While the ability for humans to host a complex microbial ecosystem is an essential property of life, the mechanisms allowing for immune tolerance of such a large microbial load are not completely understood and are currently the focus of intense research. This study shows that an important proinflammatory pathway that is commonly triggered by pathogenic bacteria upon interaction with the host is, in fact, actively repressed by the bacteria of the gut microbiome, supporting the idea that beneficial microbes themselves contribute to the immune tolerance in support of homeostasis. These findings are important for two reasons. First, many currently assume that proinflammatory signaling by lipopolysaccharide is a fundamental feature of the gut flora. This assumption influences greatly how host-microbiome interactions are theoretically modeled but also how they are experimentally studied, by using robust TLR signaling conditions to simulate commensals. Second, elucidation of the mechanisms that support host-microbe tolerance is key to the development of therapeutics for both intestinal and systemic inflammatory disorders.
KEYWORDS: microbiome, lipopolysaccharide, microbial communities, symbiosis, tolerance, Toll-like receptors
ABSTRACT
Cohabitation of microbial communities with the host enables the formation of a symbiotic relationship that maintains homeostasis in the gut and beyond. One prevailing model suggests that this relationship relies on the capacity of host cells and tissues to remain tolerant to the strong immune stimulation generated by the microbiota such as the activation of Toll-like receptor 4 (TLR4) pathways by lipopolysaccharide (LPS). Indeed, gut microbial LPS is thought to be one of the most potent activators of innate immune signaling and an important mediator of the microbiome’s influence on host physiology. In this study, we performed computational and experimental analyses of healthy human fecal samples to examine the TLR4 signaling capacity of the gut microbiota. These analyses revealed that an immunoinhibitory activity of LPS, conserved across the members of the order Bacteroidales and derived from an underacylated structural feature, silences TLR4 signaling for the entire consortium of organisms inhabiting the human gut. Comparative analysis of metagenomic data from the Human Microbiome Project and healthy-donor samples indicates that immune silencing via LPS is a microbe-intrinsic feature in all healthy adults. These findings challenge the current belief that robust TLR4 signaling is a feature of the microbiome and demonstrate that microbiome-derived LPS has the ability to facilitate host tolerance of gut microbes. These findings have broad implications for how we model host-microbe interactions and for our understanding of microbiome-linked disease.
IMPORTANCE While the ability for humans to host a complex microbial ecosystem is an essential property of life, the mechanisms allowing for immune tolerance of such a large microbial load are not completely understood and are currently the focus of intense research. This study shows that an important proinflammatory pathway that is commonly triggered by pathogenic bacteria upon interaction with the host is, in fact, actively repressed by the bacteria of the gut microbiome, supporting the idea that beneficial microbes themselves contribute to the immune tolerance in support of homeostasis. These findings are important for two reasons. First, many currently assume that proinflammatory signaling by lipopolysaccharide is a fundamental feature of the gut flora. This assumption influences greatly how host-microbiome interactions are theoretically modeled but also how they are experimentally studied, by using robust TLR signaling conditions to simulate commensals. Second, elucidation of the mechanisms that support host-microbe tolerance is key to the development of therapeutics for both intestinal and systemic inflammatory disorders.
INTRODUCTION
The human gut microbiome comprises a large and dynamic population of microorganisms representing one of the most densely populated ecosystems known (1). This microbial consortium provides many benefits to its host, including key signals that shape gastrointestinal development, immune maturation, vitamin production, extraction of otherwise indigestible carbohydrates from the diet, and pathogen resistance (2). However, this presents the challenge for the host of containing, and remaining immunologically tolerant to, a microbial load in excess of 1012 cells/ml (1). Elaborate mechanisms are required to modulate and preserve this symbiotic relationship. To date, mechanisms reported to maintain this symbiosis rely on host-driven tolerance, including the physical barriers of the oriented epithelium and mucosa, secretion of antimicrobial peptides and secreted antibodies, or negative feedback loops in NF-κB signaling (3). Several mechanisms are also at play that contain both the risk of infection and the amplitude of the immune response (4). In recent years, studies have reported examples of microorganisms themselves promoting the expansion of Foxp3+ regulatory T cells (5, 6), the suppression of tumor necrosis factor alpha (TNF-α) production (7, 8), and the maintenance of the gut epithelium (9), suggesting that the microbiota could play a role in shaping host immune responses. However, specific microbiota-derived molecular mediators of host tolerance are still largely unknown.
We recently reported that the commensal organism Bacteroides dorei produces an antagonistic form of lipopolysaccharide (LPS) that can influence the susceptibility of children to allergies and autoimmunity (10). While the ability of some LPS isoforms to inhibit Toll-like receptor 4 (TLR4) signaling has been reported (11), their broader implications with regard to gut health and disease remain unexamined. Here we directly extracted the total LPS from fecal samples from healthy adult humans and found that the LPS produced by the consortium of gut-resident microbes potently antagonizes the host TLR4 pathway. Using metagenomic sequencing, we further delineated strain level contributions to the gut LPS pool and found that numerous other members of the order Bacteroidales, which are the dominant Gram-negative bacteria in the healthy human gut microbiome (12), produce antagonistic forms of LPS, thus driving immune silencing for the entire microbial community. These findings undermine the current accepted paradigm that gut microbial communities possess a robust TLR4 signaling capacity against which the immune system needs to be heavily tolerized (13). Ours is the first report describing a phylum-wide microbiome-intrinsic mechanism actively damping innate immune activation in the healthy gut and redefines how we envision the immunological dynamics of the host-microbiota relationship.
RESULTS
Total LPS produced in the adult human gut is immunoinhibitory.
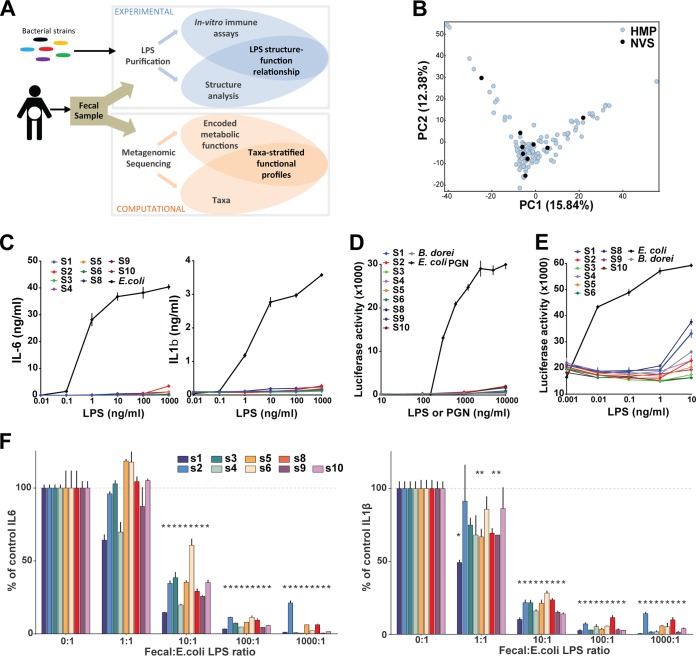
Stool samples were collected from nine healthy adults and analyzed as illustrated in Fig. 1A. The microbial composition of each sample was analyzed by metagenomic whole-genome sequencing (WGS), which revealed an overall bacterial composition that was similar to the general microbial landscape depicted in Human Microbiome Project 1 (HMP1) (14) (Fig. 1B), suggesting that these samples are representative of the general population.
FIG 1 .
Total LPS from the human gut microbiome is immunoinhibitory. (A) Schematic diagram of sample processing and analysis in this study. (B) Principal-component analysis plot comparing the HMP1 sample set (blue) and a new set of nine healthy-donor (Novartis [NVS]) samples (black). (C) Human PBMCs were stimulated in the presence of increasing amounts of fecal LPS. IL-6 and IL-1β concentrations in supernatants were measured after 20 h of culture. E. coli LPS was included as a control. (D and E) HEK-293 cells expressing an NF-κB–luciferase reporter and either hTLR2 and hCD14 (D) or hTLR4, hMD2, and hCD14 (E) were stimulated in the presence of increasing doses of fecal LPS from each donor, B. dorei LPS, or E. coli PGN. A sterile-water negative control was included in each experiment (not shown). Luciferase activity was measured after 6 h of activation. Data shown are the mean ± the standard deviation of triplicate evaluations from one out of three independent experiments. (F) PBMCs were cotreated with 1 ng/ml E. coli LPS and increasing doses of fecal LPS from the donors indicated. IL-6 and IL-1β concentrations in supernatants were measured after 20 h of culture and compared to those obtained with LPS treatment alone (0:1). Data shown are the mean ± the standard deviation of triplicates in one representative experiment. *, corrected P value of <0.05.
LPS was purified from each donor as previously described (15) and further processed as previously described by Hirschfeld et al. to remove contaminating lipoproteins and yield purified LPS (16). The immunostimulatory potency of each sample of fecal LPS was then assessed by stimulating human primary PBMCs. The production of six inflammatory cytokines, interleukin-10 (IL-10), IL-6, IL-8, TNF-α, IL-12p70, and IL-1b, was measured in parallel. All cytokines that reached detectable levels (IL-10, IL-6, IL-8, IL-1b, and TNF-α) showed similar patterns of production, in accordance with their common regulation by NF-κB activation downstream of LPS. IL-6 and IL-1b are shown as representative results. All LPS samples showed ≥2 orders of magnitude lower stimulatory potency than purified Escherichia coli LPS (no. 1, 2, 5, 8, and 10), and many (no. 3, 4, 6, and 9) were immunologically silent up to the largest dose tested on primary peripheral blood mononuclear cells (PBMCs; 1 mg/ml) (Fig. 1C). Similar results were obtained when stimulating human TLR2 (hTLR2) and hTLR4--NF-κB reporter cell lines, confirming the absence of direct signaling through these LPS-sensing signaling pathways (Fig. 1D and E). Hence, total fecal LPS has a very limited capacity to activate the TLR4--NF-κB pathway and elicit the production of inflammatory cytokines. This result is particularly striking given that the gut microbiome is thought to elicit robust TLR4 signals and that total fecal LPS is likely derived from diverse microbial origins, including species known to produce potent forms of LPS (e.g., gammaproteobacteria). We further examined the immunomodulating properties of fecal LPS by cotreating PBMCs with fecal LPS prior to stimulation with E. coli LPS to assess their ability to interfere with immune stimulation. Fecal LPS from each donor potently inhibited the cytokine (IL-6, IL-1b) production elicited by E. coli LPS. Most fecal samples reached significant inhibition at a 1:10 E. coli-to-fecal LPS ratio, while some reached significant IL-1b inhibition at a 1:1 ratio (Fig. 1F). Hence, total fecal LPS is a potent inhibitor of TLR4 stimulation.
Gut LPS derives primarily from Bacteroidales.
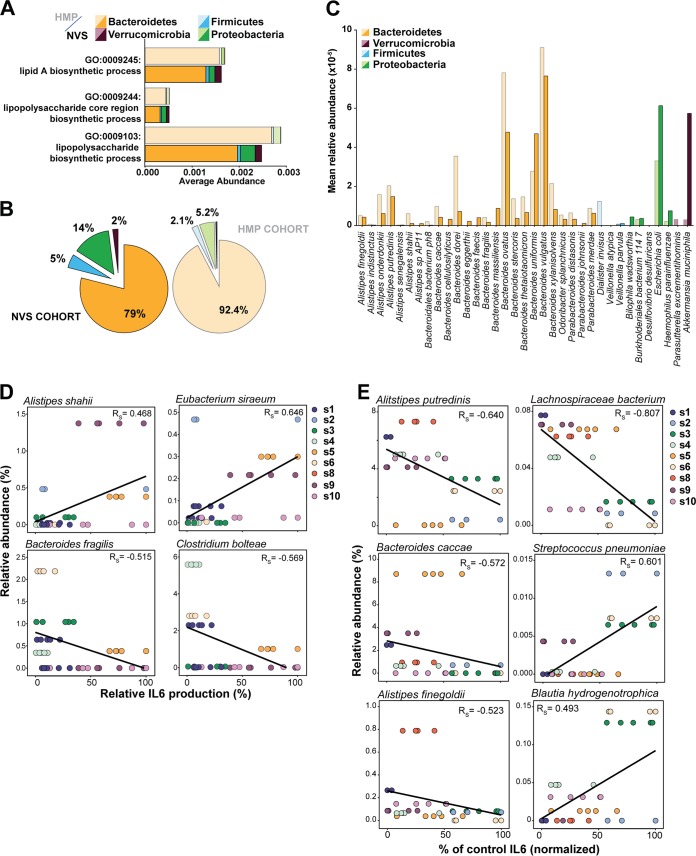
Previous studies have demonstrated that a few bacterial species known to be present in the human gut microbiome produce nonstimulatory or immunoinhibitory LPS (10, 17), which prompted us to examine which bacterial species were present in these samples that contributed to the total pool of fecal LPS. Metagenomic WGS of the fecal samples revealed that the distribution of the main phyla, as determined by using MetaPhlAn2 (18), was similar in the nine volunteers and not distinguishable from the 140 healthy-donor samples in HMP1 (Fig. 1B; see Fig. S1A in the supplemental material), suggesting that these samples are representative of the general population. We also analyzed both sample cohorts by using the HUMAnN2 algorithm (unpublished; http://huttenhower.sph.harvard.edu/HUMAnN2) to determine encoded functional pathways and inferred gene ontology (GO) functions with UniRef50 gene families. As previously described for the HMP1 cohort, the distribution of the encoded functions is similar across all samples and was also comparable within the newly collected samples (Fig. S1B). To identify which strains are contributing to LPS production in the human gut, we focused on the GO terms relevant to LPS biosynthesis (Fig. 2A and B). Both in fresh samples and in the HMP1 data set, Bacteroidetes species dominate the three main GO terms related to LPS biosynthesis. Overall, Bacteroidetes species contribute 79% of the LPS biosynthesis in healthy volunteers and 92.4% of that in HMP1 samples (Fig. 2B). In contrast, proteobacteria are minor contributors to LPS biosynthesis, with an average of 14% of the total produced LPS being of E. coli origin in volunteers; in HMP1 donors, the percentage is 5.2% (Fig. 2B; Fig. S2C). The resulting estimated average Bacteroidetes-to-E. coli LPS ratio in the gut would be between 6:1 in volunteers and 18:1 in the HMP1 cohort. The abundances of the Bacteroidales species, and therefore their likely contribution to the LPS pool, are shown in Fig. 2C. Bacteroides species contributing to LPS are diverse but follow similar trends. Notably, Bacteroides ovatus, B. uniformis, and B. vulgatus dominate LPS production in both cohorts (Fig. 2C).
FIG 2 .
Bacteroidetes bacteria are the main contributors to LPS biosynthesis in the human gut microbiome. (A) Average abundance per sample of genes related to the three main LPS biosynthesis-related functions. (B) Relative contributions of the different phyla to the total LPS-encoding capacity of the gut microbiome determined in both cohorts. The three functions of LPS biosynthesis were pooled. The darker-color set is HMP samples, and the lighter-color set is NVS samples. (C) Contribution of individual species to LPS biosynthesis functions. The average relative abundances of genes related to any of the three LPS-related GO functions are shown for individual species within each phylum. Only species detected in >50% of the donors in each data set are shown. The darker-color set is HMP samples, and the lighter-color set is NVS samples. (D) Correlation of individual-species abundance with stimulation of IL-6 production as determined in panel A. (E) Correlation of individual-species abundance with inhibition of IL-6 production as determined in panel B. Each data point represents one independent experiment. Rs, Spearman rho.
(A) Distribution of species in individual samples into the five main phyla in the new-sample (S1 to S10) and HMP1 sample sets. (B) Distribution of encoded functional pathways in individual samples of the new data set (NVS samples) or the HMP1 data set (HMP). (C) Contribution of the phylum Bacteroidetes to the total LPS-encoding capacity of the gut microbiome relative to that of other phyla. Data are presented for each individual donor. The darker-color set is HMP samples, and the lighter-color set is NVS samples. Download FIG S1, TIF file, 0.8 MB (794.7KB, tif) .
Copyright © 2017 d’Hennezel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Stimulation of PBMCs by Bacteroidales LPS. (A and B) Human PBMCs were stimulated in the presence of increasing amounts of LPS from the Bacteroidales species indicated. TNF-α and IL-1β concentrations in supernatants were measured after 20 h of culture. E. coli LPS was included as a control. Data shown are the mean ± the standard deviation of triplicates in one representative experiment. (C and D) LPS isolated from the species indicated. Human PBMCs were cotreated with zymosan and increasing doses of Bacteroidales LPS. TNF-α and IL-1β concentrations in supernatants were measured after 20 h of culture. Treatment with zymosan alone was included as a control (black bar). Data shown are the mean ± the standard deviation of triplicates in one representative experiment. Download FIG S2, TIF file, 0.6 MB (586.3KB, tif) .
Copyright © 2017 d’Hennezel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next examined whether the bacterial composition of the samples would correlate with the potency of stimulation or inhibition in these samples (Fig. 2D and E). We could not identify a strong correlation between function and composition at the phylum level and could only identify weak-to-moderate correlations between a few individual species and the stimulatory potency of individual fecal LPS samples (Fig. 2D). However, we found that the abundance of several Bacteroidales species (Alistipes putredinis, Bacteroides caccae, Alistipes finegoldii) show a moderate-to-strong correlation with the inhibition of IL-6 cytokine production (Fig. 2E), as well as TNF-α and IL-1β production (data not shown). Cytokine production and trends were similar for all of the cytokines measured. Interestingly, we also identified several Gram-positive species whose abundance correlates with functionality, notably, Lachnospiraceae bacterium 5_1_63FAA (Fig. 2E). However, these strains are present at very low abundance (<0.5% of the genomic pool), Gram-positive strains do not contribute to the LPS pool, and it remains unclear what indirect impact they may have on LPS signaling. Our results show that Bacteroidetes species are by far the most abundant contributors to LPS biosynthesis functions in the healthy human intestinal microbiota, consistent with their high abundance relative to other Gram-negative species in the gut (12, 14).
Immunosuppressive LPS in Bacteroidales species.
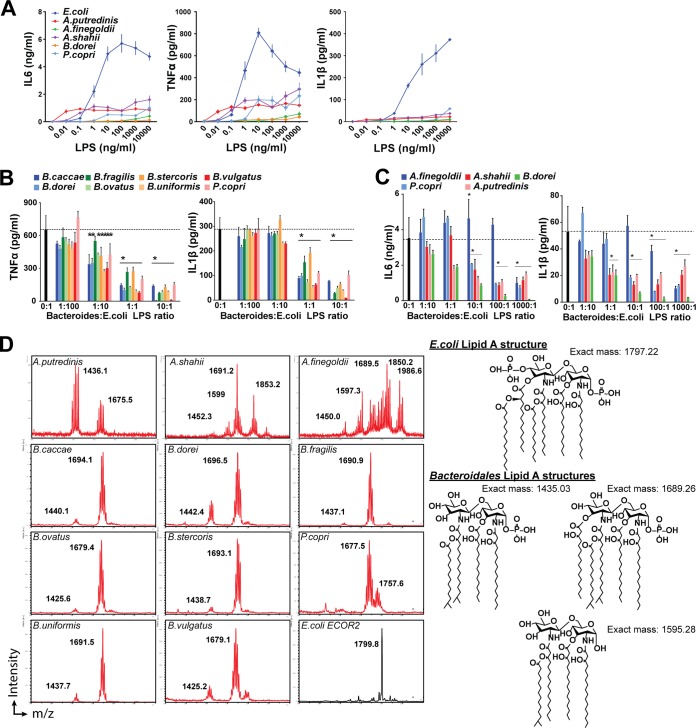
We had previously shown that LPS from several members of the order Bacteroidales were unable to activate TLR4, consistent with literature reports (10, 17, 19), and that B. dorei produces an immunoinhibitory form of LPS (10). Thus, we examined whether immunosilent LPS is produced not only by Bacteroides and Prevotella spp. but also by additional gut-associated Bacteroidales bacteria (i.e., Alistipes spp.) not examined in our previous studies and whether immunoinhibitory LPS is a common, phylum-wide feature of the order Bacteroidales. We isolated LPS from 11 Bacteroidales strains and tested its capacity to elicit inflammatory responses from primary human PBMCs. All three Alistipes spp. tested produced an LPS with low immunostimulatory capacity, similar to other members of the order Bacteroidales (Fig. 3A; Fig. S2A and B). We then tested its capacity to modulate the response of primary human PBMCs to stimulation by E. coli LPS. Across the order Bacteroidales, LPS induces a potent reduction of TNF-α or IL-6 production. Production and trends were similar for all of the cytokines measured. This included a significant suppression at a 1:10 Bacteroidales-to-E. coli LPS ratio for all of the species tested, with the notable exception of Alistipes finegoldii, which only suppresses at higher ratios (100:1 or 1,000:1) (Fig. 3B and C). Comparable suppression was seen with IL-1β (Fig. 3B and C), and unexpectedly, similar results were obtained for inhibition of zymosan-elicited cytokine production (Fig. S2C).
FIG 3 .
Bacteroidales LPS inhibits TLR stimulation. (A to C) LPS was isolated from the species indicated. (A) Human PBMCs were stimulated in the presence of increasing amounts of LPS from the Bacteroidales species indicated. IL-6, TNF-α, and IL-1β concentrations in supernatants were measured after 20 h of culture. E. coli LPS was included as a control. Data shown are the mean ± the standard deviation of triplicates in one representative experiment. (B and C) Human PBMCs were cotreated with E. coli LPS and increasing doses of LPS from Bacteroidales (B) and Alistipes (C) spp. TNF-α and IL-1β concentrations in supernatants were measured after 20 h of culture. E. coli LPS is shown as a control (dotted line). Data shown are the mean + the standard deviation of triplicate measurements in one representative of three or more independent experiments. *, corrected P value of <0.05. (C) MALDI-TOF MS analysis of the lipid A moiety of individual Bacteroidales and E. coli LPSs. m/z values are indicated for major peaks. On the right are the lipid A structures associated with the main m/z values.
Inhibitory forms of LPS have been previously described whose activity is derived from variations in the structure of the lipid A moiety (20, 21). The lipid A domain of LPS is responsible for the endotoxic properties associated with LPS due to recognition by the TLR4 complex and downstream activation of the NF-κB pathway (22). A reduction in the number of lipid A acyl chains by bacteria has been shown to modulate the recognition of LPS by TLR4 (23). Structural analysis of the lipid A domain from all of the members of the order Bacteroidales studied by matrix-assisted laser desorption ionization--time of flight mass spectrometry (MALDI-TOF MS) analysis revealed masses with a mass/charge ratio (m/z) of <1,700, consistent with the presence of underacylated lipid A structures (Fig. 3D). Specifically, all Bacteroides spp. and Prevotella copri showed a dominant peak having an m/z of 1,677.5 to 1,696.5, consistent with the [M − H]− ions of a penta-acylated form of lipid A. Also common but with variable abundance is a peak at an m/z of approximately 1,435, consistent with the [M − H]− ions of a tetra-acylated form of lipid A. Variation in determined mass is consistent with variable length in acyl chains. The spectrum of P. copri shows an additional minor peak with an m/z of 1,757.6, consistent with the addition of a phosphate group but otherwise identical to other members of the order Bacteroidales. All Alistipes spp. showed masses consistent with underacylated lipid A structures in addition to larger masses in A. shahii and A. finegoldii of unknown significance (Fig. 3D). Our analysis indicate that immunosilent and immunoinhibitory underacylated lipid A structures are conserved across the order Bacteroidales.
In a parallel effort to identify novel microbe-derived immunomodulatory molecules, we screened extracellular polymeric substances (EPS), a secreted cell-associated matrix often containing biologically active molecules (24). Given the intimate association of the commensal gut microbes with mucosal surfaces, EPS represent an unexplored niche for immunologically active factors. We extracted the EPS of 29 bacterial strains selected to comprise representatives of the four main phyla of the human gut microbiome (7) (Fig. S3A). We then assessed the capacity of these EPS extracts to modulate inflammation in primary human immune cells and showed that TLR2 (zymosan)- and TLR4 (LPS)-driven cytokine production is reduced 4- to 20-fold by extracts from several strains, mainly members of the order Bacteroidales (Fig. S3B to E). Using an in-gel LPS staining procedure (25), we determined that the EPS extracts from Bacteroides strains exhibit high LPS content (Fig. S3F). These results further confirm that LPS from members of the order Bacteroidales potently antagonizes TLR signaling and suggest that the EPS matrices of commensal organisms are a likely vehicle for immunomodulatory factors.
EPS extracts from bacterial species of the human microbiome have immunomodulatory properties. (A) Phylogenetic tree of the 120 most abundant bacterial strains in the human gut microbiome. EPS extracts were prepared from 29 strains (red). (B and C) Human monocyte-derived dendritic cells (B and C) or PBMCs (D and E) were cotreated with individual EPS extracts and zymosan (B and D) or LPS (C and E). The TNF-α concentration in supernatants was compared to that obtained with zymosan or LPS treatment alone. Data shown are the mean of triplicate measurements in one representative of three or more independent experiments. (F) EPS extracts from Gram-negative species were analyzed by SDS-PAGE, followed by Pro-Q Emerald staining for glycoproteins. Purified E. coli LPS was loaded as a control. Download FIG S3, TIF file, 2.2 MB (2.3MB, tif) .
Copyright © 2017 d’Hennezel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Immune mechanisms underlying intestinal commensalism have yet to be fully elucidated. Since the realization that our intestinal tract is host to billions of bacteria in the absence of an overwhelming immune response, the mechanisms that maintain this relationship have been the subject of intense study. Here we formally demonstrate that the total LPS produced by the human gut microbiome not only is itself nonimmunogenic but also inhibits TLR4-dependent cytokine production (Fig. 1B to F). We further show that production of immunoinhibitory forms of LPS is a common feature across members of the order Bacteroidales (Fig. 3), which are the major contributors of LPS synthesis in the human gut (Fig. 2). Previous publications have demonstrated that distinct structural features of the lipid A domain, produced by a few bacterial species, interfere with proper TLR4-MD2 signaling via competitive inhibition (17, 19). The exact mechanism of signaling inhibition by feces-derived LPS has not been demonstrated in this study but is likely to be identical to previously described mechanisms.
We had previously reported on the immunoinhibitory function of LPS produced by B. dorei and the role it likely plays in precluding proper immune education in infants genetically predisposed to type 1 diabetes, thus favoring the development of allergies and autoimmunity (10). We also observed a general overabundance of Bacteroidetes bacteria in these infants, which, in light of the findings reported here, suggests that the collective contribution of all Bacteroides spp. likely enhanced the disease susceptibility of these children. Any effort to target the gut microbial community in these infants for therapeutic purposes should therefore likely focus broadly on all Bacteroidetes bacteria, not just B. dorei.
Our findings also shed new light on a number of discoveries made in recent years that suggest a link between the inflammatory stimulation arising from the intestinal lumen and local or peripheral inflammatory disorders. Inflammatory bowel disease (IBD) has been associated with a bloom of Proteobacteria (26, 27). Interestingly, treatment with the aminoglycoside antibiotic gentamicin reduces the abundance of Proteobacteria and results in a dominance of the gut flora by Bacteroidetes, leading to protection from colitis in a murine model of IBD. Conversely, vancomycin treatment has been shown to favor an increase in Proteobacteria and does not prevent disease (28, 29). While the specific role of inflammatory LPS in the etiology and recovery of IBD remains to be elucidated, it is possible that differences in LPS immunogenicity between Bacteroidetes and Proteobacteria underlie these observations. In obesity, a common hypothesis is that intestinal LPS leaks out into the circulation, leading to subclinical, chronic inflammation in peripheral adipose tissues, altering their metabolic functions (30–32). However, our findings suggest that freely circulating LPS coming from the gut lumen tends to prevent, rather than favor, inflammation. Interestingly, obesity is associated with a decrease in Bacteroidetes species, relative to an increase in Firmicutes species, which are mostly Gram-positive bacteria (33). Thus, the intestinal LPS composition in these patients could be shifted away from anti-inflammatory Bacteroidetes LPS subtypes in favor of inflammatory LPS subtypes, possibly producing a more inflammatory LPS. Finally, the current methodologies used to quantify peripheral exposure to intestinal LPS are also sensitive to hypoacylated LPS structures; therefore, no assay currently exists that can distinguish inhibitory from inflammatory LPS in the periphery. While we do not dispute the effect of inflammatory LPS on the metabolic profile of peripheral tissue, our findings warrant caution in interpreting the significance of peripheral LPS levels in studies attempting to connect microbiome LPS and peripheral inflammation.
Most importantly, our findings challenge the current perception of the mechanisms regulating the cohabitation of the gut microbiota and the host. Commensalism is commonly thought of as an equilibrium of two powerful forces, a heavy bacterial load endowed with high inflammatory potential and a well-protected host with a tightly regulated immune system. This model has long guided efforts to elucidate the mechanisms underlying commensalism between the host and the gut microbiome. Experimentally, this has translated into the use of potent inflammatory LPS from pathogenic organisms to simulate the interaction with the microbiome in vitro and in vivo (34). In contrast, our findings show that total gut microbiome LPS is, in fact, overall immunoinhibitory. However, other bacterial factors contribute to the immunogenicity of the microbiome. Notably the production of the TLR2 ligand peptidoglycan (PGN) is ubiquitous in intestinal bacteria. While it has been shown that some pathogenic bacteria can use autolytic enzymes to alter their PGN to reduce the stimulation of TLR2, whether commensal bacteria produce altered forms of PGN or bear attenuated immune functions remains to be determined (35, 36). The inhibition of zymosan-mediated TLR2 stimulation by Bacteroides EPS and LPS we have observed also points to the possibility of a more widespread inhibition of immunogenicity by Bacteroidales that would extend to other signaling pathways beyond TLR4.
In recent years, a few reports have described contributions of individual species to specific mechanisms interfering with NF-κB signaling or actively modulating the TLR responsiveness in the gut epithelium (37, 38). Importantly, the present report is the first describing a phylum-wide, microbiome-derived mechanism that actively promotes immune tolerance of gut microbiota. Until now, the innate immune inflammatory potential of the commensal microbiota has likely been overestimated and the signaling capacity of the gut microbiome must be reassessed to accurately model the impact that resident commensal microbes have on health and disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are summarized in tables at https://figshare.com/s/5e56cc1a347ef4f1de49. All strains were started from 20% glycerol stocks stored at −80°C, plated onto brain heart infusion (BHI) agar supplemented with hemin and vitamin K (Teknova; B1093), and grown anaerobically at 37°C. Liquid cultures of all strains were started from a single colony inoculated into 1,000 ml of BHI liquid medium supplemented with 10 ml of vitamin K-hemin solution (BD; 212354), 10 ml of trace minerals (ATCC; MD-TMS), 10 ml of trace vitamins (ATCC; MD-VS), and 50 ml of fetal bovine serum (HyClone; SH30071) and grown anaerobically for at least 48 h at 37°C. A flexible anaerobic chamber (Coy Laboratory Products) containing 20% CO2, 10% H2, and 70% N2 was used for all anaerobic microbiology steps.
EPS extracts.
EPS matrix was extracted from all strains as previously described (7). Briefly, 250-ml volumes of 24-h cultures were recovered by centrifugation at 18,400 × g for 10 min (4°C). The prewashed cell pellet was suspended in 8 ml of phosphate-buffered saline by vortexing for 5 min, allowing the cell-bound EPS to dissolve. Planktonic cells were subsequently pelleted by centrifugation at 18,400 × g for 10 min (4°C). The supernatant was then carefully removed, filter sterilized with a 0.2-µm-pore-size filter, and added to 4 volumes of ice-cold absolute ethanol to precipitate the EPS. After centrifugation at 3,300 × g for 30 min, the precipitated-EPS pellet was washed with 70% ethanol, lyophilized, and then stored at −20°C. For further experiments, lyophilized EPS fractions were normalized by being dissolved in ultrapure water at the desired concentration. An average of 5 mg of lyophilized extract was recovered for each strain by using this protocol.
Human cell isolation and differentiation for immune stimulation assays.
Blood buffy coats were obtained from healthy volunteers after informed consent was obtained. The study protocol and any amendments were reviewed and approved by an independent review board (New England IRB, Newton, MA) before the start of the study. This study was conducted in accordance with the ethical principles of the Declaration of Helsinki.
PBMCs were freshly isolated from blood by Ficoll-Hypaque gradient centrifugation as previously described (39). Monocyte-derived dendritic cells were differentiated in vitro from freshly isolated human monocytes as previously described (39). Briefly, CD14+ monocytes were isolated from freshly purified PBMCs by negative selection and magnetic bead sorting (Miltenyi). Cells were then incubated in complete RPMI 1640 in the presence of 50 ng/ml recombinant human granulocyte-macrophage colony-stimulating factor and 20 ng/ml recombinant human IL-4 (R&D Systems) for 7 days. For all experiments using human donor cells, data were generated independently with at least two donors. A representative data set was selected for incorporation into the figure.
LPS purification and analysis.
To isolate the total LPS from a fecal sample, approximately 5 g of fecal material was homogenized into 10 ml of endotoxin-free water with a gentleMACS Dissociator. The resulting fecal slurry was allowed to settle for 5 min, allowing large particles to settle, and the supernatant was lyophilized for LPS purification. LPS purification from fecal material was performed with 500 mg of lyophilized material but otherwise performed as described for bacterial strains below.
For LPS isolation from bacterial strains, LPS from all strains was isolated from a 1,000-ml liquid culture grown under standard conditions for ~48 h by the hot water-phenol method as previously described (15). To remove trace amounts of endotoxin protein, phenol-purified LPS was further treated as previously described (16), with the modifications described below. Following the final ethanol precipitation, LPS was lyophilized to determine the yield with a Mettler Toledo XS105 Dual Range analytical balance (sensitivity, ≥0.1 ng) and resuspended in HyPure cell culture grade endotoxin-free water (HyClone) to a final concentration of 1 mg/ml without the addition of triethanolamine. To confirm the purity and normalization of feces-derived LPS, the final product was visualized with the Pro-Q Emerald 488 in-gel staining kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. In all cases, the Pro-Q Emerald 488 in-gel staining kit indicated a purity identical to that of LPS purified from bacterial isolates. However, given the complex molecular nature of human fecal material, our analysis does not exclude the possibility of contaminating substances in feces-derived LPS and it is not considered ultrapure.
In vitro LPS stimulation assays and competition assays.
PBMCs (105) or monocyte-derived dendritic cells (5 × 104) were incubated in the presence of LPS purified from the bacterial isolates indicated at doses ranging from 10−3 to 104 ng/ml for 18 to 20 h. For inhibition assays, cells were plated in medium. LPS purified from the strain indicated was then added, followed immediately by 100 pg/ml LPS purified from E. coli. Supernatants were collected after 18 to 20 h of culture and analyzed with the cytokine bead array human inflammation kit (BD Biosciences) in accordance with the manufacturer’s instructions. This kit analyzes the levels of IL-10, IL-6, IL-8, TNF-α, IL-12p70, and IL-1b in the same samples. Groups were compared by using a two-tailed nonhomoscedastic t test corrected for multiple testing by the Sidak-Bonferroni method with GraphPad Prism software.
Stool sample collection and DNA extraction.
Stool samples were collected from healthy volunteers after informed consent was obtained. The study protocol and any amendments were reviewed and approved by an independent review board (Western IRB, Puyallup, WA) before the start of the study. Stool samples were collected by the participants in the morning and transported to the Novartis Institute for Biomedical Research in Cambridge, MA, on the same day. Samples were then stored at −80°C until shipping to the Broad Institute for DNA extraction. DNA extractions from stool samples were carried out with the QIAamp DNA Stool minikit (QIAGEN).
Metagenome library construction.
Metagenomic whole-genome shotgun sequencing libraries were prepared as follows. Metagenomic DNA samples were quantified by Quant-iT PicoGreen dsDNA Assay (Life Technologies, Inc.) and normalized to a concentration of 50 pg/μl. Illumina sequencing libraries were prepared from 100 to 250 pg of DNA with the Nextera XT DNA Library Preparation kit (Illumina) in accordance with the manufacturer’s recommended protocol, with reaction volumes scaled accordingly. Batches of 24, 48, or 96 libraries were pooled by transferring equal volumes of each library with a Labcyte Echo 550 liquid handler. Insert sizes and concentrations for each pooled library were determined with an Agilent Bioanalyzer DNA 1000 kit (Agilent Technologies).
Sequencing and analysis of metagenomic samples.
Metagenomic whole-genome shotgun sequencing was performed essentially as previously described (10). Metagenomic libraries were sequenced on the Illumina HiSeq 2500 platform, targeting ~2.5 Gb of sequence per sample with 101-bp paired-end reads. Reads were quality controlled by trimming low-quality bases and removing reads of <60 nucleotides. Reads aligning with the human genome were identified with bowtie (40) and filtered out. Samples were profiled taxonomically with MetaPhlAn 2.0 (41) (http://huttenhower.sph.harvard.edu/MetaPhlAn2) and profiled functionally with HUMAnN2 (42) (http://huttenhower.sph.harvard.edu/HUMAnN2). HUMAnN2 maps metagenomic reads to UniRef50 (41) gene families of species identified in the MetaPhlAn2 taxonomic profiling step. Protein-coding sequences in these pangenomes have been preannotated to their respective UniRef50 families, which serve as a comprehensive, nonredundant protein sequence database. Reads that do not align with a known pangenome are separately mapped to the entirety of UniRef50 by translated search with DIAMOND (42). All hits are weighted on the basis of alignment quality and sequence length, with per-species and unclassified hits combined to produce community totals for each protein family (in addition to species-stratified totals) in numbers of reads per kilobase (RPK). RPK units were further normalized to numbers of RPK per million sample reads to account for variation in sequence depth across samples. Principal-component analysis plots were generated with the scikit-learn python package by using species abundance from MetaPhlAn2. Data are available at https://figshare.com/s/5e56cc1a347ef4f1de49 and https://www.ncbi.nlm.nih.gov/bioproject/PRJNA414479.
GO functional annotation.
We used HUMAnN2 to map UniRef50 gene families to GO terms, which were then aggregated into larger metabolic clusters with the CateGOrizer tool (43). This procedure yielded a comprehensive but manageable set of 13 nonredundant GO biological process terms for comparison of HMP1 and new samples (https://figshare.com/s/5e56cc1a347ef4f1de49).
Isolation of lipid A for MS analysis.
For analysis of crude biomass, pellets from 10-ml overnight broth bacterial cultures were washed three times in water, methanol, and chloroform (0.8:1:2). Alternatively, purified LPS (200 µg) was used directly. The material was subjected to mild acid hydrolysis at 100°C for 30 min in 12.5 mM sodium acetate buffer, pH 4.5, in the presence of 1% SDS to break the 3-deoxy-d-manno-octulosonic acid linkage, and free lipid A was recovered by two-phase Bligh-Dyer extraction. The lipid A species were analyzed with a MALDI-TOF mass spectrometer (Bruker Ultraflex) equipped with a smartbeam laser at a 2-kHz firing rate. Spectra were acquired in negative-ion linear mode. The matrix used was a saturated solution of 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1, vol/vol). Samples were dissolved in chloroform-methanol (4:1, vol/vol) and deposited on the sample plate, followed by an equal portion of matrix solution (0.5 µl).
HEK-293 NF-κB reporter cell assays.
HEK-293 cells (5 × 104) stably expressing the NF-κB-inducible Lucia luciferase reporter gene and the genes for either hTLR4, CD14, and MD2 (TLR4-HEK) or hTLR2 and CD14 (TLR2-HEK) (5 × 104) were seeded into the wells of 96-well plates and stimulated with doses of LPS purified from the strains indicated at doses ranging from 10−3 to 104 ng/ml for 6 to 8 h. For inhibition assays, cells were stimulated simultaneously with 1 ng/ml LPS purified from E. coli. Luciferase activity was measured by BrightGlo (Promega) in accordance with the manufacturer’s instructions. HEK-293 reporter cells were purchased from InvivoGen. All cell lines were tested for mycoplasma contamination with a PCR-based assay by an independent service provider.
Correlation of function and composition.
Human PBMCs were stimulated in the presence of increasing amounts of fecal LPS or cotreated with 1 ng/ml E. coli LPS and increasing doses of fecal LPS from the donors indicated. IL-6, TNF-α, and IL-1β concentrations in supernatants were measured after 20 h of culture and compared to those obtained with LPS treatment alone (0:1). Cytokine concentration upon stimulation at 1 or 10 μg/ml was obtained for LPS from individual stool samples and normalized. Inhibition potency at a ratio of 10:1 or 100:1 was obtained for individual stool samples and normalized. Five independent experiments were performed. Spearman rank correlation between the abundance of individual species and either stimulation or inhibition of IL-6, TNF-α, or IL-1β production was calculated by using pooled data from all experiments. Species that significantly correlate with function for all three cytokines measured after Benjamini-Hochberg correction are shown. Data are available at https://figshare.com/s/5e56cc1a347ef4f1de49.
ACKNOWLEDGMENTS
We thank Sena Fowler, John Annand, and Xiaoping Chen for assistance with sample preparation and experimental design, Glen Dillow for help with MS, and members of the NIBR Microbiome Hub for helpful conversation. We also thank Ramnik Xavier and Tiffany Poon of the Broad Institute for sequence production and sample management.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
E.H. and T.W.C. designed, conducted, and analyzed all experiments. S.A. and E.H. performed metagenomic data analysis. E.H., S.A., L.M., and T.W.C. assembled and wrote the manuscript. T.W.C. served as the principal investigator.
REFERENCES
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich S, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol Rev 90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 3.Rossi O, van Baarlen P, Wells JM. 2013. Host-recognition of pathogens and commensals in the mammalian intestine. Curr Top Microbiol Immunol 358:291–321. doi: 10.1007/82_2011_191. [DOI] [PubMed] [Google Scholar]
- 4.Mowat AM, Agace WW. 2014. Regional specialization within the intestinal immune system. Nat Rev Immunol 14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 5.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 6.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. 2013. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 7.Rossi O, Khan MT, Schwarzer M, Hudcovic T, Srutkova D, Duncan SH, Stolte EH, Kozakova H, Flint HJ, Samsom JN, Harmsen HJ, Wells JM. 2015. Faecalibacterium prausnitzii Strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. PLoS One 10:e0123013. doi: 10.1371/journal.pone.0123013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lahdesmaki H, Huttenhower C, Gevers D, Cullen TW, Knip M, DIABIMMUNE Study Group, Xavier RJ. 2016. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coats SR, Do CT, Karimi-Naser LM, Braham PH, Darveau RP. 2007. Antagonistic lipopolysaccharides block E. coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell Microbiol 9:1191–1202. doi: 10.1111/j.1462-5822.2006.00859.x. [DOI] [PubMed] [Google Scholar]
- 12.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. 2016. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med 8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westphal O, Jann K. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure, p 83–91. In Whistler RL (ed), Methods in carbohydrate chemistry, vol. 5. Academic Press, New York, NY. [Google Scholar]
- 16.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J Immunol 165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 17.Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, Tarelli E, Marsh PD. 2011. Temperature-dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defenses. Infect Immun 79:1187–1193. doi: 10.1128/IAI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 19.Tan Y, Zanoni I, Cullen TW, Goodman AL, Kagan JC. 2015. Mechanisms of Toll-like receptor 4 endocytosis reveal a common immune-evasion strategy used by pathogenic and commensal bacteria. Immunity 43:909–922. doi: 10.1016/j.immuni.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heij HA, Ekkelkamp S, Vos A. 1989. Lung operations in dyspneic newborn infants. Ned Tijdschr Geneeskd 133:1978–1981. [PubMed] [Google Scholar]
- 21.Coats SR, Pham TT, Bainbridge BW, Reife RA, Darveau RP. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J Immunol 175:4490–4498. doi: 10.4049/jimmunol.175.7.4490. [DOI] [PubMed] [Google Scholar]
- 22.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi N, Takayama K, Kurtz R. 1991. Diphosphoryl lipid A obtained from the nontoxic lipopolysaccharide of Rhodopseudomonas sphaeroides is an endotoxin antagonist in mice. Infect Immun 59:441–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. 2011. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog 7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A, Clemente JC, Cullender TC, Barnich N, Darfeuille-Michaud A, Vijay-Kumar M, Knight R, Ley RE, Gewirtz AT. 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, van Hylckama-Vlieg JE, Ballal SA, Morgan XC, Glickman JN, Gevers D, Huttenhower C, Garrett WS. 2014. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J 8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 31.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, Muccioli GG, Delzenne NM. 2009. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta NN, McGillicuddy FC, Anderson PD, Hinkle CC, Shah R, Pruscino L, Tabita-Martinez J, Sellers KF, Rickels MR, Reilly MP. 2010. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes 59:172–181. doi: 10.2337/db09-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters WA, Xu Z, Knight R. 2014. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett 588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caesar R, Reigstad CS, Bäckhed HK, Reinhardt C, Ketonen M, Lundén GÖ, Cani PD, Bäckhed F. 2012. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut 61:1701–1707. doi: 10.1136/gutjnl-2011-301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atilano ML, Pereira PM, Vaz F, Catalão MJ, Reed P, Grilo IR, Sobral RG, Ligoxygakis P, Pinho MG, Filipe SR. 2014. Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. Elife 3:e02277. doi: 10.7554/eLife.02277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finlay BB, McFadden G. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 37.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. 2004. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol 5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 38.Tien MT, Girardin SE, Regnault B, Le Bourhis L, Dillies MA, Coppée JY, Bourdet-Sicard R, Sansonetti PJ, Pédron T. 2006. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol 176:1228–1237. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- 39.Nair S, Archer GE, Tedder TF. 2012. Isolation and generation of human dendritic cells. Curr Protoc Immunol Chapter 7:Unit 7.32. doi: 10.1002/0471142735.im0732s99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, UniProt Consortium . 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 43.Na D, Son H, Gsponer J. 2014. Categorizer: a tool to categorize genes into user-defined biological groups based on semantic similarity. BMC Genomics 15:1091. doi: 10.1186/1471-2164-15-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Distribution of species in individual samples into the five main phyla in the new-sample (S1 to S10) and HMP1 sample sets. (B) Distribution of encoded functional pathways in individual samples of the new data set (NVS samples) or the HMP1 data set (HMP). (C) Contribution of the phylum Bacteroidetes to the total LPS-encoding capacity of the gut microbiome relative to that of other phyla. Data are presented for each individual donor. The darker-color set is HMP samples, and the lighter-color set is NVS samples. Download FIG S1, TIF file, 0.8 MB (794.7KB, tif) .
Copyright © 2017 d’Hennezel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Stimulation of PBMCs by Bacteroidales LPS. (A and B) Human PBMCs were stimulated in the presence of increasing amounts of LPS from the Bacteroidales species indicated. TNF-α and IL-1β concentrations in supernatants were measured after 20 h of culture. E. coli LPS was included as a control. Data shown are the mean ± the standard deviation of triplicates in one representative experiment. (C and D) LPS isolated from the species indicated. Human PBMCs were cotreated with zymosan and increasing doses of Bacteroidales LPS. TNF-α and IL-1β concentrations in supernatants were measured after 20 h of culture. Treatment with zymosan alone was included as a control (black bar). Data shown are the mean ± the standard deviation of triplicates in one representative experiment. Download FIG S2, TIF file, 0.6 MB (586.3KB, tif) .
Copyright © 2017 d’Hennezel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
EPS extracts from bacterial species of the human microbiome have immunomodulatory properties. (A) Phylogenetic tree of the 120 most abundant bacterial strains in the human gut microbiome. EPS extracts were prepared from 29 strains (red). (B and C) Human monocyte-derived dendritic cells (B and C) or PBMCs (D and E) were cotreated with individual EPS extracts and zymosan (B and D) or LPS (C and E). The TNF-α concentration in supernatants was compared to that obtained with zymosan or LPS treatment alone. Data shown are the mean of triplicate measurements in one representative of three or more independent experiments. (F) EPS extracts from Gram-negative species were analyzed by SDS-PAGE, followed by Pro-Q Emerald staining for glycoproteins. Purified E. coli LPS was loaded as a control. Download FIG S3, TIF file, 2.2 MB (2.3MB, tif) .
Copyright © 2017 d’Hennezel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.