ABSTRACT
DNA replication is frequently perturbed by intrinsic, as well as extrinsic, genotoxic stress. At damaged forks, DNA replication and repair activities require proper coordination to maintain genome integrity. We show here that PARI antirecombinase plays an essential role in modulating the initial response to replication stress in mice. PARI is functionally dormant at replisomes during normal replication, but upon replication stress, it enhances nascent-strand shortening that is regulated by RAD51 and MRE11. PARI then promotes double-strand break induction, followed by new origin firing instead of replication restart. Such PARI function is apparently obstructive to replication but is nonetheless physiologically required for chromosome stability in vivo and ex vivo. Of note, Pari-deficient embryonic stem cells exhibit spontaneous chromosome instability, which is attenuated by differentiation induction, suggesting that pluripotent stem cells have a preferential requirement for PARI that acts against endogenous replication stress. PARI is a latent modulator of stalled fork processing, which is required for stable genome inheritance under both endogenous and exogenous replication stress in mice.
KEYWORDS: genome stability, homologous recombination, replication stress, embryonic stem cells
INTRODUCTION
DNA replication is vulnerable to hampering by various environmental agents, as well as intrinsic cellular conditions. Impaired replication forks, if not properly processed, result in genetic instability, including DNA mutations and structural and numerical chromosome aberrations, which are detrimental to normal cellular proliferation and differentiation (1, 2). Particularly sensitive to replication perturbation are highly proliferative cells, e.g., early embryonic cells, which exhibit rapid DNA replication and cell division with a short gap (G1) phase and ineffective DNA damage checkpoints (3). Such a truncated cell cycle in early embryos is evolutionarily conserved and supposedly developmentally programmed to promptly achieve sufficient cell numbers for early embryonic morphogenesis (4, 5) but is at the same time intrinsically associated with genome instability due to high replication stress. After embryonic cell proliferation, postnatal stem cell lineages continue to self-renew and produce differentiated progeny and thus are also inherently vulnerable to replication perturbation during their lifelong proliferation (6). Such replication-associated genome instability poses potential risks of cellular transformation or cell death, which in multicellular organisms is linked to cancer predisposition, premature aging, developmental disorders, etc. (7).
To protect cells and organisms from these detrimental consequences, DNA repair mechanisms, together with cell cycle checkpoints, act to sense and restore impaired replication fork structures (8). Upon replication stalling, cell cycle checkpoint mechanisms stabilize replisomes to later resume DNA synthesis (9) and several different pathways, including replication restart by fork regression, translesion synthesis, and repriming, act to restore DNA replication at and around stalled forks (10). After prolonged replication stalling, however, replisomes collapse, followed by new origin firing (NOF) and DNA double-strand break (DSB) induction, which subsequently are repaired by homology-dependent repair or nonhomologous end-joining pathways (11). How these distinct replication recovery pathways are properly integrated at damaged forks is under active investigation.
Central to the replication fork recovery mechanisms is homology-dependent repair. At stalled replication forks, RAD51 recombinase is recruited to extended single-stranded DNA (ssDNA) stretches, where RAD51 either promotes DNA strand annealing and homologous recombination (recombinogenic function) (12) or protects newly synthesized DNA strands from exonucleolytic resection by MRE11 (non- or prerecombinogenic function) (13). RAD51 activity is intricately controlled by pro- and antirecombination factors, such as RAD52 epistasis group genes and RecQ family genes, respectively, whose functions have been well studied with regard to general DSB repair (14, 15). In contrast, much less is understood about how recombination activity is regulated at damaged replication forks in proper coordination with replication machineries (16). For instance, it is still largely unclear what specific factors and mechanisms act at damaged forks to modulate recombinogenic or nonrecombinogenic activities and how such specific regulation, if any, influences the replication fidelity and genome stability of cells. Also, the question of whether developmental-stage-specific and/or cellular-context-dependent control of replication stress response operates in multicellular organisms for proper coordination of cell proliferation, differentiation, and homeostasis remains mostly unaddressed.
Pari (also known as Parpbp) is one of the most recently identified antirecombinase genes, which is related to the Saccharomyces cerevisiae Srs2 and bacterial UvrD genes (17). Almost all other antirecombinases, such as RecQ family members (BLM, WRN, and RECQL in mammals) (15) and Srs2/UvrD-related family members (RTEL1 and FBH1 in mammals) (18, 19), are functional helicases, but PARI lacks a full repertoire of helicase motifs, discriminating it from other antirecombinase helicases. Previous studies have shown that PARI associates with PCNA, a sliding clamp for DNA polymerases (20), and it dissociates RAD51-ssDNA and PCNA-DNA polymerase δ interactions in vitro (17, 21). However, it remained to be determined how PARI actually controls DNA replication during normal, as well as damaged, fork processing. It is also unknown whether PARI has any physiological function in multicellular organisms.
In this study, we generated a Pari gene-targeted mouse line and show that Pari deficiency does not lead to immediate lethality, but upon replication stress, genome instability became evident in vivo and ex vivo. Specifically, PARI functions to reduce nascent DNA strand lengths at stalled forks and promotes DSB induction followed by NOF instead of replication restart. Such PARI function to enhance replication fork breakdown appears to be deleterious to stable replication recovery, but it is necessary for chromosome stability of proliferative cells, including somatic and germ line cells, upon replication stress in mice. Further, PARI acts to maintain the genetic stability of embryonic stem (ES) cells, but not their differentiated progeny, in the absence of exogenous replication stress, suggesting that pluripotent stem cells derived from early embryos have a developmental requirement for PARI-mediated control against endogenous replication stress to maintain their genome integrity.
RESULTS
PARI domain architecture and tissue expression in mice.
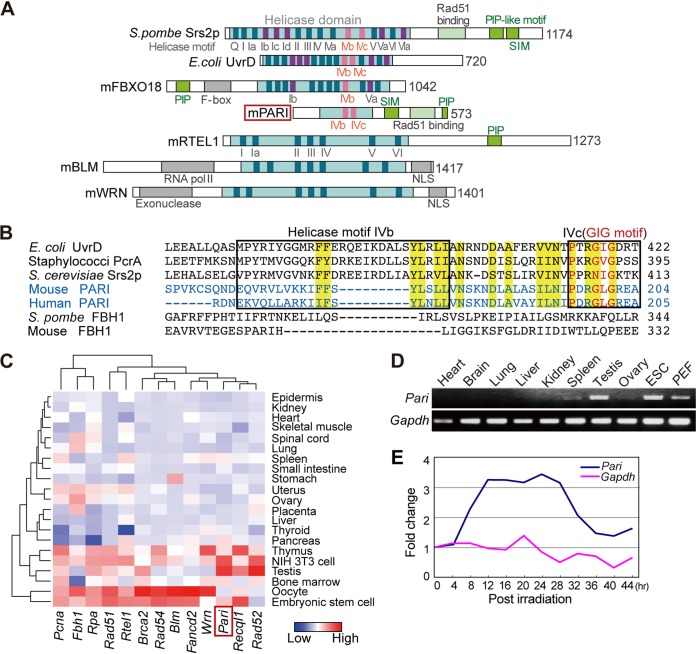
We carried out in silico screening of the mouse genome to identify putative homologues of DNA repair pathway genes in yeast and bacteria, focusing on preferential tissue expression in pluripotent stem cells and germ cells. With this criterion, 4930547N16Rik (Riken FANTOM database [22]) was shown to exhibit sequence similarity to uvrD in Escherichia coli and SRS2 in yeast. 4930547N16Rik was then found to be the orthologue of human PARI (PARPBP) during our analysis of the mouse gene in this study (17, 21, 23, 24).
Mouse PARI exhibited partial homology to helicase superfamily 1 and 2 proteins but lacked most of the conserved functional helicase motifs (Fig. 1A), suggesting that it does not possess an actual helicase activity, as was reported for human PARI. The sequence similarity of mouse PARI with other UvrD/Srs2 family members was observed in the IVb and IVc helicase motifs (25), including a PXXGXG sequence (the “GIG” motif, which binds DNA) (Fig. 1B). Mouse PARI also possesses a PCNA interaction peptide (PIP) box, a SUMO interaction motif (SIM), and a putative RAD51 binding domain, all of which are conserved in yeast Srs2 and human PARI (Fig. 1A) (17).
FIG 1.
PARI domain architecture and tissue expression in mice. (A) Protein domain architectures of PARI, Srs2, UvrD, and other selected antirecombinase helicases. Light-blue boxes, helicase domains; dark-blue boxes, highly conserved motifs common to helicase superfamilies 1 and 2; purple and pink boxes, helicase motifs uniquely found in Srs2/UvrD family members. (B) Amino acid sequence alignment of helicase motifs IVb and IVc (boxed) of mouse and human PARI and other selected Srs2/UvrD family members. The conserved GIG motif (red) is in motif IVc. Highly conserved amino acids in the Srs2-related family are highlighted in yellow. (C) Heat map representation of the mRNA expression levels of Pari and other selected recombination and replication genes in representative tissues and cells in mice. The data were extracted from a published microarray data set (GEO accession no. GSE1133). (D) RT-PCR analysis of Pari expression in selected tissues of adult mice, embryonic stem cells (ESC), and PEF. Gapdh was used as a control. (E) Quantitative RT-PCR analysis for Pari and Gapdh expression in X-ray-irradiated (20 Gy) ES cells. The data were normalized to gene expression values of control ES cells without irradiation, and fold changes of Pari and Gapdh expression are shown as means from triplicate PCRs.
The expression of Pari mRNA was detected in various tissues at basal levels in expressed sequence tag (EST) (NCBI Unigene Mm.179378) and microarray (NCBI GEO GSE1133 [26]) (Fig. 1C) data sets but was preferentially abundant in the testis, which contains highly proliferative cells. By reverse transcription (RT)-PCR, Pari mRNA was abundant in ES cells and to a lesser extent in embryonic fibroblasts, in addition to testis and spleen, among the tissues we examined (Fig. 1D). Pari expression was also regulated by DNA damage, as exemplified by its increased expression following ionizing radiation of ES cells (Fig. 1E). Radiation-induced Pari upregulation was also seen in a public transcriptome sequencing (RNA-seq) data set (PRJNA268981 [27]) of irradiated microglial cells in C56BL/6 mice (transcript per million values [n = 2]: control, 0.067 and 0.076; irradiated, 0.14 and 0.22; mean fold change, 2.5). These tissue and cell expression profiles and DNA damage-induced upregulation suggested a role of Pari in proliferative cells and DNA damage response in mice.
Pari deficiency causes preferential sensitivity to replication stress.
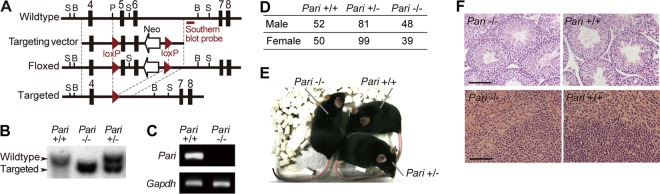
We generated a Pari gene-targeted mouse line with deletion of exons 5 and 6, which encode the conserved IVb and IVc motifs (Fig. 2A to C). Pari−/− mice were obtained in the expected Mendelian ratio and were viable and fertile (Fig. 2D and E). Under standard housing conditions, no gross pathological changes were discernible in Pari−/− mouse tissues, including testis and spleen, which expressed high levels of Pari mRNA in wild-type mice. (Fig. 2F).
FIG 2.
Generation of Pari gene-targeted mice. (A) Pari gene-targeting construct. Exons are shown as boxes, and loxP sites are shown as arrowheads. S, ScaI; B, BstXI; P, PflFI. (B) Southern blot of genomic DNA from Pari+/+, Pari+/−, and Pari−/− mice digested with BstXI. The probe position is indicated in panel A. (C) RT-PCR analysis of Pari expression in Pari+/+ and Pari−/− adult testes. Gapdh was used as a control. (D) The number of live births of each genotype obtained by crossing Pari+/− mice (44 females × 19 males; 72 litters). (E) Gross appearance of Pari+/+, Pari+/−, and Pari−/− sibling mice. (F) HE-stained sections of testes (top) and spleens (bottom) of Pari+/+ and Pari−/− mice. Scale bars, 100 μm.
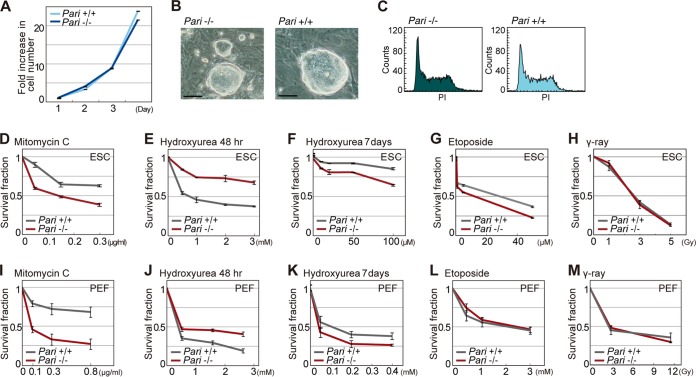
We then examined the DNA damage response in Pari-deficient cells. Pari−/− ES cells and primary embryonic fibroblasts (PEF), expanded under standard culture conditions, exhibited normal cell proliferation and cell cycle profiles compared to wild-type cells (Fig. 3A to C). However, when exogenous genotoxic agents were applied, Pari−/− ES cells and PEF both exhibited increased cellular sensitivity to hydroxyurea (HU), a replication inhibitor that depletes deoxynucleoside triphosphate (dNTP) pools, and to mitomycin C (MMC), a cross-linking agent that disturbs replication and transcription (Fig. 3D, F, I, and K). In contrast, weak or no significant responses were observed with etoposide, a topoisomerase II inhibitor, and gamma ray irradiation (Fig. 3G, H, L, and M). These results indicated that Pari deficiency provokes a preferential sensitivity to replication stress rather than general DNA damage. Of note, Pari deficiency conferred transient cellular resistance to short-term treatment with the replication inhibitor HU (Fig. 3E and J), whereas prolonged exposure resulted in increased sensitivity compared with wild-type cells (Fig. 3F and K). These results suggested that Pari deficiency temporarily suppresses cell cycle arrest or apoptosis, which are usually provoked by cell cycle checkpoints triggered by DSBs or ssDNA gaps in the short term, whereas prolonged replication stress impairs cell proliferation or survival, possibly due to accumulated genome damage in the long term.
FIG 3.
Genotoxic sensitivity of Pari-deficient cells. (A) Growth curve of Pari+/+ and Pari−/− ES cells under standard culture conditions. (B) ES cell colonies established from Pari+/+ and Pari−/− sibling blastocysts. Scale bars, 50 μm. (C) fluorescence-activated cell sorter (FACS) analysis of cell cycle profiles (DNA content stained with propidium iodide [PI]) of Pari+/+ and Pari−/− ES cells under standard culture conditions. (D to H) Cellular sensitivity of Pari+/+ and Pari−/− ES cells to mitomycin C (D), hydroxyurea (E and F), etoposide (G), and gamma rays (H). Treatment times were 24 h (D, G, and H), 48 h (E), and 7 days (F). Means ± standard errors (SE) from three independent cultures are shown. (I to M) Cellular sensitivity of Pari+/+ and Pari−/− PEF to mitomycin C (I), hydroxyurea (J and K), etoposide (L), and gamma rays (M). Treatment times were 24 h (I, L, and M), 48 h (J), and 7 days (K). Means ± SE from three independent cultures are shown.
PARI localization at replisomes and recruitment to DNA damage sites.
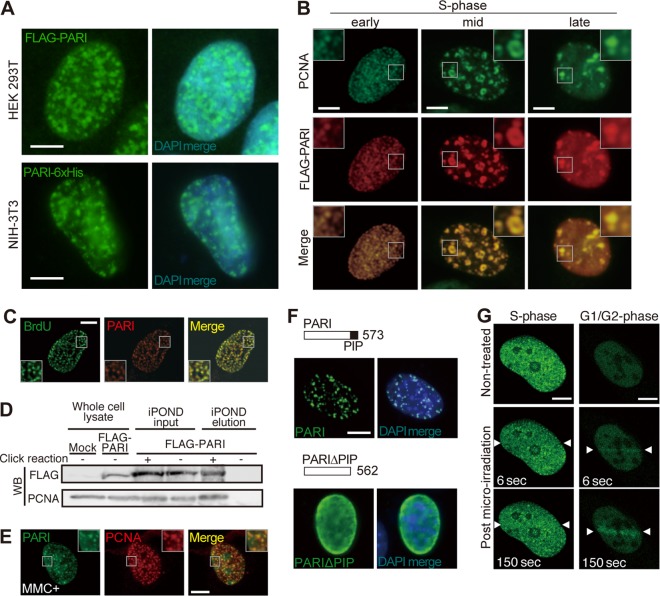
PARI interacts with PCNA, a component of replisomes, via its PIP box (17). In order to examine the subcellular localization of PARI under normal and replication stress conditions, we transiently expressed FLAG- and 6×His-tagged PARI in human 293T and mouse 3T3 cells (Fig. 4A). Both PARI constructs expressed in the two cell lines exhibited similar discrete nuclear foci, which were reminiscent of replication complexes in the S phase. These PARI foci overlapped almost completely with those of PCNA throughout the S phase (Fig. 4B) and colocalized with pulse-labeled bromodeoxyuridine (BrdU) signals (Fig. 4C), demonstrating that PARI resides at ongoing replication forks. The association of PARI with nascent replication forks was further corroborated by an iPOND (isolation of proteins on nascent DNA) analysis (28, 29) of 293T cells expressing FLAG-tagged PARI (Fig. 4D). Of note, PARI localization at replisomes (i.e., colocalization with PCNA nuclear foci) did not change in the presence of MMC (Fig. 4E), indicating that PARI stably associates with replisomes irrespective of replication stress. As expected, PARI localization at replisomes depends on the PIP box (Fig. 4F).
FIG 4.
PARI localization at replication forks and DNA damage sites. (A) HEK 293T (top) and NIH 3T3 (bottom) cells transiently transfected with pCAG-FLAG-PARI and pCAG-PARI-6×His plasmids were immunostained with anti-FLAG and anti-6×His antibodies, respectively. DAPI was used to visualize the nuclei. Scale bars, 5 μm. (B) NIH 3T3 cells transiently transfected with pCAG-FLAG-PARI immunostained for FLAG (red) and PCNA (green). Representative images of early, mid-, and late S phase cells are presented. The insets show higher-magnification views of the boxed regions. Scale bars, 5 μm. (C) NIH 3T3 cells transiently transfected with pCAG-FLAG-PARI and pulse-labeled with BrdU were stained with anti-FLAG (red) and anti-BrdU (green) antibodies. (D) iPOND analysis of 293T cells transfected with pCAG-FLAG-PARI and pulse-labeled with EdU. Nascent replication forks were captured by the click reaction, and FLAG-PARI and PCNA were detected by Western blotting (WB). The input lanes represent 2.4% of the cell lysates used for the iPOND elution. (E) NIH 3T3 cells transiently transfected with pCAG-FLAG-PARI were treated with MMC for 24 h and immunostained for FLAG (green) and PCNA (red). (F) NIH 3T3 cells transiently transfected with plasmids that express FLAG-tagged full-length PARI (top) or PIP box deletion mutant PARIΔPIP (bottom) constructs were stained with anti-FLAG antibody (green). Nuclei were counterstained with DAPI (blue). (G) U2OS cells transfected with a plasmid that expresses GFP-human PARI were laser irradiated (between arrowheads) during S phase (left) or G1/G2 phase (right), and live-cell imaging was carried out. Representative images at the indicated time points are shown. Scale bars, 5 μm.
We then conducted a laser microirradiation experiment using green fluorescent protein (GFP)-tagged human PARI in U2OS cells. In the S phase, when GFP-tagged PARI exhibited nuclear foci corresponding to replisomes, laser microirradiation did not apparently alter the localization pattern of PARI (Fig. 4G, left). In contrast, in the G1/G2 phases, when PARI was mostly diffusely distributed under unstressed conditions, rapid accumulation of PARI at laser-induced DNA damage sites was observed within a few seconds (Fig. 4G, right). This laser-induced relocalization of PARI is likely attributable to the PIP-mediated PARI-PCNA association, considering that PCNA is recruited to DNA damage sites outside the S phase (30). The reasons why PARI localization to laser-irradiated sites was not discernible in S phase cells may include the following: (i) PARI forms bright, discrete foci at replisomes, which might have masked the detection of PARI at laser-irradiated sites (31) or (ii) PCNA is not stably localized at DNA damage sites in S phase cells (30) and, accordingly, does not efficiently recruit PARI. Together, these results suggested that PARI, which primarily acts at replisomes, may also play a role outside the S phase at DNA damage sites.
PARI promotes stalled fork processing and modulates replication repair pathway choice.
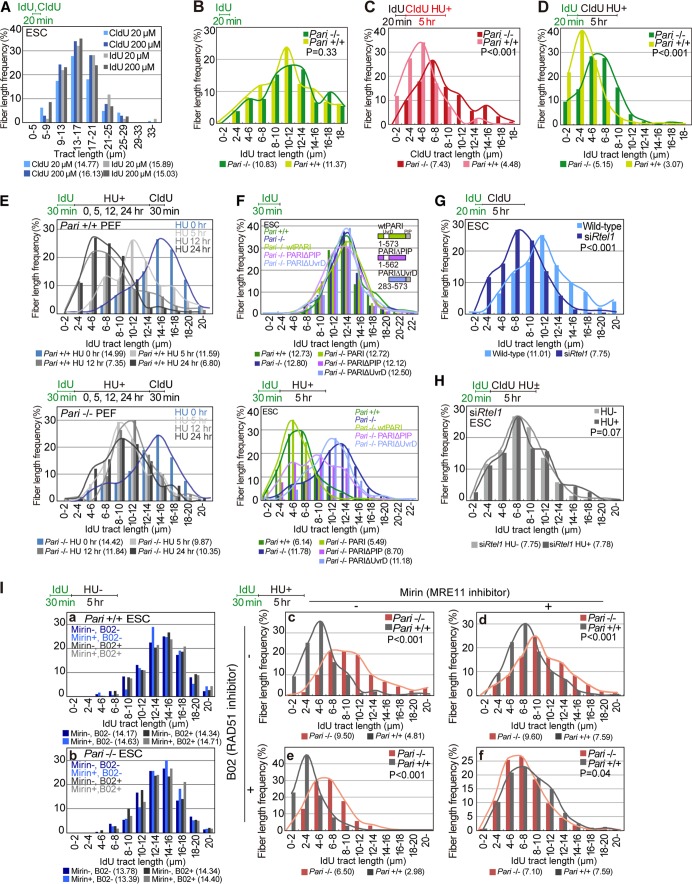
We next employed the DNA fiber assay to focus on a possible molecular function of PARI at replication forks. Pari−/− and wild-type ES cells were pulse-labeled with nucleoside analogues, iododeoxyuridine (IdU) and/or chlorodeoxyuridine (CldU), and then DNA fiber spreads were immunostained for these nucleoside analogues to measure replication tracts. In our preliminary experiments, ES cells exhibited similar rates of replication in the presence of both low and high (20 μM and 200 μM) concentrations of the nucleoside analogues (Fig. 5A). Therefore, these two concentrations were used for DNA fiber analyses (20 μM for the first label and 200 μM for the second label) in this study.
FIG 5.
PARI promotes nascent-strand processing in response to replication stress. (A) DNA fiber analyses of wild-type ES cells pulse-labeled with a low (20 μM) or high (200 μM) concentration of nucleoside analogues (IdU or CldU) for 20 min under normal culture conditions. Median tract lengths are given in parentheses below each DNA fiber graph. (B) DNA fiber analyses of replication fork progression in Pari+/+ and Pari−/− ES cells pulse-labeled with IdU (20 min) under normal culture conditions. IdU tract lengths (n > 250) are shown with P values obtained from the Mann-Whitney U test. (C and D) DNA fiber analyses of Pari+/+ and Pari−/− ES cells pulse-labeled with IdU (20 min), followed by CldU labeling in the presence of HU (5 h). CldU (C) and IdU (D) tract lengths (n > 250) are shown. (E) DNA fiber analyses of Pari+/+ and Pari−/− PEF pulse-labeled with IdU (30 min) and treated with HU for 5, 12, and 24 h or without HU treatment (0 h), followed by CldU postlabeling (30 min). IdU tract lengths (n > 300) are shown. (F) DNA fiber analyses of ES cells transiently transfected with plasmids that express FLAG-tagged full-length PARI (wtPARI), a PIP box deletion mutant (PARIΔPIP), or a UvrD homology domain deletion mutant (PARIΔUvrD). The cells were pulse-labeled with IdU (30 min) under normal culture conditions and treated with HU for 5 h (bottom) or without HU (top). IdU tract lengths (n > 300) are shown. (G) DNA fiber analyses of wild-type and Rtel1 knockdown ES cells pulse-labeled with IdU (20 min), followed by CldU labeling (5 h) under normal culture conditions. IdU tract lengths (n > 300) are shown. (H) DNA fiber analysis of Rtel1 knockdown ES cells pulse-labeled with IdU (20 min) under normal culture conditions, followed by CldU labeling in the presence or absence of HU (5 h). IdU tract lengths (n > 300) are shown. (I) Nascent-strand lengths of Pari+/+ and Pari−/− ES cells in the presence or absence of MRE11 inhibitor (30 μM mirin) and/or RAD51 inhibitor (50 μM B02) with (c to f) or without (a and b) HU for 5 h. IdU tract lengths (n > 300) are shown.
Under standard culture conditions, Pari−/− and wild-type ES cells exhibited similar rates of replication fork progression (Fig. 5B, IdU), indicating that normal replication kinetics is not disturbed by Pari deficiency. In contrast, in the presence of replication stress (HU), Pari−/− ES cells showed substantially longer replication tracts than wild-type controls (Fig. 5C, CldU). Similarly, replication tract lengths that were pulse-labeled before HU treatment were longer in Pari−/− ES cells than in wild-type cells (Fig. 5D). These observations indicated that PARI reduces nascent-strand lengths following replication stress, possibly through (i) the enhancement of nascent-strand degradation or (ii) the suppression of replication fork progression in response to replication stress. In addition to ES cells, we examined replication fork kinetics of Pari−/− and wild-type PEF and observed that nascent-strand lengths were again more stable in Pari−/− PEF than in wild-type controls in the presence of HU (Fig. 5E). Of note, wild-type ES cells and PEF were different in that it took longer for PEF to exhibit a significant difference in nascent-strand shortening (12 to 24 h) than ES cells (1 to 5 h) under replication stress (Fig. 5D and E and 6C). One explanation for this is that the kinetics of replication stress response is different between ES cells and PEF, which have distinct proliferative capacities and cell cycle structures.
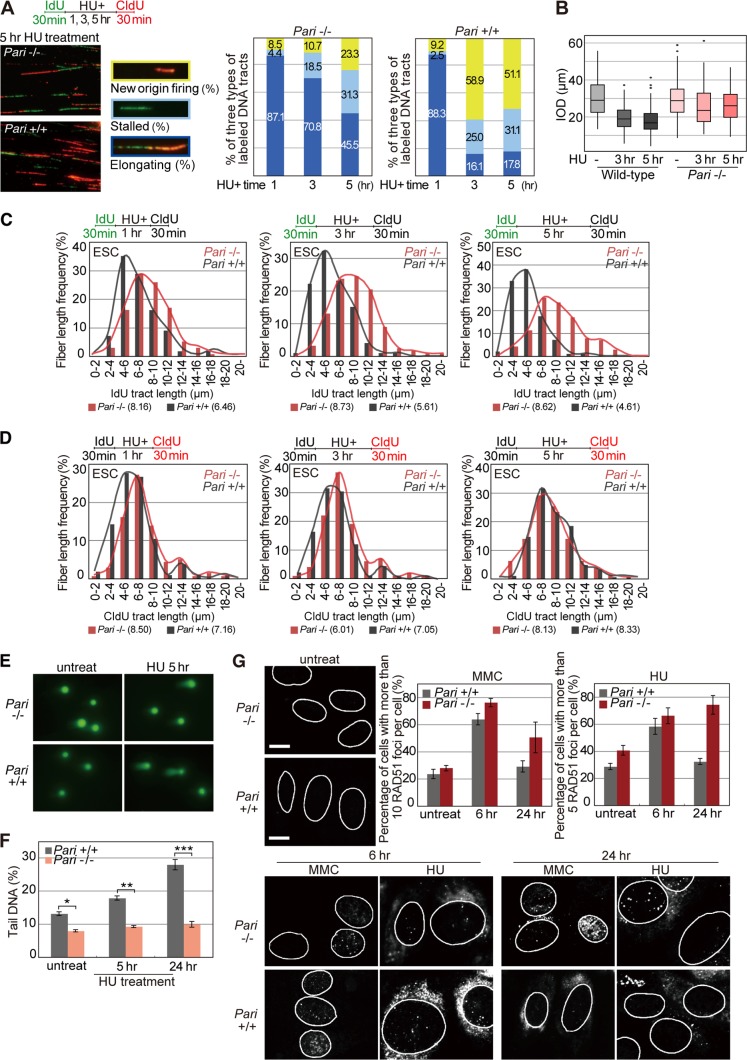
FIG 6.
PARI controls the pathway choice of replication recovery. (A) Pari+/+ and Pari−/− ES cells pulse-labeled with IdU and treated with HU for 1, 3, and 5 h, followed by CldU postlabeling. Representative images of DNA fibers of Pari+/+ and Pari−/− ES cells treated with HU for 5 h and relative frequencies of three types of labeled tracks, i.e., stalled forks (IdU only), fork elongation (IdU plus CldU), and new origin firing (CldU only), are shown. The data were obtained from >300 labeled tracts/sample. P values between Pari+/+ and Pari−/− ES cells calculated by Pearson's chi-square test were 0.65 for HU treatment for 1 h and <0.001 for HU treatment for 3 and 5 h. (B) IOD of Pari+/+ and Pari−/− ES cells pulse-labeled with IdU and treated with HU for 3 and 5 h, followed by CldU postlabeling. The data were obtained from >100 labeled tracts/sample. (C and D) ldU (C) and CldU (D) tract lengths of the same experiment shown in panel A. These data represent DNA tract lengths (n > 250) of Pari+/+ and Pari−/− ES cells before (C) and after (D) HU treatment for 1, 3, and 5 h. Median tract lengths are given in parentheses. (E and F) Neutral comet assay of Pari+/+ and Pari−/− ES cells with or without HU treatment for 5 and 24 h. Representative images (E) and tail DNA percentages measured from >100 cells/sample (F) are shown. *, P < 1E−4; **, P < 1E−8; ***, P < 1E−9 (Mann-Whitney U test). (G) Immunofluorescence staining for RAD51 of untreated Pari+/+ and Pari−/− PEF (top left) or Pari+/+ and Pari−/− PEF treated with MMC or HU for 6 and 24 h (bottom). Nuclei are outlined in white. (Top right) Percentages (±SE) of Pari+/+ and Pari−/− PEF with >10 (MMC) and >5 (HU) RAD51 foci in the nuclei. Scale bars, 10 μm.
We then carried out add back experiments in Pari−/− ES cells with full-length PARI (wtPARI), PARI lacking the PIP box (PARIΔPIP), and PARI lacking the UvrD homology domain (PARIΔUvrD) (Fig. 5F). Under standard culture conditions, the expression of these three constructs in Pari−/− ES cells did not affect replication tract lengths (Fig. 5F, top). In the presence of HU, on the other hand, wtPARI expression in Pari−/− ES cells restored nascent-strand shortening, indicating that the knockout phenotype was rescued (Fig. 5F, bottom). In contrast, PARIΔUvrD failed to restore the mutant phenotype of Pari−/− ES cells, whereas PARIΔPIP exhibited intermediate phenotype recovery (Fig. 5F, bottom), suggesting that the UvrD homology domain is necessary and the PIP box is partially required for PARI function to regulate nascent-strand shortening in response to replication stress. A plausible explanation for the intermediate phenotype of PARIΔPIP could be that PARI lacking the PIP box still binds and inhibits RAD51 (and possibly other targets) that acts to protect nascent strands (see below).
The activity of PARI, which is functionally latent at replisomes during normal replication but acts to reduce nascent-strand lengths at stalled forks, is distinct from previously reported functions of most other antirecombinases that also localize at replisomes, such as RTEL1. By RNA interference (RNAi) knockdown of Rtel1 in ES cells, replication fork speed was slowed down under unperturbed culture conditions, as previously reported (Fig. 5G) (32), but nascent-strand lengths were not further diminished in the presence of HU (Fig. 5H). Thus, PARI and RTEL1, both belonging to the Srs2-related family (17, 18), play distinct roles in normal replication fork progression and stalled fork processing.
Previous studies have shown that nascent DNA strands are resected by MRE11 nuclease following replication fork stalling (33). The degradation of newly synthesized DNA by MRE11 is alleviated by the recombination-independent function of RAD51, which covers ssDNA gaps at stalled forks (34). To see whether PARI operates in the RAD51-MRE11 pathway, we carried out DNA fiber experiments in combination with chemical inhibitors of MRE11 and RAD51, mirin (35) and B02 (36), respectively (Fig. 5I). Mirin inhibits MRE11 by binding close to the exonuclease active site, and B02 associates with RAD51 to suppress RAD51-ssDNA nucleofilament formation. Both mirin and B02 did not interfere with normal replication tracts without HU in both Pari−/− and Pari+/+ ES cells (Fig. 5I, panels a and b).
In wild-type ES cells, nascent-strand resection by HU was attenuated by MRE11 inhibition by mirin and enhanced by RAD51 inhibition by B02, in accord with previous reports (Fig. 5I, panels c to e) (34, 37). When both MRE11 and RAD51 were inhibited in wild-type ES cells by simultaneous treatment with mirin and B02, nascent-strand shortening by HU was suppressed and was similar to that of MRE11 inhibition (mirin) alone, suggesting that MRE11 acts downstream of RAD51 (Fig. 5I, panel f) (13, 34).
In contrast to wild-type ES cells, nascent-strand shortening in Pari−/− ES cells in the presence of HU was not significantly altered by MRE11 inhibition (mirin), and Pari−/− and Pari+/+ ES cells treated with mirin exhibited similar degrees of nascent-strand shortening upon HU treatment (Fig. 5I, panels c and d), indicating that MRE11 and PARI function in the same or overlapping pathways. RAD51 suppression by B02, on the other hand, enhanced nascent-strand shortening by HU in both Pari−/− and Pari+/+ ES cells (Fig. 5I, panel e). This was in accord with previous observations that RAD51 protects nascent DNA strands (13, 34) and that PARI counteracts RAD51 (17). Together, these results indicated that PARI regulates nascent-strand processing in the same pathway as MRE11 and RAD51 and were in accord with the presumed functional relationship wherein PARI suppresses RAD51, which inhibits MRE11 that executes nascent-strand degradation upon replication stress.
We then examined whether PARI affects the pathway choice of replication repair after release from replication stress. To this end, we followed the fate of stalled replication forks after HU treatment by using another DNA fiber assay that distinguishes between (i) NOF (CldU only), (ii) stalled forks without replication restart (IdU only), and (iii) replication restart from stalled forks (IdU plus CldU) (Fig. 6A). Following a short-term treatment of ES cells with HU (1 h), stalled replication forks resumed DNA synthesis with similar efficiencies (>85%) in Pari−/− and wild-type ES cells. In contrast, after prolonged HU exposure (3 and 5 h), Pari−/− and wild-type ES cells exhibited distinct responses. In wild-type cells, the efficiency of fork restart was substantially decreased (<20%), and instead, the frequency of NOF increased (>50%). In contrast, Pari−/− cells retained higher efficiencies of replication restart from stalled/collapsed forks (>70% and >45% after 3 and 5 h of HU treatment, respectively) than wild-type cells, while NOF frequencies remained low (Fig. 6A).
If PARI regulates the replication repair pathway choice between fork restart and NOF following replication stress, then the interorigin distances (IOD) should be different between Pari−/− and wild-type cells. Indeed, although IOD of Pari−/− and wild-type ES cells were similar under normal culture conditions (average, around 30 μm), replication stress (HU) induced a greater decrease in IOD of wild-type ES cells (41% reduction compared to the HU− condition) than in those of Pari−/− ES cells (12% reduction compared to the HU− condition) (Fig. 6B). Therefore, PARI regulates the pathway choice of replication recovery, i.e., it increases NOF rather than replication restart following replication perturbation. This function of PARI again is distinct from the previously reported roles of most other antirecombinases, such as BLM, which promotes replication restart from stalled forks (38). We then examined whether PARI modulates nascent-strand lengths after release from replication stress. However, replication tract lengths were similar in Pari−/− and wild-type ES cells after the removal of HU (Fig. 6D), suggesting that PARI does not play a significant role in nascent-strand processing following replication restart, unlike BLM (34), etc.
Pari deficiency leads to chromosome instability in response to replication stress in vivo and ex vivo.
In wild-type cells, prolonged replication stalling provokes replisome collapse and induces DSBs (11). These DSBs are processed by one-ended DSB repair or by two-ended DSB repair subsequent to NOF and replication fork merging (39). In Pari−/− ES cells, however, DSBs were not increased even after prolonged replication inhibition, as evidenced by the comet assay (Fig. 6E and F). DSB suppression (Fig. 6F), together with nascent-strand stabilization (Fig. 5C and D), in Pari−/− cells seemed to be a protective mechanism of damaged forks. However, it has been proposed that replication restart from stalled forks, involving template switching and homologous recombination, is a more complicated process than NOF-mediated replication recovery in mammalian cells, and illegitimate increase in such replication restart likely leads to the accumulation of unresolved recombination intermediates (40). In agreement with this notion, Pari−/− ES cells exhibited higher levels of RAD51 nuclear foci than wild-type cells after prolonged replication stress (HU and MMC for 24 h) (Fig. 6G). This, together with the suppression of DSBs (Fig. 6F), suggested that Pari−/− ES cells accumulate unresolved recombination intermediates after prolonged replication stress, which should then interfere with chromosome segregation in subsequent mitotic division.
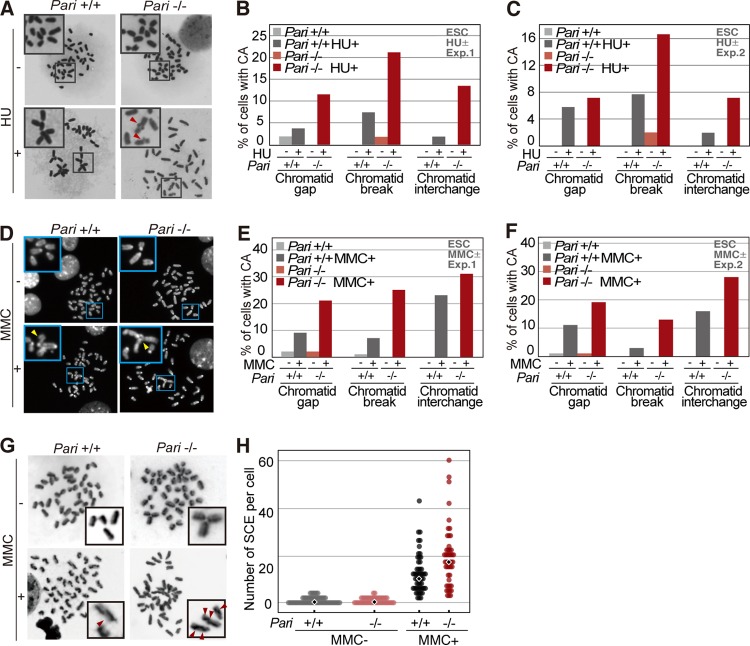
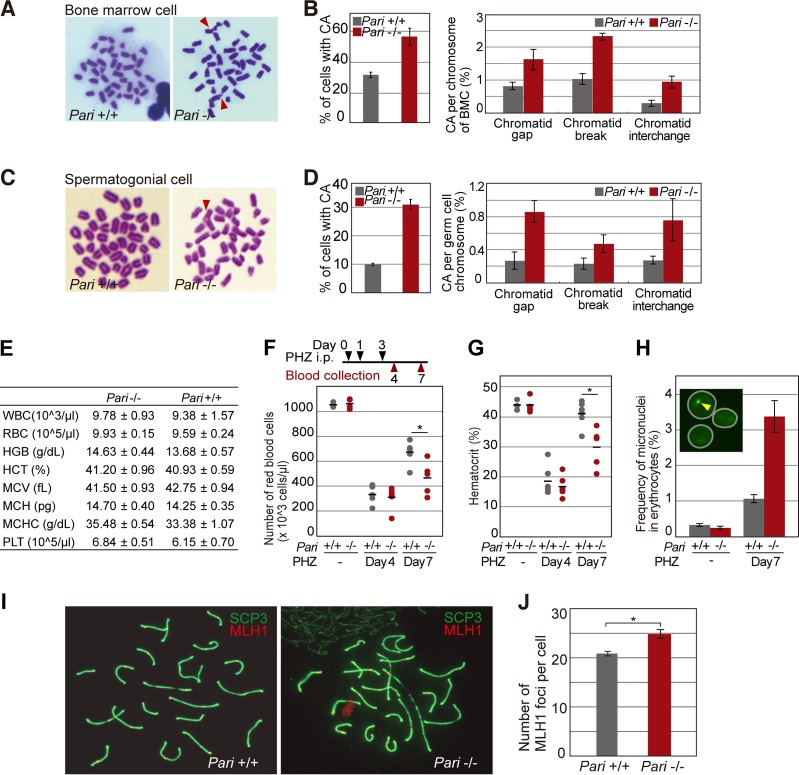
Chromosome aberrations were indeed more increased in Pari−/− ES cells than in wild-type cells in the presence of replication inhibitors (HU and MMC) (Fig. 7A to C for HU and D to F for MMC). Sister chromatid exchange (SCE), a measure of recombination repair, was also elevated in Pari−/− ES cells in response to replication stress (MMC) (Fig. 7G and H). In addition to ES cells that were expanded in vitro, we observed that chromosome aberrations were also increased in Pari−/− bone marrow cells (BMC) and spermatogonial cell populations, both of which have high proliferative capacities, after in vivo induction of replication stress by MMC in mice (Fig. 8A to D).
FIG 7.
Chromosome instability of Pari-deficient ES cells in response to replication stress. (A) Metaphase spreads (Giemsa) of Pari+/+ and Pari−/− ES cells cultured in the presence or absence of HU (2 mM). Representative images are shown, with chromosome aberrations indicated by the arrowheads. The insets are higher magnifications of the boxed regions. (B and C) Quantification (percentages) of Pari+/+ and Pari−/− ES cells at the metaphase stage with each type of chromosome aberration (chromatid gap, break, and interchange) in the presence or absence of HU. The data represent the results of two independent experiments. (D) Metaphase spreads (DAPI) of Pari+/+ and Pari−/− ES cells cultured in the presence or absence of 0.4 μg/ml MMC. Representative images are shown, with chromosome aberrations indicated by the arrowheads. The insets are higher magnifications of the boxed regions. (E and F) Quantification (percentages) of Pari+/+ and Pari−/− ES cells at the metaphase stage, with chromatid gap, break, and interchange, in the presence or absence of MMC. The data shown represent the results of two independent experiments. (G) SCE analysis of Pari+/+ and Pari−/− ES cells cultured in the presence or absence of MMC (0.4 μg/ml). (Insets) SCE sites are indicated by the arrowheads. (H) Quantification of frequencies of SCE of Pari+/+ and Pari−/− ES cells in the presence or absence of MMC from the experiment shown in panel G. The diamonds indicate median frequencies of SCE.
FIG 8.
Chromosome instability of Pari-deficient cells in response to replication stress in vivo. (A) Metaphase spreads (Giemsa) of bone marrow cells from Pari+/+ and Pari−/− mice injected with MMC (15 mg/kg). Chromosome aberrations are indicated with arrowheads. (B) Quantification (percentages) of chromosome aberrations in bone marrow cells from the experiment shown in panel A. Percentages of metaphase cells with chromosome aberrations (left) and each type of chromosome aberration (right) are shown. Means ± SE were obtained from >70 metaphases/sample from four pairs of Pari+/+ and Pari−/− mice. (C) Metaphase spreads (Giemsa) of spermatogonial cells from Pari+/+ and Pari−/− males injected with MMC (8 mg/kg). (D) Quantification (percentages) of chromosome aberrations in spermatogonial cells from the experiment shown in panel C. Percentages of metaphase cells with chromosome aberrations (left) and each type of chromosome aberration (right) are shown. Means ± SE were obtained from >70 metaphases/sample from three pairs of Pari+/+ and Pari−/− mice. (E) Hematological values in Pari+/+ and Pari−/− mice under standard housing conditions (means ± SE; n = 4). WBC, white blood cells; HGB, hemoglobin; HCT, hematocrits; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; PLT, platelets. (F and G) Numbers of red blood cells (F) and hematocrit values (G) of peripheral blood collected from Pari−/− and Pari+/+ mice treated (days 4 and 7) or not with intraperitoneal injections of PHZ (40 mg/kg) (n = 6). Each dot represents a single mouse, and the bars indicate the means. P = 0.81 (day 4) and P < 0.05 (day 7) (F) and P = 0.70 (day 4) and P < 0.01 (day 7) (G) (Mann-Whitney U test). (H) Frequencies of micronuclei in >2,000 erythrocytes in peripheral blood from Pari−/− and Pari+/+ mice treated (day 7) or not with PHZ. The data represent means ± SE (n = 3). (Inset) Representative image of erythrocytes (white circles) stained with acridine orange. The arrowhead indicates a micronucleus. (I) Surface spreads of meiotic spermatocytes from Pari+/+ and Pari−/− male mice immunostained for MLH1 (red) and SYCP3 (green). (J) Numbers of MLH1 foci (recombination nodules) on synaptonemal complexes (SYCP3) per spermatocyte from the experiment shown in panel I. Means ± SE from the results of three independent experiments are presented. Fifty spermatocytes were analyzed for each experiment. *, P < 0.001 (Mann-Whitney U test).
We then examined a possible role of Pari in vivo under physiological conditions that induce extensive cell proliferation and replication stress. As a model system to address this, we studied phenylhydrazine (PHZ)-induced acute anemia in Pari−/− and Pari+/+ mice. PHZ denatures hemoglobin and causes severe hemolytic anemia (41–43) without causing systemic defects, unlike general replication inhibitors, and thus enabled us to observe cell-type-specific (i.e., erythroid lineage) proliferation during the process of recovery from acute anemia. Pari−/− and Pari+/+ mice raised under standard conditions both showed normal hematologic indices (Fig. 8E), including red blood cell (RBC) counts and hemoglobin levels. PHZ administration (40 mg/kg of body weight on days 0, 1, and 3) induced a severe decrease in the number of RBCs and in hematocrit indices in both Pari−/− and Pari+/+ mice (Fig. 8F and G, day 4), but the subsequent recovery (Fig. 8F and G, day 7) was delayed in Pari−/− mice compared to Pari+/+ mice, suggesting that acute cell proliferation of the erythroid lineage in response to PHZ-induced anemia was impaired by Pari deficiency. In addition, we also found that the Howell-Jolly body was more frequent in Pari−/− erythrocytes than in wild-type controls following PHZ administration (Fig. 8H). The Howell-Jolly body corresponds to the micronucleus, which is derived from a chromosome(s) or its fragment(s) missegregated during cell division, and increase in the Howell-Jolly body/micronucleus frequency indicates genome damage, including replication stress. Together, these results suggested that Pari deficiency impairs the process of recovery from PHZ-induced acute anemia with an indication of chromosome segregation deficiency.
We then asked whether PARI might participate in meiosis, in which homologous recombination plays an essential role. We examined recombination nodules in meiotic spermatocytes from Pari−/− and wild-type male mice (Fig. 8I and J). Although the number of recombination nodules was slightly increased in Pari−/− spermatocytes, such a difference is occasionally seen between different strains of wild-type mice. This suggested that Pari does not play a major role, if any, in meiotic recombination control in mice. In accord with this, Pari−/− male mice exhibited normal fertility comparable to that of wild-type male mice (pregnancy rates for Pari−/− and Pari+/+ males mated with wild-type females were 0.64 and 0.67; mean litter sizes for Pari−/− and Pari+/+ mice were 6.7 and 7.3; n = 4 [Pari−/−] and 3 [Pari+/+]).
Spontaneous genome instability in Pari−/− ES cells and its attenuation by differentiation induction.
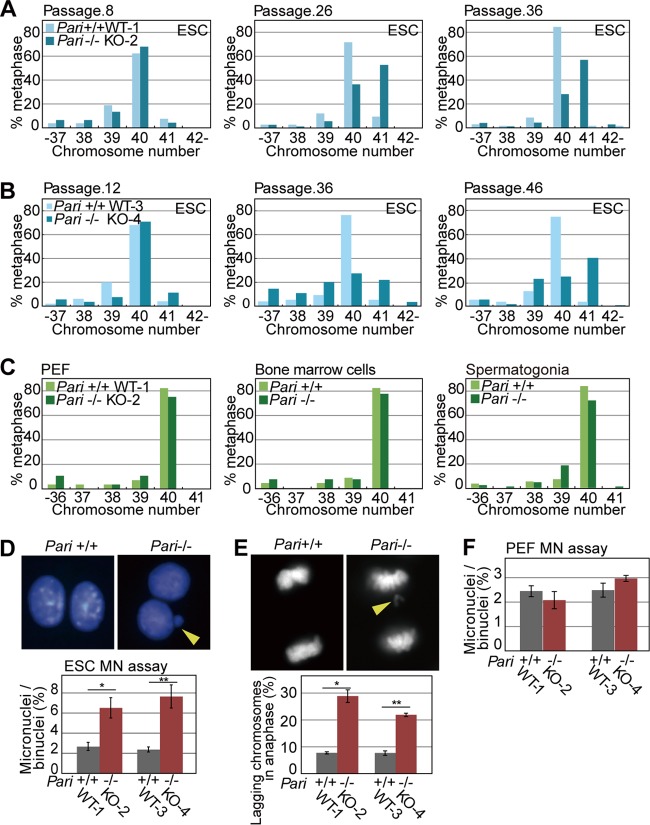
During the expansion of Pari−/− ES cells, we noticed that Pari−/− ES cells exhibited higher frequencies of spontaneous aneuploidy than wild-type controls under standard culture conditions (Fig. 9A and B). This propensity for spontaneous aneuploidy without exogenous replication stress was observed in two independently established Pari−/− ES cell lines. In contrast, other cell types we examined, including PEF expanded ex vivo and bone marrow cells and spermatogonial cells in vivo, did not exhibit such intrinsic predisposition to aneuploidy due to Pari deficiency (Fig. 9C).
FIG 9.
Spontaneous chromosome instability in Pari-deficient ES cells. (A and B) Numbers of chromosomes in Pari+/+ and Pari−/− ES cell lines at early (left), middle (middle), and late (right) passages. Pari+/+ (WT-1) and Pari−/− (KO-2) (A) and Pari+/+ (WT-3) and Pari−/− (KO-4) (B) ES cell lines were established from two independent pairs of sibling blastocysts. (C) Numbers of chromosomes in PEF (left) obtained from Pari+/+ and Pari−/− embryos and in bone marrow cells (middle) and spermatogonial cells (right) obtained from Pari+/+ and Pari−/− mice. (D) Cytokinesis block micronucleus (MN) assay of Pari+/+ and Pari−/− ES cells. Representative images (top) and frequencies of micronuclei in >500 binuclei per sample (bottom) are shown. ES cell lines established from two independent pairs of sibling blastocysts (Pari+/+ [WT-1] versus Pari−/− [KO-2] and Pari+/+ [WT-3] versus Pari−/− [KO-4], as shown in panels A and B) were used for the experiment. The data are means ± SE (n = 4). *, P < 0.05; **, P < 0.05 (Mann-Whitney U test). The arrowhead indicates a micronucleus. (E) Lagging-chromosome assay of Pari+/+ and Pari−/− ES cells. Representative images (top) and frequencies of anaphase cells with a lagging chromosome(s) (bottom) are shown. Fifty anaphase cells were examined per sample. The data are means ± SE (n = 4). *, P < 0.05; **, P < 0.05 (Mann-Whitney U test). The arrowhead indicates a lagging chromosome. (F) Cytokinesis block micronucleus assay of Pari+/+ and Pari−/− PEF. Frequencies of micronuclei in >500 binuclei per sample are shown. PEF expanded from two independent pairs of Pari+/+ and Pari−/− embryos (WT-1 versus KO-2 and WT-3 versus KO-4) were used for the experiment. The data are means ± SE (n = 4). P = 0.49 and P = 0.34 (Mann-Whitney U test).
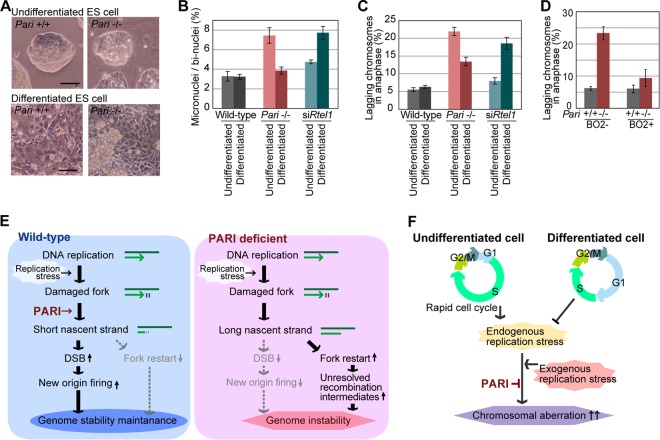
Since aneuploidy primarily arises from chromosome missegregation (44), we examined whether chromosome segregation was impaired in Pari−/− ES cells in the absence of exogenous replication stress using micronucleus formation and lagging-chromosome frequency assays (45, 46). These assays are generally more sensitive for detecting aneugenic and clastogenic effects than the chromosome aberration test, which detects only clastogenic events. In Pari−/− ES cells, spontaneous micronucleus and lagging-chromosome frequencies were both higher than in wild-type cells in the absence of exogenous replication stress (Fig. 9D and E). In contrast, no such difference was observed between Pari−/− and wild-type PEF (Fig. 9F), suggesting that ES cells are more sensitive to Pari deficiency than differentiated cells in terms of spontaneous chromosome segregation defects. Consistent with this notion, in vitro differentiation induction of Pari−/− ES cells (Fig. 10A) attenuated intrinsic chromosome segregation errors (Fig. 10B and C), indicating that PARI has a developmentally regulated function to maintain chromosome stability in undifferentiated ES cells. Of note, this preferential requirement for Pari in ES cells is not a general feature of other antirecombinases that act at replisomes. RNAi knockdown of Rtel1 resulted in an increase in chromosome segregation errors after, but not before, in vitro differentiation induction from undifferentiated ES cells (47) (Fig. 10B and C). Hence, Pari and Rtel1 play nonoverlapping and developmentally opposing functions to maintain chromosome stability in undifferentiated ES cells and their differentiated progeny, respectively.
FIG 10.
Preferential requirement for PARI function in undifferentiated ES cells to maintain chromosome stability. (A) Phase-contrast microscope images of undifferentiated Pari+/+ and Pari−/− ES cell colonies (top) and their differentiated progeny (bottom). Scale bars, 50 μm. (B) Micronucleus frequencies (percentages) of wild-type, Pari−/−, and Rtel1 knockdown (siRtel1) ES cells and their differentiated progeny; >500 binucleated cells were examined for each sample. The data are means ± SE (n = 4). (C) Frequencies of anaphase cells with a lagging chromosome(s) in wild-type, Pari−/−, and Rtel1 knockdown (siRtel1) ES cells and their differentiated progeny; 50 anaphase cells were examined for each sample. The data are means ± SE (n = 4). (D) Frequencies of lagging chromosomes in Pari+/+ and Pari−/− ES cells cultured or not with the RAD51 inhibitor B02 (50 μM) for 24 h. Fifty anaphase cells were examined per sample. The data are means ± SE (n = 4). (E) Schematic model of PARI function during DNA replication. PARI constitutively resides at replisomes but is functionally dormant during normal replication. Upon replication stress, PARI acts to reduce nascent-strand lengths through the RAD51-MRE11 pathway. PARI-DNA polymerase competition may also be involved in the nascent-strand shortening (see Discussion). Short nascent strands then promote DSB, together with NOF, and suppress the formation of unresolved recombination intermediates that interfere with proper chromosome segregation. (F) Developmental requirement for PARI function in ES cells. Undifferentiated ES cells undergo rapid self-renewal with an atypical cell cycle structure with short gap phases and inefficient checkpoints (see the introduction and Discussion). These cellular characteristics of ES cells inherently induce high endogenous replication stress and invoke an increased requirement for PARI-mediated control of stalled fork processing to ensure genome integrity.
Recent studies have shown that ES cells are under high intrinsic replication stress owing to their rapid proliferation and the lack of a G1 checkpoint (48, 49). We reasoned that spontaneous chromosome instability in Pari−/− ES cells might be a secondary consequence of intrinsic replication stress that causes illegitimate accumulation of unresolved recombination intermediates, which then interfere with normal chromosome segregation. Consistent with this, RAD51 inhibition by B02, which attenuates the Pari-deficient phenotype in the S phase (Fig. 5I), suppressed chromosome missegregation in Pari−/− ES cells (Fig. 10D). The observation indicated that chromosome instability resulting from Pari deficiency originates from the primary function of PARI to antagonize RAD51 during DNA replication, and this activity of PARI is responsible for the chromosome stability of undifferentiated ES cells.
DISCUSSION
Maintaining genetic integrity is essential for all living cells and organisms. However, genome DNA is not ensured to be stable by default but, rather, is always under attack by numerous environmental sources, as well as metabolic processes. Naturally occurring frequencies of DNA damage that arise in a mammalian cell, for example, are estimated to be tens of thousands in a day (50), necessitating highly efficient pathways of DNA repair, which, however, are not complete or accurate. Unrepaired DNA damage interferes with DNA replication, which is also frequently perturbed by intrinsic replication barriers, including DNA binding proteins, transcription machinery, and unusual DNA structures. Such impediments to DNA replication need to be resolved by proper cooperation and choice between DNA repair and replication activities, such as fork stabilization versus fork processing, depending on the types of replication damage and possibly cellular and organismal states.
In this study, we demonstrated that PARI plays a pivotal role in modulating the initial processing of stalled replication forks in mice. PARI is constitutively associated with replisomes independently of the presence or absence of replication stress, and this persistent localization of PARI at replisomes should enable PARI to perform its molecular function immediately upon replication fork perturbation (Fig. 10E). Previous biochemical studies have shown that (i) PARI displaces RAD51 from ssDNA through its antirecombinase activity (17) and (ii) PARI competes for PCNA with DNA polymerase δ at D loop recombination intermediates (21). However, the exact physiological significance of PARI during DNA replication and repair under normal, as well as stressed, conditions remained unknown. By DNA fiber analyses, we showed that PARI does not affect replication kinetics during normal cell proliferation (Fig. 5B) but reduces nascent-strand lengths (Fig. 5D) and enhances DSB (Fig. 6F) in response to replication perturbation. The nascent-strand shortening can be explained by PARI function (i) to counteract the nonrecombinogenic role of RAD51, which protects nascent strands from nucleolytic degradation (17), or (ii) to compete for PCNA with DNA polymerase δ, which extends nascent strands (21). Nascent-strand shortening enhanced by PARI should then repress replication restart from stalled forks and instead provoke NOF to complete genome duplication. PARI thereby plays a pivotal role in determining how stalled forks are processed and DNA replication is recovered upon replication stress. As mentioned above, such PARI functioning contrasts with that of other antirecombinases, which generally act to protect stalled forks. Our results show that antirecombinases exert opposing control over impaired replication forks, i.e., to enhance (PARI) or to suppress (BLM, WRN, etc. [51]) nascent-strand processing, and both activities are necessary for faithful completion of genome duplication and maintaining genetic stability.
In order to investigate the physiological function of Pari in multicellular organisms, we generated a gene-targeted mouse line. In Pari-deficient mice, chromosome instability became evident in proliferative cells, including bone marrow and spermatogenic cells, upon exposure to replication inhibitors. We also showed that Pari plays an in vivo role to ensure cell proliferation and chromosome stability during accelerated recovery from acute anemia. Such a phenotype of Pari-deficient mice is in accord with the role of PARI during DNA replication, which is latent at normal replisomes but exerts its function in response to replication stress, which arises in the presence of replication perturbation and/or during extensive cell proliferation (Fig. 10E).
We observed spontaneous chromosome instability in Pari−/− ES cells. ES cells, which are established from early preimplantation embryos, are characterized by pluripotency to differentiate into all three germ layers and have unique cell cycle properties, including a rapid cell cycle (approximately 8 to 12 h for mouse ES cells), short gap phases, and lack of the G1 checkpoint (52). These cell cycle characteristics are supposed to be developmentally programmed in early embryos, whose morphogenesis should rely on rapid and synchronized cell proliferation. Such extensive cell proliferation is inherently associated with endogenous replication stress, and indeed, ES cells exhibit elevated levels of spontaneous γH2AX signals, together with increased ssDNA breaks, in the absence of exogenous genotoxic agents (53, 54), both of which are thought to arise from intrinsic replication damage. The lack of a G1 checkpoint is possibly another cause of replication stress, since DNA damage imposed before the S phase potentially remains unrepaired and hampers DNA replication. Our results indicate that ES cells with early embryonic cell cycle profiles depend more on specific factors and mechanisms, including PARI, that act against endogenous replication stress than later differentiated progeny, which generally slow down cell proliferation rates (Fig. 10F). We propose that the functional status of PARI, and possibly other factors involved in DNA replication repair, has a significant impact on the genome stability of pluripotent stem cells, which often are expanded under highly proliferative conditions for use in both basic biological research and clinical applications.
Besides ES cells derived from early embryos, other cell types that also experience high physiological replication stress, such as cancer cells and aging stem cells, may also be prone to spontaneous chromosome instability due to Pari deficiency. In relation to this, recent studies reported that Pari expression is aberrantly upregulated in pancreatic cancer cell lines, and its RNAi knockdown reduces their cell viability (23, 24). A possible cancer-associated, as well as aging-related, phenotype(s) in Pari gene-targeted mice warrants future investigation. The mechanisms and factors underlying the genome instability associated with endogenous replication stress are still poorly understood, and their elucidation should be of fundamental importance in understanding the development, homeostasis, and diseases of long-lived organisms, including humans.
MATERIALS AND METHODS
Generation of Pari gene-targeted mice.
The Pari gene-targeting plasmid was electroporated into KY1.1 mouse ES cells (kindly provided by Junji Takeda). After G418 drug selection, three subclones identified as correctly targeted were injected into C57BL/6 × DBA/2 blastocysts. Chimeric males were mated with C57BL/6 females. To excise loxP-flanked exons 5 and 6, floxed males were mated with CMV-Cre transgenic females (55). To induce acute anemia, the mice were injected intraperitoneally with 40 mg/kg of PHZ (TCI, Japan) three times on days 0, 1, and 3. All animal experiments were performed according to the institution's ethical guidelines.
Establishment of Pari+/+ and Pari−/− ES cells.
Pari+/+ and Pari−/− sibling blastocysts were collected at 4.5 days postcoitum (dpc) from Pari+/− pregnant females mated with Pari+/− males. The zona pellucida was removed with acidic Tyrode's solution. Zona-free blastocysts were cultured in embryonic stem cell culture medium supplemented with 2i (PD032591 and CHIR99021; Axon Medchem) and LIF (Millipore) (56) on MMC-treated primary mouse embryonic fibroblasts. The inner cell masses of outgrowing blastocysts were picked up by using a glass capillary and then trypsinized and expanded to establish ES cell lines.
Pari+/+ and Pari−/− primary embryonic fibroblasts.
Pari+/+ and Pari−/− primary embryonic fibroblasts were prepared from Pari+/+ and Pari−/− sibling embryos at 13.5 dpc. Briefly, after removal of the head, limbs, and internal organs, each embryo was mechanically minced, and a culture dish was plated with the minced embryos in standard Dulbecco's modified Eagle's medium (DMEM)-based culture medium. Primary embryonic fibroblasts were passaged by trypsinization and used for experiments between passage numbers 3 and 6.
Cell culture medium.
Mouse ES cells were maintained in DMEM–F-12 medium supplemented with 15 to 20% fetal calf serum, l-glutamine, penicillin-streptomycin, sodium pyruvate, sodium bicarbonate, nonessential amino acids, LIF (Millipore), and 2i (PD032591 and CHIR99021) (56). Mouse primary embryonic fibroblasts and mouse NIH 3T3 and human HEK 293T cells were cultured in high-glucose DMEM supplemented with 10% fetal calf serum and penicillin-streptomycin at 37°C under 5% CO2.
Assay of cell sensitivity to genotoxic agents.
Pari+/+ and Pari−/− ES cells were plated in 96-well plates (5 × 103 cells/well) and cultured in the presence or absence of genotoxic agents for the times indicated in the figures. Relative cell numbers were determined by using the Cell Titer Blue assay kit (Promega) and a microplate reader (iMark; Bio-Rad). For embryonic fibroblasts, 4 × 104 cells were seeded in each well of 12-well plates. The numbers of cells were determined by using a Countess cell counter (Life Technologies). HU and etoposide were purchased from Sigma-Aldrich and MMC from Kyowa Hakko Kirin, Japan. The cells were irradiated with gamma rays from a cesium 137 source in a Gammacell 40 Exactor (Best Theratronics).
Histological examination.
Tissues were fixed in Bouin's solution, embedded in paraffin, and cut into 7-μm-thick sections. Paraffin sections were stained with hematoxylin and eosin (HE) and examined by light microscopy.
Southern blot analysis.
For Southern blot analysis, 5 μg genomic DNA was digested with restriction enzymes PflFI and BstXI. The digested DNA was separated by agarose gel electrophoresis, transferred to a Hybond N+ membrane (GE Healthcare), hybridized with [α-32P]dCTP-labeled Pari probe (Fig. 2A), and exposed to X-ray film.
RT-PCR.
Cells and tissues were lysed in TRIzol reagent (Life Technologies). Total RNA was extracted according to the manufacturer's instructions, and then, cDNA was synthesized by using a high-capacity RNA-to-cDNA kit (Life Technologies) and used as the template for RT-PCR. The PCR primers for Pari were 5′-GGGAGAGAGGCCTTCACCAACTTG-3′ and 5′-CTGTGGCCTTTGATCAGCTGCATC-3′, and those for Gapdh were 5′-ACGGCCGCATCTTCTTGTGC-3′ and 5′-CCCTTTTGGCTCCACCCTTCAAGT-3′.
Cell cycle analysis.
ES cells were trypsinized to a single-cell suspension and stained with propidium iodide in the presence of RNase (Guava cell cycle reagent kit; Millipore), and the DNA contents were analyzed by flow cytometry (Guava easyCyte flow cytometer; Millipore).
Plasmid construction and transfection.
pCAG-FLAG-PARI and pCAG-3× FLAG-PARI encode the full-length of PARI tagged with a single or triple FLAG tag, respectively. pCAG-FLAG-PARIΔPIP encodes FLAG-tagged truncated PARI lacking the C-terminal PIP box. pCAG-FLAG-PARIΔUvrD encodes FLAG-tagged truncated PARI lacking the N-terminal UvrD domain. All the plasmids were constructed by inserting cDNA fragments between ClaI and ScaI sites downstream of the CAG promoter of pCAGGS. Transient transfection was carried out using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer's instructions. Human PARI cDNA was cloned from normal human peripheral blood cells by RT-PCR. The primer pair used was 5′-TGGCTGTGTTTAATCAGAAG-3′ and 5′-TTATAGTCTAAAAAACTGAG-3′. The human PARI cDNA was subcloned into the pENTR plasmid and then fused in frame to EGFP of the destination vector by the Gateway LR reaction (Life Technologies).
Immunocytochemistry.
Cells were cultured in 8-well glass chamber slides (Nunc Lab-Tek chamber slide), fixed with 2% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X-100 in PBS, blocked with Blocking One (Nacalai Tesque, Japan), and then immunostained with primary antibodies diluted in Blocking One solution, followed by secondary antibodies conjugated with Alexa Fluor 488 or 555 (Life Technologies). The primary antibodies used were anti-FLAG polyclonal antibody (1:1,000; Sigma-Aldrich), anti-FLAG M2 monoclonal antibody (1:1,000; Sigma-Aldrich), anti-PCNA monoclonal antibody (1:500; PC10; Biolegend), anti-RAD51 (1:100; H-92; Santa Cruz), anti-BrdU antibody (1:400; BD); and anti-γH2AX (1:500; JBW301; Millipore). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole).
DNA fiber assay.
DNA fiber assays were carried out as previously described (57). Briefly, ES cells and PEF were cultured in the presence of 20 μM IdU (1st label) for the indicated periods of time, washed twice with PBS, and then cultured in the presence of 200 μM CldU (2nd label). Replication stress was induced with 2 mM HU as indicated in the figures. The cells were trypsinized and resuspended in ice-cold PBS or 70% ethanol at a concentration of 7.5 × 105 cells/ml, and then 2 μl of the cell suspension was spotted onto amino silane-coated slide glass. The cell suspension was briefly dried, and then 7 to 15 μl of lysis solution (50 mM EDTA and 0.5% SDS in 200 mM Tris-HCl, pH 7.5) was applied to the cells and gently mixed. To spread DNA fibers, the slides were tilted at 15° and air dried. DNA fibers on the slides were fixed in Carnoy's solution for 10 min and then denatured in 2.5 M HCl for 80 min. The slides were blocked with Blocking One (Nacalai Tesque) and immunostained with mouse anti-BrdU antibody (1:500; BD Biosciences; 347580) for IdU and rat anti-BrdU antibody (1:800; Abcam; ab6326) for CldU detection. The secondary antibodies used were Alexa Fluor 488- and 555-conjugated anti-mouse and -rat IgG antibodies (Life Technologies). Mirin MRE11 inhibitor and B02 RAD51 inhibitor were purchased from Sigma-Aldrich. For Rtel1 knockdown, small interfering RNAs (siRNAs) were transfected 36 h before DNA fiber experiments. Replication tract lengths were measured for 250 to 400 IdU- and/or CldU-labeled DNA fibers (1 to 3 independent experiments) using ImageJ. P values were calculated by the Mann-Whitney test.
RNA interference.
For RNA interference of Rtel1, siRNA was purchased from GE Healthcare. Nontarget siRNA was used as a control. Pari+/+ and Pari−/− ES cells were plated at a density of 1 × 106 cells per well of a 12-well plate. After 16 h, 100 fmol siRNAs was transfected with 2 μl of DharmaFect 1 reagent (Thermo Scientific) per well.
Neutral comet assay.
A neutral comet assay was carried out using a comet assay kit (Trevigen) following the manufacturer's instructions. Briefly, 500 ES cells were mixed with molten low-melting-point agarose and spread onto a comet slide. The slide was placed flat in a humid chamber at 4°C for gelation and then incubated in the lysis solution for 2 h at 4°C. Electrophoresis was carried out in neutral electrophoresis buffer (100 mM Tris-HCl and 300 mM sodium acetate at pH 9.0) for 45 min at 4°C. DNA in the gel was precipitated with 7.5 M ammonium acetate in 95% ethanol, and then the gel was dried at 37°C. The slides were stained with SYBR Gold dye (Life Technologies). Images were taken with a fluorescence microscope (BX61; Olympus) and analyzed using ImageJ software with an Open Comet plug-in (58).
Metaphase spread preparation of embryonic stem cells.
Pari+/+ and Pari−/− ES cells were cultured in the presence of 60 ng/ml colcemid for 1 h before harvest. A single-cell suspension obtained by trypsinization was treated in hypotonic solution (75 mM KCl) for 5 to 10 min, fixed with Carnoy's solution (methanol-acetic acid, 3:1), and then stored at −30°C overnight. The fixed cell suspensions were dropped onto clean glass slides and allowed to spread on a hot plate at 60°C under humidified conditions. The slides were stained with Giemsa solution and analyzed by light microscopy.
Chromosome aberration test of bone marrow cells and spermatogonial cells.
For the bone marrow chromosome aberration assay, Pari+/+ and Pari−/− mice were injected intraperitoneally with 15 mg/kg MMC 24 h and 4 mg/kg colchicine 1 h before sacrifice. Bone marrow cells were flushed out from dissected femurs with a 27-gauge needle and a 1-ml syringe and incubated in prewarmed hypotonic solution (0.9% trisodium citrate dihydrate) at 37°C for 10 to 15 min. The cells were then fixed with Carnoy's solution without resuspension for 10 min and rinsed with the fixative solution twice. Using a 26-gauge needle, the cell pellet was dispersed to a single-cell suspension. Metaphase spreads were prepared as described above. For the spermatogonial chromosome aberration test, male mice were administered 8 mg/kg MMC 24 h and 4 mg/kg colchicine 4 h before sacrifice. The tunica albuginea was removed from each testis, and the seminiferous tubules were teased apart and minced in PBS using fine forceps. The seminiferous tubule fragments were incubated in 0.5 mg/ml collagenase in PBS for 15 min at 37°C, and then spermatogonia were released into suspension by careful pipetting. The cells were rinsed with 2.2% sodium citrate (trisodium citrate) solution, followed by hypotonic treatment with 0.9% sodium citrate solution. Carnoy's fixative was added to the cells and incubated for 10 min on ice. Metaphase spreads were prepared as described above. The slides were stained with Giemsa solution and analyzed by light microscopy.
Sister chromatid exchange assay.
ES cells were cultured in the presence of 10 μM BrdU for 24 h in the dark with 0.4 μg/ml MMC added during the last 12 h and 50 ng/ml colcemid added for the last 1 h before harvest. Metaphase spreads were prepared as described above. Differential staining of sister chromatids was carried out as described previously (59). Briefly, slides were stained with Hoechst 33258, rinsed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), exposed to UV light, washed in 2× SSC, and then stained with Giemsa solution. The slides were mounted with DPX (Merck) and examined under light microscopy.
Surface spreading of meiotic chromosomes.
Seminiferous tubules of Pari+/+ and Pari−/− testes were mechanically dissected, and germ cells, including meiotic spermatocytes (as well as spermatids, etc.), were squeezed out using fine forceps. Hypotonic extraction buffer (30 mM Tris-HCl, pH 8.3, 5 mM EDTA, 1.7% sucrose, and 0.5% trisodium citrate dihydrate) was added to an equal volume of the cell suspension. The cells were then centrifuged and resuspended in 100 mM sucrose solution. A few drops of the cell suspension were spotted onto glass slides prewetted with 1% PFA and 0.15% Triton X-100 solution. The slides were then incubated in a humid chamber for 12 h at 4°C, washed with PBS, and air dried. Immunostaining was carried out with anti-SYCP3 (60), anti-MLH1 (1:100; BD Pharmingen), Alexa Fluor 488-conjugated anti-rabbit IgG, and Alexa Fluor 555-conjugated anti-mouse IgG antibodies.
Micronucleus formation assay and lagging-chromosome assay.
The cytokinesis block micronucleus formation assay was carried out as described previously (46). Briefly, cells were cultured in the presence of 4.5 μg/ml cytochalasin for 8 h (ES cells) or 20 h (PEF) in standard culture medium. After trypsinization, a single-cell suspension was treated with hypotonic solution (75 mM KCl) for 3 min and fixed with Carnoy's solution. Cells were spread on glass slides and stained with DAPI (nuclei) and fluorescein isothiocyanate (FITC) (cytoplasm). Five hundred to 1,000 binucleated cells/sample were analyzed for the presence of micronuclei under fluorescence microscopy. For the lagging-chromosome assay, cells were trypsinized and resuspended in PBS, and an equal volume of 2% PFA was added. The cell suspension was then spread on glass slides and air dried. Nuclei were stained with DAPI, and then the frequencies of lagging chromosomes in anaphase cells were scored.
In vitro differentiation of mouse ES cells.
To induce in vitro differentiation, Pari+/+ and Pari−/− ES cells were cultured in the absence of LIF, PD032591, and CHIR99021 on gelatin-coated dishes in the absence of feeder cells for 5 passages in 10 days.
Laser microirradiation experiments.
Human U2OS cells in 35-mm glass bottom culture dishes (Iwaki) were transiently transfected with the GFP-human PARI-expressing plasmid using Lipofectamine 3000 reagent (Life Technologies) according to the manufacturer's instructions. Prior to laser irradiation, the cells were treated with 10 μg/ml Hoechst 33258 for 10 min. DNA damage was introduced into the nuclei of the cells by microirradiation with a pulsed 405-nm laser (TCS-SP5; Leica), and time-lapse imaging was performed using the FRAP Wizard system (Leica). During microirradiation and imaging, the cell culture plate was maintained at 37°C and 5% CO2 using the INUH-SFB incubation system for microscopes (Tokai Hit, Japan). Cell cycle phases were determined by the subcellular localization of GFP-PARI, i.e., distinct and bright nuclear foci (S phase) or more diffuse distribution in the nucleus (G1/G2 phase).
iPOND experiments.
iPOND experiments were carried out as previously described (28, 29). Briefly, 1 × 108 293T cells were transiently transfected with the pCAG-FLAG-PARI plasmid and pulse-labeled with 20 μM EdU (Thermo Scientific) for 30 min. The cells were cross-linked with 1% PFA and 2 mM DSP (Thermo Scientific) for 10 min, and then, the fixatives were quenched with 125 mM glycine and fetal calf serum. The cross-linked cells were permeabilized with 0.25% Triton-X for 30 min, and then a click reaction was carried out using 100 μM biotin-polyethylene glycol 4-azide (Thermo Scientific) in 10 mM sodium-l-ascorbate and 5 mM CuSO4 solution. The cells were lysed in 1% SDS, 50 mM Tris-HCl, pH 8.0, and sonicated with an ultrasonicator (S220; Covaris). After centrifugation, protease inhibitor cocktail (Roche) was added to the supernatant, incubated with streptavidin-agarose resin beads (Novagen), and washed with PBS, followed by a high-stringency wash with 1 M NaCl and 0.5% SDS in 25 mM Tris-HCl, pH 8.0, solution. Then, the beads were eluted with standard SDS-PAGE loading buffer for 25 min at 95°C. SDS-PAGE and Western blotting were carried out according to standard procedures using anti-FLAG polyclonal antibody (1:2,000; Sigma-Aldrich) and anti-PCNA monoclonal antibody (1:1,000; PC10; Biolegend).
Hematological analysis.
Peripheral blood from 14- to 16-week-old mice was collected from the tail in EDTA-coated vials (Terumo, Japan), and blood cell parameters were measured with a Celltac analyzer (Nihon Kohden, Japan). To quantify micronuclei in erythrocytes, blood smears were stained with 0.007% acridine orange, and more than 2,000 erythrocytes were analyzed.
ACKNOWLEDGMENTS
We thank T. Tanaka and E. Kawase for technical assistance with gene targeting and ES cell establishment experiments. We are grateful to all the members of our laboratory for kind support and encouragement.
This study was funded by Grants-in-Aid from MEXT/JSPS, Japan, to S.C. (JP25112004).
A.L.M. conducted most of the experiments in collaboration with A.K., E.H., M.H., E.M., N.N., and S.C. G.K. and H.W. performed blastocyst injection and generated chimeric mice. M.I. performed laser microirradiation experiments. A.M. and M.T. analyzed the data on DNA replication and DNA damage response. A.L.M. and S.C. designed the research and wrote the paper.
We declare no conflict of interest.
REFERENCES
- 1.Aguilera A, Gómez-González B. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 3.Tichy ED, Stambrook PJ. 2008. DNA repair in murine embryonic stem cells and differentiated cells. Exp Cell Res 314:1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mac Auley A, Werb Z, Mirkes PE. 1993. Characterization of the unusually rapid cell cycles during rat gastrulation. Development 117:873–883. [DOI] [PubMed] [Google Scholar]
- 5.Solter D, Škreb N, Damjanov I. 1971. Cell cycle analysis in the mouse EGG-cylinder. Exp Cell Res 64:331–334. doi: 10.1016/0014-4827(71)90084-X. [DOI] [PubMed] [Google Scholar]
- 6.Adams PD, Jasper H, Rudolph KL. 2015. Aging-induced stem cell mutations as drivers for disease and cancer. Cell Stem Cell 16:601–612. doi: 10.1016/j.stem.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 9.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 10.Mourón S, Rodriguez-Acebes S, Martínez-Jiménez MI, García-Gómez S, Chocrón S, Blanco L, Méndez J. 2013. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol 20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 11.Petermann E, Helleday T. 2010. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 12.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. 2005. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol 25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helleday T. 2003. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res 532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Croteau DL, Popuri V, Opresko PL, Bohr VA. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem 83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundin C, Schultz N, Arnaudeau C, Mohindra A, Hansen LT, Helleday T. 2003. RAD51 is involved in repair of damage associated with DNA replication in mammalian cells. J Mol Biol 328:521–535. doi: 10.1016/S0022-2836(03)00313-9. [DOI] [PubMed] [Google Scholar]
- 17.Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD. 2012. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J Cell Biol 186:655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moldovan G-L, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Burkovics P, Dome L, Juhasz S, Altmannova V, Sebesta M, Pacesa M, Fugger K, Sorensen CS, Lee MY, Haracska L. 2016. The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res 44:3176–3189. doi: 10.1093/nar/gkw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y, FANTOM Consortium, RIKEN Genome Exploration Research Group Phase I and II Team . 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 23.Piao L, Nakagawa H, Ueda K, Chung S, Kashiwaya K, Eguchi H, Ohigashi H, Ishikawa O, Daigo Y, Matsuda K, Nakamura Y. 2011. C12orf48, termed PARP-1 binding protein, enhances poly(ADP-ribose) polymerase-1 (PARP-1) activity and protects pancreatic cancer cells from DNA damage. Genes Chromosomes Cancer 50:13–24. doi: 10.1002/gcc.20828. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor KW, Dejsuphong D, Park E, Nicolae CM, Kimmelman AC, D'Andrea AD, Moldovan GL. 2013. PARI overexpression promotes genomic instability and pancreatic tumorigenesis. Cancer Res 73:2529–2539. doi: 10.1158/1538-7445.AM2013-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JY, Yang W. 2006. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li MD, Burns TC, Kumar S, Morgan AA, Sloan SA, Palmer TD. 2015. Aging-like changes in the transcriptome of irradiated microglia. Glia 63:754–767. doi: 10.1002/glia.22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirbu BM, Couch FB, Cortez D. 2012. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. 2005. Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol 25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis AJ, Chi L, So S, Lee K-J, Mori E, Fattah K, Yang J, Chen DJ. 2014. BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res 42:11487–11501. doi: 10.1093/nar/gku824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. 2013. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 33.Ying S, Hamdy FC, Helleday T. 2012. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res 72:2814–2821. doi: 10.1158/1538-7445.AM2012-2814. [DOI] [PubMed] [Google Scholar]
- 34.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee J-H, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. 2008. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol 5:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. 2011. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem Biol 6:628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies SL, North PS, Hickson ID. 2007. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol 14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 39.Petermann E, Luis Orta M, Issaeva N, Schultz N, Helleday T. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gelot C, Lopez BS. 2015. Replication stress in mammalian cells and its consequences for mitosis. Genes 6:267–298. doi: 10.3390/genes6020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itano HA, Hirota K, Hosokawa K. 1975. Mechanism of induction of haemolytic anaemia by phenylhydrazine. Nature 256:665–667. doi: 10.1038/256665a0. [DOI] [PubMed] [Google Scholar]
- 42.Paulson RF, Shi L, Wu D-C. 2011. Stress erythropoiesis: new signals and new stress progenitor cells. Curr Opin Hematol 18:139. doi: 10.1097/MOH.0b013e32834521c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farres J, Llacuna L, Martin-Caballero J, Martínez C, Lozano J, Ampurdanés C, López-Contreras A, Florensa L, Navarro J, Ottina E. 2015. PARP-2 sustains erythropoiesis in mice by limiting replicative stress in erythroid progenitors. Cell Death Differ 22:1144. doi: 10.1038/cdd.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fenech M. 2000. The in vitro micronucleus technique. Mutat Res 455:81–95. doi: 10.1016/S0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 46.Fenech M. 1993. The cytokinesis-block micronucleus technique: a detailed description of the method and its application to genotoxicity studies in human populations. Mutat Res 285:35–44. doi: 10.1016/0027-5107(93)90049-L. [DOI] [PubMed] [Google Scholar]
- 47.Ding H, Schertzer M, Wu XL, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, Tam PPL, Nagy A, Lansdorp PM. 2004. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 48.Burdon T, Smith A, Savatier P. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 12:432–438. doi: 10.1016/S0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 49.Ahuja AK, Jodkowska K, Teloni F, Bizard AH, Zellweger R, Herrador R, Ortega S, Hickson ID, Altmeyer M, Mendez J. 2016. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat Commun 7:10660. doi: 10.1038/ncomms10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindahl T, Barnes DE. 2000. Repair of endogenous DNA damage. Cold Spring Harbor Symp Quant Biol 65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 51.Sidorova JM, Kehrli K, Mao F, Monnat R Jr. 2013. Distinct functions of human RECQ helicases WRN and BLM in replication fork recovery and progression after hydroxyurea-induced stalling. DNA Repair 12:128–139. doi: 10.1016/j.dnarep.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White J, Dalton S. 2005. Cell cycle control of embryonic stem cells. Stem Cell Rev 1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 53.Chuykin IA, Lianguzova MS, Pospelova TV, Pospelov VA. 2008. Activation of DNA damage response signaling in mouse embryonic stem cells. Cell Cycle 7:2922–2928. doi: 10.4161/cc.7.18.6699. [DOI] [PubMed] [Google Scholar]
- 54.Banath J, Banuelos C, Klokov D, MacPhail S, Lansdorp P, Olive P. 2009. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res 315:1505–1520. doi: 10.1016/j.yexcr.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Schwenk F, Baron U, Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23:5080. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453:519-U5. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwab RA, Niedzwiedz W. 2011. Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J Vis Exp 56:e3255. doi: 10.3791/3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gyori BM, Venkatachalam G, Thiagarajan P, Hsu D, Clement M-V. 2014. OpenComet: an automated tool for comet assay image analysis. Redox Biol 2:457–465. doi: 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolff S, Perry P. 1974. Differential Giemsa staining of sister chromatids and the study of sister chromatid exchanges without autoradiography. Chromosoma 48:341–353. doi: 10.1007/BF00290991. [DOI] [PubMed] [Google Scholar]
- 60.Chuma S, Nakatsuji N. 2001. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev Biol 229:468–479. doi: 10.1006/dbio.2000.9989. [DOI] [PubMed] [Google Scholar]