ABSTRACT
Staphylococcus aureus is a major human pathogen that imposes a great burden on the health care system. In the development of antistaphylococcal modalities intended to reduce the burden of staphylococcal disease, it is imperative to select appropriate models of S. aureus strains when assessing the efficacy of novel agents. Here, using whole-genome sequencing, we reveal that the commonly used strain Newman D2C from the American Type Culture Collection (ATCC) contains mutations that render the strain essentially avirulent. Importantly, Newman D2C is often inaccurately referred to as simply “Newman” in many publications, leading investigators to believe it is the well-described pathogenic strain Newman. This study reveals that Newman D2C carries a stop mutation in the open reading frame of the virulence gene regulator, agrA. In addition, Newman D2C carries a single-nucleotide polymorphism (SNP) in the global virulence regulator gene saeR that results in loss of protein function. This loss of function is highlighted by complementation studies, where the saeR allele from Newman D2C is incapable of restoring functionality to an saeR-null mutant. Additional functional assessment was achieved through the use of biochemical assays for protein secretion, ex vivo intoxications of human immune cells, and in vivo infections. Altogether, our study highlights the importance of judiciously screening for genetic changes in model S. aureus strains when assessing pathogenesis or the efficacy of novel agents. Moreover, we have identified a novel SNP in the virulence regulator gene saeR that directly affects the ability of the protein product to activate S. aureus virulence pathways.
IMPORTANCE Staphylococcus aureus is a human pathogen that imposes an enormous burden on health care systems worldwide. This bacterium is capable of evoking a multitude of disease states that can range from self-limiting skin infections to life-threatening bacteremia. To combat these infections, numerous investigations are under way to develop therapeutics capable of thwarting the deadly effects of the bacterium. To generate successful treatments, it is of paramount importance that investigators use suitable models for examining the efficacy of the drugs under study. Here, we demonstrate that a strain of S. aureus commonly used for drug efficacy studies is severely mutated and displays markedly reduced pathogenicity. As such, the organism is an inappropriate model for disease studies.
KEYWORDS: Newman, Staphylococcus, WGS, genome analysis, infection, pathogenesis, polymorphism, toxins, virulence
INTRODUCTION
Staphylococcus aureus is a versatile human pathogen that is capable of causing a wide spectrum of disease states that can range from mild skin and soft tissue infections to life-threatening bacteremia (1). The pathogen has steadily taken a devastating toll on the health care system for decades, and in response, many inspired efforts have been undertaken to develop therapeutics capable of thwarting the bacterium's deadly effects (2). Concomitant with the demand to develop an effective antistaphylococcal therapy is the need to select the most applicable model(s) for studying disease caused by S. aureus. S. aureus strain Newman (here referred to as Newman) is a commonly used model strain for studying S. aureus pathogenesis (3–13) and in turn has been extensively used to examine the therapeutic efficacy of agents designed to treat S. aureus infection (12, 14–20). Newman was originally isolated from a human patient in 1952 (21, 22) and has since been a fixture in the investigation of S. aureus pathogenesis due in large part to its strong virulence in both animal models and human ex vivo systems.
In addition, Newman has served as a valuable measuring stick for assessing the therapeutic efficacy of agents designed to protect the host from S. aureus infection. The importance of Newman has been demonstrated by multiple studies in animals that have shown reduced S. aureus-caused lethality in cohorts treated with a number of different therapies (12, 14–20). In our own attempts to develop a therapeutic agent to treat S. aureus infection, we discovered significant inconsistencies in the virulence profiles of Newman strains and a derivative thereof, Newman D2C. Specifically, the strain Newman in our laboratory collection, which originates from the same source that was used in determining the original genome sequence (22), differed immensely in virulence compared to the strain Newman D2C obtained from the American Type Culture Collection (ATCC). Newman D2C was originally described as a clumping-factor-positive variant of Newman strain D2 (23), although the relationship between the original Newman strain and the Newman D2 strain is not apparent. As described here, Newman D2C from the ATCC was observed to exhibit reduced virulence in mouse models of infection and in ex vivo assays with human immune cells. This is in stark contrast to the widely used Newman strain described by Baba et al. (22), which is highly virulent in mice and highly cytotoxic to human tissues (3–13, 15, 24). Unfortunately, numerous studies utilize Newman D2C as a model strain for S. aureus virulence (25–31) and, in many cases, refer to the strain as simply “Newman” (25, 27, 31, 32). The use of this strain creates confusion and leads investigators to believe that it is the same strain originally isolated by Duthie et al. in 1952 (33) and sequenced by Baba et al. (22). This error is perhaps defensible, however, as the ATCC lists the chain of custody for Newman D2C as E. Duthie (1952) to J. Hawiger (1970) to the ATCC.
In an effort to understand the differences in virulence we observed between the strain of Newman in our laboratory collection and Newman D2C obtained from the ATCC, we performed whole-genome sequencing of the two strains. In doing so, we discovered multiple mutations in the ATCC strain compared to the strain in our collection. One mutation, a single-nucleotide variant, was identified in the DNA binding domain of the response regulator gene saeR of the sae regulatory system (34, 35). The sae locus contains four open reading frames (ORFs) (saePQRS), among which saeS encodes a two-component sensor histidine kinase and saeR encodes the cognate response regulator (34). The additional genes in the locus, saeP and saeQ, encode a less-well-characterized lipoprotein and a stabilization protein for SaeS, respectively (36, 37). The sae locus consists of two promoters (P1 and P3) that drive the production of four mRNA transcripts: the T1 transcript initiates upstream of saeP and extends the length of the sae locus, the T2 transcript starts in the intergenic region upstream of saeQ, transcript T3 begins directly upstream of saeR, and T4 only extends the length of saeP (38, 39). The sae regulatory system is critical for the expression and production of a number of key S. aureus virulence proteins and is essential for pathogenesis in animal models of infection (40–46). Here, we show that the saeR mutation in strain Newman D2C renders the sae system nonfunctional and thus incapable of activating virulence gene expression. We demonstrate that the saeQRS allele from the Newman strain in our collection can rescue an saeQRS-null mutant, while the Newman D2C saeQRS allele is incapable of restoring pathogenesis in the same genetic background. Interestingly, a mutation in the saeS gene of strain Newman results in an L18P substitution in the first transmembrane domain and renders the sensor kinase constitutively active (47–49). Hence, the saeR mutation in strain Newman D2C nullifies the effects of the SaeSL18P protein (as found here in both strain Newman and Newman D2C). Moreover, this study raises concerns about the value of agents whose therapeutic efficacy has been assessed only with Newman D2C from the ATCC as a model organism and provides a cautionary note with regard to the future use of the strain in investigations that intend to examine S. aureus pathogenesis.
RESULTS
Strain Newman D2C is severely attenuated.
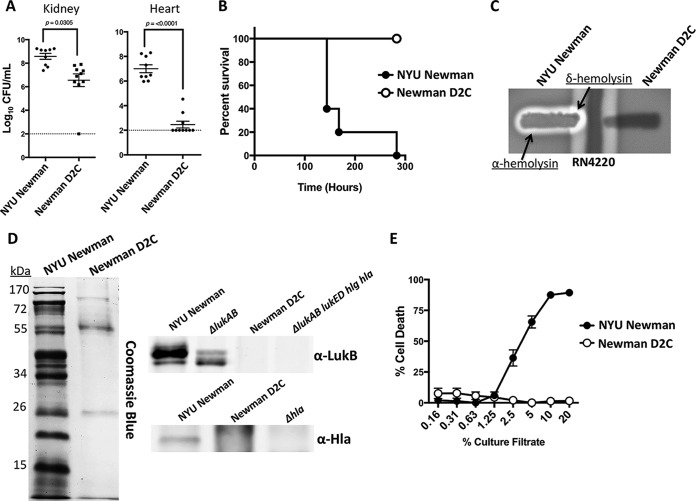
In our collaborative efforts to develop novel antistaphylococcal protein therapeutics with Janssen R&D LLC (2), it was observed that the Janssen group was unable to reproduce virulence phenotypes routinely observed with Newman infections in mice (9–11). Upon investigating the source of their strain, it was determined that, unlike the origin of our strain (22), the group had obtained a strain referred to as Newman D2C from the ATCC. The ATCC lists this strain as having originated from the same source as Newman in our laboratory collection (21), and thus, it was deemed an appropriate model for conducting studies on S. aureus pathogenesis. The Newman in our collection, which we refer to below as “NYU Newman,” originated from the strain that the genome sequence was obtained from (22). First, to corroborate the findings observed at Janssen, we examined the in vivo fitness level of Newman D2C compared to that of NYU Newman. Our results confirmed that the Newman D2C strain that Janssen acquired from the ATCC exhibited a significantly reduced ability to infect mice in multiple critical organs compared to NYU Newman (Fig. 1A). To confirm these observations, we obtained a fresh stock of Newman D2C from the ATCC and tested the strain's ability to induce lethality in mice. Consistent with the original study, we found that whereas the NYU Newman strain was acutely lethal to mice, the newly acquired Newman D2C was again found to be essentially avirulent (Fig. 1B).
FIG 1.
Newman D2C is nonpathogenic. (A) In an intravenous-infection model, S. aureus strain Newman D2C exhibited an attenuated ability to infect mice compared to that of NYU Newman (n = 9 and 10 mice/strain, respectively). The dashed lines indicate the limit of detection for the bacterial burden. Statistics were performed with Student's t test. (B) Newman D2C was nonpathogenic to mice in an assessment of lethality, while NYU Newman was acutely lethal (n = 5 mice). The points indicate individual mouse deaths. (C) Newman D2C was nonhemolytic in blood plates. (D) NYU Newman produced robust amounts of secreted proteins, whereas Newman D2C had minimal detectable proteins and no detectable levels of the toxin Hla or LukAB. (E) Culture filtrates from Newman D2C exhibited no observable cytotoxic properties against primary human PMNs, while NYU Newman was cytotoxic (n = 3 donors). The error bars indicate standard errors of the means.
In our efforts to assess the extent of Newman D2C's attenuation, we next screened for differences in hemolysis in Newman D2C compared to NYU Newman, as hemolysis is associated with S. aureus virulence (50). Using the method described by Adhikari et al. (51), which analyzes the synergistic hemolytic activity of β-hemolysin from RN4220 and δ-hemolysin from the S. aureus strain under examination, we found that Newman D2C was nonhemolytic while NYU Newman demonstrated hemolytic synergism between β-hemolysins, as well as α-hemolysin activity (Fig. 1C). Importantly, the δ-hemolysin synergism is a direct readout of Agr activity, an S. aureus regulatory system that is fundamental to the production of virulence products (46, 52–54). Thus, Newman D2C's lack of δ-hemolysin synergism serves as an indicator of Agr dysfunction in the strain.
Strain Newman is a member of the S. aureus clonal complex lineage 8 (CC8), a lineage known for secreting a multitude of cytotoxic virulence factors (46). To investigate whether there was an alteration in the levels of secreted proteins in Newman D2C, we took concentrated supernatants from in vitro-grown bacteria and examined global secretion profiles. Upon staining for all detectable secreted proteins, it was evident that Newman D2C had markedly reduced amounts of protein secretions compared to NYU Newman (Fig. 1D). To investigate whether the reduction in overall protein secretion corresponded to dampened production of key S. aureus secreted toxins, we also performed immunoblots using antibodies against either the LukB subunit of the LukAB leukotoxin (8, 55) or alpha-toxin (Hla) (56). In accordance with the exoprotein profiles, NYU Newman demonstrated clear toxin production while Newman D2C showed no apparent production of either toxin (Fig. 1D). Of note, the polyclonal anti-LukB antibody recognizes other homologous S. aureus leukotoxins (55), as indicated by the blot. As such, the lack of cross-reactivity in the Newman D2C supernatant supports the notion that a global loss in toxin production had occurred. To further support this observation, we next examined the cytotoxicity of the two Newman strains on primary human polymorphonuclear leukocytes (PMNs). Consistent with the weak exoprotein profile displayed by Newman D2C, it was observed that culture filtrates obtained from Newman D2C were severely attenuated in cytotoxicity on primary PMNs (Fig. 1E). Taken together, these data support the notion that the attenuation observed in Newman D2C from the ATCC can be attributed to either a lack of key virulence factors or a defect in the regulatory machinery necessary to secrete these toxins.
Whole-genome sequencing identifies mutations within Newman D2C.
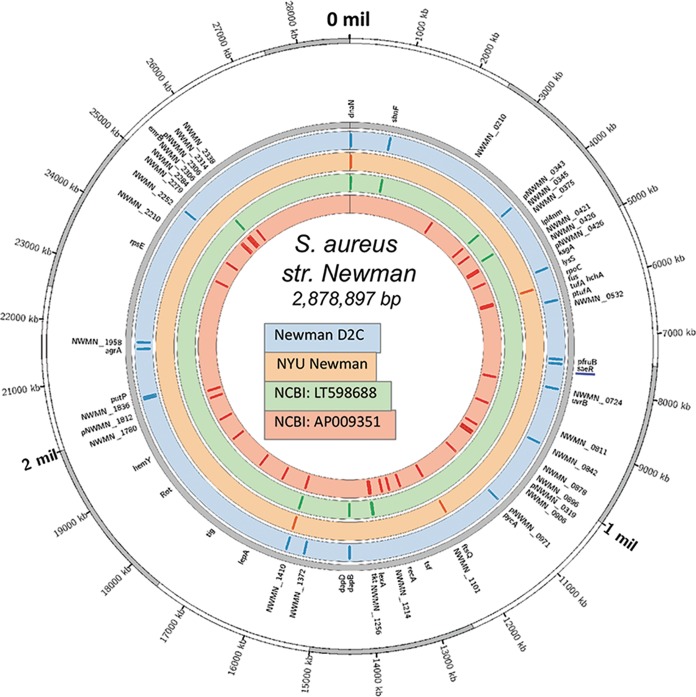
To obtain a global picture of the genomic differences between NYU Newman and Newman D2C that could account for the gross phenotypic inconsistencies observed between the two strains, we performed whole-genome sequencing and compared both genomes to the Newman reference genomes available at the National Center for Biotechnology Information (NCBI), LT598688 and AP009351 (the NYU Newman parent strain [22]). After excluding mobile genetic elements, we identified a total of 143 single-nucleotide polymorphisms (SNPs) across the 4 genomes (Fig. 2). Among these, the Newman D2C genome contained 9 unique mutations (see Table S1 in the supplemental material). Interestingly, a handful of these mutations occur in genes that are instrumental in regulating S. aureus pathogenesis, thus likely accounting for the phenotypic abnormalities observed in the strain. Namely, we uncovered the presence of a stop codon in the ORF for the universal virulence gene activator AgrA (agrA), confirming the lack of toxin production described above, and a nonsynonymous SNP in the gene for the virulence gene regulator SaeR (saeR). These two regulatory proteins have been demonstrated to impact S. aureus pathogenesis (57–60), and thus, it is not surprising that Newman D2C displays a weak virulence phenotype in animal and ex vivo models of pathogenesis.
FIG 2.
Whole-genome sequencing reveals disparities between Newman D2C and NYU Newman. Illumina HiSeq2000 sequencing demonstrated crucial mutations in Newman D2C compared to NYU Newman. Shown is a visualization of S. aureus Newman genetic diversity on a circular map of the chromosome of the Newman reference strain AP009351. The first ring from the outside shows the scale of the chromosome in nucleotides. The second ring indicates the positions of mutated genes in the genome (gray). The next four circles illustrate the four genomes used in this study (Newman D2C, NYU, NCBI LT598688, and NCBI AP009351). The colored tiles inside each circle represent the positions of mutations within each respective genome.
Restoration of AgrA function does not reestablish virulence of Newman D2C.
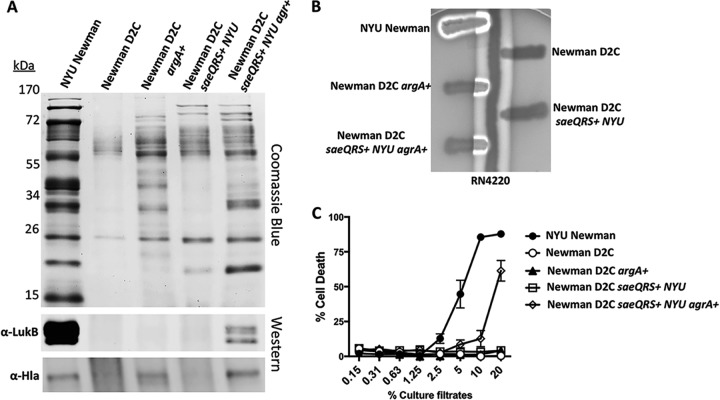
The most glaring mutation in the list of mutations that we uncovered in the Newman D2C strain was a stop codon in the ORF of the virulence gene regulator, agrA. The response regulator AgrA has been demonstrated to be critical for the production of a number of key S. aureus virulence factors (46, 53), including the transcription of the virulence gene activator RNAIII and the production of phenol-soluble modulins (PSMs) (54). As is evident in Fig. 1D, Newman D2C has a severe secretion deficiency compared to NYU Newman. To address this paucity of secretion, we wanted to determine if the addition of AgrA in trans could restore the strain's ability to secrete proteins. Upon the introduction of a wild-type agrA allele on a high-copy-number plasmid in Newman D2C, it became evident that the agr mutation accounted for only a fraction of the strain's deficiencies, as overexpression of agrA did not fully restore secretion in Newman D2C (Fig. 3A). In contrast, overexpression of agrA fully restored the production of δ-toxin by Newman D2C (Fig. 3B), a toxin controlled by AgrA (61), consistent with the functionality of the agrA overexpression plasmid (62).
FIG 3.
Addition of AgrA partially complements Newman D2C virulence deficiency. (A) Complementation of the Agr and Sae regulatory systems enhanced secretion in Newman D2C. (B) δ-Hemolysin activity was restored in Newman D2C upon the addition of AgrA. (C) Partial recovery of cytotoxicity was achieved in Newman D2C only in the presence of both functional Agr and Sae systems. The error bars indicate standard errors of the means.
We next sought to establish the less obvious role of the SNP occurring in the virulence gene regulator gene saeR. To make this assessment, we introduced the saeQRS locus from NYU Newman onto the chromosome at the SapI1 site of Newman D2C (63). We introduced the saeQRS locus, excluding saeP because subsequent complementation experiments utilized our previously published saeQRS mutant in the NYU Newman background; this mutant has no Sae function and can be complemented with saeRS in trans (64). The integration of saeQRS had a modest impact on the levels of protein production in Newman D2C (Fig. 3A). However, the tandem introduction of agrA and saeQRS with NYU Newman sequences bestowed the greatest enhancement in protein production for Newman D2C (Fig. 3A). Moreover, the synergism between AgrA and SaeR was highlighted by the production of specific S. aureus secreted virulence factors. Where Hla could be visualized upon the expression of functional agrA, expression of functional agrA and saeQRS resulted in greater production of the toxin. Furthermore, LukB and other leukocidins were detectable only when both response regulators were present in Newman D2C (Fig. 3A).
We next assessed the ability of the Newman D2C complement strains to induce hemolysis. Interestingly, the only detectable hemolytic activity that was restored was the function of δ-hemolysin in the presence of AgrA (Fig. 3B). This is a logical result, as it has been well documented that Agr contributes to the activation of this particular hemolysin (65). Of note, Newman D2C has a mutation in the dihydrodipicolinate reductase gene, dapB, a gene that lies adjacent to dapA, which has been previously shown to be crucial for hemolysis in Newman (66). Thus, this could explain the lack of full α-hemolysin activity despite the reconstitution of Agr and Sae.
Given Newman D2C's heightened level of protein secretion in the presence of functional AgrA and SaeR, we next wanted to determine whether this resulted in restoration of cytotoxicity. Again, supporting the synergism between these two regulators, human PMNs were susceptible to intoxication by culture filtrates from Newman D2C only when the strain was complemented with both NYU Newman agrA and saeQRS (Fig. 3C). However, this increase in cytotoxicity was still substantially lower than that of NYU Newman, indicating that the other mutations in Newman D2C are likely important for complete restoration of cytotoxicity.
The SNP in the Newman D2C saeR allele renders the SaeRS system nonfunctional.
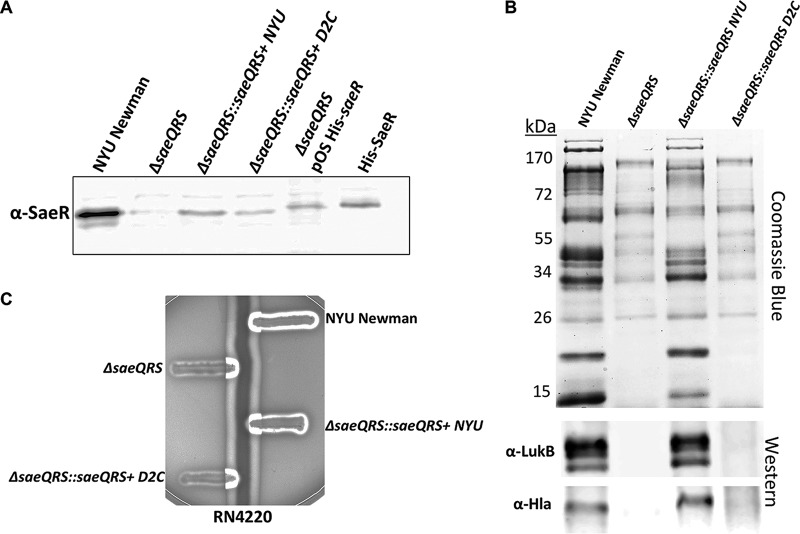
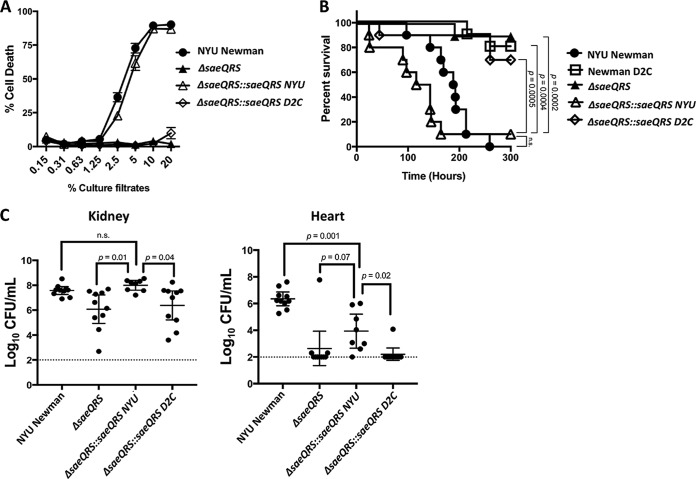
We next wanted to further dissect the significance of the SNP occurring in the ORF of the saeR allele (C595T, resulting in the amino acid change R199 to C199) in Newman D2C. While our complementation studies in Newman D2C (Fig. 3) certainly suggested that the NYU Newman saeR allele conferred greater virulence-activating power, we did not clarify the extent to which the Newman D2C saeR allele was attenuated. To address this question, we integrated copies of the saeQRS locus from either Newman D2C (ΔsaeQRS::saeQRS D2C) or NYU Newman (ΔsaeQRS::saeQRS NYU) into the SapI1 locus on the chromosome of a ΔsaeQRS mutant in the NYU Newman background. Importantly for functionality studies, we first confirmed that both ΔsaeQRS::saeQRS NYU and ΔsaeQRS::saeQRS D2C were capable of producing SaeR protein (Fig. 4A). To assess the extent of attenuation in SaeR activity from Newman D2C, we first examined exoproteome profiles from the ΔsaeQRS complemented strains. Interestingly, we found that while the saeQRS NYU allele was able to substantially restore protein secretion in the ΔsaeQRS mutant, the saeQRS D2C allele provided no restoration of protein secretion (Fig. 4B). Moreover, upon evaluating toxin production, we witnessed a reconstitution of LukB and Hla production in ΔsaeQRS::saeQRS NYU but not in ΔsaeQRS::saeQRS D2C (Fig. 4B). While we did not see a recovery of hemolysis upon the integration of the saeQRS NYU allele into Newman D2C (Fig. 3B), we did, as expected, observe a reconstitution of hemolytic activity upon the integration of the saeQRS NYU allele into ΔsaeQRS (Fig. 4C). The saeQRS D2C allele was unable to restore hemolysis, further highlighting the weakened activity of the system in the Newman D2C strain.
FIG 4.
A unique SNP in saeR from Newman D2C renders the protein product nonfunctional. (A) Western blot demonstrating that a ΔsaeQRS mutant complemented with saeQRS from either Newman D2C or NYU Newman produced SaeR protein. SaeR produced from a plasmid and purified SaeR served as positive controls; these proteins carried a histidine tag. (B) Only the saeQRS allele from NYU Newman was capable of restoring protein secretion in a complemented ΔsaeQRS mutant. (C) The Newman D2C saeQRS allele could not reconstitute hemolysis in a ΔsaeQRS mutant, while the NYU Newman saeQRS allele could restore hemolysis.
The Newman D2C saeR allele confers no pathogenic activity.
To assess whether the inability to produce secreted proteins at wild-type levels impacted pathogenesis, we determined whether ΔsaeQRS::saeQRS D2C could elicit cytotoxic properties. Further substantiating the observation that the Newman D2C SaeR protein could not activate the expression of secreted proteins, we found that only the saeQRS NYU allele was capable of restoring cytotoxicity in the ΔsaeQRS mutant (Fig. 5A). Our in vitro assessment of protein production and ex vivo assays with human PMNs suggested that the saeR allele in Newman D2C produces an inactive protein. To verify this notion, we tested the pathogenicity of the ΔsaeQRS complement strains in a mouse model of bacteremia. We observed that the saeQRS NYU allele restored lethality to the saeQRS mutant while ΔsaeQRS::saeQRS D2C behaved very similarly to the saeQRS-null mutant in that it was essentially nonpathogenic (Fig. 5B). While statistically insignificant, the slightly increased lethality observed in the ΔsaeQRS::saeQRS NYU strain compared to that of wild-type NYU Newman was likely due to the placement of the saeQRS at the Sap1I site (67). Further confirming the lack of functionality of the Newman D2C saeQRS allele, we found that the bacterial burden in critical organs was significantly reduced in mice that had been infected with the ΔsaeQRS::saeQRS D2C strain compared to those infected with the ΔsaeQRS::saeQRS NYU strain (Fig. 5C). While the saeQRS-null mutant complemented with the saeQRS NYU allele was able to regain wild-type levels of bacterial burden in the kidney, full restoration of the bacterial burden was not achieved in the heart (Fig. 5C). This is again likely due to the expression dynamic of placing the saeQRS locus at the Sap1 site on the chromosome.
FIG 5.
SNP C595T in Newman D2C abolishes pathogenesis. (A) The NYU Newman saeR allele restored cytotoxicity to an ΔsaeQRS mutant, whereas the saeQRS allele from Newman D2C provided no restoration of cytotoxicity (n = 6 donors). Intoxications were repeated with unique transductants for both ΔsaeQRS::saeQRS D2C and ΔsaeQRS::saeQRS NYU, and the same results were achieved (data not shown). (B) Mice infected intravenously were susceptible to acute lethality only in the presence of the NYU Newman saeQRS allele, as neither Newman D2C, ΔsaeQRS, nor ΔsaeQRS::saeQRS D2C had acutely lethal effects on the mice (n = 10 mice/strain). The points indicate individual mouse deaths. Infections were done in sets of 5 mice, and each infection was performed with a unique transductant for ΔsaeQRS::saeQRS D2C and ΔsaeQRS::saeQRS NYU. Statistical analysis for survival was performed using the Gehan-Breslow-Wilcoxon test with P values adjusted for multiple comparisons. (C) Mice infected intravenously exhibited reduced bacterial burdens in critical organs when infected with either ΔsaeQRS or ΔsaeQRS::saeQRS D2C compared to that of NYU Newman or ΔsaeQRS::saeQRS NYU (n = 10 mice/strain). Dotted lines represent the limit of detection for bacterial burden in these organs. Statistical analysis was performed using one-way analysis of variance (ANOVA) with post hoc Holm-Sidak multiple-comparison tests where appropriate. The error bars indicate standard errors of the means.
DISCUSSION
In order to develop an effective therapy to combat the deadly consequences of invasive S. aureus infection, models that appropriately demonstrate the bacterium's pathogenic potential must be utilized. Here, we reveal that the commonly used S. aureus strain Newman D2C from the ATCC, used in multiple studies of antistaphylococcal therapeutics and vaccines (25–30), contains numerous mutations that render the strain essentially nonpathogenic. Remarkably, Newman D2C contains both a stop mutation in the virulence gene activator gene, agrA, and a novel polymorphism in the virulence gene regulator gene, saeR, that abolishes SaeR function. The importance of these two mutations is highlighted by the partial recovery of virulence that we observed in Newman D2C upon complementation of both of these pathways. Additionally, it is clearly evident that the remaining mutations in Newman D2C are significant, as we were unable to achieve full restoration of virulence with both functional Agr and Sae systems. Complete analysis of the remaining mutations in Newman D2C (see Table S1 in the supplemental material) would be required to fully understand the extent of Newman D2C's pathogenic shortcomings.
Here, we show that a previously unidentified polymorphism occurring in saeR eliminates the functionality of the protein product. This loss of function can likely be attributed to both the location of the SNP on the chromosome and the nature of the amino acid alteration that occurs. Specifically, amino acid 199 in SaeR from NYU Newman is arginine (R199), while in Newman D2C, the arginine has been replaced with cysteine. The mutation is significant because this residue in SaeR has been shown to be a crucial residue in the protein's DNA binding domain (68). In an effort to identify the DNA binding domain of SaeR, Fan et al. demonstrated that upon replacing the arginine 199 residue with alanine, S. aureus loses the ability to produce alpha toxin and to induce hemolysis (68). Moreover, purified SaeR containing the R→A199 substitution exhibited reduced binding to the P1 promoter region of the sae operon, while wild-type SaeR maintained its high binding affinity (68). Thus, the loss of function in SaeR from Newman D2C can likely be attributed to the protein product's inability to bind critical promoter regions associated with virulence gene expression.
This study also emphasizes the importance of scrupulously examining genomic-sequence data when assessing bacterial virulence. Here, we show that strain Newman D2C from the ATCC, a strain that has been used in numerous studies that sought to assess S. aureus pathogenesis in both in vivo and ex vivo models of infection (25–30), is in fact nonpathogenic. The chain of custody of strain Newman D2C at the ATCC is listed as E. Duthie (1952) to J. Hawiger (1970) to the ATCC, and it was originally described as a clumping-factor-positive variant of Newman strain D2 (23), although the relationship between the original Newman strain and the Newman D2 strain is not apparent. Unfortunately, Newman D2C has been the centerpiece of multiple studies on S. aureus pathogenesis and antistaphylococcal therapeutics and vaccines that have substantially influenced the field's understanding of both S. aureus biology and assessments of the efficacy of antistaphylococcal agents (25–30) and is often referred to simply as strain Newman. It should be noted that the references listed here are by no means a comprehensive list of all the publications that have used Newman D2C as their model organism, as that would be quite extensive. While the conclusions from the majority of studies using Newman D2C may be sound, the data described here bring the results of some studies into question. This is particularly problematic for studies that have assessed the efficacy of therapeutic agents, as the threshold for positive data is likely lowered with models using Newman D2C. While it may be impractical to continually sequence bacterial stocks of commonly used strains, a range of basic tests are available for S. aureus to evaluate the integrity of the bacterium's virulence. Some of these assessments are described here, including examining for hemolysis, profiling of protein secretions, and determining ex vivo cytotoxicity. As efforts continue to identify and characterize novel antibacterial agents and vaccines, these studies provide a cautionary tale in ensuring that the integrity of the virulence of the model organisms employed is maintained and is consistent between laboratories.
MATERIALS AND METHODS
Bacterial culture conditions.
The S. aureus strains used in this study were grown at 37°C on tryptic soy agar (TSA) or broth (TSB). When appropriate, the strains were grown in the presence of either tetracycline (4 μg/ml) or chloramphenicol (10 μg/ml). Escherichia coli DH5α was used for cloning and propagation of plasmids. E. coli culturing was done in Luria-Bertani broth supplemented with ampicillin (100 μg/ml). Liquid cultures were grown in 5 ml of growth medium in 15-ml conical tubes incubated at a 45° angle with shaking at 180 rpm. For all experiments involving the growth of S. aureus bacteria, overnight cultures were diluted 1:100 in fresh TSB.
Genome sequencing, assembly, and annotation.
We prepared sequencing libraries from DNA extracted from Newman D2C and NYU Newman as previously described (69). Whole-genome sequencing was performed using an Illumina HiSeq2000 with 100-base paired-end reads. The paired-end Illumina reads were mapped against the S. aureus Newman reference genomes AP009351.1 and LT598688 using the Burrows-Wheeler Aligner (BWA) (70). AP009351.1 was described by Baba et al. and is the parental strain of NYU Newman (22). The LT598688 sequence was deposited by Monk et al. (49). The BWA outputs were analyzed and annotated using SAMtools (71), GATK (72), and ANNOVAR (73). SNPs in genes annotated as integrases, transposases, resolvases, maturases, or phages were removed from the analysis. Other mobile genetic elements, including SaPI5, phiSA2usa, phiSA3usa, SCCmecIV, and ACME, were identified using IslandPath-DIMOB and PHASTSNPs (74, 75). Genomes and mutations were visualized using Circos (76).
Construction of bacterial strains.
All the strains and plasmids used in this study are listed in Tables 1 and 2. Complementation of saeQRS NYU or saeQRS D2C in the chromosome of ΔsaeQRS or Newman D2C was performed with the pJC1306 suicide plasmid (63), which stably integrates DNA into the SaPI1 site, leading to a single-copy chromosomal insertion. The only difference between the NYU Newman saeQRS locus and the Newman D2C saeQRS locus occurs at the C595T polymorphism in saeR, and as such, the same primer set was used to amplify the saeQRS loci from both strains. Primers VJT1796 (5′ CCCCCCCCTGCAGTCAATTTCTGAGTTAAACT 3′) and VJT1797 (5′ CCCCCCCGGATCCTTATGACGTAATGTCTAATT 3′) amplified a product that extends from the 5′ intergenic region upstream of saeQ to the 3′ end of the saeS ORF. Within this genetic fragment, a transcriptional promoter exists in saeQ's intragenic region (39). These products were then digested with BamHI and PstI, cloned into pJC1306, and transformed into E. coli DH5α. Sanger sequencing was used to verify that the C595T polymorphism in saeR was present in the pJC1306/saeQRS D2C plasmid, and wild-type saeR was similarly confirmed in pJC1306/saeQRS NYU. To generate the integrated strains, we first transformed pJC1306 saeQRS (NYU or D2C) or pJC1306 (empty vector) plasmids into RN4220 carrying pRN7023, which contains an integrase and thus allows integration of pJC1306 into the SaPI1 site (63). The SapI1 locus from RN4220 was then transduced with phage Φ80 into either ΔsaeQRS NYU Newman, Newman D2C, or NYU Newman. Strains harboring the plasmid pOSplgt-agrA were transformed via electroporation.
TABLE 1.
Bacterial strains used in this study
| Strain | Background | Description | Designation | Reference |
|---|---|---|---|---|
| VJT 3.81 | NYU Newman | Wild-type NYU Newman | NYU Newman | 21 |
| VJT 50.23 | Newman D2C | Newman D2C (10833) | Newman D2C | 21, 23 |
| VJT 57.16 | NYU Newman | NYU Newman carrying the empty pJC1306 vector at the SapI1 site | NYU Newman | This study |
| VJT 57.15 | Newman D2C | Newman D2C carrying the empty pJC1306 vector at the SapI1 site | Newman D2C | This study |
| VJT 2.60 | RN4220 | Wild-type RN4220 | RN4220 | 77 |
| VJT 3.76 | NYU Newman | Newman carrying the bursa aurealis transposon in the hla gene | hla::bursa | 78 |
| VJT 8.91 | NYU Newman | Newman with a clean deletion of the lukAB locus | ΔlukAB | 8 |
| VJT 57.17 | NYU Newman | ΔsaeQRS::spec carrying the empty pJC1306 vector at the SapI1 site | ΔsaeQRS | This study |
| VJT 57.10 | NYU Newman | ΔsaeQRS::spec carrying pJC1306 expressing saeQRS NYU in the SaPI1 site | ΔsaeQRS::saeQRS NYU | This study |
| VJT 57.09 | NYU Newman | ΔsaeQRS::spec carrying pJC1306 expressing saeQRS D2C in the SaPI1 site | ΔsaeQRS::saeQRS D2C | This study |
| VJT 50.43 | NYU Newman | NYU Newman carrying the empty pOS1plgt vector | NYU Newman | This study |
| VJT 50.45 | Newman D2C | Newman D2C carrying the empty pOS1plgt vector | Newman D2C | This study |
| VJT 50.48 | Newman D2C | Newman D2C carrying the pOS1plgt agrA vector | Newman D2C argA+ | This study |
| VJT 57.32 | Newman D2C | Newman D2C carrying the empty pOS1plgt vector and pJC1306 expressing saeQRS NYU in the SaPI1 site | Newman D2C::saeQRS NYU | This study |
| VJT 57.33 | Newman D2C | Newman D2C carrying the pOS1plgt agrA vector and pJC1306 expressing saeQRS NYU in the SaPI1 site | NewmanD2C::saeQRS NYU agrA+ | This study |
| VJT 31.57 | NYU Newman | ΔlukED:hlgACB::tet::lukAB::spec::hla::ermC | ΔΔΔΔ | 11 |
| VJT 16.99 | NYU Newman | ΔsaeQRS::spec | ΔsaeQRS | 64 |
TABLE 2.
Plasmids used in this study
| Name | Description | Resistancea | Reference |
|---|---|---|---|
| pOS1-Plgt | lgt promoter in an empty vector | Cm | 79 |
| pOS1-Plgt-agrA | lgt promoter driving agr expression | Cm | 62 |
| pJC1306 | Single-copy integration vector that inserts at the SaPI1 site | Tet | 63 |
| pJC1306/saeQRS NYU | Plasmid pJC1306 expressing saeQRS NYU from its native promoter | Tet | This study |
| pJC1306/saeQRS D2C | Plasmid pJC1306 expressing saeQRS D2C from its native promoter | Tet | This study |
| pOS1-Plgt-saeR | lgt promoter driving saeR expression | Cm | This study |
Cm, chloramphenicol; Tet, tetracycline.
Exoprotein analysis.
Proteins were concentrated from 5-h culture supernatants using 10% (vol/vol) trichloroacetic acid (TCA) precipitation as described previously (46). Protein visualization was achieved by separating samples using SDS-12% PAGE and staining with Coomassie brilliant blue. To assess the production of specific products, proteins were resolved with 12% SDS-PAGE, transferred to a nitrocellulose membrane, and then probed with the indicated primary antibody. Alex Fluor 680–anti-rabbit antibody was used as a secondary antibody, and the membranes were visualized using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Cell lysate preparation.
Five-hour S. aureus subcultures were centrifuged, and the cell pellets were washed one time in 1× PBS. Following the wash, the pellets were resuspended in 1 ml of TSM (50 mM Tris, 0.5M d-sucrose, 10 mM MgCl2, pH 7.5) and 10 μl of 2-mg/ml lysostaphin and then incubated at 37°C for 10 min. Following incubation, samples were centrifuged and the pellets were resuspended in 250 μl of 1× PBS. The samples were then moved to tubes containing 0.1-mm glass beads and disrupted using an MP Biomedicals FastPrep-24 homogenizer. The resulting samples were then centrifuged, and the soluble fraction was resuspended in 2× SDS sample buffer.
Ethics statement.
Buffy coats were obtained from anonymous blood donors with informed consent from the New York Blood Center. Because all of the samples were collected anonymously prior to their delivery, the New York University Langone Medical Center (NYULMC) Institutional Review Board determined that our study was exempt from further ethics approval requirements.
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of NYULMC. All experiments were performed according to NIH guidelines, the Animal Welfare Act, and U.S. federal law.
Cytotoxicity assays.
To evaluate the cytotoxicity of the secreted factors produced from each strain, primary human PMNs were intoxicated with culture filtrates obtained from 5-h S. aureus subcultures. Prior to intoxication, culture filtrates were diluted 2-fold in a 96-well plate (20 to 0.15%). PMNs were isolated as described by Reyes-Robles et al. (11) and normalized to 200,000 cells per 80 μl RPMI (10 mM HEPES plus 0.1% human serum albumin). PMNs (80 μl) were then pipetted into each well, and the supernatant-PMN mixtures were incubated in a 37°C-5% CO2 incubator for 1 h. To assess toxicity, 10 μl of CellTiter 96 Aqueous One solution (CellTiter; Promega) was added to the 96-well plate, and the mixture was incubated at 37°C in 5% CO2 for 1.5 h. PMN viability was assessed with a PerkinElmer EnVision 2103 multilabel reader at an absorbance of 492 nm.
Murine sepsis model.
Three-hour subcultures of S. aureus were washed, resuspended in 1× phosphate-buffered saline, measured for cell density (optical density at 600 nm [OD600]), normalized, and then plated for CFU. The initial experiment comparing tissue burdens in mice infected with NYU Newman and Newman D2C (purchased by and obtained from Janssen R&D LLC) used 5 × 107 CFU; the remaining tissue colonization experiments were conducted with 2.5 × 107 CFU. All mouse survival infections were conducted with 5 × 107 CFU. Retro-orbital infections (100-μl inoculum) were performed on 5-week-old female ND4 Swiss-Webster mice (Envigo) that had been anesthetized intraperitoneally with 300 μl of Avertin (2,2,2-tribromoethanol dissolved in tert-amyl alcohol and diluted to a final concentration of 2.5% [vol/vol] in sterile saline). To examine for tissue colonization at 96 h postinfection, the mice were euthanized with CO2, and the indicated organs were harvested as described previously (9). For acute/survival experiments, mice administered retro-orbital infections were monitored every 4 to 6 h for signs of morbidity (hunched posture, lack of movement, paralysis, and inability to acquire food or water), at which time the animals were euthanized and survival curves were plotted.
Accession number(s).
All the genomic data are available at NCBI under the following accession numbers: NYU Newman, CP023390.1; ATCC Newman, CP023391.1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Janssen Research and Development LLC under the auspices of an exclusive License and Research Collaboration Agreement with NYU, as well as by NIH-NIAID Institutional Research Training Grant T32 AI007180 (W.E.S.) and NIH-NIAID R01 AI103268 and HHSN272201400019C (B.S. and V.J.T.). V.J.T. is a Burroughs Wellcome Fund Investigator in the pathogenesis of infectious diseases.
V.J.T. is an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. B.J.M., P.T.B., J.F., and A.S.L. are employees of Janssen Research and Development and declare competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00476-17.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Sause WE, Buckley PT, Strohl WR, Lynch AS, Torres VJ. 2016. Antibody-based biologics and their promise to combat Staphylococcus aureus infections. Trends Pharmacol Sci 37:231–241. doi: 10.1016/j.tips.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 4.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 5.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. 2009. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med 206:2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumont AL, Nygaard TK, Watkins RL, Smith A, Kozhaya L, Kreiswirth BN, Shopsin B, Unutmaz D, Voyich JM, Torres VJ. 2011. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol 79:814–825. doi: 10.1111/j.1365-2958.2010.07490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonzo F, Benson MA, Chen J, Novick RP, Shopsin B, Torres VJ. 2012. Staphylococcus aureus leukocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol Microbiol 83:423–435. doi: 10.1111/j.1365-2958.2011.07942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonzo F III, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, Torres VJ. 2013. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493:51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes-Robles T, Alonzo F III, Kozhaya L, Lacy DB, Unutmaz D, Torres VJ. 2013. Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell Host Microbe 14:453–459. doi: 10.1016/j.chom.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thammavongsa V, Missiakas DM, Schneewind O. 2013. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342:863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes-Robles T, Lubkin A, Alonzo F III, Lacy DB, Torres VJ. 2016. Exploiting dominant-negative toxins to combat Staphylococcus aureus pathogenesis. EMBO Rep 17:780. doi: 10.15252/embr.201670010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Liu H, Zhu K, Gong S, Dramsi S, Wang YT, Li J, Chen F, Zhang R, Zhou L, Lan L, Jiang H, Schneewind O, Luo C, Yang CG. 2014. Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase. Proc Natl Acad Sci U S A 111:13517–13522. doi: 10.1073/pnas.1408601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAdow M, Kim HK, Dedent AC, Hendrickx AP, Schneewind O, Missiakas DM. 2011. Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog 7:e1002307. doi: 10.1371/journal.ppat.1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farinelli G, Jofra Hernandez R, Rossi A, Ranucci S, Sanvito F, Migliavacca M, Brombin C, Pramov A, Di Serio C, Bovolenta C, Gentner B, Bragonzi A, Aiuti A. 2016. Lentiviral vector gene therapy protects XCGD mice from acute Staphylococcus aureus pneumonia and inflammatory response. Mol Ther 24:1873–1880. doi: 10.1038/mt.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauch S, Gough P, Kim HK, Schneewind O, Missiakas D. 2014. Vaccine protection of leukopenic mice against Staphylococcus aureus bloodstream infection. Infect Immun 82:4889–4898. doi: 10.1128/IAI.02328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HK, Kim HY, Schneewind O, Missiakas DM. 2011. Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J 25:3605–3612. doi: 10.1096/fj.11-187963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagnoli F, Fontana MR, Soldaini E, Mishra RPN, Fiaschi L, Cartocci E, Nardi-Dei V, Ruggiero P, Nosari S, De Falco MG, Lofano G, Marchi S, Galletti B, Mariotti P, Bacconi M, Torre A, Maccari S, Scarselli M, Rinaudo CD, Inoshima N, Savino S, Mori E, Rossi-Paccani S, Baudner B, Pallaoro M, Swennen E, Petracca R, Brettoni C, Liberatori S, Norais N, Monaci E, Wardenburg JB, Schneewind O, O'Hagan DT, Valiante NM, Bensi G, Bertholet S, De Gregorio E, Rappuoli R, Grandi G. 2015. Vaccine composition formulated with a novel TLR7-dependent adjuvant induces high and broad protection against Staphylococcus aureus. Proc Natl Acad Sci U S A 112:3680–3685. doi: 10.1073/pnas.1424924112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomer L, Emolo C, Thammavongsa V, Kim HK, McAdow ME, Yu W, Kieffer M, Schneewind O, Missiakas D. 2016. Antibodies against a secreted product of Staphylococcus aureus trigger phagocytic killing. J Exp Med 213:293–301. doi: 10.1084/jem.20150074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol 6:95–107. [DOI] [PubMed] [Google Scholar]
- 22.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawiger J, Niewiarowski S, Gurewich V, Thomas DP. 1970. Measurement of fibrinogen and fibrin degradation products in serum by staphylococcal clumping test. J Lab Clin Med 75:93–108. [PubMed] [Google Scholar]
- 24.Attia AS, Cassat JE, Aranmolate SO, Zimmerman LJ, Boyd KL, Skaar EP. 2013. Analysis of the Staphylococcus aureus abscess proteome identifies antimicrobial host proteins and bacterial stress responses at the host-pathogen interface. Pathog Dis 69:36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavakis T, Hussain M, Kanse SM, Peters G, Bretzel RG, Flock JI, Herrmann M, Preissner KT. 2002. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med 8:687–693. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 26.Liang HP, Kerschen EJ, Hernandez I, Basu S, Zogg M, Botros F, Jia S, Hessner MJ, Griffin JH, Ruf W, Weiler H. 2015. EPCR-dependent PAR2 activation by the blood coagulation initiation complex regulates LPS-triggered interferon responses in mice. Blood 125:2845–2854. doi: 10.1182/blood-2014-11-610717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen KJ, Mattsson AH, Pilely K, Asferg CA, Ciofu O, Vitved L, Koch C, Kemp M. 2016. Proteome-wide antigen discovery of novel protective vaccine candidates against Staphylococcus aureus infection. Vaccine 34:4602–4609. doi: 10.1016/j.vaccine.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Kumar P, Kretzschmar B, Herold S, Nau R, Kreutzfeldt M, Schutze S, Bahr M, Hein K. 2015. Beneficial effect of chronic Staphylococcus aureus infection in a model of multiple sclerosis is mediated through the secretion of extracellular adherence protein. J Neuroinflammation 12:22. doi: 10.1186/s12974-015-0241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura CY, Hollands A, Tran DN, Olson J, Dahesh S, von Kockritz-Blickwede M, Thienphrapa W, Corle C, Jeung SN, Kotsakis A, Shalwitz RA, Johnson RS, Nizet V. 2012. A new pharmacological agent (AKB-4924) stabilizes hypoxia inducible factor-1 (HIF-1) and increases skin innate defenses against bacterial infection. J Mol Med 90:1079–1089. doi: 10.1007/s00109-012-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramaniam R, Barnes PF, Fletcher K, Boggaram V, Hillberry Z, Neuenschwander P, Shams H. 2014. Protecting against post-influenza bacterial pneumonia by increasing phagocyte recruitment and ROS production. J Infect Dis 209:1827–1836. doi: 10.1093/infdis/jit830. [DOI] [PubMed] [Google Scholar]
- 31.Cashman JD, Jackson JK, Mugabe C, Gilchrist S, Ball K, Tredwell S, Burt HM. 2013. The use of tissue sealants to deliver antibiotics to an orthopaedic surgical site with a titanium implant. J Orthop Sci 18:165–174. doi: 10.1007/s00776-012-0325-6. [DOI] [PubMed] [Google Scholar]
- 32.Brouillette E, Lacasse P, Shkreta L, Belanger J, Grondin G, Diarra MS, Fournier S, Talbot BG. 2002. DNA immunization against the clumping factor A (ClfA) of Staphylococcus aureus. Vaccine 20:2348–2357. doi: 10.1016/S0264-410X(02)00100-7. [DOI] [PubMed] [Google Scholar]
- 33.Duthie ES. 1952. Variation in the antigenic composition of staphylococcal coagulase. J Gen Microbiol 7:320–326. doi: 10.1099/00221287-7-3-4-320. [DOI] [PubMed] [Google Scholar]
- 34.Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett 177:15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- 35.Giraudo AT, Martinez GL, Calzolari A, Nagel R. 1994. Characterization of a Tn925-induced mutant of Staphylococcus aureus altered in exoprotein production. J Basic Microbiol 34:317–322. doi: 10.1002/jobm.3620340507. [DOI] [PubMed] [Google Scholar]
- 36.Jeong DW, Cho H, Jones MB, Shatzkes K, Sun F, Ji Q, Liu Q, Peterson SN, He C, Bae T. 2012. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol Microbiol 86:331–348. doi: 10.1111/j.1365-2958.2012.08198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong DW, Cho H, Lee H, Li C, Garza J, Fried M, Bae T. 2011. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol 193:4672–4684. doi: 10.1128/JB.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinhuber A, Goerke C, Bayer MG, Doring G, Wolz C. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J Bacteriol 185:6278–6286. doi: 10.1128/JB.185.21.6278-6286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol 190:3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benton BM, Zhang JP, Bond S, Pope C, Christian T, Lee L, Winterberg KM, Schmid MB, Buysse JM. 2004. Large-scale identification of genes required for full virulence of Staphylococcus aureus. J Bacteriol 186:8478–8489. doi: 10.1128/JB.186.24.8478-8489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun 74:4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol 188:7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR, Whitney AR, Otto M, DeLeo FR. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis 199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis 201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun F, Li C, Jeong D, Sohn C, He C, Bae T. 2010. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol 192:2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapman JR, Balasubramanian D, Tam K, Askenazi M, Copin R, Shopsin B, Torres VJ, Ueberheide BM. 2017. Using quantitative spectrometry to understand the influence of genetics and nutritional perturbations on the virulence potential of Staphylococcus aureus. Mol Cell Proteomics 16:S15–S28. doi: 10.1074/mcp.O116.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adhikari RP, Novick RP. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949–959. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- 48.Beenken KE, Mrak LN, Zielinska AK, Atwood DN, Loughran AJ, Griffin LM, Matthews KA, Anthony AM, Spencer HJ, Skinner RA, Post GR, Lee CY, Smeltzer MS. 2014. Impact of the functional status of saeRS on in vivo phenotypes of Staphylococcus aureus sarA mutants. Mol Microbiol 92:1299–1312. doi: 10.1111/mmi.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monk IR, Howden BP, Seemann T, Stinear TP. 2017. Spontaneous secondary mutations confound analysis of the essential two-component system WalKR in Staphylococcus aureus. Nat Commun 8:14403. doi: 10.1038/ncomms14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foster T. 1996. Staphylococcus, p 6 In Baron S. (ed), Medical microbiology, 4th ed University of Texas Medical Branch at Galveston, Galveston, TX. [Google Scholar]
- 51.Adhikari RP, Arvidson S, Novick RP. 2007. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan. Infect Immun 75:4534–4540. doi: 10.1128/IAI.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 53.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaan AN, van Strijp JAG, Torres VJ. 2017. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol 15:435–447. doi: 10.1038/nrmicro.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berube BJ, Bubeck Wardenburg J. 2013. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins 5:1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet 202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 58.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun 61:3879–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giraudo AT, Raspanti CG, Calzolari A, Nagel R. 1994. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol 40:677–681. doi: 10.1139/m94-107. [DOI] [PubMed] [Google Scholar]
- 60.Rampone H, Martinez GL, Giraudo AT, Calzolari A, Nagel R. 1996. In vivo expression of exoprotein synthesis with a Sae mutant of Staphylococcus aureus. Can J Vet Res 60:237–240. [PMC free article] [PubMed] [Google Scholar]
- 61.Janzon L, Lofdahl S, Arvidson S. 1989. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol Gen Genet 219:480–485. doi: 10.1007/BF00259623. [DOI] [PubMed] [Google Scholar]
- 62.Benson MA, Lilo S, Wasserman GA, Thoendel M, Smith A, Horswill AR, Fraser J, Novick RP, Shopsin B, Torres VJ. 2011. Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol Microbiol 81:659–675. doi: 10.1111/j.1365-2958.2011.07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. 2014. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76:1–7. doi: 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benson MA, Lilo S, Nygaard T, Voyich JM, Torres VJ. 2012. Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J Bacteriol 194:4355–4365. doi: 10.1128/JB.00706-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Traber K, Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol Microbiol 59:1519–1530. doi: 10.1111/j.1365-2958.2006.04986.x. [DOI] [PubMed] [Google Scholar]
- 66.Burnside K, Lembo A, de Los Reyes M, Iliuk A, Binhtran NT, Connelly JE, Lin WJ, Schmidt BZ, Richardson AR, Fang FC, Tao WA, Rajagopal L. 2010. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balasubramanian D, Ohneck EA, Chapman J, Weiss A, Kim MK, Reyes-Robles T, Zhong J, Shaw LN, Lun DS, Ueberheide B, Shopsin B, Torres VJ. 2016. Staphylococcus aureus coordinates leukocidin expression and pathogenesis by sensing metabolic fluxes via RpiRc. mBio 7:e00818-16. doi: 10.1128/mBio.00818-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan X, Zhang X, Zhu Y, Niu L, Teng M, Sun B, Li X. 2015. Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 71:1768–1776. doi: 10.1107/S1399004715010287. [DOI] [PubMed] [Google Scholar]
- 69.Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li M, Cheung GY, Hu J, Wang D, Joo HS, Deleo FR, Otto M. 2010. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis 202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langille MG, Hsiao WW, Brinkman FS. 2008. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 9:329. doi: 10.1186/1471-2105-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. [DOI] [PubMed] [Google Scholar]
- 78.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A 101:12312–12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.