; cat. no., ab6789; Abcam) was added and incubated for 1 h at room temperature. The protein bands were visualized using an enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.). ImageJ 1.49 (National Institutes of Health, Bethesda, MD, USA) was used to quantify of the western blots. This assay was repeated three times.
Luciferase reporter assay
293T cells were seeded at 70% confluence into 24-well plates. Cells were co-transfected with PmirGLO-ADAM9-3′ UTR Wt or Mut, and miR-154 mimics or NC using Lipofectamine 2000 according to the manufacturer's protocol. At 48 h after transfection, cells were harvested and the luciferase activities were measured by a Dual-Luciferase Reporter assay system (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol. The experiment was performed independently in triplicate.
Statistical analysis
All data are presented as mean ± standard deviation. Statistical analysis was performed with a Student's t-test or a one-way analysis of variance using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). The Student-Newman-Keuls test was used as a post hoc test following ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-154 is downregulated in breast cancer tissues and cell lines
To investigate the role of miR-154 in human breast cancer, its expression was analyzed in breast cancer and corresponding non-tumor breast tissue samples. Using RT-qPCR, it was identified that miR-154 expression levels were significantly reduced in breast cancer tissues compared with the corresponding non-tumor breast tissues (Fig. 1A; P<0.05). miR-154 expression levels were then analyzed in the breast cancer cell lines MCF-7, SKBR3 and MDA-MB-231, and in the MCF-10A normal mammary epithelial cell line. Compared with MCF-10A, miR-154 was downregulated in the breast cancer cell lines (Fig. 1B; P<0.05). These results suggested that the downregulation of miR-154 may be associated with the initiation and progression of breast cancer.
Figure 1.
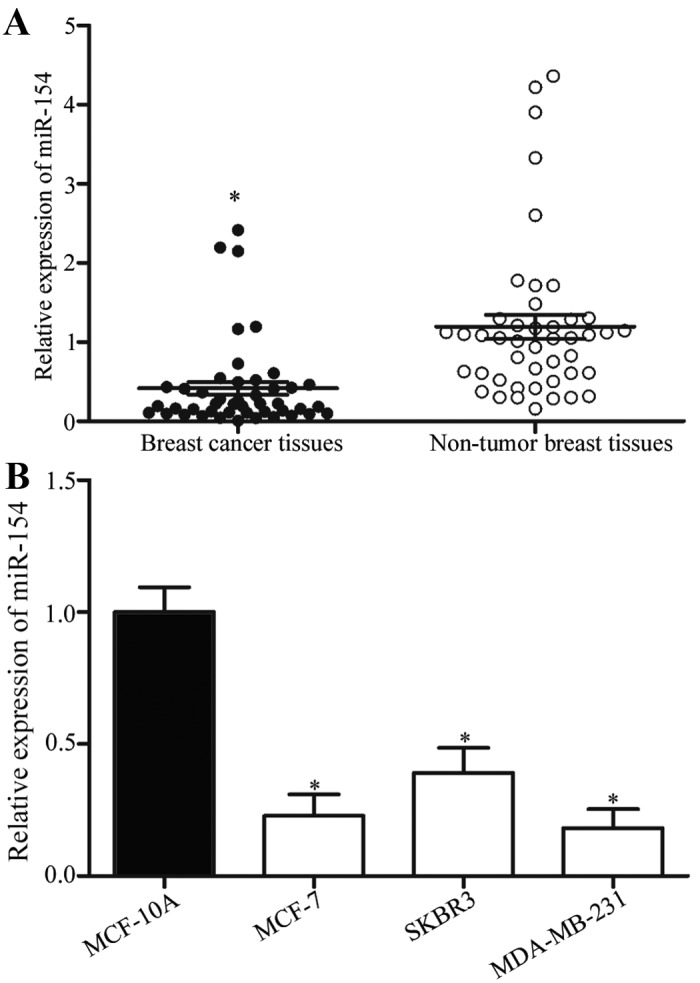
miR-154 expression is downregulated in breast cancer. (A) Relative expression levels of miR-154 in breast cancer tissue and corresponding non-tumor breast tissue samples were determined using RT-qPCR. *P<0.05 compared with non-tumor tissue. (B) Relative expression levels of miR-154 in breast cancer cell lines (MCF-7, SKBR3 and MDA-MB-231) and the MCF-10A normal mammary epithelial cell line were analyzed with RT-qPCR. *P<0.05 compared with MCF-10A cells. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-154 inhibits the proliferation of breast cancer cells
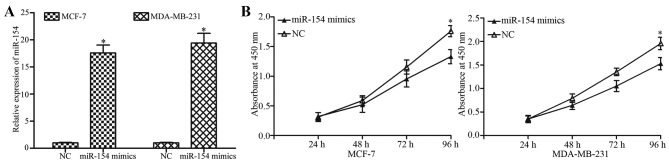
To investigate the effect of miR-154 on breast cancer cell proliferation, MCF-7 and MDA-MB-231 cells were transfected with an miR-154 mimic or NC. The overexpression of miR-154 in the cells transfected with the mimic was confirmed by RT-qPCR (Fig. 2A; P<0.05). The extent of proliferation in the miR-154 mimic- or NC-treated cells was detected by with a CCK8 assay. Transfection with the miR-154 mimic significantly decreased the proliferation of MCF-7 and MDA-MB-231 cells compared with NC-transfected cells (Fig. 2B; P<0.05) at 96 h.
Figure 2.
miR-154 inhibits the proliferation of breast cancer cells. (A) Levels of miR-154 in MCF-7 and MDA-MB-231 cells were detected by RT-qPCR following transfection with an miR-154 mimic or NC. (B) Proliferation assay revealed a significant inhibition of proliferation in cells transfected with an miR-154 mimic compared with cells transfected with the NC. *P<0.05 compared with NC. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control.
miR-154 inhibits the migration and invasion of breast cancer cells
To determine whether miR-154 was associated with the regulation of breast cancer metastasis, transwell migration and invasion assays were performed. Compared with the NC, transfection with the miR-154 mimic significantly decreased the migration ability of MCF-7 and MDA-MB-231 cells (Fig. 3; P<0.05). Furthermore, miR-154 overexpression significantly repressed the capacity for invasion of MCF-7 and MDA-MB-231 cells, compared with the NC (Fig. 3; P<0.05). Taken together, the results indicated that miR-154 acted as a tumor suppressor in breast cancer cells.
Figure 3.
miR-154 inhibits the migration and invasion of breast cancer cells. Transwell migration and invasion assays were performed to evaluate the migration and invasion abilities following transfection with an miR-154 mimic or NC in MCF-7 and MDA-MB-231 cells (magnification, ×200). *P<0.05 compared with NC. miR, microRNA; NC, negative control.
ADAM9 is a direct target of miR-154
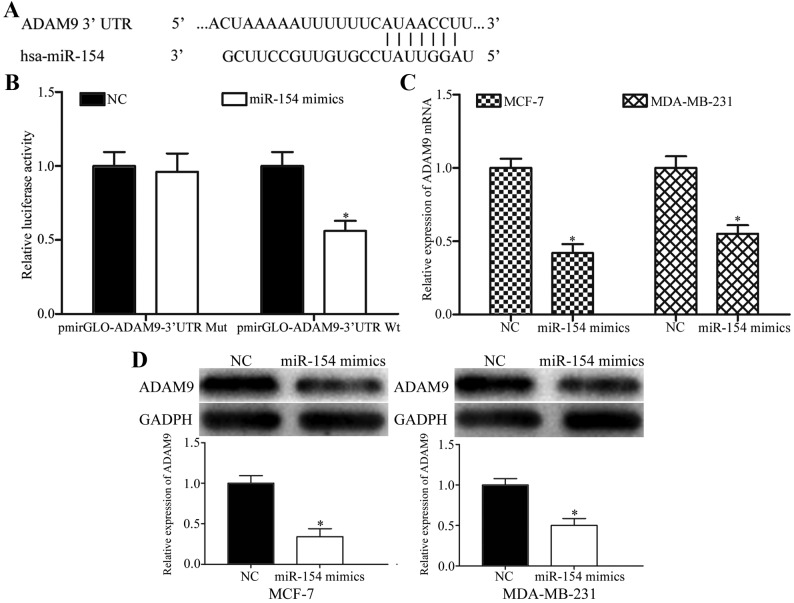
To investigate the molecular mechanism by which miR-154 inhibited breast cancer cell proliferation, migration and invasion, potential target genes of miR-154 were predicted with miRanda and TargetScan. As shown in Fig. 4A, the 3′UTR of ADAM9 contained a putative miR-154 binding site.
Figure 4.
ADAM9 is a direct target gene of miR-154. (A) Schematic of a putative miR-154 binding site in the 3′UTR of ADAM9. (B) Luciferase reporter assays exhibited a reduction in the relative luciferase activities of PmirGLO-ADAM9-3′UTR Wt following transfection with an miR-154 mimic. (C) Reverse transcription-quantitative polymerase chain reaction analysis of ADAM9 mRNA expression levels in MCF-7 and MDA-MB-231 cells transfected with an miR-154 mimic or NC. (D) Western blot analysis of ADAM9 protein expression levels in MCF-7 and MDA-MB-231 cells transfected with an miR-154 mimic or NC. *P<0.05 compared with NC. ADAM9, ADAM metallopeptidase domain 9; miR, microRNA; 3′UTR, 3’-untranslated region; Wt, wild-type; Mut, mutant.
Luciferase reporter assays were performed to explore whether ADAM9 was a direct target of miR-154. 293T cells were transfected with luciferase reporter vectors, along with an miR-154 mimic or NC. The miR-154 mimic significantly decreased the luciferase activities of PmirGLO-ADAM9-3′ UTR Wt compared with the NC (Fig. 4B; P<0.05), but had no effect on the PmirGLO-ADAM9-3′UTR Mut, suggesting that miR-154 could directly bind to the 3′UTR of ADAM9.
To determine whether ADAM9 expression was regulated by miR-154, RT-qPCR and western blotting were performed to detect the ADAM9 expression levels in MCF-7 and MDA-MB-231 cells transfected with the miR-154 mimic or NC. Restoration of miR-154 expression suppressed the ADAM9 mRNA expression levels in MCF-7 and MDA-MB-231 cells transfected with miR-154 mimics, compared with the NC (Fig. 4C; P<0.05). Western blotting demonstrated that the ectopic expression of miR-154 resulted in a significant decrease in ADAM9 protein expression (Fig. 4D; P<0.05). Taken together, ADAM9 was identified as a novel direct target of miR-154 in breast cancer.
Restoration of ADAM9 abrogates tumor suppression by miR-154 in breast cancer cells
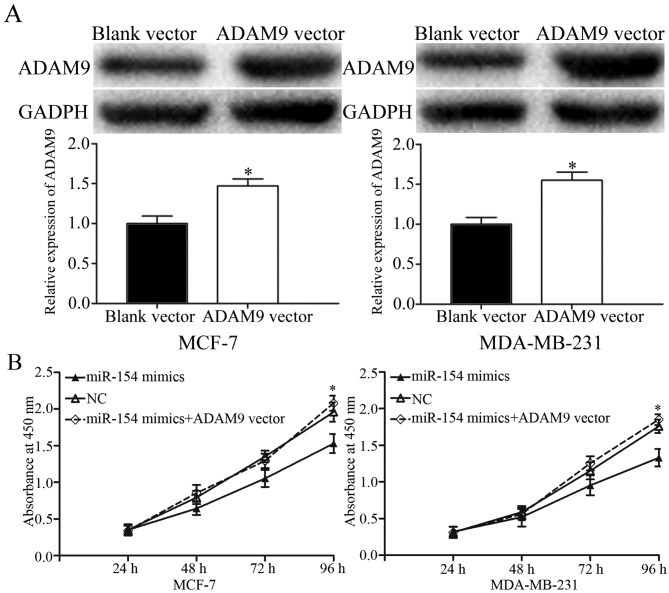
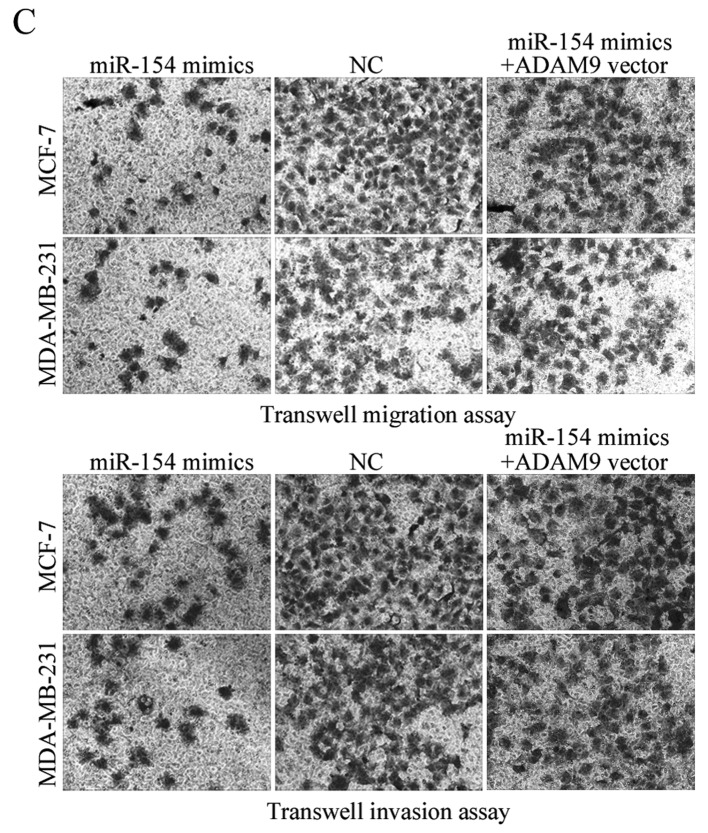
To determine whether ADAM9 acted as a functional target for miR-154, a gain-of-function analysis was performed by the transfection of an ADAM9 vector or control into the miR-154-overexpressing MCF-7 and MDA-MB-231 cells. Western blotting revealed that ADAM9 was significantly upregulated in MCF-7 and MDA-MB-231 cells transfected with the ADAM9 vector compared with the control (Fig. 5A; P<0.05). Proliferation assays, and transwell migration and invasion assays, were performed. The data showed that ADAM9 overexpression significantly abrogated the tumor suppressive effects of miR-154 on breast cancer cell proliferation (Fig. 5B; P<0.05), migration and invasion (Fig. 5C). These results provided further support that ADAM9 was a downstream functional target of miR-154.
Figure 5.
Restoration of ADAM9 expression abrogates the suppressive effects of miR-154 on breast cancer cells. (A) Western blot analysis of ADAM9 protein expression levels in MCF-7 and MDA-MB-231 cells transfected with an ADAM9 vector or a blank vector control. *P<0.05 compared with blank vector. (B) ADAM9 overexpression abrogated the inhibition of proliferation in MCF-7 and MDA-MB-231 cells transfected with an miR-154 mimic. *P<0.05 compared with miR-154 mimic alone. (C) ADAM9 reversed the inhibition of migration and invasion in MCF-7 and MDA-MB-231 cells transfected with an miR-154 mimic (magnification, ×200). ADAM9, ADAM metallopeptidase domain 9; miR, microRNA; NC, negative control.
Discussion
Preventing tumor growth and metastasis is the central problem in the treatment of breast cancer (24). The abnormal expression of miRNAs has been identified in various types of human cancer; miRNAs have been demonstrated to serve a significant role in the progression and carcinogenesis of cancer (25). In breast cancer, a number of miRNAs have been reported to affect growth and metastasis. For example, miR-362-5p may target CYLD lysine 63 deubiquitinase to inhibit the proliferation, migration and invasion of breast cancer cells (26). Zhang et al (27) reported that miR-147 repressed the growth and metastasis of breast cancer through the Akt/mechanistic target of rapamycin pathway. Therefore, miRNAs may be suitable for development as therapeutic targets to prevent breast cancer growth and metastasis.
In the present study, the expression and role of miR-154 in breast cancer was investigated. It was identified that miR-154 was significantly downregulated in breast cancer tissues and cell lines. Functional studies demonstrated that miR-154 overexpression suppressed the proliferation, migration and invasion of breast cancer cells. These results indicated that miR-154 acted as a tumor suppressor, and therefore, the low expression of miR-154 may contribute to breast cancer initiation and development.
miR-154 was previously identified as dysregulated in a number of cancer types, and its expression has been associated with clinical outcomes. For example, in colorectal cancer, miR-154 was previously identified as downregulated in tumor tissue, and its low expression was associated with an increased tumor size, a positive lymph node metastasis status and an advanced clinical stage (28). In addition, multivariate analysis identified low miR-154 expression as an independent predictor of reduced survival time (28). Lin et al (29) reported that miR-154 expression levels were lower in non-small-cell lung cancer relative to normal lung tissue, and that low miR-154 expression was associated with metastasis, a larger tumor size and an advanced TNM stage. The results of these studies collectively indicated that miR-154 may be a prognostic marker in a number of types of cancer.
Alterations in miR-154 expression have been demonstrated to contribute to the initiation and progression of various types of cancer. Lin et al (29) identified that miR-154 overexpression suppressed non-small-cell lung cancer cell proliferation, colony formation, invasion and migration, and induced apoptosis and G0/G1 cell cycle arrest in vitro. Restoration of miR-154 expression also decreased the growth of non-small-cell lung cancer cell xenografts in vivo (29). Xin et al (30) reported that miR-154 inhibited the proliferation, colony formation, migration and invasion of colorectal cancer cells. In prostate cancer, enforced miR-154 expression decreased the proliferation of prostate cancer cells in vitro (31). In accord with the present study, these studies suggest that miR-154 acts as a tumor suppressor and may be a suitable therapeutic target in these types of cancer.
miRNAs serve major roles in a wide range of physiological and pathological processes through negative regulation of their target mRNAs (13,14). Cyclin D2 (31), high mobility group AT-hook 2 (32) and toll-like receptor 2 (30) were previously validated as target genes of miR-154. In the present study, ADAM9 was identified as a direct and functional target of miR-154 in breast cancer. Bioinformatics analysis identified a putative miR-154 binding site in the 3′UTR of ADAM9. miR-154 mimic transfection markedly decreased the luciferase activity of a luciferase reporter vector containing the ADAM9 3′UTR sequence. miR-154 mimic transfection also suppressed ADAM9 mRNA and protein levels in breast cancer cells. The restoration of ADAM9 expression abrogated the suppression of cell proliferation, invasion and migration by miR-154 in breast cancer cells. These data demonstrated that miR-154 inhibited breast cancer cell proliferation, migration and invasion through the miR-154/ADAM9 axis.
A disintegrin and metalloproteinases (ADAMs), members of the metzincin superfamily of matrix metalloproteinases, have been demonstrated to contribute to a range of biological functions, including fertilization, adhesion, migration and proteolysis (33,34). ADAM9, a member of the ADAM family, has been demonstrated to be upregulated in a range of types of human cancer, including renal cell cancer (35), prostate cancer (36), breast cancer (37), hepatocellular carcinoma (38), and pancreatic cancer (39). Previous studies have also identified miRNAs that may interact with ADAM9. Wang et al (40) identified that miR-126 targeted ADAM9 to inhibit osteosarcoma proliferation and invasion. Zhang et al (41) reported that miR-33a repressed breast cancer cell growth and metastasis through the negative regulation of ADAM9. Further studies that explore additional novel targets for miR-154 and other miRNAs that can regulate ADAM9 in breast cancer will facilitate a complete understanding of the molecular mechanisms underlying the initiation and progression of the disease.
In conclusion, the data presented in the present study indicated that miR-154 may be a tumor-suppressing gene in breast cancer. Transfection with an miR-154 mimic inhibited the proliferation, migration and invasion of breast cancer cells. Therefore, the restoration of miR-154 expression may be a effective therapeutic strategy for the treatment of breast cancer in the future.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Fu SW, Lee W, Coffey C, Lean A, Wu X, Tan X, Man YG, Brem RF. miRNAs as potential biomarkers in early breast cancer detection following mammography. Cell Biosci. 2016;6:6. doi: 10.1186/s13578-016-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter P. ‘Westernizing’ women's risks? Breast cancer in lower-income countries. N Engl J Med. 2008;358:213–216. doi: 10.1056/NEJMp0708307. [DOI] [PubMed] [Google Scholar]
- 4.Beaglehole R, Yach D. Globalisation and the prevention and control of non-communicable disease: The neglected chronic diseases of adults. Lancet. 2003;362:903–908. doi: 10.1016/S0140-6736(03)14335-8. [DOI] [PubMed] [Google Scholar]
- 5.Parkin DM, Fernández LM. Use of statistics to assess the global burden of breast cancer. Breast J. 2006;12(Suppl 1):S70–S80. doi: 10.1111/j.1075-122X.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 6.Badar F, Faruqui ZS, Ashraf A, Uddin N. Third world issues in breast cancer detection. J Pak Med Assoc. 2007;57:137–140. [PubMed] [Google Scholar]
- 7.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura S, Yagata H, Ohno S, Yamaguchi H, Iwata H, Tsunoda N, Ito Y, Tokudome N, Toi M, Kuroi K, Suzuki E. Multi-center study evaluating circulating tumor cells as a surrogate for response to treatment and overall survival in metastatic breast cancer. Breast Cancer. 2010;17:199–204. doi: 10.1007/s12282-009-0139-3. [DOI] [PubMed] [Google Scholar]
- 9.Gong Y, He T, Yang L, Yang G, Chen Y, Zhang X. The role of miR-100 in regulating apoptosis of breast cancer cells. Sci Rep. 2015;5:11650. doi: 10.1038/srep11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 11.Alonso DF, Ripoll GV, Garona J, Iannucci NB, Gomez DE. Metastasis: Recent discoveries and novel perioperative treatment strategies with particular interest in the hemostatic compound desmopressin. Curr Pharm Biotechnol. 2011;12:1974–1980. doi: 10.2174/138920111798377076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson I, Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- 13.Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. doi: 10.12688/f1000research.2-136.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gromak N. Intronic microRNAs: A crossroad in gene regulation. Biochem Soc Trans. 2012;40:759–761. doi: 10.1042/BST20120023. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 16.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 17.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pichler M, Calin GA. MicroRNAs in cancer: From developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 20.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis; Proc Natl Acad Sci USA; 2008; pp. 13556–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Ma S, Zhao G, Yang L, Zhang P, Yi Q, Cheng S. miR-708/LSD1 axis regulates the proliferation and invasion of breast cancer cells. Cancer Med. 2016;5:684–692. doi: 10.1002/cam4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Jaarsveld MT, Helleman J, Berns EM, Wiemer EA. MicroRNAs in ovarian cancer biology and therapy resistance. Int J Biochem Cell Biol. 2010;42:1282–1290. doi: 10.1016/j.biocel.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Ni F, Gui Z, Guo Q, Hu Z, Wang X, Chen D, Wang S. Downregulation of miR-362-5p inhibits proliferation, migration and invasion of human breast cancer MCF7 cells. Oncol Lett. 2016;11:1155–1160. doi: 10.3892/ol.2015.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Zhang HE, Liu Z. MicroRNA-147 suppresses proliferation, invasion and migration through the AKT/mTOR signaling pathway in breast cancer. Oncol Lett. 2016;11:405–410. doi: 10.3892/ol.2015.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kai Y, Qiang C, Xinxin P, Miaomiao Z, Kuailu L. Decreased miR-154 expression and its clinical significance in human colorectal cancer. World J Surg Oncol. 2015;13:195. doi: 10.1186/s12957-015-0607-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Lin X, Yang Z, Zhang P, Shao G. miR-154 suppresses non-small cell lung cancer growth in vitro and in vivo. Oncol Rep. 2015;33:3053–3060. doi: 10.3892/or.2015.3895. [DOI] [PubMed] [Google Scholar]
- 30.Xin C, Zhang H, Liu Z. miR-154 suppresses colorectal cancer cell growth and motility by targeting TLR2. Mol Cell Biochem. 2014;387:271–277. doi: 10.1007/s11010-013-1892-3. [DOI] [PubMed] [Google Scholar]
- 31.Zhu C, Shao P, Bao M, Li P, Zhou H, Cai H, Cao Q, Tao L, Meng X, Ju X, et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol. 2014;32:31.e9–e16. doi: 10.1016/j.urolonc.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379:69–75. doi: 10.1007/s11010-013-1628-4. [DOI] [PubMed] [Google Scholar]
- 33.Blobel CP. ADAMs: Key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 34.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fritzsche FR, Wassermann K, Jung M, Tölle A, Kristiansen I, Lein M, Johannsen M, Dietel M, Jung K, Kristiansen G. ADAM9 is highly expressed in renal cell cancer and is associated with tumour progression. BMC Cancer. 2008;8:179. doi: 10.1186/1471-2407-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritzsche FR, Jung M, Tölle A, Wild P, Hartmann A, Wassermann K, Rabien A, Lein M, Dietel M, Pilarsky C, et al. ADAM9 expression is a significant and independent prognostic marker of PSA relapse in prostate cancer. Eur Urol. 2008;54:1097–1106. doi: 10.1016/j.eururo.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 37.O'Shea C, McKie N, Buggy Y, Duggan C, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Expression of ADAM-9 mRNA and protein in human breast cancer. Int J Cancer. 2003;105:754–761. doi: 10.1002/ijc.11161. [DOI] [PubMed] [Google Scholar]
- 38.Tao K, Qian N, Tang Y, Ti Z, Song W, Cao D, Dou K. Increased expression of a disintegrin and metalloprotease-9 in hepatocellular carcinoma: Implications for tumor progression and prognosis. Jpn J Clin Oncol. 2010;40:645–651. doi: 10.1093/jjco/hyq030. [DOI] [PubMed] [Google Scholar]
- 39.Grützmann R, Lüttges J, Sipos B, Ammerpohl O, Dobrowolski F, Alldinger I, Kersting S, Ockert D, Koch R, Kalthoff H, et al. ADAM9 expression in pancreatic cancer is associated with tumour type and is a prognostic factor in ductal adenocarcinoma. Br J Cancer. 2004;90:1053–1058. doi: 10.1038/sj.bjc.6601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Wang X, Guo Q, Wang G, Han X, Li X, Shi ZW, He W. MicroRNA-126 overexpression inhibits proliferation and invasion in osteosarcoma cells. Technol Cancer Res Treat. 2016;15:NP49–NP59. doi: 10.1177/1533034615601563. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Zhang Y, Ding W, Lin Y, Huang Z, Luo Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6:881–889. doi: 10.1007/s13238-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]