Abstract

Tropoelastin, as the monomer unit of elastin, assembles into elastic fibers that impart strength and resilience to elastic tissues. Tropoelastin is also widely used to manufacture versatile materials with specific mechanical and biological properties. The assembly of tropoelastin into elastic fibers or biomaterials is crucially influenced by key submolecular regions and specific residues within these domains. In this work, we identify the functional contributions of two rarely occurring negatively charged residues, glutamate 345 in domain 19 and glutamate 414 in domain 21, in jointly maintaining the native conformation of the tropoelastin hinge, bridge and foot regions. Alanine substitution of E345 and/or E414 variably alters the positioning and interactive accessibility of these regions, as illustrated by nanostructural studies and detected by antibody and cell probes. These structural changes are associated with a lower propensity for monomer coacervation, cross-linking into morphologically and functionally atypical hydrogels, and markedly impaired and abnormal elastic fiber formation. Our work indicates the crucial significance of both E345 and E414 residues in modulating specific local structure and higher-order assembly of human tropoelastin.
Keywords: tropoelastin, glutamate, hinge region, small-angle X-ray scattering, solution structure
Introduction
Tropoelastin is the monomer unit of elastin, the main component of elastic fibers in the extracellular matrix that confer strength and resilience to elastic tissues such as skin, lungs, and vasculature.1 Apart from this native function, the mechanical, biological, and assembly properties of tropoelastin have also enabled its fabrication into diverse biomaterials.2
Tropoelastin assembly into higher-order structures is strongly influenced at each stage by distinct contributions from its submolecular regions. Previous studies have identified the pivotal roles of the N-terminal coil,3 the central hinge and bridge,4−7 and the C-terminal foot8,9 regions during the coacervation, microfibrillar deposition and cross-linking stages of elastic fiber formation. Within these regions, there are specific residues that directly participate in intra- and intermolecular contacts,10−12 or maintain the local and global conformation essential for functional assembly.3,6 Such residues potentially modulate tropoelastin behavior via charge-based interactions.
Tropoelastin contains an abundance of residues with positively charged side chains at physiological pH, which are proposed to facilitate elastogenesis in a number of ways. Positively charged residues can promote tropoelastin coacervation by binding negatively charged glycosaminoglycans.13−15 The C-terminal RKRK cluster mediates tropoelastin tethering to cell surface receptors.16,17 The C-terminus also modulates tropoelastin deposition onto microfibrils via interactions with microfibril-associated glycoproteins.8,10,18,19 Specific lysine residues are directly involved in tropoelastin cross-linking into mature elastic fibers.12,20 The role of charged residues also extends beyond electrostatic or ionic interactions with elastogenic components. The substrate recognition of these residues relies heavily on their conformation,12,21 which can likewise be stabilized by the charged moieties. In support, a well-conserved arginine residue (R515) in the tropoelastin bridge region has been shown to maintain the native tertiary shape required for self-assembly.6
In contrast to the numerous positively charged residues, negatively charged residues occur at only three sites within human tropoelastin. We have previously identified the significance of the aspartate 72 (D72) residue in stabilizing the N-terminal coil region3 and facilitating elastogenic assembly. The roles of the other two negatively charged sites, glutamate 345 (E345) in domain 19 and glutamate 414 (E414) in domain 21, are still uncharacterized. Domains 19 and 21 are proximate to the bridge region formed by domains 25–26, and are located within a segment enriched for cross-links.12,20 In particular, domain 19 is reported to participate in cross-linking even at low cross-linker concentrations, which points to this region as the initial point of alignment during tropoelastin coacervation.5 Consistent with this is the high propensity of domain 19 for α helix formation, which positions lysine residues for cross-linking.22,23 Domain 19 has been identified to form cross-links with domain 25 and domain 10,21 which forms the basis of head-to-tail elastin assembly.24
Domain 21 is uniquely positioned preceding another hydrophilic domain 23 due to the constitutive splicing of domain 22 in human tropoelastin.25 The juxtaposition of domains 21 and 23 forms a hinge region26 as evidenced by nuclear magnetic resonance,23 small-angle X-ray scattering,5 and cross-linking experiments. This hinge region is structurally flexible27 and critically influences the dynamics of the tropoelastin molecule central to the assembly process.7
The positioning of E345 and E414 within structurally and functionally important regions suggests likely modulatory roles for tropoelastin shape and assembly. Interestingly, negatively charged residues also occur rarely in other mammalian tropoelastin,28 including the chimpanzee, baboon, cow, dog, pig, and rat sequences, and are predominantly clustered around the central domains 17–25 in proximity to the human 345 and 414 positions. In this work, we aim to investigate the significance of these negatively charged residues by constructing tropoelastin variants with mutations at one or both sites: E345A, where the E345 residue has been replaced with an alanine; E414A, where the E414 residue has been replaced with an alanine; and E345A+E414A, where both E345 and E414 have been substituted by alanines (Figure 1). The structure and functionality of these mutants will be assessed relative to those of the wild-type (WT) protein, specifically in terms of coacervation, cross-linking into hydrogel materials, and elastic fiber assembly.
Figure 1.
Domain structures of WT, E345A, E414A, and E345A+E414A tropoelastin. Hydrophobic domains are represented by black boxes, whereas hydrophilic domains are represented by white boxes. The mutation/s in each construct are indicated.
Materials and Methods
Tropoelastin Production
Mutant tropoelastin sequences were constructed by site-targeted mutagenesis of the pET-3d plasmid containing the WT sequence (Genscript) and validated by plasmid sequencing (Australian Genome Research Facility). The recombinant wild-type (WT, corresponding to amino acid residues 27–724 of GenBank entry AAC98394) and mutant proteins were expressed from transformed Escherichia coli BL21 cells, purified as previously described,29 and confirmed by SDS-PAGE and mass spectrometry (Figure S1).
Mass Spectrometry
WT and mutant tropoelastin constructs (5 mg/mL in water) were digested with 0.05 mg/mL Lys-C at 25 °C overnight. The samples were subjected to comparative matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry using a QSTAR XL mass spectrometer. A mass/charge window of 800–5000 was applied, and the resulting peaks were assigned by comparison with expected monoisotopic peptide masses from a theoretical Lys-C digest of singly charged peptides containing up to one missed cleavage. The mass peaks from the WT and mutant tropoelastin samples were overlaid, and peptide mass shifts corresponding to the mutation were identified.
Far-UV Circular Dichroism (CD)
CD spectra of tropoelastin constructs (0.15 mg/mL in 10 mM phosphate and 150 mM NaF) were recorded on a Jasco J-815 spectrometer equipped with a Peltier-controlled sample chamber. Samples were scanned with a bandwidth of 1.0 at 20 nm/min. Each spectrum was averaged from five scans, buffer-corrected, and smoothed using 3 point adjacent averaging. Secondary structure composition was estimated from the CD spectrum using the CONTINLL and CDSSTR methods30 with a reference set of 37 soluble proteins.
Small-Angle X-ray Scattering (SAXS)
SAXS data of tropoelastin constructs (10 mg/mL in phosphate buffered saline (PBS) with 2 mM dithiothreitol) were collected on the European Molecular Biology Laboratory beamline X33 at the DORISIII light source facilities at Hamburger Synchrotronstrahlungslabor/Deutsches Elektronen-Synchrotron (HASYLAB/DESY). Data were acquired using 4 × 30 s exposures and a 2.4 m sample-to-detector distance to cover a momentum transfer interval 0.008 < q < 0.54 Å–1. The modulus of the momentum transfer is defined as q = 4∏sin θ/λ, where 2θ is the scattering angle, and λ is the wavelength. The q range was calibrated using silver behenate powder based on diffraction spacings of 58.38 Å. The scattering images obtained were spherically averaged using in-house software and buffer scattering intensities subtracted using PRIMUS.31 Radius of gyration (Rg) values were calculated using AUTORG.32 Particle shapes were generated ab initio using GASBOR.33 Multiple GASBOR runs were performed to generate 10 similar shapes that were combined and filtered to produce an averaged model using the DAMAVER software package.34 The bead model was then visualized in UCSF Chimera.35
Enzyme-Linked Immunosorbent Assay (ELISA)
Wells were coated with up to 30 μg/mL of each tropoelastin construct at 4 °C overnight (n = 6). Unbound protein was removed with three PBS washes and nonspecific antibody binding was blocked with 3% (w/v) BSA for 1 h at room temperature. Excess BSA was washed off with PBS, and bound tropoelastin was detected with 1:2000 mouse antielastin BA4 antibody and 1:5000 goat antimouse IgG conjugated with horseradish peroxidase for 1 h at room temperature. Antibody-bound tropoelastin was visualized by incubation with ABTS solution (1.04 mg/mL ABTS, 0.05% (v/v) H2O2, 10 mM CH3COONa, 5 mM Na2HPO4, pH 5) at 37 °C for 1 h. Sample absorbances at 405 nm were read with a plate reader, and subtracted by the absorbance of BSA-blocked wells without tropoelastin. To compare the exposure of the C-terminus, another ELISA was performed using 1:5000 rabbit anti-C-terminal peptide antibody and 1:5000 goat antirabbit horseradish peroxidase-conjugated IgG as the primary and secondary antibody, respectively.
Cell Attachment
Cell culture wells were incubated in up to 30 μg/mL WT or mutant tropoelastin at 4 °C overnight. Wells were washed three times with PBS to remove unbound protein, blocked with heat-denatured BSA for 1 h at room temperature, and washed with PBS. Human dermal fibroblasts (GM3348, from Coriell Research Institute) grown in DMEM with 10% (v/v) fetal bovine serum were harvested with 0.25% (v/v) trypsin-EDTA at 37 °C for 3 min. The cells were centrifuged at 800 g for 5 min and resuspended in serum-free DMEM. Wells were seeded at 1.56 × 105 cells/cm2. To estimate the percentage of cell attachment, cells were diluted in DMEM to 10, 20, 50, 80, and 100% of the cell density used for the sample wells and added to uncoated and unblocked wells. Cells were allowed to attach at 37 °C in 5% CO2 for 1 h, and washed with two PBS washes. Cells were fixed with 3% (v/v) formaldehyde in PBS for 20 min, washed three times with PBS, and stained with 0.1% (w/v) crystal violet in 0.2 M MES, pH 5.0 at room temperature for 1 h. Excess stain was removed with four washes of reverse osmosis water, and the crystal violet was solubilized with 10% (w/v) acetic acid. Absorbance was measured at 570 nm using a plate reader and fitted to a linear regression, which was used to convert sample absorbances into percentage of cell attachment.
Coacervation Assay
Tropoelastin constructs at 10 mg/mL in PBS (10 mM phosphate, 150 mM NaCl, pH 7.4) were placed in quartz cuvettes and monitored in a Shimadzu UV-1601 spectrophotometer heated to a set temperature by a Julabo F4 recirculation water bath. Light scattering was examined by measuring the absorbance at 300 nm over 600 s at 20–60 °C. Between each temperature shift, the sample was cooled at 4 °C until turbidity was visibly reduced. The tropoelastin species were assessed according to the time taken to reach maximum turbidity at each temperature, as well as the temperature at which maximum sample turbidity was achieved.
Particle Size Analysis
The particle sizes of tropoelastin solutions at 20–60 °C were determined via dynamic light scattering using a Malvern Zetasizer Nano (Malvern Instruments). Tropoelastin solutions (10 mg/mL in PBS) were equilibrated for 5 min at the set temperature. Three runs of measurements, each with at least 12 data acquisitions, were taken and averaged to obtain the relative percentages of particle sizes present in each tropoelastin solution.
Hydrogel Construction
Tropoelastin constructs (100 mg/mL in PBS) were mixed with 10 mM of the chemical cross-linker bis(sulfosuccinimidyl)suberate (BS3) at 4 °C, and transferred in 200 μL volumes into LabTek Chamber Slides. The tropoelastin solutions were incubated at 37 °C for 16 h, washed in PB (10 mM phosphate, pH 7.4), and lyophilized.
Scanning Electron Microscopy (SEM)
Lyophilized hydrogels were sputter-coated with a 20 nm gold layer. Sample imaging was performed using the Zeiss EVO/Qemscan electron microscope at the Australian Centre for Microscopy and Microanalysis, University of Sydney.
Microcomputed Tomography (micro-CT)
Three-dimensional X-ray structures of hydrogels were determined with a SkyScan 1072 microcomputed tomography system. Samples were scanned with a 60 kV X-ray beam at a resolution of 3.23 μm. The resulting X-ray projection images were converted into a stack of cross sections with the cone-beam reconstruction program NRecon 1.4.4. (SkyScan) and rendered into a three-dimensional structure with VGStudio Max 1.2.1 (Volume Graphics GmbH). Images of the 3D structures, as well as horizontal and vertical sections, were taken with the same software. Hydrogel porosity was estimated from the cross-section images using CTan (SkyScan) and averaged across triplicate samples.
Hydrogel Swelling
Freeze-dried and preweighed hydrogels were swelled in water for 24 h at 37 °C, 25 °C, and 4 °C. Between each temperature shift, excess water was removed and the hydrogels were weighed. The amount of liquid absorbed per gram of hydrogel was recorded at each temperature.
Immunofluorescence Staining of Elastic Fibers
Fibroblasts were seeded on glass coverslips at a density of 18,400 cells/cm2. After 10 days, 20 μg/mL tropoelastin was added to the culture medium. At 1, 4, 7, and 10 days after tropoelastin addition, cells were fixed with 4% (w/v) paraformaldehyde for 20 min and quenched with 0.2 M glycine. The cells were incubated with 0.2% (v/v) Triton X-100 for 6 min, blocked with 5% bovine serum albumin at 4 °C overnight, and stained with 1:500 mouse antielastin BA4 antibody for 1.5 h and 1:100 FITC-conjugated antimouse IgG antibody for 1 h. The coverslips were mounted onto glass slides with ProLong Gold antifade reagent with DAPI. Slides were imaged with an Olympus FluoView FV1000 confocal microscope under identical laser settings. Z-stacks were taken from areas distributed across each sample and converted to maximum projection images.
Confocal images of elastic fibers were analyzed using ImageJ. To compare fiber fluorescence, a threshold was set to exclude background and saturated pixel intensities. The average intensity of pixels within this threshold was measured for each image and averaged for each sample. To compare fiber number, two perpendicular reference lines were consistently drawn through the center of each image. The number of fibers intersecting either reference line was counted and averaged for each sample. The area occupied by cell nuclei was used to indicate similar cell numbers in all samples.
Statistical Analyses
All data were reported as mean ± standard error (n = 3 unless otherwise indicated). Analysis of variance was performed using GraphPad Prism (GraphPad Software). Statistical significance was set at p < 0.05, and indicated in the figures as “ns” (p ≥ 0.05), * (p < 0.05), ** (p < 0.01), or *** (p < 0.001).
Results
Alanine Substitution of E345 and/or E414 Alters the Solution Shape of Tropoelastin
The WT and mutant tropoelastin constructs possessed comparable overall secondary structure, characterized by similar far-UV CD spectral features including a large negative minimum at 200 nm, which corresponds to disordered hydrophobic regions, and a slight negative shoulder at 220 nm, which is assigned to the α-helical structure of cross-linking domains36−38 (Figure 2A). These features computationally translated to a similar secondary structure composition, consisting predominantly of unordered regions (48–50%), with a small percentage of alpha-helices (7–10%), beta-sheets (18–20%), turns (12–13%), and polyproline-2 helices (9–11%) (Figure 2B).
Figure 2.
Structure of WT, E345A, E414A, and E345A+E414A tropoelastin. (A) Far-UV CD spectra and (B) secondary structure composition of tropoelastin constructs. Ahel, α helix; BSht, beta-sheet; PP2, polyproline-2 helix; UNR, unordered. (C) Solution structures of tropoelastin constructs obtained from SAXS. The models were aligned to show spatial overlap of common features (N, N-terminus; H, hinge region; BR, bridge region; C, C-terminus). Scale bar: 5 nm.
The solution shapes of the E345A, E414A and E345A+E414A mutants obtained by small-angle X-ray scattering also exhibited the same characteristic features as WT tropoelastin (Figure 2C). Each structure displayed an elongated N-terminal coil leading to a hinge (spur) region, and connected by a bridge region to the C-terminal foot. Although the scattering curves showed similar global features between the WT and mutants, the low q data displayed differences suggestive of subtle shape changes (Figure S2). These differences were manifested as alterations in the Rg between the WT and mutants (WT, 6.1 ± 0.08 nm; E345A, 5.84 ± 0.08 nm; E414A, 6.63 ± 0.17 nm; E345A+E414A, 6.08 ± 0.08 nm). Consistent with the trend of the Rg values, the hinge, bridge and foot regions appeared condensed in E345A, extended in E414A, and most similar to the native structure in E345A+E414A. These results suggest central and C-terminal conformational changes associated with mutation/s at E345 or E414, which encompass but are not confined to the expected locations of these sites, and which are not linked to global changes in protein secondary structure composition.
Antibody and Cell Probes Detect Central and C-Terminal Conformational Changes in the Mutants
To confirm the structural differences observed in the solution shapes of the tropoelastin mutants, we used antibodies to probe the accessibility of specific domains on substrate-bound constructs. The amount of bound protein detected by the BA4 antielastin antibody increased proportionally to the coating concentration until saturation at ∼10 μg/mL tropoelastin. This antibody recognizes multiple epitopes, but primarily the central domain 24, within the tropoelastin molecule. At subsaturation concentrations of tropoelastin, BA4 antibody binding to the E345A, E414A, and E345A+E414A constructs was significantly reduced by up to 58 ± 11%, 78 ± 7%, and 58 ± 17% compared to WT, suggesting decreased availability of this central region of the mutant proteins (Figure 3A).
Figure 3.

Antibody and cell probing of tropoelastin local conformation. Exposure of the (A) central and (B) C-terminal tropoelastin regions as detected by antibodies targeted against these regions. (C) Attachment of human dermal fibroblasts to WT, E345A, E414A, and E345A+E414A tropoelastin.
In the same manner, relative exposure of the tropoelastin C-terminal regions was compared using an antibody directed against domain 36 (Figure 3B). Even at the maximum tropoelastin coating concentration, the mutant constructs displayed significantly decreased antibody binding. Compared to WT, the C-terminal accessibility of E345A, E414A, and E345A+E414A was reduced by 19 ± 3, 33 ± 2, and 23 ± 5%, respectively.
In addition to the antibodies, human dermal fibroblast cells were also used to probe for conformational changes in the tropoelastin constructs. These cells are known to bind to specific sequences within the central and C-terminal segments.17,39 All WT and mutant tropoelastin supported attachment of cells in a dose-dependent manner until saturation at 10 μg/mL. At tropoelastin concentrations supporting maximum cell attachment, WT facilitated the adhesion of 82 ± 9% of seeded cells. In contrast, all mutant constructs exhibited significantly reduced cell interactions. Compared to the WT construct, E345A, E414A, and E345A+E414A supported the attachment of 61 ± 4%, 58 ± 0.5%, and 50 ± 0.5% of seeded cells, respectively. These antibody and cell binding results support the presence of structural shifts in the central and C-terminal regions associated with E345 and E414 mutations.
Tropoelastin with E345 and E414 Mutations Display Impaired Coacervation
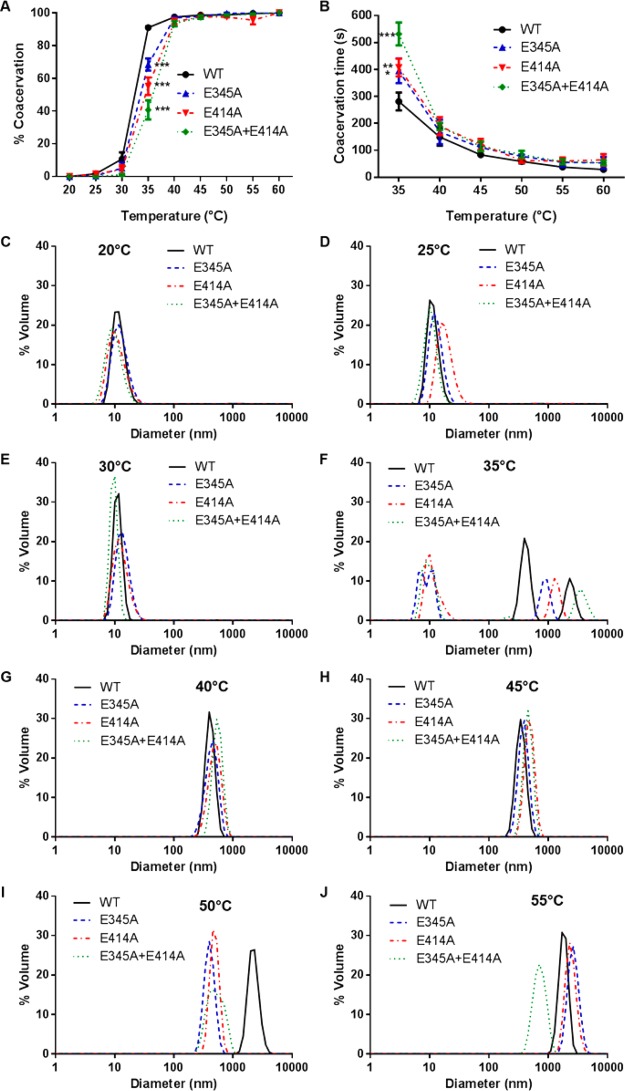
All tropoelastin constructs displayed temperature-dependent coacervation (Figure 4A). As each sample reached a critical temperature, a sharp increase in turbidity was detected and interpreted as a rapid rise in coacervation level. Negligible increases in sample turbidity were observed beyond this transition temperature, which was distinct for each tropoelastin construct. Full coacervation was achieved at 35 °C by WT, but only at 40 °C by E345A, E414A and E345A+E41A. At the WT transition temperature of 35 °C, E345A and E414A displayed comparable self-association that was significantly decreased relative to WT, whereas E345A+E414A exhibited an even greater reduction in coacervation compared to either E345A or E414A.
Figure 4.
Coacervation of WT, E345A, E414A, and E345A+E414A tropoelastin. (A) Extent of coacervation at each temperature, expressed as a percentage of the maximum coacervation achieved by each tropoelastin construct. (B) Time taken by each sample to reach maximum coacervation at each temperature. (C–J) Particle size distribution of tropoelastin solutions at (C) 20, (D) 25, (E) 30, (F) 35, (G) 40, (H) 45, (I) 50, and (J) 55 °C.
The time required for coacervation to occur was also temperature-dependent for all tropoelastin constructs (Figure 4B). Coacervation time decreased exponentially with increasing temperature. A difference in the coacervation time of the constructs was observed at temperatures below the transition temperature of the mutants at 40 °C. At 35 °C, the Glu-to-Ala variants required a significantly longer time to aggregate (E345A, 395 ± 46 s; E414A, 407 ± 33 s; E345A+E414A, 532 ± 42 s) compared to WT (281 ± 33 s). Among the tropoelastin mutants, E345A+E414A coacervated more slowly than either E345A or E414A.
The differences in the coacervation profiles of the WT and mutant species were confirmed by analysis of solution particle sizes over a range of temperatures (Figure 4C–J). At and below 30 °C, all tropoelastin constructs were in the form of ∼10 nm monomers in solution. At 35 °C, 67% of WT species associated into 255–615 nm particles, 33% into larger 1.4–3.6 μm aggregates, and no monomers remained; in contrast, 66% of E345A, 60% of E414A, and 62% of E345A+E414A remained as monomers. At 40 °C, all mutant species had coacervated into 255–825 nm particles similarly to WT. The same trend was observed at 45 °C. WT tropoelastin further aggregated into 1.3–4.8 μm assemblies at 50 °C, whereas E345A and E414A formed similar-sized particles only at 55 °C. Notably, the E345A+E414A coacervates did not attain this end size even at 55 °C. These results clearly point to the lower propensity of E345A, E414A, and most severely, E345A+E414A, for temperature-dependent self-assembly.
Tropoelastin Mutated at E345 and/or E414 Forms Atypical Hydrogels
Addition of a 6-fold molar excess of the chemical cross-linker BS3 to tropoelastin solutions allowed the fabrication of hydrogels from each monomer construct. SDS-PAGE analysis of the aqueous solution left after tropoelastin polymerization revealed the absence of monomer species in all samples (Figure S3), indicating the complete cross-linking of WT and mutant constructs into the elastin materials.
The WT and mutant hydrogels demonstrated distinct differences in their surface composition as revealed by SEM (Figure 5). The top surface of the WT hydrogel appeared as a flat layer with large ∼100 μm pores. In contrast, the top surfaces of the E345A, E414A, and E345A+E414A hydrogels were marked by an abundance of ∼10 μm globules that are variably interlinked. On the E345A hydrogel, these spherules appeared as discrete entities connected by very fine fibers. On the E414A hydrogel, the particles were interspersed among sheet-like fragments containing a number of ∼10 μm pores. On the E345A+E414A hydrogel, the globules were joined by thick fibers which appeared to arise from the coalescence of the globules themselves. Unlike the top surfaces, the bottom surfaces of the elastin hydrogels shared a similar morphology, consisting of a smooth sheetlike layer with pores ranging from 20 to 100 μm in size.
Figure 5.
SEM imaging of the top and bottom surfaces of hydrogels produced by chemical cross-linking of WT, E345A, E414A, or E345A+E414A tropoelastin. Scale bar: 20 μm.
Micro-CT imaging also revealed discernible differences in the structural composition of the WT and mutant hydrogels (Figure 6A). The WT hydrogel consisted of a filamentous network interspersed with numerous pores that were visible across the top surface and throughout the cross-section of the material. In contrast, this predominantly porous network structure was not observed in any of the mutant hydrogels. The E345A, E414A, and E345A+E414A hydrogels were denser than the WT material, and were morphologically distinct from each other. The E345A hydrogel appeared more compact than the E414A or E345A+E414A constructs, as evidenced by a reduced thickness despite having a mass comparable to the other hydrogels. The E414A hydrogel comprised of more loosely packed clusters of wispy structures that were easily dislodged during sample handling. The E345A+E414A hydrogel exhibited a fibrous structure reminiscent of the WT material; however, its structure was more compact and characterized by pores smaller than those observed in the WT.
Figure 6.
(A) Three-dimensional reconstruction of WT, E345A, E414A, and E345A+E414A hydrogels by micro-CT imaging, showing a top-down and cross-sectional view of each material. Scale bar: 0.8 mm. (B) Porosity of hydrogels calculated from micro-CT sections. (C) Swelling of hydrogels in water at 4, 25, and 37 °C.
The most apparent difference between the WT and mutant hydrogels was the abundance of large pore structures in the former that were absent in the mutant constructs. Calculations of hydrogel porosity from micro-CT cross-sections estimated the WT material to be 87 ± 1% porous, and the mutant hydrogels to be significantly less porous at 76 ± 2% for E345A, 76 ± 1% for E414A, and 76 ± 1% for E345A+E414A (Figure 6B). The porosities of the mutant hydrogels were similar to each other.
The WT and mutant elastin hydrogels exhibited profound but differential swelling after being submerged in water for a 24 h period at various temperatures (Figure 6C). At 37 °C, WT hydrogels absorbed water 59 ± 5 times their dry weight. In contrast, water influx into the mutant hydrogels was significantly reduced. The E345A, E414A, and E345A+E414A hydrogels swelled 2.4 ± 0.4, 3.4 ± 0.1, and 4.7 ± 0.3 fold less compared to the WT, respectively. This trend was consistent at all tested temperatures. There were no significant differences among the swelling properties of the mutant hydrogels.
E345 and E414 Tropoelastin Mutations Severely Impede Elastic Fiber Assembly
The ability of the tropoelastin constructs to form elastic fibers in a cellular environment was determined by the addition of WT, E345A, E414A, and E345A+E414A to the culture medium of human dermal fibroblasts (Figure 7). WT spherules were arranged in a linear, fiber-like formation day 1 after tropoelastin addition, and progressively formed thin elastic fibers by day 4, which developed into an extensive, well-defined fiber network by day 7. In contrast, the Glu-to-Ala mutants underwent elastogenic pathways that were impaired to varying degrees. E345A did not produce definitive fibers throughout the 10-day postaddition period, and remained as randomly dispersed punctate species in the extracellular space, which were gradually cleared from the cell environment during media changes. E414A and E345A+E414A spherules were clustered in a globular organization by day 1, and formed short segments of elastic fibers by day 4, but did not progress further into a networked structure even by day 10. The elastic fibers detected by immunostaining represent those formed solely from exogenous tropoelastin, as evidenced by the absence of fibers in the control sample with no added tropoelastin.
Figure 7.
Elastic fiber assembly of WT, E345A, E414A, or E345A+E414A tropoelastin at 1, 4, 7, and 10 days after addition into human dermal fibroblast cultures. Elastic fibers were stained with the mouse BA4 antielastin primary antibody and a FITC-conjugated goat antimouse secondary antibody. Controls consist of fully stained samples with no exogenous tropoelastin, and samples in which WT tropoelastin was added but not stained, or stained only with the secondary antibody, or stained with a nonspecific mouse IgG and the FITC-conjugated antimouse secondary antibody. The faintly fluorescent fibers observed in the staining controls were due to the autofluorescence of elastic fibers. Cell nuclei in the control samples were stained with DAPI. Scale bar: 20 μm.
Moreover, the elastic fibers formed by E414A and E345A+E414A, respectively, displayed a 39 ± 1% and 31 ± 0.4% reduction in immunofluorescence, and a 50 ± 5% and 42 ± 3% decrease in abundance compared to the WT fibers, despite comparable cell numbers in all samples (Figure 8). E345A did not form elastic fibers, and therefore could not be analyzed. The differences in the elastogenic ability of the WT and mutant tropoelastin constructs were consistently observed at 4, 7, and 10 days after tropoelastin addition, pointing to the significant elastogenic impairment associated with the E345A and E414A tropoelastin mutations.
Figure 8.
Properties of elastic fibers formed by WT, E414A, and E345A+E414A tropoelastin. (A) Elastin-specific fluorescence and (B) abundance of elastic fibers. (C) Cell numbers as measured by the area occupied by cell nuclei per field of view.
Discussion
Key tropoelastin residues with charged side chains at physiological pH have been shown to modulate local and global protein structure,3,6 as well as a number of elastogenic assembly events.8,10,12−15,18−20 While human and other mammalian tropoelastin contain an abundance of positively charged amino acids, negatively charged residues occur infrequently, on average at three positions within each sequence,28 which are clustered either in the N-terminal domain 6 or in the central domains 19–25. We have previously identified the functional roles of the aspartate in domain 6 of human tropoelastin.3 In this work, we sought to characterize the significance of the glutamates in domain 19 and 21, a region tightly associated with tropoelastin structural flexibility26 and functional assembly.5,11,21
The secondary structure of the WT and Glu-to-Ala mutant tropoelastin, estimated from CD spectra using established algorithms,30 consists mainly of unordered regions and a small percentage of alpha helices, beta sheets, turns, and polyproline-2 helices. This trend was comparable to findings obtained from nuclear magnetic resonance40 and Raman spectroscopic data.41,42 The high amount of unordered regions and corresponding low level of helical structures are consistent with the flexibility of tropoelastin.7 The presence of beta turns occurs in hydrophobic domains and is proposed to be responsible for the elasticity of the elastin polymer.43 Although there is no evidence of significant changes in the overall secondary structure composition of E345A, E414A, and E345A+E414A, our results do not preclude the possibility of shifts in local secondary structure. For instance, the tropoelastin domain 19 which contains E345 has a greater propensity for helix formation than any other cross-linking domain.22,23 Likewise, domain 21, which contains E414 and forms part of a flexible hinge region, also has a high helical content.7 Because charged residues are known to stabilize helical structures,44 the substitution of E345 or E414 with a neutral alanine can potentially disrupt helix formation and affect local molecular structure.
In light of this, the solution structures of the mutant tropoelastin species were determined by SAXS. The constructs exhibited general structural features similar to those previously described in human tropoelastin, including an N-terminal elastic coil region, a hinge region spanning domains 21/23, a bridge region containing domains 25–26, and a C-terminal foot region.24 However, the E345A and E414A shapes revealed displacement of their hinge, bridge and foot regions from the WT, whereas the E345A+E414A model more closely resembled the native structure, suggesting potential compensatory effects of the double mutation.
Nevertheless, orientation differences in the central and C-terminal regions of E345A, E414A, and E345A+E414A were confirmed by reduced antibody and cell binding to the mutants compared to the WT, particularly at low tropoelastin concentrations. The BA4 antibody predominantly targets the hydrophobic VGVAPG pentapeptide in domain 24,45 and to some extent, other similar sequences with a xGxxPG or xGxPGx motif.46 The anti-C-terminal peptide antibody is custom designed against the RKRK motif in domain 36. These sequences were unmodified in the mutant tropoelastin species, which allows the extent of antibody detection to be correlated to the relative conformational exposure of these domains. In the same manner, fibroblasts are known to bind tropoelastin via domains 17–1839 and domain 36.17,47 Because E345 and E414 fall outside these regions, the decreased cell attachment is not due to the mutational disruption of cell binding sites, but likely arises from the differential availability and engagement of these regions to cell receptors implicated in fibroblast-tropoelastin adhesion, such as glycosaminoglycans18 and integrins.39 In addition, the reduced exposure of domain 24 in the mutant variants also likely hinders interactions with the elastin binding protein.48 Decreased antibody and cell binding supports the spatial displacement or partial obscuring of the central and C-terminal domains in E345A, E414A, and E345A+E414A tropoelastin.
The propagation of structural changes from domain 19 and 21, where the E345 and E414 sites are respectively located, to the broader central and C-terminal regions is likely due to a turn formed by the adjoining domains 21 and 23.26 This hinge positions domain 19 in close proximity with domain 25 as part of a symmetrical loop around this region,24 which supports the formation of a well-characterized intramolecular cross-link between the two domains21 (Figure 9). This conformation potentially allows the E345 residue in domain 19 to contact one of several positively charged residues in domains 25 and 26, such as K507, K511, or R515. A native salt bridge involving E345 may promote the availability and specificity of proximal lysines, such as K507, for participation in cross-link formation.12 Alternatively, removal of the E345 site and its stabilizing interactions may allow the cross-linking of a typically unavailable lysine residue, or enable aberrant intramolecular contacts that bias against the cross-linking of native lysines. In the E345A mutant, non-native interactions between the free lysine or arginine in domains 25–26 with an alternative site such as the E414 residue would result in an apparent lengthening of the hinge region and a dramatic shortening of the bridge region, resulting in the contraction of the C-terminal foot region toward the central mass of the E345A molecule.
Figure 9.

Model of proposed interactions involving the tropoelastin E345 and E414 residues. Domains 18–26 encompassing the hinge and bridge regions are represented schematically, adapted from Baldock et al. (2011).24 The hinge region is formed by domains 21 and 23. The proposed tetra-functional cross-link occurring between domains 19 and 2521,24 is represented by red dotted lines. Potential contacts formed by the E345 and E414 residues are indicated by green dashed lines.
The tropoelastin hinge, formed by the adjoining domains 21 and 23, is predicted to be stabilized by a salt bridge between the E414 residue in domain 21 and the K441 residue in domain 2327 (Figure 9). Abolishment of the E414 site may therefore destabilize the hinge and contribute to a structural change in this region. In addition, the positively charged residue normally bound to E414 may then interact with an upstream site such as E345, which is consistent with the high local flexibility of the hinge region,5,49 and result in the elongation of the bridge region and extension of the foot region.
We propose that in the absence of both E345 and E414 residues, the available positively charged sites do not form aberrant local structures that contract or extend the bridge region. However, the elimination of stabilizing interactions within the tropoelastin hinge may increase the torsional flexibility of this region and modify its equilibrium conformation,27 resulting in the differential presentation of specific central and C-terminal domains as observed in E345A+E414A. In the absence of local stabilization, however, the global features of E345A+E414A are still preserved, likely due to structural contributions from the rest of the molecule. Our findings suggest that both E345 and E414 are tandemly involved in maintaining the local tropoelastin hinge and bridge structures.
Given the structural modifications in the mutant constructs, their capacity for functional assembly was examined. Coacervation represents the first crucial stage in elastogenesis and greatly impacts upon tropoelastin incorporation into elastic fibers.50 Similar to studies on various tropoelastin isoforms,4,51,52 the WT and mutant tropoelastin species displayed a sharp transition from the monomer to coacervate stage over a narrow temperature range (<10 °C), consistent with the process being entropy-driven. However, E345A, E414A, and E345A+E414A initially coacervated at a higher temperature compared to the WT, and also required a higher temperature to form end-sized assemblies.53 Studies have shown a strong inverse correlation between coacervation temperature and the number of hydrophobic domains in tropoelastin.54−58 However, this model does not explain the higher transition temperature of the tropoelastin mutants compared to WT, as all constructs possess the same number and length of hydrophobic domains. An increased coacervation temperature therefore conceptually reflects an apparently less hydrophobic monomer, potentially due to less solvent-exposed hydrophobic regions consistent with the subtle structural differences between the WT and mutant constructs.
In addition to thermodynamic differences, kinetic differences were also observed in the coacervation of WT and mutant constructs. In all constructs, coacervation rate increased at higher temperatures. Coacervation rate is likewise linked to protein hydrophobicity, as greater cooperativity among a larger number of hydrophobic segments improves coacervation efficiency.4 The conformational changes in the hinge and bridge regions of E345A, E414A, and E345A+E414A may obscure or displace neighboring large hydrophobic domains such as domains 20, 24 and 26, resulting in decreased cooperative interactions during coacervation.
The ability of tropoelastin to be cross-linked strongly reflects its propensity to be incorporated into elastic fibers. Cross-linking of the tropoelastin constructs with BS3, which targets lysine residues within a maximum distance of 11.4 Å,59 approximates in vivo cross-linking by lysyl oxidase12,60 and identifies regions aligned by coacervation.5 Hydrogels produced from WT and mutant tropoelastin displayed strikingly different morphological and functional properties. The porous nature of the WT hydrogel surface was similar to previous descriptions of synthetic elastin.61 In contrast, the mutant hydrogel surfaces consisted of globular clusters, with sizes consistent with partially cross-linked nascent elastin prior to their condensation into fibrous structures.53,62 These nascent elastin globules particularly suggest that E345A, E414A, and E345A+E414A are less able to form mature cross-linked structures characteristic of normal elastin. The globules were linked by coalesced structures—fine fibrils in E345A, sheetlike fragments in E414A, and thick fibers which seemed intermediate between a fibrillar and a flat structure in E345A+E414A—to form a closed network. These morphologically distinct assemblies further indicate differences in the extent and/or nature of cross-linking among the mutant tropoelastin constructs.
Micro-CT reconstruction of the WT hydrogel revealed a fibrous and porous network consistent with the filamentous nature of natural elastin,63−65 which markedly contrasted with the compact, less porous structures of the mutant hydrogels. Hydrogel porosity is thought to be determined by the kinetics of separation into polymer-rich and polymer-lean phases.66 Variations in this phase separation, and therefore in hydrogel porosity, may lead to differences during cross-linking.
The differences in WT and mutant hydrogel porosities likely account for their differential swelling in water. Water absorption by WT hydrogels to ∼60 times the protein mass is consistent with the reported swelling ability of cross-linked elastin67,68 and elastin–mimetic peptides.69 Hydrogel swelling is defined by interactions between the solvent and the polymer. The influx of solvent stretches the junctions of the hydrogel70,71 and decreases the mobility of the flexible hydrophobic segments within the rigid cross-linked domains.72 This is balanced by the entropic increase associated with the mixing of solvent and bound water within the polymer.71 The significantly reduced water absorption of mutant hydrogels may therefore be due to changes in polymer-associated hydration brought about by the non-native conformation of the cross-linked material. The reduced swelling of mutant hydrogels may also be simply reflective of their reduced porosity as characterized by a more compact structure, fewer or smaller channels, and/or less interconnectivity between pores.
The aberrant cross-linking of E345A, E414A, and E345A+E414A is unlikely to be directly due to the elimination of negatively charged residue/s, since cross-linking involves specific lysine residues12 that remain present in these constructs. The observed conformational shifts in the mutant hinge and bridge regions, which contain important cross-linking domains, likely displace these sites of contact and detrimentally affect native cross-link formation.5
Tropoelastin incorporation into elastic fibers in a cellular environment was determined by the addition of purified constructs to the culture medium of dermal fibroblasts. Even at the earliest stages of elastogenesis, the mutant constructs already displayed lower propensity for the WT linear organization, and instead remained randomly dispersed, e.g. E345A, or clustered in a globular arrangement, e.g. E414A and E345A+E414A. The muted early stage fiber assembly of the mutants suggests differential presentation of intermolecular interacting domains, consistent with the impairment observed during the coacervation and cross-linking stages that requisitely precede elastic fiber formation.
Elastic fibers formed by the mutant constructs not only are inefficiently assembled but also appear morphologically abnormal. WT elastic fibers displayed extensive branched structures consistent with the architecture of the skin elastic network,14,73 whereas the E414A and E345A+E414A fibers were disjointed, scarcer, and less fluorescent. The decreased staining of mutant elastic fibers by the BA4 antibody contrasts against the comparable detection of all tropoelastin constructs above 10 μg/mL previously observed in the ELISA experiments. This result indicates that fewer mutant monomers may be deposited into elastic fibers and are subsequently removed from the cell environment. Alternatively, the BA4 epitopes equally accessible in the WT and mutant monomers may become differentially exposed in the assembled elastic fibers, which strongly suggests that the E414A and E345A+E414A molecules are atypically arranged within the elastic fiber. The structural modifications to the E414A and E345A+E414A hinge, bridge and C-terminal regions coincide with the submolecular segments specifically involved in native head-to-tail protein assembly.7,21,24 Inefficient or abnormal association of the mutant species, compounded by atypical cross-linking as demonstrated by their hydrogel properties, would hinder expansion of the elastic fiber network and account for the markedly reduced number of mutant fibers compared to WT. In addition, because cell anchorage of elastic fibers is thought to mechanically regulate the elastic network architecture,14,62,74 the decreased fibroblast adhesion of the mutant tropoelastin constructs may also contribute to the non-native fiber morphology and reduced fiber assembly.75
The inability of E345A to form elastic fibers was unexpected in light of its comparable functionality with E414A and E345A+E414A in previous cell adhesion, coacervation and cross-linking assays. E345A potentially self-associates in a manner that is incompatible with fibrillar assembly, as contraction of its bridge and C-terminal domains may sterically prevent head-to-tail monomer contact. Alternatively, E345A may have impaired interactions with other essential elastogenic components, such as microfibrillar proteins and lysyl oxidase enzymes, which prevents its stable incorporation into elastic fibers. The improved elastogenic ability of E345A+E414A compared to E345A suggests functional compensation by the loss of both glutamate residues.
We have identified key contributions of the E345 and E414 residues to the structure and functional assembly of human tropoelastin. While such negatively charged residues also occur rarely in other mammalian tropoelastin sequences, those of species such as chicken, lizard, frog and fish (e.g., zebrafish, fugu) do not contain negatively charged residues at all.76 However, these sequences also contain other large sections of insertions and deletions, particularly in domains 18–24, which diverge from the human isoform. These significant sequence differences suggest that mechanisms for tropoelastin structural stabilization may also vary between species, and can be independent of charge-based intramolecular interactions.
Conclusions
We propose that the tropoelastin hinge and bridge regions are stabilized by charge interactions between E345 or E414 and proximal lysines or arginines. These intramolecular contacts likely strongly influence the proximity and interactional accessibility of tropoelastin domains. Removal of either E345 or E414 may support aberrant intramolecular interactions that alter the length of the hinge and the adjoining bridge, which consequently affects the position of the C-terminal foot. The abolishment of both E345 and E414 may prevent the formation of abnormal interactions and preserve the normal bridge length, but can nevertheless destabilize the local hinge and bridge structures. These conformational changes likely perturb the relative positions of hydrophobic and cross-linking domains essential for intermolecular interactions, resulting in significantly impaired coacervation, hydrogel cross-linking, and elastic fiber assembly. These findings indicate the importance of both E345 and E414 in modulating human tropoelastin structure and functional assembly.
Acknowledgments
The authors acknowledge use of the facilities as well as scientific and technical assistance at the Australian Centre for Microscopy and Microanalysis. The authors also thank DESY for SAXS beamtime and Manfred Roessle and Anne Tuukkanen for assistance in using beamline X33 during data collection.
Glossary
Abbreviations
- ELISA
enzyme-linked immunosorbent assay
- BS3
bis(sulfosuccinimidyl)suberate
- CD
circular dichroism
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- micro-CT
microcomputed tomography
- PBS
phosphate buffered saline
- SAXS
small-angle X-ray scattering
- SEM
scanning electron microscopy
- WT
wild-type human tropoelastin
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsbiomaterials.6b00564.
Figure S1, mass spectrometry validation of E345A, E414A, and E345A+E414A tropoelastin; Figure S2, SAXS measurements of WT, E345A, E414A, and E345A+E414A tropoelastin, showing Log I vs q, P(r) vs distance, and Guinier plots; Figure S3, SDS-PAGE analysis of the aqueous solution left after tropoelastin cross-linking into hydrogels (PDF)
Author Contributions
C.B., G.C.Y., and A.S.W. performed and analyzed SAXS measurements of tropoelastin constructs. S.G.W. performed and analyzed mass spectrometry of tropoelastin samples. G.C.Y. and A.S.W. designed, performed, and analyzed all other experiments. G.C.Y. and A.S.W. wrote the manuscript. All authors have given approval to the final version of the manuscript.
A.W. acknowledges funding from the Australian Research Council DP120103911, DP130103693 and National Health & Medical Research Council APP1075357. C.B. acknowledges funding from the Biotechnology and Biological Sciences Research Council (BB/L00612X/1). The Wellcome Trust Centre for Cell-Matrix Research, University of Manchester, is supported by core funding from the Wellcome Trust (088785/Z/09/Z).
The authors declare the following competing financial interest(s): AW is the Scientific Founder of Elastagen Pty Ltd.
Supplementary Material
References
- Wise S. G.; Weiss A. S. Tropoelastin. Int. J. Biochem. Cell Biol. 2009, 41, 494–497. 10.1016/j.biocel.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Yeo G. C.; Aghaei-Ghareh-Bolagh B.; Brackenreg E. P.; Hiob M. A.; Lee P.; Weiss A. S. Fabricated Elastin. Adv. Healthcare Mater. 2015, 4 (16), 2530–2556. 10.1002/adhm.201400781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G. C.; Baldock C.; Wise S. G.; Weiss A. S. A negatively charged residue stabilizes the tropoelastin N-terminal region for elastic fiber assembly. J. Biol. Chem. 2014, 289 (50), 34815–26. 10.1074/jbc.M114.606772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. A.; Vrhovski B.; Weiss A. S. Domain 26 of tropoelastin plays a dominant role in association by coacervation. J. Biol. Chem. 2000, 275 (37), 28449–28454. 10.1074/jbc.M004265200. [DOI] [PubMed] [Google Scholar]
- Dyksterhuis L. B.; Baldock C.; Lammie D.; Wess T. J.; Weiss A. S. Domains 17–27 of tropoelastin contain key regions of contact for coacervation and contain an unusual turn-containing crosslinking domain. Matrix Biol. 2007, 26 (2), 125–135. 10.1016/j.matbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Yeo G. C.; Baldock C.; Tuukkanen A.; Roessle M.; Dyksterhuis L. B.; Wise S. G.; Matthews J.; Mithieux S. M.; Weiss A. S. Tropoelastin bridge region positions the cell-interactive C terminus and contributes to elastic fiber assembly. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (8), 2878–2883. 10.1073/pnas.1111615108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G. C.; Tarakanova A.; Baldock C.; Wise S. G.; Buehler M. J.; Weiss A. S. Subtle balance of tropoelastin molecular shape and flexibility regulates dynamics and hierarchical assembly. Sci. Adv. 2016, 2, e1501145. 10.1126/sciadv.1501145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BrownAugsburger P.; Broekelmann T.; Rosenbloom J.; Mecham R. P. Functional domains on elastin and microfibril-associated glycoprotein involved in elastic fibre assembly. Biochem. J. 1996, 318, 149–155. 10.1042/bj3180149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel B. A.; Wachi H.; Davis E. C.; Mecham R. P. Domains in tropoelastin that mediate elastin deposition in vitro and in vivo. J. Biol. Chem. 2003, 278 (20), 18491–18498. 10.1074/jbc.M212715200. [DOI] [PubMed] [Google Scholar]
- Brown P. L.; Mecham L.; Tisdale C.; Mecham R. P. The cysteine residues in the carboxy terminal domain of tropoelastin form an intrachain disulfide bond that stabilizes a loop structure and positively charged pocket. Biochem. Biophys. Res. Commun. 1992, 186, 549–555. 10.1016/S0006-291X(05)80843-5. [DOI] [PubMed] [Google Scholar]
- Dyksterhuis L. B.; Weiss A. S. Homology models for domains 21–23 of human tropoelastin shed light on lysine crosslinking. Biochem. Biophys. Res. Commun. 2010, 396 (4), 870–873. 10.1016/j.bbrc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Wise S. G.; Mithieux S. M.; Raftery M. J.; Weiss A. S. Specificity in the coacervation of tropoelastin: solvent exposed lysines. J. Struct. Biol. 2005, 149 (3), 273–281. 10.1016/j.jsb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Gheduzzi D.; Guerra D.; Bochicchio B.; Pepe A.; Tamburro A. M.; Quaglino D.; Mithieux S.; Weiss A. S.; Pasquali Ronchetti I. Heparan sulphate interacts with tropoelastin, with some tropoelastin peptides and is present in human dermis elastic fibers. Matrix Biol. 2005, 24 (1), 15–25. 10.1016/j.matbio.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Kozel B. A.; Rongish B. J.; Czirok A.; Zach J.; Little C. D.; Davis E. C.; Knutsen R. H.; Wagenseil J. E.; Levy M. A.; Mecham R. P. Elastic fiber formation: A dynamic view of extracellular matrix assembly using timer reporters. J. Cell. Physiol. 2006, 207, 87–96. 10.1002/jcp.20546. [DOI] [PubMed] [Google Scholar]
- Tu Y. D.; Weiss A. S. Glycosaminoglycan-mediated coacervation of tropoelastin abolishes the critical concentration, accelerates coacervate formation, and facilitates spherule fusion: Implications for tropoelastin microassembly. Biomacromolecules 2008, 9 (7), 1739–1744. 10.1021/bm7013153. [DOI] [PubMed] [Google Scholar]
- Akhtar K.; Broekelmann T. J.; Song H. W.; Turk J.; Brett T. J.; Mecham R. P.; Adair-Kirk T. L. Oxidative modifications of the C-terminal domain of tropoelastin prevent cell binding. J. Biol. Chem. 2011, 286 (15), 13574–13582. 10.1074/jbc.M110.192088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax D. V.; Rodgers U. R.; Bilek M. M.; Weiss A. S. Cell adhesion to tropoelastin is mediated via the C-terminal GRKRK motif and integrin alphaVbeta3. J. Biol. Chem. 2009, 284, 28616–28623. 10.1074/jbc.M109.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekelmann T. J.; Kozel B. A.; Ishibashi H.; Werneck C. C.; Keeley F. W.; Zhang L.; Mecham R. P. Tropoelastin interacts with cell-surface glycosaminoglycans via its COOH-terminal domain. J. Biol. Chem. 2005, 280 (49), 40939–40947. 10.1074/jbc.M507309200. [DOI] [PubMed] [Google Scholar]
- Sato F.; Wachi H.; Ishida M.; Nonaka R.; Onoue S.; Urban Z.; Starcher B. C.; Seyama Y. Distinct steps of cross-linking, self-association, and maturation of tropoelastin are necessary for elastic fiber formation. J. Mol. Biol. 2007, 369 (3), 841–851. 10.1016/j.jmb.2007.03.060. [DOI] [PubMed] [Google Scholar]
- Mithieux S. M.; Wise S. G.; Raftery M. J.; Starcher B.; Weiss A. S. A model two-component system for studying the architecture of elastin assembly in vitro. J. Struct. Biol. 2005, 149 (3), 282–289. 10.1016/j.jsb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Brown-Augsburger P.; Tisdale C.; Broekelmann T. J.; Sloan C.; Mecham R. P. Identification of an elastin cross-linking domain that joins three peptide chains. Possible role in nucleated assembly. J. Biol. Chem. 1995, 270 (30), 17778–17783. 10.1074/jbc.270.30.17778. [DOI] [PubMed] [Google Scholar]
- Tamburro A. M.; Bochicchio B.; Pepe A. Dissection of human tropoelastin: Exon-by-exon chemical synthesis and related conformational studies. Biochemistry 2003, 42 (45), 13347–13362. 10.1021/bi034837t. [DOI] [PubMed] [Google Scholar]
- Tamburro A. M.; Pepe A.; Bochicchio B. Localizing alpha-helices in human tropoelastin: Assembly of the elastin ″puzzle″. Biochemistry 2006, 45 (31), 9518–9530. 10.1021/bi060289i. [DOI] [PubMed] [Google Scholar]
- Baldock C.; Oberhauser A. F.; Ma L.; Lammie D.; Siegler V.; Mithieux S. M.; Tu Y.; Chow J. Y. H.; Suleman F.; Malfois M.; Rogers S.; Guo L.; Irving T. C.; Wess T. J.; Weiss A. S. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (11), 4322–4327. 10.1073/pnas.1014280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani H.; Hirano E.; Knutsen R. H.; Shifren A.; Wagenseil J. E.; Ciliberto C.; Kozel B. A.; Urban Z.; Davis E. C.; Broekelmann T. J.; Mecham R. P. Alternative splicing and tissue-specific elastin misassembly act as biological modifiers of human elastin gene frame shift mutations associated with dominant cutis laxa. J. Biol. Chem. 2012, 287, 22055–22067. 10.1074/jbc.M111.327940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M.; Cirulis J. T.; Lee S.; Keeley F. W. Structural determinants of cross-linking and hydrophobic domains for self-assembly of elastin-like polypeptides. Biochemistry 2005, 44 (43), 14367–14375. 10.1021/bi0510173. [DOI] [PubMed] [Google Scholar]
- Djajamuliadi J.; Kagawa T. F.; Ohgo K.; Kumashiro K. K. Insights into a putative hinge region in elastin using molecular dynamics simulations. Matrix Biol. 2009, 28, 92–100. 10.1016/j.matbio.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Piontkivska H.; Zhang Y.; Green E.; Elnitski L. Multi-species sequence comparison reveals dynamic evolution of the elastin gene that has involved purifying selection and lineage-specific insertions/deletions. BMC Genomics 2004, 5 (1), 31. 10.1186/1471-2164-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. J.; Weiss A. S. Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur. J. Biochem. 1999, 266, 308–314. 10.1046/j.1432-1327.1999.00891.x. [DOI] [PubMed] [Google Scholar]
- Sreerama N.; Woody R. W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287 (2), 252–260. 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Konarev P. V.; Volkov V. V.; Sokolova A. V.; Koch M. H. J.; Svergun D. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. 10.1107/S0021889803012779. [DOI] [Google Scholar]
- Petoukhov M. V.; Konarev P. V.; Kikhney A. G.; Svergun D. I. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 2007, 40 (s1), s223–s228. 10.1107/S0021889807002853. [DOI] [Google Scholar]
- Svergun D. I.; Petoukhov M. V.; Koch M. H. J. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001, 80, 2946–2953. 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V. V.; Svergun D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003, 36, 860–864. 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25 (13), 1605–12. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Urry D. W.; Starcher B.; Partridge Sm Coacervation of solubilized elastin effects a notable conformational change. Nature 1969, 222 (5195), 795–796. 10.1038/222795a0. [DOI] [PubMed] [Google Scholar]
- Starcher B. C.; Saccomani G.; Urry D. W. Coacervation and ion-binding studies on aortic elastin. Biochim. Biophys. Acta, Protein Struct. 1973, 310 (2), 481–486. 10.1016/0005-2795(73)90132-3. [DOI] [PubMed] [Google Scholar]
- Tamburro A. M.; Guantieri V.; Daga-Gordini D.; Abatangelo G. Conformational transitions of alpha-elastin. Biochim. Biophys. Acta, Protein Struct. 1977, 492, 370–376. 10.1016/0005-2795(77)90088-5. [DOI] [PubMed] [Google Scholar]
- Lee P.; Bax D. V.; Bilek M. M. M.; Weiss A. S. A novel cell adhesion region in tropoelastin that mediates attachment to integrin alphaVbeta5. J. Biol. Chem. 2014, 289, 1467–1477. 10.1074/jbc.M113.518381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyerla J. R.; Torchia D. A. Molecular mobility and structure of elasitn deducted from the solvent and temperature dependence of 13C magnetic resonance relaxation data. Biochemistry 1975, 14, 5175–5183. 10.1021/bi00694a024. [DOI] [PubMed] [Google Scholar]
- Frushour B. G.; Koenig J. L. Raman scattering of collagen, gelatin, and elastin. Biopolymers 1975, 14, 379–391. 10.1002/bip.1975.360140211. [DOI] [PubMed] [Google Scholar]
- Prescott B.; Renugopalakrishnan V.; Thomas G. J. Raman spectrum and structure of elastin in relation to type-II β-turns. Biopolymers 1987, 26, 934–936. 10.1002/bip.360260612. [DOI] [PubMed] [Google Scholar]
- Urry D. W. What is elastin; what is not. Ultrastruct. Pathol. 1983, 4, 227–251. 10.3109/01913128309140793. [DOI] [PubMed] [Google Scholar]
- Forood B.; Feliciano E. J.; Nambiar K. P. Stabilization of alpha-helical structures in short peptides via end capping. Proc. Natl. Acad. Sci. U. S. A. 1993, 90 (3), 838–842. 10.1073/pnas.90.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn D. S.; Griffin G. L.; Senior R. M.; Mecham R. P. Characterization of biologically-active domains on elastin - identification of a monoclonal antibody to a cell recognition site. Biochemistry 1986, 25 (18), 5172–5176. 10.1021/bi00366a028. [DOI] [PubMed] [Google Scholar]
- Grosso L. E.; Scott M. Peptide sequences selected by BA4, a tropoelastin-specific monoclonal antibody, are ligands for the 67-kilodalton bovine elastin receptor. Biochemistry 1993, 32 (48), 13369–13374. 10.1021/bi00211a052. [DOI] [PubMed] [Google Scholar]
- Rodgers U. R.; Weiss A. S. Integrin alpha(v)beta(3) binds a unique non-RGD site near the C-terminus of human tropoelastin. Biochimie 2004, 86 (3), 173–178. 10.1016/j.biochi.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Senior R. M.; Griffin G. L.; Mecham R. P.; Wrenn D. S.; Prasad K. U.; Urry D. W. Val-Gly-Val-Ala-Pro-Gly, a repeating peptide in elastin, is chemotactic for fibroblasts and monocytes. J. Cell Biol. 1984, 99 (3), 870–874. 10.1083/jcb.99.3.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiznieks L. D.; Weiss A. S. Flexibility in the solution structure of human tropoelastin. Biochemistry 2007, 46 (27), 8196–8205. 10.1021/bi700139k. [DOI] [PubMed] [Google Scholar]
- Cox B. A.; Starcher B.; Urry D. W. Coacervation of tropoelastin results in fiber formation. J. Biol. Chem. 1973, 249 (3), 997–998. [PubMed] [Google Scholar]
- Vrhovski B.; Jensen S. A.; Weiss A. S. Coacervation characteristics of recombinant human tropoelastin. Eur. J. Biochem. 1997, 250, 92–98. 10.1111/j.1432-1033.1997.00092.x. [DOI] [PubMed] [Google Scholar]
- Toonkool P.; Jensen S. A.; Maxwell A. L.; Weiss A. S. Hydrophobic domains of human tropoelastin interact in a context-dependent manner. J. Biol. Chem. 2001, 276 (48), 44575–44580. 10.1074/jbc.M107920200. [DOI] [PubMed] [Google Scholar]
- Clarke A. W.; Arnspang E. C.; Mithieux S. M.; Korkmaz E.; Braet F.; Weiss A. S. Tropoelastin massively associates during coacervation to form quantized protein spheres. Biochemistry 2006, 45 (33), 9989–9996. 10.1021/bi0610092. [DOI] [PubMed] [Google Scholar]
- Miao M.; Bellingham C. M.; Stahl R. J.; Sitarz E. E.; Lane C. J.; Keeley F. W. Sequence and structure determinants for the self-aggregation of recombinant polypeptides modeled after human elastin. J. Biol. Chem. 2003, 278 (49), 48553–48562. 10.1074/jbc.M308465200. [DOI] [PubMed] [Google Scholar]
- Luan C. H.; Parker T. M.; Prasad K. U.; Urry D. W. Differential Scanning Calorimetry Studies of Nacl Effect on the Inverse Temperature Transition of Some Elastin-Based Polytetrapeptides, Polypentapeptides, and Polynonapeptides. Biopolymers 1991, 31 (5), 465–475. 10.1002/bip.360310502. [DOI] [PubMed] [Google Scholar]
- Urry D. W. The change in Gibbs free energy for hydrophobic association. Derivation and evaluation by means of inverse temperature transitions. Chem. Phys. Lett. 2004, 399 (1), 177–183. 10.1016/S0009-2614(04)01565-9. [DOI] [Google Scholar]
- Urry D. W.; Luan C. H.; Parker T. M.; Gowda D. C.; Prasad K. U.; Reid M. C.; Safavy A. Temperature of polypeptide inverse temperature transition depends on mean residue hydrophobicity. J. Am. Chem. Soc. 1991, 113 (11), 4346–4348. 10.1021/ja00011a057. [DOI] [Google Scholar]
- Urry D. W.; Trapane T. L.; Prasad K. U. Phase-structure transitions of the elastin polypentapeptide-water system within the framework of composition-temperature studies. Biopolymers 1985, 24 (12), 2345–2356. 10.1002/bip.360241212. [DOI] [PubMed] [Google Scholar]
- Cuatrecasas P.; Parikh I. Adsorbents for Affinity Chromatography - Use of N-Hydroxysuccinimide Esters of Agarose. Biochemistry 1972, 11 (12), 2291–2299. 10.1021/bi00762a013. [DOI] [PubMed] [Google Scholar]
- Mithieux S. M.; Weiss A. S., Elastin. In Advances in Protein Chemistry; Academic Press: New York, 2005; Vol. 70, pp 437–461. [DOI] [PubMed] [Google Scholar]
- Mithieux S. M.; Rasko J. E. J.; Weiss A. S. Synthetic elastin hydrogels derived from massive elastic assemblies of self-organized human protein monomers. Biomaterials 2004, 25 (20), 4921–4927. 10.1016/j.biomaterials.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Kozel B. A.; Ciliberto C. H.; Mecham R. P. Deposition of tropoelastin into the extracellular matrix requires a competent elastic fiber scaffold but not live cells. Matrix Biol. 2004, 23, 23–34. 10.1016/j.matbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Pasquali-Ronchetti I.; Baccarani-Contri M. Elastic fiber during development and aging. Microsc. Res. Tech. 1997, 38, 428–435. . [DOI] [PubMed] [Google Scholar]
- Pasquali-Ronchetti I.; Baccarani-Contri M.; Fornieri C.; Mori G.; Quaglino D. Structure and composition of the elastin fibre in normal and pathological conditions. Micron 1993, 24 (1), 75–89. 10.1016/0968-4328(93)90016-T. [DOI] [Google Scholar]
- Gotte L.; Giro M. G.; Volpin D.; Horne R. W. The ultrastructural organization of elastin. J. Ultrastruct. Res. 1974, 46, 23–33. 10.1016/S0022-5320(74)80019-5. [DOI] [PubMed] [Google Scholar]
- Annabi N.; Mithieux S. M.; Weiss A. S.; Dehghani F. The fabrication of elastin-based hydrogels using high pressure CO2. Biomaterials 2009, 30, 1–7. 10.1016/j.biomaterials.2008.09.031. [DOI] [PubMed] [Google Scholar]
- Mark J. E. Dependence of swelling of elastin on elongation, and its importance in fluorescence probe analysis. Biopolymers 1976, 15 (9), 1853–1856. 10.1002/bip.1976.360150919. [DOI] [PubMed] [Google Scholar]
- Gosline J. M. Temperature-dependent swelling of elastin. Biopolymers 1978, 17 (3), 697–707. 10.1002/bip.1978.360170312. [DOI] [PubMed] [Google Scholar]
- Trabbic-Carlson K.; Setton L. A.; Chilkoti A. Swelling and mechanical behaviors of chemically cross-linked hydrogels of elastin-like polypeptides. Biomacromolecules 2003, 4 (3), 572–580. 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- Flory P. J.; Rehner J. J. Statistical mechanics of cross-Linked polymer networks II. Swelling. J. Chem. Phys. 1943, 11 (11), 521–526. 10.1063/1.1723792. [DOI] [Google Scholar]
- Nickerson M. T.; Farnworth R.; Wagar E.; Hodge S. M.; Rousseau D.; Paulson A. T. Some physical and microstructural properties of genipin-crosslinked gelatin-maltodextrin hydrogels. Int. J. Biol. Macromol. 2006, 38 (1), 40–44. 10.1016/j.ijbiomac.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Debelle L.; Tamburro A. M. Elastin: molecular description and function. Int. J. Biochem. Cell Biol. 1999, 31 (2), 261–272. 10.1016/S1357-2725(98)00098-3. [DOI] [PubMed] [Google Scholar]
- Vrhovski B.; Weiss A. S. Biochemistry of tropoelastin. Eur. J. Biochem. 1998, 258, 1–18. 10.1046/j.1432-1327.1998.2580001.x. [DOI] [PubMed] [Google Scholar]
- Kielty C. M.; Sherratt M. J.; Shuttleworth C. A. Elastic fibres. J. Cell Sci. 2002, 115, 2817–2828. [DOI] [PubMed] [Google Scholar]
- Kelleher C. M.; Silverman E. K.; Broekelmann T. J.; Litonjua A. A.; Hernandez M.; Sylvia J. S.; Stoler J.; Reilly J. J.; Chapman H. A.; Speizer F. E.; Weiss S. T.; Mecham R. P.; Raby B. A. A functional mutation in the terminal exon of elastin in severe, early-onset chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2005, 33, 355–362. 10.1165/rcmb.2005-0206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley F. W., The Evolution of Elastin. In Evolution of Extracellular Matrix, Keeley F. W., Mecham R. P., Eds.; Springer: Berlin, 2013; pp 73–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.