Abstract
Microglia are resident macrophages of the central nervous system that contribute to homeostasis and neuroinflammation. Although known to play an important role in brain development, their exact function has not been fully described. Here, we show that in contrast to healthy adult and inflammation‐activated cells, neonatal microglia show a unique myelinogenic and neurogenic phenotype. A CD11c+ microglial subset that predominates in primary myelinating areas of the developing brain expresses genes for neuronal and glial survival, migration, and differentiation. These cells are the major source of insulin‐like growth factor 1, and its selective depletion from CD11c+ microglia leads to impairment of primary myelination. CD11c‐targeted toxin regimens induced a selective transcriptional response in neonates, distinct from adult microglia. CD11c+ microglia are also found in clusters of repopulating microglia after experimental ablation and in neuroinflammation in adult mice, but despite some similarities, they do not recapitulate neonatal microglial characteristics. We therefore identify a unique phenotype of neonatal microglia that deliver signals necessary for myelination and neurogenesis.
Keywords: CD11c, IGF1, microglia, myelinogenesis
Subject Categories: Immunology, Neuroscience
Introduction
Microglia are resident myeloid cells of the central nervous system (CNS) that originate from yolk sac precursors and colonize the brain very early during embryonic life (Ginhoux et al, 2010; Schulz et al, 2012; Kierdorf et al, 2013). They are autonomously maintained through proliferation (Askew et al, 2017) and are not replaced from blood‐derived precursors during the lifetime of the host, at least under normal circumstances (reviewed in Ginhoux et al, 2013; Prinz & Priller, 2014). The role of microglia has traditionally been studied in the context of immune responses in the diseased CNS, and they have been implicated in neuroinflammatory and neurodegenerative diseases. Although their importance in homeostasis and brain development is recognized (Biber et al, 2014; Matcovitch‐Natan et al, 2016), their exact role in these processes remains incompletely defined. Evidence supporting the view that microglia are crucial players in neurodevelopment includes that mice lacking the CSF1R receptor for CSF1 or IL34, which is critical for microglia maintenance, show abnormal brain development (Elmore et al, 2014). Moreover, they are involved in synaptic pruning (Paolicelli et al, 2011; Schafer et al, 2012; Kettenmann et al, 2013; Zhan et al, 2014), they modulate axonal outgrowth and cortical interneuron positioning (Squarzoni et al, 2014), and they support survival of layer V cortical neurons during postnatal development by producing neuroprotective insulin‐like growth factor 1 (IGF1) (Ueno et al, 2013). IGF1 has also been shown to be essential for primary myelination, but the microglial contribution to this process is relatively unstudied.
In many neuroinflammatory (Wlodarczyk et al, 2014, 2015) and degenerative (Butovsky et al, 2006) conditions, a normally rare subpopulation of microglia that expresses the integrin complement receptor CD11c increases in proportion and number. We have previously shown in an animal model for multiple sclerosis, experimental autoimmune encephalomyelitis (EAE), that while CD11c+ microglia are effective antigen‐presenting cells for T‐cell proliferation, they are a poor source of pro‐inflammatory cytokines (Wlodarczyk et al, 2014) and that they differ from infiltrating DC and CD11c− microglia with respect to expression of many genes (Wlodarczyk et al, 2015). Interestingly, we showed that CD11c+ microglia uniquely express Igf1 during EAE (Wlodarczyk et al, 2015), suggesting that they may be neuroprotective.
Here, we show that neonatal microglia differ dramatically in their gene expression profile from microglia in healthy adults and mice with EAE, showing a neurogenic signature. CD11c+ microglia greatly expand during postnatal development (PN3‐5) and then dramatically contract as mice age to adulthood. They express a characteristic neurosupportive gene profile, equipping them to play a fundamental role in the developing CNS. Moreover, they are appropriately located for delivery of signals necessary for neuronal development and primary myelination. Importantly, we show that Igf1 deficiency in CD11c+ microglia leads to an impairment in primary myelination. Three separate CD11c‐targeted toxin regimens all resulted in a similar selective transcriptional outcome in neonatal mice, with a response distinct from that of adult microglia. Furthermore, whereas CD11c+ microglia are among the cells that repopulate the adult brain after microglial ablation, they do not show myelinogenic or neurogenic signatures. Thus, we identify neonatal microglia, and especially the CD11c+ subset, as key tissue macrophages for CNS development.
Results
CD11c+ microglia emerge during postnatal development
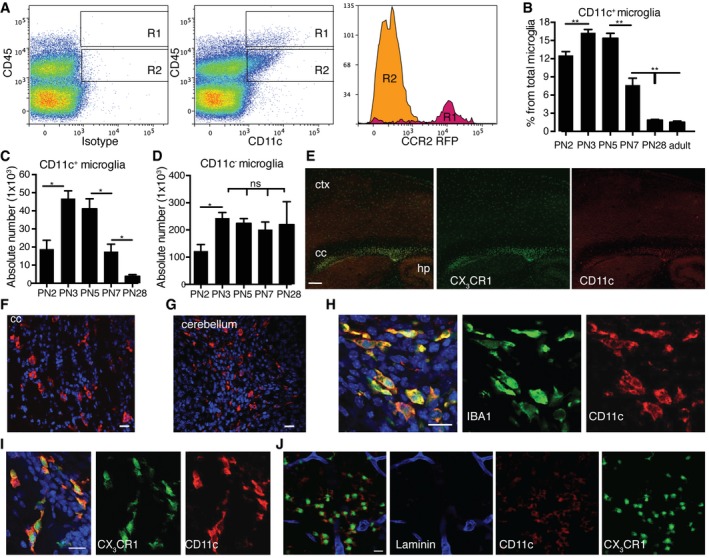
During neuroinflammation, CD11c+ microglia are the major source of Igf1 (Wlodarczyk et al, 2015), a gene critical for neurodevelopment, myelination, and neurogenesis. Thus, we asked whether this microglial subset is present during postnatal development. Mononuclear cells were isolated from perfused brains from PN2, PN3, PN7, PN28, and adult (8–12 weeks old) B6 and CCR2rfp/+ mice and analyzed by flow cytometry for CD11c expression. Microglia are defined by a lower level of cell surface CD45 than blood‐derived leukocytes, expression of fractalkine receptor CX3CR1, and lack of CCR2 chemokine receptor (Mizutani et al, 2012). We used relative CD45 levels (Remington et al, 2007) and CCR2 expression to discriminate between blood‐derived leukocytes (CD45high CCR2+) and resident microglia (CD45dimCCR2−). There were very few CD45high CD11c+ cells (0.2% live gate) in brain isolates at indicated time points. Nevertheless, nearly 85% of them were CCR2‐positive (Fig 1A). In contrast, more than 99% of CD45dim CD11b+ CD11c+ cells (microglia) were CCR2‐negative (Fig 1A). The proportion of CD11c+ microglia, as a percentage of total microglia, increased significantly from PN2 (12%) to PN3 (17%) and then sharply decreased by PN7 (8%), falling to < 3% in young (PN28) and adult animals (Fig 1B). Absolute numbers of CD11c+ microglia significantly increased from PN2 to PN3‐5. After PN5, the numbers of CD11c+ microglia dramatically decreased, reaching only 50 and 10% of PN3‐5 levels at PN7 and PN28, respectively (Fig 1C). Numbers of CD11c– microglia were also elevated in neonatal CNS, increasing from PN2 to PN3. However, unlike CD11c+ microglia, their numbers were stable from PN3 throughout adulthood (Fig 1D). Immunofluorescent stainings of perfused PN4‐5 B6 and CX3CR1GFP/+ murine brains showed that CD11c+ microglia cells were not homogenously distributed throughout the brain, but localized mainly in the corpus callosum and cerebellar white matter, in contrast to other areas of the brain where they were virtually absent (Fig 1E–G). All of the CD11c‐positive cells co‐stained with the microglial marker IBA1 (Fig 1H) and co‐localized with CX3CR1 (Fig 1I). Importantly, laminin staining revealed that they were localized in the brain parenchyma and not in blood vessels or in the perivascular space (Fig 1J).
Figure 1. CD11c+ microglia emerge during postnatal neurodevelopment.
-
ARepresentative flow cytometry profiles of five individual brain suspensions prepared from PN5 mice showing RFP expression driven by Ccr2 promoter on CD45highCD11c+ cells (R1) and lack of expression on CD45lowCD11c+ microglia (R2).
-
B–DFlow cytometry analysis showing CD11c+ microglia presented as a percentage from total microglia (B), absolute numbers of CD11c+ microglia (C), and CD11c− microglia (D) at different time points PN2 (n = 4), PN3 (n = 6), PN5 (n = 6), PN7 (n = 6), PN28 (n = 9), and adult mouse brain (n = 16).
-
ERepresentative low‐power micrographs showing patches of CD11c‐stained cells (red) co‐localizing with GFP driven by Cx3cr1 promoter (green) in corpus callosum (cc) and single CX3CR1‐positive cells in cortex (ctx) and hippocampus (hp) in PN4‐5 brains (n = 3). Scale bar = 200 μm.
-
F–IRepresentative confocal microscopic micrographs showing CD11c‐stained cells (red) in corpus callosum (F) and cerebellum (G) as well as co‐localization of CD11c marker (red) and IBA1 (green) (H) or CX3CR1 (green) (I) in PN4‐5 brains (n = 3). Scale bars = 15 μm.
-
JConfocal microscopic analysis of two individual brains showing CD11c (red), CX3CR1 (green) double‐positive cells localized in the parenchyma, outside of the laminin‐stained blood vessels (blue) (n = 2). Scale bar = 15 μm.
Neonatal CD11c+ microglia are a critical source of IGF1 for primary myelination
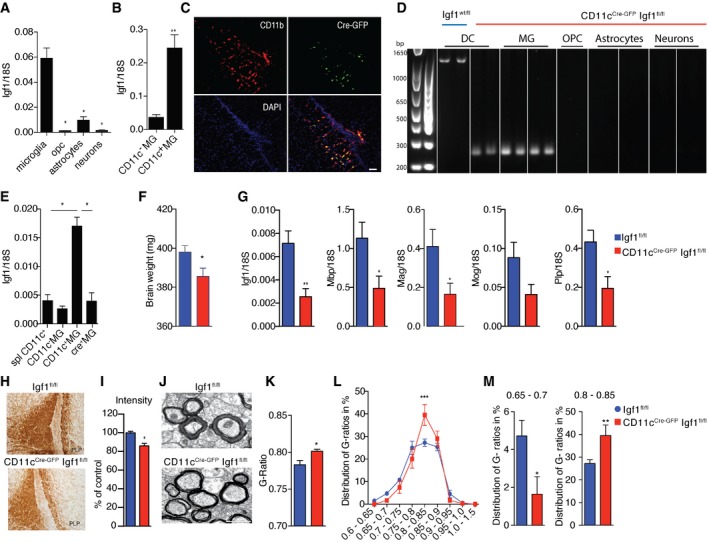
Next, we were interested whether CD11c+ microglia, so abundant in areas of primary myelination, are a source of Igf1 that is critical for this process. We compared levels of Igf1 expression in MACS‐sorted neurons, astrocytes, oligodendrocyte precursor cells (OPCs), and CD11b+ cells (which include mostly microglia) from unperfused PN4‐7 brains. Although all cell populations expressed detectable levels of Igf1, CD11b+ cells showed at least eightfold higher expression of this gene (Fig 2A). Igf1 transcripts were further compared in CD11c+ and CD11c− microglial populations. CD11c+ microglia expressed significantly higher levels of Igf1 (sevenfold) than their CD11c− counterparts (Fig 2B).
Figure 2. Neonatal CD11c+ microglia are a major source of myelinogenic Igf1 .
-
A, BExpression of Igf1 relative to 18S rRNA in MACS‐sorted microglia, OPC, astrocytes and neurons (n = 4) (A) as well as FACS‐sorted CD11c+ and CD11c− microglia (n = 6) (B) from brains of PN4‐7 mice.
-
CRepresentative micrographs showing patches of Cre‐GFP, CD11b double‐positive cells in corpus callosum from PN4‐5 CD11c Cre‐GFP Igf1fl/fl brains (n = 3), Scale bar = 50 μm.
-
DGenomic PCR analysis of Cre recombination in MACS‐sorted splenic dendritic cells (DC) from Igf1wt/fl and microglia, OPC, astrocytes, and neurons from CD11cCre‐GFP Igf1fl/fl. Wild‐type Igf1 gene is detected as an ˜1‐kb band; Cre‐induced recombination is detected as an ˜0.2‐kb band, while the Igf1/flox locus cannot be amplified under the assay condition (Liu et al, 1998).
-
EExpression of Igf1 relative to 18S rRNA in FACS‐sorted splenic CD11c+ cells (n = 5), CD11c+ microglia (n = 5), and CD11c− microglia (n = 5) from Igf1fl/fl mice and Cre+ microglia (n = 4, each n represents a pool of 2 brains) from CD11cCre‐GFP Igf1fl/fl mice.
-
FBar graph showing weights of brains from PN21 Igf1fl/fl (blue) (n = 8) and CD11cCre‐GFP Igf1fl/fl (red) (n = 4) mice.
-
GExpression of Igf1, Mog, Plp, and Mbp relative to 18S rRNA in brain tissue from PN21 CD11cCre‐GFP Igf1fl/fl and Igf1fl/fl mice (F) n = 6.
-
H, IRepresentative micrographs (H) and quantification of PLP staining intensity (I) in corpus callosum of CD11cCre‐GFP Igf1fl/fl (red) (n = 4) and Igf1fl/fl (blue) (n = 6) PN21 brains.
-
J–MRepresentative electron microscopy micrographs (J), mean G‐ratios (K), and distribution of G‐ratios (L, M) in corpus callosum from Igf1fl/fl (blue) (n = 8) and CD11c CD11cCre‐GFP Igf1fl/fl (red) (n = 6) PN21 brains. Scale bar = 1 μm.
To confirm the importance of CD11c+ microglia‐derived IGF1 on primary myelination, we used a CD11c‐Cre‐GFP driver to delete the Igf1 gene specifically in CD11c+ cells. In line with data presented in Fig 1, Cre‐GFP positive cells were localized mainly in corpus callosum (Fig 2C) and cerebellum. We confirmed that all of these cells co‐stained with CD11b (Fig 2C). PCR analysis of genomic DNA in sorted OPC, astrocytes, neurons, and microglia from CD11cCre‐GFP Igf1fl/fl PN7 mice revealed Cre‐induced recombination only in microglia (Fig 2D). We additionally showed lack of recombination in astrocytes from PN21 CD11cCre‐GFP Igf1fl/fl mice, and we showed recombination in microglia but not in neurons, OPC, or astrocytes from PN7 CD11cCre‐GFP Igf1fl/WT heterozygous mice (Fig EV1). This is in line with results from Goldmann et al (2013), who showed that in a CD11cCre: Rosa EYFP reporter mouse EYFP was exclusively expressed by Iba1 + microglia in CNS parenchyma and that there was no ectopic expression outside the myeloid lineage in these mice. Flow cytometry analysis showed that CD11c+ but not CD11c− microglia or CD45 negative cells were Cre‐GFP positive (not shown). Igf1 expression was significantly reduced in sorted Cre+ CD11c+ microglia, reaching levels of CD11c− microglia and splenic CD11c+ cells (Fig 2E). Even though the efficiency of Cre recombination in CD11c+ microglia was only close to 40% (Fig 2D), we observed lower brain weight (Fig 2F), significant decrease in Igf1, Plp, Mag, and Mbp, and slight downregulation of Mog gene expression (Fig 2G) in PN21 CD11cCre‐GFP Igf1fl/fl brains in comparison with Igf1‐intact Igf1fl/fl littermate controls. This was accompanied by less intense PLP staining (Fig 2H) and significantly higher myelin G‐ratio (Fig 2I–K) in corpus callosum. Conversely, we observed higher representation of less myelinated fibers (G‐ratio 0.8–0.85) and significantly less frequent sufficiently myelinated axons (G‐ratio = 0.65–0.7) in corpus callosum of CD11cCre‐GFP Igf1fl/fl than in littermate controls (Fig 2L and M). Altogether, our data point to an important role for IGF1‐producing CD11c+ microglia in primary myelination.
Figure EV1. Cre recombination in microglia.
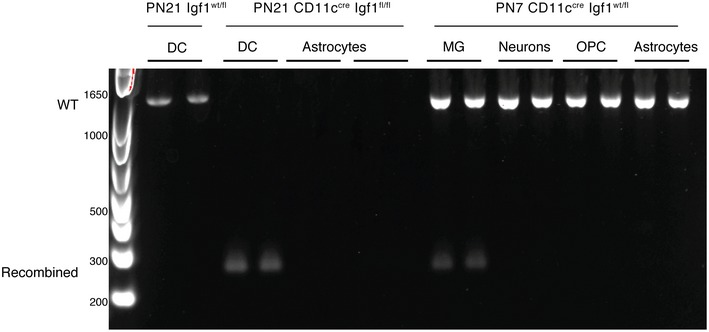
Genomic PCR analysis of Cre recombination in MACS‐sorted splenic dendritic cells (DC) from PN21 Igf1wt/fl; DC, astrocytes from PN21 CD11cCre‐GFP Igf1fl/fl (homozygous) as well as microglia, neurons, OPC, and astrocytes from CD11cCre‐GFP Igf1wt/fl (heterozygous). Wild‐type Igf1 gene is detected as an ~1‐kb band; Cre‐induced recombination is detected as an ~0.2‐kb band, while the Igf1/flox locus cannot be amplified under the assay condition (Liu et al, 1998).
Distinct gene signatures in microglia subsets during development and EAE
We have previously shown that numbers and proportions of CD11c+ microglia dramatically increase during EAE, and the data suggested that they might play a protective role during the disease (Wlodarczyk et al, 2014, 2015). To assess whether microglial subsets from developing CNS and mice with symptomatic EAE are similar, we compared transcriptomes of sorted CD11c+ and CD11c− microglia from PN4‐6 CNS and symptomatic EAE (grades 3‐5) as well as of total naïve adult microglia (Dataset EV1). Microglial markers (Aif1, Itgam, Cx3cr1, Csf1r) and signature genes (Butovsky et al, 2014; Bennett et al, 2016) (Spi1, Irf8, Olfml3, Hexb, Fcrls, Tgfbr1, P2ry12, Siglech, Tmem119) were similarly expressed in both CD11c+ and CD11c− neonatal and EAE microglia populations as in adult microglia (not shown). CD11c+ microglia from neonates and mice with EAE strongly upregulated Itgax expression, confirming high purity of sorted cells and validating the RNA‐seq assay (not shown). Moreover, we identified 20 genes (Itgax, Gpnmb, Spp1, Igf1, Colec12, Ccl5, Ak4, Lox, Mmp12, Cpeb1, Ntn1, Clec7a, Saa3, Ahnak2, Fabp5, Hpse, Gm26902, Cspg4, Fam20c, and Stra6 l) that were associated with the CD11c+ microglia population.
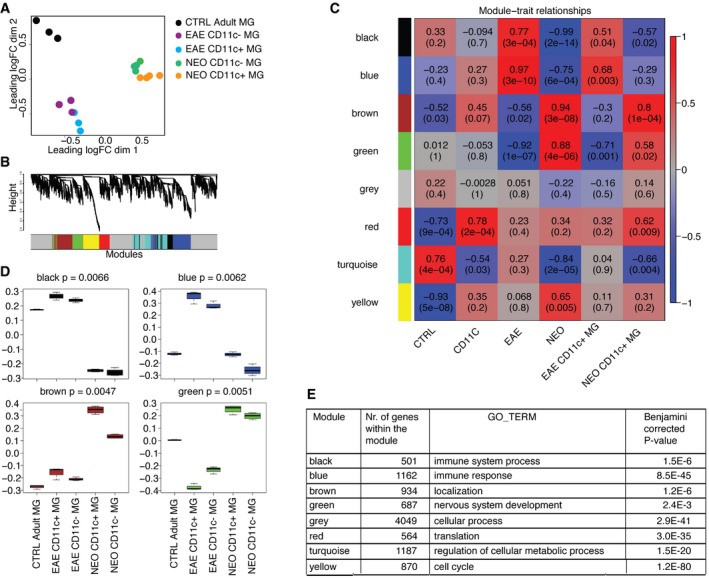
A multidimensional scaling (MDS) plot showed that neonatal, naïve adult microglia, and microglia from EAE formed three separate and distinct global gene expression clusters, while CD11c‐positive and negative subpopulations clustered relatively close together for each condition (Fig 3A). The major difference was therefore related to developmental age rather than subset phenotype.
Figure 3. Transcriptome analysis of neonatal, EAE, and adult microglia.
-
AMultidimensional scaling shows that neonatal, EAE subpopulations of microglia and adult microglia have distinct transcriptional profiles. Colors indicate six different groups of samples: orange represents neonatal CD11c+ microglia (n = 4), green neonatal CD11c− microglia (n = 4), blue EAE CD11c+ microglia (n = 3), purple EAE CD11c− microglia (n = 3), and black adult microglia (n = 3). Each n represents a pool of 10–15 mice from five individual EAE immunizations and four individual cell sorts of neonatal and naïve adult microglia.
-
B, CCo‐expression networks were generated for 12,691 genes of the transcriptome dataset. Average linkage hierarchical clustering was applied to the topological overlap matrix and branches of highly correlating genes were formed, which were cut and assigned a color (B). For each module, the Module Eigengene (ME) was calculated, which represents the expression profile of the module. The ME values were correlated with binary variables (Spearman's correlation) that represent control, CD11c+, EAE, neonatal, and CD11c+ microglia in neonates and EAE. Within each table cell, upper values represent correlation coefficients between ME and the variable, while lower values in brackets correspond to Student asymptotic P‐value (C).
-
DA boxplot containing the distribution of the black, blue, brown, and green ME values across the samples. The boxes contain the first and third quartiles; center line indicates the median and whiskers indicate minimum and maximum values. Kruskal–Wallis test was applied to determine whether ME values were significantly different between the groups.
-
ETable showing number of genes within the module and the top GO term for each module with Benjamini‐corrected P‐value.
In order to identify gene expression profiles associated with microglia from the different conditions, weighted gene co‐expression network analysis (WGCNA) was applied to the RNA‐seq data. In WGCNA, genes are clustered according to co‐expression and a network with seven co‐expression modules was identified (Fig 3B). From each module, the Module Eigengene (ME) was calculated, which is the first principal component and functions as a representative of the module. The ME values were correlated with variables that represent control adult microglia (“control”), CD11c+ microglia population (“CD11c”), EAE microglia populations (“EAE”), neonatal microglia populations (“neonatal”), and CD11c+ microglia from EAE (“EAE CD11c+”) and neonates (“neonatal CD11c+”) (Fig 3C) are depicted as a box plot per condition (Fig 3D).
With the exception of the gray module (that contained unclustered genes), all modules correlated significantly (P < 0.005) with some of the indicated variables. The ME of the yellow module was negatively correlated with “control” (−0.93; downregulated) and positively with “neonatal” (between 0.068 and 0.65). The top gene ontology (GO) enrichment category associated with the yellow module was the “cell cycle” term (Fig 3E). This suggested that microglia from neonates more abundantly expressed cell cycle genes, which is in line with the observed microglia proliferation during neurodevelopment (Matcovitch‐Natan et al, 2016). In contrast, the turquoise module was negatively correlated with “neonatal” variable (−0.84) and positively correlated with “control” (0.76). This module was enriched for GO term “regulation of cellular metabolic process”. The red module was not only negatively correlated with the “control”, but also positively correlated with the “CD11c” variable (−0.73 versus 0.78). This suggested an activation‐related increase in expression, which is more pronounced in CD11c‐positive microglia. This red module was enriched for a “translation” GO term (Fig 3E), suggesting that neonatal and EAE microglia and in particular CD11c microglia were more translationally active.
Interestingly, four modules (black, blue, green, and brown) showed clear opposite correlations with the “EAE” and “neonatal” variables (Fig 3C). Where the blue and black modules increased their expression in EAE microglia, the expression of these modules was reduced in neonatal microglia (Fig 3D). The blue and black modules were enriched for “immune system process” and “immune response” categories. In contrast, ME values of the brown and green modules negatively correlated (Fig 3C) and were decreased (Fig 3D) in EAE microglia and positively correlated (Fig 3C) and increased (Fig 3D) in neonatal microglia. The green and brown modules were enriched for “nervous system development” and “localization” GO terms, respectively (Fig 3E). Additionally, the brown module also significantly correlated with the CD11c+ neonatal microglia variable, suggesting that the CD11c+ population in neonatal brains most abundantly expressed genes related to the “localization” GO term (Fig 3E). Taken together, these findings suggest that microglia under EAE conditions became immune‐activated, while microglia in neonatal brains displayed a CNS‐supportive and neurogenic phenotype.
Distinct gene expression patterns in neonatal subsets of microglia and adult microglia
In order to further investigate differences between neonatal and adult microglia, the transcriptomes of CD11c+ and CD11c− microglia from PN4‐6 as well as of total adult microglia were compared. MDS plot showed that neonatal CD11c+, neonatal CD11c−, and adult microglia formed three separate and distinct global gene expression clusters (Fig 4A). From 12,691 genes expressed, 1,104 genes were increased and 1,001 decreased in neonatal CD11c− microglia compared to adult microglia. In neonatal CD11c+ microglia, 1,633 genes were increased and 1,131 decreased, compared to adult microglia, and 641 were increased and 163 decreased, compared to CD11c− microglia (Fig 4B).
Figure 4. Distinct gene expression profiles in neonatal and adult microglial subsets.
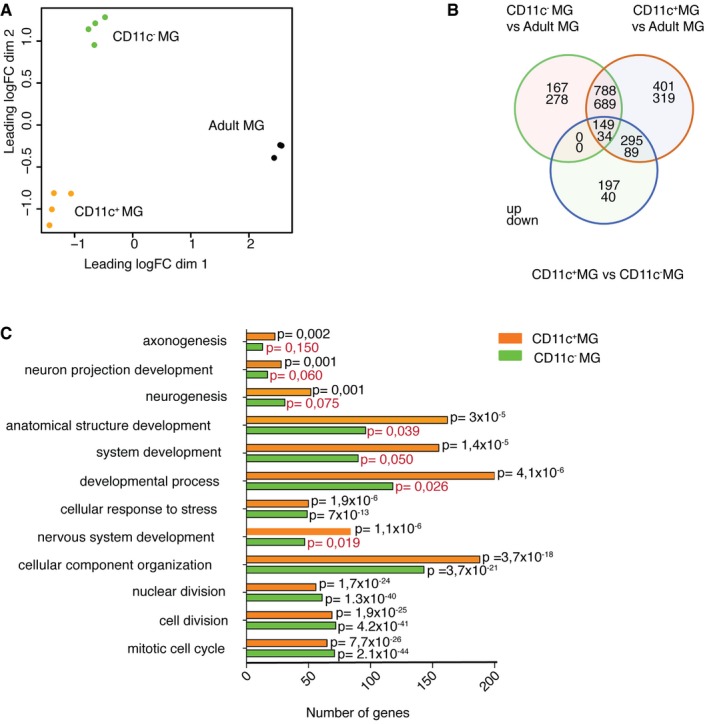
- Multidimensional scaling shows that neonatal subpopulations of microglia and adult microglia have distinct transcriptional profiles. Colors indicate three different groups of samples: orange represents neonatal CD11c+ microglia (n = 4), green neonatal CD11c− microglia (n = 4), and black adult microglia (n = 3). Each n represents a pool of 10–15 mice from four individual cell sorts.
- Venn diagram showing differentially expressed genes by neonatal CD11c+ microglia, neonatal CD11c− microglia, and adult microglia. Numbers of genes differentially expressed comparing neonatal CD11c− versus adult microglia, neonatal CD11c+ versus adult microglia, and neonatal CD11c+ versus neonatal CD11c− microglia are indicated.
- GO enrichment analysis of upregulated genes (logFC ≥ 1.5; FDR ≤ 0.01) in neonatal CD11c+ microglia (orange bars) and CD11c− microglia (green bars) versus adult microglia identified significant (P‐values ≤ 0.01 after Benjamini correction) enrichment for 55 GOTERM_BP_ALL biological processes categories, 12 of which are shown on the graph. Benjamini‐corrected P‐values are indicated on the bar graph; P ≥ 0.01 are marked with red font.
GO enrichment analysis on genes differentially expressed (logFC ≥ 1,5; FDR ≤ 0.01) in neonatal CD11c+ and CD11c− microglia compared to adult microglia revealed significant enrichment (P ≤ 0.01 after Benjamini correction) for 55 GOTERM_BP_ALL biological processes categories. Among those, some were associated with “cell cycle”, “mitosis”, and “cell division”, suggesting that both neonatal microglia populations were in cell cycle or proliferating. Categories related to developmental processes including “nervous system development”, “neurogenesis”, “axonogenesis”, and “neuron projection development” were significantly enriched in neonatal CD11c+ microglia, but not in the CD11c− population (Fig 4C).
Neonatal microglia express neuroectodermal genes
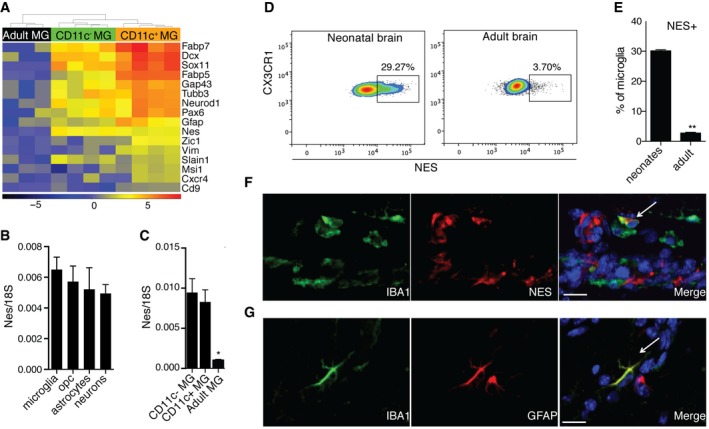
Neonatal microglia expressed neuroectodermal markers such as Nes, Gfap, Pax6, Fabp5, Fabp7, Gap43, Vim, Dcx, Sox11, and NeuroD1 (Fig 5A). Nestin gene expression was compared in neonatal microglia, OPCs, astrocytes, and neurons from PN4‐7 brains. All the cell populations expressed equal levels of nestin transcripts (Fig 5B). Nestin transcripts were also compared between neonatal CD11c− and CD11c+ microglia, and adult microglia. While both neonatal microglia subsets expressed similar levels of nestin message, significantly less nestin transcripts were detected in adult microglia (Fig 5C). Consistent with these observations, flow cytometry analysis showed that around 30% of neonatal but only 3.7% of adult microglia expressed nestin (Fig 5D). Immunostaining revealed co‐localization of nestin (Fig 5E) and GFAP (Fig 5F) with IBA1‐positive cells in the neonatal brain.
Figure 5. Neuroectodermal marker expression in neonatal microglia.
-
AA heatmap showing neuroectodermal gene expression in neonatal versus adult microglia. Scale represents log2 fold change normalized CPM expression values.
-
B, CExpression of nestin in MACS‐sorted microglia (n = 4), OPC (n = 4), astrocytes (n = 4), and neurons (n = 3) from brains of PN3‐7 mice (B) as well as FACS‐sorted CD11c+ microglia (n = 5), CD11c− microglia (n = 5) from brains of PN4‐7 mice, and total microglia from adult brain (C).
-
D, ERepresentative flow cytometry profiles (D) and bar graph (E) showing nestin expression in microglia from brains of PN5 mice (n = 5) and adult mice (n = 6).
-
F, GConfocal microscopic analysis showing co‐localization of nestin (red) (F) or GFAP (red) (G) with IBA1 (green) (n = 3). Arrows point to IBA1, NES (F) and IBA1, GFAP (G) double positive cells. Scale bars = 15 μm.
Neonatal CD11c+ microglia are a major source of neurogenic signals
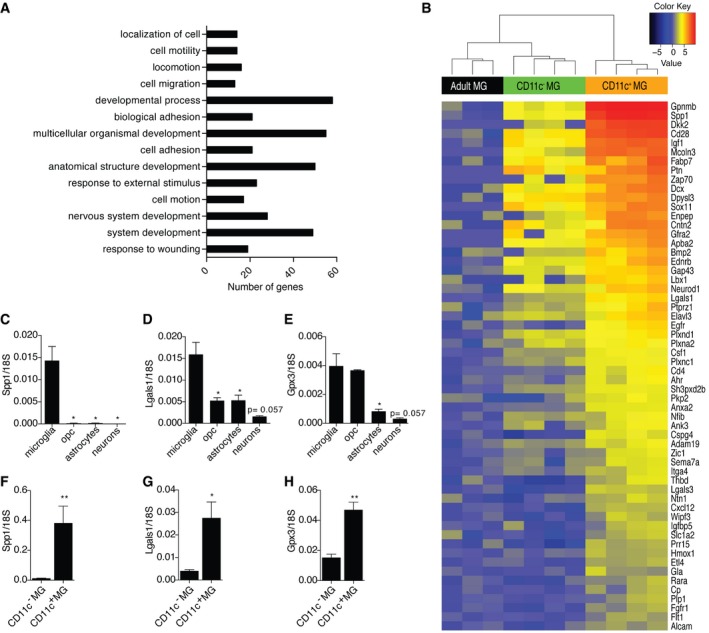
To further understand the difference between neonatal CD11c+ and CD11c− microglia populations, GO enrichment analysis was performed on genes that were significantly more abundant (logFC ≥ 1,5; FDR ≤ 0.01) in neonatal CD11c+ microglia compared to both neonatal CD11c− microglia and adult microglia. 251 genes fulfilled these criteria and showed significant enrichment (P ≤ 0.01 after Benjamini correction) for 14 GOTERM_BP_ALL biological processes categories. Interestingly, nearly 25% of the genes were annotated to the ontology term “developmental process” and “multicellular organismal development”, 20% to anatomical “structure development” and “system development”, 12% to “nervous system development”, and < 10% to terms such as “cell motion”, “cell migration”, “cell adhesion”, “response to external stimulus”, and “response to wounding” (Fig 6A). Similar analysis for neonatal CD11c− microglia showed only 14 genes that fulfilled set criteria and the only significant enrichment was found for “immune effector process” category.
Figure 6. Neonatal CD11c+ microglia are a major source of neurogenic signals.
-
AOntological analysis of upregulated genes (logFC ≥ 1.5; FDR ≤ 0.01) in neonatal CD11c+ microglia in comparison with both neonatal CD11c− microglia and adult microglia identified significant (P ≤ 0.01 after Benjamini correction) enrichment for 14 GOTERM_BP_ALL biological processes categories.
-
BA heatmap showing upregulated genes involved in system development in neonatal CD11c+ microglia versus neonatal CD11c− and adult microglia. Scale represents log2 fold change normalized CPM expression values.
-
C–HExpression of Spp1, Lgals1, and Gpx3 relative to 18S rRNA in MACS‐sorted microglia (n = 4), OPC (n = 4), astrocytes (n = 4), and neurons (n = 3) (C, D, E) as well as FACS‐sorted CD11c+ (n = 5) and CD11c− microglia (n = 5) (F, G, H) from brains of PN4‐7 mice.
Importantly, oligodendrocyte supportive genes such as Spp1 and Igf1 were most abundant in CD11c+ microglia. They significantly expressed Bmp2, a gene for a protein that promotes astrocyte differentiation, Csf1 that induces microglial proliferation as well as genes involved in axonal guidance such as Cxcl12, Ntn1, Sema7a, and Plxna2. Moreover, CD11c+ microglia expressed Gpx3 and Ptn that promote neurite outgrowth, Lgals1 promoting axonal growth, neuroprotective Gpnmb, Adam19 known to shed neuregulin isoforms (Fig 6B), and tissue remodeling metalloproteinases: Mmp12, Mmp19, and Mmp24 (not shown).
Gene expression levels of selected gene candidates encoding secreted proteins involved in neurodevelopment, including Csf1, Spp1, Cxcl12, Gpx3, Ntn1, and Lgals1, were compared in neonatal microglia, OPCs, astrocytes, and neurons, sorted from PN4‐7 brains. These genes were expressed in all of the sorted cell populations. Microglia were the major source of Spp1 and Lgals1 (Fig 6C and D), and they also expressed significantly higher levels of Gpx3 than astrocytes and neurons (Fig 6E). Cxcl12 was equally expressed by all of investigated cell populations. Ntn1 was expressed predominantly by OPCs and astrocytes and Csf1 mainly by neurons, OPCs, and astrocytes (data not shown). Spp1, Lgals1, and Gpx3 transcripts were further compared in CD11c+ and CD11c− microglial populations. CD11c+ microglia expressed much higher levels of Spp1 (30‐fold), Lgals1 (sevenfold), and Gpx3 (threefold) than their CD11c− counterparts (Fig 6F–H).
CD11c‐targeted toxin regimens resulted in Itgax upregulation in neonatal brain
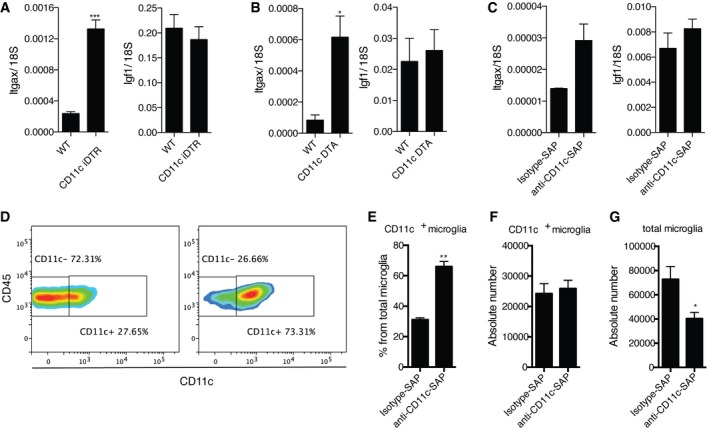
Having shown that CD11c+ microglia are a major source of many neurodevelopmentally important genes as well as being a critical source of IGF1 for myelinogenesis, we then asked the functional consequence of removal of these cells for developing brain. CD11c‐iDTR mice that express diphtheria toxin receptor under control of a CD11c/Itgax promoter and WT littermates were injected with diphtheria toxin (DT) at days PN1 and PN3. Flow cytometry analysis, 48 h after the last DT injection, showed no difference in the percentage of CD11c+ microglia compared to WT littermates (data not shown). Notably however, Itgax expression was significantly elevated in the brains of these mice, while Igf1 expression did not change (Fig 7A). Similar findings were obtained in PN7 CD11c‐DTA mice that express diphtheria toxin‐A subunit under a CD11c/Itgax promoter—Itgax expression was significantly upregulated and Igf1 expression was not altered (Fig 7B) in transgenic mice compared to WT littermates. In a separate approach, we injected saporin toxin conjugated to anti‐CD11c antibodies into the corpus callosum and cerebellum of PN3 mice. Similar to the two other depletion strategies, we observed upregulation of Itgax and unchanged Igf1 (Fig 7C) expression in comparison with isotype control‐treated mice. Interestingly, we also observed a dramatic increase in CD11c+ microglia as a percentage of total microglia (Fig 7D and E) with unchanged absolute numbers (Fig 7F). A concomitant decrease in total microglia (Fig 7G) may be explained by CD11c− microglia having also expressed itgax and therefore becoming CD11c+ in response to this stimulus. Taken together, the similarity in response of CD11c+ microglia to three distinct toxin regimens underlines their unique status in the developing CNS.
Figure 7. Effect of CD11c‐directed depletion strategies.
-
A–CExpression of Itgax and Igf1 in PN5 brains of CD11c iDTR mice (n = 6) and WT littermates (n = 3) treated with DTx at days 1 and 3 after birth (A); PN7 brains of CD11c DTA mice (n = 5) and WT littermates (n = 3) (B); PN5 WT brains I.C. injected with saporin toxin conjugated with anti‐CD11c (n = 6) or isotype control antibody (n = 5) (C).
-
D–GRepresentative flow cytometry profiles (D) and flow cytometry analysis showing percentage from total microglia (E) and absolute numbers of CD11c+ microglia (F) as well as absolute numbers of total microglia (G) in PN5 WT brains I.C. injected with saporin toxin conjugated with anti‐CD11c (n = 6) or isotype control antibody (n = 5).
Clusters of repopulating microglia in adulthood contain CD11c+ microglia
In contrast to developing CNS, where depletion of microglia has never exceeded 50% (Ueno et al, 2013), near‐total ablation has been shown for adult mice (Bruttger et al, 2015). Recently, some of us demonstrated that 7 days after microglial ablation from adult CNS, using a CX3CR1CreER iDTR model, micro‐clusters containing highly proliferating microglia were present throughout the CNS (Bruttger et al, 2015). We further analyzed these repopulating cells by RNA‐seq and showed overexpression of Itgax (CD11c) and other genes associated with CD11c+ microglia such as Igf1, Spp1, and Gpnmb, in comparison with microglia from naïve adult mice (Fig 8A). Moreover, some clusters of repopulating microglia contained CD11c+ cells (Fig 8B). These cells were also observed outside the clusters, likely migrating to and colonizing the tissue (Fig 8C). Interestingly, virtually all microglia in the clusters were nestin‐positive (not shown). Next, we determined the overlap between the WGCNA modules and up‐ and downregulated genes of the repopulating microglia. Genes that were upregulated in repopulating microglia, in comparison with control microglia, showed significant overlap for red (“translation”) and blue modules (“immune system processes”), while downregulated genes for the blue module (Fig 8D). These data suggest that after ablation, repopulating adult microglia, similar to neonates, express nestin and CD11c but do not show a neurogenic gene expression profile. Thus, the novel neuro‐ and myelinogenic CD11c+ microglial phenotype that we have described is unique to neonatal CNS.
Figure 8. Clusters of repopulating microglia in adult mice contain CD11c+ microglia.
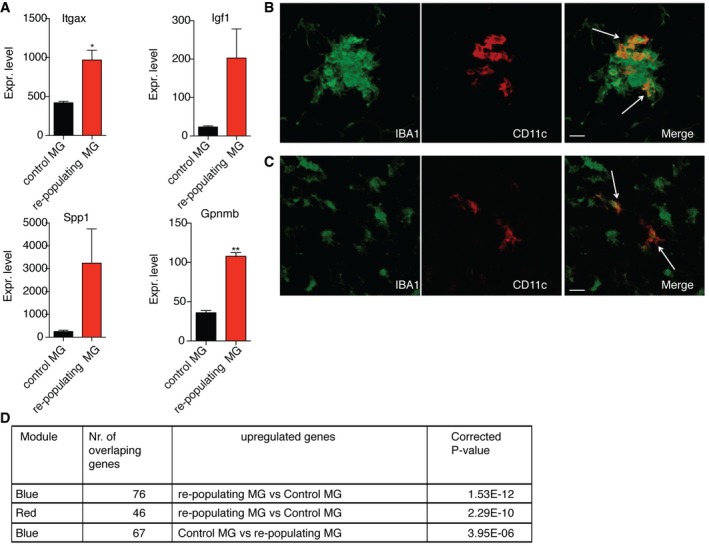
-
AGene expression values of Itgax, Igf1, Spp1, and Gpnmb in control microglia (n = 2) and repopulating microglia (n = 3) obtained from RNA‐seq analysis. Each n represents a pool of two individual mice. P‐values were determined by Student's t‐test. ns, not significant; *P < 0.05; **P < 0.01.
-
B, CConfocal microscopic analysis showing co‐localization of CD11c (red) and IBA1 (green) within (B) and outside the repopulating microglia cluster (C) in brain stem of adult CX3CR1CreER iDTR mouse 7 days after DTx treatment (n = 2). Arrows point to IBA1, CD11c double positive cells. Scale bars = 15 μm.
-
DTable showing significant correlation of the up‐ and downregulated genes from repopulating microglia with two modules (red and blue) from Fig 3.
Discussion
We have identified a unique, myelinogenic phenotype of neonatal microglia. They are distinct from adult, EAE, and repopulating microglia. We have shown that a CD11c+ microglial subset significantly increases in early postnatal brain and then dramatically contracts as animals mature to adulthood. These microglia are a major cellular source of Igf1, Spp1, Lgals1, Gpx3, and other genes associated with neurogenic and myelinogenic processes in the neonatal brain, which is supported by effects of selective Igf1 ablation from CD11c+ microglia on myelin gene expression and myelination in 3‐week‐old mice. These findings identify neonatal microglia, and especially the CD11c+ subset, as key players in neurodevelopment.
Microglia include functionally distinct subsets that can be distinguished by expression of the integrin CD11c, also known as complement receptor‐4 (CR4). CD11c‐expressing microglia have been found in several neuroinflammatory and neurodegenerative conditions possibly responding to CNS damage (Butovsky et al, 2006; Remington et al, 2007). We have previously shown that this subset of microglia expands in cuprizone‐demyelinated corpus callosum (Remington et al, 2007; Wlodarczyk et al, 2015), in EAE (Wlodarczyk et al, 2014), and in neuromyelitis optica (NMO)‐like pathology (Wlodarczyk et al, 2015). Now we show that the great majority of CD11c+ cells in the neonatal brain are microglia. Similar to microglia populations from EAE (Wlodarczyk et al, 2014, 2015), they express IBA1, CX3CR1 and, in contrast to blood‐derived cells, do not express CCR2. Microglia have been recently shown to undergo temporal developmental stages characterized by different gene expression profiles (Matcovitch‐Natan et al, 2016) and to mature in the second postnatal week (Bennett et al, 2016). Our data, in agreement with these studies, show that the gene expression profile of neonatal microglia significantly differed from adult homeostatic microglia and in addition from that seen during EAE, showing myelin‐ and neurogenic in neonates versus an immune response signature in EAE. In line with the fact that neonatal mice have been shown to be resistant or to have delayed onset of EAE in comparison with adult mice (Smith et al, 1999; Cravens et al, 2013), our findings suggest that the neurosupportive capacity of microglia decreases with aging.
A role for microglia in neurodevelopment is increasingly accepted. Microglia have been reported to prune or sculpt developing synaptic connections, and a role for complement has been proposed (Stevens et al, 2007; Schafer et al, 2012). Microglia are defined by expression of CR3 (CD11b) and we show important anti‐inflammatory and myelinogenic roles for CR4/CD11c‐expressing microglia. We have not examined the role of complement, but instead show dramatic and provocative correlation between expression of this receptor with myelinogenic capabilities, about which very little has been reported. We demonstrate that CD11c+ microglia expressed the majority of myelinogenic Igf1 in the developing brain. IGF1 is necessary for neurodevelopment both in humans and in mice. Rare cases of IGF1 deficiency in humans are characterized by growth alteration, microcephaly, sensorineural deafness, and delayed psychomotor development (reviewed in Netchine et al, 2011). Deficiency of Igf1 and its receptor in mice is usually postnatally lethal and those mice that survive to adulthood show microcephaly associated with increased neuronal death and myelination impairment (Beck et al, 1995). Expression of myelin‐specific proteins as well as oligodendrocyte numbers in the developing corpus callosum is significantly reduced in Igf1‐deficient mice (Ye et al, 2002). In contrast, mice with transgenic overexpression of Igf1 have larger brains, increased populations of neurons and oligodendrocytes as well as enhanced myelin production (reviewed in de la Monte & Wands, 2005). Neurons, OPCs, and microglia have all been reported to express Igf1 (Liu et al, 1994; Mueller et al, 2013; Suh et al, 2013; Ueno et al, 2013) but their relative importance in postnatal development has not been defined so far. Microglial Igf1 was recently shown to support layer V cortical neuron survival during postnatal development (Ueno et al, 2013). We have now shown that even partial depletion of Igf1 in CD11c+ microglia leads to reduction in brain weight, decrease in Plp, Mbp, Mag, and Mog expression in brain, and is associated with higher frequency of less myelinated fibers in corpus callosum. We showed that highly myelinating areas such as the developing corpus callosum and cerebellum are colonized specifically by CD11c+ microglia, which further supports that through their high Igf1 expression these cells influence myelin formation within these structures. Thus, as the major source of Igf1, the CD11c+ subset of microglia plays a critical role in primary myelination and neuronal support in the neonatal CNS.
We show that neonatal microglia express many genes that are important for brain formation. Although genes associated with developmental processes were expressed in both subsets of neonatal microglia, this was much more pronounced in the CD11c+ population. CD11c+ microglia may contribute to shaping axonal organization in the CNS through expression of well‐described axonal guidance signals such as Cxcl12, Ntn1, Plxna2, and Sema7a. They may stimulate neuronal and neurite outgrowth and astrocytogenesis through expression of Ptn (Garcia‐Gutierrez et al, 2014), Gpx3 (Buchser et al, 2012), and Bmp2 (Nakashima et al, 1999), respectively, and affect remodeling processes in the developing brain tissue by expression of metalloproteinases Mmp12, Mmp19, and Mmp24. CD11c+ microglia are therefore seen to be involved in promoting and directing organization of the developing CNS.
Microglia colonize the neuroepithelium very early during neurodevelopment (Ginhoux et al, 2010) and are autonomously maintained through proliferation (Ajami et al, 2007). Both CD11c+ and CD11c− populations of microglia proliferated equivalently in response to neuroinflammation (Wlodarczyk et al, 2015). The mechanism by which the number of CD11c+ microglia is regulated during brain maturation is obviously of key importance. Neonatal CD11c+ microglia are themselves a source of Csf1, which suggests that these cells may represent a self‐renewing population, since microglial development and maintenance is highly dependent on CSF1R signaling (Elmore et al, 2014), and mice deficient in CSF1R almost completely lack microglia (Ginhoux et al, 2010). Neonatal microglia upregulate genes involved in cell division and their number increases rapidly during the first postnatal days. Our findings show that CD11c+ microglia survive and show a unique response to antibody‐directed as well as cell‐intrinsic and cell‐extrinsic transgenic toxicity, and indirectly suggest that besides proliferation, they may also be reconstituted from the CD11c− microglia population.
Interestingly, both of the neonatal microglia populations that we studied, and therefore all neonatal microglia, express nestin at a similar level as other brain cell populations such as OPCs, astrocytes, and neurons. Nestin expression by neonatal microglia was transient and significantly decreased in adulthood. Microglia have been shown to express Nes and Vim also in naïve adult rat brain (Takamori et al, 2009), and nestin‐expressing microglia have been shown to act as multipotential stem cells that give rise to neurons, astrocytes, or oligodendrocytes in vitro (Yokoyama et al, 2004). Recently, nestin expression has been demonstrated in repopulating adult microglia after their genetic ablation (Bruttger et al, 2015) and in inflammatory‐activated microglia (Krishnasamy et al, 2017), pointing to nestin‐positive microglia progenitors with high proliferative potential (Elmore et al, 2014). Here, we show that the clusters of such repopulating microglia are rich in CD11c+ Nes+ microglia. However, they do not show a neonatal‐like gene signature, identifying the neonatal neurogenic phenotype of microglia as unique and transient.
The CD11c+ microglial response is not restricted to neurodevelopment. Despite their immune response gene signature and being less myelinogenic/neurogenic than neonatal microglia, evidence suggests that they may be involved in regenerative processes in adulthood. CD11c+ microglia express Igf1 (Wlodarczyk et al, 2015) and Lgals1 during EAE in adult animals, although this expression is significantly lower than in neonatal CD11c+ microglia (Wlodarczyk, unpublished observations). Galectin‐1 encoded by Lgals1 promotes neurite outgrowth and has been reported to promote axonal regeneration (Horie et al, 1999; McGraw et al, 2004; Quinta et al, 2014). It also suppresses microglial activation (Starossom et al, 2012), promotes apoptosis of activated T cells, and has immunoregulatory properties in models of autoimmune diseases (Camby et al, 2006) including amelioration of EAE (Starossom et al, 2012). In the adult brain, only a small pool of CD11c+ microglia remains, being available when needed for immunoregulation, regeneration, and remyelination, for instance in response to brain damage. Increase in this subset in EAE as well as in cuprizone‐mediated acute demyelination and NMO‐like pathology (Remington et al, 2007; Wlodarczyk et al, 2014, 2015) argues for stimulus‐dependent response. Neonatal population dynamics were recapitulated in the adult brain, where the expanded population of cuprizone‐induced CD11c+ cells contracted dramatically upon withdrawal of the demyelinating stimulus (Remington et al, 2007). This suggests a more general response to loss of homeostasis, which will be further studied.
Taken together, we identify neonatal CD11c+ microglia as a potent and inducible source of developmental proteins for neurogenesis, and myelinogenesis in the developing CNS.
Materials and Methods
Mice
C57BL/6j bom female mice aged 7–8 weeks were obtained from Taconic Europe A/S; CX3CR1GFP/GFP and CCR2RFP/RFP, CD11cCre‐GFP (Stranges et al, 2007) and Igf1fl/fl were obtained from The Jackson Laboratory and maintained as a breeding colony (CX3CR1GFP/GFP and CCR2RFP/RFP were crossed with B6 mice to obtain heterozygotes) in the Biomedical Laboratory, University of Southern Denmark (Odense). All experiments performed in Denmark were approved by the Danish Animal Experiments Inspectorate (approval number 2014‐15‐0201‐00369). CX3CR1CreER, CD11cCre, DTA, and iDTR mice were housed in specific pathogen‐free conditions and used in accordance with the guidelines of the Central Animal Facility Institution (ZVTE, University of Mainz). Neonatal mice (p2‐p21) used for experiments were of mixed sex.
EAE model
Seven‐ to ten‐week‐old female mice were immunized by injecting subcutaneously 100 μl of an emulsion containing 300 μg of myelin oligodendrocyte glycoprotein (MOG)p35–55 (TAG Copenhagen A/S, Frederiksberg, Denmark) in incomplete Freund's adjuvant (DIFCO, Albertslund, Denmark) supplemented with 400 μg H37Ra Mycobacterium tuberculosis (DIFCO). Bordetella pertussis toxin (300 ng; Sigma Aldrich, Brøndby, Denmark) in 200 μl of PBS was injected intraperitoneally at day 0 and day 2. Animals were monitored daily from day 5 and scored on a 7‐point scale as follows: 0, no symptoms; 1, partial loss of tail tonus; 2, complete loss of tail tonus; 3, difficulty walking; 4, paresis in both hind legs; 5, paralysis in both hind legs; and 6, front limb weakness. Due to ethical consideration, mice were sacrificed when they reached grade 6 or 24 h after hind legs paralysis.
Microglia depletion strategies
For CD11c+ microglia depletion, CD11c‐iDTR mice and WT littermates were injected subcutaneously with 25 ng/g body weight of DT 1 and 3 days after birth and were sacrificed 5 days after birth.
C57BL/6j bom PN3 pups were randomized into two groups and injected intracerebrally into two areas of the brain (coordinates: 2.2 mm anterior, 1.0 mm ventral and 1.0 posterior, 1.2 ventral) with 1.3 μg of biotinylated anti‐CD11c antibody (N418; eBioscience) or biotinylated anti‐hamster IgG (Jackson ImmunoResearch) conjugated to streptavidin‐saporin (ATS) prepared according to the manufacturer's protocol. Pups were sacrificed 2 days after injection.
For microglia depletion in CX3CR1CreER iDTR mice, first 2 mg tamoxifen was administered subcutaneously (s.c.) twice on postnatal days 12 and 14, and then at age of 8 weeks, mice were injected intraperitoneally (i.p.) with 500 ng diphtheria toxin (DT; Merck Millipore) three times, with a 1‐day interval between each injection (Bruttger et al, 2015).
Magnetic‐activated cell sorting (MACS)
To isolate cells from brain PN3‐7, mice were sacrificed, brain tissue was collected, and single cell suspensions were generated using Neural Tissue Dissociation Kit (P) (Miltenyi Biotec) for OPC, CD11b+ cells (microglia), and astrocyte isolation, or Neural Tissue Dissociation Kit ‐Postnatal Neurons (Miltenyi Biotec) for neuron isolation. Cells were isolated by magnetic separation using CD140a (PDGFRα) MicroBead Kit (Miltenyi Biotec), CD11b (Microglia) MicroBeads (Miltenyi Biotec), anti‐ACSA‐2 MicroBead Kit (Miltenyi Biotec), or Neuron Isolation Kit (Miltenyi Biotec). All steps were done according to the manufacturer's protocols.
Fluorescence‐activated cell sorting (FACS) and flow cytometry
To isolate microglia from brain, PN2‐PN5, PN7, PN28, and 8‐ to 12‐week‐old mice were anaesthetized with 200 mg/kg of pentobarbital and intracardially perfused with ice‐cold PBS. Brain tissue was collected and a single cell suspension was generated by forcing through a 70‐mm cell strainer (BD Biosciences). Mononuclear cells were collected after centrifugation on 37% Percoll (GE Healthcare Biosciences AB). They were first incubated with anti‐Fc receptor (Clone 2.4G2; 1 mg/ml; BD Pharmingen) and Syrian hamster IgG (50 mg/ml; Jackson Immuno Research Laboratories Inc.) in PBS, 2% fetal bovine serum (FBS), then with anti‐CD45 (Clone 30‐F11; Biolegend), anti‐CD11b (Clone M1/70; Biolegend), and biotin‐conjugated anti‐CD11c (Clone HL3; BD Pharmingen) antibodies in PBS, 1% FBS, and finally with streptavidin‐APC (Biolegend). Cell populations were gated based on isotype‐matched control antibodies as CD45dim CD11b+ CD11c− (CD11c− microglia), CD45dim CD11b+ CD11c+ (CD11c+ microglia) and sorted on a FACSAria™ III cell sorter (BD Biosciences) or data were collected on an LSRII™ flow cytometer (BD Biosciences) and analyzed using FACSDiva™ software version 6.1.2 (BD Biosciences). For the analysis of nestin expression after extracellular staining (as above), cells were fixed and permeabilized using BD Cytofix/Cytoperm™ according to the manufacturer's protocol and stained with anti‐nestin and corresponding isotype control (Clone 307501; R&D Systems).
RNA extraction, quantitative RT–PCR (RT–qPCR)
FACS‐sorted CD11c+ and CD11c− microglia were placed in RLT buffer (Qiagen) and total RNA was extracted using RNeasy columns as per the manufacturer's protocol (Qiagen). MACS‐sorted neurons, OPCs, CD11b+ cells, and astrocytes were placed in QIAzol Lysis Reagent and total RNA was extracted using miRNeasy Micro Kit according to the manufacturer's protocol (Qiagen). Reverse transcription was performed with M‐MLV reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Quantitative real‐time PCR (qPCR) was performed with 1 μl cDNA in a 25 μl reaction volume containing Maxima® Probe/ROX qPCR Master mix (Fermentas), TaqMan® PreAmp Master Mix Kits for Cxcl12: Mm00445553_m1, Ntn1: Mm00500896_m1, Lgals1: Mm00839408_g1, Gpx3: Mm00492427_m and Nes: Mm00450205_m1, Spp1: Mm00436767_m1, Mog: Mm00447824_m1, Plp: Mm01297210_m1, Mbp: Mm01266402_m1, Mag: Mm00487538_m1. For Igf1, Maxima® SYBR Green/ROX qPCR Master Mix (2X) Probe/ROX qPCR Master Mix (Fermentas) with forward and reverse primers (800 nM; from ATGC) were used. Igf1 primer sequences were as follows: For: CCG AGG GGC TTT TAC TTC AAC AA; Rev: CGG AAG CAA CAC TCA TCC ACA A. PCR were done on an ABI Prism 7300 Sequence Detection System (Applied Biosystems). Results were expressed relative to 18S rRNA (2ΔCT method) as endogenous control (TaqMan® Ribosomal RNA control Reagents kit; Applied Biosystems). cDNA was diluted 1/500 for 18S rRNA analysis.
Genomic PCR
DNA was extracted by DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's protocol, and quantitated using NanoDrop (ThermoFisher). To detect Cre‐induced recombination, PCR on 90 ng DNA was performed as described previously (Liu et al, 1998) using ID‐3: CACTAAGGAGTCTGTATTTGGACC; ES‐1: AGCCTCTCAACTAAGACAATA primers.
RNA sequencing
RNA quality check was done using an Agilent bioanalyzer, and only high‐quality samples were used (RIN > 7) for sequencing. Sequence libraries were prepared using the Illumina Truseq RNA sample preparation kit. RNA sequencing was performed at the Genome Analysis Facility of the University Medical Center Groningen with an Illumina Hiseq 2500. Single‐read 50‐bp sequencing was done, aligned to the ensemble reference genome using the Star 2.3.1 l aligner (Dobin et al, 2013), allowing for two mismatches. Samtools 0.1.19 was used to sort the aligned reads (Roussos et al, 2012). Quantification was done using HT‐seq count 0.5.4 (Anders et al, 2015). Between 6 and 11 million uniquely aligned reads per sample were generated. Count data were loaded in R and analyzed with the EdgeR BioConductor Package (Robinson et al, 2010). Differential gene expression was done between groups of interest (Dataset EV1). Heatmaps were generated with heatmap2 function of package gplots. Functional annotation was done using DAVID (Huang et al, 2009).
Detailed procedures for RNA‐seq of repopulating microglia including data analysis are described in Bruttger et al (2015).
Weighted gene co‐expression network analysis (WGCNA)
WGCNA (Langfelder & Horvath, 2008) was applied to the count per million (CPM) expression data. A co‐expression network was constructed with a beta value of 20. Genes were clustered into branches of highly expressed genes, and modules were identified with the tree cut algorithm with the additional PAM stage. Six binary variables were generated that were used to calculate the module trait relationships in which all groups were set to zero with the exception of particular groups of interest: control (1's for microglia obtained from healthy control brain), CD11c (1's for both EAE CD11c and neonatal CD11c), EAE (1's for CD11c negative and microglia obtained from EAE brains), neonatal (1's for CD11c negative and microglia obtained from neonatal brains), CD11c EAE, and CD11c neonatal. In addition, a between‐group Kruskal–Wallis test was applied to the Module Eigengene values, to detect overall differential expression. Modules were functionally annotated using DAVID (Huang et al, 2009).
Histology
Seventy‐micrometer or twelve‐micrometer sections from 4% PFA‐fixed, frozen brains, or spinal cords of perfused mice were cut on a cryostat and stored in de Olmos cryoprotectant solution (Khorooshi & Owens, 2013) at −14°C or in −20°C on Superfrost Plus slides (Thermo Scientific), respectively. Sections were washed in PBS and incubated for 10 min in ice‐cold acetone or 30 min with 10% methanol, 10% H2O2 in PBS to block endogenous peroxidase. After repeated rinses with 0.2% Triton X‐100 in PBS (PBST), they were incubated for 1 h in 3% BSA in PBS to block unspecific binding. Next, sections were incubated overnight at 4°C with corresponding primary antibodies: rabbit anti‐IBA1 (Cat. No. 019‐19741 Wako), rat anti‐nestin antibody (7A3; Abcam), hamster anti‐CD11c (N418; Bio‐Rad), anti‐myelin PLP (Cat. No ab105784; Abcam), and rabbit anti‐laminin (Cat. No. CL54851AP; Cedarlane). Following primary antibody incubation, the sections were washed with PBST and incubated for 1 h with the appropriate secondary antibody: goat anti‐Armenian hamster IgG‐HRP (Cat. No sc‐2904; Santa Cruz Biotechnology), donkey anti‐rabbit 488 (Cat. No A21206; Invitrogen), donkey anti‐rat Alexa 594 (Cat. No A21209; Invitrogen), and goat anti‐rabbit Alexa 647 (Cat. No A21245; Invitrogen). Following secondary antibody incubation, the sections were washed with PBS and incubated for 4 min with TSA™ Plus Cy3 System (PerkinElmer).
After immunofluorescent labeling, nuclei were visualized by DAPI staining and the sections were mounted with Fluorescence Mounting Medium (DAKO). Visualization of GFP was done as previously described (Khorooshi et al, 2015). The sections were visualized on an Olympus FV1000MPE confocal microscope and analyzed by FV10‐ASW 4.06 software.
Electron microscopy
Animals were sacrificed and transcardially perfused with PBS followed by a fixative containing 4% paraformaldehyde and 1.5% glutaraldehyde. After postfixation of the brains in the same fixative, sagittal sections were acquired using a vibrating microtome (Leica Microsystems, Wetzlar, Germany) with a thickness of 75 μm. The sections were stained with 0.5% osmium tetroxide in PBS for 30 min and subsequently rinsed in PBS followed by dehydration with 30, 50 and 70% ethanol. Afterward, the sections were treated with 1% uranyl acetate in 70% ethanol for 1 h, before the tissue was further dehydrated by using 80, 90, 96, 100% ethanol and finally propylene oxide. The sections were transferred in Durcupan (Sigma Aldrich, Steinheim, Germany) and embedded in between coated microscope slides and cover slips before the polymerization process at 56°C for 48 h. Regions of interest were located by light microscopy, marked, and transferred on blocks of resin for a second polymerization step. Finally, the embedded tissue was cut into semi‐thin sections and areas for ultrastructural analysis were cut into 55‐nm ultra‐thin sections using an ultra‐microtome (Leica Microsystems, Wetzlar, Germany) and transferred on Formvar‐coated grids and stained with lead citrate for 6 min. The analysis was performed using Zeiss SIGMA electron microscope (Zeiss NTS, Oberkochen, Germany).
Four different regions of the corpus callosum from each sample were analyzed. G‐ratios of transversally sectioned axons were calculated using ImageJ software and compared between groups. The analysis was performed by an investigator who was blinded to the experiment.
Statistical analysis
No statistical methods were used to predetermine sample sizes, and exact group numbers were determined by animal availability. We ensured that the sample sizes were similar to those generally employed in the field. For each EAE experiment, at least 16 mice were immunized (av. 50% of mice show EAE symptoms required to enter the experiment). In order to collect sufficient amount of RNA for RNA‐seq analysis, each sample consisted of 10–15 pooled mice.
To obtain unbiased data, analysis of the samples was performed blinded to the genotype. Only after finalization of all quantitative measurements, the samples genotypes were decoded.
All experiments were repeated at least twice, and data are presented as means ± SEM. Statistical significance was assessed using the two‐tailed Mann–Whitney U‐test (GraphPad Prism 6) unless specified otherwise. Data met assumptions for the used tests. Normal distribution was assessed by Shapiro–Wilk normality test. P‐values < 0.05 were considered significant.
Data availability
The datasets of RNA‐seq from adult, EAE, and neonatal microglia are deposited in Gene Expression Omnibus (GEO): GSE78809, and will become available via the Glia Open Access Database (Holtman et al, 2015): www.goad.education. The datasets of RNA‐seq of repopulating microglia are described in Bruttger et al (2015) and deposited in GEO: GSE68376.
Author contributions
AWl, TO, AWa, and BJLE designed the work. AWl, AB‐B, IK, RK, NY, NAM, JJB‐B, JB, KK, and MK performed the experiments. AWl, IRH, MK, EWGMB, BJLE, and JB analyzed the data. AWl and TO wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Review Process File
Source Data for Figure 2
Acknowledgements
This work was supported by grants from Lundbeckfonden, Danish Council for Independent Research, Danish MS Society, and Warwara Larsens Fond. We thank Dina Arengoth, Pia Nyborg Nielsen, Kirstine Nolling Jensen, and Nieske Brouwer for expert technical assistance and Inger Andersen and Lars Vitved for help with cell sorting. The bioimaging experiments reported in this paper were performed at DaMBIC, a bioimaging research core facility, at the University of Southern Denmark. DaMBIC was established by an equipment grant from the Danish Agency for Science Technology and Innovation and by internal funding from the University of Southern Denmark.
See also: ML Bennett & BA Barres (November 2017)
References
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM (2007) Local self‐renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 10: 1538–1543 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew K, Li K, Olmos‐Alonso A, Garcia‐Moreno F, Liang Y, Richardson P, Tipton T, Chapman MA, Riecken K, Beccari S, Sierra A, Molnar Z, Cragg MS, Garaschuk O, Perry VH, Gomez‐Nicola D (2017) Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep 18: 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck KD, Powell‐Braxton L, Widmer HR, Valverde J, Hefti F (1995) Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin‐containing neurons. Neuron 14: 717–730 [DOI] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG, Barres BA (2016) New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA 113: E1738–E1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biber K, Owens T, Boddeke E (2014) What is microglia neurotoxicity (Not)? Glia 62: 841–854 [DOI] [PubMed] [Google Scholar]
- Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, Mack M, Pinteaux E, Muller W, Zipp F, Binder H, Bopp T, Prinz M, Jung S, Waisman A (2015) Genetic cell ablation reveals clusters of local self‐renewing microglia in the mammalian central nervous system. Immunity 43: 92–106 [DOI] [PubMed] [Google Scholar]
- Buchser WJ, Smith RP, Pardinas JR, Haddox CL, Hutson T, Moon L, Hoffman SR, Bixby JL, Lemmon VP (2012) Peripheral nervous system genes expressed in central neurons induce growth on inhibitory substrates. PLoS ONE 7: e38101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Koronyo‐Hamaoui M, Kunis G, Ophir E, Landa G, Cohen H, Schwartz M (2006) Glatiramer acetate fights against Alzheimer's disease by inducing dendritic‐like microglia expressing insulin‐like growth factor 1. Proc Natl Acad Sci USA 103: 11784–11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL (2014) Identification of a unique TGF‐beta‐dependent molecular and functional signature in microglia. Nat Neurosci 17: 131–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camby I, Le Mercier M, Lefranc F, Kiss R (2006) Galectin‐1: a small protein with major functions. Glycobiology 16: 137R–157R [DOI] [PubMed] [Google Scholar]
- Cravens PD, Kieseier BC, Hussain R, Herndon E, Arellano B, Ben LH, Timmons BC, Castro‐Rojas C, Hartung HP, Hemmer B, Weber MS, Zamvil SS, Stuve O (2013) The neonatal CNS is not conducive for encephalitogenic Th1 T cells and B cells during experimental autoimmune encephalomyelitis. J Neuroinflammation 10: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, Green KN (2014) Colony‐stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82: 380–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Gutierrez P, Juarez‐Vicente F, Wolgemuth DJ, Garcia‐Dominguez M (2014) Pleiotrophin antagonizes Brd2 during neuronal differentiation. J Cell Sci 127: 2554–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330: 841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Lim S, Hoeffel G, Low D, Huber T (2013) Origin and differentiation of microglia. Front Cell Neurosci 7: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, Luedde T, Heikenwalder M, Jung S, Prinz M (2013) A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 16: 1618–1626 [DOI] [PubMed] [Google Scholar]
- Holtman IR, Noback M, Bijlsma M, Duong KN, van der Geest MA, Ketelaars PT, Brouwer N, Vainchtein ID, Eggen BJ, Boddeke HW (2015) Glia Open Access Database (GOAD): a comprehensive gene expression encyclopedia of glia cells in health and disease. Glia 63: 1495–1506 [DOI] [PubMed] [Google Scholar]
- Horie H, Inagaki Y, Sohma Y, Nozawa R, Okawa K, Hasegawa M, Muramatsu N, Kawano H, Horie M, Koyama H, Sakai I, Takeshita K, Kowada Y, Takano M, Kadoya T (1999) Galectin‐1 regulates initial axonal growth in peripheral nerves after axotomy. J Neurosci 19: 9964–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Kirchhoff F, Verkhratsky A (2013) Microglia: new roles for the synaptic stripper. Neuron 77: 10–18 [DOI] [PubMed] [Google Scholar]
- Khorooshi R, Owens T (2013) Detection and cellular localization of phospho‐STAT2 in the central nervous system by immunohistochemical staining. Methods Mol Biol 967: 179–188 [DOI] [PubMed] [Google Scholar]
- Khorooshi R, Morch MT, Holm TH, Berg CT, Dieu RT, Draeby D, Issazadeh‐Navikas S, Weiss S, Lienenklaus S, Owens T (2015) Induction of endogenous Type I interferon within the central nervous system plays a protective role in experimental autoimmune encephalomyelitis. Acta Neuropathol 130: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, Muller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F et al (2013) Microglia emerge from erythromyeloid precursors via Pu.1‐ and Irf8‐dependent pathways. Nat Neurosci 16: 273–280 [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Weng YC, Thammisetty SS, Phaneuf D, Lalancette‐Hebert M, Kriz J (2017) Molecular imaging of nestin in neuroinflammatory conditions reveals marked signal induction in activated microglia. J Neuroinflammation 14: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yao DL, Bondy CA, Brenner M, Hudson LD, Zhou J, Webster HD (1994) Astrocytes express insulin‐like growth factor‐I (IGF‐I) and its binding protein, IGFBP‐2, during demyelination induced by experimental autoimmune encephalomyelitis. Mol Cell Neurosci 5: 418–430 [DOI] [PubMed] [Google Scholar]
- Liu JL, Grinberg A, Westphal H, Sauer B, Accili D, Karas M, LeRoith D (1998) Insulin‐like growth factor‐I affects perinatal lethality and postnatal development in a gene dosage‐dependent manner: manipulation using the Cre/loxP system in transgenic mice. Mol Endocrinol 12: 1452–1462 [DOI] [PubMed] [Google Scholar]
- Matcovitch‐Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben‐Yehuda H, David E, Zelada Gonzalez F, Perrin P, Keren‐Shaul H, Gury M, Lara‐Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo‐Vivas E, Itzkovitz S et al (2016) Microglia development follows a stepwise program to regulate brain homeostasis. Science 353: aad8670 [DOI] [PubMed] [Google Scholar]
- McGraw J, McPhail LT, Oschipok LW, Horie H, Poirier F, Steeves JD, Ramer MS, Tetzlaff W (2004) Galectin‐1 in regenerating motoneurons. Eur J Neurosci 20: 2872–2880 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE (2012) The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J Immunol 188: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte SM, Wands JR (2005) Review of insulin and insulin‐like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer's disease. J Alzheimers Dis 7: 45–61 [DOI] [PubMed] [Google Scholar]
- Mueller AM, Nassery A, Conlon H, Liu X, Jun E, Yoon BH, Cristofanilli M, Sadiq SA (2013) Effects of intraventricular methotrexate administration on Cuprizone‐induced demyelination in mice. Front Mol Neurosci 6: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Taga T (1999) Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS Lett 457: 43–46 [DOI] [PubMed] [Google Scholar]
- Netchine I, Azzi S, Le Bouc Y, Savage MO (2011) IGF1 molecular anomalies demonstrate its critical role in fetal, postnatal growth and brain development. Best Pract Res Clin Endocrinol Metab 25: 181–190 [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333: 1456–1458 [DOI] [PubMed] [Google Scholar]
- Prinz M, Priller J (2014) Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci 15: 300–312 [DOI] [PubMed] [Google Scholar]
- Quinta HR, Pasquini JM, Rabinovich GA, Pasquini LA (2014) Glycan‐dependent binding of galectin‐1 to neuropilin‐1 promotes axonal regeneration after spinal cord injury. Cell Death Differ 21: 941–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington LT, Babcock AA, Zehntner SP, Owens T (2007) Microglial recruitment, activation, and proliferation in response to primary demyelination. Am J Pathol 170: 1713–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian V (2012) A system‐level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen Psychiatry 69: 1205–1213 [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B (2012) Microglia sculpt postnatal neural circuits in an activity and complement‐dependent manner. Neuron 74: 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo‐Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F (2012) A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336: 86–90 [DOI] [PubMed] [Google Scholar]
- Smith ME, Eller NL, McFarland HF, Racke MK, Raine CS (1999) Age dependence of clinical and pathological manifestations of autoimmune demyelination. Implications for multiple sclerosis. Am J Pathol 155: 1147–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont‐Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, Garel S (2014) Microglia modulate wiring of the embryonic forebrain. Cell Rep 8: 1271–1279 [DOI] [PubMed] [Google Scholar]
- Starossom SC, Mascanfroni ID, Imitola J, Cao L, Raddassi K, Hernandez SF, Bassil R, Croci DO, Cerliani JP, Delacour D, Wang Y, Elyaman W, Khoury SJ, Rabinovich GA (2012) Galectin‐1 deactivates classically activated microglia and protects from inflammation‐induced neurodegeneration. Immunity 37: 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, Barres BA (2007) The classical complement cascade mediates CNS synapse elimination. Cell 131: 1164–1178 [DOI] [PubMed] [Google Scholar]
- Stranges PB, Watson J, Cooper CJ, Choisy‐Rossi CM, Stonebraker AC, Beighton RA, Hartig H, Sundberg JP, Servick S, Kaufmann G, Fink PJ, Chervonsky AV (2007) Elimination of antigen‐presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity 26: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh HS, Zhao ML, Derico L, Choi N, Lee SC (2013) Insulin‐like growth factor 1 and 2 (IGF1, IGF2) expression in human microglia: differential regulation by inflammatory mediators. J Neuroinflammation 10: 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori Y, Mori T, Wakabayashi T, Nagasaka Y, Matsuzaki T, Yamada H (2009) Nestin‐positive microglia in adult rat cerebral cortex. Brain Res 1270: 10–18 [DOI] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T (2013) Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci 16: 543–551 [DOI] [PubMed] [Google Scholar]
- Wlodarczyk A, Lobner M, Cedile O, Owens T (2014) Comparison of microglia and infiltrating CD11c(+) cells as antigen presenting cells for T cell proliferation and cytokine response. J Neuroinflammation 11: 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarczyk A, Cédile O, Jensen KN, Jasson A, Mony JT, Khorooshi R, Owens T (2015) Pathologic and protective roles for microglial subsets and bone marrow‐ and blood‐derived myeloid cells in central nervous system inflammation. Front Immunol 6: 463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Li L, Richards RG, DiAugustine RP, D'Ercole AJ (2002) Myelination is altered in insulin‐like growth factor‐I null mutant mice. J Neurosci 22: 6041–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Yang L, Itoh S, Mori K, Tanaka J (2004) Microglia, a potential source of neurons, astrocytes, and oligodendrocytes. Glia 45: 96–104 [DOI] [PubMed] [Google Scholar]
- Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, Vyssotski AL, Bifone A, Gozzi A, Ragozzino D, Gross CT (2014) Deficient neuron‐microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci 17: 400–406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Dataset EV1
Review Process File
Source Data for Figure 2
Data Availability Statement
The datasets of RNA‐seq from adult, EAE, and neonatal microglia are deposited in Gene Expression Omnibus (GEO): GSE78809, and will become available via the Glia Open Access Database (Holtman et al, 2015): www.goad.education. The datasets of RNA‐seq of repopulating microglia are described in Bruttger et al (2015) and deposited in GEO: GSE68376.


