Abstract
Enteroviral infections have been associated with the development of type 1 diabetes (T1D), a chronic inflammatory disease characterized by autoimmune destruction of insulin-producing pancreatic beta cells. Cultured human islets, including the insulin-producing beta cells, can be infected with coxsackievirus B4 (CVB4) and thus are useful for understanding cellular responses to infection. We performed quantitative mass spectrometry analysis on cultured primary human islets infected with CVB4 to identify molecules and pathways altered upon infection. Corresponding uninfected controls were included in the study for comparative protein expression analyses. Proteins were significantly and differentially regulated in human islets challenged with virus compared with their uninfected counterparts. Complementary analyses of gene transcripts in CVB4-infected primary islets over a time course validated the induction of RNA transcripts for many of the proteins that were increased in the proteomics studies. Notably, infection with CVB4 results in a considerable decrease in insulin. Genes/proteins modulated during CVB4 infection also include those involved in activation of immune responses, including type I interferon pathways linked to T1D pathogenesis and with antiviral, cell repair, and inflammatory properties. Our study applies proteomics analyses to cultured human islets challenged with virus and identifies target proteins that could be useful in T1D interventions.
Keywords: coxsackievirus, immune response, inflammation, innate immunity, insulin, mass spectrometry, pancreatic islets, proteomics, type 1 diabetes
To understand how enteroviruses contribute to the etiology of type 1 diabetes, we performed proteomic analyses to characterize coxsackievirus B4–specific alterations in cultured primary human islets.
The pathogenesis of type 1 diabetes (T1D) is characterized by immune-mediated damage of insulin-producing beta cells of pancreatic islets. Clinical reports and epidemiological data support that enteroviral infections may accelerate the autoimmune disease process (see Hyöty [1] for review). Pancreatic tissue from patients with recent-onset T1D reveals enteroviral RNA sequences and evidence of viral proteins in islets, consistent with the possibility that low-grade infection in pancreatic islets may contribute to disease progression [2], which is of interest given implications on prevention strategies for T1D. In addition, enteroviral infections, including those with coxsackievirus B (CVB), have been associated infrequently with fulminant T1D, characterized by rapid onset of insulin-deficient hyperglycemia and linked to a strong inflammatory immune response that preferentially destroys beta cells [3, 4]. In such circumstances, viral infection likely plays a more direct etiologic role toward disease.
Cultured primary human islets provide a platform for studying enteroviral replication and host responses in a controlled experimental setting. Cytotoxic effects can be observed by microscopy, and beta cell function and innate immune responses can be evaluated in RNA and protein harvested from islets. Previous studies reveal permissiveness of human islets to infection with various enteroviruses, including poliovirus, coxsackievirus A, CVB, and echovirus [5–11]. Following enterovirus infection of cultured primary human islets, beta cells exhibit defects in glucose-stimulated insulin secretion [6, 7]. Viral replication efficiency, cytotoxicity, and beta cell tropism may all contribute to virus-induced beta cell dysfunction, but the mechanisms of insulin suppression following infection are incompletely understood.
Cytokine production and gene expression changes in infected primary human islets provide insights into the diabetogenic potential of viruses. Infection of cultured primary human islets with CVB4 induces type I interferon (IFN) production [12], and gene expression studies with CVB-infected islets show induction of CXCL10 and other IFN-stimulated genes (ISGs) [8, 13, 14]. ISGs are considerably enhanced in insulitic islets from living donors with recent-onset T1D [15]. Notably, initial infection or repeat infection could establish a setting for more chronic immune-mediated damage over time. Identifying markers of CVB4 infection in islets, including specific IFN signatures, could be useful for developing new diagnostic, treatment, or prevention options (see review by Jean-Baptiste et al. [16]).
Although such immune responses have been characterized in whole islets, defining the contributions of individual cell types to the overall response is difficult. Understanding both genes involved in the antiviral response and cell types that mediate responses could provide insights into autoimmunity directed at beta cells. Newly available sources of human beta cells mitigate some limitations of cultured primary human islets such as limited availability and variability in donor genetics. The EndoC-βH1 cell line, an immortalized clonal human beta cell line, is an experimental model that provides the advantage of monotypic insulin-producing beta cells [17] and eliminates the paracrine effects of other cell types during infection. Because EndoC-βH1 cells are a homogenous population, they can help define cell-intrinsic effects of virus on human beta cell gene expression and function.
The primary objective of our study was to characterize protein modulation in cultured primary human islets following challenge with CVB4 strain JVB, with the goal of identifying proteins and pathways that may play a role in the etiology of T1D. We selected this strain given our previous findings in diabetes models with human islets [18]. We examined specific gene expression changes in cultured primary human islets following CVB4 infection over a time course for comparison with proteomics findings. To define the specific impact of CVB4 in human beta cells, we also evaluated gene expression changes in EndoC-βH1 cells during infection. In all cases, we found decreases in insulin following CVB4 but not in uninfected controls. We identify additional genes/proteins modulated during CVB4 infection that may participate in T1D pathogenesis.
1. Materials and Methods
A. Virus and Stimulants
CVB4 strain JVB (American Type Culture Collection, Manassas, VA) was propagated in HeLa cells, and purified using a 30% sucrose cushion [19]. CVB3–enhanced green fluorescent protein was a gift from J. L. Whitton [20]. For some experiments, cells were challenged with polyinosinic-polycytidylic acid (poly I:C) at 100 μg/mL (InvivoGen, San Diego, CA) with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA).
B. Human Islets
Primary human islets from adult donors without diabetes were obtained from Prodo Laboratories, Inc. (Aliso Viejo, CA) and used according to protocols approved by the Institutional Review Board of the University of Massachusetts Medical School. A total of 150 to 200 islet equivalents was cultured in CMRL-1066 media (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin. Cultured islets were infected with 1 × 106 plaque-forming units of CVB/100 islet equivalents by incubating islets in a minimal volume of media at 37°C for 1 hour. Mock-infected islets were treated with media alone. Complete medium was added, and cells were cultured at 37°C in 5% CO2.
C. EndoC-βH1 Cells
EndoC-βH1 cells (gift from Dr. R. Scharfmann) were cultured as described [17]. Virus was added to cells at a multiplicity of infection (MOI) of 10 for 1 hour in a minimal volume to allow for adsorption. Cells were washed with phosphate-buffered saline, and fresh medium was added.
D. Microscopy and Immunofluorescence
EndoC-βH1 cells were cultured on coverslips and infected with CVB4 (MOI of 10) for 24 hours. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature. See Supplemental Materials and Methods (19.2MB, docx) for details on wide-field microscopy and staining.
E. Flow Cytometry
EndoC-βH1 cells were trypsinized to obtain a single-cell suspension at the indicated time points. Dead cells were stained using LIVE/DEAD™ Fixable Blue Stain (1:1000; Invitrogen, Carlsbad, CA) for 20 minutes. Cells were fixed and permeabilized using BD Cytofix/Cytoperm reagents (BD Biosciences, San Jose, CA) and then stained with mouse antibody to VP1, clone 5-D8/1 (1:1000 of 71 mg/L; catalog number M7064, RRID:AB_2118128; DakoCytomation) and Alexa Fluor 488 goat antibody to mouse immunoglobulin G (1:1000; catalog number A-11029, RRID:AB_2534088; Thermo Fisher Scientific). Flow cytometry was performed with an LSRII (BD Biosciences) equipped with Trigon and Blue 488-nm lasers running BD FACS Diva software (version 8.0.1; BD Biosciences) and analyzed using FlowJo (version 10.1r5; Tree Star).
F. Extraction and Processing of Proteins for Mass Spectrometry
Cultured mock-treated and CVB4-infected primary human islets from adult donors without diabetes and EndoC-βH1 cells were harvested and washed three times with phosphate-buffered saline to remove residual fetal bovine serum and other potential contaminants from the culture medium. Pelleted cells were solubilized in 50% trifluoroethanol in 50 mM triethyl ammonium bicarbonate and sonicated as described [21]. The liquid chromatography-tandem mass spectrometry (MS) methods are listed in the Supplemental Materials and Methods (19.2MB, docx) .
G. Bioinformatic Analysis of Differentially Expressed Proteins
We used Ingenuity Pathway Analysis (IPA; see Ingenuity Systems, http://www.ingenuity.com; and Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) to characterize the molecular functions of differentially expressed proteins between the mock- and CVB4-infected islets and EndoC-βH1 cells.
H. Gene Expression Profiling
TRIzol reagent (Thermo Fisher Scientific) was added to cells for total RNA extraction. We used NanoString (NanoString Technologies, Inc., Seattle, WA) CodeSets containing probes for 123 genes, including 7 housekeeping genes (ACTB, G6PD, GAPDH, GUSB, HPRT1, PPP1CA, and TUBB) and CVB as previously described [18]. A total of 100 ng RNA was hybridized, processed per the manufacturer’s procedure, and analyzed on an nCounter Digital analyzer. Data were normalized using the NanoStringNorm R package. Replicate values from each experiment were averaged and used for downstream analysis. Heat maps reflect the gene expression patterns of genes with at least twofold change by either CVB or poly I:C compared with mock-treated samples. K-means clustering was performed on the data.
2. Results
A. CVB4 Infection of Primary Human Islets Activates Specific Cellular and Molecular Pathways in Protein Expression Profiles
We challenged islets from three independent human donors (see Supplemental Table 1 (19.2MB, docx) for details) with CVB4 and compared them to uninfected islets by MS at 48 hours postinfection (hpi), a time point previously used to examine changes in gene expression in CVB4-infected human islets [18]. Using software algorithms to identify and quantify differential protein expression, we identified >2000 proteins, of which 55 proteins were significantly differentially regulated in at least two of the three donors. In addition to viral proteins that constitute the CVB4 polyprotein (CXB4 in Table 1), 36 host proteins were significantly upregulated and 19 were significantly downregulated in the infected islets compared with the mock-infected controls. Table 1 summarizes the expression profiles of the differentially regulated proteins, and the fold expression values represent the mean expression of the proteins in the islets from the three donors. Supplemental Table 2 (19.2MB, docx) includes the −log(P value) for the differentially expressed proteins, the fold differences in the expression, and protein description information for each individual donor.
Table 1.
Differential Protein Expression Between CVB4- vs Mock-Infected Primary Human Islets at 48 hpi
| Protein/Gene | Fold Difference | Protein/Gene | Fold Difference |
|---|---|---|---|
| CXB4 | ↑8.089 | PC | ↑1.284 |
| MX1 | ↑7.199 | SLC25A4 | ↑1.284 |
| ISG15 | ↑5.817 | SLC25A5 | ↑1.207 |
| IFIT3 | ↑5.138 | IDH3B | ↑1.185 |
| CXCL1 | ↑4.112 | MCCC1 | ↑1.108 |
| CXCL8 | ↑4.014 | HIBCH | ↑1.078 |
| IFIT1 | ↑3.571 | SAMHD1 | ↑1.066 |
| HIC1 | ↑3.484 | PPOX | ↑1.062 |
| TAP1 | ↑3.051 | EIF2AK2 | ↑1.041 |
| ICAM1 | ↑2.833 | RBM14 | ↓−1.020 |
| IFIT2 | ↑2.769 | UCHL1 | ↓−1.027 |
| HLA-B | ↑2.7624 | SEC24C | ↓−1.132 |
| OAS3 | ↑2.390 | RAP1GAP2 | ↓−1.249 |
| TAPBP | ↑2.286 | PDCD5 | ↓−1.275 |
| STAT1 | ↑2.183 | GPD1 | ↓−1.277 |
| MRPS36 | ↑2.146 | NACA | ↓−1.312 |
| HLA-C | ↑1.840 | PCP4 | ↓−1.338 |
| DDX58 | ↑1.830 | GHRL | ↓−1.404 |
| TYMP | ↑1.793 | PLCXD3 | ↓−1.421 |
| PML | ↑1.651 | VAT1L | ↓−1.463 |
| LGALS9 | ↑1.625 | RPL27 | ↓−1.503 |
| GBP1 | ↑1.563 | PRSS2 | ↓−1.689 |
| HLA-A | ↑1.477 | PPP1R1A | ↓−1.752 |
| EHD4 | ↑1.417 | UBQLN2 | ↓−1.891 |
| WARS | ↑1.379 | ATP6AP1 | ↓−1.970 |
| APOL2 | ↑1.378 | INS | ↓−2.152 |
| LAP3 | ↑1.337 | WIBG | ↓−2.315 |
| COX7C | ↑1.312 | REG3A | ↓−3.103 |
We evaluated the molecular processes/functions and disease processes modulated in primary human islets following CVB4 challenge vs mock infection by analyzing differentially expressed proteins using IPA. The Top Canonical Pathways, Top Diseases and Bio Functions, Molecular and Cellular Functions, and Physiological System Development and Function predicted for the proteins/genes that are differentially regulated in CVB4-infected islets and their respective probability values are summarized in Table 2.
Table 2.
Summary of IPA of CVB4-Infected Islets vs Mock-Infected Islets
| Top canonical pathways | ||
| Name | P value | Overlap |
| Interferon signaling | 7.45E-10 | 16.7% (6/36) |
| Antigen presentation pathways | 7.35E-08 | 13.2% (5/38) |
| T1D signaling | 1.28E-05 | 4.7% (5/106) |
| Activation of IRF by cytosolic PRRs | 2.57E-05 | 6.7% (4/60) |
| Th1 pathway | 3.44E-05 | 3.8% (5/130) |
| Top upstream regulators | ||
| Upstream regulator | P value of overlap | Predicted activation |
| IFNG | 1.12E-22 | Activated |
| IFNL1 | 6.98E-22 | Activated |
| MAPK1 | 9.78E-19 | Inhibited |
| IFNA2 | 4.17E-16 | Activated |
| IFNB1 | 3.95E-14 | Activated |
| Top diseases and bio functions | ||
| Diseases and disorders | ||
| Name | P value | Number of molecules |
| Antimicrobial response | 2.85E-03–4.91E-12 | 11 |
| Inflammatory disease | 2.19E-02–4.91E-12 | 28 |
| Dermatological diseases and conditions | 1.14E-02–2.04E-09 | 21 |
| Organismal injury and abnormalities | 2.19E-02–2.04E-09 | 53 |
| Immunological disease | 2.18E-02–1.79E-08 | 32 |
| Molecular and cellular functions | ||
| Name | P value | Number of molecules |
| Cell signaling | 1.08E-02–2.20E-07 | 14 |
| Cell death and survival | 2.13E-02–3.53E-07 | 23 |
| Cellular growth and proliferation | 1.98E-02–1.05E-06 | 26 |
| Cellular movement | 2.25E-02–2.94E-05 | 17 |
| Carbohydrate metabolism | 1.70E-02–3.26E-05 | 4 |
| Physiological system development and function | ||
| Name | P value | Number of molecules |
| Cardiovascular system development and function | 1.85E-02–2.17E-05 | 10 |
| Organismal development | 1.85E-02–2.17E-05 | 12 |
| Tissue development | 1.85E-02–6.40E-05 | 10 |
| Tissue morphology | 1.98E-02–6.92E-03 | 7 |
| Hematological system development and function | 1.98E-02–1.21E-04 | 11 |
| Top regulator effect networks | ||
| ID regulators | Diseases and functions | Consistency score |
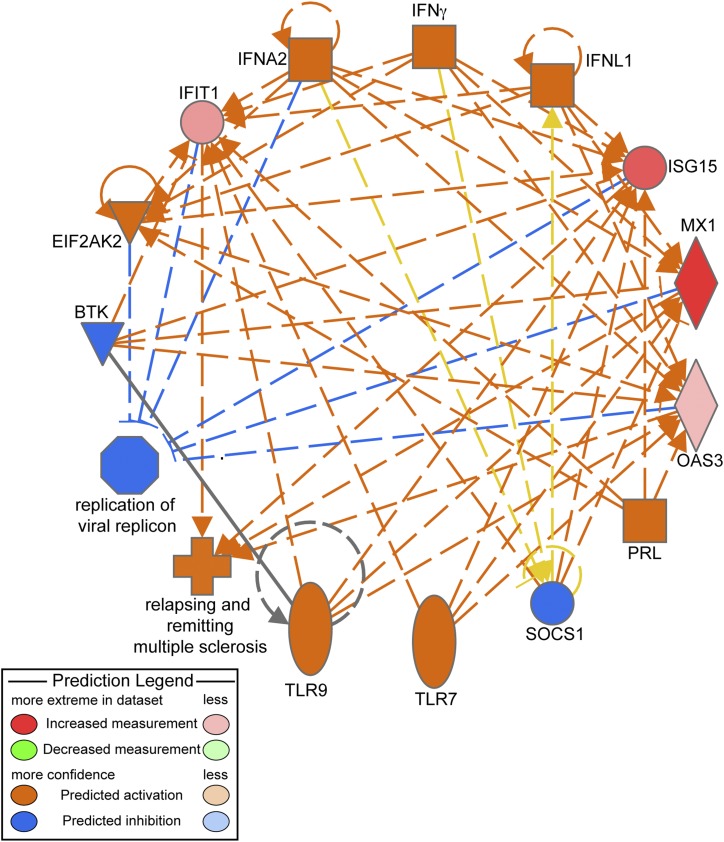
| BTK, EIF2AK2, IFNA2, IFNG, IFNL1, PRL, SOCS1, TLR7, TLR9 | Relapsing-remitting multiple sclerosis and replication of virus replicon | 33.5 |
| JAK1, NLRC5, TGM2 | Viral infection | 2.774 |
| MAPK1 | Viral infection | −1.664 |
| IFNA2 | Cell death | −2.333 |
| IL1RN | Cell death | −2.828 |
| Top networks | ||
| ID-associated network functions | Score | |
| Antimicrobial response, inflammatory response, infectious diseases | 40 | |
| Cellular growth and proliferation, development, cellular development, and lymphoid tissue structure | 14 | |
| Cell signaling, cancer, organismal injury, and abnormalities | 8 | |
| Developmental disorder, hereditary disorder, metabolic disease | 2 | |
| Cellular assembly and organization, gastrointestinal disease, and hepatic system disease | 2 | |
Abbreviations: IRF, IFN-regulatory factor; PRR, pattern recognition receptor; Th1, T helper 1.
Many of the significantly upregulated proteins participate in the host antiviral response, as anticipated, and have been shown by others to be induced at the level of gene expression in cultured human islets challenged with enteroviruses [8, 13, 14]. MX1/MxA, which is more than sevenfold upregulated in CVB4-infected islets compared with mock-infected islets, possesses broad antiviral activity. The ubiquitin-like protein ISG15 plays a critical role in the innate immune response to viral infection. Multiple additional IFN-induced antiviral RNA-binding proteins, including IFIT1, IFIT2, IFIT3, STAT1, and OAS3, are upregulated in CVB4-infected islets. TAP1, also known as antigen peptide transporter 1, is 3.5-fold upregulated in CVB4-infected cells vs controls. TAP1 is involved in the transport of antigens from the cytoplasm to the endoplasmic reticulum (ER) for association with major histocompatibility complex (MHC) class I molecules and acts as a molecular scaffold for the final stage of MHC class I folding by binding with the peptides [22]. TAPBP (tapasin) was also upregulated and is involved in the association of MHC class I with TAP1 and in the assembly of MHC class I with peptide [23]. Other genes/proteins involved in antigen presentation, including HLA-A, -B, and -C, were also increased.
The most significantly downregulated proteins in the CVB4-infected islets are regenerating islet-derived protein 3-α (REG3A), WIBG, and INS. The decrease in INS is consistent with our previous finding of decreased INS at the transcriptional level following CVB4 infection of human islets [18] and is in agreement with findings that intracellular insulin levels decrease in CVB-infected human pancreatic cultures [24].
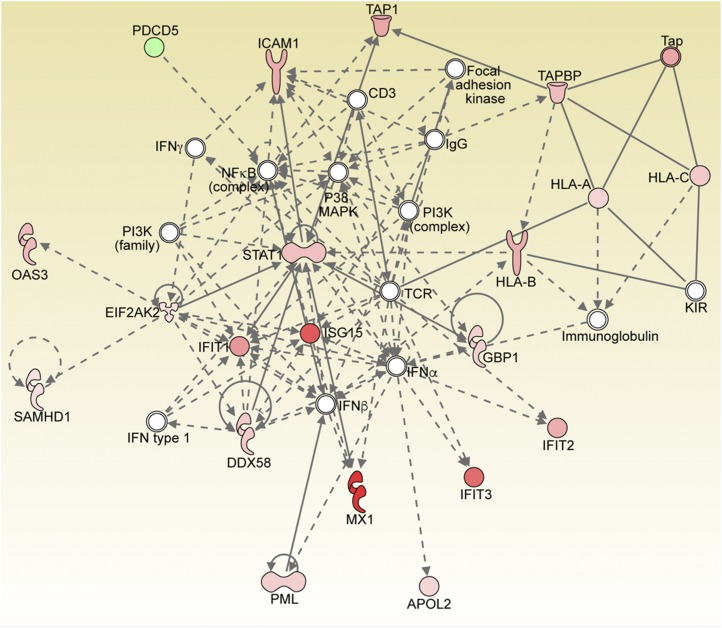
We performed regulator effects analysis using IPA to elucidate the causes and effects of differentially expressed proteins upon infection of islets. “Regulator Effects” explains how predicted activated or inhibited upstream regulators might cause increases or decreases in phenotypic or functional outcomes downstream. These causal hypotheses take the form of directionally coherent networks formed from the merger of Upstream Regulator networks with Downstream Effects networks [25]. IPA was used to investigate the regulation of the top networks associated with the profile of differentially expressed proteins obtained from quantitative proteomic analyses. Table 2 also summarizes the top five regulator effect networks and their associated networks, the corresponding diseases and functions, and respective scores obtained from IPA. The consistency score is a measurement used to rank or prioritize the most useful networks. The protein network with the highest consistency score of 33.5 includes BTK, EIF2AK2, IFNA2, IFNG, IFNL1, PRL, SOCS1, TLR7, and TLR9, which are all involved in autoimmunity. The associated activated networks are relapsing-remitting multiple sclerosis, which is predicted to be activated, and the replication of virus replicon, which is inhibited (Fig. 1). The top network corresponds to pathways related to antimicrobial response, inflammatory response, and infectious diseases (significance score of 40) (Fig. 2). The central molecule node of the network upregulated in CVB4-infected islets is STAT1. The top network analysis includes the following proteins, all of which were significantly upregulated: HLA-A, HLA-B, HLA-C, TAP1, TAP, ICAM, OAS3, SAMHD1, ISG, PML, DDX58, and GBP1. The other nodes in the network include the nuclear factor κB complex, a master regulator of the immune response, P38 mitogen-activated protein kinase (MAPK), and the phosphatidylinositol 3-kinase complex.
Figure 1.
IPA Top Regulator Effect with the highest consistent score of 33.5 (Table 2) showing inhibited and activated diseases and functions and the associated molecules. Proteins shown in orange are activated, whereas those in blue are inhibited in the CVB4-infected islets. Orange dashed lines depict activation, whereas blue dashed lines depict inhibition. The predictions indicate that pathways associated with the “replication of viral replicon” are inhibited upon CVB4 infection, whereas pathways associated with “relapsing and remitting multiple sclerosis” (associated with the inflammatory immune response) are activated. Yellow dashed lines depict findings inconsistent with the state of downstream molecules. Gray lines show effects not predicted.
Figure 2.
IPA Top Network generated from differentially regulated proteins between CVB4- and mock-infected primary human islets. Proteins shown in color are significantly differentially expressed based on the statistical analysis (red, upregulated; green, downregulated). A solid line represents a direct interaction between the two gene products, and a dotted line represents an indirect interaction. STAT1 is significantly upregulated and is one of the central nodes in the network.
B. CVB4 Infection Modulates Gene Expression in Cultured Human Islets
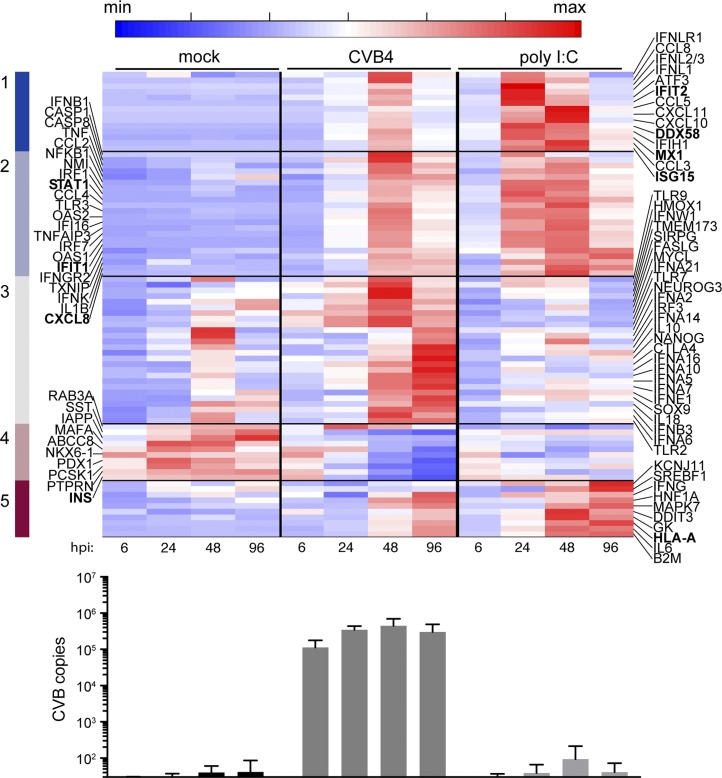
To correlate potential changes in gene expression with our proteomics analyses, we examined islet RNA expression over a time course using a NanoString platform. Cultured islets were independently studied from four different human donors, including donors used in the proteomics study (Supplemental Table 1 (19.2MB, docx) ). Cytotoxicity was examined and infection monitored (Supplemental Fig. 1 (19.2MB, docx) ). We focused on genes with at least a twofold change with CVB4 infection or poly I:C treatment compared with mock treatment. We clustered the expression pattern using a k-means clustering algorithm. In the heat map of Fig. 3, clusters 1 and 2 (dark blue and light blue) contain genes induced upon treatment, including inflammatory response genes and ISGs, whereas cluster 4 (light maroon) contains genes downregulated, such as INS and PDX1; GCG is unaffected by CVB4 infection, consistent with our previous report [18]. Type I IFN genes are among the first to increase in expression following CVB4 infection. IFNL3 gene expression is induced, consistent with recent studies on CVB triggering primary human islets to produce type III IFN [9]. CVB gene expression is sustained over the time course (Fig. 3, bottom panel).
Figure 3.
Gene expression in cultured primary human islets following challenge with CVB4 or poly I:C. The heat map depicts the five clusters of genes that have at least twofold change compared with mock in the average of the four replicates. The color bar highlights five clusters obtained after k-means clustering. Genes in boldface are also found in Table 1. Averages for four independent human donors were used. Time points are indicated. Average CVB copies per condition, measured by NanoString, are shown in the bottom panel; error bars represent the standard deviation.
We also challenged cultured human islets with poly I:C, a double-stranded RNA mimetic and MDA5 agonist, and compared specific gene induction patterns induced by poly I:C vs CVB4 over time (Fig. 3). Although some genes are induced by both, cluster 3 (gray) includes genes more robustly induced by CVB4 compared with poly I:C. Conversely, cluster 5 (dark maroon) includes genes more robustly induced by poly I:C than by CVB4. We found that many antiviral host responses are induced by poly I:C, particularly at 48 hpi, but unlike for CVB4, INS gene expression does not decrease with poly I:C challenge, which suggests that CVB4 may specifically influence INS independent from dsRNA signaling and type I IFN induction.
C. EndoC-βH1 Cells Exhibit High Cytotoxicity Following Infection With CVB4
To help define beta cell–specific factors that may affect the development of autoimmune T1D in the context of viral perturbation, we challenged EndoC-βH1 cells with CVB4. EndoC-βH1 cells exhibit considerable cytotoxicity by 48 hpi following infection with CVB4 (MOI of 1) (Supplemental Fig. 2A (19.2MB, docx) ). To visualize viral protein and insulin, CVB4-infected (MOI of 10) EndoC-βH1 cells at 6 hpi were stained with anti-VP1– (green) and anti-insulin–specific (red) antibodies. VP1 was readily detected, and colocalization with insulin was present (yellow) (Supplemental Fig. 2B (19.2MB, docx) ). Although EndoC-βH1 cells that appear to have escaped infection display a similar staining intensity for insulin as mock-treated cells, from image analysis alone, it is unclear if individual CVB4-infected cells have reduced insulin compared with uninfected (VP1-negative) cells (Supplemental Fig. 2C (19.2MB, docx) ). Challenge with CVB4 (MOI of 10) resulted in 62% of VP1-positive cells at 6 hpi. CVB4-infected EndoC-βH1 cells have a similar proportion of dead cells compared with mock-treated cultures at 6 hpi. By 24 hpi, 29% of cells were VP1 positive (Supplemental Fig. 2D (19.2MB, docx) ), and 74% dead cells were observed (Supplemental Fig. 2E (19.2MB, docx) ).
D. Gene Expression and Proteomics Were Evaluated in CVB4-Infected EndoC-βH1 Cells
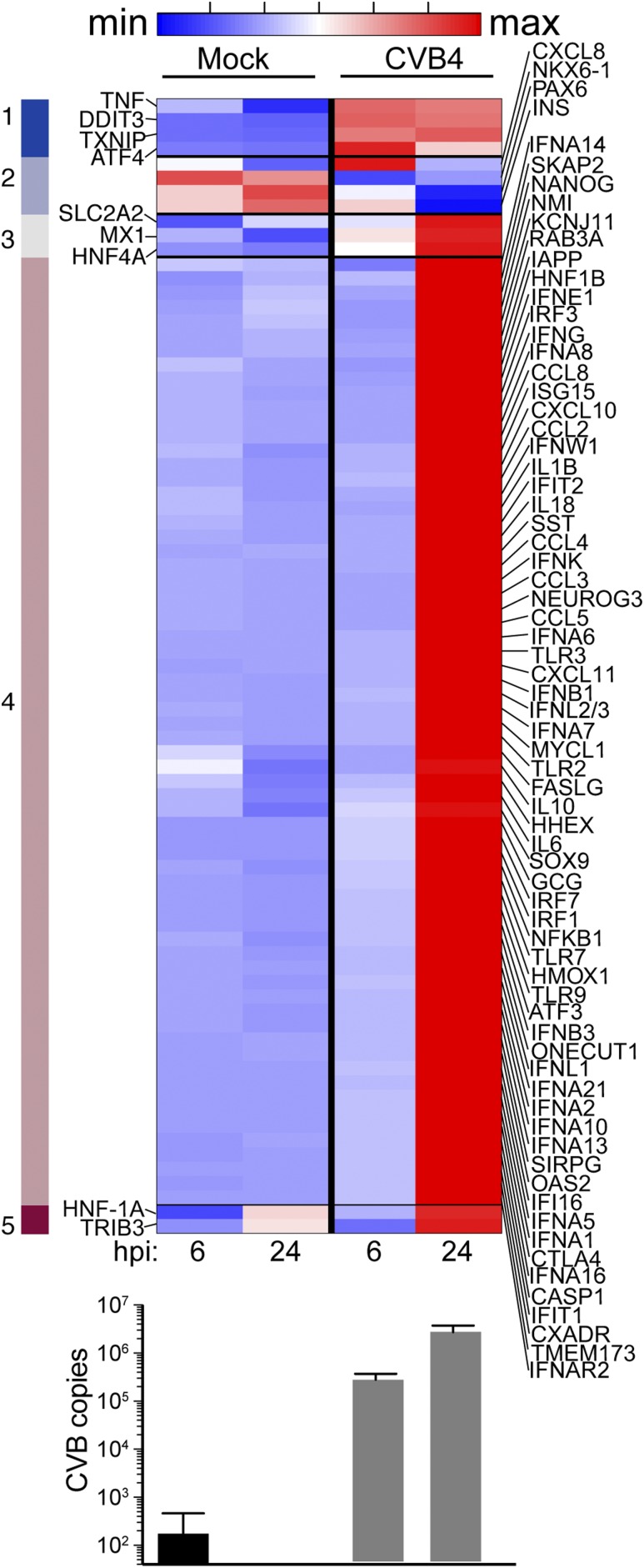
To compare the gene expression effects of CVB4 infection between human islets and EndoC-βH1 cells, we performed three independent EndoC-βH1 infections using CVB4 (MOI of 10). Gene expression was assessed at 6 and 24 hpi using the same NanoString CodeSet as for the primary human islets. We performed similar clustering as for human islets and observed robust gene induction at 24 hpi, particularly for ISGs (Fig. 4). INS gene expression was decreased in CVB4-challenged cells compared with mock-treated cells at 24 hpi. CVB gene expression increased by 10-fold between 6 and 24 hpi, indicative of viral replication (Fig. 4, bottom panel). The gene expression data of CVB4-infected EndoC-βH1 cells recapitulate some features observed in human islets infected with CVB4 and confirm that human beta cells are indeed capable of antiviral responses following infection. Of note, we processed EndoC-βH1 cells for proteomics analysis following infection with CVB4 (MOI of 5) at 24 hpi in two independent experiments. However, infected samples revealed no increases in either viral proteins or proteins expected to be induced based on gene expression data, presumably due to excessive cell death (data not shown).
Figure 4.
Gene expression in EndoC-βH1 cells following challenge with CVB4 (MOI of 10) compared with mock. The heat map shows five clusters obtained after k-means clustering of genes having at least a twofold change with CVB4 compared with mock. Clusters 1, 3, 4, and 5 have genes induced with CVB4. Cluster 2 (light blue) has genes downregulated with CVB4, including INS. Averages for three independent experiments were used. Time points are indicated. Average CVB copies per condition, measured by NanoString, are shown in the bottom panel; error bars represent the standard deviation.
3. Discussion
Understanding a mechanistic role of enteroviral infection in T1D is a priority. The role of virus infection in the setting of autoimmunity is still under investigation, but it has become increasingly clear that an islet insult leading to local inflammation is likely central in initiating the autoimmune process. Using a proteomics approach, we identified host factors modulated during acute CVB4 infection that could prove useful in identifying individuals at risk for progressing to T1D as well as therapeutic targets for prevention and intervention. We found that the insulin level is decreased in human islets following viral challenge and characterized the modulation of islet proteins not previously reported to be associated with viruses and T1D. Beta cell death following CVB4 infection likely contributes to some of the observed decreases in insulin [5] and will be important to quantify in future studies.
We confirmed the upregulation of several antiviral response factors including MxA, ISG15, and HLA class I antigens, as discussed above. HIC1 (hypermethylated in cancer 1 protein) is a transcriptional repressor that is upregulated in CVB4-infected human islets. HIC1 downregulates SIRT1, a protein involved in regulating p53/TP53-dependent apoptotic DNA-damage responses [26]. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target of HIC1 [27]. CXCR7 and other chemokine receptors are suggested to be T1D determinants, especially in autoimmunity and beta cell destruction [28]. HIC1 has also been demonstrated to modulate transcriptional stimulation of the genes regulated by canonical Wnt/beta-catenin signaling [29].
A high correlation between the protein and gene expression is observed between MS and gene expression analyses using protein and RNA, respectively, extracted from islets. The proteins/genes significantly modulated in both analyses include MX1, ISG15, CXCL8, IFIT1, IFIT2, STAT1, DDX58, HLA-A (all increased), and INS (decreased). Furthermore, many of the proteins increased in our study reportedly have significantly increased transcripts at 48 hpi by RNA-Seq in primary cultured human islets challenged with CVB3 strain Nancy [14]. These include APOL2, DDX58, EIF2AK2, GBP1, IFIT1, IFIT2, IFIT3, ISG15, LAP3, MX1, OAS3, PML, SAMHD1, STAT1, TAP1, TYMP, and WARS or ~10% of the genes reported to be increased [14]. Differences in the expression profiles of proteins and RNA transcripts in the CVB4-infected islets can be partially explained by molecular events such as translational efficiency, alternative splicing, trafficking and localization, posttranslational modifications, secretion, and proteolytic degradation, all of which affect protein abundance independently of transcripts. Discrepancies could be explained further by human islet donor variability and differences in viral serotypes used, as CVB3 Nancy and CVB4 JVB are ~80% identical by nucleotide sequence. Regardless, the orthogonal validation of differential expression of the genes using independent but complementary technologies validate the robustness of our data.
A number of proteins besides insulin were downregulated in CVB4-infected islets compared with control. Following CVB4 challenge, REG3A was the most significantly decreased. REG3A contains a peptide that may stimulate islet neogenesis and insulin production [30, 31], and its downmodulation following islet infection is reported in this study, although, interestingly, we previously described its upregulation in pancreata from autoantibody-positive subjects [32]. Decreased WIBG expression was also observed. The significance of this downregulation is unknown; however, a critical regulator of the exon junction complex, WIBG is postulated to bind to RNA and may be involved in positive regulation of translation [33]. ATP6AP1 (V-type proton ATPase subunit S1) is a vacuolar ATPase belonging to a family of proteins responsible for acidifying a variety of intracellular compartments in eukaryotic cells. Its downregulation (−1.970) may correlate with the observed decrease in INS because it is predicted to be involved in insulin receptor recycling [34]. Ubiquilin-2 (UBQLN2), −1.891 downregulated, is a critical molecule in the regulation of cellular protein degradation pathways including the ubiquitin-proteasome system, autophagy and the ER-associated protein degradation pathway [35–38]. The downregulation (−1.752) of protein phosphatase 1 regulatory subunit 1A (PPP1R1A) correlates with previous reports proposing that the protein is a real-time biomarker of beta cell destruction [39]. Downregulation of PRSS2 (trypsin-2), a trypsinogen, may reflect acinar cell presence in the islet preparations; given that human acinar cells are not typically infected by CVB4, this may occur secondary to changes in the local culture environment.
Many proteins modulated in CVB4-infected islets were reported in proteomic analysis of islets from individuals with enterovirus-associated fulminant diabetes [40], including WARS, STAT1, HLA-C, TYMP, APOL2, and SAMHD1. WARS, also known as tryptophan–transfer RNA ligase, has been linked to the regulation of angiogenesis and distinct cytokine activities [41]; like indoleamine 2,3-dioxygenase, it is preferentially induced by IFN-γ [42]. STAT1 mediates cellular responses to IFNs, cytokines and other growth factors, and STAT1 signaling ultimately activates the transcription of ISGs, which drive the cell to an antiviral state. TYMP (thymidine phosphorylase) catalyzes the reversible phosphorolysis of thymidine and possesses angiogenic activity in vivo and chemotactic activity on endothelial cells in vitro [43]. APOL2 binds to high-density lipoproteins and is involved in transportation of lipids, but its role in the context of enteroviral infection remains unknown. SAMHD1 (deoxynucleoside triphosphate triphosphohydrolase) is a restriction nuclease that blocks early-stage virus replication in myeloid cells [44]. SAMHD1 has a protective role in preventing self-activation of innate immunity, including type I IFN responses, by cell-intrinsic components in Aicardi-Goutières syndrome [45].
The permissiveness of EndoC-βH1 cells to CVB infection enables investigation of the virus-host relationship within human beta cells and potential identification/characterization of early biomarkers of viral infection and beta cell function. Indeed, type I IFN stimulation of EndoC-βH1 cells are reported to increase ER stress markers [46]. Although we observed ISG induction and downregulation of INS gene expression following infection of these cells, the impact on insulin secretion at the protein level requires further evaluation. Although high toxicity confounded proteomics assessment, examination of earlier time points or with a lower MOI could be useful for assessing beta cell–specific protein changes following CVB4 infection. The high cytotoxicity observed in the EndoC-βH1 cells could be due to a lack of a protective barrier in the islet microenvironment. Alternatively, the cells may lack survival signals from surrounding cell types present in primary islets.
Despite the increasing evidence correlating enterovirus infections with the development of T1D, establishing a causal relationship remains elusive. In this study, we provide additional insight on how enteroviruses may suppress human beta cells and identify potential markers for the initiation and progression of T1D. Although we used a standard sample size for our MS analyses, some proteins of potential interest did not meet the criteria for statistical significance that perhaps would have been identified with a larger sample size (i.e., our study was limited by low power). Nevertheless, our current studies confirm the upregulation/activation of specific molecules and pathways that were previously described in clinical samples of patients with enterovirus-associated fulminant diabetes. These molecules are potential therapeutic targets for intervention against T1D. Further studies are required to elucidate the role of these molecules in the etiology of T1D and may perhaps be gained through single-cell RNA sequencing of virus-infected islets, which could further facilitate the development of improved therapeutic or preventive treatments to halt the development of autoimmunity against beta cells.
Acknowledgments
We thank Melanie Trombly for assistance with manuscript preparation.
Financial Support: This work was supported by National Institutes of Health (NIH) Grant R01 AI092105 and R01 AI116920 (to J.P.W.), Human Islet Research Network NIH/National Institute of Diabetes and Digestive and Kidney Grant UC4 DK104166 (to J.L.N., J.O.N., and M.A.M.); and NIH Grants R01 DK105837 and UC4 DK104218 (to D.M.H. and R.B.). This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative type 1 diabetes research project sponsored by the Juvenile Diabetes Research Foundation (JDRF).
Author Contributions: J.O.N. designed the study, performed and analyzed the MS experiments, and wrote the manuscript. G.R.G., L.G., M.A.M., A.J., R.B., and D.M.H. contributed to the design and interpretation of the experiments and contributed to the discussion. G.R.G. performed experiments (all infections, NanoString, microscopy, and flow cytometry). P.V. performed statistical analysis on gene expression studies, interpreted the data, and contributed to the discussion. J.P.W. and J.L.N. contributed to the original idea and design of the experiments and wrote, revised, and edited the manuscript. All authors read and approved the manuscript and gave informed consent for publication.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ATP6AP1
- V-type proton ATPase subunit S1
- CVB
- coxsackievirus B
- CXCR
- chemokine (CXC motif) receptor
- ER
- endoplasmic reticulum
- hpi
- hours postinfection
- HIC1
- hypermethylated in cancer 1 protein
- IFN
- interferon
- IPA
- Ingenuity Pathway Analysis
- ISG
- interferon-stimulated gene
- MAPK
- mitogen-activated protein kinase
- MHC
- major histocompatibility complex
- MOI
- multiplicity of infection
- MS
- mass spectrometry
- poly I:C
- polyinosinic-polycytidylic acid
- REG3A
- regenerating islet-derived protein 3-α
- T1D
- type 1 diabetes.
References and Notes
- 1.Hyöty H. Viruses in type 1 diabetes. Pediatr Diabetes. 2016;17(Suppl 22):56–64. [DOI] [PubMed] [Google Scholar]
- 2.Krogvold L, Edwin B, Buanes T, Frisk G, Skog O, Anagandula M, Korsgren O, Undlien D, Eike MC, Richardson SJ, Leete P, Morgan NG, Oikarinen S, Oikarinen M, Laiho JE, Hyöty H, Ludvigsson J, Hanssen KF, Dahl-Jørgensen K. Detection of a low-grade enteroviral infection in the islets of Langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2015;64(5):1682–1687. [DOI] [PubMed] [Google Scholar]
- 3.Imagawa A, Hanafusa T. Fulminant type 1 diabetes--an important subtype in East Asia. Diabetes Metab Res Rev. 2011;27(8):959–964. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka S, Aida K, Nishida Y, Kobayashi T. Pathophysiological mechanisms involving aggressive islet cell destruction in fulminant type 1 diabetes. Endocr J. 2013;60(7):837–845. [DOI] [PubMed] [Google Scholar]
- 5.Roivainen M, Rasilainen S, Ylipaasto P, Nissinen R, Ustinov J, Bouwens L, Eizirik DL, Hovi T, Otonkoski T. Mechanisms of coxsackievirus-induced damage to human pancreatic beta-cells. J Clin Endocrinol Metab. 2000;85(1):432–440. [DOI] [PubMed] [Google Scholar]
- 6.Roivainen M, Ylipaasto P, Savolainen C, Galama J, Hovi T, Otonkoski T. Functional impairment and killing of human beta cells by enteroviruses: the capacity is shared by a wide range of serotypes, but the extent is a characteristic of individual virus strains. Diabetologia. 2002;45(5):693–702. [DOI] [PubMed] [Google Scholar]
- 7.Frisk G, Diderholm H. Tissue culture of isolated human pancreatic islets infected with different strains of coxsackievirus B4: assessment of virus replication and effects on islet morphology and insulin release. Int J Exp Diabetes Res. 2000;1(3):165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulte BM, Lanke KH, Piganelli JD, Kers-Rebel ED, Bottino R, Trucco M, Huijbens RJ, Radstake TR, Engelse MA, de Koning EJ, Galama JM, Adema GJ, van Kuppeveld FJ. Cytokine and chemokine production by human pancreatic islets upon enterovirus infection. Diabetes. 2012;61(8):2030–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ylipaasto P, Smura T, Gopalacharyulu P, Paananen A, Seppänen-Laakso T, Kaijalainen S, Ahlfors H, Korsgren O, Lakey JR, Lahesmaa R, Piemonti L, Oresic M, Galama J, Roivainen M. Enterovirus-induced gene expression profile is critical for human pancreatic islet destruction. Diabetologia. 2012;55(12):3273–3283. [DOI] [PubMed] [Google Scholar]
- 10.Ylipaasto P, Klingel K, Lindberg AM, Otonkoski T, Kandolf R, Hovi T, Roivainen M. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia. 2004;47(2):225–239. [DOI] [PubMed] [Google Scholar]
- 11.Elshebani A, Olsson A, Westman J, Tuvemo T, Korsgren O, Frisk G. Effects on isolated human pancreatic islet cells after infection with strains of enterovirus isolated at clinical presentation of type 1 diabetes. Virus Res. 2007;124(1-2):193–203. [DOI] [PubMed] [Google Scholar]
- 12.Chehadeh W, Kerr-Conte J, Pattou F, Alm G, Lefebvre J, Wattré P, Hober D. Persistent infection of human pancreatic islets by coxsackievirus B is associated with alpha interferon synthesis in beta cells. J Virol. 2000;74(21):10153–10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anagandula M, Richardson SJ, Oberste MS, Sioofy-Khojine AB, Hyöty H, Morgan NG, Korsgren O, Frisk G. Infection of human islets of Langerhans with two strains of Coxsackie B virus serotype 1: assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway. J Med Virol. 2014;86(8):1402–1411. [DOI] [PubMed] [Google Scholar]
- 14.Domsgen E, Lind K, Kong L, Hühn MH, Rasool O, van Kuppeveld F, Korsgren O, Lahesmaa R, Flodström-Tullberg M. An IFIH1 gene polymorphism associated with risk for autoimmunity regulates canonical antiviral defence pathways in Coxsackievirus infected human pancreatic islets. Sci Rep. 2016;6:39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg M, Krogvold L, Kuric E, Dahl-Jørgensen K, Skog O. Expression of interferon-stimulated genes in insulitic pancreatic iIslets of patients recently diagnosed with type 1 diabetes. Diabetes. 2016;65(10):3104–3110. [DOI] [PubMed] [Google Scholar]
- 16.Jean-Baptiste VSE, Xia CQ, Clare-Salzler MJ, Horwitz MS. Type 1 diabetes and type 1 interferonopathies: localization of a type 1 common thread of virus infection in the pancreas. EBioMedicine. 2017;22:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravassard P, Hazhouz Y, Pechberty S, Bricout-Neveu E, Armanet M, Czernichow P, Scharfmann R. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher GR, Brehm MA, Finberg RW, Barton BA, Shultz LD, Greiner DL, Bortell R, Wang JP. Viral infection of engrafted human islets leads to diabetes. Diabetes. 2015;64(4):1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber M, Watson KA, Selinka HC, Carthy CM, Klingel K, McManus BM, Kandolf R. Cleavage of RasGAP and phosphorylation of mitogen-activated protein kinase in the course of coxsackievirus B3 replication. J Virol. 1999;73(5):3587–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuer R, Mena I, Pagarigan R, Slifka MK, Whitton JL. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol. 2002;76(9):4430–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG II, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4(6):2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eggensperger S, Tampé R. The transporter associated with antigen processing: a key player in adaptive immunity. Biol Chem. 2015;396(9-10):1059–1072. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Paulsson KM, Chen S, Sjögren HO, Wang P. Tapasin is required for efficient peptide binding to transporter associated with antigen processing. J Biol Chem. 2000;275(3):1581–1586. [DOI] [PubMed] [Google Scholar]
- 24.Yoon JW, Onodera T, Jenson AB, Notkins AL. Virus-induced diabetes mellitus. XI. Replication of coxsackie B3 virus in human pancreatic beta cell cultures. Diabetes. 1978;27(7):778–781. [DOI] [PubMed] [Google Scholar]
- 25.Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. [DOI] [PubMed] [Google Scholar]
- 27.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guérardel C, Ramsey K, Monté D, Bégue A, Tschan MP, Stephan DA, Leprince D. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1). J Biol Chem. 2009;284(31):20927–20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fallahi P, Corrado A, Di Domenicantonio A, Frenzilli G, Antonelli A, Ferrari SM. CXCR3, CXCR5, CXCR6, and CXCR7 in Diabetes. Curr Drug Targets. 2016;17(5):515–519. [DOI] [PubMed] [Google Scholar]
- 29.Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006;25(11):2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levetan CS, Upham LV, Deng S, Laury-Kleintop L, Kery V, Nolan R, Quinlan J, Torres C, El-Hajj RJ. Discovery of a human peptide sequence signaling islet neogenesis. Endocr Pract. 2008;14(9):1075–1083. [DOI] [PubMed] [Google Scholar]
- 31.Choi JH, Lee MY, Kim Y, Shim JY, Han SM, Lee KA, Choi YK, Jeon HM, Baek KH. Isolation of genes involved in pancreas regeneration by subtractive hybridization. Biol Chem. 2010;391(9):1019–1029. [DOI] [PubMed] [Google Scholar]
- 32.Burch TC, Morris MA, Campbell-Thompson M, Pugliese A, Nadler JL, Nyalwidhe JO. Proteomic analysis of disease stratified human pancreas tissue indicates unique signature of type 1 diabetes. PLoS One. 2015;10(8):e0135663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14(12):1173–1179. [DOI] [PubMed] [Google Scholar]
- 34.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33(Database issue):D428–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6(2):409–419. [DOI] [PubMed] [Google Scholar]
- 36.Xia Y, Yan LH, Huang B, Liu M, Liu X, Huang C. Pathogenic mutation of UBQLN2 impairs its interaction with UBXD8 and disrupts endoplasmic reticulum-associated protein degradation. J Neurochem. 2014;129(1):99–106. [DOI] [PubMed] [Google Scholar]
- 37.N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10(2):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim TY, Kim E, Yoon SK, Yoon JB. Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem Biophys Res Commun. 2008;369(2):741–746. [DOI] [PubMed] [Google Scholar]
- 39.Jiang L, Brackeva B, Ling Z, Kramer G, Aerts JM, Schuit F, Keymeulen B, Pipeleers D, Gorus F, Martens GA. Potential of protein phosphatase inhibitor 1 as biomarker of pancreatic β-cell injury in vitro and in vivo. Diabetes. 2013;62(8):2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishida Y, Aida K, Kihara M, Kobayashi T. Antibody-validated proteins in inflamed islets of fulminant type 1 diabetes profiled by laser-capture microdissection followed by mass spectrometry. PLoS One. 2014;9(10):e107664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci USA. 2002;99(1):173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleckner J, Martensen PM, Tolstrup AB, Kjeldgaard NO, Justesen J. Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine. 1995;7(1):70–77. [DOI] [PubMed] [Google Scholar]
- 43.Usuki K, Saras J, Waltenberger J, Miyazono K, Pierce G, Thomason A, Heldin CH. Platelet-derived endothelial cell growth factor has thymidine phosphorylase activity. Biochem Biophys Res Commun. 1992;184(3):1311–1316. [DOI] [PubMed] [Google Scholar]
- 44.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. [DOI] [PubMed] [Google Scholar]
- 45.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marroqui L, Dos Santos RS, Op de Beeck A, Coomans de Brachène A, Marselli L, Marchetti P, Eizirik DL. Interferon-α mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia. 2017;60(4):656–667. [DOI] [PubMed] [Google Scholar]