ABSTRACT
Mechanistic target of rapamycin complex 1 (MTORC1) is a critical negative regulator of general autophagy. We hypothesized that MTORC1 may specifically regulate autophagic clearance of damaged mitochondria. To test this, we used cells lacking tuberous sclerosis complex 2 (TSC2−/− cells), which show constitutive MTORC1 activation. TSC2−/− cells show MTORC1-dependent impaired autophagic flux after chemical uncoupling of mitochondria, increased mitochondrial-protein aging, and accumulation of p62/SQSTM1-positive mitochondria. Mitochondrial autophagy (mitophagy) was also deficient in cells lacking TSC2, associated with altered expression of PTEN-induced putative kinase 1 (PINK1) and PARK2 translocation to uncoupled mitochondria, all of which were recovered by MTORC1 inhibition or expression of constitutively active forkhead box protein O1 (FoxO1). These data prove the necessity of intact MTORC1 signaling to regulate two synergistic processes required for clearance of damaged mitochondria: (i) general autophagy initiation and (ii) PINK1/PARK2-mediated selective targeting of uncoupled mitochondria to the autophagic machinery.
KEYWORDS: MTORC1, PINK1, TSC2, autophagy, mitophagy, rapamycin
INTRODUCTION
Macroautophagy, referred to here as autophagy, is an essential process for cellular homeostasis. Autophagy is a defense mechanism against acute stress, such as starvation or infection (1, 2). It is also fundamental under basal conditions, where it carries out cellular quality control by removing damaged proteins or organelles (3).
Multiple signaling pathways regulate this process, which ultimately leads to the engulfment of the cargo to be degraded in a double membrane compartment (autophagosome) that fuses with lysosomes to form autolysosomes with degradative capacity.
One of the best-described nodes for autophagy regulation is mechanistic target of rapamycin complex 1 (MTORC1). Inhibition of MTORC1 is a potent proautophagic stimulus (4, 5), in part due to its interaction with the complex formed by UNC-51-like kinase 1/2 (ULK1/2), ATG13, and FIP200 (6–8). We and others have shown that chronic hyperactivation of MTORC1, by loss of tuberous sclerosis complex proteins 1/2 (TSC1/2) (9, 10), causes accumulation of autophagic substrates (11) and autophagic impairment upon nutrient starvation (12, 13). However, whether autophagy induction is also impaired in cells lacking TSC2 (TSC2−/− cells) by other cellular stressors has been less well studied. For example, cargo-selective autophagy can also occur under nutrient-rich conditions. In addition, TSC2/MTORC1 may be important in the regulation of the specific autophagy of mitochondria (mitophagy) (12, 14–16), but the underlying molecular mechanism is not clear. Here, we describe the essential role of TSC2/MTORC1 in general autophagy induction after acute uncoupling of oxidative phosphorylation by carbonyl cyanide m-chlorophenyl hydrazone (CCCP), as well as its importance in the regulation of the PTEN-induced putative kinase 1 (PINK1)/parkin (PARK2) axis that enables the targeting of uncoupled mitochondria for autophagic degradation.
RESULTS
TSC2-deficient cells show impaired CCCP-induced autophagic flux.
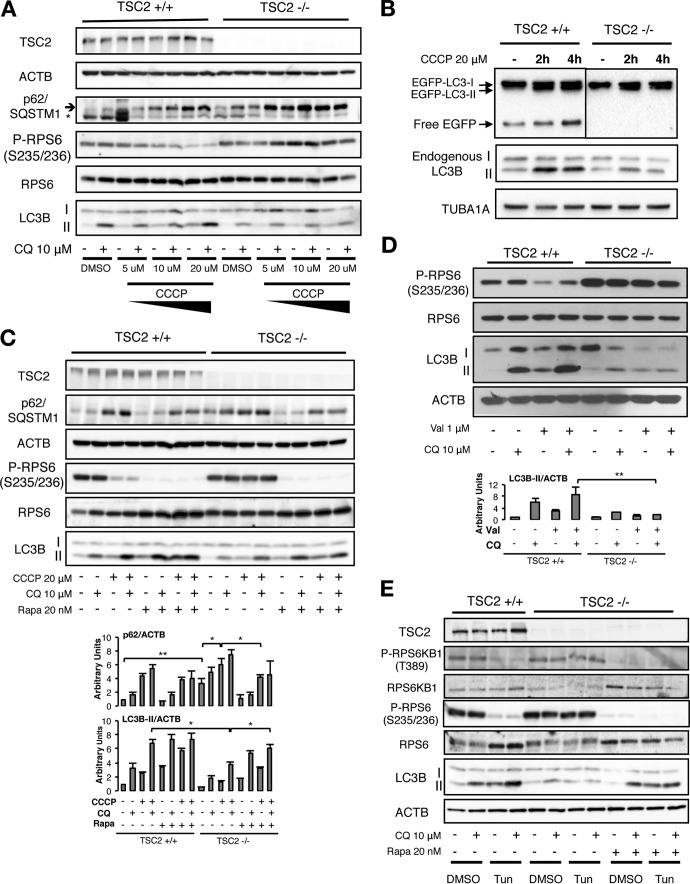
Knockout of TSC2 in pancreatic β cells causes MTORC1 hyperactivation and a parallel accumulation of damaged mitochondria with collapsed mitochondrial membrane potential (Δψm) (12). Based on this result, we hypothesized that TSC2/MTORC1 is critical for normal mitochondrial turnover. We used CCCP to chemically uncouple oxidative phosphorylation and induce mitophagy (17, 18). TSC2−/− and control cells did not show differences in CCCP sensitivity, lacking mitochondrial uptake of the Δψm-sensitive probe MitoTracker at any of the time points and CCCP doses used in the study (data not shown; see Fig. S1 in the supplemental material). Increasing doses of CCCP reduced MTORC1 activity, as indicated by less ribosomal protein S6 (RPS6) phosphorylation, in control but not TSC2−/− cells (Fig. 1A). Next, we monitored autophagic flux by treatment with the lysosomotropic autophagic inhibitor chloroquine (CQ) and found that CCCP stimulation resulted in increased microtubule-associated protein 1 light chain 3 beta II (LC3B-II) in control compared to TCS2−/− cells, which also showed relatively increased levels of the autophagic adaptor protein p62 sequestosome 1 (p62/SQSTM1) (Fig. 1A). Similarly, we observed impaired autolysosome activity in basal or CCCP-stimulated TSC2−/− cells stably expressing enhanced green fluorescent protein (EGFP)-LC3 (Fig. 1B). Rapamycin treatment enhanced autophagic flux in response to CCCP in both control and TSC2−/− cells (Fig. 1C), indicating the key role of MTORC1 for autophagic induction after loss of Δψm. Similar results were observed with another chemical uncoupler, valinomycin, which also reduced RPS6 phosphorylation and induced autophagy, but only in cells with intact TSC2 (Fig. 1D).
FIG 1.
MTORC1-dependent autophagy induction by CCCP. (A) Western blots from TSC2+/+ or TSC2−/− MEFs treated with different doses of the mitochondrial uncoupler CCCP for 15 h in the presence or absence of 10 μM CQ. (B) Western blots from TSC2+/+ or TSC2−/− MEFs expressing the fusion protein EGFP-LC3 stimulated with CCCP for the indicated times. As EGFP is more resistant to lysosomal hydrolases than LC3B, the appearance of free EGFP is indicative of autolysosome activity. (C) Western blots from TSC2+/+ or TSC2−/− MEFs stimulated with CCCP with or without CQ in the presence or absence of 20 nM rapamycin (Rapa), with densitometric quantification of p62/SQSTM and LC3-II. (D) Western blots from TSC2+/+ or TSC2−/− MEFs stimulated for 15 h with 1 μM valinomycin (Val) with or without CQ in the presence or absence of Rapa, with densitometric quantification of LC3-II. The values represent means and standard deviations (SD); n = 3 independent experiments. (E) Western blots from TSC2+/+ or TSC2−/− MEFs stimulated for 15 h with 2 μg/ml tunicamycin (Tun) with or without CQ in the presence or absence of Rapa. The values represent means and SD; n = 3 independent experiments. *, P < 0.05; **, P < 0.01 compared to the indicated control.
Mitochondrial uncoupling impairs cellular ATP production capability and induces an AMP-activated protein kinase (AMPK) stress response (19, 20). As AMPK activation has been shown to inhibit MTORC1 by direct phosphorylation of TSC2 and the MTORC1 scaffold protein regulatory-associated protein of MTOR (RAPTOR) (21, 22), we hypothesized that CCCP-associated MTORC1 inhibition could be AMPK dependent. However, although CCCP treatment indeed induced phosphorylation of AMPK substrates, such as acetyl coenzyme A (acetyl-CoA)-carboxylase (ACAC) Ser79 or RAPTOR Ser792 (see Fig. S2A in the supplemental material), CCCP was still able to block MTORC1 activity and induce autophagic flux in AMPK α1/α2 double-knockout (DKO) mouse embryonic fibroblasts (MEFs), which can be further increased by rapamycin (see Fig. S2B in the supplemental material). Thus, we conclude that CCCP-induced inhibition of MTORC1 activity and stimulation of autophagy is TSC2 dependent but AMPK independent.
As a similar autophagic defect was observed in TSC2−/− cells after nutrient starvation (13), we hypothesized that nutrient availability could fully explain our phenotype. However, treatment with tunicamycin (an endoplasmic reticulum [ER] stressor) in nutrient-rich medium potently inhibited MTORC1 signaling and induced autophagy and autophagic flux in control but not TSC2-deficient cells (Fig. 1E), an effect reversed by rapamycin. These data suggest that normal MTORC1 inhibition is critical for autophagy stimulation in response to stimuli beyond nutrient- or energy-related stress.
MTORC1 regulates mitochondrial aging and accumulation of mitochondrial autophagic cargo.
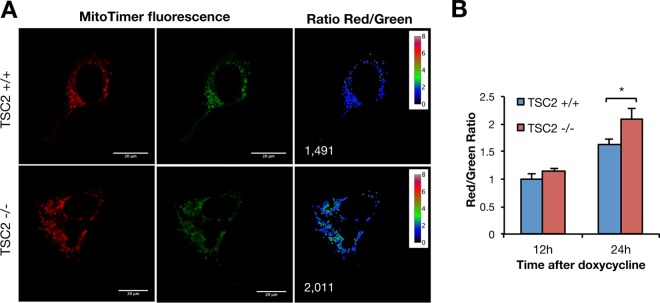
Higher basal levels of p62/SQSTM1 in TSC2−/− cells were further increased with CCCP stimulation (Fig. 1A and C). Thus, we hypothesized that MTORC1 inhibition might be necessary for loss of Δψm-induced mitochondrial turnover. Indeed, we found abundant perinuclear p62/SQSTM1 and translocase of outer mitochondrial membrane 20 (TOMM20) double-positive mitochondrial structures in TSC2−/− cells (see Fig. S3A in the supplemental material). Rapamycin treatment was able to reduce basal p62/SQSTM1 in whole-cell lysates, as well as its colocalization with mitochondria in TSC2−/− cells (see Fig. S3B). These data point to the accumulation of mitochondrial autophagic substrates as a result of chronic MTORC1 activation. To further explore this hypothesis, we utilized MitoTimer (23), which consists of mitochondrial-matrix-targeted Timer, a fluorescent protein that shifts its fluorescence over time (from green to red), to uncover mitochondrial-protein aging. After a doxycycline-induced pulse of MitoTimer, we found that TSC2−/− cells showed an increased red-to-green ratio 24 h after MitoTimer induction (Fig. 2). This result revealed increased mitochondrial-protein aging and suggested inefficient autophagic turnover of mitochondrial proteins.
FIG 2.
TSC2-deficient cells show increased mitochondrial aging. (A) Representative images of TSC2+/+ and TSC2−/− MEFs, stably expressing rtTA3, transfected with the doxycycline-inducible MitoTimer probe and then pulsed with doxycycline prior to visualization after 24 h. Ratiometric images representing the red/green ratio (MitoTimer fluorescence shifts from green to red with time) are also shown; the value of the ratio is represented as intensity and color coded as indicated in the scales. The numbers represent the specific ratios of the images shown. (B) Quantification of the red/green ratios 12 and 24 h after doxycycline pulse. The values represent means and SD (n = 3 independent experiments). *, P < 0.05 compared to the indicated control.
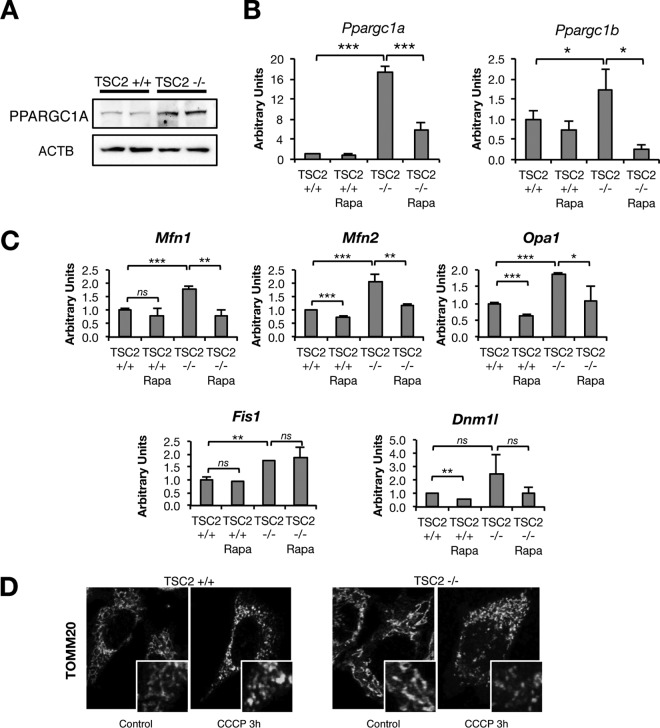
MTORC1 has also been shown to increase peroxisome proliferative activated receptor gamma coactivator 1 alpha (PPARGC1A) to regulate mitochondrial biogenesis (24, 25), which in turn is sufficient to increase expression of mitofusin 2 (MFN2), leading to mitochondrial elongation (26, 27). As alteration of mitochondrial complexity has been shown to affect autophagic clearance of damaged mitochondria (28), we evaluated the influence of MTORC1 in mitochondrial complexity as an explanation for altered mitochondrial turnover. Consistent with the above-described data, TSC2−/− cells showed a rapamycin-sensitive increase in Ppargc1a and Ppargc1b mRNA and PPARGC1A protein levels (Fig. 3A and B and data not shown). Surprisingly, genes involved in fusion (Mfn1 and Mfn2, encoding mitofusin 1/2), fission (Dnm1l and Fis1), and reorganization of cristae (Opa1, encoding optic atrophy 1) were all increased in TSC2−/− cells and partially regulated by rapamycin treatment (Fig. 3C), leading to an overall unchanged ratio of fission/fusion genes by MTORC1 hyperactivation or inhibition. Further, the rapid mitochondrial fragmentation that occurs after CCCP exposure was unchanged in TSC2−/− cells (Fig. 3D). These data suggest that an alteration in mitochondrial complexity is unlikely to explain the differences in mitochondrial-protein aging and CCCP-induced accumulation of autophagic substrates in TSC2−/− cells.
FIG 3.
MTORC1-dependent expression of genes involved in mitochondrial dynamics. (A) Western blots from TSC2+/+ and TSC2−/− MEF lysates. (B and C) qPCR analysis of TSC2+/+ or TSC2−/− MEFs treated with Rapa or vehicle for 15 h. (D) TOMM20 staining in TSC2+/+ and TSC2−/− MEFs stimulated with CCCP for 3 h. The values represent means and SD (n = 3 to 5 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared to the indicated control.
Role of TSC2/MTORC1 in CCCP-induced mitophagy.
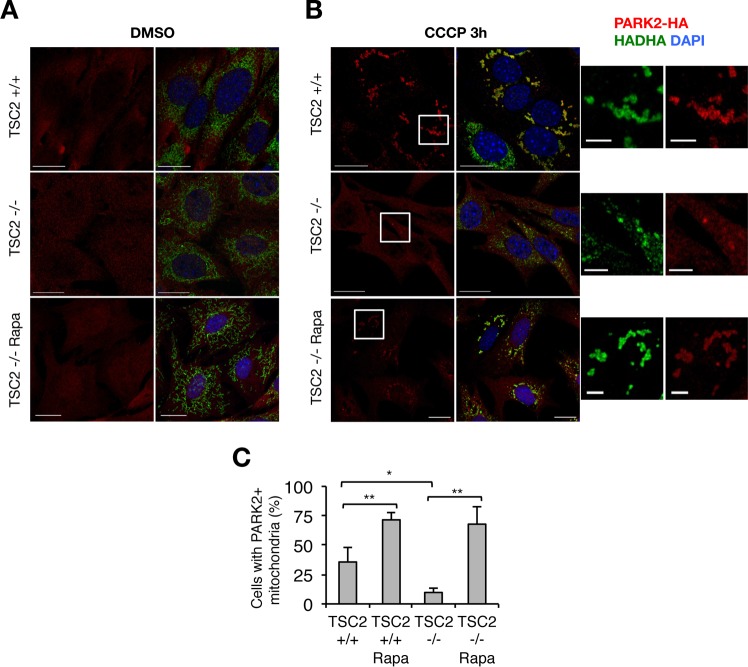
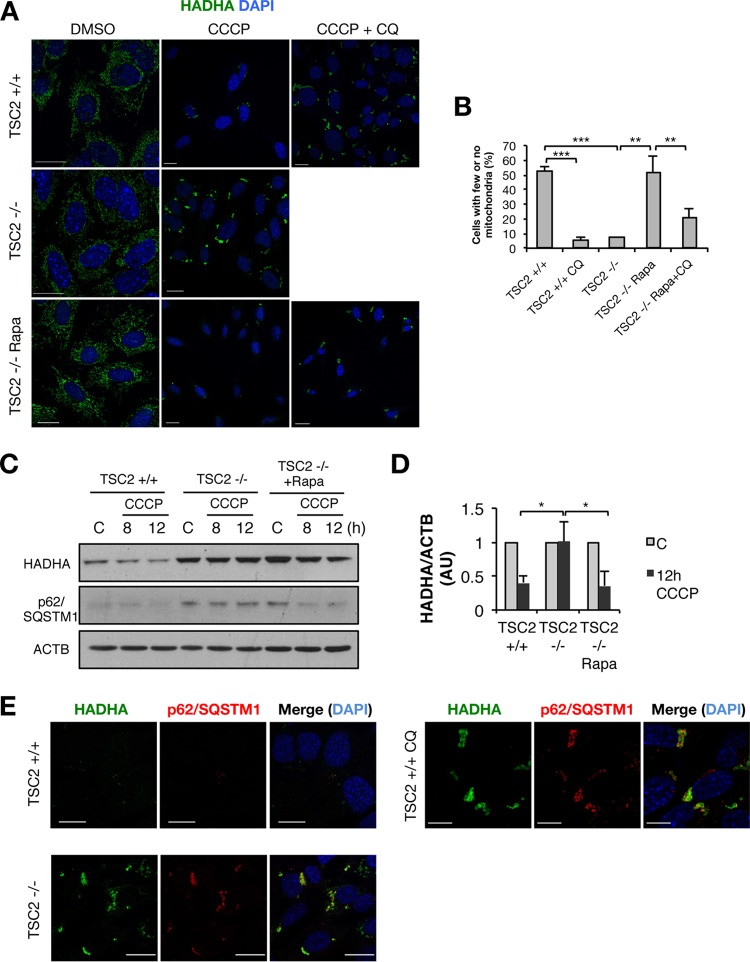
As CCCP-induced general autophagy is impaired in TSC2−/− cells, we hypothesized that mitophagy might be similarly impaired. Mitochondria with collapsed Δψm are rapidly targeted by the autophagic machinery and degraded in autolysosomes in a manner dependent on the translocation of the E3 ubiquitin ligase PARK2 to mitochondria (18). For this experiment, we took advantage of the complete dependence on mitophagy for degradation of mitochondrial matrix proteins, such as hydroxyacyl-CoA dehydrogenase (HADHA), whereas outer mitochondrial membrane (OMM) proteins, such as TOMM20, are degraded by the proteasome after mitochondrial damage in an autophagy-independent manner (29). Exogenous PARK2 expression is required to experimentally detect mitophagy in fibroblast models (30–34). We generated TSC2−/− and control cell lines stably expressing hemagglutinin (HA)-tagged PARK2; in resting cells, PARK2 localizes in the cytoplasm, and no PARK2-positive mitochondria were observed in control cells or TSC2−/− or rapamycin-treated cells (Fig. 4A). CCCP or valinomycin stimulation induced PARK2 localization to mitochondria in control cells, but to a much lesser degree in TSC2−/− cells (Fig. 4B and C; see Fig. S4A in the supplemental material), which could be rescued by rapamycin pretreatment (Fig. 4B and C). Thus, mitochondrial degradation after prolonged CCCP treatment is impaired in TSC2−/− cells or if autophagy is inhibited with chloroquine (Fig. 5A and B). Consistent with our confocal microscopy results, we observed accumulation of both p62/SQSTM1 and the mitochondrial matrix protein HADHA by Western blotting in CCCP-treated TSC2−/− cells, which was reversed by concomitant rapamycin treatment (Fig. 5C and D). A similar accumulation of p62/SQSTM1-positive mitochondrial structures in basal TSC2−/− cells, as well as in chloroquine-treated control cells, was further confirmed by confocal imaging (Fig. 5E), indicating poor clearance of mitochondria that are targeted as an autophagic substrate. Interestingly, some TSC2−/− cells showed PARK2 in mitochondria after prolonged CCCP treatment, which points to a relative as opposed to absolute defect in PARK2 translocation (see Fig. S4B in the supplemental material).
FIG 4.
Impaired PARK2 mitochondrial translocation in TSC2-deficient cells. (A and B) Representative images of PARK2-HA localization in TSC2+/+ and TSC2−/− MEFs with or without 3 h of CCCP exposure and/or 15 h of Rapa pretreatment. Bars, 20 μm; bars in magnified images, 50 μm. DMSO, dimethyl sulfoxide; DAPI, 4′,6-diamidino-2-phenylindole. (C) Quantification of cells with PARK2-positive mitochondria in CCCP-treated cells from the experiment shown in panel B. The values represent means and SD (n = 3 independent experiments). *, P < 0.05; **, P < 0.01 compared to the indicated control.
FIG 5.
Impaired mitophagy in TSC2-deficient cells. (A) PARK2-HA-expressing TSC2+/+ and TSC2−/− MEFs were stimulated with CCCP with or without CQ and/or Rapa for 20 h and then assayed for mitochondria by staining of the mitochondrial matrix protein HADHA. (B) Quantification of the percentages of cells with few or no mitochondria. (C) Western blots from PARK2-HA-expressing TSC2+/+ and TSC2−/− MEFs stimulated with CCCP with or without Rapa for the indicated times. (D) Densitometric analysis of HADHA/ACTB, represented as change from the basal value. AU, arbitrary units. (E) Representative confocal images of mitochondria (HADHA) and p62/SQSTM1 in PARK2-HA-expressing TSC2+/+ and TSC2−/− MEFs stimulated with CCCP for 20 h. The values represent means and SD (n = 3 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared to the indicated control.
MTORC1 inhibition facilitates PINK1 expression.
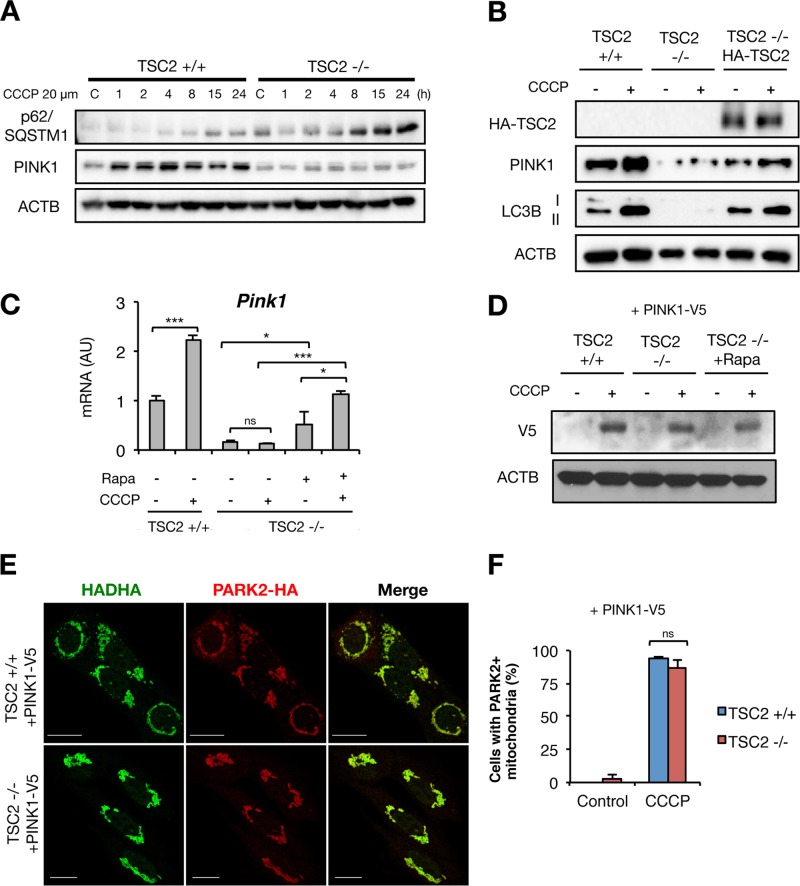
We next assessed whether altered PINK1, known to be sensitive to Δψm and critical to regulating PARK2 recruitment to the OMM (35), could explain inefficient PARK2 mitochondrial recruitment in TSC2−/− cells. As expected, CCCP treatment resulted in rapid full-length PINK1 accumulation in control cells, but this accumulation was blunted in TSC2−/− cells (Fig. 6A). TSC2 reconstitution into TSC2−/− cells was able to recover PINK1 accumulation after CCCP treatment (Fig. 6B). This regulatory effect of MTORC1 is likely transcriptionally mediated, as TSC2−/− cells showed a dramatic reduction of basal and CCCP-stimulated Pink1, which was partially salvaged by rapamycin cotreatment (Fig. 6C). Finally, to test whether exogenous PINK1 could rescue the PARK2 translocation defect in TSC2−/− cells, we generated cells stably expressing PINK1-V5 (Fig. 6D). We observed that both control and TSC2−/− cells showed abundant PARK2-positive labeled mitochondria upon CCCP exposure (Fig. 6E and F), suggesting that the PINK1 deficiency in cells with MTORC1 hyperactivity expression is the likely cause of defective PARK2 mitochondrial translocation.
FIG 6.
MTORC1-dependent PINK1 expression. (A and B) Western blots from TSC2+/+ and TSC2−/− MEFs stimulated with CCCP for the indicated times (A) or with reconstituted TSC2 (B). (C) qPCR in TSC2+/+ and TSC2−/− MEFs pretreated with Rapa for 24 h and then exposed to CCCP for 15 h. (D) Western blots from TSC2+/+ and TSC2−/− MEFs stably expressing PINK1-V5 and treated with CCCP for 3 h. (E) PARK2 localization in TSC2+/+ and TSC2−/− MEFs stably expressing PARK2-HA and PINK1-V5 and treated with CCCP for 3 h. (F) Quantification of cells with PARK2-positive mitochondria. The values represent means and SD (n = 3 to 5 independent experiments). *, P < 0.05; ***, P < 0.001; ns, not significant compared to the indicated control.
Constitutively active FoxO restores PINK1 expression and PARK2 translocation.
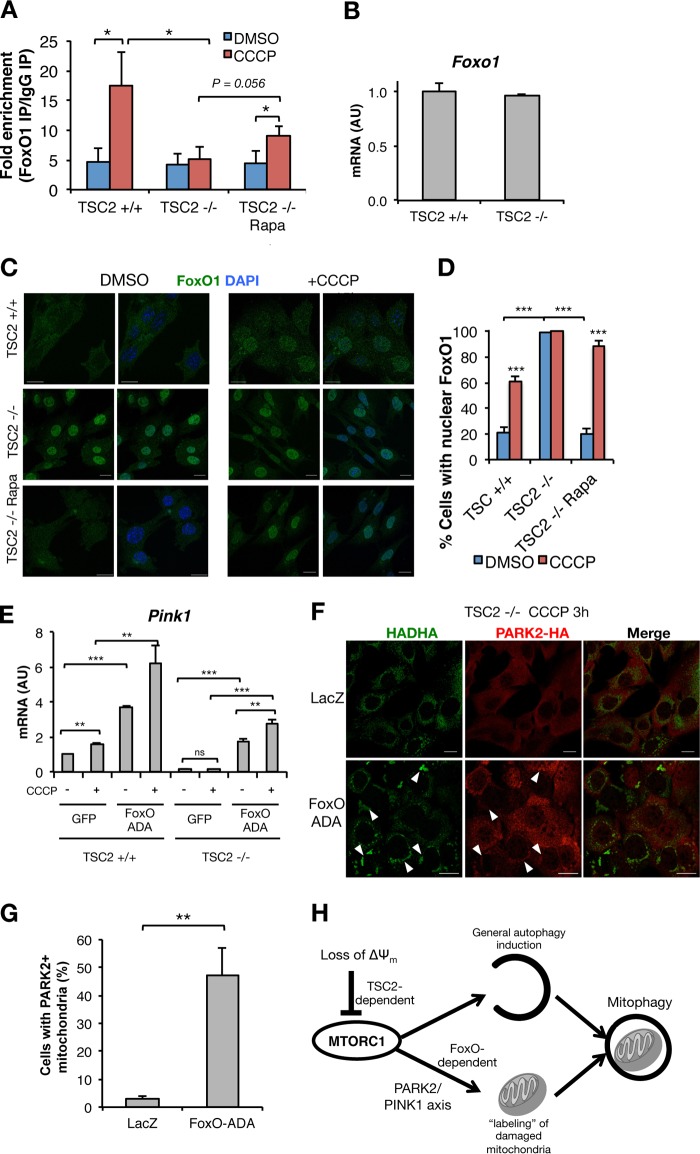
We next turned our attention to the mechanism of reduced Pink1 in TSC2−/− cells. Unrestrained activity of MTORC1/S6K in TSC2−/− cells generates a well-described negative-feedback loop on phosphatidylinositol 3-kinase (PI3K)–Akt signaling (36). The insulin-repressible family of forkhead box protein O (FoxO) transcription factors have been shown to induce Pink1 expression (37, 38). Consistently, we found that FoxO1 occupancy on the Pink1 promoter increased in response to CCCP treatment in control but not TSC2−/− cells, which was partially recovered after MTORC1 inhibition (Fig. 7A). This suggests that FoxO1 function may be altered with MTORC1 hyperactivity; indeed, while Foxo1 expression was unchanged (Fig. 7B), we noted constitutive nuclear FoxO1 localization in TSC2−/− cells as opposed to the normal cytosolic-nuclear shuttling in response to CCCP-induced stress in TSC2+/+ and rapamycin-treated TSC2−/− cells (Fig. 7C and D). Next, we determined if exogenous FoxO1 is sufficient to rescue Pink1 expression in TSC2−/− cells. For this experiment, we transduced cells with a constitutively active (i.e., insulin-insensitive) FoxO1-ADA mutant in an effort to reconstitute “normal” FoxO function in TSC2−/− cells (39) and found that FoxO1-ADA enhanced basal and CCCP-induced Pink1 mRNA in both control and TSC2−/− cells (Fig. 7E). FoxO-ADA also rescued CCCP-induced mitochondrial PARK2 translocation in TSC2−/− cells (Fig. 7F and G), suggesting that chronic disruption of normal insulin-FoxO signaling in TSC2−/− cells causes reduced Pink1 expression and thus impaired PARK2 translocation to mitochondria.
FIG 7.
FoxO-ADA rescues Pink1 expression and PARK2 translocation to mitochondria. (A) ChIP, using anti-FoxO1 or IgG control, in TSC2+/+ and TSC2−/− MEFs stimulated with CCCP or vehicle for 15 h with or without Rapa pretreatment, followed by qPCR for the Pink1 promoter region. (B) qPCR in TSC2+/+ and TSC2−/− MEFs. (C and D) Representative images of FoxO1 localization (C) and percentages of cells with nuclear FoxO1 (D) in TSC2+/+ and TSC2−/− MEFs stimulated with CCCP or vehicle for 15 h, with or without Rapa pretreatment. (E) qPCR in TSC2+/+ and TSC2−/− MEFs transduced with adenovirus encoding GFP or FoxO-ADA, with or without 15 h of exposure to CCCP. (F and G) Representative images of PARK2 mitochondrial localization (arrowheads) (F) and percentages of cells with PARK2+ mitochondria (G) in TSC2+/+ and TSC2−/− MEFs stably expressing PARK2-HA and transduced with LacZ or FoxO-ADA and then treated for 3 h with CCCP. The values represent means and SD (n = 3 to 5 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant compared to the indicated control. (H) Model in which loss of ΔΨm leads to TSC2-dependent MTORC1 inhibition, fundamental to coordinating two synergistic MTORC1-dependent processes—autophagy initiation and labeling of the cargo to be degraded, enabling its recognition by the autophagic machinery—to execute the clearance of damaged mitochondria.
DISCUSSION
Although MTORC1 has been shown to be a critical inhibitor of autophagy, our study proves that normal TSC2/MTORC1 action is required for not just general autophagy induction but also mitophagy after acute uncoupling of oxidative phosphorylation by CCCP. These data may be relevant to a host of pathophysiologic metabolic and neoplastic conditions that have been linked to both abnormal MTORC1 signaling (40) and mitochondrial function (41) or uncoupling (42). Although we did not explore the signals upstream of TSC2/MTORC1, which might be required for loss of Δψm-mediated MTORC1 inhibition, including BNIP3L (42, 43) and REDD1 (44), we found that TSC2 is critically required for CCCP-induced MTORC1 inhibition and autophagy induction, independent of AMPK.
TSC2 ablation has also been reported to increase mitochondrial biogenesis (24, 25). Autophagic turnover of mitochondria is also dependent on the mitochondrial dynamics and complexity of the mitochondrial network (28). We did find evidence of increased mitochondrial complexity in β cell-specific TSC2−/− mice (12), but on the whole, TSC2-deficient MEFs showed balanced mitochondrial fusion and fission and no defect in CCCP-induced mitochondrial fragmentation. Enhanced expression of Mfn1 and Mfn2 in MTORC1-hyperactive cells is consistent with previous reports that link MTORC1 to the master regulator of mitochondrial biogenesis, PPARGC1A (24), also found to regulate the expression of mitochondrial fusion genes (27, 45). However, we did not expect concomitant MTORC1-dependent expression of other genes, such as Dnm1l, Opa1, and Fis1, similarly observed in PPARGC1A/B-deficient models, which display coordinated downregulation of all mitochondrial complexity genes (46). Further, although we did not explicitly focus on mitochondrial transport, also shown to regulate mitochondrial turnover (47) and potentially regulated by TSC in neurons (16), we found no differences in perinuclear clustering of PARK2-positive mitochondria. Nevertheless, coupled with increased mitochondrial-protein aging in TSC2−/− cells, as revealed by a mitochondrial time-sensitive probe, we conclude that MTORC1 may act as a master regulator of the mitochondrial life span by integrating mitochondrial biogenesis, complexity, and clearance by mitophagy.
Damaged mitochondria with collapsed Δψm accumulate full-length PINK1 and recruit PARK2, which leads to recognition and processing by the autophagic machinery (32, 34, 35, 48). We showed that MTORC1 inhibition enhances PARK2 translocation in control cells while CCCP-mediated PARK2 translocation to mitochondria and mitophagy are severely impaired in TSC2−/− cells but recovered by rapamycin treatment. Interestingly, this effect recapitulates the previously observed MTORC1 dependence of clearance of dysfunctional mitochondria resulting from mitochondrial DNA (mtDNA) mutations by the autophagic machinery (14) and is also consistent with recent observations on the beneficial effects of MTORC1 inhibition on mitochondrial myopathies in mice (49). Our data also point to severe impairment of PARK2-dependent mitophagy by MTORC1 hyperactivation, but not complete inhibition. PARK2 translocation to mitochondria and accumulation of p62/SQSTM1-positive mitochondrial clusters were evident in TSC2−/− cells with chronic CCCP treatment long after full mitochondrial clearance in control cells. These results resemble the effects observed from the expression of several pathogenic PARK2 mutants (33), which also results in a highly delayed translocation of PARK2 after loss of Δψm.
We found that MTORC1 hyperactivity represses Pink1 expression and that reconstituted PINK1 is sufficient to rescue PARK2 translocation in TSC2−/− cells. These data are consistent with the critical role of PINK1 in PARK2 recruitment for mitochondria (35). We hypothesize that loss of Pink1 in TSC2−/− cells may result from compromised insulin signaling in the chronic MTORC1 hyperactive state (36). Thus, whereas CCCP led to FoxO1 nuclear translocation and Pink1 expression in control cells, consistent with the role of FoxO transcription factors to shuttle from cytosol to nucleus in stress to activate expression of cellular protection machinery (50), TSC2−/− cells showed simultaneous constitutive FoxO1 nuclear confinement yet incompetent binding to the Pink1 promoter. This odd pairing suggests that in the absence of functional insulin-Akt signaling in TSC2−/− cells, FoxO1 might undergo posttranslational modifications that render it ineffectual despite nuclear localization, as transactivation of Pink1 can be rescued by exogenous FoxO1. These findings highlight the importance of intact insulin-FoxO signaling for mitochondrial homeostasis and are in agreement with reports that link mitochondrial dysfunction with either chronic insulin resistance (51) or FoxO deficiency (52).
In summary, our data describe a mechanistic link between MTORC1 and mitochondrial turnover and highlight the need for proper regulation of autophagy for aspects of cellular homeostasis, such as preservation of mitochondrial quality, well beyond the oft-documented necessity of autophagy to allow cell survival under nutrient stress. MTORC1 integrates the molecular cues that control not only autophagy initiation but also the PINK1/PARK2 axis that primes the damaged organelle for cargo-specific autophagy of mitochondria (Fig. 7H). These data suggest that therapeutic interventions that regulate mitochondrial turnover may prove useful in the vast array of metabolic and neoplastic disorders related to inappropriate MTORC1 activation.
MATERIALS AND METHODS
Antibodies and chemicals.
We used antibodies directed against ACAC (number 3662), phosphorylated ACAC (P-ACAC) (number 3661), AMPK (number 2352), P-AMPK (number 2531), LC3B (number 4108), RAPTOR (number 4978), P-RAPTOR (number 2083), RPS6 (number 2217), phosphorylated ribosomal protein S6 kinase polypeptide 1 (P-RPS6KB1) (number 9205), RPS6KB1 (number 9202), TSC2 (number 3612), and TUBA1A (number 2144), all from Cell Signaling Technology; HADHA (number ab54477; Abcam); TOMM20 (number sc-11415) and green fluorescent protein (GFP) (number sc-9996) from Santa Cruz Biotechnology; SQSTM1 (number GP62; Progen); PINK1 (number BC100-494; Novus Biologicals); P-RPS6 (number MA5-15140; Thermo Scientific); HA (number 12CA5; Roche); actin beta (ACTB) (number A5316; Sigma); and V5 (number R960-25; Thermo Fisher). The antibody dilutions used are specified in Table S1 in the supplemental material. We also used rapamycin (Merck; number 553210) and blasticidin S-hydrochloride, chloroquine, CCCP, doxycycline, tunicamycin, and valinomycin (Sigma; numbers 15205, C6628, C2759, D9891, T7765, and V0627, respectively).
Cell culture and stable cell lines.
TSC2+/+ and TSC2−/− MEFs were kindly provided by David Kwitakowski (Harvard Medical School, Boston, MA). AMPK-DKO and control MEFs were provided by Arkaitz Carracedo (bioGune, Spain). Cells stably expressing PARK2-HA were generated by infection with retrovirus generated using Phoenix cells and pMXs-IP HA-Parkin (Addgene; number 38248) (29), a gift from Noboru Mizushima (University of Tokyo, Tokyo, Japan). Stable cells were selected with puromycin (5 μg/ml) for 2 weeks, subcloned, and assayed for expression of PARK2-HA. Three different clones with similar expression levels were pooled for subsequent work. For PINK1-V5 stable expression, lentiviral particles were generated with pLenti6-DEST PINK1-V5 wild type (WT) (Addgene; number 13320) (53), a gift from Mark Cookson (NIH), and packaging plasmids, using 293T cells, as previously described (54). The cells were infected with PINK1-V5 lentivirus and selected with blasticidin (10 μg/ml) for 2 weeks, and a pool of resistant cells was used for subsequent experiments.
Adenovirus infection and lipofection.
Adenovirus encoding murine FoxO1-ADA (T24A S253D S316A) and various controls (GFP or LacZ) were previously described (55, 56). EGFP-LC3 (Addgene; number 11546) plasmid (57) was a gift from Karla Kirkegaard (Stanford School of Medicine). Cells were transfected with Lipofectamine 3000 (Thermo Fisher; L3000015), with experiments performed 24 h after transfection unless otherwise indicated.
Western blotting and quantitative PCR.
Cell lysates were resolved by SDS-PAGE, followed by Western blotting and visualization using an ECL Western blotting detection kit (GE Healthcare Bio-Sciences; RPN2106). Densitometric quantification of blots was performed with NIH ImageJ (https://imagej.nih.gov/ij/), using the Analyze/Gels tools and the rolling-ball algorithm for background subtraction (radius set to 25 pixels). RNA was isolated with TRIzol (Invitrogen; 15596-026), cDNA was obtained with a high-capacity cDNA reverse transcription kit (Applied Biosystems; 4368813), and quantitative PCR (qPCR) was performed using Power SYBR master mix (Applied Biosystems; 4367659) as described previously (58). The primer sequences used are detailed in Table S2 in the supplemental material.
Immunofluorescence and confocal imaging.
For immunofluorescence assays, cells were seeded on glass coverslips. For MitoTracker experiments, cells were incubated for 30 min in serum-free Dulbecco's modified Eagle's medium (DMEM) with 500 nmol/liter MitoTracker Orange CMTMRos (Thermo Fisher; M-7510). After fixation and permeabilization, standard techniques were used for multiple staining procedures. For confocal microscopy imaging, Axio Observer Z1 with an LSM 700 scanning module (Zeiss) was used. Images were collected using a 63× Zeiss Plan-Apochromat oil objective (numerical aperture = 1.4). All images were obtained in a 1,024- by 1,024-pixel format. For microscope operation and image gathering, ZEN2 (Zeiss) software was used. The images were processed with ZEN2, and no further processing was performed. For quantification of single cell events (presence of PARK2-positive mitochondria and degradation of all mitochondria), at least 300 cells per condition were assayed in at least three independent experiments.
MitoTimer experiments.
TSC2+/+ and TSC2−/− stable cell lines expressing a tetracycline-dependent reverse transcriptional transactivator were generated. Lentiviruses were generated with pLenti cytomegalovirus (CMV) rtTA3 BLAST (Addgene; number 26429), a gift from Eric Campeau (University of Massachusetts Medical School), and packaging plasmids, using 293T cells, as previously described (54). The cells were subjected to blasticidin selection (10 μg/ml) for 2 weeks. The cells were transfected with pTRE-Tight-MitoTimer (Addgene; number 50547) (23), a gift from Roberta Gottlieb (Cedars-Sinai Heart Institute), using Lipofectamine 3000 (Thermo Fisher; L3000015). Twenty-four hours after transfection, the cells were exposed for 1 h to doxycycline (1 μg/ml), extensively washed with complete medium, and then fixed for 12 to 24 h prior to confocal imaging as described above. Images were processed with Fiji (http://fiji.sc/Fiji). Background was subtracted by using a rolling-ball algorithm (50-pixel radius). To eliminate the pixel shift between red and green channel images, a registering plugin was used (TurboReg). A threshold was set on the “numerator” image (red channel) to cover the mitochondrial region (all other pixels were discarded), and ratiometric images were generated by the division of pixel intensities (red by green), using the RatioPlus plugin. The mean intensity of the resulting images was used for quantification of the red/green ratio. The ratiometric images shown as examples were obtained after thresholding the intensity of the pixels shown between 0 and 8 and applying the Rainbow RGB look-up table. Twenty randomly selected individual cells were quantified per condition in three independent experiments. The Fiji macros for image processing and analysis are available on request.
Chromatin immunoprecipitation (ChIP).
Cells grown to confluence in three 100-mm2 cell culture dishes were fixed with 1% formaldehyde for 10 min, and chromatin was isolated with a Magna ChIP A kit (Merck-Millipore; 17-610) according to the instructions provided by the manufacturer. DNA was sheared by sonication, using 24 cycles of 30 s on, 30 s off (high setting) in a Bioruptor sonicator (Diagenode) on ice. Twenty-five micrograms of sheared chromatin was used for immunoprecipitation, with 2 μg Anti-FOXO1 (Abcam; number ab39670) or 2 μg of normal rabbit IgG (Cell Signaling; number 2729). The immunoprecipitated DNA was assayed by qPCR with primers for the Pink1 promoter (5′-AGATCTAAGCCCCGGAACCT-3′; 5′-GTTCACACTGCCCTGGCTAT-3′) region previously determined to contain a FoxO-binding element (37). The results are presented as fold enrichment above control (IgG) calculated with the following formula: 2(ΔCT IP − ΔCT control).
Supplementary Material
ACKNOWLEDGMENTS
A.B. acknowledges a grant from the Russell Berrie Foundation. This work is supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) (22590981 to Y.K.); the Ministry of Economy, Industry and Competitiveness, Spain (MINECO) (SAF2011-22555 and SAF2014-51795-R to M.B.); and the National Institutes of Health (NIH) (DK103818 to U.B.P.).
We declare that we have no conflicts of interest regarding the contents of this article.
A.B., A.G.-A., and C.G. researched data and contributed to the experimental design and discussion. S.-I.A., Y.K., U.B.P., and M.B. contributed to discussion and review of the project. A.B. and U.B.P. wrote the manuscript. All the authors reviewed the results and approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00441-17.
REFERENCES
- 1.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DCO, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. 2010. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 2.Russell RC, Yuan H-X, Guan K-L. 2014. Autophagy regulation by nutrient signaling. Cell Res 24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murrow L, Debnath J. 2013. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noda T, Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 5.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 6.Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S-I, Natsume T, Takehana K, Yamada N, Guan J-L, Oshiro N, Mizushima N. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, Kundu M, Kim D-H. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol 4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. 2003. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest 112:1223–1233. doi: 10.1172/JCI200317222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkhitko A, Myachina F, Morrison TA, Hindi KM, Auricchio N, Karbowniczek M, Wu JJ, Finkel T, Kwiatkowski DJ, Yu JJ, Henske EP. 2011. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci U S A 108:12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartolomé A, Kimura-Koyanagi M, Asahara S-I, Guillén C, Inoue H, Teruyama K, Shimizu S, Kanno A, García-Aguilar A, Koike M, Uchiyama Y, Benito M, Noda T, Kido Y. 2014. Pancreatic β-cell failure mediated by mTORC1 hyperactivity and autophagic impairment. Diabetes 63:2996–3008. doi: 10.2337/db13-0970. [DOI] [PubMed] [Google Scholar]
- 13.Ng S, Wu Y-T, Chen B, Zhou J, Shen H-M. 2011. Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy 7:1173–1186. doi: 10.4161/auto.7.10.16681. [DOI] [PubMed] [Google Scholar]
- 14.Gilkerson RW, de Vries RLA, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, Schon EA. 2012. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet 21:978–990. doi: 10.1093/hmg/ddr529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taneike M, Nishida K, Omiya S, Zarrinpashneh E, Misaka T, Kitazume-Taneike R, Austin R, Takaoka M, Yamaguchi O, Gambello MJ, Shah AM, Otsu K. 2016. mTOR hyperactivation by ablation of tuberous sclerosis complex 2 in the mouse heart induces cardiac dysfunction with the increased number of small mitochondria mediated through the down-regulation of autophagy. PLoS One 11:e0152628. doi: 10.1371/journal.pone.0152628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL, Conrad C, Rothberg JM, Lipton JO, Kölker S, Hoffmann GF, Han M-J, Polleux F, Sahin M. 2016. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep 17:1053–1070. doi: 10.1016/j.celrep.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundu M, Lindsten T, Yang C-Y, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. 2008. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra D, Tanaka A, Suen D-F, Youle RJ. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLeod LE, Proud CG. 2002. ATP depletion increases phosphorylation of elongation factor eEF2 in adult cardiomyocytes independently of inhibition of mTOR signalling. FEBS Lett 531:448–452. doi: 10.1016/S0014-5793(02)03582-2. [DOI] [PubMed] [Google Scholar]
- 20.Kwon K-Y, Viollet B, Yoo OJ. 2011. CCCP induces autophagy in an AMPK-independent manner. Biochem Biophys Res Commun 416:343–348. doi: 10.1016/j.bbrc.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 21.Inoki K, Zhu T, Guan K-L. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 22.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. 2008. AMPK phosphorylation of Raptor mediates a metabolic checkpoint. Mol Cell 30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez G, Thornton C, Stotland A, Lui D, Sin J, Ramil J, Magee N, Andres A, Quarato G, Carreira RS, Sayen MR, Wolkowicz R, Gottlieb RA. 2013. MitoTimer: a novel tool for monitoring mitochondrial turnover. Autophagy 9:1852–1861. doi: 10.4161/auto.26501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 25.Koyanagi M, Asahara S-I, Matsuda T, Hashimoto N, Shigeyama Y, Shibutani Y, Kanno A, Fuchita M, Mikami T, Hosooka T, Inoue H, Matsumoto M, Koike M, Uchiyama Y, Noda T, Seino S, Kasuga M, Kido Y. 2011. Ablation of TSC2 enhances insulin secretion by increasing the number of mitochondria through activation of mTORC1. PLoS One 6:e23238. doi: 10.1371/journal.pone.0023238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soriano FX, Liesa M, Bach D, Chan DC, Palacín M, Zorzano A. 2006. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes 55:1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 27.Liesa M, Borda-d'Agua B, Medina-Gómez G, Lelliott CJ, Paz JC, Rojo M, Palacín M, Vidal-Puig A, Zorzano A. 2008. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS One 3:e3613. doi: 10.1371/journal.pone.0003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshii SR, Kishi C, Ishihara N, Mizushima N. 2011. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakovic A, Shurkewitsch K, Seibler P, Grünewald A, Zanon A, Hagenah J, Krainc D, Klein C. 2013. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J Biol Chem 288:2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrané J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 34.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. 2010. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. 2004. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mei Y, Zhang Y, Yamamoto K, Xie W, Mak TW, You H. 2009. FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation. Proc Natl Acad Sci U S A 106:5153–5158. doi: 10.1073/pnas.0901104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta A, Molkentin JD, Paik J-H, DePinho RA, Yutzey KE. 2011. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem 286:7468–7478. doi: 10.1074/jbc.M110.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C-C, Jeon S-M, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, Park Y, Hay N. 2010. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell 18:592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson SC, Rabinovitch PS, Kaeberlein M. 2013. mTOR is a key modulator of ageing and age-related disease. Nature 493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding W- X, Ni H-M, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, Yin X-M. 2010. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem 285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Wang Y, Kim E, Beemiller P, Wang C-Y, Swanson J, You M, Guan K-L. 2007. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem 282:35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 44.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cartoni R, Léger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener J-L, Luthi F, Dériaz O, Zorzano A, Gobelet C, Kralli A, Russell AP. 2005. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J Physiol 567:349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin OJ, Lai L, Soundarapandian MM, Leone TC, Zorzano A, Keller MP, Attie AD, Muoio DM, Kelly DP. 2014. A role for peroxisome proliferator-activated receptor γ coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ Res 114:626–636. doi: 10.1161/CIRCRESAHA.114.302562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soubannier V, McLelland G-L, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM. 2012. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol 22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou Y-S, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivelä R, Pessia A, Velagapudi V, Suomalainen A. 2017. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab 26:419–428.e5. doi: 10.1016/j.cmet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Frescas D, Valenti L, Accili D. 2005. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem 280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 51.Cheng Z, Guo S, Copps K, Dong X, Kollipara R, Rodgers JT, DePinho RA, Puigserver P, White MF. 2009. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med 15:1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim-Muller JY, Zhao S, Srivastava S, Mugabo Y, Noh H-L, Kim YR, Madiraju SRM, Ferrante AW, Skolnik EY, Prentki M, Accili D. 2014. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metab 20:593–602. doi: 10.1016/j.cmet.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. 2005. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A 102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartolomé A, Guillén C, Benito M. 2012. Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic β cell death. Autophagy 8:1757–1768. doi: 10.4161/auto.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pajvani UB, Shawber CJ, Samuel VT, Birkenfeld AL, Shulman GI, Kitajewski J, Accili D. 2011. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nat Med 17:961–967. doi: 10.1038/nm.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim K, Qiang L, Hayden MS, Sparling DP, Purcell NH, Pajvani UB. 2016. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun 7:10255. doi: 10.1038/ncomms10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson WT, Giddings TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sparling DP, Yu J, Kim K, Zhu C, Brachs S, Birkenfeld AL, Pajvani UB. 2016. Adipocyte-specific blockade of gamma-secretase, but not inhibition of Notch activity, reduces adipose insulin sensitivity. Mol Metab 5:113–121. doi: 10.1016/j.molmet.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.