Abstract
Context:
Hyperinsulinemia is often observed in obese people, owing to their insulin resistance accompanied by visceral fat accumulation, but the frequency of hyperinsulinemia in nonobese people is not well known. Mutations in the insulin receptor gene are known to cause insulin resistance and hyperinsulinemia in type A insulin resistance syndrome, Rabson-Mendenhall syndrome, and Donohue syndrome. However, insulin receptor gene abnormalities have not been investigated in asymptomatic hyperinsulinemic subjects.
Purpose:
The aim of the current study was to investigate the prevalence of hyperinsulinemia in nonobese Japanese subjects and to examine the involvement of insulin receptor gene mutations.
Methods:
We enrolled 11,046 subjects who received health checkups. From these, we extracted nonobese subjects (body mass index <25 kg/m2) who exhibited hyperinsulinemia (serum fasting immunoreactive insulin ≥15 µU/mL). Genetic analysis was performed for the insulin receptor gene in 11 nonobese subjects with hyperinsulinemia.
Results:
The prevalence of hyperinsulinemia without apparent diabetes in nonobese subjects was 0.4% (33/8630). In the 11 analyzed subjects, two novel heterozygous nonsense mutations were detected [c.2106 T>G (p.Y702X) and c.2779-2780 GC>A]. The prevalence of insulin receptor gene mutations was 18.2% (2/11).
Conclusions:
To our knowledge, this is the first report of the prevalence of hyperinsulinemia in nonobese healthy subjects. We identified two novel mutations in the insulin receptor gene. These findings indicate that mutations in the insulin receptor gene may be related to fasting hyperinsulinemia, and insulin receptor gene screening may be useful for determining the cause of unexplained hyperinsulinemia.
Keywords: glucose intolerance, healthy subjects, hyperinsulinemia, insulin receptor gene, nonobese
In nonobese healthy subjects, the prevalence of hyperinsulinemia without apparent diabetes is 0.4%. The prevalence of insulin receptor gene mutations in 11 subjects with hyperinsulinemia is 18.2%.
Hyperinsulinemia is often observed in obese people, owing to their insulin resistance accompanied by visceral fat accumulation. However, in nonobese people, hyperinsulinemia is seldom identified, except in cases of pancreatic β-cell tumors (insulinoma), insulin or insulin receptor autoantibodies, or certain diseases causing insulin resistance. Because most diseases causing insulin resistance and insulinoma are characterized by specific abnormalities other than hyperinsulinemia, few apparently healthy patients are identified as having hyperinsulinemia.
Mutations in the insulin receptor (IR) gene are known to produce clinical abnormalities, such as insulin resistance and hyperinsulinemia, and are associated with heterogeneous phenotypes [1, 2]. Donohue syndrome and Rabson-Mendenhall syndrome usually have homozygous or compound heterozygous mutations in the IR gene, and patients with these diseases have severe insulin resistance together with various symptoms, such as growth retardation, occasional hypoglycemia from infancy, intrauterine growth retardation, and low birth weight [3–6]. In contrast, patients with type A insulin resistance syndrome usually have heterozygous mutations in the IR gene and present with mild insulin resistance, acanthosis nigricans, and glucose intolerance but rarely have hypoglycemia [7, 8]. In contrast to these findings, we have recently reported two independent cases of adult-onset hyperinsulinemic hypoglycemia linked to mutations in the IR gene [9]. Because the phenotypes of the two cases were different from those seen in typical type A insulin resistance syndrome, we concluded that these were novel phenotypes related to IR gene abnormalities. Moreover, we observed an adult case with a novel IR gene mutation who presented with hyperinsulinemia but not obesity (unpublished data). Thus, we speculated that there might be a certain number of patients with latent hyperinsulinemia related to IR gene mutations in nonobese subjects.
The mechanism of hyperinsulinemia is considered as follows: mutation in the insulin receptor decreases the expression or function of insulin receptor and induces insulin resistance, resulting in compensative excessive insulin secretion from pancreatic β-cells. As hyperinsulinemia usually poses a burden to insulin-producing cells and could cause exhaustion of the cells, hyperinsulinemia with an IR gene mutation might increase the risk of diabetes mellitus. Thus, it is important to identify hyperinsulinemia in apparently healthy subjects and to introduce lifestyle modifications to prevent the onset of diabetes mellitus. In this study, we investigated the prevalence of hyperinsulinemia in nonobese Japanese subjects who participated in the health checkup program and examined insulin receptor gene mutations in nonobese subjects with hyperinsulinemia.
1. Materials and Methods
A. Subjects and Screening for Hyperinsulinemia
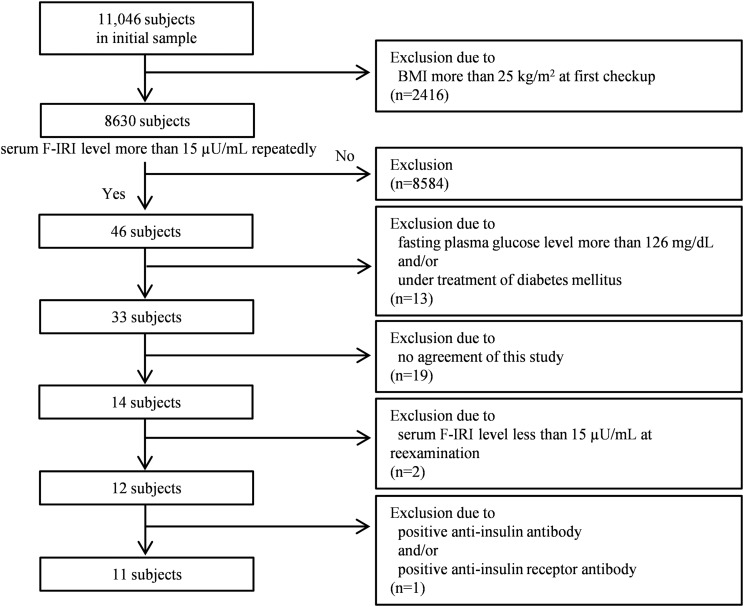
In Fig. 1, the flowchart shows the subject selection process. In this study, we enrolled 11,046 subjects who had received health checkups at the Physical Checkup Center in Sumitomo Hospital more than twice between 1 January 2010 and 31 December 2015. In Japan, people often voluntarily schedule their medical checkups at a physical checkup center within a hospital to evaluate their health. The subjects in this study were not admitted patients at our hospital, and none of them had apparent diseases that required hospital admission. All participants signed informed consent to provide medical information and blood samples before undergoing the medical checkup, and they had the right to refuse the use of their data. We first extracted 8630 subjects with a body mass index (BMI) of <25 kg/m2 and checked their serum fasting immunoreactive insulin (F-IRI) levels. Forty-six subjects were identified to repeatedly have serum F-IRI levels >15 µU/mL. Next, we excluded 13 subjects who either had fasting plasma glucose levels >126 mg/dL or were under treatment for diabetes. Thus, the prevalence of hyperinsulinemia in nonobese subjects without apparent diabetes was ~0.4% (33/8630). Then, we excluded 19 subjects who did not consent to further examination and 2 subjects with serum F-IRI levels <15 µU/mL at reexamination. Furthermore, we excluded 1 subject with positive anti-insulin antibody. Finally, we extracted 11 subjects and performed genetic analysis of the IR gene on these subjects.
Figure 1.
Flow diagram for the subjects included in this study.
B. Measurements
Plasma glucose concentrations were measured with the glucose oxidase method, and insulin and C-peptide levels were measured with enzyme immunoassay kits. Hemoglobin A1c (HbA1c) National Glycohemoglobin Standardization Program equivalent values (%) were calculated with the following formula: HbA1c (%) = 1.02 × HbA1c (Japan Diabetes Society, %) + 0.25 (%) [10]. Measurement of serum anti-insulin antibodies was outsourced to SRL, Inc. (Tokyo, Japan), and the radioreceptor assay of serum anti-insulin receptor antibodies was outsourced to BML, Inc. (Tokyo, Japan). The latter assay is based on the detection of antibodies that inhibit [125I] insulin binding in human IM-9 lymphocytes expressing the insulin receptor. Insulin resistance was determined by using the homeostasis model assessment of insulin resistance (HOMA-IR), and values were obtained with the following formula: HOMA-IR = fasting insulin (IU/mL) × fasting plasma glucose (mmol/L)/22.5 [11].
C. Evaluation of Hyperinsulinemia
The general reference range of serum F-IRI levels is 2 to 15 µU/mL. Iwahashi et al. [12], in a study of nonobese (BMI <25 kg/m2) Japanese subjects, have shown that the mean serum F-IRI level is 4.3 ± 2.6 μU/mL in subjects with normal glucose tolerance, 5.1 ± 2.7 μU/mL in subjects with impaired glucose tolerance, and 5.7 ± 1.9 μU/mL in patients with diabetes mellitus. In another study, the mean serum F-IRI level even in subjects with abdominal obesity with a BMI <25 kg/m2 was 5.4 ± 2.5 μU/mL [13]. Therefore, these data indicate that a serum F-IRI level of 15 μU/mL is more than three standard deviations higher than normal in nonobese people. Thus, in the current study, we defined hyperinsulinemia as a serum F-IRI level >15 µU/mL.
D. 75-g Oral Glucose Tolerance test and Reactive Hypoglycemia
Glucose tolerance was assessed by an oral glucose tolerance test (OGTT) after an overnight fast. Blood samples were collected at 0, 30, 60, 90, and 120 minutes in all cases and at 180, 240, and 300 minutes in cases 1, 2, 4, 6, 7, 8, and 10. In cases 5 and 11, we were unable to obtain consent to perform OGTT. In the OGTT results, reactive hypoglycemia was diagnosed if the minimum plasma glucose level was <70 mg/dL (3.9 mmol/L) [14]. Diabetes mellitus and impaired glucose tolerance were classified according to the criteria of the World Health Organization [15].
E. Polymerase Chain Reaction Direct DNA Sequencing
Genomic DNA extraction from leukocytes was outsourced to SRL, Inc. Each exon of the IR gene was amplified using primers reported previously [16, 17]. Ex-Taq DNA polymerase (Takara Shuzo Co., Biomedical Group, Shiga, Japan) was used to amplify exons 2 to 22, and AmpliTaq Gold 360 Master Mix (Applied Biosystems, Foster City, CA) was used to amplify exon 1. Amplified polymerase chain reaction (PCR) products were electrophoresed, and the size of each product was confirmed. PCR products were sequenced after purification with a QIA quick PCR Purification kit (Qiagen, Hilden, Germany). The sequencing reaction was carried out using an ABI Prism dye terminator cycle sequencing kit (Applied Biosystems), and the products were analyzed on an ABI gene analyzer 1100 system, according to the protocol supplied by the manufacturer (Applied Biosystems). Genetic analysis in this study was performed with the approval of the Research Ethics Committee at Osaka University. In all subjects, written informed consent was obtained for analysis of the IR gene after genetic counseling had been performed. In the genetic counseling, we explained to prospective subjects what we might learn from the genetic test, the merits and demerits of knowing the results, and that the subjects had the right to decide whether they wished to know the results. The results of the tests were explained to the subjects if they wished.
F. Restriction Fragment Length Polymorphism Analysis
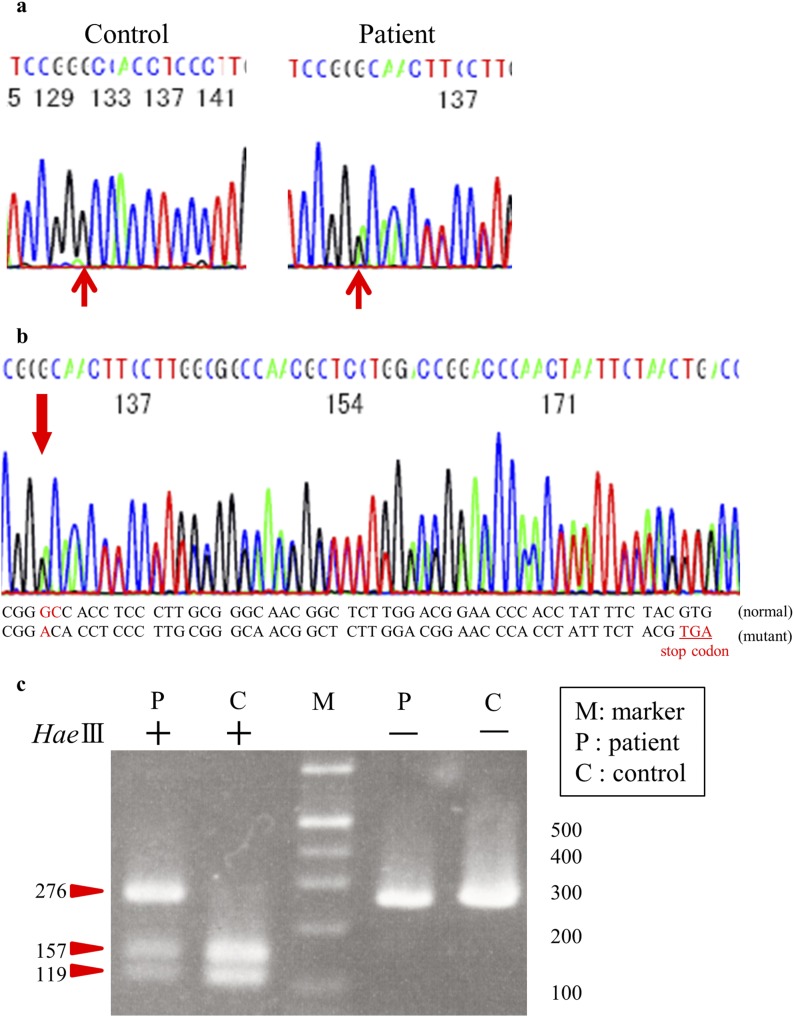
To determine whether the novel mutation (c.2779-2780 GC>A) found in exon 14 in case 5 was located on one allele and whether the other allele was normal, we performed the following restriction fragment length polymorphism study. PCR products were digested with HaeIII (Takara Bio, Shiga, Japan) at 37°C for 2 hours, separated on a 3% agarose gel, and stained with ethidium bromide to visualize the fragments using ultraviolet light.
2. Results
A total of 11,046 subjects were enrolled in the current study, from which we extracted 8630 nonobese (BMI <25 kg/m2) subjects. The prevalence of hyperinsulinemia without apparent diabetes in nonobese subjects was 0.4% (33/8630). Finally, we extracted 11 subjects and performed genetic analysis of the IR gene in these 11 subjects.
A. Clinical Characteristics of Subjects
The clinical features of the 11 subjects whose genetic analyses were performed are presented in Table 1. Because 2 subjects (cases 5 and 11) did not agree to the OGTT, 9 of the 11 subjects’ OGTT were performed as planned (Table 2). Glucose intolerance was prevalent in 6 of 9 (66.7%) subjects. Regarding the details of 6 subjects with glucose intolerance, 5 of 6 had impaired glucose tolerance, and 1 of 6 was diabetic. Reactive hypoglycemia was prevalent in 4 of 9 (44.4%) subjects.
Table 1.
Clinical Characteristics of the 11 Subjects Whose Genetic Analyses Were Performed in This Study
| Case No. | Age, y | Sex | BMI, kg/m2 | HbA1c, % | FPG, mg/dL | F-IRI, µU/mL | HOMA-IR | Adiponectin, µg/mL |
|---|---|---|---|---|---|---|---|---|
| 1 | 74 | Female | 22.6 | 6.1 | 101 | 15.6 | 3.9 | 6.9 |
| 2 | 65 | Male | 23.8 | 5.9 | 109 | 19.0 | 5.1 | 4.1 |
| 3 | 75 | Female | 19.8 | 5.5 | 90 | 15.3 | 3.4 | 8.4 |
| 4 | 54 | Male | 24.7 | 5.6 | 97 | 23.5 | 5.6 | 6.7 |
| 5 | 55 | Male | 20.8 | 6.0 | 111 | 27.8 | 7.6 | 8.2 |
| 6 | 62 | Male | 23.0 | 5.6 | 89 | 17.2 | 3.8 | 3.6 |
| 7 | 61 | Male | 24.8 | 5.8 | 99 | 15.1 | 3.7 | 6.2 |
| 8 | 66 | Male | 23.2 | 5.8 | 110 | 18.3 | 5.0 | 7.3 |
| 9 | 40 | Female | 24.1 | 5.4 | 85 | 20.0 | 4.2 | 5.4 |
| 10 | 43 | Female | 24.1 | 5.6 | 99 | 16.6 | 4.1 | 4.4 |
| 11 | 72 | Male | 24.7 | 6.0 | 102 | 18.0 | 4.5 | 4.9 |
Abbreviation: FPG, fasting plasma glucose.
Table 2.
OGTT Data of the Subjects Whose Genetic Analyses Were Performed, Excluding Case 5 and Case 11
| Characteristic |
Time, min |
Glycemic Status | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 30 | 60 | 120 | 180 | 240 | 300 | ||
| Case 1 | Diabetes | |||||||
| Glucose (mg/dL) | 103 | 222 | 270 | 236 | 135 | 87 | 46a | |
| IRI (µU/mL) | 17.7 | 62.4 | 108.2 | 210.8b | 133.7 | 79.1 | 7.7 | |
| Case 2 | IGT | |||||||
| Glucose (mg/dL) | 114 | 174 | 184 | 163 | 73a | 77 | 90 | |
| IRI (µU/mL) | 27.5 | 118 | 178.3 | 178.4b | 25.5 | 10.1 | 10.5 | |
| Case 3 | NGT | |||||||
| Glucose (mg/dL) | 91a | 154 | 134 | 114 | 93 | |||
| IRI (µU/mL) | 9.1 | 158.7 | 232.8b | 42.5 | 35.7 | |||
| Case 4 | IGT | |||||||
| Glucose (mg/dL) | 96 | 121 | 178 | 150 | 127 | 50a | 80 | |
| IRI (µU/mL) | 15.9 | 68.3 | 111.6 | 113.3b | 108.5 | 14.5 | 8.1 | |
| Case 6 | NGT | |||||||
| Glucose (mg/dL) | 96 | 214 | 188 | 83 | 72a | 81 | 84 | |
| IRI (µU/mL) | 24.1 | 423.0b | 387.0 | 110.3 | 22.5 | 13.1 | 24.0 | |
| Case 7 | IGT | |||||||
| Glucose (mg/dL) | 108 | 214 | 243 | 182 | 92 | 62a | 81 | |
| IRI (µU/mL) | 30.3 | 207.5 | 269.8 | 279.1b | 101.3 | 32.4 | 20.9 | |
| Case 8 | IGT | |||||||
| Glucose (mg/dL) | 96 | 171 | 193 | 176 | 92 | 52a | 75 | |
| IRI (µU/mL) | 11.0 | 68.9 | 86.5 | 196.5b | 71.9 | 9.5 | 9.1 | |
| Case 9 | IGT | |||||||
| Glucose (mg/dL) | 87a | 152 | 130 | 148 | 121 | |||
| IRI (µU/mL) | 8.8 | 79.0b | 55.4 | 72.8 | 53.1 | |||
| Case 10 | NGT | |||||||
| Glucose (mg/dL) | 100 | 164 | 159 | 113 | 75a | 88 | 91 | |
| IRI (µU/mL) | 19.7 | 180.9 | 198.9b | 142.7 | 28.7 | 15.7 | 17.8 | |
Abbreviations: IGT, impaired glucose tolerance; IRI, immunoreactive insulin; NGT, normal glucose tolerance.
Minimum glucose level.
Peak immunoreactive insulin level.
B. Genetic Analysis of the IR Gene in the Subjects
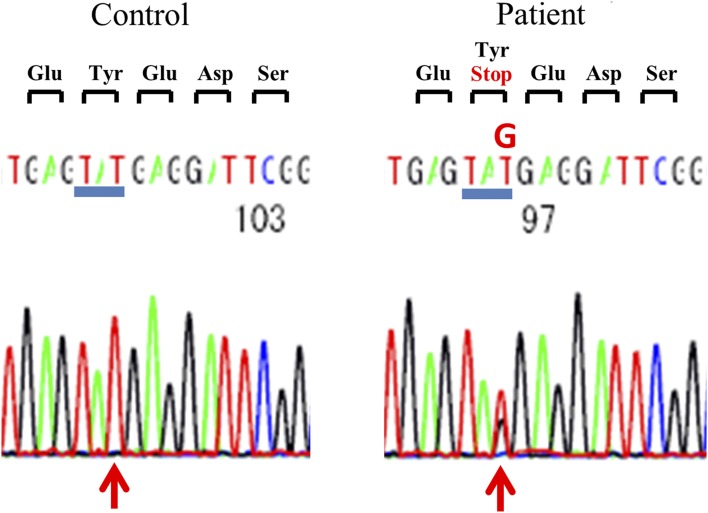
To investigate whether the IR genes in the 11 subjects had mutations, we conducted genetic analysis of all 22 exons and the intron-exon boundaries of the insulin receptor gene. Table 3 shows the results of genetic analysis of the IR gene. In exons 1, 2, 4, 5, 7, 9, 15, 16, and 18 to 22, no novel variants or single-nucleotide polymorphisms (SNPs) were detected. Previously reported SNPs—including rs891087 and rs2860179 in exon 3; rs2860178 and rs7252268 in exon 6; rs2059806 in exon 8; rs3745548 in exon 10; rs2252673 in exon 11; rs2229430, rs2229434, and rs13306451 in exon12; rs2229431 in exon 13; and rs1799817 in exon 17—were detected. There was not a large difference in the frequency of these SNPs between subjects in this study and healthy Japanese subjects in the database of the 1000 Genomes Project (International Genome Sample Resource). Novel heterozygous mutations were detected in 2 of 11 subjects (in cases 4 and 5). In case 4, a novel heterozygous missense mutation was identified in which Tyr 702 was replaced with a stop codon [c.2106T>G (p.Y702X); Fig. 2]. In case 5, a novel heterozygous mutation (c.2779-2780 GC>A) was detected, which led to a frame shift and a subsequent premature stop codon at downstream codon 17 (Fig. 3a and 3b). No other novel variants were found in any of the exons sequenced in these 11 study subjects.
Table 3.
Results of Genetic Analyses of the Insulin Receptor Gene in 11 Nonobese Subjects With Hyperinsulinemia
| Exon No. |
Case No. |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| 1 | — | — | — | — | — | — | — | — | — | — | — |
| 2 | — | — | — | — | — | — | — | — | — | — | — |
| 3 | — | rs891087 rs2860179 | — | — | — | — | — | — | — | — | — |
| 4 | — | — | — | — | — | — | — | — | — | — | — |
| 5 | — | — | — | — | — | — | — | — | — | — | — |
| 6 | rs2860178 rs7252268 | — | — | rs2860178 | rs2860178 rs7252268 | — | — | — | — | — | — |
| 7 | — | — | — | — | — | — | — | — | — | — | — |
| 8 | rs2059806 | — | — | rs2059806 | rs2059806 | — | — | — | — | — | — |
| 9 | — | — | — | — | — | — | — | — | — | — | — |
| 10 | rs3745548 | — | rs3745548 | c.2106 T>G (p.Tyr702X) | — | rs3745548 | rs3745548 | rs3745548 | rs3745548 | rs3745548 | rs3745548 |
| 11 | — | rs2252673 | — | rs2252673 | rs2252673 | — | rs2252673 | — | — | rs2252673 | rs2252673 |
| 12 | — | — | — | rs2229430 rs2229434 rs13306451 | — | — | — | — | — | — | rs2229430 rs2229434 rs13306451 |
| 13 | rs2229431 | rs2229431 | — | — | — | — | — | — | — | — | — |
| 14 | — | — | — | — | c.2779-2780 GC>A | — | — | — | — | — | — |
| 15 | — | — | — | — | — | — | — | — | — | — | — |
| 16 | — | — | — | — | — | — | — | — | — | — | — |
| 17 | — | — | — | rs1799817 | rs1799817 | rs1799817 | rs1799817 | rs1799817 | — | rs1799817 | — |
| 18 | — | — | — | — | — | — | — | — | — | — | — |
| 19 | — | — | — | — | — | — | — | — | — | — | — |
| 20 | — | — | — | — | — | — | — | — | — | — | — |
| 21 | — | — | — | — | — | — | — | — | — | — | — |
| 22 | — | — | — | — | — | — | — | — | — | — | — |
“—” indicates no mutation or no polymorphism.
Figure 2.
Nucleotide sequence of the insulin receptor gene in case 4 is shown in the right panel and compared with that from the control in the left.
Figure 3.
Case 5 sequence analysis of the insulin receptor gene. (a) Nucleotide sequence of the insulin receptor gene in case 5 is shown in the right panel and compared with that from the control in the left. (b) The position of GC converted to A is indicated with an arrow. This mutation causes a 1-bp deletion leading to a frame shift and a subsequent premature stop codon. (c) Restriction fragment length polymorphism study. PCR products of exon 14 from case 5 were digested with HaeIII. Three bands were observed: 276-bp, 157-bp, and 119-bp bands, thus indicating the presence of both the normal allele and mutant allele.
C. Case Reports and Genetic Analysis of the IR Gene in the Two Cases
Case 4 was a 54-year-old man with a BMI of 24.2 kg/m2. No signs of acanthosis nigricans or hypertrichosis were noted. Liver function tests, renal function tests, and serum electrolytes were within the normal range at first checkup. An OGTT showed a high serum insulin level (113.3 μU/mL at 2 hours; Table 2). During the test, he had no symptoms of hypoglycemia, but the plasma glucose was 50 mg/dL at 4 hours in the OGTT. As a result of genetic analysis of the IR gene, a novel heterozygous nonsense mutation was identified with c.2106T>G (p.Y702X; Fig. 2). In addition, seven SNPs were detected in the IR gene (rs2860178, rs2059806, rs2252673, rs2229430, rs2229434, rs13306451, and rs1799817).
Case 5 was a 55-year-old man with a BMI of 20.8 kg/m2. No appearance of acanthosis nigricans or hypertrichosis was noted. Liver function tests, renal function tests, and serum electrolytes were within the normal range at the first checkup. Because we did not obtain consent for performing the OGTT, we could not conduct the OGTT in this case. Genetic analysis of the IR gene revealed a novel heterozygous mutation (c.2779-2780 GC>A). This mutation caused a 1-bp deletion (Fig. 3a), leading to a frame shift and a subsequent premature stop codon (Fig. 3b). In addition, five SNPs were detected in the IR gene (rs2860178, rs7252268, rs2059806, rs2252673, and rs1799817). To determine whether the novel mutation (c.2779-2780 GC>A) found in exon 14 in case 5 was truly located on one allele and that the other allele was normal, we performed a restriction fragment length polymorphism study. When the PCR products of exon 14 were digested with HaeIII, the product of a normal allele should have been digested to 157- and 119-bp bands, whereas the product of a mutated allele would not be digested with HaeIII, thereby resulting in a single 276-bp band. Thus, the appearance of 276-, 157-, and 119-bp bands (Fig. 3c) indicated that the mutation was on one allele and that the other allele was normal.
3. Discussion
In this study, we found that the prevalence of hyperinsulinemia without apparent diabetes in nonobese subjects was 0.4% (33/8630). Although this rate was not as high as that in obese subjects, which was 8.5% (206/2416) in our subjects, our identification of apparently healthy nonobese individuals with hyperinsulinemia was unexpected. In addition, we found that 66.7% (6/9) of the subjects presented with glucose intolerance. Furthermore, we also found that approximately 44% (4/9) of the subjects presented with reactive hypoglycemia in the OGTT, thus indicating potential latent hypoglycemia; hence, we recommend that hyperinsulinemic patients be evaluated for symptoms of hypoglycemia in their daily life. Therefore, it would be useful to measure serum insulin levels during health checkups to screen for hyperinsulinemia and to provide preventive health guidance for diabetes mellitus, even in nonobese people.
In the 11 hyperinsulinemic subjects, we identified two novel IR gene mutations, one of which was a heterozygous stop codon in case 4, and the other was a 1-bp deletion leading to a subsequent premature stop codon in case 5. These mutations should result in nonsense-mediated messenger RNA (mRNA) decay (NMD). NMD is a widely known mRNA quality control mechanism by which the mRNA with premature stop codon is degraded, preventing the production of truncated abnormal proteins [18]. Thus, we considered that the two novel mutations lead to the decreased function of insulin receptor by NMD. In fact, Kadowaki et al. [19] have previously reported a case of Donohue syndrome by compound heterozygous mutations in c.2095C>T (p.Q699X) at exon 10 and c.1459A>G (p.K487E) at exon 6. Furthermore, the proband’s father had a heterozygous mutation (p.Q699X) and presented with severe hyperinsulinemia. Because the mutation c.2106 T>G (p.Y702X), identified in our case 4, is located very near p.Q699X, it is reasonable to consider that in both cases, hyperinsulinemia is associated with heterozygous mutations in the IR gene. In addition, Kadowaki et al. [20] have also reported a patient with Donohue syndrome who was a compound heterozygote for two cis-acting dominant mutations in the IR gene; the paternal allele, which had a mutation in c.2770C>T (p.R924X), decreased the levels of insulin receptor mRNA. Because the mutation (c.2779-2780 GC>A) identified at exon 14 in the IR gene in our case 5, which caused a 1-bp deletion leading to a frame shift and a subsequent premature stop codon at exon 14, is also close to c.2770C>T, it is reasonable to speculate that in case 5, the levels of insulin receptor mRNA might also be decreased, thus causing insulin receptor dysfunction. When the function of insulin receptor is decreased, insulin signaling is lowered and insulin resistance occurs. Insulin resistance usually results in compensatory hyperinsulinemia for keeping blood glucose normal as well as impairing the clearance of insulin in the target organs like liver, muscle, and adipose tissues. Further analysis would be needed to elucidate the functional changes caused by these two novel nonsense mutations and how they are linked to hyperinsulinemia.
Although the two cases with identified mutations in IR gene did not present a substantial lipid metabolism abnormality, liver dysfunction, or renal dysfunction, they showed some of the highest fasting insulin levels and HOMA-IR among the 11 subjects. It may be due to the severity of decreased function of insulin receptor in the two cases. In addition, their plasma adiponectin levels were also relatively higher among the 11 subjects whose genetic analyses were performed (Table 1). A previous report has shown that plasma adiponectin is elevated in patients with IR gene abnormalities in contrast to other forms of severe insulin resistance [21]. That report has indicated that an adiponectin level >7 μg/mL in severe insulin resistance has a 97% positive predictive value for insulin receptor gene mutations, whereas a level <5 μg/mL has a 97% negative predictive value. In the current study, 3 of the 11 subjects had an adiponectin level >7 μg/mL, 4 subjects had <5 μg/mL, and 4 subjects had >5 μg/mL and ≤7 μg/mL. All subjects with an adiponectin level <5 μg/mL in this study had no mutations in the IR gene, and this result was concordant with findings from the previous report. In three subjects with an adiponectin level >7 μg/mL, however, only one subject had a mutation in the IR gene. This discrepancy in frequency might have been caused by a difference in the severity of insulin resistance. In the previous report, patients with severe insulin resistance were defined as having fasting insulin >21.6 μU/mL or peak insulin in an oral glucose tolerance test >216.0 μU/mL in nondiabetic patients. Otherwise, in our study, hyperinsulinemia was defined as a fasting insulin level >15 µU/mL. In addition, in our study, one subject with an adiponectin level of 6.7 μg/mL (>5 μg/mL and ≤7 μg/mL) had a mutation in the IR gene. Because the subject had severe insulin resistance [fasting insulin 23.5 μU/mL (>21.6 μU/mL)], we recommend that IR gene screening be performed even in subjects with severe insulin resistance and an adiponectin level >5 μg/mL and ≤7 μg/mL.
Among the 11 apparently healthy nonobese subjects with hyperinsulinemia, we found two cases with IR gene mutations. The two cases had no signs of acanthosis nigricans or hypertrichosis, which are usually seen in type A insulin resistance syndrome. This result confirmed the existence of some patients with latent hyperinsulinemia related to IR gene mutations, as we had expected. Furthermore, one subject who consented to the OGTT presented with glucose intolerance and reactive hypoglycemia, similarly to the cases that we have recently reported [9]. These results support the existence of a novel subtype related to IR gene abnormalities, as we have proposed previously. Because the frequency of IR gene abnormalities in nonobese hyperinsulinemia was as high as 18% (2/11), we recommend IR gene screening for nonobese people with unexplained hyperinsulinemia. Given that the prevalence of hyperinsulinemia in nonobese people was 0.4%, the prevalence of hyperinsulinemia with mutations in the insulin receptor gene in nonobese Japanese might be ~0.07% (0.4 × 0.18). Because they are at high risk of developing diabetes and their offspring might also be involved by the mutations, it would be important to extract these subjects. In general, hyperinsulinemia with genetic mutations ought to be observed from youth, and thus it might be enough to measure serum fasting insulin levels only once during their lifetime. In Japan, it is common to measure blood glucose and HbA1c in an annual health checkup program every year, and it costs the participants about a half the price of measuring insulin levels; thus, we think it is not wasteful to measure fasting insulin levels at least once during their lifetime (e.g., at 40 years old).
We also detected SNPs that have been previously reported in these 11 subjects, but there was not a large difference in the frequency of these SNPs between the subjects in this study and healthy Japanese subjects. This finding suggests that the SNPs are not correlated with hyperinsulinemia and do not appear to be the cause of hyperinsulinemia. The causes of hyperinsulinemia in the rest of the nine cases, other than cases 4 and 5, were unclear and hence require further study.
In conclusion, to our knowledge, this is the first study to detect the prevalence of hyperinsulinemia in nonobese healthy subjects and we identified two novel mutations in the insulin receptor gene. Furthermore, the prevalence of insulin receptor gene mutations was even higher. These findings indicate that mutations in the insulin receptor gene may be related to fasting hyperinsulinemia, and insulin receptor gene screening may be useful for understanding unexplained hyperinsulinemia.
Acknowledgments
The authors are grateful to the participants in this study.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BMI
- body mass index
- F-IRI
- fasting immunoreactive insulin
- HbA1c
- hemoglobin A1c
- HOMA-IR
- homeostasis model assessment of insulin resistance
- IR
- insulin receptor
- mRNA
- messenger RNA
- NMD
- nonsense-mediated messenger RNA decay
- OGTT
- oral glucose tolerance test
- PCR
- polymerase chain reaction
- SNP
- single-nucleotide polymorphism.
References and Notes
- 1.Glaser B, Kesavan P, Heyman M, Davis E, Cuesta A, Buchs A, Stanley CA, Thornton PS, Permutt MA, Matschinsky FM, Herold KC. Familial hyperinsulinism caused by an activating glucokinase mutation. N Engl J Med. 1998;338(4):226–230. [DOI] [PubMed] [Google Scholar]
- 2.Otonkoski T, Jiao H, Kaminen-Ahola N, Tapia-Paez I, Ullah MS, Parton LE, Schuit F, Quintens R, Sipilä I, Mayatepek E, Meissner T, Halestrap AP, Rutter GA, Kere J. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am J Hum Genet. 2007;81(3):467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor SI. Lilly Lecture: molecular mechanisms of insulin resistance. Lessons from patients with mutations in the insulin-receptor gene. Diabetes. 1992;41(11):1473–1490. [DOI] [PubMed] [Google Scholar]
- 4.Musso C, Cochran E, Moran SA, Skarulis MC, Oral EA, Taylor S, Gorden P. Clinical course of genetic diseases of the insulin receptor (type A and Rabson-Mendenhall syndromes): a 30-year prospective. Medicine (Baltimore). 2004;83(4):209–222. [DOI] [PubMed] [Google Scholar]
- 5.Højlund K, Hansen T, Lajer M, Henriksen JE, Levin K, Lindholm J, Pedersen O, Beck-Nielsen H. A novel syndrome of autosomal-dominant hyperinsulinemic hypoglycemia linked to a mutation in the human insulin receptor gene. Diabetes. 2004;53(6):1592–1598. [DOI] [PubMed] [Google Scholar]
- 6.Huang Z, Li Y, Tang T, Xu W, Liao Z, Yao B, Hu G, Weng J. Hyperinsulinaemic hypoglycaemia associated with a heterozygous missense mutation of R1174W in the insulin receptor (IR) gene. Clin Endocrinol (Oxf). 2009;71(5):659–665. [DOI] [PubMed] [Google Scholar]
- 7.Kahn CR, Flier JS, Bar RS, Archer JA, Gorden P, Martin MM, Roth J. The syndromes of insulin resistance and acanthosis nigricans: insulin-receptor disorders in man. N Engl J Med. 1976;294(14):739–745. [DOI] [PubMed] [Google Scholar]
- 8.Krook A, O’Rahilly S. Mutant insulin receptors in syndromes of insulin resistance. Baillieres Clin Endocrinol Metab. 1996;10(1):97–122. [DOI] [PubMed] [Google Scholar]
- 9.Kuroda Y, Iwahashi H, Mineo I, Fukui K, Fukuhara A, Iwamoto R, Imagawa A, Shimomura I. Hyperinsulinemic hypoglycemia syndrome associated with mutations in the human insulin receptor gene: report of two cases. Endocr J. 2015;62(4):353–362. [DOI] [PubMed] [Google Scholar]
- 10.Committee on the Standardization of Diabetes Mellitus-Related Laboratory Testing of Japan Diabetes Society (JDS) International clinical harmonization of glycated hemoglobin in Japan: From Japan Diabetes Society to National Glycohemoglobin Standardization Program values. Diabetol Int. 2012;3(1):8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. [DOI] [PubMed] [Google Scholar]
- 12.Iwahashi H, Okauchi Y, Ryo M, Noguchi M, Morita S, Kishida K, Okita K, Ohira T, Funahashi T, Nakamura T, Imagawa A, Shimomura I. Insulin-secretion capacity in normal glucose tolerance, impaired glucose tolerance, and diabetes in obese and non-obese Japanese patients. J Diabetes Investig. 2012;3(3):271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki R, Yano Y, Yasuma T, Onishi Y, Suzuki T, Maruyama-Furuta N, Gabazza EC, Sumida Y, Takei Y. Association of waist circumference and body fat weight with insulin resistance in male subjects with normal body mass index and normal glucose tolerance. Intern Med. 2016;55(11):1425–1432. [DOI] [PubMed] [Google Scholar]
- 14.Harris S. Hyperinsulinism and dysinsulinism. JAMA. 1924;83(10):729–733. [Google Scholar]
- 15.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. [DOI] [PubMed] [Google Scholar]
- 16.Seino S, Seino M, Bell GI. Human insulin-receptor gene: partial sequence and amplification of exons by polymerase chain reaction. Diabetes. 1990;39(1):123–128. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki T, Kadowaki H, Rechler MM, Serrano-Rios M, Roth J, Gorden P, Taylor SI. Five mutant alleles of the insulin receptor gene in patients with genetic forms of insulin resistance. J Clin Invest. 1990;86(1):254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karousis ED, Nasif S, Mühlemann O. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA. 2016;7(5):661–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadowaki T, Bevins CL, Cama A, Ojamaa K, Marcus-Samuels B, Kadowaki H, Beitz L, McKeon C, Taylor SI. Two mutant alleles of the insulin receptor gene in a patient with extreme insulin resistance. Science. 1988;240(4853):787–790. [DOI] [PubMed] [Google Scholar]
- 20.Kadowaki T, Kadowaki H, Taylor SI. A nonsense mutation causing decreased levels of insulin receptor mRNA: detection by a simplified technique for direct sequencing of genomic DNA amplified by the polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87(2):658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semple RK, Cochran EK, Soos MA, Burling KA, Savage DB, Gorden P, O’Rahilly S. Plasma adiponectin as a marker of insulin receptor dysfunction: clinical utility in severe insulin resistance. Diabetes Care. 2008;31(5):977–979. [DOI] [PubMed] [Google Scholar]