Abstract
Sorting, transport, and autophagic degradation of proteins in endosomes and lysosomes, as well as the division of these organelles, depend on scission of membrane‐bound tubulo‐vesicular carriers. How scission occurs is poorly understood, but family proteins bind these membranes. Here, we show that the yeast PROPPIN Atg18 carries membrane scission activity. Purified Atg18 drives tubulation and scission of giant unilamellar vesicles. Upon membrane contact, Atg18 folds its unstructured CD loop into an amphipathic α‐helix that inserts into the bilayer. This allows the protein to engage its two lipid binding sites for PI3P and PI(3,5)P2. PI(3,5)P2 induces Atg18 oligomerization, which should concentrate lipid‐inserted α‐helices in the outer membrane leaflet and drive membrane tubulation and scission. The scission activity of Atg18 is compatible with its known roles in endo‐lysosomal protein trafficking, autophagosome biogenesis, and vacuole fission. Key features required for membrane tubulation and scission by Atg18 are shared by other PROPPINs, suggesting that membrane scission may be a generic function of this protein family.
Keywords: autophagy, endosomes, lysosomes, membrane fission, membrane traffic
Subject Categories: Autophagy & Cell Death, Membrane & Intracellular Transport
Introduction
Endosomal and lysosomal compartments permanently produce tubulo‐vesicular carriers that transport material between them while maintaining their identity in terms of content, pH, and lipid composition. While numerous components involved in the formation of these carriers have been identified, such as the retromer complex and sorting nexins (Seaman et al, 1998; Rojas et al, 2007; Temkin et al, 2011), scission of these carriers from their donor compartments remains poorly understood. Also the biogenesis of autophagosomes requires growth of the phagophore and delivery of lipids to it, probably by membrane flux (Tooze et al, 2014). Among other factors, growth of the isolation membrane depends on the transmembrane protein Atg9, which localizes to it but also to other compartments. A current hypothesis on Atg9 function therefore stipulates that Atg9 might shuttle in vesicles between the phagophore membrane and other compartments, which may serve as lipid donors for the phagophore. These include the Golgi, endosomes, the plasma membrane, and ER exit sites (Reggiori et al, 2004; Mari et al, 2010; Puri et al, 2013). According to this hypothesis, autophagosome biogenesis might require membrane fission reactions on both the phagophore and the lipid donor organelles. Critical lipid factors required for the biogenesis of autophagosomes and for membrane fission reactions on endo‐lysosomal organelles are the phosphoinositides PI3P, PI(5)P, and PI(3,5)P2 (Obara et al, 2008a,b; Vicinanza et al, 2015).
The GTPase dynamin is the prototypical mechano‐chemical membrane fission protein (Antonny et al, 2016). However, other membrane fission factors exist. In the context of retromer‐mediated recycling, for example, actin promotes membrane fission on endosomes via the action of the WASH complex, but retromer trafficking in yeast requires also the SNX‐BAR protein Mvp1 and the dynamin‐like GTPase Vps1, which constricts retromer‐tubules (Derivery et al, 2009; Gomez & Billadeau, 2009; Chi et al, 2014). It is unknown whether actin promotes retromer fission solely by providing mechanical stress, or also by promoting lipid phase separation, which alone can suffice to drive membrane fission (Allain et al, 2004; Römer et al, 2010). Also shallow insertion of amphipathic helices into a membrane leaflet can drive fission, as illustrated by the fission of endocytic vesicles at the plasma membrane by endophilin‐A2 (Renard et al, 2015). Similarly, the small GTPases Arf1 and Sar1 were proposed to use amphipathic helix insertion to promote scission of vesicles for retrograde and anterograde transport on the ER and the Golgi, respectively (Bielli et al, 2005; Lee et al, 2005; Beck et al, 2008). However, reconstitution and real‐time visualization of protein‐mediated membrane fission events on liposomes has only been achieved for dynamin (Ferguson & De Camilli, 2012; Morlot & Roux, 2013).
PROPPINs are a family of conserved proteins (Proikas‐Cezanne et al, 2015). Eukaryotes express multiple PROPPIN isoforms, three in yeasts (Atg18, Atg21, and Hsv2) and four in mammalian cells (WIPI 1 through 4, with several splice variants), which localize to a variety of cellular membranes (Dove et al, 2004; Jeffries et al, 2004; Stromhaug et al, 2004; Polson et al, 2010; Proikas‐Cezanne & Robenek, 2011). PROPPINs interact with core components of the autophagy machinery, with signaling components regulating PI3P production, and are important for the growth of phagophores into autophagosomes (Reggiori et al, 2004; Proikas‐Cezanne & Robenek, 2011; Dooley et al, 2014; Bakula et al, 2017). In the endo‐lysosomal system of mammals and yeast, inactivation of PROPPINs affects retrograde traffic (Dove et al, 2004; Jeffries et al, 2004; McCartney et al, 2014). Furthermore, the PROPPIN Atg18 accelerates fission of yeast vacuoles, which are the lysosomal compartment of these cells (Dove et al, 1997; Zieger & Mayer, 2012). Key features shared by PROPPINs are their two lipid interaction sites with a capacity to bind PI3P, PI(3,5)P2, and PI(5)P, phosphoinositides that are particularly enriched on endosomes, lysosomes, and autophagosomes (Schink et al, 2013; McCartney et al, 2014; Vicinanza et al, 2015). A hydrophobic loop is located between these two lipid binding sites. It must face the membrane surface when the protein binds PI3P and PI(3,5)P2 (Baskaran et al, 2012; Krick et al, 2012; Watanabe et al, 2012; Busse et al, 2015). In Pichia pastoris Atg18, the loop can be phosphorylated, which reduces the affinity of the protein for PI(3,5)P2‐containing membranes (Tamura et al, 2013). Despite the availability of crystal structures and a wealth of information on the interaction of PROPPINs with lipids and other proteins, the molecular function of PROPPINs has remained incompletely understood.
We used the lysosome‐like yeast vacuoles as a model to study this function. Fission of vacuoles into multiple smaller organelles can be triggered in vivo by a mild salt shock, which activates phosphorylation of PI3P by the kinase Fab1 and leads to a sudden increase in PI(3,5)P2 (Dove et al, 1997; Cooke et al, 1998; Gary et al, 1998; Bonangelino et al, 2002). As a consequence, vacuoles first form multiple invaginations that require PI3P. The remaining tubular structures between the invaginations fission into smaller vesicles (Peters et al, 2004; Zieger & Mayer, 2012). Scission of these vesicles requires PI(3,5)P2 and is accelerated by (but not entirely dependent on) Atg18 (Bonangelino et al, 2002; Zieger & Mayer, 2012). Atg18 is also a negative regulator of the PI3P‐5‐kinase Fab1, and yeast cells lacking Atg18 (atg18Δ) have even higher PI(3,5)P2 levels than the wild type (Dove et al, 2004; Efe et al, 2007). Although an elevated level of PI(3,5)P2 usually leads to a hyper‐fragmentation of the vacuole (Efe et al, 2007), atg18Δ cells show a single large vacuole that undergoes fission more slowly than in wild‐type cells (Zieger & Mayer, 2012). We hence tested the hypothesis that Atg18 might be an effector protein that executes PI(3,5)P2‐triggered membrane scission on vacuoles.
Results
Yeast PROPPINs are required for vacuole fission in vivo
atg18Δ cells show only a moderate delay in osmotically induced vacuole fragmentation (Zieger & Mayer, 2012; Michaillat & Mayer, 2013). Since yeast expresses two potentially redundant Atg18 homologs, Atg21 and Hsv2, we generated triple knockout atg18Δ/atg21Δ/hsv2Δ (ΔΔΔ) cells. Wild‐type cells mostly had 1–2 vacuoles on rich media, which were often multi‐lobed and showing an invagination. By contrast, atg18Δ and ΔΔΔ cells showed a single large and round vacuole (Fig 1A) (Dove et al, 2004; Efe et al, 2007; Krick et al, 2008). Within < 15 min upon a moderate hypertonic shock, virtually all wild‐type cells contained several smaller vacuoles and more than 75% of them had > 7. By contrast, around 60% of ΔΔΔ cells and 40% of atg18Δ cells had retained a single and almost none showed > 7 (Fig 1A and B). In atg18Δ cells, the fraction of cells with one large vacuole declined down to 20% during the subsequent 45 min of incubation, indicating that fragmentation proceeded slowly, whereas vacuoles in ΔΔΔ cells expanded back into a single large organelle. The phenotype of ΔΔΔ cells could be rescued by plasmid‐based expression of Atg18‐GFP, but not by expressing Atg18FGGG‐GFP, a mutant affecting the two binding sites of the protein for PI3P and PI(3,5)P2 (Efe et al, 2007; Baskaran et al, 2012; Krick et al, 2012; Watanabe et al, 2012) (Fig 1A and B; Appendix Fig S1). Thus, Atg18‐GFP is sufficient to support salt‐induced vacuole fission.
Figure 1. Redundancy of PROPPINs in vacuole fission.
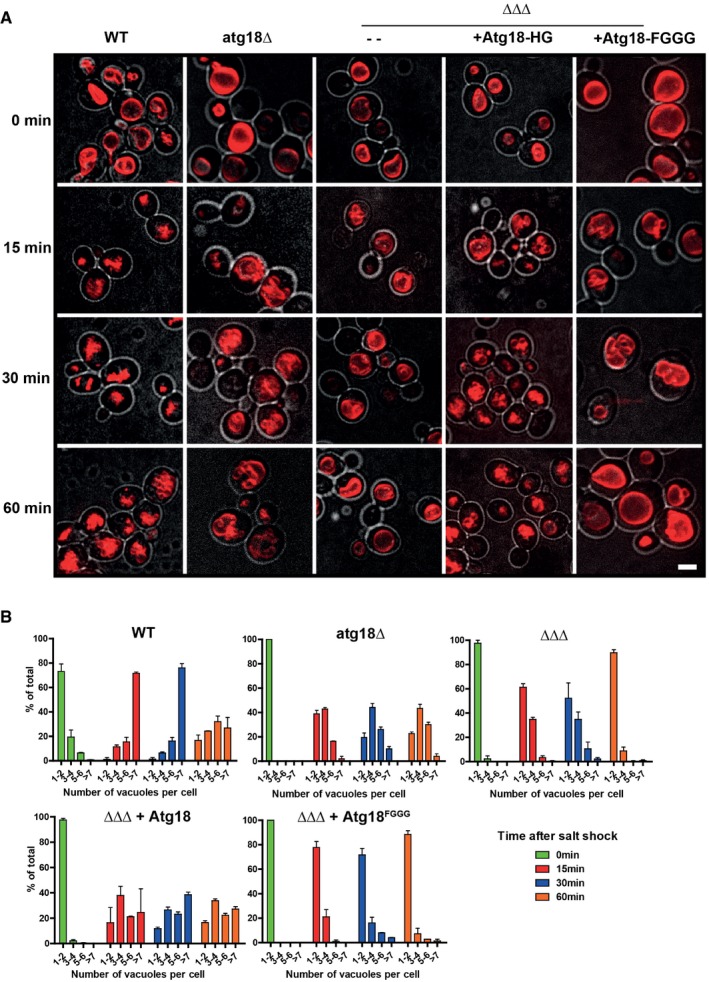
- Cells (wt, ∆atg18 or ∆atg18/∆atg21/∆hsv2 (ΔΔΔ), with or without a plasmid expressing wt or a mutant form of Atg18) were grown in YPD; vacuoles were stained with FM4‐64 and imaged by confocal microscopy. At least 10 confocal sections spaced at 300 nm were assembled into maximum projections. Images show cells before (0 min) and 15, 30, and 60 min after induction of vacuolar fragmentation by addition of 0.4 M NaCl. Red: FM4‐64. Gray: DIC. Scale bar 5 μm.
- The number of vacuoles per cell was quantified for the experiment in (A); 150 cells from three independent experiments were counted for each condition. Values represent the mean, error bars the SEM.
On rich YPD medium, most of the Atg18‐GFP resided in the cytoplasm and in one or two brightly fluorescent spots, which probably represent pre‐autophagosomal structures (Suzuki et al, 2013). Within < 2 min after an osmotic shock, Atg18‐GFP was depleted from the cytosol and strongly concentrated on the vacuole membrane (Fig 2A and C), where it accumulated at vacuole–vacuole interfaces and vertices, that is, at bona fide sites of membrane fission (Fig 2D and E; Movie EV1) (Tamura et al, 2013). This recruitment was not seen for Atg18FGGG‐GFP (Fig 2A and C). Atg18‐GFP was not recruited to vacuoles of vps34Δ or fab1Δ cells, which lack PI3P or PI(3,5)P2, respectively (Fig 2B and C; Efe et al, 2007). The rapid recruitment of Atg18 is consistent with the reported kinetics of PI(3,5)P2 accumulation after an osmotic shock, which reaches its maximum within 5–15 min after salt addition (Dove et al, 1997; Gary et al, 1998, 2002; Bonangelino et al, 2002; Duex et al, 2006).
Figure 2. Localization of Atg18‐GFP during salt‐induced vacuole fission.
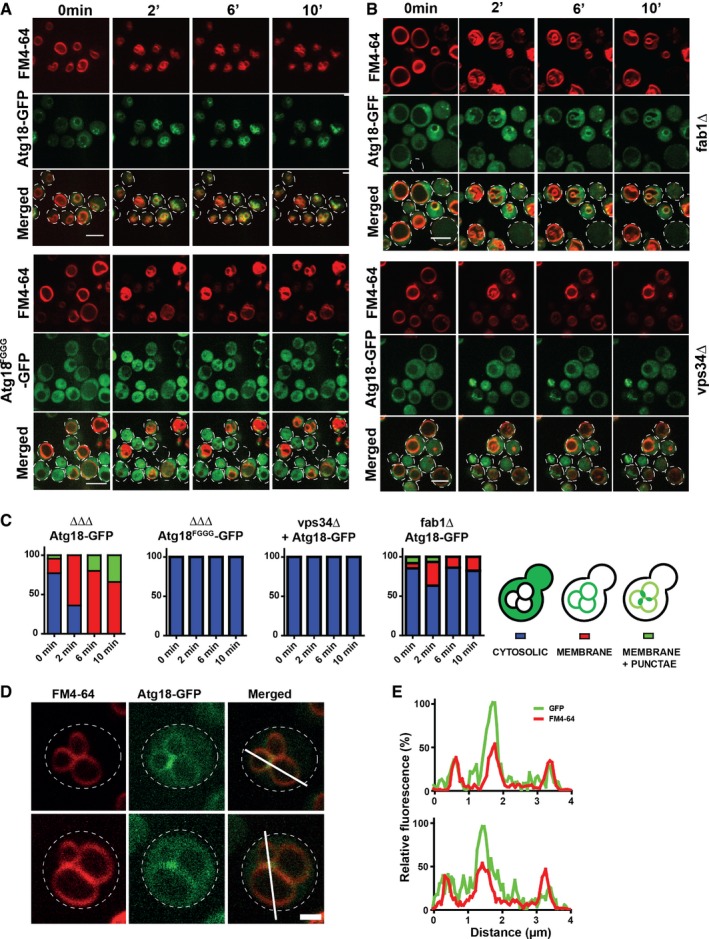
- Confocal z‐stacks were taken before and 2, 6, and 10 min after addition of 0.4 M NaCl. Stacks were processed into maximum projections using ImageJ. The cell outlines are marked with dotted lines. Scale bar: 5 μm.
- Same experiment as in (A), but in a fab1Δ or vps34Δ background. Scale bar: 5 μm.
- The cells were categorized as indicated, and the phenotypes of 150 cells were quantified.
- Localization of Atg18‐GFP 5 min after salt shock. Experiment as in (A). Scale bar: 1 μm.
- Line scan analysis of fluorescence intensities along the white lines shown in (D). Red: FM4‐64. Green: Atg18‐GFP. Scale bar: 1 μm.
The unstructured CD loop of Atg18 can undergo lipid‐triggered transformation into an amphipathic α‐helix
We analyzed the primary and predicted secondary structure of Atg18 in order to further elucidate the mechanism by which Atg18 could mediate membrane fission. Blade 6 of the β‐propeller of the Atg18‐homolog Hsv2 contains an unstructured hydrophobic loop (CD loop) located between the two lipid binding sites (Baskaran et al, 2012; Krick et al, 2012; Watanabe et al, 2012). This loop is also present in other PROPPINs, suggesting that it may be crucial for PROPPIN function (Fig 3A). We analyzed the loop sequences of a wide variety of PROPPINs with the program Heliquest (Gautier et al, 2008). Although the lengths and primary sequences of CD loops from different species vary substantially, we found that they all contain a stretch of at least 18 amino acids that has the potential to fold into an amphipathic α‐helix (Fig 3B). This conservation at the level of secondary structure suggests that the formation of amphipathic helices might be an important functional trait of the CD loops. This is intriguing because shallow insertions of amphipathic α‐helixes into bilayers can promote membrane deformation and scission (Boucrot et al, 2012). In order to test the propensity of the loop to form an α‐helix, we synthesized a peptide corresponding to the putative amphipathic α‐helix of Atg18. Circular dichroism spectroscopy showed that the peptide formed a random coil in aqueous buffer. When incubated in increasing concentrations of the hydrophobic compound hexafluoroisopropanol (HFIP), it folded into an α‐helix as the hydrophobicity of the buffer increased (Fig 4A). This conformational change occurred also in entirely aqueous buffer when the peptide was incubated together with small unilamellar vesicles (SUVs) (Fig 4B). Thus, lipid contact induces a conformational transition of the CD loop peptide from a random coil into an amphipathic α‐helix.
Figure 3. Amphipathic α‐helix formation.
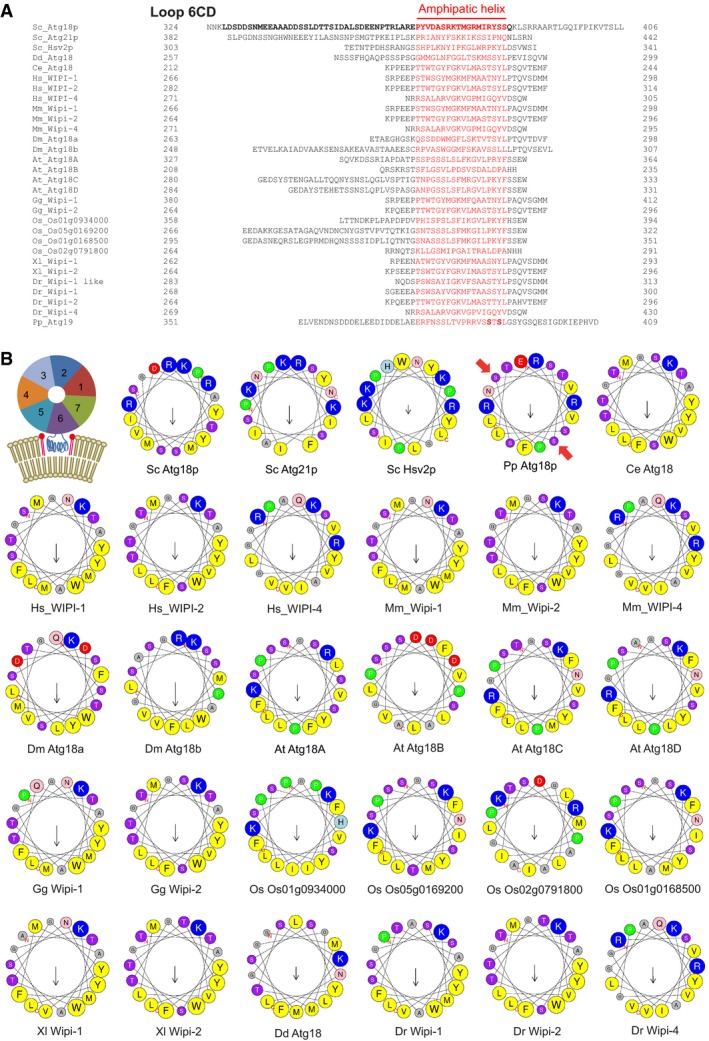
- The CD loop sequences of PROPPINs from a wide variety of organisms. Potential amphipathic helices were predicted using the online tool Heliquest and are plotted in red. Amino acids removed in Atg18DLoop (aa 328–384) are printed in bold. In the loop of Pichia pastoris Atg18, bold print mark phosphorylation sites relevant for its membrane insertion (Tamura et al, 2013). Dr: Danio rerio, Dd: Dictyostelium discoideum.
- Helical wheel projections of the red sequences from (A). A schematic representation of the 7‐bladed beta‐propeller PROPPIN structure is also shown, illustrating the localization of the putative amphipathic α‐helix (blue) of the CD loop between the two lipid binding sites for PI3P/PI(3,5)P2. Red arrows mark phosphorylation sites in the loop of Pichia pastoris Atg18, which are relevant for its membrane insertion (Tamura et al, 2013). Color code for residues: yellow, hydrophobic; purple, serine and threonine; blue, basic; red, acidic; pink, asparagine and glutamine; gray, alanine and glycine; green, proline; light blue, histidine.
Figure 4. Lipid‐triggered folding of the amphipathic helix of CD loop peptides and its requirement for vacuole scission.
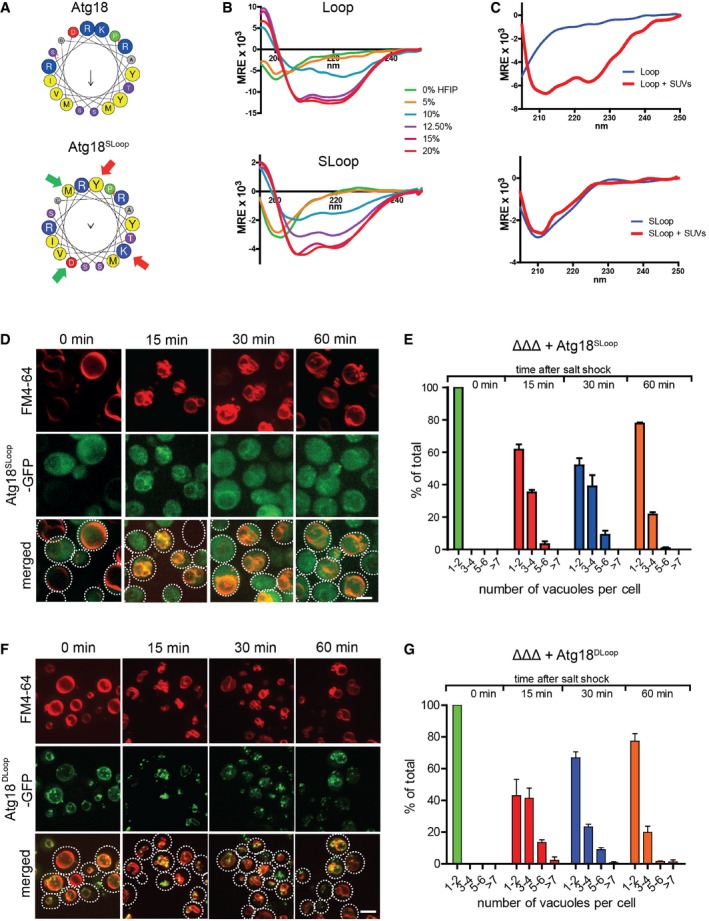
-
ACD loop folding in hydrophobic buffer. Peptides were synthesized that correspond to the predicted amphipathic helix from Saccharomyces cerevisiae Atg18 (TRLAREPYVDASRKTMGRMIRYSSQ) or to a sequence with reduced tendency to form amphipathic helices, generated by a swap of two amino acid pairs (indicated by green and red arrows; SLoop). Helical wheel projections of the sequences are shown.
-
B, CTheir secondary structure was analyzed by circular dichroism spectroscopy in the presence of increasing concentrations of hexafluoro‐2‐propanol (HFIP), in the presence of 6 mM small unilamellar vesicles (SUVs, 70% egg PC and 30% PS, total phospholipid concentration approx. 6 mM), or of or control buffer only (blue). Spectra are shown for (B) the wild‐type peptide, and (C) the SLoop peptide.
-
DVacuole morphology of ΔΔΔ cells expressing Atg18Sloop‐GFP, imaged before and after osmotic shock as in Fig 1A. Scale bar: 5 μm.
-
ENumber of vacuoles per cell in (D), assayed as in Fig 1B. n = 3.
-
FVacuole morphology of ΔΔΔ cells expressing Atg18DLoop‐GFP, imaged as in (D).
-
GNumber of vacuoles per cell in (E) assayed as in Fig 1B. n = 3.
We also synthesized a scrambled loop (SLoop) mutant, in which two pairs of hydrophobic/hydrophilic amino acids in the region of the predicted amphipathic helix were swapped (Fig 4A). The peptide has identical amino acid composition and similar overall hydrophobicity as the “wild‐type” peptide but lost its potential to form an amphipathic α‐helix. While the SLoop peptide still folded into an α‐helix in HFIP buffer, it lost the capacity to fold into an α‐helix when incubated with SUVs (Fig 4C).
Vacuole scission requires the amphipathic α‐helical nature of the CD loop peptide
We tested the relevance of the amphipathic character of this lipid‐induced α‐helix in vivo by generating Atg18 carrying this scrambled sequence in its CD loop (Atg18SLoop). Furthermore, we designed a mutant lacking the largest part of the CD loop (aa 328–384; Atg18DLoop) (Fig 3A). We expressed Atg18SLoop‐GFP and Atg18DLoop‐GFP in ΔΔΔ cells. As judged from the fluorescence signals and from Western blots (Appendix Fig S1), both constructs were expressed at similar levels as Atg18wt‐GFP. Atg18DLoop‐GFP did not support vacuole fission upon a hypertonic shock (Fig 4D and E), although it efficiently bound the vacuole membrane. Interestingly, Atg18DLoop‐GFP concentrated on vacuoles even in the absence of an osmotic shock (Fig 4D), which was much less the case for Atg18wt‐GFP (Fig 2A). This suggests that membrane association of Atg18DLoop‐GFP cannot be controlled and that the CD loop acts as a negative regulator of membrane binding, which is consistent with the presence of phosphorylation sites in the loop region (Tamura et al, 2013). Atg18SLoop‐GFP was recruited to vacuoles upon hypertonic shock, though less efficiently than Atg18wt‐GFP, but it did not at all rescue salt‐induced vacuole scission (Fig 4F and G). This suggests that the lipid‐triggered folding of part of the CD loop into an amphipathic helix is necessary for vacuole scission.
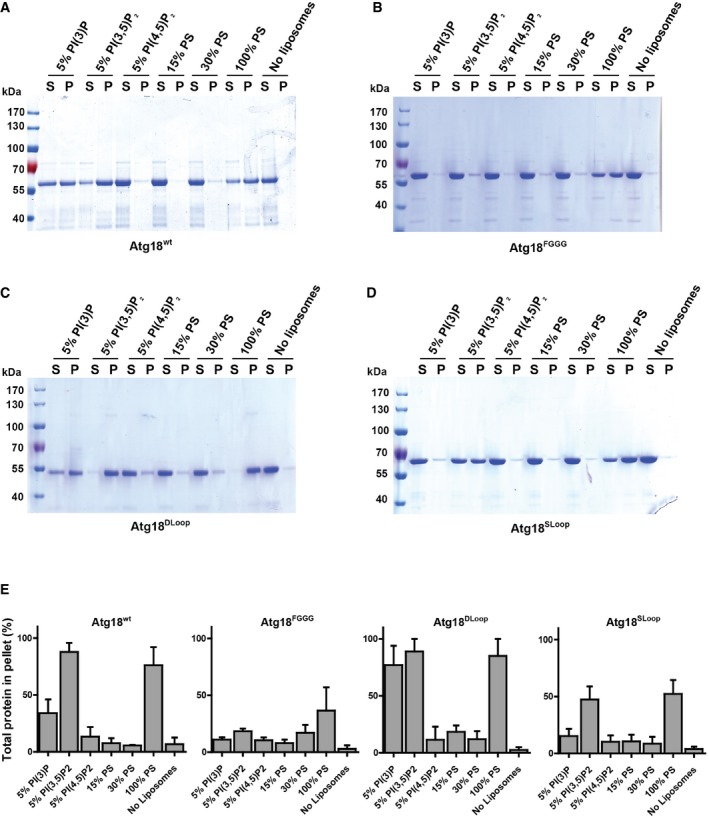
In order to study Atg18 activity in detail, we recombinantly expressed Atg18wt and the mutant forms Atg18FGGG, Atg18DLoop, and Atg18SLoop in Escherichia coli, purified the proteins, and studied their effect on unilamellar vesicles. First, we tested membrane binding of the various Atg18 mutant forms in a liposome centrifugation assay. In line with previous binding studies (Dove et al, 2004; Baskaran et al, 2012; Busse et al, 2015), purified Atg18wt bound to PI3P as well as to PI(3,5)P2‐containing liposomes (Fig 5A and E). Atg18wt bound better to 5% PI(3,5)P2 than to 5% PI3P. Atg18FGGG bound PI(3,5)P2 and PI3P liposomes very poorly (Fig 5B and E). In contrast to Atg18Sloop, which hardly bound to PI3P liposomes but showed significant binding to PI(3,5)P2 liposomes, Atg18DLoop bound efficiently to both liposome types (Fig 5C–E). This suggests that the loop might contribute to the differentiation between PI(3,5)P2 and PI3P, but we did not pursue this aspect further in the framework of this study. Since lipid binding involves two lipid binding sites and two blades of Atg18 (Baskaran et al, 2012; Krick et al, 2012; Watanabe et al, 2012; Busse et al, 2015), its preservation suggests that the protein can reach its native β‐propeller conformation independently of the CD loop. None of the Atg18 variants bound to liposomes containing 5% PI(4,5)P2 or 15% or 30% phosphatidylserine. All variants bound to liposomes containing 100% phosphatidylserine, probably by non‐specific electrostatic interactions. These binding experiments are compatible with the in vivo observations on Atg18 association with vacuoles (Fig 4D and F). They show that membrane binding of Atg18 via its two lipid binding sites is not sufficient for vacuole scission since Atg18DLoop retained its lipid binding activity but did not support vacuole scission in vivo. Furthermore, a CD loop that is present yet has lost the capacity to form an amphipathic α‐helix, which is expected to interfere with its membrane insertion, prevents membrane binding of the protein, possibly by steric interference with lipid binding. This suggests that the CD loop may have to fold into an amphipathic α‐helix and insert into the membrane in order to allow the two lipid binding sites to become engaged.
Figure 5. Binding of Atg18Sloop and Atg18Dloop to small unilamellar vesicles.
-
A–DBinding of recombinant Atg18 variants to liposomes. SUVs were made of EPC with 5% of the indicated phosphoinositides, or of EPC and the indicated fraction of PS. Cholesterol had been added to 20 mol% of the total phospholipid amount. Liposomes were incubated with the purified proteins for 30 min at room temperature (25°C) and centrifuged, and the supernatants (S) and pellets (P) analyzed by SDS–PAGE and Coomassie staining. Binding is shown for (A) Atg18wt; (B) Atg18FGGG; (C) Atg18DLoop; (D) Atg18SLoop.
-
EThe bands from (A–D) were quantified from three independent experiments. The mean with SEM is shown.
Differential requirements for Atg18 function in vacuole fission and autophagy
Atg18 and Atg21 are also important factors in autophagosome biogenesis. We assayed the impact of CD loop mutations on autophagy using the maturation of a soluble, cytosolic pro‐alkaline phosphatase (Pho8Δ60) by vacuolar phosphatases (Noda & Klionsky, 2008). Whereas Atg18FGGG reduced the autophagic activity that is induced through nitrogen starvation (SD‐N medium) by 70%, Atg18SLoop provided similar activity as wild type (Fig 6A). Also simultaneous expression of Atg18Sloop and Atg21Sloop in atg18Δ atg21Δ double mutants rescued starvation‐induced autophagic activity to 75% of the wild‐type level (Fig 6B).
Figure 6. Different functional aspects of Atg18 displayed in vacuole fission and autophagy.
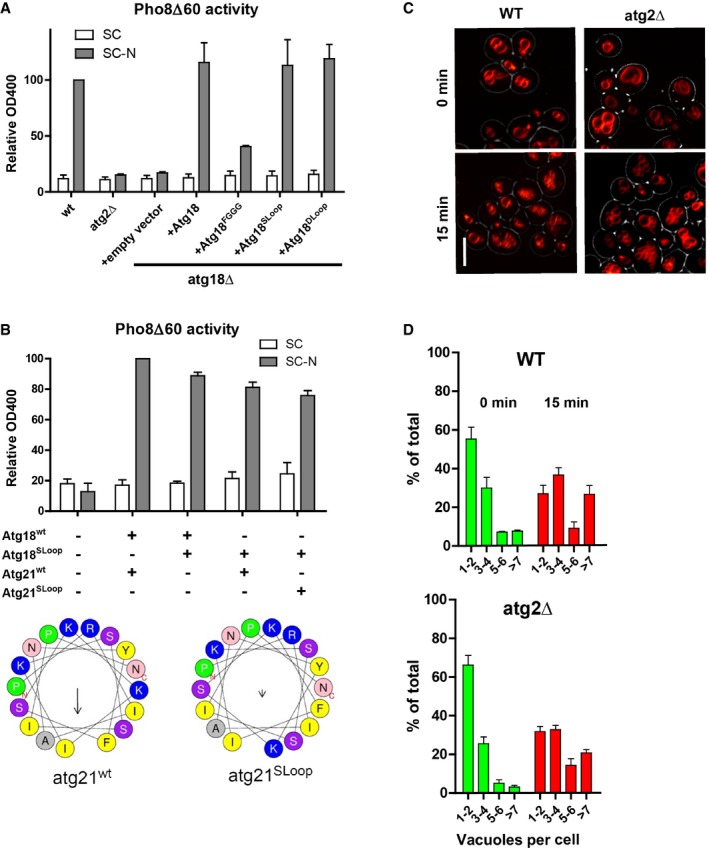
- Autophagy measured with the pho8∆60 assay in atg18Δ cells complemented with wt, FGGG, SLoop, or DLoop alleles of ATG18. Error bars represent SEM of 3 independent experiments.
- Same assay as in (A), but in atg18Δ atg21Δ cells complemented with wt or SLoop versions of ATG18 and ATG21. Atg21SLoop contains K426F and F429K substitutions. Helical wheel projections for both Atg21 helices are given below. The arrows indicate the magnitude of the hydrophobic moment. Error bars represent SEM of 3 independent experiments.
- Vacuole fragmentation in atg2Δ cells, assayed 15 min after a moderate hypertonic shock as in Fig 1. Scale bar: 5 μm.
- Number of vacuoles per cell in (C), assayed as in Fig 1B. n = 3.
Atg18 interacts with Atg2, a crucial factor for autophagy (Tsukada & Ohsumi, 1993). This interaction recruits Atg2 to the isolation membrane. It is mediated through a binding site at the membrane‐distal face of Atg18 (Obara et al, 2008b; Watanabe et al, 2012). Upon exposure to a moderate hypertonic shock, as used above, atg2Δ cells showed a similar degree of vacuole fission as the wild type (Fig 6C and D). Thus, Atg18 displays different functional aspects in vacuole fission, where its binding partner Atg2 is dispensable but its amphipathic CD loop helix is crucial, and in autophagy, where the Atg2–Atg18 interaction is necessary, but the amphipathic character of the Atg18 CD loop is of moderate importance.
Atg18 drives tubulation and scission of giant unilamellar vesicles
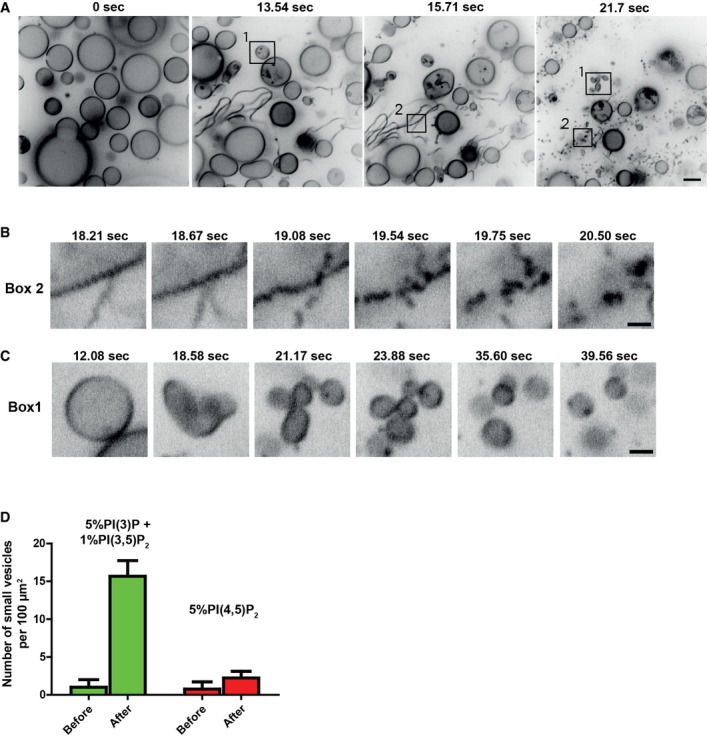
We tested the effects of purified, recombinant Atg18 on pure lipid membranes by live confocal microscopy of giant unilamellar vesicles (GUVs). The GUVs had been generated from synthetic lipid mixtures containing PI3P (5%) and PI(3,5)P2 (1%) at concentrations and in ratios close to those observed in vivo during vacuole fragmentation (Dove et al, 1997, 2004; Gary et al, 1998; Bonangelino et al, 2002; Duex et al, 2006). 1% rhodamine‐phosphatidyl ethanolamine served as a fluorescent tracer. GUVs were left to sediment at the bottom of a 96‐well plate. Atg18 was added to a final concentration of 0.9 μM. This is not far from its cytosolic concentration, which can be approximated to be between 0.04 and 0.2 μM, based on published values on Atg18 abundance [345–1,560 molecules per cell (Ghaemmaghami et al, 2003; Kulak et al, 2014)], the median volume of a yeast cell [42 fl (Uchida et al, 2011)], and an excluded volume of the membrane‐bound organelles of around 40% of total cell volume (Uchida et al, 2011). When Atg18 was added to sedimented GUVs containing PI3P and PI(3,5)P2, the GUVs tubulated extensively (Fig 7A and D; Movie EV6) and most of them underwent scission into smaller vesicles. Larger tubules often pearled prior to scission (Fig 7B). GUVs could also convert directly into a cluster of smaller buds (Fig 7C). These clusters underwent scission because the vesicular products diffused away from each other. Scission was not observed when Atg18 was added to GUVs containing 5% PI(4,5)P2 (Fig 7D; Movie EV7).
Figure 7. Time lapse analysis of the effects of Atg18 on giant unilamellar vesicles.
-
ATime lapse analysis of Atg18 added to liposomes. 10 μl of 10 μM purified recombinant Atg18wt was added to 100 μl buffer containing GUVs with 5% PI3P and 1% PI(3,5)P2. The sample was imaged on a spinning disk confocal microscope. Most of the tubules underwent scission into small vesicles (see Movie EV6). Scale bar 5 μm.
-
B, CAreas in boxes 1 and 2 of (A) are shown at higher magnification in (B and C). Scale bars: 2.5 μm.
-
DQuantification of fission activity. For each movie, the whole frame was divided into squares of 10 per 10 μm and the number of small vesicles per 100 μm2 counted. The mean of three independent experiments is shown with SEM.
Since the formation of tubular structures during vacuole fission depends on PI3P but not on PI(3,5)P2 (Zieger & Mayer, 2012), we tested whether Atg18 might exhibit different effects when incubated with liposomes containing only one of the two phosphoinositides. We incubated Atg18 with GUVs containing only PI3P or PI(3,5)P2. In time‐lapse experiments, addition of Atg18wt to GUVs containing PI3P drove massive tubulation. About 30 s to 1 min after the onset of tubulation, GUVs frequently collapsed into a network of long flexible tubules (Fig 8A, [Link], [Link]), suggesting that the tubulating activity of the protein could generate sufficient membrane tension to force explosion of the GUV. In the presence of PI(3,5)P2 only, tubulation and collapse of GUVs occurred as well, but the tubules quickly retracted to the surface of the GUV and became immobile clusters there (Fig 8A, [Link], [Link]). Incubation of fluorescently labeled Atg18wt with GUVs showed that Atg18 localized to tubules in the presence of PI3P and to dense collapsed regions in the presence of PI(3,5)P2 (Fig 8B and D). These were not observed with PI(4,5)P2. Atg18SLoop had no effect on GUVs (Fig 8C). Incubation of Atg18DLoop with GUVs containing PI3P or PI(3,5)P2 did not lead to membrane tubulation either (Fig 8C and E). However, it induced adherence of the liposome membranes over larger areas, suggesting that the vesicles might associate via protein–protein interactions.
Figure 8. Effect of Atg18 mutants on GUVs of different lipid composition.
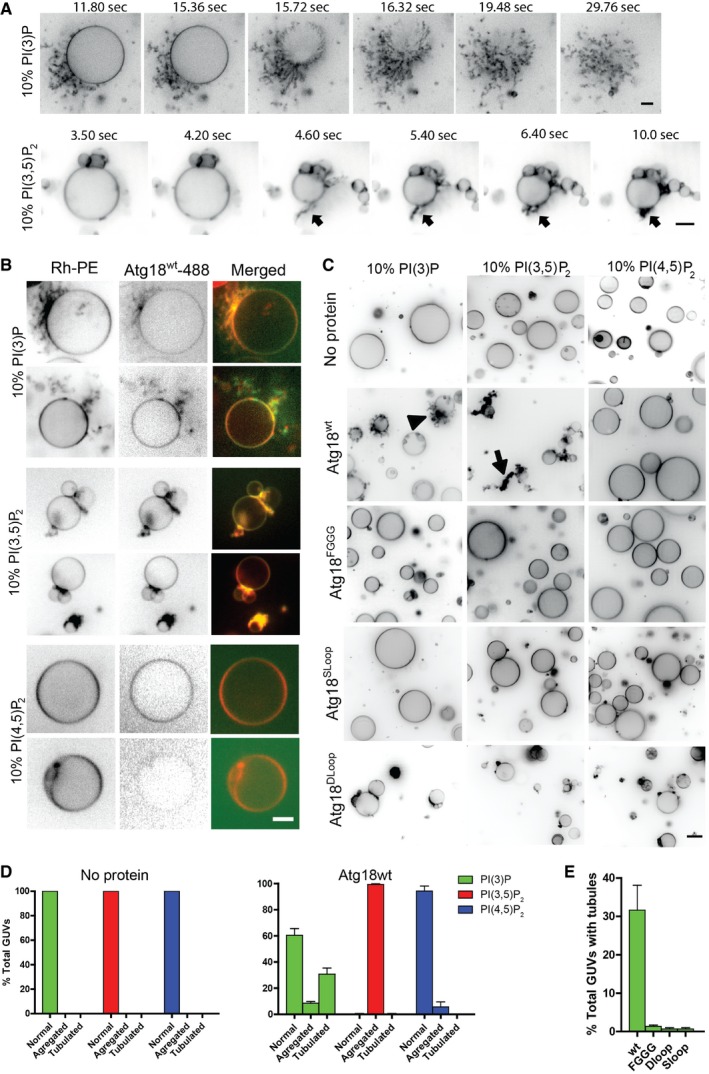
- 1 μM Atg18 was added to wells containing sedimented GUVs with 10% PI3P or PI(3,5)P2 while imaging a single plane by spinning disk microscopy. Membrane tension exerted by Atg18 binding suffices to induce liposome collapse, which allows to better judge the mobility of the tubular or clustered membranes that remain. Black arrow points to a PI(3,5)P2 induced tubule collapsing to the GUV surface. Images are extracted from [Link], [Link], [Link], [Link]. Scale bar 5 μm.
- Atg18 was covalently labeled with the green fluorescent dye Alexa488 TFP (ThermoFisher) and used in experiments as in (A), with rhodamine‐phosphatidylethanolamine‐labeled (Rh‐PE) GUVs containing the indicated phosphoinositides. Scale bar 5 μm.
- Effect of 1 μM of purified recombinant Atg18 variants on GUVs containing 10 mol% of the indicated phosphoinositides. Vesicles are shown after 1–3 min of incubation. Scale bar: 7 μm. Arrow and arrowhead point to the structures quantified in (D).
- Quantification of the experiments in (C). The percentage of GUVs showing an enrichment of Atg18 flexible tabulated (arrowhead) or stiff aggregated structures (arrow) was determined. Shown are the mean and SEM from three independent experiments.
- Quantification of the effect of different Atg18 version on the formation of tubules in (C). Shown is the mean from three independent experiments with SD.
PI(3,5)P2 induces Atg18 oligomerization
The rigidity of the PI(3,5)P2‐induced tubules on collapsed liposomes and the mutual adherence of GUVs over larger parts of their surface pointed to possible self‐interaction of Atg18. We employed chemical cross‐linking assays to test this. After incubation of Atg18 with liposomes and the cleavable cross‐linker DTSSP, high molecular weight forms of Atg18 could be observed by SDS–PAGE (Fig 9A and B). They migrated close to the stacking gel, suggesting that they are larger than the separation range of the gel system, that is, > 300 kDa, whereas monomeric Atg18 migrated at 55 kDa. The high‐MW forms did not appear if the cross‐linker was cleaved by the reduction agent DTT or if DTSSP was omitted from the incubation with the liposomes (Fig 9A). Increasing the concentration of liposomes increased the amount of high molecular weight product (Fig 9B) and the lipid binding mutant Atg18FGGG did not form cross‐linked products (Fig 9A). The cross‐linked product depended on the presence of PI(3,5)P2. PI3P and PI(4,5)P2 were less efficient in inducing cross‐linking (Fig 9A, Appendix Fig S2), but they also recruited Atg18 less well to the membrane (Fig 5E). PS liposomes, by contrast, bound Atg18 with similar efficiency as PI(3,5)P2 liposomes (Fig 5E), but they did not promote cross‐linking (Fig 9A). This suggests that PI(3,5)P2 might promote self‐interaction of Atg18 on liposomes.
Figure 9. Oligomerization of Atg18.
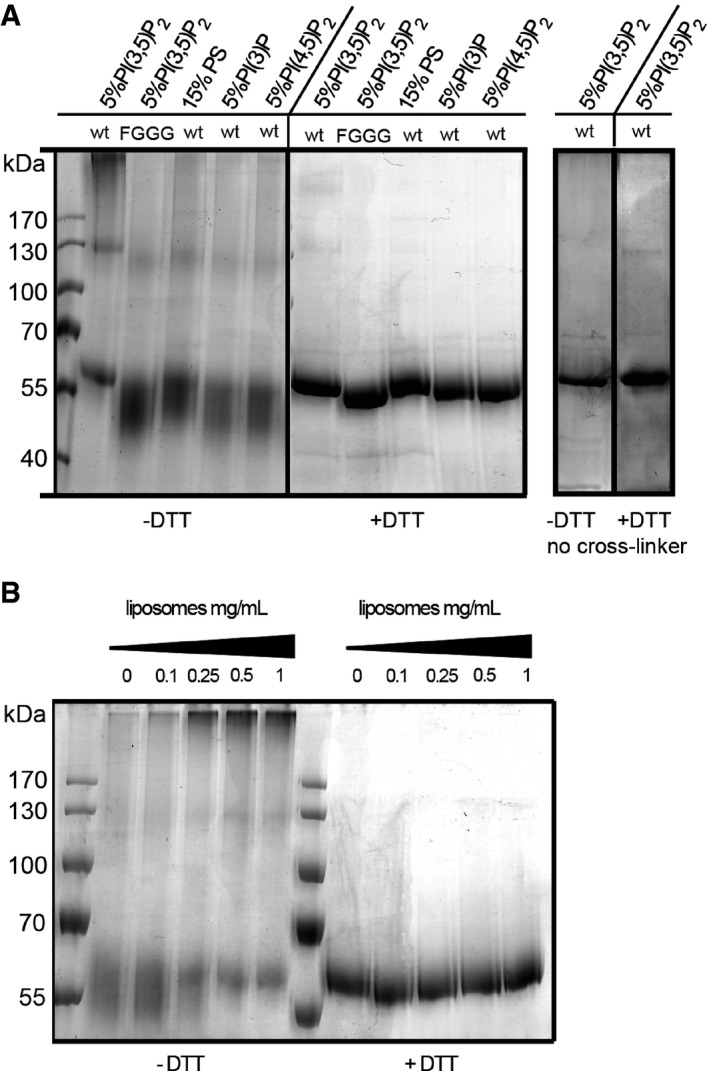
- Cross‐linking of Atg18. Purified recombinant Atg18wt or Atg18FGGG (1.5 μM) was incubated with liposomes containing 5% PI3P, PI(3,5)P2, PI(4,5)P2, or 15% PS as a control for negative charge. After addition of a cleavable cross‐linker or buffer only, liposomes were pelleted, dissolved in sample buffer, incubated in the presence or absence of 100 mM dithiothreitol (DTT) for 5 min at room temperature, and analyzed by SDS–PAGE and Coomassie staining. Cross‐link products migrate at the upper limit of the separating gel.
- Cross‐linking experiment with recombinant Atg18wt as in (A), performed in the presence of increasing concentrations of liposomes containing 5% PI(3,5)P2.
Discussion
Our study provides evidence for a function of the PROPPIN Atg18 in membrane fission. Vacuole fission activity of Atg18 depends on the presence of the CD loop and its capacity to fold into an amphipathic α‐helix. The positioning of this loop between the two phosphoinositide binding sites is a general feature of the PROPPIN family (Proikas‐Cezanne et al, 2004). Its insertion into the lipid phase may be necessary in order to allow the protein to approach the bilayer sufficiently to fully engage both lipid binding sites. Although the primary sequences of the CD loops are highly divergent, their potential to fold into an amphipathic α‐helix is well conserved. This suggests that the formation of an amphipathic helix in the CD loop and its membrane insertion is a generic feature of PROPPINs. Then, also other members of this family might display membrane fission activity. This notion is strengthened by the fact that inactivation of other PROPPINS affects cellular processes of which membrane fission is an essential part, such as retrograde trafficking from endosomes and lysosomes (Dove et al, 2004; Jeffries et al, 2004).
During autophagosome biogenesis, an isolation membrane bends into a cup‐shaped structure, but membrane bending activity has not yet been identified for any of the known autophagy proteins. Atg18 carries such activity. The interactions of its mammalian homolog WIPI2b with Atg5, Atg12, and Atg16L1, core components necessary for growth of the isolation membrane, suggest that it may be well‐placed to promote phagophore bending (Dooley et al, 2014; Juris et al, 2015; Proikas‐Cezanne et al, 2015). Furthermore, inactivation of mammalian WIPI2 or Atg18 arrests phagophore growth (Reggiori et al, 2004; Mari et al, 2010; Orsi et al, 2012; Suzuki et al, 2013). While this arrest can reflect the functions of Atg18 or WIPI proteins in recruiting and regulating core proteins for autophagosome assembly (Reggiori et al, 2004; Obara et al, 2008b; Dooley et al, 2014), a membrane bending and scission activity might also contribute to this phenotype. It might be particularly relevant in relation to Atg9, which interacts with Atg18 (Reggiori et al, 2004). Atg9 is a transmembrane protein that localizes to the isolation membrane and to several cellular membranes that may serve as its lipid sources, such as on endosomes, the Golgi, or at ER exit sites (Reggiori et al, 2004; Mari et al, 2010; Puri et al, 2013; Shibutani & Yoshimori, 2014; Tooze et al, 2014). One model of autophagy therefore stipulates that Atg9‐containing vesicles deliver lipids to the phagophore, probably by membrane fusion (Mari et al, 2010; Moreau et al, 2011; Orsi et al, 2012). If phagophore expansion indeed required cycling of Atg9‐containing vesicles between the phagophore and lipid donor membranes, the accumulation of Atg9 on the rim of the isolation membrane that is observed in atg18Δ mutants, or upon depletion of the mammalian WIPI proteins (Reggiori et al, 2004; Mari et al, 2010; Orsi et al, 2012; Yamamoto et al, 2012; Suzuki et al, 2013; Dooley et al, 2014), could result from stalled scission of Atg9 carriers from this site. Lack of Atg18 or WIPI protein activity might also rupture Atg9 cycling by stalling formation of Atg9 carriers at lipid donor membranes.
We observed functional differences between Atg18 function in vacuole fission and in autophagy. Atg18 interacts tightly with Atg2, and this interaction is crucial for autophagy. The resulting recruitment of Atg2 to the phagophore may be a major function of Atg18 in autophagy (Kobayashi et al, 2012). Since atg2Δ mutants fragment their vacuoles like wild type, this functional aspect of Atg18 may be dispensable for vacuole fission. The opposite applies to the amphipathic character of the CD loop, which appears to be less crucial for autophagy than for vacuole scission. This, however, should not be taken as evidence that Atg18 is not involved in membrane fission during autophagosome biogenesis. Proteins can facilitate membrane fission in numerous, not mutually exclusive ways (Johannes et al, 2014). Interesting examples are provided by the recent discoveries that simple crowding of proteins, or the frictional barrier to lipid diffusion that they create, can suffice to facilitate fission (Simunovic et al, 2017; Snead et al, 2017). Also mechanical stresses can alleviate the requirement for helix insertions in fission reactions (Renard et al, 2015). This can render membrane fission by endophilin A2 or epsin independent of the membrane insertion of their hydrophobic helices. Such effects and/or the interaction of Atg18 with Atg2 might also render the amphiphilic nature of the Atg18 CD loop less critical for autophagy. Detailed future analyses will have to test whether this is the case.
We propose a working model in which membrane contact of Atg18 transforms its CD loop into an amphipathic α‐helix, which reversibly inserts into the bilayer and allows Atg18 to be anchored there. Insertions of amphipathic α‐helices into the outer membrane leaflet bend the bilayer. This effect should be amplified by the concentration and potential oligomerization of Atg18 that is triggered through PI3P and PI(3,5)P2, the lipids required for vacuole fission. Increased recruitment favors bending and scission through crowding effects (Snead et al, 2017), and a tendency to form oligomeric assemblies of Atg18 could drastically increase the density of inserted CD loops. Crystal structures and the predicted length of the amphipathic helix allow to estimate their footprints on the membrane at 15 nm2 for the whole protein and at 1.4–3.5 (nm)2 for the amphipathic α‐helix (considering total folding and insertion of the hydrophobic loop for the maximal value). Coverage of a membrane patch with Atg18 would then increase the surface of its outer leaflet by 9–23%, which by itself can suffice to form membrane tubules of 35–50 nm diameter (Campelo et al, 2008).
Membrane fission by Atg18 might require constriction, similarly as by dynamin. In this model, the membrane tubules obtained by Atg18 loop insertion would have to be constricted further. This could be promoted by membrane scaffolding through Atg18 oligomerization, or by accessory constriction factors. In this respect, it is notable that vacuole fission and retrograde trafficking both require not only Atg18 but also the dynamin‐like GTPase Vps1 (Peters et al, 2004; Chi et al, 2014; Arlt et al, 2015). Vps1 lacks the PH domain through which dynamins interact with the membrane. It promotes membrane constriction but not fission (Smaczynska‐de Rooij et al, 2010) and might hence act in concert with Atg18.
Additional mechanisms, by which Atg18 could promote fission, are the above‐mentioned crowding and friction effects (Simunovic et al, 2017; Snead et al, 2017), but also lipid phase separation and the line tension resulting from it. Lipid phase separations can drive fission reactions, and they can be promoted or induced by proteins binding to one of the lipid phases. This is exemplified by the fission of Shiga‐toxin carriers, which is driven by actin‐induced lipid phase separation (Allain et al, 2004; Roux et al, 2005; Römer et al, 2010). Also vacuoles undergoing membrane fission show striking lipid phase separation. They segregate a liquid‐ordered phase that is readily detectable by freeze‐fracture EM because it is virtually free of intramembranous particles (bona fide transmembrane proteins) (Müller et al, 2000; Takatori et al, 2016). This phase, which is often found at sites where the membrane tubulates and will undergo fission, is strongly enriched for PI(3,5)P2 and PI3P (Zieger & Mayer, 2012; Takatori et al, 2016). Similar segregation of PI(3,5)P2 and PI3P‐enriched liquid‐ordered phases, with which the mammalian Atg18 homolog WIPI‐1 preferentially associates, occurs on mammalian endo‐lysosomal compartments (Takatori et al, 2016).
An interesting aspect of the fission activity of Atg18 concerns the driving force for the reaction. Whereas the prototypic fission catalyst dynamin is directly fueled by GTP hydrolysis and also Sar1 and Arf1 are GTPases (Pucadyil & Schmid, 2009), neither an enzymatic activity nor a nucleotide binding site has been found or predicted for Atg18 or other PROPPINs. However, Atg18 membrane recruitment and oligomerization requires the ATP‐consuming synthesis of PI3P and PI(3,5)P2, and subsequent recycling of Atg18 into its soluble cytosolic form is expected to occur upon hydrolysis of PI(3,5)P2. The system can thus be energized indirectly, via the energy invested into the synthesis of its lipid ligand.
Taken together, the tubulation and scission activity that we observed for pure Atg18 in a liposome system requires molecular features that are conserved in the PROPPIN family. Such a scission activity is compatible with known effects of PROPPIN mutations on endo‐lysosomal trafficking, autophagy, and vacuole fission (Dove et al, 2004; Jeffries et al, 2004). Therefore, we propose that membrane bending and fission activity represents a general feature of PROPPINs.
Materials and Methods
Materials
Lipids were purchased from Avanti Polar Lipids (USA): Egg L‐a‐phosphatidylcholine (EPC); 1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐L‐serine sodium salt (DOPS); 1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐(1′‐myo‐inositol‐3′‐phosphate) (PI3P); 1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐(1′‐myo‐inositol‐3′,5′‐bisphosphate) (P(3,5)P2); 1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐(1′‐myo‐inositol‐4′,5′‐bisphosphate) (PI(4,5)P2); cholesterol; 1,2‐dioleoyl‐sn‐glycero‐3‐phosphoethanolamine‐N‐(lissamine rhodamine B sulfonyl) (Rhodamine‐PE). All lipids were dissolved in chloroform. Phosphatidylinositol phosphates were dissolved in chloroform/methanol/water (20:10:1).
Strains and plasmids
BJ3505 yeast cells were grown in HC media as described previously (Zieger & Mayer, 2012). Genes were deleted by replacing a complete open reading frame with a marker cassette (Janke et al, 2004) (see Appendix Table S1 for a list of strains used in this study and Appendix Table S2 for a list of PCR primers used in this study). Atg18‐GFP was expressed from pRS316‐Atg18‐3HA‐GFP (Atg18‐HG), which was a kind gift of Dr Y. Ohsumi (Tokyo Institute of Technology). To generate Atg18SLoop‐GFP and Atg18DLoop‐GFP, pRS316‐Atg18‐3HA‐GFP was cut with BspEI and BglII and a synthetic gene fragment (Eurofins MWG operon) containing the corresponding sequence with the scrambled loop or the Delta loop mutations was inserted by ligation. To generate Atg18FGGG‐GFP, Atg18FGGG was amplified from pJE184 (GFP‐Atg18‐ALP with a 285GG286 mutation in Atg18 (Efe et al, 2007), kind gift from Dr. Scott Emr, Cornell University), digested with BglII and BspEI, and ligated into pRS316‐Atg18‐3HA‐GFP. pRS316‐Atg18 and mutants were generated from the corresponding pRS316‐Atg18‐3HA‐GFP plasmid from which the 3HA‐GFP was removed. For removal of the tag, a sequence containing the C‐terminus and part of the 3′‐UTR of Atg18 was amplified from the genome and inserted by gap repair into a SphI and NotI digested vector. pRS314‐Atg21 was generated by amplifying a fragment containing the promotor and coding sequence and the terminator of Atg21 from the genome. The purified PCR fragments were the inserted into pRS314 by gap repair. For pRS314‐Atg21Sloop, a DNA fragment containing the Sloop mutation was ordered (Eurofins) and inserted into pRS314‐Atg21 with ClaI and SexAI. All constructs were verified by DNA sequencing.
Live microscopy
Vacuoles were stained with FM4‐64. An overnight preculture in HC medium was used to inoculate a 10 ml culture. Cells were then grown in HC to an OD600 between 0.6 and 1.0. The culture was diluted to an OD600 of 0.4, and FM4‐64 was added to a final concentration of 10 μM from a 10 mM stock in DMSO. Cells were labeled for 60 min with FM4‐64, washed three times in fresh media, and then incubated for 60 min in media without FM4‐64 before imaging. For the quantification of vacuole fragmentation, NaCl was added to FM4‐64 stained cells (OD600 between 0.8 and 1.0) at a final concentration of 0.4 M and cells were imaged at 0, 15, 30, and 60 min after salt addition. Just before imaging, cells were concentrated by a brief low‐speed centrifugation and placed on a glass microscopy slide overlaid with a thin glass coverslip. Z‐stacks with a spacing of 0.3 μm were taken, assembled into maximum projections, and the number of vacuoles per cell was counted.
Peptide synthesis and circular dichroism spectroscopy
Peptides were synthesized by the protein and peptide synthesis facility of our department on a CS Bio 336 synthesizer and purified by preparative HPLC on a C18 column (Waters). Peptide purity was assessed by mass spectrometry (Applied Biosystem) and Analytical HPLC (Agilent). Purity was 97.7%. Lyophilized peptide was reconstituted in liposome CD buffer (50 mM NaCl, 20 mM NaHPO4 pH 6.8, 1 mM MgCl2) at 10 mg/ml and used at 1 mg/ml for CD acquisition. CD spectra were acquired in a Jasco J‐815 spectrometer using a 10‐mm‐wide quartz cell.
Protein purification
All steps were performed on ice or at 4°C, unless stated otherwise. Atg18 wt and mutant DNA were amplified from the corresponding pRS316 plasmids and cloned into a pEXP5‐NT/TOPO vector (Invitrogen). Plasmids were transformed into E. coli BL21. A 50‐ml preculture was used to inoculate 2 l of LB media (37°C). Cells were grown to an OD600 of 0.8–0.9. The cultures were then cooled to 16°C on ice, and IPTG (Roche) was added to a final concentration of 0.2 mM. Cells were shaken overnight at 16°C, pelleted, washed once in ice cold lysis buffer (500 mM NaCl, 50 mM Tris pH 7.4, 10 mM KPi), and resuspended in one pellet volume of lysis buffer + complete EDTA‐free protease inhibitor cocktail (Roche). Cells were lysed in a French press (One shot cell disruptor, Constant Systems LTD, Daventry, UK). Triton X‐100 was added to a final concentration of 1% and DNAse I to 0.1 mg/ml. The lysate was then incubated on a rotating wheel for 20 min at 4°C. Insoluble material was pelleted at 30,000 × g for 30 min in a Beckman JA25.50 rotor at 4°C. Supernatants were filtered through a 0.2 μm filter (Sarstedt, Germany), imidazole was added to a final concentration of 10 mM, and lysates were passed three times over 1 ml of Ni‐NTA resin (Qiagen). The resin was washed with 30 ml of lysis buffer containing 10 mM imidazole, and Atg18 was eluted with 250 mM imidazole in lysis buffer (pH 7.4). Bradford‐positive fractions were pooled. The his‐tag was cleaved using AcTEV protease. The protease was added directly to the eluted protein, and this solution was dialyzed overnight against TEV cleavage buffer (500 mM NaCl, 50 mM Tris pH 7.4, 10 mM KPi, 1 mM DTT, 0.5 mM EDTA). The protein was then dialyzed against lysis buffer for 3–4 h. Un‐cleaved protein and AcTEV were removed by passing the solution over 0.4 ml Ni‐NTA resin, washing with 20 ml of lysis buffer and eluting with lysis buffer containing 250 mM imidazole. The protein‐containing fractions were pooled and concentrated using an Amicon ultra column (30 kDa cutoff, Merck Millipore) and further purified by FPLC on a Superose‐6 column (GE Healthcare). For this, the column was equilibrated with lysis buffer and 0.5 ml of the concentrated protein was injected. Buffer flow was set to 0.4 ml/min, and 0.5 ml fractions were collected. Fractions analyzed by 10% SDS–PAGE and Coomassie staining. Fractions corresponding to Atg18 were pooled and dialyzed overnight against lysis buffer containing 50% glycerol. The protein was stored in aliquots at −20°C.
Liposome preparation and imaging
Giant unilamellar vesicles and SUVs contained phospholipids in the following ratios: 93% EPC + 5% PI3P + 1% PI(3,5)P2 or 94% EPC + 5% PI(4,5)P2 (Fig 7) or 89% EPC + 10% PI3P or 89% EPC + 10% PI(3,5)P2, or 89% EPC + 10% PI(4,5)P2 (Fig 8). All GUVs were supplemented with 1% Rhodamine‐PE. Cholesterol was added to 20 mole% of the phospholipids. SUVs were made as described previously (Baskaran et al, 2012). Briefly, lipids were diluted in chloroform in a glass tube and the solvent evaporated under nitrogen flux while gently vortexing the tube. Tubes were placed in vacuum for 1 h in order to remove traces of solvents. Liposome buffer (50 mM NaCl, 20 mM HEPES pH 6.8, 1 mM MgCl2) was added (final lipid concentration 5 mg/ml), and the tubes were placed in a water bath for 60 min at 37°C to hydrate the lipid film. For CD spectroscopy experiments, CD buffer was used (50 mM NaCl, 20 mM NaH2PO4 pH 6.8, 1 mM MgCl2). After 30–60 s of vigorous vortexing, the liposome suspension was transferred to an Eppendorf tube and frozen and thawed three times in liquid nitrogen. SUVs were used the same day or kept at −20°C and used within a week. GUVs were made by electro‐formation as described (Angelova et al, 1992). Briefly, lipid mix in chloroform was deposited on indium‐titan oxide glass slides and dried for 60 min at 55°C to evaporate all solvents. GUVs were electroformed at 1 V and 10 Hz for 60 min at 55°C in a 220 mM sucrose solution containing 1 mM Tris pH 6.8 and 0.2 mM EDTA (~220 mOsm). GUVs were then removed from the chamber and placed in an Eppendorf tube until use. GUVs were used within 1–2 h after formation. For imaging, 10 μl of GUV suspension was added to 100 μl of GUV buffer (50 mM NaCl, 20 mM HEPES pH 6.8, 1 mM MgCl2, 130 mM sucrose, ~250 mOsm) in a 96‐well plate pretreated with a BSA solution (1 mg/ml) for 30 min. GUVs were left to sediment for 20 min. For live imaging, 10 μl of GUVs was left to sediment in 100 μl GUV buffer for 20 min in BSA‐coated 96‐well plates and imaged by confocal spinning disk microscopy. The reaction was initiated by adding 10 μl of 10 μM Atg18 in hypertonic GUV buffer (50 mM NaCl, 20 mM HEPES pH 6.8, 1 mM MgCl2, 377 mM sucrose, ~500 mOsm), resulting in a slight increase in osmotic value of the suspension from 250 to 270 mOsm. This increase allows the spherical GUVs to lose water and adjust their surface‐to‐volume ratio to the lower values that are necessary for converting a sphere into a partially tubular structure. The sedimented GUVs were imaged by spinning disk confocal microscopy.
Liposome binding and cross‐linking assay
For the binding assay, 1.5 μM protein and 1 mg/ml SUVs were incubated in liposome buffer for 30 min at room temperature in a total volume of 50 μl. SUVs were then centrifuged at 21,000 × g on a table top centrifuge for 30 min at 4°C. Pellet and supernatant fraction were analyzed by SDS–PAGE and Coomassie staining. The cross‐linking assay was performed under the same conditions but in a total volume of 120 μl. After 30 min, 0.5 mM of a cleavable cross‐linker (DTSSP, sigma) was added and, 10 min later, the reaction was quenched by addition of 10 μl 1 M glycine. After 5 min, samples were split into two 60 μl fractions and 20 μl 4× non‐reducing sample buffer (−DTT) was added to half of the samples, while the other half was treated with reducing sample buffer (+DTT). Samples were analyzed by SDS gel and Coomassie staining.
SDS–PAGE and Western blotting
Inconsistent Atg18 migration was observed using precast commercial gels. SDS gels were therefore freshly prepared and used the same day. They had the following composition: 10% Protogel (30% w/v acrylamide, 0.8% bisacrylamide (37.5:1 solution), National diagnostics, Atlanta, USA), 380 mM Tris–HCl pH 8.8, 0.1% w/v SDS (Applichem, Darmstadt, Germany), 0.06% v/v TEMED (Applichem), 0.06% w/v APS (Applichem) for the running gel and 5% Protogel, 165 mM Tris–HCl pH 6.8, 0.1% w/v SDS, 0.08% v/v TEMED, 0.04% w/v APS for the stacking gel. Running buffer for SDS–PAGE was 190 mM glycine (Applichem), 25 mM Tris‐base (Applichem), 0.5% SDS. Final sample buffer composition was as follows: 50 mM Tris‐Cl pH 6.8, 2% SDS, 10% glycerol, 0.02% bromophenol blue. Samples were mixed with an equivalent volume of 2× sample buffer and incubated at room temperature for 5 min before being loaded. Sample boiling leads to Atg18 aggregation. Gels were blotted overnight at a constant current of 30 mA using a Mini Trans‐Blot® Cell (Bio‐Rad, USA).
For the preparation of total cell lysates, cells were grown as for live imaging and 2 OD600 equivalents were collected and precipitated with TCA (final concentration 6%) and put on ice for at least 5 min. Cells were then pelleted by centrifugation, washed twice with cold acetone, and dried (5–10 min on a heating block at 30°C). Cells were resuspended in 100 μl of lysis buffer (50 mM Tris pH 7.5, 5 mM EDTA, 100 mM NaCl, 1% SDS, 1 mM PMSF, 0.01% bromophenol blue), and 50 μl of glass beads was added. Cells were broken by vortexing for 10 min at 4°C and beads, and cell debris were pelleted by centrifugation on a table top centrifuge (18,000× g, 2 min). 0.5 OD600 equivalent (25 μl) was loaded per lane.
Pho8∆60 assay
The pho8∆60 assay was performed as described in Noda & Klionsky (2008).
Author contributions
NG, AR, and AM designed the research. NG carried out research. BF and HL provided technical expertise for CD spectroscopy. NG, BF, HL, AR, and AM analyzed the data. NG and AM wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Review Process File
Acknowledgements
We thank Catherine Servis for the synthesis of peptides and Scott Emr and Yoshinori Ohsumi for strains and plasmids. This work was supported by ERC Grant no. 233458 and SNF grant 144258 (to A.M.).
The EMBO Journal (2017) 36: 3274–3291
References
- Allain J‐M, Storm C, Roux A, Ben Amar M, Joanny J‐F (2004) Fission of a multiphase membrane tube. Phys Rev Lett 93: 158104 [DOI] [PubMed] [Google Scholar]
- Angelova MI, Soléau S, Méléard P, Faucon JF, Bothorel P (1992) AC field controlled formation of giant fluctuating vesicles and bending elasticity measurements In The structure and conformation of amphiphilic membranes, Lipowsky R, Richter D, Kremer K. (eds), pp 178–182. Berlin, Heidelberg: Springer; [Google Scholar]
- Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, Ford M, Frolov VA, Frost A, Hinshaw JE, Kirchhausen T, Kozlov MM, Lenz M, Low HH, McMahon H, Merrifield C, Pollard TD, Robinson PJ, Roux A, Schmid S (2016) Membrane fission by dynamin: what we know and what we need to know. EMBO J 35: 2270–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Reggiori F, Ungermann C (2015) Retromer and the dynamin Vps1 cooperate in the retrieval of transmembrane proteins from vacuoles. J Cell Sci 128: 645–655 [DOI] [PubMed] [Google Scholar]
- Bakula D, Müller AJ, Zuleger T, Takacs Z, Franz‐Wachtel M, Thost A‐K, Brigger D, Tschan MP, Frickey T, Robenek H, Macek B, Proikas‐Cezanne T (2017) WIPI3 and WIPI4 β‐propellers are scaffolds for LKB1‐AMPK‐TSC signalling circuits in the control of autophagy. Nat Commun 8: 15637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH (2012) Two‐site recognition of phosphatidylinositol 3‐phosphate by PROPPINs in autophagy. Mol Cell 47: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Sun Z, Adolf F, Rutz C, Bassler J, Wild K, Sinning I, Hurt E, Brügger B, Béthune J, Wieland F (2008) Membrane curvature induced by Arf1‐GTP is essential for vesicle formation. Proc Natl Acad Sci USA 105: 11731–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M (2005) Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol 171: 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS (2002) Osmotic stress‐induced increase of phosphatidylinositol 3,5‐bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol 156: 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon HT, Kozlov MM (2012) Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell 149: 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse RA, Scacioc A, Krick R, Pérez‐Lara A, Thumm M, Kühnel K (2015) Characterization of PROPPIN‐phosphoinositide binding and role of Loop 6CD in PROPPIN‐membrane binding. Biophys J 108: 2223–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F, McMahon HT, Kozlov MM (2008) The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J 95: 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi RJ, Liu J, West M, Wang J, Odorizzi G, Burd CG (2014) Fission of SNX‐BAR‐coated endosomal retrograde transport carriers is promoted by the dynamin‐related protein Vps1. J Cell Biol 204: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ (1998) The stress‐activated phosphatidylinositol 3‐phosphate 5‐kinase Fab1p is essential for vacuole function in S. cerevisiae . Curr Biol 8: 1219–1222 [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A (2009) The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell 17: 712–723 [DOI] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA (2014) WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12‐5‐16L1. Mol Cell 55: 238–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH (1997) Osmotic stress activates phosphatidylinositol‐3,5‐bisphosphate synthesis. Nature 390: 187–192 [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA (2004) Svp1p defines a family of phosphatidylinositol 3,5‐bisphosphate effectors. EMBO J 23: 1922–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS (2006) Phosphoinositide 5‐phosphatase Fig 4p is required for both acute rise and subsequent fall in stress‐induced phosphatidylinositol 3,5‐bisphosphate levels. Eukaryot Cell 5: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Botelho RJ, Emr SD (2007) Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5‐bisphosphate. Mol Biol Cell 18: 4232–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P (2012) Dynamin, a membrane‐remodelling GTPase. Nat Rev Mol Cell Biol 13: 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD (1998) Fab1p is essential for PtdIns(3)P 5‐kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol 143: 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD (2002) Regulation of Fab1 phosphatidylinositol 3‐phosphate 5‐kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell 13: 1238–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G (2008) HELIQUEST: a web server to screen sequences with specific alpha‐helical properties. Bioinformatics 24: 2101–2102 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W‐K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD (2009) A FAM21‐containing WASH complex regulates retromer‐dependent sorting. Dev Cell 17: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno‐Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004) A versatile toolbox for PCR‐based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jeffries TR, Dove SK, Michell RH, Parker PJ (2004) PtdIns‐specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol Biol Cell 15: 2652–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Wunder C, Bassereau P (2014) Bending ‘on the rocks’–a cocktail of biophysical modules to build endocytic pathways. Cold Spring Harb Perspect Biol 6: a016741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juris L, Montino M, Rube P, Schlotterhose P, Thumm M, Krick R (2015) PI3P binding by Atg21 organises Atg8 lipidation. EMBO J 34: 955–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Suzuki K, Ohsumi Y (2012) Autophagosome formation can be achieved in the absence of Atg18 by expressing engineered PAS‐targeted Atg2. FEBS Lett 586: 2473–2478 [DOI] [PubMed] [Google Scholar]
- Krick R, Henke S, Tolstrup J, Thumm M (2008) Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy 4: 896–910 [DOI] [PubMed] [Google Scholar]
- Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kühnel K (2012) Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β‐propeller protein family. Proc Natl Acad Sci USA 109: E2042–E2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic‐sample processing applied to copy‐number estimation in eukaryotic cells. Nat Methods 11: 319–324 [DOI] [PubMed] [Google Scholar]
- Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R (2005) Sar1p N‐terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122: 605–617 [DOI] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F (2010) An Atg9‐containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 190: 1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AJ, Zhang Y, Weisman LS (2014) Phosphatidylinositol 3,5‐bisphosphate: low abundance, high significance. BioEssays 36: 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaillat L, Mayer A (2013) Identification of genes affecting vacuole membrane fragmentation in Saccharomyces cerevisiae . PLoS One 8: e54160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC (2011) Autophagosome precursor maturation requires homotypic fusion. Cell 146: 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot S, Roux A (2013) Mechanics of dynamin‐mediated membrane fission. Annu Rev Biophys 42: 629–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O, Sattler T, Flötenmeyer M, Schwarz H, Plattner H, Mayer A (2000) Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J Cell Biol 151: 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Klionsky DJ (2008) The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 451: 33–42 [DOI] [PubMed] [Google Scholar]
- Obara K, Noda T, Niimi K, Ohsumi Y (2008a) Transport of phosphatidylinositol 3‐phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae . Genes Cells 13: 537–547 [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y (2008b) The Atg18‐Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3‐phosphate and exerts an essential function. J Biol Chem 283: 23972–23980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23: 1860–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Baars TL, Buhler S, Mayer A (2004) Mutual control of membrane fission and fusion proteins. Cell 119: 667–678 [DOI] [PubMed] [Google Scholar]
- Polson HEJ, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, Tooze SA (2010) Mammalian Atg18 (WIPI2) localizes to omegasome‐anchored phagophores and positively regulates LC3 lipidation. Autophagy 6: 506–522 [DOI] [PubMed] [Google Scholar]
- Proikas‐Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A (2004) WIPI‐1alpha (WIPI49), a member of the novel 7‐bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation‐induced autophagy. Oncogene 23: 9314–9325 [DOI] [PubMed] [Google Scholar]
- Proikas‐Cezanne T, Robenek H (2011) Freeze‐fracture replica immunolabelling reveals human WIPI‐1 and WIPI‐2 as membrane proteins of autophagosomes. J Cell Mol Med 15: 2007–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas‐Cezanne T, Takacs Z, Dönnes P, Kohlbacher O (2015) WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci 128: 207–217 [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL (2009) Conserved functions of membrane active GTPases in coated vesicle formation. Science 325: 1217–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC (2013) Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154: 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ (2004) The Atg1‐Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre‐autophagosomal structure. Dev Cell 6: 79–90 [DOI] [PubMed] [Google Scholar]
- Renard H‐F, Simunovic M, Lemière J, Boucrot E, Garcia‐Castillo MD, Arumugam S, Chambon V, Lamaze C, Wunder C, Kenworthy AK, Schmidt AA, McMahon HT, Sykes C, Bassereau P, Johannes L (2015) Endophilin‐A2 functions in membrane scission in clathrin‐independent endocytosis. Nature 517: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, Kametaka S, Haft CR, Bonifacino JS (2007) Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6‐phosphate receptors. Mol Cell Biol 27: 1112–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer W, Pontani L‐L, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, Johannes L (2010) Actin dynamics drive membrane reorganization and scission in clathrin‐independent endocytosis. Cell 140: 540–553 [DOI] [PubMed] [Google Scholar]
- Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B (2005) Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J 24: 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink KO, Raiborg C, Stenmark H (2013) Phosphatidylinositol 3‐phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. BioEssays 35: 900–912 [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD (1998) A membrane coat complex essential for endosome‐to‐Golgi retrograde transport in yeast. J Cell Biol 142: 665–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, Yoshimori T (2014) A current perspective of autophagosome biogenesis. Cell Res 24: 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Manneville J‐B, Renard H‐F, Evergren E, Raghunathan K, Bhatia D, Kenworthy AK, Voth GA, Prost J, McMahon HT, Johannes L, Bassereau P, Callan‐Jones A (2017) Friction mediates scission of tubular membranes scaffolded by BAR proteins. Cell 170: 172–184.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczynska‐de Rooij II, Allwood EG, Aghamohammadzadeh S, Hettema EH, Goldberg MW, Ayscough KR (2010) A role for the dynamin‐like protein Vps1 during endocytosis in yeast. J Cell Sci 123: 3496–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead WT, Hayden CC, Gadok AK, Zhao C, Lafer EM, Rangamani P, Stachowiak JC (2017) Membrane fission by protein crowding. Proc Natl Acad Sci USA 114: E3258–E3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug PE, Reggiori F, Guan J, Wang C‐W, Klionsky DJ (2004) Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell 15: 3553–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo‐Kakuta C, Yamamoto H, Ohsumi Y (2013) Fine mapping of autophagy‐related proteins during autophagosome formation in Saccharomyces cerevisiae . J Cell Sci 126: 2534–2544 [DOI] [PubMed] [Google Scholar]
- Takatori S, Tatematsu T, Cheng J, Matsumoto J, Akano T, Fujimoto T (2016) Phosphatidylinositol 3,5‐bisphosphate‐rich membrane domains in endosomes and lysosomes. Traffic 17: 154–167 [DOI] [PubMed] [Google Scholar]
- Tamura N, Oku M, Ito M, Noda NN, Inagaki F, Sakai Y (2013) Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide‐binding activity. J Cell Biol 202: 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P, Lauffer B, Jäger S, Cimermancic P, Krogan NJ, von Zastrow M (2011) SNX27 mediates retromer tubule entry and endosome‐to‐plasma membrane trafficking of signalling receptors. Nat Cell Biol 13: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Abada A, Elazar Z (2014) Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb Perspect Biol 6: a018358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy‐defective mutants of Saccharomyces cerevisiae . FEBS Lett 333: 169–174 [DOI] [PubMed] [Google Scholar]
- Uchida M, Sun Y, McDermott G, Knoechel C, Le Gros MA, Parkinson D, Drubin DG, Larabell CA (2011) Quantitative analysis of yeast internal architecture using soft X‐ray tomography. Yeast 28: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC (2015) PI(5)P regulates autophagosome biogenesis. Mol Cell 57: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN (2012) Structure‐based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem 287: 31681–31690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo‐Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieger M, Mayer A (2012) Yeast vacuoles fragment in an asymmetrical two‐phase process with distinct protein requirements. Mol Biol Cell 23: 3438–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Review Process File


