ABSTRACT
Bacterial cell wall dynamics have been implicated as important determinants of cellular physiology, stress tolerance, and virulence. In Streptococcus mutans, the cell wall is composed primarily of a rhamnose-glucose polysaccharide (RGP) linked to the peptidoglycan. Despite extensive studies describing its formation and composition, the potential roles for RGP in S. mutans biology have not been well investigated. The present study characterizes the impact of RGP disruption as a result of the deletion of rgpF, the gene encoding a rhamnosyltransferase involved in the construction of the core polyrhamnose backbone of RGP. The ΔrgpF mutant strain displayed an overall reduced fitness compared to the wild type, with heightened sensitivities to various stress-inducing culture conditions and an inability to tolerate acid challenge. The loss of rgpF caused a perturbation of membrane-associated functions known to be critical for aciduricity, a hallmark of S. mutans acid tolerance. The proton gradient across the membrane was disrupted, and the ΔrgpF mutant strain was unable to induce activity of the F1Fo ATPase in cultures grown under low-pH conditions. Further, the virulence potential of S. mutans was also drastically reduced following the deletion of rgpF. The ΔrgpF mutant strain produced significantly less robust biofilms, indicating an impairment in its ability to adhere to hydroxyapatite surfaces. Additionally, the ΔrgpF mutant lost competitive fitness against oral peroxigenic streptococci, and it displayed significantly attenuated virulence in an in vivo Galleria mellonella infection model. Collectively, these results highlight a critical function of the RGP in the maintenance of overall stress tolerance and virulence traits in S. mutans.
IMPORTANCE The cell wall of Streptococcus mutans, the bacterium most commonly associated with tooth decay, is abundant in rhamnose-glucose polysaccharides (RGP). While these structures are antigenically distinct to S. mutans, the process by which they are formed and the enzymes leading to their construction are well conserved among streptococci. The present study describes the consequences of the loss of RgpF, a rhamnosyltransferase involved in RGP construction. The deletion of rgpF resulted in severe ablation of the organism's overall fitness, culminating in significantly attenuated virulence. Our data demonstrate an important link between the RGP and cell wall physiology of S. mutans, affecting critical features used by the organism to cause disease and providing a potential novel target for inhibiting the pathogenesis of S. mutans.
KEYWORDS: Streptococcus mutans, rhamnose, RGP, cell wall, acid tolerance
INTRODUCTION
Streptococcus mutans is the primary etiologic agent of dental caries, the most prevalent bacterial disease worldwide (1, 2). The Gram-positive aerotolerant lactic acid bacterium resides on the tooth surface, in concert with hundreds of different microorganisms within the multispecies biofilm known as dental plaque (3, 4). Oral hygiene dictates whether the odontopathogen can dominate in these biofilms, with a key contributing factor being consistent dietary carbohydrate intake. S. mutans utilizes a variety of mechanisms for efficient sugar uptake, including ATP-binding cassette transport systems and the phosphoenolpyruvate:sugar phosphotransferase system (PTS) (5, 6). Through the homofermentative breakdown of these sugars, S. mutans generates significant concentrations of lactic acid as an end product, so as to reduce local pH levels within the dental plaque microenvironment to values lower than 5.0 and, importantly, below the threshold for demineralization of the tooth enamel (7). While these conditions render the biofilm inhospitable to the majority of organisms, S. mutans is capable of persisting due to its ability to dynamically adapt to acid stress, a trait referred to as aciduricity.
The aciduric property of S. mutans is defined by a number of mechanisms that become induced when the bacterium encounters acid stress, and these are collectively referred to as the acid tolerance response (ATR). S. mutans utilizes the ATR to maintain a more alkaline cytosolic pH relative to its environmental conditions. A principal component of this response is the upregulation of genes associated with the membrane-bound proton-translocating F1Fo ATPase. Through the hydrolysis of ATP, protons are extruded from the cell, and enzyme activity is significantly increased during acid stress (8). Additionally, previous work from our group has identified fundamental changes that occur in the fatty acid (FA) composition of the plasma membrane that are required for acid adaptation in S. mutans (9). Under conditions of acid stress, the bacterium incorporates a greater proportion of unsaturated FAs than saturated FAs into its membrane, a process dependent on the FA biosynthesis enzyme FabM (10). The deletion of fabM resulted in strains that were highly susceptible to acid stress, incapable of inducing ATR processes, and significantly attenuated in caries formation using an in vivo model of infection in rats (10, 11). It is obvious that proper regulation of the physiological state of S. mutans is necessary for ATR induction, and ultimately, virulence. While this has been appreciated specifically for changes in the membrane, S. mutans physiology is also highly dependent on the overall composition of the cell wall, for which very little is known with regard to stress tolerance.
Proper maintenance of the bacterial cell wall has important ramifications for the way cells respond to their environment. In Gram-positive bacteria, the contribution of wall teichoic acids (WTAs) to both stress response and overall physiology has been extensively studied. The disruption of these surface-associated polyanionic sugar alcohol structures results in improper cell shape and division, enhanced cellular autolysis, reduced adherence and biofilm growth, and an increased susceptibility to stress, antimicrobial agents, and host defense mechanisms (12–17). S. mutans (like many other streptococcal species) does not produce WTAs and instead generates an antigenically distinct cell wall polysaccharide (CWP) termed the rhamnose-glucose polysaccharide (RGP). These structures are composed of a polyrhamnose backbone with glucose side chains, and following extracellular transport, they become covalently linked to the peptidoglycan (PG) (18). The process by which the RGP is constructed has been well established; however, the specific roles for these structures remain understudied (18–20). In examinations of related streptococcal species, it has been demonstrated that the rhamnose-containing CWPs dominate and that their formation is often essential, or at least critically required, for many important cellular processes. Inactivation of the genes encoding the rhamnosyltransferases responsible for capsule formation in Streptococcus pneumoniae proved to be lethal, highlighting the essentiality of these enzymes (21, 22). Similarly, the rhamnosyltransferase enzymes of Streptococcus pyogenes have also been shown to be essential, while the disruption of side-chain formation on the CWP of S. pyogenes resulted in significantly reduced virulence following infection in both mice and rabbits (23, 24). In S. mutans, the loss of RGP resulted in reduced bacteriophage adherence and an increased susceptibility to certain cell wall-targeting antibiotics, while recent studies have suggested that these structures may influence biofilm formation (25–27). Beyond this, however, specific roles for these abundant CWPs have not been investigated, and the contribution of RGP to the critical virulence features of S. mutans is unknown.
Previous work in our laboratory resulted in the generation of a gene deletion library spanning the genome of S. mutans (28). The loss of genes involved in RGP synthesis caused a considerable reduction in the overall fitness of the bacterium. The present study sought to investigate the importance of this cell wall component as a means of better understanding the aciduric abilities of S. mutans, and by extension, the effects on the virulence capabilities of the bacterium. We demonstrate that disruption of RGP synthesis via the deletion of a single rhamnosyltransferase gene, rgpF, resulted in heightened sensitivity to a variety of stress conditions, ultimately resulting in an inability to undergo stress adaptation. Furthermore, biofilm cultures of the ΔrgpF mutant strain were significantly less robust than the parent strain, suggesting a role for the RGP structures in adherence. In addition, the competitive fitness of S. mutans was reduced upon the deletion of rgpF, with the deletion strain displaying growth inhibition caused by peroxigenic streptococci. Finally, the virulence of the ΔrgpF mutant was significantly attenuated using the Galleria mellonella in vivo infection model. Our findings suggest a critical role for RGP in the maintenance of S. mutans homeostasis and provide a potential novel target for inhibiting the overall ability of the bacterium to mount a stress response and cause disease.
RESULTS
Disruption of RGP synthesis in S. mutans results in reduced fitness.
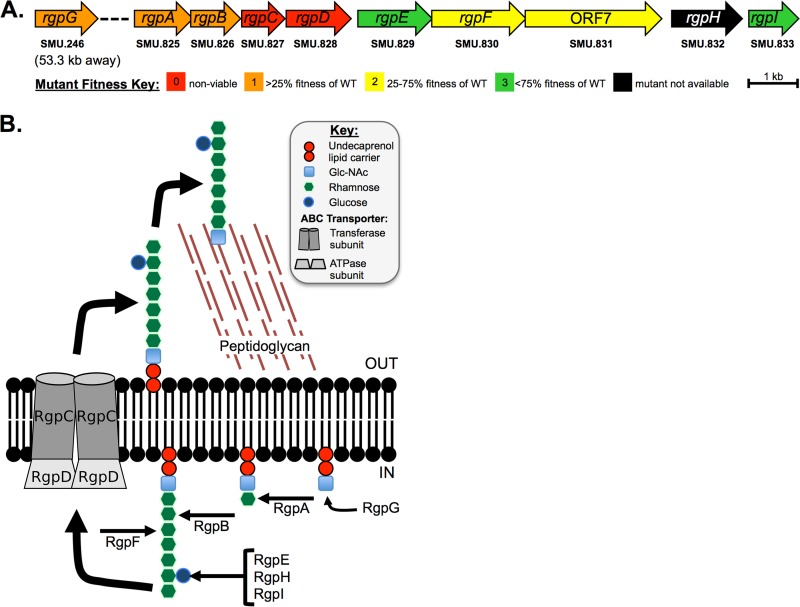
Mutant strains carrying deletions for each gene involved in the synthesis of RGP in S. mutans were constructed previously in an effort to generate a genome-wide gene deletion library in the organism (28). The cost of losing each rgp gene was assayed under a variety of conditions, and their resulting growth was then assigned a score representative of the individual mutant's overall fitness. Figure 1A displays the gene map of the rgp locus with the respective growth phenotype for each gene deletion mutant. Genes thought to be responsible for the assembly of the core polyrhamnose backbone of RGP (rgpG, rgpA, rgpB, and rgpF; SMU.246, SMU.825, SMU.826, and SMU.830, respectively) demonstrated the greatest reduction in overall fitness (20, 28). Attempts to delete rgpC (SMU.827) and rgpD (SMU.828), annotated as the transferase subunit and ATPase subunit, respectively, of an ATP-binding cassette transporter proposed to be the mechanism by which mature RGP is transported out of the cell, appeared to be lethal mutations in S. mutans (18, 28). Overall fitness was minimally affected by the deletion of rgpE (SMU.829) and rgpI (SMU.833), each of which have been implicated in linking glucose as a side chain onto the polyrhamnose backbone (20, 29).
FIG 1.
Gene map and enzymatic pathway leading to rhamnose-glucose polysaccharide (RGP) synthesis. (A) The gene loci responsible for synthesis of RGP are displayed. The overall fitness cost for losing each individual gene is color-coded and based on a combined score from growth phenotypes under various conditions compared to the wild-type parent strain, UA159, as described by Quivey et al. (28). Briefly, a score of “0” indicated no viable organisms recovered (thus, presumably an essential gene); “1” was equivalent to <25% growth compared to the WT; “2” indicates between 25% and 75% growth compared to WT; and “3” indicates >75% growth compared to that of the WT. (B) A depiction of the enzymatic pathway for RGP formation. The initiation of RGP synthesis relies on the transfer of N-acetylglucosamine (GlcNAc) onto a membrane-linked undecaprenol lipid carrier, facilitated by RgpG. RgpA, RgpB, and RgpF are rhamnosyltransferases that likely function in a stepwise fashion, each respectively adding the first three l-rhamnose units onto the lipid-linked GlcNAc, with further elongation of the polyrhamnose backbone carried out by alternating activities of RgpB and RgpF (18–20). Side chains are formed by glucose linkages, carried out by the glucosyltransferase enzymes RgpE, RgpH, and RgpI (18, 29). The mature structure is then transferred to the extracellular space by the ABC transporter, made up of RgpC/RgpD (18).
The ΔrgpF mutant has heightened sensitivity to acid stress.
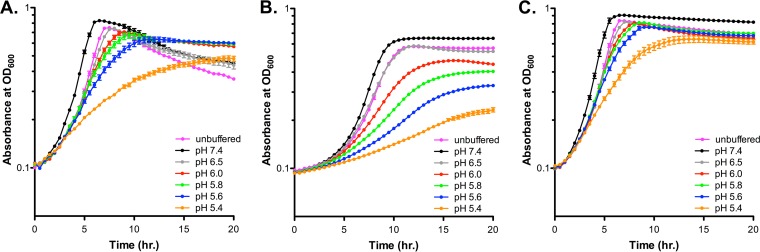
To expand our examination into the fitness defects mentioned above, the growth rate of the ΔrgpF mutant was further analyzed using growth medium that had been titrated to various pH values ranging from 7.4 to 5.4. Although the growth rates for S. mutans UA159 (Fig. 2A) and the rgpF+ complement strain (Fig. 2C) were impacted in a pH-dependent manner, the maximal growth of the two strains was similar to one another regardless of pH value. However, the growth rate of the ΔrgpF mutant strain was not only hindered under neutral pH conditions, but it also displayed a distinct acid sensitivity beginning at pH 6.0 that progressively inhibited the growth of the culture at each successive decreasing pH value (Fig. 2B). The calculated doubling times for each strain (Table 1) revealed that the growth rate of the ΔrgpF mutant was significantly reduced compared to UA159 in all growth scenarios (P ≤ 0.001). The growth rates of the rgpF+ complement strain were restored to parental levels, and in fact enhanced, compared to those of UA159, despite comparable levels of rgpF expression between the two strains (see Fig. S1A in the supplemental material). It is possible that these differences arise from the ectopic insertion of the complementing locus away from potential cis-acting elements within the core rgp locus. In turn, this may affect RgpF function or RGP formation, possibly influencing the growth of the complemented rgpF+ strain. When growth rates were measured from cultures grown in tryptone-yeast extract (TY) medium plus 1% (wt/vol) glucose, without pH control (Fig. S1B), no differences were observed between UA159 and the rgpF+ strain, while the rate of ΔrgpF mutant growth remained significantly reduced (P ≤ 0.001).
FIG 2.
Growth rate of the ΔrgpF mutant is impaired. Cultures of S. mutans UA159 (A), ΔrgpF (B), and rgpF+ (C) strains were grown in BHI medium or BHI medium titrated to pH values between 5.4 and 7.4. Growth was assessed by measuring optical density at 600 nm in a Bioscreen C plate reader (as described in Materials and Methods). Data are represented as mean ± standard deviation (SD) (n = 10). The calculated doubling times from graphs are displayed in Table 1.
TABLE 1.
Doubling times of S. mutans UA159, ΔrgpF mutant, and rgpF+ complementa
| pH of medium | Doubling time (min) |
|||||
|---|---|---|---|---|---|---|
| UA159 |
ΔrgpF strain |
rgpF+ strain |
||||
| Mean | SD | Mean | SD | Mean | SD | |
| Unbuffered | 142.1 | 1.9 | 290.7b,c | 12.0 | 134.5 | 3.9 |
| 7.4 | 123.6 | 5.9 | 243.9b,c | 10.6 | 97.7b | 8.1 |
| 6.5 | 186.3 | 5.7 | 309.5b,c | 19.2 | 149.1b | 3.7 |
| 6.0 | 198.8 | 11.5 | 496.5b,c | 14.6 | 161.7b | 4.3 |
| 5.8 | 220.8 | 9.2 | 732.0b,c | 29.8 | 174.9b | 6.5 |
| 5.6 | 277.5 | 11.5 | 1,034.5b,c | 23.2 | 197.0b | 12.8 |
| 5.4 | 645.7 | 54.8 | 1,962.4b,c | 83.1 | 286.4b | 14.4 |
Data are derived from growth curves shown in Fig. 2. Doubling times were calculated as described in Materials and Methods.
P ≤ 0.001 versus UA159.
P ≤ 0.001 versus rgpF+ strain.
The ΔrgpF mutant strain is incapable of adapting to stress conditions.
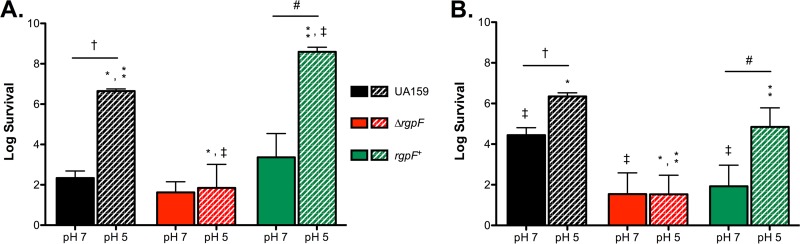
The acid sensitivity observed for the ΔrgpF strain suggested an impairment in the overall acid tolerance response (ATR) of the mutant strain. To assess the ability of the ΔrgpF mutant to mount a response, the S. mutans UA159, ΔrgpF and rgpF+ strains were grown in continuous culture to steady state at pH values of 7 and 5 and challenged with an acid shock (pH 2.5) for 60 min. As expected, the survival of UA159 was significantly enhanced following growth at pH 5 versus pH 7, indicating induction of ATR and conferred resistance to acid challenge in these cells. The ΔrgpF mutant strain, however, remained highly susceptible to acid challenge, while adaptation was restored in the rgpF+ complement strain (Fig. 3A).
FIG 3.
The ΔrgpF mutant has heightened sensitivity to stress. Cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains were grown in continuous culture to steady state at pH values of 7 and 5 and challenged with either 0.1 M glycine (pH 2.5) (A) or 0.05% H2O2 (B). Surviving organisms were enumerated after a 60-min exposure, and data are expressed as mean log survival (versus starting inoculum) ± SD. Data were derived from three independent chemostat cultures, each performed in duplicate. Statistical significance (P ≤ 0.02) was determined by pairwise comparison using Student's t test and is indicated by comparison of like symbols.
Previous work from our laboratory and from others has demonstrated that increased resistance to oxidative stress is coupled with ATR mechanisms (30–32). To test the response of the ΔrgpF mutant strain to oxidative stress, assays similar to those described above were performed, using instead H2O2 (final concentration of 0.05%) as the test agent. Again, both the UA159 and rgpF+ strains demonstrated significant increases in survival following growth at pH 5 versus pH 7, indicating conferred tolerance. The ΔrgpF mutant strain was significantly more susceptible to oxidative stress than UA159, regardless of the culture pH (Fig. 3B). Collectively, these results indicate that a loss of RGP impairs the ability of S. mutans to fully adapt to stress conditions routinely encountered in the oral cavity.
Disruption of RGP in S. mutans causes membrane impairment.
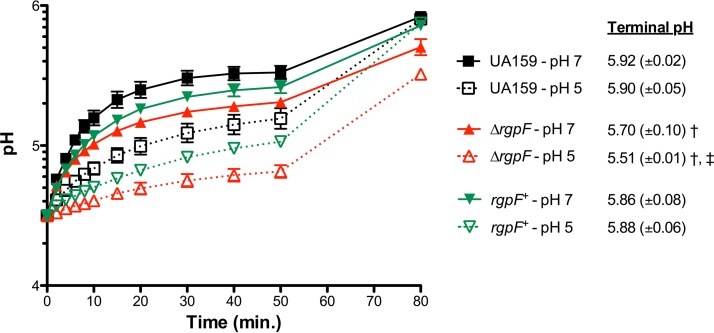
To explain the stress sensitivity and lack of adaptation phenotypes observed in the ΔrgpF mutant strain, we turned our attention to interrogating the integrity of the cell membrane. A fundamental outcome of the ATR in S. mutans is the maintenance of a greater cytosolic pH than the external acidic environment created by the organism, allowing for enhanced survival. Critical to this maintenance of internal pH are changes associated with the membrane, in both general structure and activity of enzymes embedded within (9, 33). To test this, cells from steady-state cultures grown at pH 7 and pH 5 were assayed for membrane permeability by measuring the rate of proton movement into the cell (Fig. 4). A decrease in proton flow was observed over 50 min in the UA159 strain grown to steady state at pH 5 compared to that in the pH 7-grown cells. This observation likely reflects previous findings from our group demonstrating that acid-adapted S. mutans strains contain a greater proportion of unsaturated (versus saturated) fatty acids in their plasma membranes, which may influence membrane fluidity (9, 10). Proton flow through the membrane of the ΔrgpF mutant strain was reduced in cultures grown to steady state at pH 7 compared to UA159 under similar conditions. However, an almost complete collapse of proton movement was observed with cells from the ΔrgpF mutant culture grown to steady state at pH 5. These observations were further punctuated by the terminal pH values measured at the 80-min time point, after the cytosolic pH was allowed to equilibrate with the external pH. In experiments with UA159, the terminal pH value for cultures grown to steady state at pH 7 was 5.92, and it was 5.90 for growth at pH 5. These values were significantly lower following pH equilibration of the ΔrgpF mutant samples, where cells grown to steady state at pH 7 had a terminal pH value of 5.70, while the value for cells grown to steady state at pH 5 was 5.51 (P ≤ 0.05). These data provide insight into the pH conditions of the cytosol, as a more alkaline internal pH would elevate the equilibrated pH value. Conversely, the lower terminal pH reading observed for the ΔrgpF strain suggests that the mutant has a much more acidic cytosol than the parent strain. In turn, this affects the readout of the assay, since the higher intracellular proton concentration displaces the proton gradient across the membrane. For the rgpF+ complement, proton permeability was moderately restored, with terminal pH values in accordance with those measured in UA159 (5.86 and 5.88 for pH 7 and pH 5, respectively).
FIG 4.
Proton permeability is disrupted in the ΔrgpF mutant. Cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains were grown in continuous culture to steady state at pH values of 7 and 5. At the 50-min time point, butanol was added to samples (10% [vol/vol] final concentration) to lyse cells. Terminal pH values listed represent mean pH recordings (±SD) at the 80-min time point, where the pH of the ΔrgpF mutant was significantly lower (†, P ≤ 0.02) than that of both UA159 and the rgpF+ complement under the same conditions. The terminal pH values for the ΔrgpF mutant strain were also significantly different between pH 7 and pH 5 (‡, P ≤ 0.02). Data were derived from three independent chemostat cultures, each performed in duplicate and represented as the mean values ± SD. Statistical significance was determined by a pairwise comparison using Student's t test.
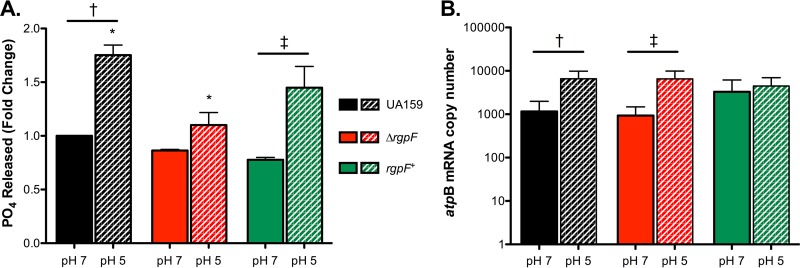
Another approach to measuring membrane stability is to examine the activities of membrane-bound enzymes. Relevant to the ATR in S. mutans, the membrane-bound, proton-translocating F1Fo ATPase uses the hydrolysis of ATP as energy to extrude protons from the cell, and this process is critical to the ability of the bacterium to regulate cytosolic pH (34). Accordingly, when ATPase enzyme activity was assayed for S. mutans UA159, a significant increase was observed in cells grown to steady state at pH 5 versus pH 7 (Fig. 5A), consistent with previous reports (33). No significant changes in ATPase activity were observed in cultures of the ΔrgpF strain grown to steady state at pH 7 and pH 5. Further, the activity measured in the ΔrgpF mutant strain at pH 5 was significantly reduced compared to that in UA159 (P ≤ 0.05). The induction of ATPase enzyme activity at pH 5 was faithfully restored in the rgpF+ complement strain. When gene expression for the catalytic subunit of the ATPase complex was analyzed by quantitative real-time PCR (qRT-PCR), atpB was upregulated in response to steady-state growth at pH 5 in all strains tested, although not significantly in the rgpF+ strain (Fig. 5B).
FIG 5.
ATPase activity is reduced in the ΔrgpF mutant. (A) Cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains were grown in continuous culture to steady state at pH values of 7 and 5. The release of inorganic phosphate was measured as a readout for membrane-bound ATPase activity. Data were normalized to activity levels for UA159 grown to steady state at pH 7 (assigned a value of 1) and are represented as mean values ± SD from three independent chemostat cultures. Statistical significance was determined by pairwise comparison using Student's t test († and ‡, P ≤ 0.05; *, P ≤ 0.02). Note the values for the ΔrgpF mutant are not statistically significant in a comparison of the activities in pH 7- and pH 5-grown cultures. (B) Transcription of atpB was determined by qRT-PCR using RNA isolated from cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains grown in continuous culture to steady state at pH values of 7 and 5. Samples from three independent chemostat cultures were measured in triplicate and are represented as mean values ± SD. Statistical significance was determined by pairwise comparison using Student's t test († and ‡, P ≤ 0.001).
Loss of rgpF attenuates virulence in S. mutans.
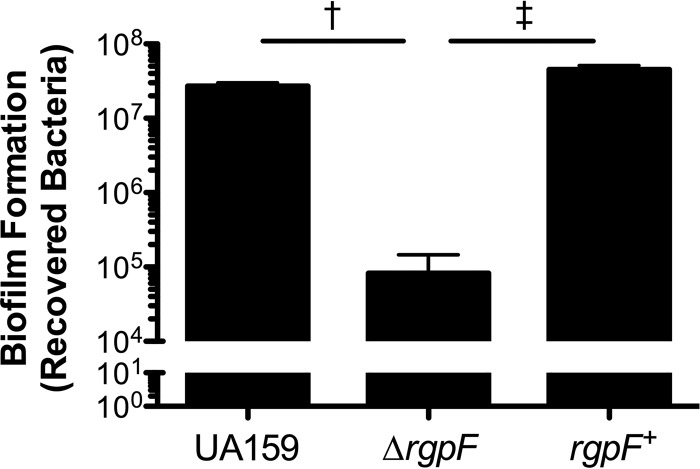
While stress tolerance and acid adaptation contribute to the persistence of S. mutans within the oral cavity, adherence to the tooth surface and subsequent dominance within the multispecies biofilm that develops are critical to the pathogenicity of the organism. To test the capacity of the ΔrgpF mutant strain to form model biofilms, hydroxyapatite discs were coated with human saliva in order to mimic the glycoprotein pellicle that forms on the surface of teeth. Bacteria were cultured on saliva-coated hydroxyapatite (sHA) discs in the presence of sucrose for 5 days and then the total number of bacteria was determined (Fig. 6). Similar quantities were recovered for both the UA159 and rgpF+ strains (2.7 × 107 and 4.5 × 107 CFU · ml−1, respectively), while the number of cells recovered from biofilm cultures of the ΔrgpF strain (8.2 × 104 CFU · ml−1) was decreased by 2.5 log (P ≤ 0.001).
FIG 6.
The ΔrgpF mutant forms less-robust biofilms. Cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains were grown on sHA discs in the presence of TY medium plus 1% (wt/vol) sucrose for 5 days. Adherent cells were removed from discs and enumerated to determine the total bacteria. Data are represented as mean CFU values ± SD. Statistical significance was determined by pairwise comparison using Student's t test, comparing the ΔrgpF strain to both UA159 and the rgpF+ complement († and ‡, P ≤ 0.001).
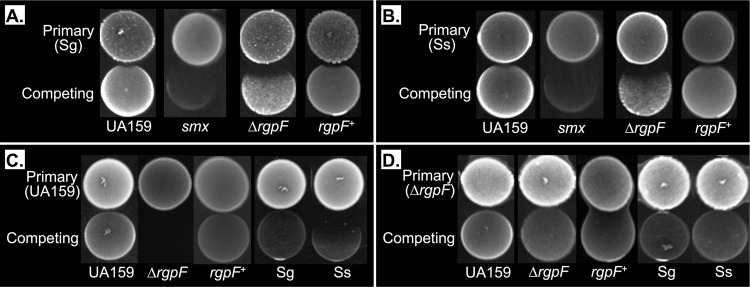
Among the challenges S. mutans faces within the multispecies dental plaque biofilm is reactive oxygen species in the form of H2O2 produced by the peroxigenic streptococci S. gordonii and S. sanguinis (35). To determine the effects of H2O2 production on ΔrgpF mutant outgrowth, S. gordonii DL-1 (Fig. 7A) or S. sanguinis 10904 (Fig. 7B) was spotted onto agar plates as a primary culture and allowed to grow; then, competitor strains of S. mutans were spotted immediately adjacent to the primary spot. The outgrowth of both UA159 and the rgpF+ complement was minimally affected; however, a much larger zone of growth inhibition was consistently observed for the ΔrgpF mutant strain. An S. mutans strain carrying a mutation in the base excision repair enzyme Smx was also highly susceptible to growth inhibition caused by the peroxigenic streptococci and included as a positive control for H2O2 sensitivity (36). Assays were repeated under anaerobic conditions, and outgrowth was fully restored for all susceptible strains (Fig. S2A and B).
FIG 7.
Deletion of rgpF results in loss of competitive fitness in S. mutans. Competitive spot plating was performed as described in Materials and Methods using as primary spots S. gordonii DL-1 (Sg) (A) and S. sanguinis 10904 (Ss) (B) as oxidative stress agents, and S. mutans UA159 (C) and the S. mutans ΔrgpF mutant (D) as acidic stress agents. The S. mutans smx mutant strain was included as a positive oxidative stress-sensitive control. The strain carries a mutation in smx (SMU.1649) encoding a base excision repair enzyme (36).
In contrast, S. mutans relies on its acidogenesis to gain an advantage over competing organisms. A similar competition assay was used to measure the ability of the ΔrgpF mutant strain to withstand and produce acidic end products from glycolysis. Here, S. mutans strains were used as primary spots. Both S. gordonii and S. sanguinis had diminished outgrowth when plated adjacent to S. mutans UA159, with a greater degree of inhibition observed for S. sanguinis (Fig. 7C). Outgrowth of the ΔrgpF mutant strain was completely inhibited by UA159, while growth was restored in the rgpF+ complement strain. When the ΔrgpF mutant was used as the primary spot (Fig. 7D), no growth inhibition was observed for any of the competing strains. Assays were then repeated on agar medium that had been buffered to neutral pH (Fig. S2C and D). All growth susceptibilities were fully restored, suggesting that the observed zones of clearance were caused by acid production, further highlighting the acid sensitivity of the ΔrgpF mutant strain.
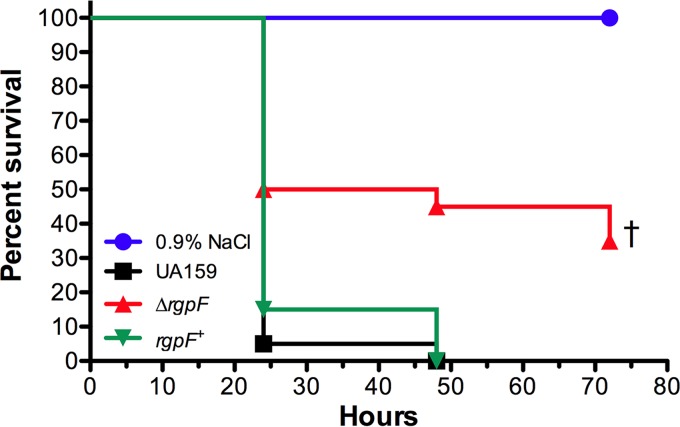
Lastly, infection of larvae of the greater waxworm G. mellonella was used to determine the impact of the deletion of rgpF on S. mutans virulence. The insect relies on specialized phagocytic cells called hemocytes to generate superoxide and combat microbial infections (37). Following the injection of S. mutans UA159 and the rgpF+ complement, the larvae demonstrated rapid melanization, with greater than 80% larval kill observed by 24 h postinfection (Fig. 8). In contrast, the survival of larvae was significantly enhanced (P ≤ 0.001) when injected with the ΔrgpF mutant strain. These data further support the observed oxidative stress sensitivity of ΔrgpF and demonstrate an overall attenuation of virulence for the mutant in an in vivo model system.
FIG 8.
Virulence of the S. mutans rgpF deletion mutant strain is attenuated in a G. mellonella infection model. Larvae of the greater wax worm Galleria mellonella were injected with 107 CFU of S. mutans UA159, the ΔrgpF strain, the rgpF+ strain, or the 0.9% NaCl control. Kaplan-Meier survival plots were used to demonstrate rate of larvae kill. Assays were performed in triplicate with 20 larvae injected per group, and the data shown are representative of a typical experiment. Statistical significance (†, P ≤ 0.001) was determined by pairwise comparison using Student's t test, comparing the ΔrgpF mutant to both UA159 and the rgpF+ complement.
DISCUSSION
While the importance of WTAs has been demonstrated for many organisms, these structures are not ubiquitous among Gram-positive bacteria. Notably, S. mutans lacks the machinery for WTA synthesis but does produce the RGP, which is similarly surface associated and abundant in the cell wall. This distinctive polysaccharide structure is composed of repeating units of l-rhamnose to form a core backbone, onto which glucose is linked as side chains. Historically, the RGP structure has been used as a means of serotype distinction within the strain family, with grouping based on the specific linkage used to anchor glucose onto the backbone (38, 39). Formation of the RGP has been investigated, and the stepwise activities of the enzymes involved are summarized in Fig. 1B (20, 29). The deletion of rgpG, rgpA, rgpB, and rgpF in our study produced mutants with severely attenuated fitness, highlighting the importance of the mature RGP structure, while also demonstrating that it is dispensable for viability. This is in stark contrast to a recent concurrent study, where the authors describe the disruption of rgpG in S. mutans and reported no differences in growth rate or stress sensitivity (27). While the reason for these discrepancies is yet unclear, our data are in agreement with studies conducted in related streptococcal species, as well as the general consensus of reports on the importance of WTA and other CWP structure formation (23, 24, 40–49). Interestingly, our attempts to produce single-deletion mutants of either rgpC or rgpD failed, suggesting their essentiality under the conditions tested. These genes produce the enzymatic subunits making up the ABC transporter system thought to be required for extracellular transport of the mature RGP structure (18). A similar phenomenon has been described for deletion of the transport system of WTA in S. aureus, which was proven to be a conditionally lethal event, as simultaneous disruption of WTA synthesis rescued the mutants (50). It is possible that although RGP export is disrupted in a ΔrgpC or ΔrgpD mutant, the structure's synthesis proceeds unabated, leading to accumulation of the polymer and sequestration of the universal undecaprenol phosphate required for cell wall synthesis, ultimately poisoning the cell.
The profound effects on S. mutans fitness consequent to the deletion of rgpF led us to investigate the stress response of the mutant strain, with specific attention on stress factors routinely encountered in the oral cavity. Experiments using acidified brain heart infusion (BHI) medium to support ΔrgpF mutant propagation resulted in retarded growth in comparison to the parent strain, highlighting the sensitivity of this strain to low pH. In addition, the mutant strain was highly susceptible to acid challenge when cultures were grown to steady state at pH 5, demonstrating an inability to appropriately acid adapt. Oxidative stress tolerance has also been shown to be part of the ATR in S. mutans, and the ΔrgpF mutant strain was exquisitely sensitive to H2O2 challenge, regardless of growth conditions, further exemplifying its heightened sensitivity to stress (30, 31). The specific mechanisms that contribute to induction of the ATR involve fundamental changes in S. mutans physiology, particularly in efforts to maintain a more alkaline cytosol as the microenvironment it inhabits becomes acidified. As such, these changes are central to the virulence profile of S. mutans. Our lab and others have reported on a number of regulators and biosynthetic systems that are required for induction of the ATR (31, 32, 51). Notably, it has been speculated that changes associated with the plasma membrane of S. mutans confer a degree of membrane fluidity that allows for an appropriate proton gradient across the membrane and more efficient alignment of membrane-bound proteins specifically utilized during acid adaptation (9, 52). Similar observations have been made during the heat shock response of Bacillus subtilis, as well as regarding the ability of Lactobacillus casei to tolerate acid stress (53, 54). Here, we show that proton permeability is impacted in UA159 following growth at pH 5 versus pH 7, with reduced proton flow into the cells and an elevated ΔpH following acid adaptation. Conversely, in the ΔrgpF mutant strain, proton flow into the cell proceeded at a more reduced rate, and terminal pH recordings were also significantly lower than in the parent strain, demonstrative of a disturbed proton gradient across the membrane. This may be caused by a reduced ability of the mutant strain to effectively regulate intracellular pH and, as a consequence, the ΔrgpF mutant strain maintains a more acidic cytosol, which would, in turn, negatively impact the proton gradient. The ability to trap protons close to the membrane and influence the gradient has been attributed to WTA in S. aureus due to the presence of phosphate groups within the structure's backbone; however, this is unlikely to be the case for S. mutans, since the RGP is uncharged (55).
In concert with membrane permeability, the proton-translocating F1Fo ATPase functions primarily to extrude protons from the cell at the expense of ATP (34). During acid adaptation, this mechanism is thought to be the principal means by which S. mutans maintains a more alkaline cytosol than its acidic outside environment (56). Failure of the ΔrgpF strain to activate this complex expounds upon the membrane impairment phenotype of this mutant and may likely explain, at least in part, the observed collapse of proton gradient across the membrane, allowing unchecked accumulation of protons within the cell. A hallmark of the S. mutans ATPase is robust induction at low pH (8, 33). It is possible that disruption of RGP prevents the optimal conformation of specific membrane-bound proteins, such as the ATPase complex. In support of this hypothesis, the gene expression of atpB, which encodes the catalytic subunit of the ATPase, was upregulated under acid adaptation conditions, even in the ΔrgpF mutant strain. This signifies an appropriate response of the mutant strain to acid stress at the transcriptional level but a failure of the protein complex to become induced. These data suggest that in the ΔrgpF strain, the disruption of this particular stress response mechanism occurs posttranslationally. While increases in membrane fluidity have been shown to positively influence ATPase activity in mammalian systems, this process has not been specifically interrogated in bacteria (57–59). Experiments are under way to investigate cellular morphology and physiology under various rgp-deficient backgrounds to help address this question.
Adherence to the tooth surface and robust biofilm growth are fundamental requirements for S. mutans cariogenicity. We used a model in vitro system where a hydroxyapatite substrate was coated with salivary glycoproteins to mimic the tooth surface and pellicle. Bacterial yield was significantly less robust from the ΔrgpF mutant biofilm cultures than from both the parent and complement strains. While the reduced growth rate of the ΔrgpF mutant strain may contribute to this finding, a more likely explanation, based on observations of the biofilm cultures, is that the ΔrgpF strain is impaired in its ability to adhere to the model sHA surfaces. Most of these cells remained in planktonic culture throughout the assay and were also more readily disrupted from discs during the washing steps (compared to both the parent strain and rgpF+ complement). It is yet unclear if the RGP structure itself can contribute to S. mutans adherence or if the disruption of its formation affects orientation or activity of important anchoring proteins in the cell wall, similar to the hypothesis proposed here for the ATPase enzyme. Although not shown specifically in S. mutans, it is known that the depletion of CWP in other Gram-positive bacteria leads to mislocalization of cell wall proteins and reduced adherence (14, 40, 60). We are currently investigating how particular proteins interact with the cell wall in the various rgp deletion backgrounds, and future experiments will address RGP-mediated adherence in an animal model that more faithfully resembles caries progression.
Competitive interactions between the hundreds of organisms inhabiting dental plaque biofilm ultimately dictate the pathogenic propensity of these communities. The oral peroxigenic streptococci S. gordonii and S. sanguinis can produce bactericidal levels of H2O2, inhibiting S. mutans growth (61). Both available oxygen and low-glucose concentration have been shown to positively influence H2O2 generation from these organisms in a pyruvate oxidase-dependent manner (35, 61, 62). For S. mutans to survive and dominate, the bacterium must not only withstand the toxic products produced by neighbors but also gain a competitive advantage via its own acidogenesis. These interactions were severely diminished in the ΔrgpF mutant, in that this strain was more sensitive to H2O2 production and was incapable of inhibiting the outgrowth of the peroxigenic streptococci; further, the growth of the ΔrgpF strain was arrested by acid production arising from the S. mutans parent strain. Mutacin production by S. mutans may act antagonistically against other bacteria; however, the contribution of these peptides on growth inhibition of the ΔrgpF mutant strain was not investigated in this study. Previous reports have demonstrated that both mutacin I and mutacin IV of S. mutans are influenced by cell density; thus, it cannot be ruled out that the release of these peptides may have affected the outgrowth of competing strains in our study (63). Nonetheless, the inhibitory effects of S. mutans UA159 described above were completely abolished when the assays were performed using agar medium that was buffered to neutral pH, indicating that the phenomenon was primarily driven by acidogenesis. Likewise, outgrowth of the ΔrgpF strain during peroxigenic competition assays was similarly restored under anaerobic conditions, demonstrating that the mutant strain is specifically influenced by the H2O2 generated by the competing peroxigenic bacteria. These data further exemplify the stress sensitivities of the ΔrgpF mutant strain and also suggest that disruption of RGP formation may ablate the competitive fitness of S. mutans.
Perturbation of RGP synthesis via rgpF deletion also resulted in attenuated virulence and a reduced rate of larval death using the greater wax worm (G. mellonella) model of virulence. These insects use specialized innate immune cells called hemocytes to phagocytose invading microorganisms and kill by oxidative burst, much like mammalian neutrophils (37). Their utility has been demonstrated for virulence studies of numerous Gram-positive bacteria, including S. mutans, as a means of determining susceptibilities to antimicrobial peptides and tolerance of oxidative stress (64–66). It is noteworthy that the RGP structures of S. mutans have been implicated in a variety of mechanisms required for the more invasive pathology of the organism, infective endocarditis. The invasion of endothelial cells by S. mutans has been shown to be serotype specific, while the RGP structure itself has been suggested to trigger platelet aggregation and protect cells from phagocytosis by leukocytes (67–69). The fact that the ΔrgpF mutant strain induced significantly less mortality in G. mellonella, to our knowledge, represents the first time these surface polysaccharides of S. mutans have been directly tied to virulence in an in vivo system, and these findings may have important implications for better understanding the role of RGP on the pathogenic capabilities of the organism.
In conclusion, the present study has clearly demonstrated that the disruption of RGP synthesis greatly impairs the overall fitness of S. mutans. The failure of the ΔrgpF mutant to effectively tolerate both acid and oxidative stresses, coupled with a demonstrative loss of virulence in the mutant strain, establishes the importance of RGP in S. mutans pathogenicity. Proper understanding of the role of RGP in S. mutans biology may elucidate novel therapeutic strategies while also providing a broader appreciation of cell wall biology in Gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains and culture.
Streptococcus mutans strain UA159, a clinical isolate of serotype c, was used as the parent strain in the present study (70). Single deletions of each gene involved in RGP synthesis (rgpA to rgpI) were constructed by replacement of the target open reading frames by an erythromycin resistance cassette, as previously described (28).
A genetic complement strain was constructed in the ΔrgpF mutant background to confirm that the defects observed with this mutant were indeed due to a specific loss of the gene. The rgpF open reading frame was PCR amplified from UA159 genomic DNA with primers (forward, [5′-ATTTAAAAATAGATCTGACCATTCCTACAAAAAT-3′] and reverse [5′-GAGCTCGAATAGATCTTTCAATTGTTTCATGACT-3′]) designed to contain a BglII restriction site (underlined). The resulting amplicon contained 15-bp overhangs on each end, complementary to the integration site of the cloning vector pSUGK-Bgl (bold font above), which had been similarly linearized with BglII (New England BioLabs, Ipswich, MA) (30). The linearized vector and the rgpF amplicon were combined using the In-Fusion HD cloning kit (Clontech Laboratories, Inc., Mountain View, CA), in accordance with the manufacturer's recommendations. The ligation reaction mixture was transformed into Escherichia coli Stellar competent cells (Clontech Laboratories), and transformants were selected on LB agar medium containing 50 μg · ml−1 kanamycin. Positive transformants were verified by colony PCR and nucleotide sequencing using rgpF-specific primers (forward [5′-CATAGATCTGACCATTCCTACAAAAAT-3′] and reverse [5′-CATAGATCTTTCAATTGTTTCATGACT-3′]). A plasmid was isolated from a positive construct, pSUGKrgpF, and used to transform the ΔrgpF mutant, the S. mutans strain carrying the deletion, for integration into the gtfA (SMU.881) locus using previously published methods (30). Transformants were selected on BHI agar medium containing 1 mg · ml−1 kanamycin. Positive transformants were verified using colony PCR, and one successfully transformed strain was named rgpF+.
Bacterial culture.
Bacterial strains were grown in brain heart infusion (BHI) medium (BD/Difco, Franklin Lakes, NJ) at 37°C in a 5% (vol/vol) CO2–95% air atmosphere.
S. mutans UA159, the ΔrgpF mutant, and the rgpF+ complement were also grown under continuous culture conditions using a BioFlo 2000 fermenter (New Brunswick Scientific, Edison, NJ) in TY medium (3% tryptone, 0.1% yeast extract, 0.5% KOH, and 1 mM H3PO4) containing 1% glucose, as described previously (71). Cultures of all 3 strains were grown at a constant dilution rate of 0.144 h−1 under glucose-limiting (2.3 mM) conditions. Culture pH was continually maintained via the addition of 2 N KOH and verified by an indwelling pH probe (Mettler Toledo, Billerica, MA). Cultures were grown for 10 generations to steady state at pH 7 and pH 5, and aliquots from both conditions were removed for downstream analyses.
Growth curves.
Growth rates were determined using a BioScreen C plate reader (Growth Curves USA, Piscataway, NJ). Overnight cultures of the S. mutans UA159, ΔrgpF, and rgpF+ strains grown in BHI medium were subcultured 1:20 (UA159 and rgpF+ strain) or 1:10 (ΔrgpF strain) into fresh BHI medium and incubated at 37°C in a 5% (vol/vol) CO2–95% air atmosphere until cultures reached an optical density at 600 nm (OD600) of 0.3. A 10-μl aliquot was used to inoculate wells of a microtiter plate containing 300 μl of test medium. Cultures were grown in either BHI medium or BHI medium titrated to pH values of 7.4, 6.5, 6.0, 5.8, 5.6, or 5.4 in order to assess acid sensitivity. Assays were performed at 37°C, and the OD600 was continually read at 30-min intervals following 10 s of shaking at medium amplitude. Generation times were calculated using the formula 0.3/[(N − N0)/(T − T0)], where N represents the mean OD600 value at the end of exponential phase and N0 represents the mean OD600 at the beginning of exponential phase. T and T0 refer to the times in minutes that correspond to the OD600 values for N and N0, respectively. Statistical significance (P ≤ 0.001) was determined by pairwise comparison using Student's t test.
Stress sensitivity assays.
Bacterial susceptibility to acid and hydrogen peroxide (H2O2) shock was determined following steady-state growth. Samples from cultures of S. mutans UA159, ΔrgpF and rgpF+ strains grown at pH 7 and pH 5 were harvested and collected by centrifugation at 4°C and 2,272 × g for 10 min. Pellets were resuspended in either 0.1 M glycine (pH 2.5) (acid stress) or BHI medium with addition of 0.05% H2O2 (oxidative stress). Immediately, a 0.1-ml sample was removed, serially diluted 10-fold, plated onto BHI agar medium, and incubated at 37°C in a 5% (vol/vol) CO2–95% air atmosphere for enumeration of the starting inoculum. Bacteria were exposed to stress conditions (either acid or H2O2) for 60 min, and surviving organisms were enumerated. The data are represented as log survival after 60 min compared to the inoculum. All assays were performed in duplicate from three independent chemostat cultures.
Proton permeability.
The capacity of cells to restrict proton movement across their membrane was assayed as previously described (34). Briefly, cells were collected from steady-state cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains grown at pH 7 and pH 5, washed once in wash buffer (5 mM MgCl2), and then resuspended in starvation buffer (20 mM phosphate buffer [pH 7.2] containing 1 mM MgCl2 and 50 mM KCl) at a volume corresponding to 5 mg cell dry weight · ml−1. Samples were incubated for 120 min at 37°C to starve cells and eliminate stores of ATP. After starvation, cells were pelleted by centrifugation at 4°C and 2,272 × g for 10 min and resuspended in a solution of 50 mM KCl and 1 mM MgCl2 to a cell density of 20 mg cell dry weight · ml−1 (or one-fourth the volume of starvation). The pH of the cell suspension was rapidly adjusted to 4.5 (time = 0) and continually monitored over time. At 50 min, butanol was added to cell suspensions (10% [vol/vol]) in order to disrupt cells, allowing for equilibration of cytosolic pH with that of the solution, and a terminal pH reading was measured at the 80-min time point. Assays were performed in duplicate from three independent chemostat cultures.
ATPase activity assay.
The activity levels of the membrane-bound proton-translocating F1Fo ATPase were determined by measuring the release of inorganic phosphate from bacterial membrane preparations following the addition of ATP, using previously described techniques (52, 72). In brief, 50-ml aliquots were removed from steady-state cultures of S. mutans UA159, ΔrgpF, and rgpF+ strains at pH 7 and pH 5. Cells were pelleted by centrifugation at 4°C and 2,272 × g for 10 min, washed once in membrane buffer (75 mM Tris [pH 7.0], 10 mM MgSO4), and then resuspended in 1 ml of membrane buffer. Cells were permeabilized by the addition of 100 μl of toluene and two freeze-thaw cycles using an ethanol-dry ice bath and a 37°C water bath, respectively. Permeabilized cells were collected by microcentrifugation at 16,100 × g at room temperature for 10 min. The toluene-containing supernatant was discarded, and pellets were resuspended in membrane buffer, aliquoted, and stored at −80°C. To perform the assay, a 300-μl aliquot of permeabilized cells was added to 9 ml of ATPase buffer (50 mM Tris maleate [pH 6.0], 10 mM MgSO4). To this, ATP (0.5 M [pH 6.0]) was added to begin the reaction, and a sample was immediately removed for time zero. After 60 min, samples were removed and added to 20% trichloroacetic acid (TCA) to stop the reaction. TCA was then cleared by centrifugation (4°C, 2,272 × g for 10 min), and 1 ml of the resulting supernatants was transferred to tubes containing 1.5 ml of H2O and 0.5 ml of acid molybdate solution (1.25 g · dl−1 in 2.5 N sulfuric acid; Sigma Chemical Company, St. Louis, MO). For the colorimetric reaction, 125 μl of Fiske-Subbarow solution (Sigma Chemical Company) was added to each tube. After a 10-min incubation at room temperature, absorbance was measured at OD660, and the amount of inorganic phosphate released was determined by comparison to a phosphate standard curve. Data were normalized to the total protein concentration measured from each sample using the bicinchoninic acid kit (Sigma Chemical Company). Assays were performed using samples derived from three independent chemostat cultures.
Biofilm formation.
The ability of the ΔrgpF mutant to form model biofilms was assessed as previously reported (73). Briefly, bacterial cultures were grown in TY medium plus 1% (wt/vol) sucrose in a 24-well microtiter plate where saliva-coated hydroxyapatite (sHA) discs were vertically suspended within the wells. Plates were incubated at 37°C in a 5% (vol/vol) CO2–95% air atmosphere. Each day, discs were transferred to wells of a new plate containing fresh medium, for a total of 5 days. Discs were dip-washed into sterile phosphate-buffered saline (PBS) three times to remove loosely adhered cells, and biofilms were then recovered from discs by scraping with a sterile spatula. Clumps of cells were disrupted by gentle sonication. Cells were then serially diluted 10-fold and plated onto BHI agar medium for enumeration. Data are represented as the mean CFU recovered from three independent experiments, each performed in duplicate.
Competition spot assays.
To determine the competitive fitness of the ΔrgpF mutant, an agar spotting assay was employed, similar to previously published methods (61). For these, the oral commensal bacteria Streptococcus gordonii DL1 and Streptococcus sanguinis 10904, members of the peroxigenic streptococcal group, were used as primary cultures in peroxigenic competition assays to determine the susceptibility of S. mutans to H2O2 production. Acidogenic competition assays were performed using S. mutans UA159 and the ΔrgpF mutant as primary cultures. For primary spots, bacteria were grown overnight in BHI medium at 37°C in a 5% (vol/vol) CO2–95% air atmosphere, and then subcultured into fresh BHI medium until cultures reached an OD600 of 0.4. An 8.0-μl aliquot was then spotted onto prewarmed BHI agar and incubated overnight at 37°C in a 5% (vol/vol) CO2–95% air atmosphere. Concurrently, competing bacteria were similarly grown overnight and subcultured, and an 8.0-μl aliquot was spotted immediately adjacent to the primary spot. Plates were returned to 37°C in a 5% (vol/vol) CO2–95% air atmosphere and incubated overnight. An S. mutans smx mutant strain (SMU.1649, responsible for the production of the base-excision repair enzyme Smx) was included as a positive control for oxidative stress susceptibility (36). The data are shown as representative images of outgrowth inhibition from three independent experiments performed in triplicate.
Galleria mellonella infection.
G. mellonella killing assays were performed as previously described (30, 64). Larval insects were purchased from Vanderhorst Wholesale, Inc. (St. Marys, OH), stored at 4°C in the dark, and used within 7 days of shipment. Uniformly pigmented larvae, ranging from 200 to 300 mg in weight, were used for subsequent infection. Overnight cultures of S. mutans UA159 and the ΔrgpF and rgpF+ strains were normalized to an OD600 value of 1.0 in 10 ml and then resuspended to a final volume of 1.0 ml to use as inocula for injections. Twenty larvae per treatment group were injected with 5 μl of bacteria into the hemocoel via the last proleg. A group injected with 0.9% NaCl was used as a negative control in each experiment. After injection, larvae were incubated at 37°C, monitored, and scored as dead when they displayed no thigmotaxis. Data are represented as percent survival using Kaplan-Meier killing curves, where significance (P ≤ 0.05) was determined by log rank testing with Prism version 4.0 (GraphPad Software, La Jolla, CA).
Quantitative real-time PCR.
RNA was extracted from three independent cultures grown to steady-state conditions at pH 7 and pH 5, according to previously described methods (74, 75). The high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA) was used to generate cDNA from RNA samples using random primers. Primers specific to atpB were used (atpB-RT-forward [5′-ATGCTGGTGAAGTTGTTATGGC-3′] and atpB-RT-reverse [5′-GCAGAAAGGCTATTGGTGTCG-3′]) with Power SYBR green master mix (Applied Biosystems), and the reactions were carried out in a StepOnePlus real-time PCR system (Applied Biosystems). The mRNA copy number was quantified based on a standard curve of PCR products for specific gene targets.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH/NIDCR DE-13683 and DE-17425 (to R.G.Q.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00497-17.
REFERENCES
- 1.Clarke JK. 1924. On the bacterial factor in the aetiology of dental caries. Br J Exp Pathol 5:141–147. [Google Scholar]
- 2.Benjamin RM. 2010. Oral health: the silent epidemic. Public Health Rep 125:158–159. doi: 10.1177/003335491012500202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephan RM. 1944. Intra-oral hydrogen ion concentrations associated with dental caries activity. J Dent Res 23:257–266. doi: 10.1177/00220345440230040401. [DOI] [Google Scholar]
- 4.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abranches J, Chen YYM, Burne RA. 2003. Characterization of Streptococcus mutans strains deficient in EIIABMan of the sugar phosphotransferase system. Appl Environ Microbiol 69:4760–4769. doi: 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker JL, Faustoferri RC, Quivey RG Jr. 2017. Acid-adaptive mechanisms of Streptococcus mutans-the more we know, the more we don't. Mol Oral Microbiol 32:107–117. doi: 10.1111/omi.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicks J, Garcia-Godoy F, Flaitz C. 2004. Biological factors in dental caries enamel structure and the caries process in the dynamic process of demineralization and remineralization (part 2). J Clin Pediatr Dent 28:119–124. doi: 10.17796/jcpd.28.2.617404w302446411. [DOI] [PubMed] [Google Scholar]
- 8.Sutton SV, Marquis RE. 1987. Membrane-associated and solubilized ATPases of Streptococcus mutans and Streptococcus sanguis. J Dent Res 66:1095–1098. doi: 10.1177/00220345870660060201. [DOI] [PubMed] [Google Scholar]
- 9.Quivey RG Jr, Faustoferri R, Monahan K, Marquis R. 2000. Shifts in membrane fatty acid profiles associated with acid adaptation of Streptococcus mutans. FEMS Microbiol Lett 189:89–92. doi: 10.1111/j.1574-6968.2000.tb09211.x. [DOI] [PubMed] [Google Scholar]
- 10.Fozo EM, Quivey RG Jr. 2004. The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J Bacteriol 186:4152–4158. doi: 10.1128/JB.186.13.4152-4158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fozo EM, Scott-Anne K, Koo H, Quivey RG Jr. 2007. Role of unsaturated fatty acid biosynthesis in virulence of Streptococcus mutans. Infect Immun 75:1537–1539. doi: 10.1128/IAI.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell J, Singh AK, Santa Maria JP Jr, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol 6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Gotz F. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 14.Holland LM, Conlon B, O'Gara JP. 2011. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157:408–418. doi: 10.1099/mic.0.042234-0. [DOI] [PubMed] [Google Scholar]
- 15.Boyd DA, Cvitkovitch DG, Bleiweis AS, Kiriukhin MY, Debabov DV, Neuhaus FC, Hamilton IR. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J Bacteriol 182:6055–6065. doi: 10.1128/JB.182.21.6055-6065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho F, Atilano ML, Pombinho R, Covas G, Gallo RL, Filipe SR, Sousa S, Cabanes D. 2015. l-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog 11:e1004919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Wang H, Labroli M, Koseoglu S, Zuck P, Mayhood T, Gill C, Mann P, Sher X, Ha S, Yang SW, Mandal M, Yang C, Liang L, Tan Z, Tawa P, Hou Y, Kuvelkar R, DeVito K, Wen X, Xiao J, Batchlett M, Balibar CJ, Liu J, Xiao J, Murgolo N, Garlisi CG, Sheth PR, Flattery A, Su J, Tan C, Roemer T. 2016. TarO-specific inhibitors of wall teichoic acid biosynthesis restore beta-lactam efficacy against methicillin-resistant staphylococci. Sci Transl Med 8:329ra332. doi: 10.1126/scitranslmed.aad7364. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita Y, Tsukioka Y, Tomihisa K, Nakano Y, Koga T. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J Bacteriol 180:5803–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita Y, Shibata Y, Nakano Y, Tsuda H, Kido N, Ohta M, Koga T. 1999. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J Bacteriol 181:6556–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata Y, Yamashita Y, Ozaki K, Nakano Y, Koga T. 2002. Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect Immun 70:2891–2898. doi: 10.1128/IAI.70.6.2891-2898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xayarath B, Yother J. 2007. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol 189:3369–3381. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James DB, Yother J. 2012. Genetic and biochemical characterizations of enzymes involved in Streptococcus pneumoniae serotype 2 capsule synthesis demonstrate that Cps2T (WchF) catalyzes the committed step by addition of beta1-4 rhamnose, the second sugar residue in the repeat unit. J Bacteriol 194:6479–6489. doi: 10.1128/JB.01135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Sorge NM, Cole JN, Kuipers K, Henningham A, Aziz RK, Kasirer-Friede A, Lin L, Berends ETM, Davies MR, Dougan G, Zhang F, Dahesh S, Shaw L, Gin J, Cunningham M, Merriman JA, Hutter J, Lepenies B, Rooijakkers SHM, Malley R, Walker MJ, Shattil SJ, Schlievert PM, Choudhury B, Nizet V. 2014. The classical Lancefield antigen of group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe 15:729–740. doi: 10.1016/j.chom.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Beek SL, Le Breton Y, Ferenbach AT, Chapman RN, van Aalten DM, Navratilova I, Boons GJ, McIver KS, van Sorge NM, Dorfmueller HC. 2015. GacA is essential for group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD). Mol Microbiol 98:946–962. doi: 10.1111/mmi.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata Y, Yamashita Y, van der Ploeg JR. 2009. The serotype-specific glucose side chain of rhamnose-glucose polysaccharides is essential for adsorption of bacteriophage M102 to Streptococcus mutans. FEMS Microbiol Lett 294:68–73. doi: 10.1111/j.1574-6968.2009.01546.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob Agents Chemother 46:3756–3764. doi: 10.1128/AAC.46.12.3756-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De A, Liao S, Bitoun JP, Roth R, Beatty WL, Wu H, Wen ZT. 2017. Deficiency of RgpG causes major defects in cell division and biofilm formation, and deficiency of LCP proteins leads to accumulation of cell wall antigens in culture medium by Streptococcus mutans. Appl Environ Microbiol doi: 10.1128/AEM.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quivey RG Jr, Grayhack EJ, Faustoferri RC, Hubbard CJ, Baldeck JD, Wolf AS, MacGilvray ME, Rosalen PL, Scott-Anne K, Santiago B, Gopal S, Payne J, Marquis RE. 2015. Functional profiling in Streptococcus mutans: construction and examination of a genomic collection of gene deletion mutants. Mol Oral Microbiol 30:474–495. doi: 10.1111/omi.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozaki K, Shibata Y, Yamashita Y, Nakano Y, Tsuda H, Koga T. 2002. A novel mechanism for glucose side-chain formation in rhamnose-glucose polysaccharide synthesis. FEBS Lett 532:159–163. doi: 10.1016/S0014-5793(02)03661-X. [DOI] [PubMed] [Google Scholar]
- 30.Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG Jr. 2012. Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol 78:1215–1227. doi: 10.1128/AEM.06890-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker JL, Abranches J, Faustoferri RC, Hubbard CJ, Lemos JA, Courtney MA, Quivey R Jr. 2015. Transcriptional profile of glucose-shocked and acid-adapted strains of Streptococcus mutans. Mol Oral Microbiol 30:496–517. doi: 10.1111/omi.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Len AC, Harty DW, Jacques NA. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiol 150:1353–1366. doi: 10.1099/mic.0.26888-0. [DOI] [PubMed] [Google Scholar]
- 33.Kuhnert WL, Zheng G, Faustoferri RC, Quivey RG Jr. 2004. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J Bacteriol 186:8524–8528. doi: 10.1128/JB.186.24.8524-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender GR, Sutton SV, Marquis RE. 1986. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun 53:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faustoferri RC, Hahn K, Weiss K, Quivey RG Jr. 2005. Smx nuclease is the major, low-pH-inducible apurinic/apyrimidinic endonuclease in Streptococcus mutans. J Bacteriol 187:2705–2714. doi: 10.1128/JB.187.8.2705-2714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K. 2005. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun 73:4161–4170. doi: 10.1128/IAI.73.7.4161-4170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard DG, Coligan JE, Speed SE, Gray BM. 1981. Carbohydrate fingerprints of streptococcal cells. J Clin Microbiol 13:89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano K, Nomura R, Nakagawa I, Hamada S, Ooshima T. 2005. Role of glucose side chains with serotype-specific polysaccharide in the cariogenicity of Streptococcus mutans. Caries Res 39:262–268. doi: 10.1159/000084831. [DOI] [PubMed] [Google Scholar]
- 40.Caliot É, Dramsi S, Chapot-Chartier MP, Courtin P, Kulakauskas S, Pechoux C, Trieu-Cuot P, Mistou MY. 2012. Role of the group B antigen of Streptococcus agalactiae: a peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog 8:e1002756. doi: 10.1371/journal.ppat.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meredith TC, Swoboda JG, Walker S. 2008. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J Bacteriol 190:3046–3056. doi: 10.1128/JB.01880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 43.Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dengler V, Meier PS, Heusser R, Kupferschmied P, Fazekas J, Friebe S, Staufer SB, Majcherczyk PA, Moreillon P, Berger-Bachi B, McCallum N. 2012. Deletion of hypothetical wall teichoic acid ligases in Staphylococcus aureus activates the cell wall stress response. FEMS Microbiol Lett 333:109–120. doi: 10.1111/j.1574-6968.2012.02603.x. [DOI] [PubMed] [Google Scholar]
- 45.Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, McGuinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T. 2013. Discovery of wall teichoic acid inhibitors as potential anti-MRSA beta-lactam combination agents. Chem Biol 20:272–284. doi: 10.1016/j.chembiol.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sewell EW, Brown ED. 2014. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot (Tokyo) 67:43–51. doi: 10.1038/ja.2013.100. [DOI] [PubMed] [Google Scholar]
- 48.Labroli MA, Caldwell JP, Yang C, Lee SH, Wang H, Koseoglu S, Mann P, Yang SW, Xiao J, Garlisi CG, Tan C, Roemer T, Su J. 2016. Discovery of potent wall teichoic acid early stage inhibitors. Bioorg Med Chem Lett 26:3999–4002. doi: 10.1016/j.bmcl.2016.06.090. [DOI] [PubMed] [Google Scholar]
- 49.Bhavsar AP, Erdman LK, Schertzer JW, Brown ED. 2004. Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J Bacteriol 186:7865–7873. doi: 10.1128/JB.186.23.7865-7873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasquina L, Santa Maria JP Jr, McKay Wood B, Moussa SH, Matano LM, Santiago M, Martin SE, Lee W, Meredith TC, Walker S. 2016. A synthetic lethal approach for compound and target identification in Staphylococcus aureus. Nat Chem Biol 12:40–45. doi: 10.1038/nchembio.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Len AC, Harty DW, Jacques NA. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339–1351. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- 52.Cross B, Garcia A, Faustoferri R, Quivey RG. 2016. PlsX deletion impacts fatty acid synthesis and acid adaptation in Streptococcus mutans. Microbiology 162:662–671. doi: 10.1099/mic.0.000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cybulski LE, Albanesi D, Mansilla MC, Altabe S, Aguilar PS, de Mendoza D. 2002. Mechanism of membrane fluidity optimization: isothermal control of the Bacillus subtilis acyl-lipid desaturase. Mol Microbiol 45:1379–1388. doi: 10.1046/j.1365-2958.2002.03103.x. [DOI] [PubMed] [Google Scholar]
- 54.Wu C, Zhang J, Wang M, Du G, Chen J. 2012. Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J Ind Microbiol Biotechnol 39:1031–1039. doi: 10.1007/s10295-012-1104-2. [DOI] [PubMed] [Google Scholar]
- 55.Biswas R, Martinez RE, Gohring N, Schlag M, Josten M, Xia G, Hegler F, Gekeler C, Gleske AK, Gotz F, Sahl HG, Kappler A, Peschel A. 2012. Proton-binding capacity of Staphylococcus aureus wall teichoic acid and its role in controlling autolysin activity. PLoS One 7:e41415. doi: 10.1371/journal.pone.0041415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandre H, Mathieu B, Charpentier C. 1996. Alteration in membrane fluidity and lipid composition, and modulation of H(+)-ATPase activity in Saccharomyces cerevisiae caused by decanoic acid. Microbiology 142:469–475. [DOI] [PubMed] [Google Scholar]
- 58.Cid-Hernández M, Ramirez-Anguiano AC, Ortiz GG, Morales-Sanchez EW, Gonzalez-Ortiz LJ, Velasco-Ramirez SF, Pacheco-Moises FP. 2015. Mitochondrial ATPase activity and membrane fluidity changes in rat liver in response to intoxication with buckthorn (Karwinskia humboldtiana). Biol Res 48:17. doi: 10.1186/s40659-015-0008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee AG. 2004. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta 1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Vergara-Irigaray M, Maira-Litran T, Merino N, Pier GB, Penades JR, Lasa I. 2008. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiol 154:865–877. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng LY, Itzek A, Chen ZY, Kreth J. 2011. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci 3:82–89. doi: 10.4248/IJOS11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kreth J, Merritt J, Zhu L, Shi W, Qi F. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol Lett 265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 64.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC Jr, Eliopoulos GM. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis 199:532–536. doi: 10.1086/596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buckley AA, Faustoferri RC, Quivey RG Jr. 2014. β-Phosphoglucomutase contributes to aciduricity in Streptococcus mutans. Microbiol 160:818–827. doi: 10.1099/mic.0.075754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abranches J, Miller JH, Martinez AR, Simpson-Haidaris PJ, Burne RA, Lemos JA. 2011. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect Immun 79:2277–2284. doi: 10.1128/IAI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chia JS, Lin YL, Lien HT, Chen JY. 2004. Platelet aggregation induced by serotype polysaccharides from Streptococcus mutans. Infect Immun 72:2605–2617. doi: 10.1128/IAI.72.5.2605-2617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuda H, Yamashita Y, Toyoshima K, Yamaguchi N, Oho T, Nakano Y, Nagata K, Koga T. 2000. Role of serotype-specific polysaccharide in the resistance of Streptococcus mutans to phagocytosis by human polymorphonuclear leukocytes. Infect Immun 68:644–650. doi: 10.1128/IAI.68.2.644-650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ajdić D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quivey RG Jr, Faustoferri RC, Clancy KA, Marquis RE. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett 126:257–261. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- 72.Phan TN, Nguyen PT, Abranches J, Marquis RE. 2002. Fluoride and organic weak acids as respiration inhibitors for oral streptococci in acidified environments. Oral Microbiol Immunol 17:119–124. doi: 10.1046/j.0902-0055.2001.00103.x. [DOI] [PubMed] [Google Scholar]
- 73.Duarte S, Klein MI, Aires CP, Cury JA, Bowen WH, Koo H. 2008. Influences of starch and sucrose on Streptococcus mutans biofilms. Oral Microbiol Immunol 23:206–212. doi: 10.1111/j.1399-302X.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- 74.Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol 188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baker JL, Derr AM, Karuppaiah K, MacGilvray ME, Kajfasz JK, Faustoferri RC, Rivera-Ramos I, Bitoun JP, Lemos JA, Wen ZT, Quivey RG Jr. 2014. Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol 196:2166–2177. doi: 10.1128/JB.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.