ABSTRACT
Much genetic diversity within a bacterial community is likely obscured by microdiversity within operational taxonomic units (OTUs) defined by 16S rRNA gene sequences. However, it is unclear how variation within this microdiversity influences ecologically relevant traits. Here, we employ a multifaceted approach to investigate microdiversity within the dominant leaf litter bacterium, Curtobacterium, which comprises 7.8% of the bacterial community at a grassland site undergoing global change manipulations. We use cultured bacterial isolates to interpret metagenomic data, collected in situ over 2 years, together with lab-based physiological assays to determine the extent of trait variation within this abundant OTU. The response of Curtobacterium to seasonal variability and the global change manipulations, specifically an increase in relative abundance under decreased water availability, appeared to be conserved across six Curtobacterium lineages identified at this site. Genomic and physiological analyses in the lab revealed that degradation of abundant polymeric carbohydrates within leaf litter, cellulose and xylan, is nearly universal across the genus, which may contribute to its high abundance in grassland leaf litter. However, the degree of carbohydrate utilization and temperature preference for this degradation varied greatly among clades. Overall, we find that traits within Curtobacterium are conserved at different phylogenetic depths. We speculate that similar to bacteria in marine systems, diverse microbes within this taxon may be structured in distinct ecotypes that are key to understanding Curtobacterium abundance and distribution in the environment.
KEYWORDS: Actinobacteria, Curtobacterium, Microbacteriaceae, drought, ecotypes, glycoside hydrolases, nitrogen addition
IMPORTANCE
Despite the plummeting costs of sequencing, characterizing the fine-scale genetic diversity of a microbial community—and interpreting its functional importance—remains a challenge. Indeed, most studies, particularly studies of soil, assess community composition at a broad genetic level by classifying diversity into taxa (OTUs) defined by 16S rRNA sequence similarity. However, these classifications potentially obscure variation in traits that result in fine-scale ecological differentiation among closely related strains. Here, we investigated “microdiversity” in a highly diverse and poorly characterized soil system (leaf litter in a southern Californian grassland). We focused on the most abundant bacterium, Curtobacterium, which by standard methods is grouped into only one OTU. We find that the degree of carbohydrate usage and temperature preference vary within the OTU, whereas its responses to changes in precipitation are relatively uniform. These results suggest that microdiversity may be key to understanding how soil bacterial diversity is linked to ecosystem functioning.
INTRODUCTION
Currently, most studies assessing the responses of bacterial communities to environmental change rely on broad taxonomic designations, for instance, by using operational taxonomic units (OTUs) based on the nucleotide sequence similarity of the 16S rRNA gene (1). While this classification of bacterial diversity can capture broad taxonomic shifts, it provides limited genetic resolution at this loosely defined species level (2–4) by obscuring important genetic diversity within the OTU (5–7)—so-called microdiversity (8, 9). Given that most studies investigate microbial composition using 16S rRNA gene-defined OTUs (specifically, at the 97% level), a large gap in our understanding is the extent of microdiversity in natural communities and its relationship to variation in bacterial traits.
Growing evidence indicates that the genetic variation encompassed by bacterial microdiversity corresponds to variation in a wide range of functional traits (10). At fine genetic scales (11, 12), microbes with distinct physiological traits may partition niche space within the environment (13, 14). For example, extensive work in marine systems has demonstrated that microdiversity within a 16S rRNA gene-defined taxon encompasses distinct ecotypes, or lineages that respond differently to variation in the environment over space and time (14–17). However, our ability to characterize ecotypes at fine taxonomic levels is still largely dependent on cultured organisms because of the need to link genomic variation to phenotypic variation (18). Also, while metagenomic sequencing has advanced the identification of uncultivated organisms (19), the functional role of microdiversity has rarely been considered in soils, as we lack cultured representatives of microbes that are abundant in soil.
Diverse bacterial and fungal communities on leaf litter, the top layer of soil, play a key role in the carbon cycle. Litter decomposition mediates the loss of carbon through respiration to the atmosphere or its storage as organic matter in soil (20). The Loma Ridge global climate experiment (LRGCE) in southern California was established to test how future changes in precipitation and nitrogen availability may alter semiarid grassland and coastal sage scrub ecosystems. In grasslands at the LRGCE, the litter microbial community is dominated by bacteria (21), suggesting that bacteria perform the bulk of grassland litter decomposition. Over a 2-year period, the leaf litter community responded weakly but significantly to treatment manipulations (22, 23). At the 97% OTU level, a Curtobacterium OTU (phylum Actinobacteria, family Microbacteriaceae) was the most abundant taxon within the bacterial community (23). An analysis of Curtobacterium sequences from around the globe revealed the genus to be a cosmopolitan terrestrial taxon, with isolates derived primarily from plant and soil habitats (24). Further, genomic sequencing of Curtobacterium strains isolated from leaf litter indicated that the genus has a high genomic potential to decompose polymeric carbohydrates such as starch, cellulose, and xylan that are abundant in leaf litter (24).
Our previous work demonstrated that isolates belonging to Curtobacterium harbored extensive genomic diversity despite being clustered within a single OTU as defined by 16S rRNA (24). Here, we ask the following questions. (i) What is the extent of Curtobacterium microdiversity in a natural leaf litter bacterial community? (ii) Does this microdiversity encompass genetic and physiological variation in ecologically relevant traits? To address these questions, we used a combination of environmental field data and physiological lab assays to assess the distribution of traits within Curtobacterium and their phylogenetic conservatism. First, we examined the response of Curtobacterium microdiversity to manipulations of precipitation and nitrogen availability by using cultured isolates to inform metagenomic data. Moisture limitation, in particular, is likely a major stressor on litter bacteria in Southern California, which experiences long dry seasons with little rainfall. Second, we assayed both the genomic potential and metabolic capacity of isolates to depolymerize cellulose and xylan. As leaf litter is primarily composed of these polysaccharides, access to this primary carbon supply in this environment may be a highly advantageous trait.
RESULTS
Curtobacterium abundance and microdiversity.
We characterized Curtobacterium abundance and its microdiversity at the Loma Ridge global climate experiment (LRGCE) using 48 metagenomic sequence libraries from litter samples collected over a 2-year period. To estimate its relative abundance within the bacterial community, we created a custom pipeline using a curated reference database of 3,019 genomes representing 1,464 bacterial genera, including 16 Curtobacterium genomes (see Fig. S1 in the supplemental material). We calculated taxonomic abundance by using a phylogenetic classification of the metagenomic reads against the reference phylogeny, which we constructed using single-copy marker genes (25). Using our pipeline, we identified Actinobacteria and Microbacteriaceae as the most abundant phylum (46.3%) and family (28.2%), respectively. We detected similar relative abundances using MG-RAST annotations of the marker genes (Text S1; Table S1), but this approach did not detect any Curtobacterium. Therefore, we used the new pipeline to investigate finer taxonomic levels. This analysis revealed that Curtobacterium was the most abundant genus observed over the 2-year period within the leaf litter community, comprising an average of 7.8% of the bacterial community. However, even with our pipeline, we were unable to characterize 31.2% of the marker genes at or below the genus level.
Comparison of bacterial community analyses. Download TEXT S1, DOCX file, 0.1 MB (117.2KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reference database constructed from >3,000 genomes representing the entire Bacteria domain (two genomes per genus). Multilocus phylogenetic analysis is unrooted and based on a concatenated alignment of 29 single-copy marker genes. Download FIG S1, TIF file, 3.1 MB (3.1MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Average taxonomic relative abundance of the bacterial communities across the metagenomic samples from 2010 to 2012 at the LRGCE. The values are average percent taxonomic relative abundance ± 1 standard deviation (SD). Download TABLE S1, DOCX file, 0.1 MB (62.7KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
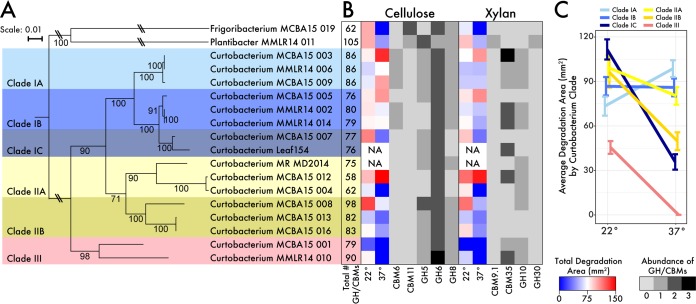
On the basis of the full-length 16S rRNA gene, the Curtobacterium genomes (14 of which were cultured isolates from leaf litter [24] and two other publically available genomes) clustered in the same OTU defined at the 97% sequence identity level. We therefore identified genomic clusters within the Curtobacterium OTU using a phylogenetic analysis of 29 single-copy marker genes and grouped the isolates into six well-supported clades (Fig. 1A). These clades were supported by nucleotide (average nucleotide identity [ANI]) and amino acid (average amino acid identity [AAI]) similarity (Table S2). Specifically, isolates shared >97% AAI within clades for the 29 marker genes. Across the whole genome, isolates within clades were more similar in ANI and AAI than isolates between clades, which had a minimum pairwise similarity of 83.2% ANI and 78.9% AAI across all Curtobacterium isolates.
FIG 1 .
Phylogeny and traits of Curtobacterium strains. (A) Multilocus phylogenetic analysis using a concatenated alignment of 29 single-copy marker genes. Bar, 0.01 amino acid substitutions per position. (B) Genomic and physiological metrics of carbohydrate utilization. The total number of GH/CBM families targeting all potential carbohydrate substrates is shown in the first column. The physiological ability to degrade cellulose and xylan is shown in blue or red, while the genomic potential (presence of GH/CBM families) to degrade either cellulose or xylan is represented in gray or black. Strains that were not assayed (NA) for carbon degradation are indicated. (C) Average degradation area (±1 standard deviation [SD] [error bar]) of the substrates by Curtobacterium clade at each temperature.
Genomic characterizations of all Curtobacterium isolates within individual clades. Download TABLE S2, DOCX file, 0.1 MB (55.9KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We then classified the metagenomic marker gene reads assigned to Curtobacterium in the taxonomic analysis onto the six identified clades. Only a tiny fraction (0.27% of the total bacterial community) of the bacteria identified as Curtobacterium failed to classify into one of the six clades, suggesting that our isolates encompassed most of the genomic diversity of Curtobacterium at the LRGCE. Across all samples, Curtobacterium was dominated by two clades (Table S1); clades IA and III averaged 3.0% and 2.4% of the marker gene sequences, respectively (Fig. 2C). Together, the remaining Curtobacterium clades (clades IB, IC, IIA, and IIB) composed >2% of the bacterial community, but separately, each of the four clades represented <0.6% of the bacterial community (Table S1).
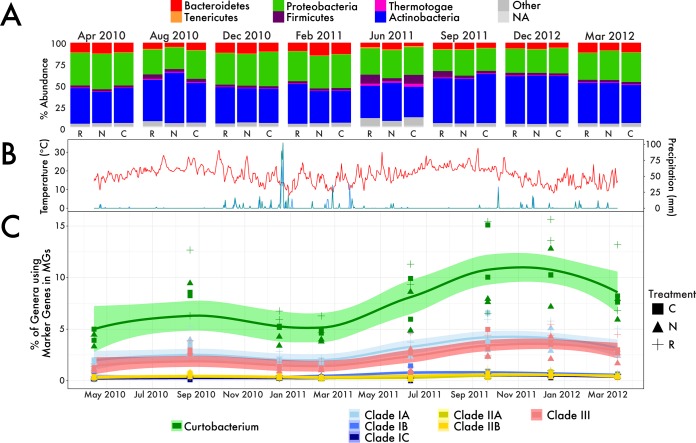
FIG 2 .
Bacterial community composition in the Loma Ridge field site over 2 years. (A) Relative abundances of the six most abundant phyla; replicates were averaged for each treatment and time point. The treatments were the addition of nitrogen (N), reduced precipitation (R), and control treatment (C). NA, not available. (B) Temperature and precipitation at Loma Ridge collected from May 2010 to March 2012. (C) Relative abundance of total Curtobacterium and each individual clade over time and by treatment. Smoothed averages (lines) were calculated from locally weighted smoothing (LOESS) with confidence intervals (colored areas). MGs, metagenomic sequences.
Response to the global change treatments.
Within the global change experiment, the composition of the microbial community varied seasonally by sampling date such that some bacterial phyla, including Actinobacteria, were strongly correlated with background precipitation (Fig. S2A) as previously reported (26). Indeed, at the phylum level (Fig. 2A), bacterial composition varied significantly over time (Bray-Curtis similarity; P < 0.002 by permutational multivariate analysis of variance [PERMANOVA]) and responded marginally to the global change treatments of reduced precipitation (drought) and added nitrogen (P = 0.061), with no significant interaction between the two factors. However, the global change treatments explained only 1.9% of the variation in phylum composition, whereas time (date of collection over a 2-year period) explained 65.1%. In particular, during a prolonged hot, dry season in the second year (Fig. 2B), the bacterial community became dominated by Actinobacteria (Fig. 2A).
Multidimensional scaling (MDS) plot depicting the difference in microbial community relative abundances at the phylum level (A) and clade level within Curtobacterium (B). Symbols represent sampling date with darker colors indicating winter/spring months and warmer colors indicating summer/fall months. Download FIG S2, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Much of the response of Actinobacteria to the global change treatments was due to Curtobacterium. The relative abundance of all Curtobacterium increased by 20.2% in the drought treatment and decreased by 17.2% in the nitrogen treatment relative to the control plots (P < 0.05 by PERMANOVA; Table S3). Similar to the phylum-level response, the time of sampling explained the greatest amount of variation in Curtobacterium abundance, accounting for 52.6% of the variation, while the treatment accounted for only 5.0%. Curtobacterium abundance was strongly associated with seasonal precipitation (Fig. S3A), increasing in relative abundance during the dry seasons and accounting for more than 10% of all leaf litter bacteria in the second, drier year of the study (Fig. 2B and C). Curtobacterium abundance, however, was not correlated with the mean temperature in the field 2 weeks prior to sampling (Fig. S3B).
Average relative abundance of Curtobacterium in the bacterial community at LRGCE as a function of the total precipitation (A) and the mean temperature (B) in the 2 weeks prior to sampling. Average relative abundance (± 1 SD) is shown. Symbols represent sampling date with darker colors indicating winter/spring months and warmer colors indicating summer/fall months. Download FIG S3, TIF file, 27 MB (27.6MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of Curtobacterium clades by treatment averaged across the metagenomic libraries from the LRGCE. Values are relative percent abundance ± 1 SD. Download TABLE S3, DOCX file, 0.1 MB (53.2KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next we tested whether microdiversity within Curtobacterium (and in particular, the six identified clades) varied in their responses to the global change treatments. All Curtobacterium clades responded similarly to drought, increasing in abundance relative to the control and, with the exception of clade IC, responded negatively to the increased nitrogen treatment (Table S3). Furthermore, Curtobacterium clade composition varied significantly over time (P < 0.001 by PERMANOVA; Fig. S2B), with clades IA and III increasing in relative abundance during the drier, second year of the study (Fig. 2C).
Carbohydrate degradation traits within Curtobacterium. (i) Genomic characterization.
To analyze the genomic potential for carbohydrate degradation, we characterized the glycoside hydrolase (GH) and carbohydrate binding module (CBM) protein families within and among Curtobacterium clades. The abundance of total GH and CBM (GH/CBM) genes varied among all genomes, ranging from 58 to 98 GH/CBM copies. The total distribution of GH/CBM genes varied significantly with phylogenetic distance such that more closely related genomes carried more similar copy numbers (RELATE test; ρ = 0.45 and P < 0.05; Fig. S4). Clades IA, IIB, and III encoded the highest abundance of GH/CBM genes (an average of 86, 87.7, and 84.5 genes), which differed significantly (F5,10 = 5.3027 by analysis of variance [ANOVA]; P < 0.05) from clade IIA (65 genes), whereas clades IB and IC encoded intermediate numbers of these genes (76.5 and 78.3, respectively; Fig. 1B).
Multidimensional scaling (MDS) plot depicting total GH/CBM gene composition for Curtobacterium isolates. Each symbol represents a genomic isolate and is colored according to the assigned clade. Download FIG S4, TIF file, 16.3 MB (16.7MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we considered the GH/CBM gene diversity that is thought to be responsible for degradation of the most abundant carbohydrates in the leaf litter at the LRGCE, cellulose and hemicellulose (specifically, xylan) (22). Overall, the numbers of both cellulose- and xylan-related GH/CBMs were significantly correlated with phylogenetic distance (RELATE test; ρ = 0.57 and P < 0.01 and ρ = 0.26 and P < 0.05, respectively). All Curtobacterium genomes contained at least one copy of a GH or CBM protein family that targeted either cellulose or xylan. However, some strains (e.g., MCBA15013 and MCBA15016, both from clade IIB) had an elevated abundance of GH/CBM genes targeting cellulose, with an apparent absence of genes targeting xylan. Clades IA and IB were the only clades to contain both GH and CBM genes targeting each substrate (Fig. 1B).
(ii) Phenotypic characterization.
The presence of GH/CBM genes within a genome suggests only the potential for substrate utilization. Therefore, we conducted substrate assays in the laboratory to confirm the ability of each isolate to degrade cellulose and xylan at 22°C. We performed these assays at this temperature, as the optimum growth for the genus is thought to range from 20 to 26°C (27, 28). All but one of the strains (MCBA15001) degraded both cellulose and xylan over a 4-day period, including the four isolates that did not carry known xylan-targeting genes (Fig. 1B). Indeed, the size of an isolate’s zone of depolymerization was not correlated with the abundance of genes targeting either cellulose (F1,14 = 1.24 by phylogenetic independent contrast [PIC] analysis; P > 0.05) or xylan (F1,14 = 0.15 by PIC analysis; P > 0.05).
The degradation patterns of the Curtobacterium strains also depended greatly on the temperature of the assay. When isolates were assayed at 37°C, the expected maximum temperature for growth in Curtobacterium (28), four strains exhibited an increase in degradation capability, including two strains from clade IA, while three strains were unable to degrade either substrate at 37°C (Fig. 1B). The total area of the zone of depolymerization varied significantly by temperature (F1,43 = 4.67 by analysis of covariance [ANCOVA]; P < 0.05) and clade (F5,43 = 4.74; P < 0.01), with a significant interaction between them (F5,43 = 2.46; P < 0.05), whereas the assay substrate had no effect on the depolymerization area (F1,43 = 0.95; P > 0.05). When averaged across Curtobacterium clades, only clade IA saw an average increase in depolymerization area when strains were grown at 37°C compared to 22°C (Fig. 1C). Most clades maintained some level of degradation capability at the higher temperature except for clade III, which failed to depolymerize either cellulose or xylan at 37°C (Fig. 1C).
DISCUSSION
In this study, we investigated the extent of genomic microdiversity of Curtobacterium in the field and the relationship between this diversity and the bacterium’s functional traits. To our knowledge, this study is the first to do so in a dominant soil bacterium. As in aquatic and host-associated ecosystems (6, 8, 14, 29–31), microdiversity within this abundant bacterium is extensive. Cooccurring strains within the same Curtobacterium OTU have an average nucleotide identity (ANI) of as low as 83%, far below the traditional species boundary (32). Our results support the growing understanding that traditional taxonomic assignments (i.e., OTUs) are insufficient to resolve ecologically distinct microorganisms (33, 34). Indeed, extensive Curtobacterium microdiversity persists in grassland leaf litter and encompasses variation in several ecologically relevant traits, including its ability to degrade abundant carbohydrates as well as temperature preferences for this degradation. Thus, binning 16S rRNA sequences obscures detection and interpretation of ecologically important trait variance.
Trait variability within soil bacterial OTUs has been described previously, suggesting that local adaptation and coexistence are probable among closely related strains (35–38). However, the combination of lab assays on cultured representative isolates in conjunction with metagenomic data allowed us to compare the physiological findings to their representation in the environment, as well as test the response to environmental change in the context of the whole community. Further, this combination enabled us to quantify and interpret metagenomic data of ecologically relevant microdiversity that would otherwise not be detected (Table S1) due to the absence of genomic representation in public databases. Indeed, particularly for terrestrial soil systems, the genomic reference databases often lack the resolution to detect fine-scale taxonomic groups, as defined by an ANI of >95% (39), or result in mischaracterization of taxonomic groups altogether (40).
The results of this study are also consistent with the idea that bacterial traits are often conserved at different phylogenetic depths (14, 41). Complex quantitative traits like an organism’s response to drought have been proposed to be more phylogenetically conserved (10, 41). Here, we observed that a response to dry conditions (both in the drought treatment and the dry seasons) appears to be generally consistent among Curtobacterium clades, suggesting that biological and physiological traits responsible for moisture response are ecologically cohesive (42) within this taxon. Thus, the response of Curtobacterium to future drought would likely be apparent at the OTU level, although certain clades may be more abundant than others. However, given that some of the clades were relatively rare within the community, further investigation is still needed to confirm this interpretation.
In contrast, traits that rely on one gene or a few genes such as carbon utilization are thought to be more shallowly conserved (43), as they may be more prone to horizontal gene transfer. Using physiological assays, we confirmed the genomic potential for Curtobacterium to degrade polymeric carbohydrates, which are likely central to their success within the leaf litter community. Although all Curtobacterium clades could depolymerize both xylan and cellulose, the degree of carbohydrate utilization varied among and within clades, suggesting that carbohydrate utilization is finely conserved. Such intricate differences in carbohydrate degradation traits among Curtobacterium may contribute to the persistence of this microdiversity within the leaf litter community. However, the genomic potential for carbohydrate utilization (number and composition of GH/CBMs) did not predict observed phenotypic variation in the lab, highlighting the difficulty in using gene annotations to predict ecological roles.
Carbohydrate degradation was also temperature dependent, regardless of the substrate. Further, this dependency varied among clades, revealing that Curtobacterium microdiversity also incorporates variation in temperature preference. Broadly, this result supports the idea that bacterial temperature preference can be relatively finely conserved (41), in agreement with rapid temperature adaptation observed in the lab (44). More specifically, it suggests differential physiological tradeoffs between temperature and carbohydrate utilization (45) among clades. Such variation in this tradeoff might explain the coexistence of these closely related clades, particularly for the two most abundant clades, clades IA and III. Despite similar environmental responses to drought and seasonal fluctuations, these clades exhibited opposite responses to temperature with respect to carbohydrate utilization (Fig. 1C). While temperature preference has previously been shown to drive shifts in ecotype abundance within marine systems (16), we did not observe a correlation between clade abundance and temperature at this one site. However, further investigation is needed across a wider temperature range to test whether temperature drives the geographic distribution of Curtobacterium clades.
In conclusion, the microdiversity within a single Curtobacterium OTU in this grassland leaf litter encompasses variation in traits involved in carbon degradation and temperature preference. Classic ecological theory would suggest that this trait variation allows microdiversity to occupy distinct ecological niches (46), although further work is needed to identify distinct Curtobacterium ecotypes in the environment. At the same time, Curtobacterium appears to be consistent in its response to changes in precipitation, suggesting that variability in moisture conditions are unlikely to explain the maintenance of this microdiversity. Thus, similar to marine bacteria (8, 9), our work highlights that the depth of trait conservatism (14) may help to understand the response of soil bacteria to changing environments.
MATERIALS AND METHODS
Field site.
The Loma Ridge global change experiment (LGRCE) (in Irvine, California, USA [33° 44′ N, 117° 42′ W]) (47) was established in 2007 with precipitation and nitrogen manipulations in areas of deciduous shrubland (coastal sage scrub) and annual grasses. For this study, we sampled only in the grassland plots, which are dominated by Avena, Bromus, and Lolium (22). We used a subset of the plots that included reduced precipitation treatment (50% reduction in annual precipitation), added nitrogen treatment (20 to 40 kg N/ha), and a control treatment, as previously described (22).
We collected leaf litter from these plots by sampling each season from May 2010 to March 2012 across three treatments: control, reduced precipitation (drought), and added nitrogen (8 time points × 3 treatments × 2 replicates). As described previously, metagenomic libraries were created from these samples by extracting DNA from ground litter, prepared using a TruSeq library kit (Illumina, San Diego, CA), and sequenced on an Illumina HiSeq2000 instrument. Samples were pooled from eight plots from each treatment to form the two replicate libraries at each time point (for more information, see reference 26). The sequence libraries are available on MG-RAST under the project identifiers (IDs) 4511045 to 4511050, 4511060 to 4511065, 4511111 to 4511116, 4511134 to 4511153, and 4511178 to 4511193. We excluded two libraries (Drought April 2010 and Nitrogen August 2010) due to low sequence count. Temperature and precipitation data were recorded at a nearby weather tower (22).
Curated marker gene reference database.
We developed a reference genomic database to characterize phylogenetic marker genes from the metagenomic sequences of the microbial community. This approach is similar to PhyloSift (48), except we performed a more robust search to compensate for the lack of genomic references to characterize soil microbial communities. We downloaded 79,838 genomes from the PATRIC database (49) with RAST (50) annotations on 9 December 2016. We screened all genomes for annotations of 29 conserved, single-copy phylogenetic marker genes (25) and discarded failed genomes, most of which were draft genomes with >1,000 contigs. The remaining genomes were manually curated by assigned nomenclature to include two genomes per genus. We used complete genomes and genomes isolated from soil ecosystems when they were available. The 3,159 resulting genomes were combined with 14 Curtobacterium genome sequences isolated from two grassland leaf litter sites (24), including four strains isolated during the time of metagenomic sampling from the LRGCE.
We curated the downloaded genomes to ensure that all genomes were properly assigned to the correct taxon. Individual marker genes from each genome were aligned using ClustalO v1.2.0 (51) and used to construct a 15,963-bp concatenated alignment for phylogenetic analysis using FastTree 2 (52). The resulting reference phylogeny guided the construction of each individual marker gene tree to maintain relative node structure across trees. For each marker gene tree, we performed a maximum likelihood bootstrap analysis using RAxML v8.0.0 (53) under the PROTGAMMAWAGF model for 100 replicates. If a genome was named incorrectly or showed a problematic alignment for any of the individual marker gene trees (i.e., genome terminal branch length was >5), the entire genome was removed (a total of 154 genomes were removed), and all trees were regenerated.
The NCBI taxonomy database (54) was downloaded on 17 June 2016. The taxonomic information of the remaining 3,019 genomes was added locally to the NCBI database using the PATRIC genome IDs. The individual marker gene trees and taxonomic information were all used to generate reference packages for the program pplacer v1.1.alpha17 (55). Reference packages were subsequently used to characterize the microbial community (available at https://github.com/alex-b-chase/LRGCE).
Metagenomic analyses.
To evaluate the taxonomic diversity of the bacterial community as well as finer-scale diversity within Curtobacterium at the LRGCE, we reanalyzed the metagenomic libraries previously described (26). Metagenomes were retrieved from the metagenomics analysis server (MG-RAST) (56) after sequences had been processed for quality control and coding regions were predicted by FragGeneScan (57). We performed an initial filter using BLASTP (58) against our custom database with an E value of 1 × 10−5. We applied a secondary filter using HMMER v3.1b2 (59) with an E value of 1 × 10−10 to achieve a higher specificity. We grouped the filtered reads for each library by each marker gene and aligned them using ClustalO v1.2.0 (51) to the corresponding marker gene reference package (see above). Aligned metagenomic reads were “placed” onto the reference phylogeny using pplacer v1.1.alpha17 (55), keeping at most 20 placements, and a posterior probability for final placement on the reference tree was calculated. Finally, we created single branch abundance matrices yielding an abundance distribution ranging from phyla to individual genomes. All abundances were normalized by the total number of marker genes present.
Comparison of the curated pipeline to other methods.
To validate the taxonomic results generated by our custom pipeline (Text S1), we compared our taxonomic abundances using two alternative approaches: (i) the MG-RAST pipeline using a read-based analysis and (ii) a de novo coassembly of all metagenomic libraries using paired-end reads.
First, to generate the MG-RAST taxonomic profiles, we downloaded the Kyoto Encyclopedia of Genes and Genomes (KEGG) database annotations for each library from the MG-RAST API (56) and calculated relative abundances across all annotated reads. Next, we standardized the MG-RAST output by filtering the MD5 IDs corresponding to the 29 marker genes and regenerated standardized taxonomic abundance profiles. All gene sequences retrieved from MG-RAST were assigned to the closest hit genus in the MG-RAST database using an E value of 1 × 10−5.
Second, we conducted a genome-centric analysis by performing a de novo coassembly of all of the paired-end shotgun metagenomic libraries using MEGAHIT (60). We used an iterative k-step ranging from k = 27 to 111 and discarded all assembled contigs of <3,000 bp. Read coverage for each assembled contig was calculated using bbwrap.sh within the suite of tools available via BBMap v35.66 (61). Taxonomic assignments for all assembled contigs were generated using MegaBLAST against the NCBI nucleotide database (January 2015 version) with an E value of 1 × 10−5.
Genomic comparisons of the isolates.
To validate that all Curtobacterium genomes, including two publically available Curtobacterium genomes, clustered within the same OTU, we used Barrnap (http://www.vicbioinformatics.com/software.barrnap.shtml) to predict rRNA genes and clustered the 16S rRNA gene using UCLUST (62). We then examined the relationship among all 16 Curtobacterium genomes using 29 single-copy phylogenetic marker genes (25). Each conserved gene was independently aligned using ClustalO v1.2.0 (51) and used to create a concatenated alignment for phylogenetic analyses. We constructed a maximum likelihood phylogenetic tree using RAxML v8.0.0 (53) under the PROTGAMMAWAGF model for 100 replicates. For convenience, we designated six monophyletic clades based on the results from the phylogenetic analyses. To confirm these designations, we calculated pairwise average amino acid identity (AAI) across the 29 marker genes across all genomes.
Next, we confirmed that our clade designations were in accordance with additional genomic characterizations. Specifically, we calculated pairwise whole-genome average nucleotide identity (ANI) and AAI (63) and computed the core genome for each clade by generating groups of orthologous proteins with MCL (64). Genes identified as orthologous groups within clades were subsequently used to calculate the AAI of all clade-specific core genes. All genomic analyses were performed using the suite of tools available in the Microbial Genomes Atlas (MiGA) (https://github.com/bio-miga/miga).
To analyze each genome for its potential to degrade carbohydrates, genomic open reading frames (ORFs) were generated by the RAST annotation pipeline (50) and searched using HMMER against the Pfam-A v30.0 database (65). We then used a subset of identified protein families, representing glycoside hydrolase (GH) and carbohydrate binding module (CBM) proteins to identify the genomic potential to degrade carbohydrates of each isolate (24, 66). GH and CBM (GH/CBM) gene composition profiles for each isolate were subsequently used to generate a Bray-Curtis similarity matrix to produce a nonmetric multidimensional scaling (MDS) ordination plot.
Physiological analyses of the isolates.
In the laboratory, we characterized the 14 Curtobacterium isolates for their ability to utilize two polysaccharides, cellulose and xylan, at two temperatures. All isolates were grown from −80°C freezer stocks for 24 to 48 h in LB liquid medium at room temperature (22°C). Isolates were spun down at a relative centrifugal force (RCF) of 13,500 for 4 min, and the LB supernatant was discarded. Pelleted cultures were washed with 0.9% saline solution three times and resuspended in 10 ml of M63 minimal medium with 0.5% (wt/vol) dextrose and allowed to grow for 24 h. All cultures were then diluted to an optical density at 600 nm (OD600) of 0.1 to ensure equal cell density across isolates. We used 10 µl of the cultures grown (in triplicates) to inoculate solid M63 medium containing 0.5% (wt/vol) carboxymethyl cellulose (CMC) (catalog no. 150560; MP Biomedicals) or xylan (catalog no. X0502; Sigma) and placed at 22°C (optimum temperature for growth [27, 28]) and 37°C (maximum temperature for growth [28]). Depolymerization of each substrate was classified after 4 days by measuring the zones of transparent growth around the inoculum as previously described (67) with Gram’s iodine stain (68). We analyzed the zones of depolymerization around inoculated colonies on ImageJ (https://imagej.nih.gov/ij/) to calculate the total area of carbohydrate degradation. An Escherichia coli strain was included as a negative control for all physiological assays.
Statistical analyses.
To test the effects of environmental treatment manipulations on the distribution of bacterial communities and Curtobacterium clade composition, we used a permutational multivariate analysis of variance (PERMANOVA) (69). The statistical model included plot treatment (control, drought, or N addition) and date of collection as fixed effects. We tested the effects of time and treatment by generating Bray-Curtis similarity matrices at the phylum and clade taxonomic levels. Subsequent PERMANOVA analyses used a type III partial sum of squares for 999 permutations of residuals under a reduced model. Similarity matrices were also used to generate nonmetric multidimensional scaling (MDS) ordination plots. All multivariate statistical analyses were conducted using PRIMER6 with the PERMANOVA+ function (Primer-E Ltd., Ivybridge, UK).
We analyzed the distribution of GH/CBM genes within and among Curtobacterium clades. To test for differences in the total abundance of GH/CBM proteins across clades, we used a one-way analysis of variance (ANOVA). For the ANOVA analysis, we used a Tukey’s honestly significant difference test to detect the difference in the total abundance of GH/CBM genes across clades. To test for correlations between the abundance of GH/CBM proteins, with respect to cellulose and xylan, and phylogenetic distance, we calculated a Spearman’s rank correlation coefficient using a RELATE test. Further, we performed a phylogenetic independent contrast (PIC) analysis to test whether the abundance of GH/CBM genes was related to an isolate’s phenotypic ability to degrade cellulose or xylan in the laboratory. Finally, to determine the factors driving degradation, we used a multiple regression model including the following variables: temperature, clade designation, and carbon substrate. Starting with a three-way analysis of covariance (ANCOVA), we implemented a backward selection process (70). If the model returned nonsignificant interactions, the interaction was removed, and the model was regenerated to decrease the chance of spurious relationships (71). All analyses were performed in the R software environment.
Coverage profiles of all coassembled contigs as a function of their GC content. Taxonomic assignments are colored by the top 10 most abundant bacterial genera (listed in descending order). Download FIG S5, TIF file, 4.3 MB (4.4MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Michaeline Nelson and Steven Allison for comments on earlier versions and Alberto Lopez for assistance in physiological assays. We thank Kristin Dolan and Renaud Berlemont for allowing us to use their isolates and metagenomic data, respectively. We thank Michael Goulden for establishing and maintaining the LRGCE.
Support for this project was provided by a U.S. Department of Education Graduate Assistance of National Need (GAANN) fellowship (A.B.C.), a UC MEXUS-CONACYT Postdoctoral Research Fellowship (Z.G.-L.), the National Science Foundation (DEB-1457160), and the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under awards DE-SC-SC0016410 and DE-PS02-09ER09-25. Work at Lawrence Berkeley National Laboratory (E.L.B. and U.K.) was supported by the Department of Energy under contract DE-AC02-05CH11231 with the University of California.
Footnotes
Citation Chase AB, Karaoz U, Brodie EL, Gomez-Lunar Z, Martiny AC, Martiny JBH. 2017. Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio 8:e01809-17. https://doi.org/10.1128/mBio.01809-17.
REFERENCES
- 1.Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D, Konstantinidis KT. 2014. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole JR, Konstantinidis K, Farris RJ, Tiedje JM. 2010. Microbial diversity and phylogeny: extending from rRNAs to genomes, p 1–20. In Liu W-T, Jansson JK (ed), Environmental molecular microbiology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. 2015. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 6.Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- 7.Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol 10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Moore LR, Rocap G, Chisholm SW. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 9.Jaspers E, Overmann J. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol 70:4831–4839. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larkin AA, Martiny AC. 2017. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ Microbiol Rep 9:55–70. doi: 10.1111/1758-2229.12523. [DOI] [PubMed] [Google Scholar]
- 11.Polz MF, Hunt DE, Preheim SP, Weinreich DM. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc Lond B Biol Sci 361:2009–2021. doi: 10.1098/rstb.2006.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL. 2015. A single genus in the gut microbiome reflects host preference and specificity. ISME J 9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreås L, Reysenbach A-L, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 14.Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW. 2009. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol 11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- 15.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 16.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 17.Becraft ED, Cohan FM, Kühl M, Jensen SI, Ward DM. 2011. Fine-scale distribution patterns of Synechococcus ecological diversity in microbial mats of Mushroom Spring, Yellowstone National Park. Appl Environ Microbiol 77:7689–7697. doi: 10.1128/AEM.05927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J, Yang F, Stepanauskas R, Cardenas E, Garoutte A, Williams R, Flater J, Tiedje JM, Hofmockel KS, Gelder B, Howe A. 2016. Strategies to improve reference databases for soil microbiomes. ISME J 11:829–834. doi: 10.1038/ismej.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons SL, DiBartolo G, Denef VJ, Goltsman DSA, Thelen MP, Banfield JF. 2008. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol 6:e177. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adair EC, Parton WJ, Del Grosso SJ, Silver WL, Harmon ME, Hall SA, Burke IC, Hart SC. 2008. Simple three‐pool model accurately describes patterns of long‐term litter decomposition in diverse climates. Glob Chang Biol 14:2636–2660. [Google Scholar]
- 21.Alster CJ, German DP, Lu Y, Allison SD. 2013. Microbial enzymatic responses to drought and to nitrogen addition in a southern California grassland. Soil Biol Biochem 64:68–79. doi: 10.1016/j.soilbio.2013.03.034. [DOI] [Google Scholar]
- 22.Allison SD, Lu Y, Weihe C, Goulden ML, Martiny AC, Treseder KK, Martiny JBH. 2013. Microbial abundance and composition influence litter decomposition response to environmental change. Ecology 94:714–725. doi: 10.1890/12-1243.1. [DOI] [PubMed] [Google Scholar]
- 23.Matulich KL, Weihe C, Allison SD, Amend AS, Berlemont R, Goulden ML, Kimball S, Martiny AC, Martiny JBH. 2015. Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J 9:2477–2489. doi: 10.1038/ismej.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chase AB, Arevalo P, Polz MF, Berlemont R, Martiny JBH. 2016. Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front Microbiol 7:1874. doi: 10.3389/fmicb.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D, Jospin G, Eisen JA. 2013. Systematic identification of gene families for use as “markers” for phylogenetic and phylogeny-driven ecological studies of bacteria and archaea and their major subgroups. PLoS One 8:e77033. doi: 10.1371/journal.pone.0077033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlemont R, Allison SD, Weihe C, Lu Y, Brodie EL, Martiny JBH, Martiny AC. 2014. Cellulolytic potential under environmental changes in microbial communities from grassland litter. Front Microbiol 5:639. doi: 10.3389/fmicb.2014.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Springer-Verlag 2012. Bergey’s manual of systematic bacteriology. The Actinobacteria, Part A and B. 2nd ed, vol 5 Springer-Verlag, New York, NY. [Google Scholar]
- 28.Evtushenko L, Takeuchi M. 2006. The family Microbacteriaceae, p 1020–1098. In Dworkin W, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 3 Springer, New York, NY. [Google Scholar]
- 29.Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NCK, Patil PB, Juge N, Mackenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J. 2011. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet 7:e1001314. doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. 2012. Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553. doi: 10.1371/journal.pone.0037553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams TC, Blackman ER, Morrison SS, Gibas CJ, Oliver JD. 2014. Transcriptome sequencing reveals the virulence and environmental genetic programs of Vibrio vulnificus exposed to host and estuarine conditions. PLoS One 9:e114376. doi: 10.1371/journal.pone.0114376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-R LM, Konstantinidis KT. 2014. Bypassing cultivation to identify bacterial species. Microbe 9:111–118. doi: 10.1128/microbe.9.111.1. [DOI] [Google Scholar]
- 34.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Res 43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schloter M, Lebuhn M, Heulin T, Hartmann A. 2000. Ecology and evolution of bacterial microdiversity. FEMS Microbiol Rev 24:647–660. doi: 10.1111/j.1574-6976.2000.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 36.Wielbo J, Marek-Kozaczuk M, Kubik-Komar A, Skorupska A. 2007. Increased metabolic potential of Rhizobium spp. is associated with bacterial competitiveness. Can J Microbiol 53:957–967. doi: 10.1139/W07-053. [DOI] [PubMed] [Google Scholar]
- 37.Choudhary DK, Johri BN. 2011. Ecological significance of microdiversity: coexistence among casing soil bacterial strains through allocation of nutritional resource. Indian J Microbiol 51:8–13. doi: 10.1007/s12088-011-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Schlatter DC, Kinkel LL. 2014. Global biogeography of Streptomyces antibiotic inhibition, resistance, and resource use. FEMS Microbiol Ecol 88:386–397. doi: 10.1111/1574-6941.12307. [DOI] [PubMed] [Google Scholar]
- 39.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. 2016. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez A, Vázquez-Baeza Y, Pettengill JB, Ottesen A, McDonald D, Knight R. 2016. Avoiding pandemic fears in the subway and conquering the platypus. mSystems 1:e00050-16. doi: 10.1128/mSystems.00050-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martiny JBH, Jones SE, Lennon JT, Martiny AC. 2015. Microbiomes in light of traits: a phylogenetic perspective. Science 350:aac9323. doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 42.Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. 2010. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 43.Martiny AC, Treseder K, Pusch G. 2013. Phylogenetic conservatism of functional traits in microorganisms. ISME J 7:830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett AF, Lenski RE, Mittler JE. 1992. Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution 46:16–30. doi: 10.1111/j.1558-5646.1992.tb01981.x. [DOI] [PubMed] [Google Scholar]
- 45.Schimel J, Balser TC, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 46.Chase JM, Leibold MA. 2003. Ecological niches: linking classical and contemporary approaches. University of Chicago Press, Chicago, IL. [Google Scholar]
- 47.Potts DL, Suding KN, Winston GC, Rocha AV, Goulden ML. 2012. Ecological effects of experimental drought and prescribed fire in a southern California coastal grassland. J Arid Environ 81:59–66. doi: 10.1016/j.jaridenv.2012.01.007. [DOI] [Google Scholar]
- 48.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. 2014. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Federhen S. 2012. The NCBI taxonomy database. Nucleic Acids Res 40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsen FA, Kodner RB, Armbrust EV. 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rho M, Tang H, Ye Y. 2010. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 61.Bushnell B. 2016. BBMap short read aligner. University of California, Berkeley, CA: https://sourceforge.net/projects/bbmap/. [Google Scholar]
- 62.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez-R LM, Konstantinidis KT. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1 https://doi.org/10.7287/peerj.preprints.1900v1. [Google Scholar]
- 64.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berlemont R, Martiny AC. 2013. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79:1545–1554. doi: 10.1128/AEM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pold G, Billings AF, Blanchard JL, Burkhardt DB, Frey SD, Melillo JM, Schnabel J, Van Diepen LTA, Deangelis KM. 2016. Long-term warming alters carbohydrate degradation potential in temperate forest soils. Appl Environ Microbiol 82:6518–6530. doi: 10.1128/AEM.02012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. 2008. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr Microbiol 57:503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- 69.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Austral Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 70.Mac Nally R. 2002. Multiple regression and inference in ecology and conservation biology: further comments on identifying important predictor variables. Biodivers Conserv 11:1397–1401. doi: 10.1023/A:1016250716679. [DOI] [Google Scholar]
- 71.Harrell FE., Jr. 2015. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis, 2nd ed. Springer-Verlag, Cham, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of bacterial community analyses. Download TEXT S1, DOCX file, 0.1 MB (117.2KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reference database constructed from >3,000 genomes representing the entire Bacteria domain (two genomes per genus). Multilocus phylogenetic analysis is unrooted and based on a concatenated alignment of 29 single-copy marker genes. Download FIG S1, TIF file, 3.1 MB (3.1MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Average taxonomic relative abundance of the bacterial communities across the metagenomic samples from 2010 to 2012 at the LRGCE. The values are average percent taxonomic relative abundance ± 1 standard deviation (SD). Download TABLE S1, DOCX file, 0.1 MB (62.7KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic characterizations of all Curtobacterium isolates within individual clades. Download TABLE S2, DOCX file, 0.1 MB (55.9KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multidimensional scaling (MDS) plot depicting the difference in microbial community relative abundances at the phylum level (A) and clade level within Curtobacterium (B). Symbols represent sampling date with darker colors indicating winter/spring months and warmer colors indicating summer/fall months. Download FIG S2, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Average relative abundance of Curtobacterium in the bacterial community at LRGCE as a function of the total precipitation (A) and the mean temperature (B) in the 2 weeks prior to sampling. Average relative abundance (± 1 SD) is shown. Symbols represent sampling date with darker colors indicating winter/spring months and warmer colors indicating summer/fall months. Download FIG S3, TIF file, 27 MB (27.6MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of Curtobacterium clades by treatment averaged across the metagenomic libraries from the LRGCE. Values are relative percent abundance ± 1 SD. Download TABLE S3, DOCX file, 0.1 MB (53.2KB, docx) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multidimensional scaling (MDS) plot depicting total GH/CBM gene composition for Curtobacterium isolates. Each symbol represents a genomic isolate and is colored according to the assigned clade. Download FIG S4, TIF file, 16.3 MB (16.7MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coverage profiles of all coassembled contigs as a function of their GC content. Taxonomic assignments are colored by the top 10 most abundant bacterial genera (listed in descending order). Download FIG S5, TIF file, 4.3 MB (4.4MB, tif) .
Copyright © 2017 Chase et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.