Abstract
Context:
Autosomal-recessive mutations in the growth hormone receptor (GHR) are the most common causes for primary growth hormone insensitivity (GHI) syndrome with classical GHI phenotypically characterized by severe short stature and marked insulin-like growth factor (IGF)-I deficiency. We report three families with dominant-negative heterozygous mutations in the intracellular domain of the GHR causing a nonclassical GHI phenotype.
Objective:
To determine if the identified GHR heterozygous variants exert potential dominant-negative effects and are the cause for the GHI phenotype in our patients.
Results:
All three mutations (c.964dupG, c.920_921insTCTCAAAGATTACA, and c.945+2T>C) are predicted to result in frameshift and early protein termination. In vitro functional analysis of variants c.964dupG and c.920_921insTCTCAAAGATTACA (c.920_921ins14) suggests that these variants are expressed as truncated proteins and, when coexpressed with wild-type GHR, mimicking the heterozygous state in our patients, exert dominant-negative effects. Additionally, we provide evidence that a combination therapy of recombinant human growth hormone (rhGH) and rhIGF-I improved linear growth to within normal range for one of our previously reported patients with a characterized, dominant-negative GHR (c.899dupC) mutation.
Conclusion:
Dominant-negative GHR mutations are causal of the mild GHI with substantial growth failure observed in our patients. Heterozygous defects in the intracellular domain of GHR should, therefore, be considered in cases of idiopathic short stature and IGF-I deficiency. Combination therapy of rhGH and rhIGF-I improved growth in one of our patients.
Keywords: dominant-negative GHR mutations, GH/IGF-I therapy
In patients with mild GHI, heterozygous, truncating GHR mutations exerted dominant-negative effects. A combined therapy of rhGH plus rhIGF-I is an effective treatment option.
The human growth hormone (GH) receptor (GHR) mediates the growth and metabolic effects of GH through a cascade of downstream signaling leading to the transcription of multiple genes, including insulin-like growth factor (IGF)-I. The GHR consists of an extracellular domain that binds GH, a short transmembrane domain, and an intracellular domain involved in transducing signals upon binding of GH. The receptor lacks intrinsic kinase activity and recruits Janus-family tyrosine kinase 2 (JAK2) for activation of downstream signaling. Binding of GH to the dimeric cell-surface receptor induces conformational changes in the receptor resulting in transphosphorylation of the associated JAK2 [1]. Phosphorylated JAK2, in turn, phosphorylates the seven intracellular tyrosine residues of GHR, including those that are critical for activation of the signal transducer and activator of transcription 5B (STAT5B) in humans [2]. Phosphorylated STAT5B dimerizes, translocates to the nucleus, and regulates the transcription of target genes, including IGF1, IGFBP3, and IGFALS [3–5].
Mutations of GHR (MIM 600946) are the most common cause for primary GH insensitivity (GHI) syndrome, phenotypically characterized by severe short stature [heights as low as −10 standard deviations (SDs) below normal], dysmorphic facial features, and marked IGF-I deficiency. To date, more than 80 different mutations in GHR have been identified in more than 300 case reports [6–9]. The overwhelming majority of the reported mutations are autosomal-recessive (deletions, point or splice mutations) and are located in sequences encoding the extracellular domain of the receptor [9]. Very few proven clinically important heterozygous mutations in GHR have been reported, of which only four have been shown to exert dominant-negative effects [10–15]. We now report the identification of three heterozygous variants in the intracellular domain of the GHR in three unrelated patients who present with postnatal growth failure, IGF deficiency, and a relatively normal facial phenotype. We demonstrate that these variants also exerted dominant-negative effects; importantly, therapeutic options are available for these patients, and we show that long-term treatment with a combination of recombinant human GH (rhGH) and rhIGF-I improved linear growth to within the normal range for our previously described patient who carried the GHR c.899dupC dominant-negative mutation.
1. Case Reports
Patient 1 (P1) is a girl initially evaluated at age 3 years, 4 months. Height was 84 cm (−3.09 SD) and weight 11.3 kg (−2.36 SD). She was born at 39 weeks of gestation with a weight of 3.5 kg (+0.23 SD) and length of 49.5 cm (−0.29 SD). Her metabolic panel, complete blood count, karyotype, celiac panel, and thyroid function tests were normal. She had a normal serum GH level (4.3 ng/mL; normal, <10), low serum IGF-I level (<25 ng/mL; normal, 49 to 290) and low normal IGF binding protein 3 (IGFBP-3) (1.1 mg/L; normal, 0.9 to 4.5). GH stimulation testing showed a peak GH level of 14.1 ng/mL (after clonidine), 23.7 ng/mL (after arginine), and 24.7 ng/mL (after glucagon). During a subsequent evaluation at the age of 7 years, 6 months, P1’s physical examination revealed a small midface, mild frontal bossing, bluish sclerae, and delayed dental age. An IGF-I generation test was performed after GH was given for a week. Baseline IGF-I was 24 ng/mL (normal, 39 to 198); after GH stimulation, IGF-I was 44 ng/mL (normal, 39 to 198). Her basal IGFBP-3 level was 1.2 mg/L (normal, 1.5 to 3.4), with no further increase after GH stimulation. She had normal values of acid labile subunit (ALS), ruling out ALS deficiency. Her GH binding protein (GHBP) was mildly elevated at 1991 pmol/L (normal, 267 to 1638).
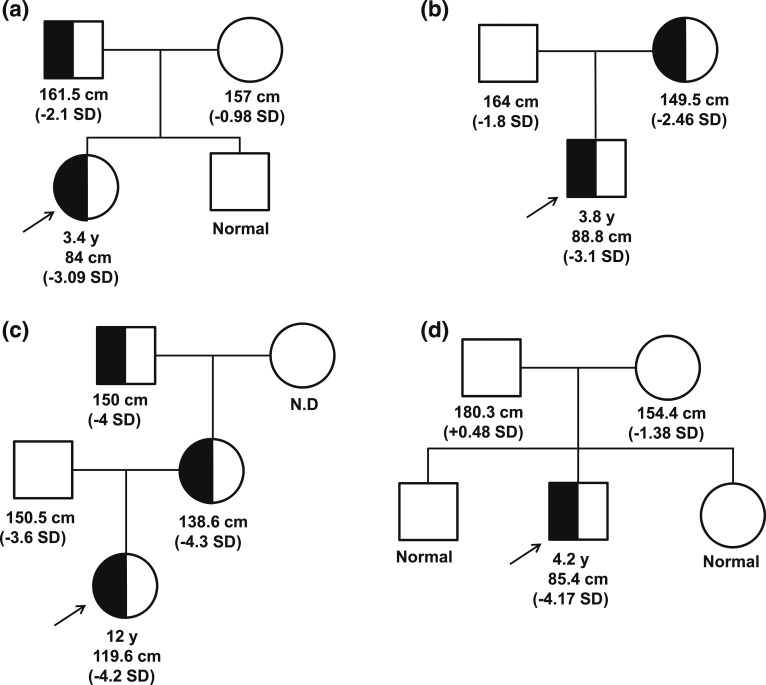
Her mother is of Hispanic ethnicity with normal height (157 cm, −0.98 SD) and had menarche at 12 years of age. Her brother was of normal stature. Her father (Northern European ethnicity) was modestly short [Fig. 1(a)] with a height of 161.5 cm (−2.1 SD) and had delayed puberty (around 17 years of age). Short stature was prevalent on the father’s side of the family. Her paternal grandmother’s height is 150 cm (−2.07 SD); paternal great aunts, 142 cm (−3.23 SD) and 150 cm (−2.07 SD); and paternal great uncle, 145 cm (−4.4 SD). The height of the paternal uncle is normal (170 cm, −0.93 SD).
Figure 1.
Heterozygous GHR variant segregation in affected families. (a) P1, c.964dupG; (b) P2, c.920_921ins14; (c) P3, c.945+2T>C; and (d) P4, de novo c.899dupC [15]. Arrow, proband.
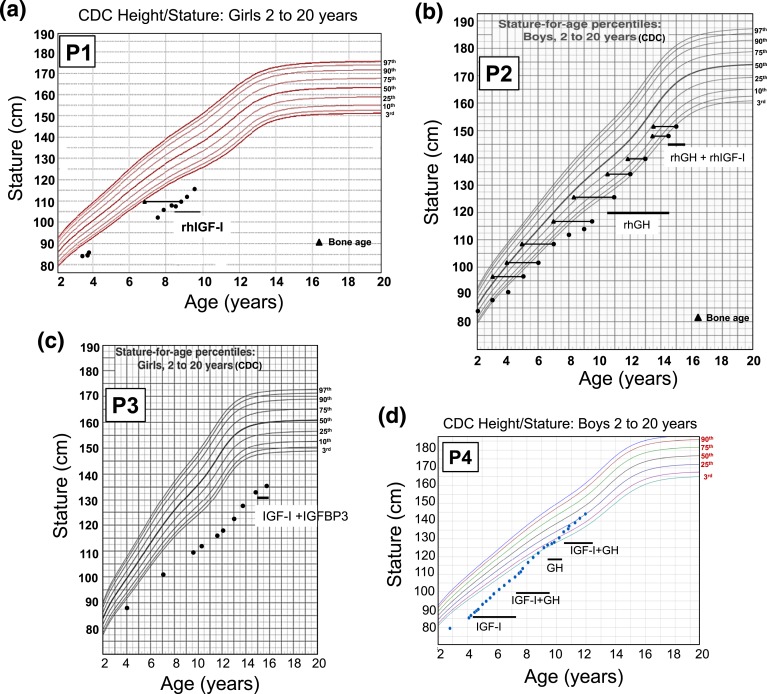
P1 was started on rhIGF-I therapy at the age of 8 years, 8 months, with incremental dosing over the course of 6 weeks to 120 µg/kg/dose twice daily. Her height at the start of rhIGF-I treatment was 109.9 cm (−3.89 SD) and, at the age of 9 years, 6 months, was 116 cm (−3.24 SD). Her annualized growth velocity on rhIGF-I improved to 8.1 cm/y compared with 4 cm/y before therapy [Fig. 2(a)]. The family also reported increased energy and muscle strength with treatment.
Figure 2.
The growth charts of the patients carrying heterozygous GHR variants. (a) P1, c.964dupG; (b) P2, c.920_921ins14; (c) P3, c.945+2T>C; and (d) P4, c.899dupC [15]. The time frame of rhIGF-I, rhGH, and rhIGF-I + IGFBP3 treatments are indicated on the growth curves. CDC, Centers for Disease Control and Prevention.
Patient 2 (P2) is a Spanish boy with a height of 88.8 cm (−3.1 SD) and a weight of 11.3 kg (−2.0 SD) at age 3.8 years. He had normal thyroid function; serum IGF-I was low normal at 69 ng/mL (normal, 55 to 297). He was born at 37 weeks with weight of 2.6 kg (−0.7 SD), birth length of 48 cm (−0.3 SD), and head circumference of 31.5 cm (−1.2 SD). His body proportions were normal and he had no dysmorphic features. At the age of 2 years, he was diagnosed with celiac disease; his autoantibodies normalized with good adherence to diet.
Growth velocity was 4 to 6 cm per year with a bone age delayed at least 2.5 years. IGF-I concentrations were repeatedly low. At age 6 years, P2 underwent a GH stimulation test (clonidine); he had a peak GH response of 17.7 mU/L. At age 9.6 years, P2 underwent an IGF-I generation test with GH given for 5 days (0.35 µg/kg/d); it elicited a poor response. Baseline IGF-I was <25 ng/mL (normal, 55 to 297) and upon GH stimulation, IGF-I remained below normal (34 ng/mL; range, 55 to 297). Basal and stimulated IGFBP-3 levels were 1.28 mg/L and 1.99 mg/L, respectively (normal, 1.5 to 7.4).
The parents were not consanguineous and his father was of normal stature (164 cm; −1.8 SD). His mother was modestly short [Fig. 1(b)] with a height of 149.5 cm (−2.46 SD).
rhGH treatment, started at 10.6 years (0.035 µg/kg/d), resulted in an increased growth velocity to 8 cm/yea but IGF-I remained persistently low (maximum value, 87 ng/mL). At age 12 years, P2’s height was at −2.5 SD, with bone age delayed by 1.5 years. He was noted to be in puberty at 13.5 years (testes, 6 mL and 8 mL). At 14.5 years, low-dose rhIGF-I (40 µg/kg/d) was added to his treatment regimen. At 15 years (testes, 12 mL), his height was 154 cm (−2.02 SD), weight 38 kg (−1.99 SD), and bone age 14.5 years [Fig. 2(b)].
Patient 3 (P3) was a 12-year-old Spanish girl from a nonconsanguineous marriage. She presented with a substantially short stature (119.6 cm, −4.30 SD) and normal facial features. She had a high basal GH (57.6 ng/mL), low IGF-I (43 ng/mL; normal, 126 to 1188 ng/mL), low IGFBP-3 (0.87 mg/L; normal, 2.0 to 9.3), and low ALS (7.4 mg/L; normal, 14 to 29). Her GHBP was elevated (35,634 pmol/L; normal, 534 to 5785). Further investigation revealed the presence of a substantially short stature in three other family members [Fig. 1(c)]: the patient’s father (150.5 cm, –3.6 SD), mother (138.6 cm, –4.3 SD), and maternal grandfather (150 cm, –4.0 SD).
The patient was started on a then-available combination therapy of rhIGF-I/rhIGFBP-3 (1 mg/kg/d) at age 15, 1 month after menarche. Treatment was well tolerated but response was poor, although only given for a total of 10 months at quite an advanced age. Her height at the beginning of the treatment was 135 cm and at the end of treatment was 137.8 cm [Fig. 2(c)]. The treatment was discontinued at age 15 years, 10 months.
Patient 4 (P4), previously reported [15], had a height of 85.4 cm (−4.17 SD) and weight of 11.4 kg (−3.66 SD) at age 4.2 years, bone age of 3.0 years, normal GH and GHBP concentrations, a low IGF-I of 16 ng/mL (normal, 54 to 178), and a low IGFBP-3 of 0.7 mg/L (normal, 0.8 to 3.0). He carried a de novo, dominant-negative, heterozygous GHR c.899dupC mutation [Fig. 1(d)] [10]. Treatment with rhIGF-I (120 µg/kg twice daily) was initiated, and his pretreatment growth velocity of 4.2 cm/y (age, 4.2 years) improved to 8.0 cm/y after 1 year of treatment, and height SD was −3.30 at age 5.3 years [15]. After 3 years of rhIGF-I therapy, rhGH (0.28 mg/kg/wk) was added to the regimen and rhIGF-I dosing adjusted to 90 µg/kg/dose, a combination that further increased growth velocity to >10 cm/y. When treatment with rhIGF-I was interrupted for a year (because of unavailability of the drug), growth velocity dropped to <7 cm/y while continuing rhGH therapy. The resumption of the coadministration therapy (at age 10.75 years) increased growth velocity to >10 cm/year again [Fig. 2(d)], and at age 11.25 years, his height was in the normal range (139.2 cm, −0.81 SD). He was noted to be in puberty at age 11.7 years (testicular volumes, 8 mL bilaterally), with bone ages of 12.5 years (phalanges) and 11.5 years (metacarpals).
The clinical, biochemical, and treatment profiles of all four patients are summarized in Table 1.
Table 1.
Summary of Clinical, Biochemical, and Treatment Profiles of Patients With Dominant-Negative Mutations in the GHR
|
P1 |
P2 |
P3 |
P4 |
|||
|---|---|---|---|---|---|---|
| c.964dupG/WT | c.920_921ins14/WT | c.945+2T>C/WT | c.899dupC/p.R229H | |||
| Mutation | Duplication | Insertion | Splicing | Duplication | ||
| Age, y | 3.4 | 3.8 | 12 | 4.2 | ||
| Sex | Female | Male | Female | Male | ||
| Height, cm (SD) | 84 (−3.09) | 88.8 (−3.1) | 119.6 (−4.3) | 85.4 (−4.17) | ||
| Weight, kg (SD) | 11.3 (−2.36) | 11.3 (−2.0) | NA | 11.4 (−3.66) | ||
| Ethnicity | Northern European/Hispanic | Spanish | Spanish | Pakistan | ||
| Birth week | 39 | 37 | NA | NA | ||
| Birth weight, kg (SD) | 3.56 (+0.23) | 2.6 (−0.7) | NA | 3.38 | ||
| Birth length, cm (SD) | 49.5 (−0.29) | 48 (−0.3) | NA | 48 | ||
| Paternal height, cm (SD) | 161.5 (−2.1) | 164 (−1.8) | 150.5 (−3.6) | 180.3 (+0.48) | ||
| Maternal height, cm (SD) | 157 (−0.98) | 149.5 (−2.46) | 138.6 (−4.3) | 154.4 (−1.38) | ||
| Biochemistry | ||||||
| GH basal, ng/mL | 4.3 | NA | 56.6 | 8 | ||
| GH stimulated | 14.1 ng/mL (clonidine) | 17.7 mU/L (clonidine) | NA | 37 ng/mL (arginine) | ||
| IGF-I, ng/mL (RR) | <25 (49–290) | 69 (55–297) | 43 (126–1188) | 16 (54–178) | ||
| IGFBP-3, mg/L (RR) | 1.1 (0.9–4.5) | NA | 0.87 (2.0–9.3) | 0.7 (0.8–3.0) | ||
| ALS, mg/L (RR) | 4.6 (2.3–11) | NA | 7.4 (14–29) | 2.6 (1.9–10) | ||
| GHBP, pmol/L (RR) | 1991 (267–1638) | NA | 35,634 (534–5785) | 1379 (267–1638) | ||
| Treatment | ||||||
| Treatment | rhIGF-I | rhGH | rhGH + rhIGF-I | rhIGF-I + rhIGFBP-3 | rhIGF-I | rhGH + rhIGF-I |
| Dose | 120 µg/kg bid | 0.035 mg/kg/d | 0.035 mg/kg/d (rhGH) + 40 µg/kg/d (rhIGF-I) | 1 mg/kg/da | 120 µg/kg bid | 0.04 mg/kg/d (rhGH) + 90 µg/kg bid (rhIGF-I) |
| Age, y | 8.8 | 12.08 | 14.8b | 15 | 4.7 | 11.7b |
| IGF-1, ng/mL (RR) | 206 (57–305) | 87 (80–690) | 309 (180–820) | NA | 211 (36–202) | 326 (86–393) |
| IGFBP3, mg/L (RR) | NA | 3.4 (2.1–9.2) | 3.73 (>3.1) | NA | 0.6 (1–4.7) | 0.9 (2–4.8) |
| ALS, mg/L (RR) | NA | NA | NA | NA | NA | 3.3 (5.6–6) |
| Reference | This study | This study | This study | Aisenberg et al. (2010) | ||
| Derr et al. (2011), this study | ||||||
Abbreviations: bid, twice daily; NA, not available; RR, reference range.
rhIGF-I + rhIGFBP3 combination therapy that is no longer available.
Tanner II-III, testes 8 mL.
2. Materials and Methods
A. Sample Procurement
Informed consent was obtained from the families to participate and provide samples (DNA, whole blood, skin biopsy for establishing primary fibroblasts), in compliance with the Institutional Review Board at Cincinnati Children’s Hospital Medical Center (Cincinnati, Ohio), SIEMENS, and Seattle Children’s laboratory. Genomic DNA was isolated from peripheral blood of patients and their parents as previously described [15]. Whole exome sequencing was performed on DNA samples from P1, her unaffected brother, and her parents, as previously described [16]. For P2 and P3, targeted Sanger sequencing of the GHR coding exons (NM_000163.4), exons 2 through 10, was performed, as previously described [15]. For P3, total RNA was extracted from peripheral leukocytes and GHR messenger RNA (mRNA) was amplified by reverse transcription polymerase chain reaction (RT-PCR) to determine integrity of exon 9 splicing events.
B. Regenerating the GHR Variants for Functional Studies
The plasmid pcDNA carrying the full-length human GHR (kind gift from Dr. Richard Ross) was used as template to regenerate variants c.964dupG and c.920_921ins14. The variants were generated using the QuikChange II XL Site-Directed Mutagenesis Kit from Agilent, as previously described [10]. The c.920_921ins14 variant was generated in three sequential steps. In the first two steps, five nucleotides were inserted; in the last step, four nucleotides were inserted resulting in insertion of 14 nucleotides in total. The primers for the site-directed mutagenesis were designed using the Quick change primer design program from Agilent. The sequence of the primers used is shown in Table 2.
Table 2.
Primers Used to Generate the Human GHR Variants, c.964dupG and c.920_921ins14
| Primer Name | Primer Sequence |
|---|---|
| Primers used to generate the hGHR c.964dupG variant | |
| hGHR c.964dupG forward | 5′ GGAAGGAAAATTAGAGGAGGGTGAACACAATCTTAGCCATT 3′ |
| hGHR c.964dupG reverse | 5′ AATGGCTAAGATTGTGTTCACCCTCCTCTAATTTTCCTTCC 3′ |
| Primers used to generate the hGHR c.920_921ins14 variant | |
| hGHR c.920_921ins14 (step 1) forward | 5′ CCCCAGTTCCAGTTCCAAAGATTAATCTCAAGGAATCGATCCA 3′ |
| hGHR c.920_921ins14 (step 1) reverse | 5′ TGGATCGATTCCTTGAGATTAATCTTTGGAACTGGAACTGGGG 3′ |
| hGHR c.920_921ins14 (step 2) forward | 5′ CAGTTCCAGTTCCAAAGATTAATCTCAAAGATAGGAATCGATCCAG 3′ |
| hGHR c.920_921ins14 (step 2) reverse | 5′ CTGGATCGATTCCTATCTTTGAGATTAATCTTTGGAACTGGAACTG 3′ |
| hGHR c.920_921ins14 (step 3) forward | 5′CCAGTTCCAAAGATTAATCTCAAAGATTACAAGGAATCGATCCGATC 3′ |
| hGHR c.920_921ins14 (step 3) reverse | 5′ GATCTGGATCGATTCCTTGTAATCTTTGAGATTAATCTTTGGAACTGG 3′ |
Nucleotides inserted are underlined.
C. Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at 37°C in 5% CO2. For in vitro functional analyses, HEK293 cells were transfected with the vector (pcDNA3.1), wild-type (WT), and/or the mutant GHR variants using Polyjet transfection reagent (SignaGen Laboratories). After 24 hours of transfection, the cells were serum starved in Dulbecco’s modified Eagle medium supplemented with 0.1% bovine serum albumin for 8 or 16 hours and then treated with rhGH as indicated. All transfection experiments were performed at least three independent times.
D. Immunoblot Analysis
For immunoblot analyses, transfected cells were treated with 100 ng/mL rhGH for 20 minutes; cell lysates were prepared and processed as previously described [10]. Protein concentrations of the lysates were determined by the Bradford assay and equal quantities of protein were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. After electrophoresis, the proteins were transferred onto nitrocellulose membranes, blocked using 5% bovine serum albumin or 5% nonfat milk in 1X tris(hydroxymethyl)aminomethane-buffered saline with Tween 20 (TBST) and incubated with the appropriate primary and secondary antibodies (fluorescent-labeled or horseradish peroxidase–conjugated). Following incubation, the blots probed with fluorescent-labeled secondary antibodies were washed in 1X TBST and membranes were scanned using the Odyssey IR Imaging System (LICOR Biosciences). The blots probed with the horseradish peroxidase–conjugated secondary antibodies were washed in 1X TBST and developed using the super signal maximum sensitivity detection system (Thermo Scientific, catalog no. 37075).
E. Luciferase Reporter Assays
HEK293 cells were transfected with a total input DNA of 2.0 µg, including 1 µg luciferase (pGHRE-LUC) [10] and vector pcDNA3.1 or specified human GHR (hGHR) constructs (WT, GHR c.964dupG, and c.920_921ins14) as indicated. After 24 hours of transfection, cells were serum starved for ~8 hours and treated with 10 ng/mL rhGH for 18 hours. Cell lysates were collected, and luciferase activity was determined using the luciferase assay system from Promega per manufacturers’ protocol. The assay was performed at least three independent times.
3. Results
A. Identification and Segregation of the GHR Variants
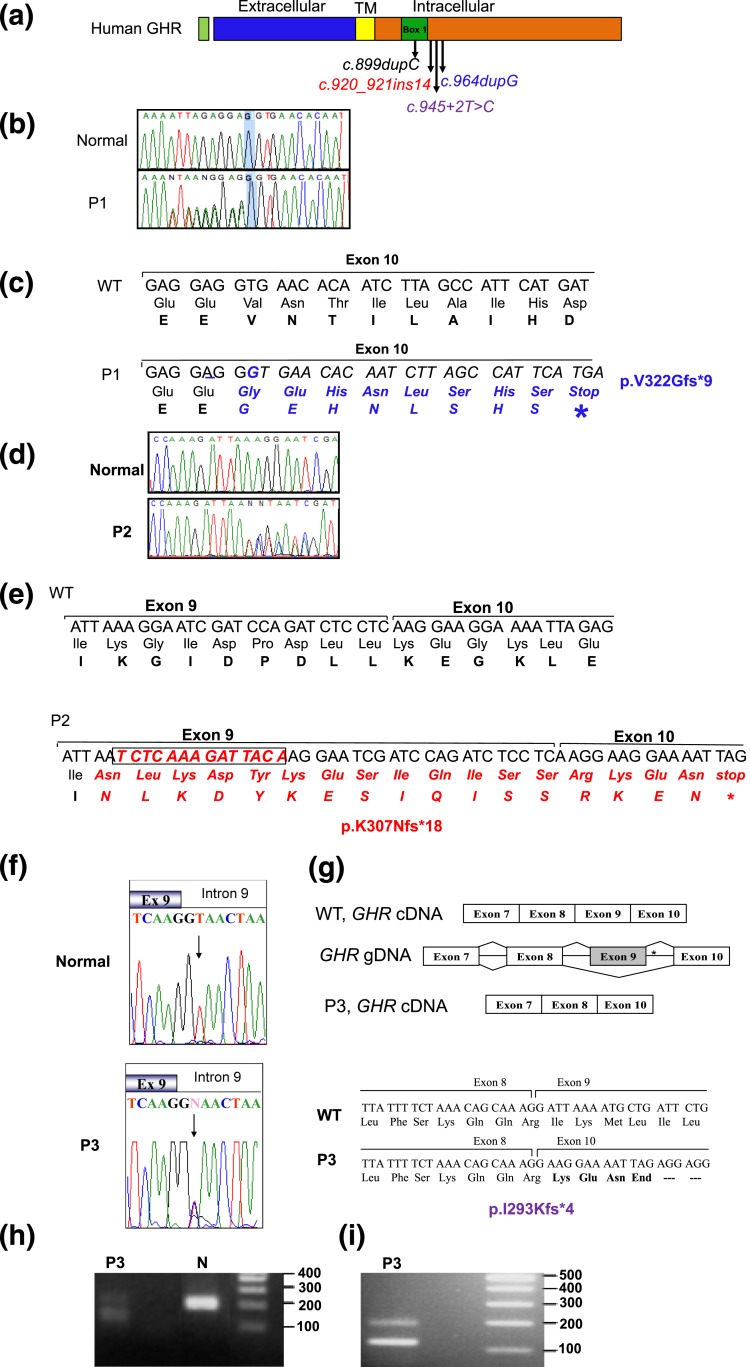
The clinical profiles of our patients who presented with progressive growth failure, normal or high GH, and low IGF-I suggest that mutations in the GHR, or in components downstream of the receptor, could be causal of their short stature. All three candidate variants subsequently identified were in GHR and located in sequences encoding the intracellular domain of the GHR [Fig. 3(a)]. Whole exome sequencing data from P1, her parents, and brother were stringently filtered for autosomal dominant variants because the father was also short and the parents were not known to be related. The top candidate was a heterozygous variant, c.964dupG in exon 10 of GHR (NM_000163.4), which was confirmed by Sanger sequencing [Fig. 3(b) and 3(c)] to cosegregate with the growth phenotype in the family [Fig. 1(a)]. For P2 and P3, targeted Sanger sequencing of GHR revealed a heterozygous insertion of 14 nucleotides in exon 9 (c.920_921ins14) [Fig. 3(d) and 3(e)] and a heterozygous c.945+2T>C (invariant dinucleotide of the donor splice site of intron 9) [Fig. 3(g) and Fig. 3(h)], respectively. Each of these variants segregated appropriately with the growth phenotype in the family [Fig. 1(a–c)]. Interestingly, the father of P3 who was also significantly short statured, was WT for GHR, and had serum IGF-I levels within the low normal range (93 ng/mL; reference range, 90 to 360); the molecular cause for his short stature remains unknown.
Figure 3.
Heterozygous GHR variants identified in affected patients. (a) Schematic representation of the hGHR protein showing the various domains and the location of the heterozygous mutations, c.899dupC, c.920_921ins14, c.945+2T>C, and c.964dupG. TM, transmembrane. (b) Electropherograms of the WT GHR and P1 carrying the c.964dupG variant. The reverse primer was used for sequencing and the electropherogram presented is the complement, forward sequences. Heterozygosity is, therefore, visualized 5′ to the highlighted G nucleotide. (c) DNA sequence (5′ to 3′) and corresponding amino acids showing the duplication of the nucleotide G (underlined) in the c.964dupG variant and the resulting frameshift and early termination (stop). (d) Electropherograms of the forward WT GHR and P2 showing the c.920_921ins14 variant. (e) DNA sequence (5′ to 3′) and corresponding amino acid sequence showing the insertion of 14 bp at nucleotide 921 of GHR cDNA (boxed) in the c.920_921ins14 variant, the resulting frameshift, and stop. (f) Electropherograms of the exon 9–intron 9 junction from WT GHR and P3 carrying the c.945+2T>C variant (arrow); the mother, but not the father, carried the variant, as did the maternal grandfather (data not shown). (g) Schematic, c.945+2T>C splicing out of exon 9. *Position of the mutation. The cDNA sequence and translated products of WT (GHR) and the c.945+2T>C variant are shown (bottom) along with the splice junctions. (h) DNA gel of GHR mRNA RT-PCR products from a normal control and P3, amplified using primers from exon 8 to 10. The expected product size is 187 bp (control). The presence of a smaller 117-bp band in the proband confirms deletion of exon 9. (i) A larger sample (20 µL) of the RT-PCR product better demonstrates the presence of two products (187 bp and 117 bp) carried by P3.
For variants c.964dupG and c.920_921ins14, the duplication of a nucleotide (c.964dupG) or insertion of 14 nucleotides (c.920_921ins14) results in frameshift and predicted early protein termination (p.V322Gfs*9 and p.K307Nfs*18, respectively). The c.945+2T>C variant is predicted to destroy the donor splice site in intron 9 and would result in the splicing out of exon 9, which encodes the Box1 region essential for JAK2 association. The absence of exon 9 was confirmed by RT-PCR amplification of the segment of GHR mRNA sequences encoded by exons 8 through 10 [Fig. 3(h) and 3(i)]. The amplified GHR complementary DNA (cDNA) product produced a 187-bp band in the normal control sample and two bands (187 bp and 117 bp) in a sample from P3 [Fig. 3(h) and 3(i)]. Sequencing of the 117-bp fragment confirmed the absence of exon 9 [Fig. 3(g)]. Skipping exon 9 would result in a frameshift, generation of three amino acids, and a premature stop codon, p.I293Kfs*4.
B. Variants hGHR:c.964dupG and hGHR: c.920_921ins14 are Readily Expressed but Cannot Activate GH-induced STAT5 Signaling
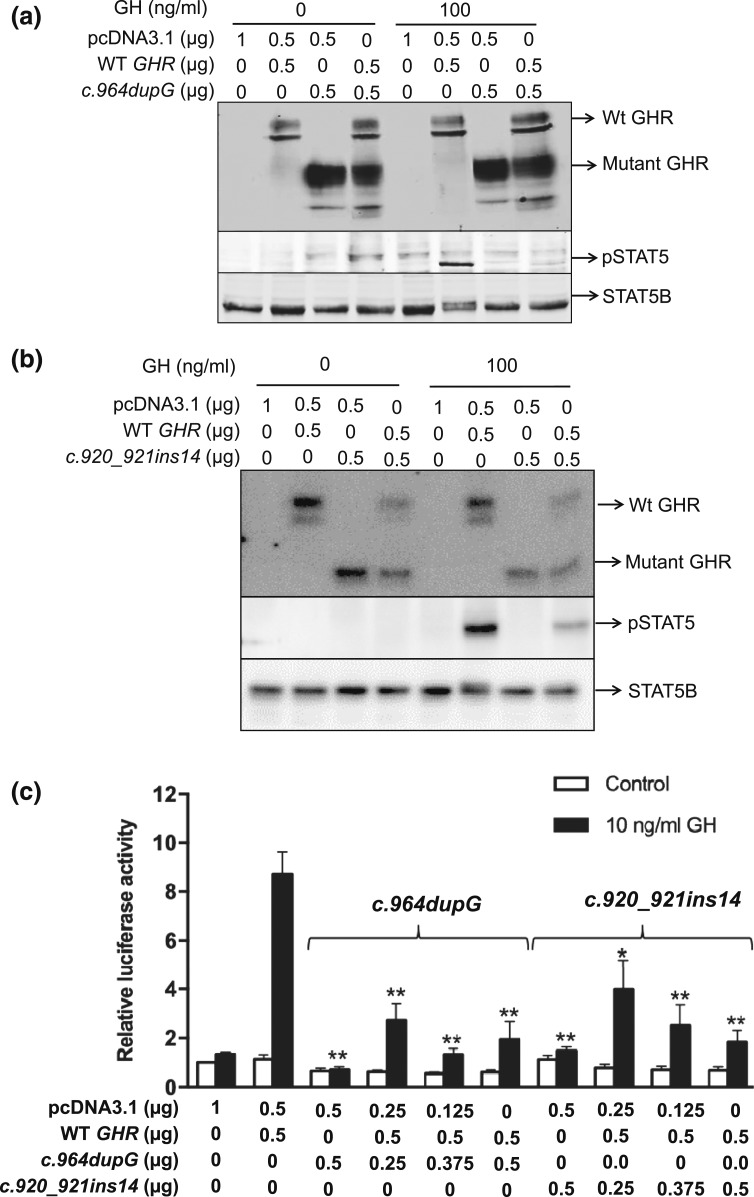
All three variants are predicted to result in truncation of the intracellular domain of the GHR, with retained transmembrane domain sequences (amino acid residues 265 through 288) presumed to permit anchoring of the truncated receptors to the membrane. The truncated GHR peptides, if expressed, should also be capable of forming heterodimers with WT GHR, possibly interfering with downstream signaling as have been described for our (and others) previously identified dominant-negative GHR variants [10–12, 14]. For our c.945+2T>C variant, the splicing out of exon 9 is identical to the two previously reported heterozygous splicing defects (c.876-1G>C and c.945+1G>A), which also resulted in excision of exon 9 [12, 14]. Because loss of exon 9 generates a mature truncated protein of 277 amino acids that has been already demonstrated to have dominant-negative effects when coexpressed with the WT GHR [12, 14], we focused on functionally evaluating the GHR frameshift variants, c.964dupG and c.920_921ins14. The regenerated variants were analyzed in reconstituted HEK293 cells. In the homozygous state, both hGHR:c.964dupG and hGHR: c.920_921ins14 variants were robustly expressed as truncated, ~65 kDa, peptides, and, as might be expected, failed to activate STAT5B signaling in response to GH treatment [Fig. 4(a) and 4(b)] .
Figure 4.
GH-induced STAT5B activities in HEK293 cells transiently transfected with the WT and mutated GHR variants regenerated by site-directed mutagenesis. Input hGHR constructs are indicated. The fully glycosylated GHR, truncated GHR, PAK STAT5 (pSTAT5), and total STAT5B are indicated by arrows. (a) Immunoblot analysis of GHR, STAT5B, and GH-induced STAT5B activation in cells transfected with the c.964dupG variant. (b) Immunoblot analysis of GHR, STAT5B, and GH-induced STAT5B activation in cells transfected with the c.920_921ins14 variant. (c) GH-induced transcriptional activities as measured by relative pGHRE-LUC activities. The data are mean ± standard error of the mean of at least three experiments. The results of mutant only (c.964dupG or c.920_921ins14) or WT + mutant cotransfections were compared with WT. *P < 0.01, **P < 0.001.
C. GH-Induced Phosphorylation of STAT5B and Transcriptional Activities are Significantly Reduced When the Variants are Coexpressed With WT GHR
To determine whether the strongly expressed truncated hGHR variants could affect normal functioning of WT hGHR, each of the variants was cotransfected with WT hGHR in a 1:1 ratio to mimic the heterozygous state seen in our patients. GH-induced STAT5B phosphorylation was significantly reduced when either variant was coexpressed with WT hGHR [Fig. 4(a) and 4(b)]. To determine if the observed reduction in STAT5B phosphorylation had an effect on GH-induced transcriptional activities, WT hGHR (0.5 µg) was cotransfected with increasing concentrations of the hGHR variants, hGHR:c.964dupG or hGHR: c.920_921ins14 (0.25–0.5 µg), and the pGHRE-LUC reporter. Luciferase activities were significantly reduced when the mutant was coexpressed with the WT GHR [Fig. 4(c)].
Altogether, our results support the hypothesis that the heterozygous mutants identified in our patients exert dominant-negative effects.
4. Discussion
We report the identification of three heterozygous, dominant-negative GHR variants, c.964dupG, c.920_921ins14, and c.945+2T>C, in three unrelated patients who present with substantial postnatal growth retardation, partial GHI, and IGF-I deficiency. Our cases increase the number of known dominant-negative GHR defects to seven [11, 13–15], but such mutations may be much more prevalent because, in comparison with classical GHI, the milder phenotype seen in affected patients may be difficult to diagnose. Indeed, such patients are likely to be classified as having idiopathic short stature because they lack the phenotype classically associated with autosomal-recessive GHR mutations. Of note, serum GHBP concentrations were high normal, or even markedly elevated (P3), supporting in vivo expression of both normal and mutant GHR. Importantly, we provide evidence that a combination of rhGH and rhIGF-I appears to be a viable option for successful, longitudinal treatment of such patients.
For our previously described patient carrying the heterozygous c.899dupC mutation (P4; height SD, −4.17), initial treatment with rhIGF-I had improved growth velocity (8 cm/y), but the subsequent addition of rhGH to his treatment regimen further increased his growth velocity to >10 cm/y, allowing his growth profile to reach the normal range [Fig. 2(d)]. Interestingly, during an interruption to his combination therapy (when only rhGH was available), a reduction in his growth velocity was noted, further supporting the benefit of the combination therapy approach (at least for this patient). Based on the successful combination therapy for P4, P2 (carrying the c.920_921ins14 mutation) is now being treated with a similar combination treatment regimen, and, although growth has improved to near the normal range, the true efficacy remains somewhat uncertain because the patient is also in puberty. It is possible that the improved growth could be attributed to the pubertal state. P1 is currently on rhIGF-I therapy only, which improved her growth velocity, as was observed initially for P4. It remains to be determined if the addition of rhGH for P1 could be as helpful as for P4.
Although administration of rhIGF-I is the first treatment of choice for patients with GHI and IGF deficiency, rhIGF-I will only transiently increase serum IGF-I and, through a negative feedback mechanism, inhibit GH secretion and downstream signaling. On the other hand, rhGH therapy, if patients are even partially responsive, such as P2 was, not only will increase circulating IGF-I but also IGFBP-3 and ALS concentrations, which can prolong the half-life of IGF-I. For P2, who was treated long-term with rhGH, serum IGF-I was only modestly increased although serum IGFBP-3 was normalized (Table 1), and the addition of rhIGF-I significantly increased serum IGF-I concentrations, as might be expected (Table 1). For patient P4, who was treated with a combination of rhGH and rhIGF-I, serum IGF-I was in the upper range of normal but serum IGFBP-3, surprisingly, remain consistently below normal, whereas serum ALS, which was normal before treatment, also did not increase as might be expected (Table 1). These biochemistries suggested either a lack of response to rhGH, or that rhIGF-I, through the negative feedback loop to the pituitary, inhibited subsequent GH-induced signaling. Optimal growth velocities for P4, however, were observed with the combination therapy, suggesting beneficial effects of including rhGH despite the biochemistries. Altogether, our data, albeit limited, correlated normalized serum IGF-I concentrations with improved growth in patients with dominant-negative GHR mutations. Interestingly, a recent, 3-year, multicenter, open-label trial demonstrated that a combination therapy of rhGH/rhIGF-I also significantly augmented growth in children with short stature, GH sufficiency and IGF-I deficiency [17]. Hence, for coadministration therapy approaches, frequent monitoring of IGF-I, IGFBP-3, and ALS may be necessary to ensure that normalized serum IGF-I has been achieved. Of note, rhGH and rhIGF-I currently need to be administered via separate injections because the combined product has not been made available for clinical use.
It is of note that one of our patients (P3, c.945+2T>C) had serum GHBP markedly exceeding the normal range. This was not observed in previously characterized patients who carried similar GHR exon 9 splicing mutations [12, 14]. Serum GHBP levels up to 100-fold above normal have been detected only in patients carrying heterozygous, GHR exon 8 splicing mutations, as we had previously reported [18]. Loss of exon 8, which encodes the GHR transmembrane domain, would result in truncated GHR unable to anchor to the cell surface [18] and the higher than normal GHBP has been hypothesized to sequester endogenous circulating GH, thereby reducing the availability of GH for binding to the GHR. However, these patients had normal serum IGF-I, IGFBP-3, and ALS levels, suggesting one copy of WT GHR may be sufficient for normal responsiveness to their elevated, endogenous GH [18], although short stature was still prevalent. For P3, although it remains unclear why her serum GHBP was so high, her serum IGF-I, IGFBP-3, and ALS levels were well below the normal ranges, suggesting an adequate proportion of expressed truncated GHR is anchored and acted in the predicted dominant-negative manner.
A modest height difference of at least 1 SD was observed between P1 (at age 3.4 years) and her father [Fig. 1(a)], both of whom carried the same heterozygous dominant-negative GHR c.964dupC; the GHR c.964dupC status of paternal family members who were short statured (height SD, −2.07 to −4.4) are not known. Variations in height among subjects carrying the same mutation are commonly observed. We previously reported, for example, that subjects carrying the homozygous GHR c.618+792A>G splicing mutation, had height SDs from −3.3 to −6.0 associated with mild to severe GHI phenotypes, and, for this splicing defect, the range could be explained, in part, by variations in splicing efficiencies [19, 20]. Mechanism(s) to explain the height variations in subjects carrying the same dominant-negative GHR frameshift mutation remain to be fully elucidated, although penetrance of the heterozygous mutation is likely to be a key contributing factor.
For two of the three GHR variants (c.964dupG and c.920_921ins14) reported, the JAK2 binding domain, Box1, is intact, whereas Box1 is lost in the c.945+2T>C variant. All three truncations also remove the intracellular tyrosines that are required for recruiting and docking of STAT5B [21]. Our functional studies for variants c.964dupG and c.920_921ins14 in HEK293 cells support the hypothesis that these variants have dominant-negative effects and are most likely the cause of GHI in our patients. When either one of the variants was coexpressed with WT GHR (to recapitulate the heterozygous state in our patients), there was a substantial reduction in phosphorylation of STAT5 [Fig. 4(a) and 4(b)]. Moreover, the STAT5B-mediated transcriptional activities were reduced when the mutant was coexpressed with the WT GHR, supportive of dominant-negative effects [Fig. 4(c)]. For the c.945+2T>C variant, the loss of exon 9, confirmed by RT-PCR of GHR mRNA, generated a truncated protein identical to the two previously reported dominant-negative splicing variants, c.876-1G>C and c.945+1G>A [12–14].
In summary, our patients carrying dominant-negative, heterozygous GHR mutations presented with milder phenotypes compared with patients with classical homozygous, autosomal-recessive GHI. The evidence suggests that this is due, in part, to the presence of predicted 25% WT GHR homodimers. When WT GHR forms a heterodimer with the truncated mutant GHR, downstream signaling and transcriptional activities are significantly blunted, supporting dominant-negative effects exerted by the mutant variants. A combined therapy with rhGH plus rhIGF-I appears to be an effective treatment option, with some response seen in patients receiving rhIGF-I or rhGH alone. The possibility of heterozygous mutations in the GHR intracellular domain should be considered in patients presenting with low IGF-I concentrations and growth phenotypes less severe than seen in classical GHI syndrome.
Acknowledgments
Acknowledgments
This work was supported by funding from the National Institutes of Health (NIH) Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Grant 1K23HD073351 (to A.D.) and NIH NICHD Grant R01HD078592 (to V.H.).
Disclosure Summary: P.F.B. is a consultant and is on the advisory boards for Novo Nordisk, Sandoz (Novartis), Versartis, and IPSEN; J.A. received lecture fees from NovoNordisk and previously from IPSEN; R.G.R. consults for OPKO, Versartis, Ascendis, Genexine, Ammonite, Sandoz, Ferring, and NovoNordisk. The remaining authors have nothing to disclose.
Footnotes
- ALS
- acid labile subunit
- cDNA
- complementary DNA
- GH
- growth hormone
- GHBP
- growth hormone binding protein
- GHI
- growth hormone insensitivity
- GHR
- growth hormone receptor
- hGHR
- human growth hormone receptor
- IGF
- insulin-like growth factor
- JAK2
- Janus-family tyrosine kinase 2
- mRNA
- messenger RNA
- P
- patient
- rhGH
- recombinant human growth hormone
- RT-PCR
- reverse transcription polymerase chain reaction
- SD
- standard deviation
- STAT5B
- signal transducer and activator of transcription 5B
- TBST
- tris(hydroxymethyl)aminomethane-buffered saline with Tween 20
- WT
- wild-type.
References and Notes
- 1.Gent J, van Kerkhof P, Roza M, Bu G, Strous GJ. Ligand-independent growth hormone receptor dimerization occurs in the endoplasmic reticulum and is required for ubiquitin system-dependent endocytosis. Proc Natl Acad Sci USA. 2002;99(15):9858–9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milward A, Metherell L, Maamra M, Barahona MJ, Wilkinson IR, Camacho-Hübner C, Savage MO, Bidlingmaier M, Clark AJ, Ross RJ, Webb SM. Growth hormone (GH) insensitivity syndrome due to a GH receptor truncated after Box1, resulting in isolated failure of STAT 5 signal transduction. J Clin Endocrinol Metab [published correction appears in J Clin Endocrinol Metab 2009;94(7):2674]. 2004;89(3):1259–1266. [DOI] [PubMed] [Google Scholar]
- 3.Davey HW, Xie T, McLachlan MJ, Wilkins RJ, Waxman DJ, Grattan DR. STAT5b is required for GH-induced liver IGF-I gene expression. Endocrinology. 2001;142(9):3836–3841. [DOI] [PubMed] [Google Scholar]
- 4.Woelfle J, Billiard J, Rotwein P. Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J Biol Chem. 2003;278(25):22696–22702. [DOI] [PubMed] [Google Scholar]
- 5.Ooi GT, Cohen FJ, Tseng LY, Rechler MM, Boisclair YR. Growth hormone stimulates transcription of the gene encoding the acid-labile subunit (ALS) of the circulating insulin-like growth factor-binding protein complex and ALS promoter activity in rat liver. Mol Endocrinol. 1997;11(7):997–1007. [DOI] [PubMed] [Google Scholar]
- 6.Savage MO, Attie KM, David A, Metherell LA, Clark AJ, Camacho-Hübner C. Endocrine assessment, molecular characterization and treatment of growth hormone insensitivity disorders. Nat Clin Pract Endocrinol Metab. 2006;2(7):395–407. [DOI] [PubMed] [Google Scholar]
- 7.Diniz ET, Jorge AA, Arnhold IJ, Rosenbloom AL, Bandeira F. [Novel nonsense mutation (p.Y113X) in the human growth hormone receptor gene in a Brazilian patient with Laron syndrome]. Arq Bras Endocrinol Metabol. 2008;52(8):1264–1271. [DOI] [PubMed] [Google Scholar]
- 8.Laron Z. Laron syndrome (primary growth hormone resistance or insensitivity): the personal experience 1958-2003. J Clin Endocrinol Metab. 2004;89(3):1031–1044. [DOI] [PubMed] [Google Scholar]
- 9.David A, Hwa V, Metherell LA, Netchine I, Camacho-Hübner C, Clark AJ, Rosenfeld RG, Savage MO. Evidence for a continuum of genetic, phenotypic, and biochemical abnormalities in children with growth hormone insensitivity. Endocr Rev. 2011;32(4):472–497. [DOI] [PubMed] [Google Scholar]
- 10.Derr MA, Aisenberg J, Fang P, Tenenbaum-Rakover Y, Rosenfeld RG, Hwa V. The growth hormone receptor (GHR) c.899dupC mutation functions as a dominant negative: insights into the pathophysiology of intracellular GHR defects. J Clin Endocrinol Metab. 2011;96(11):E1896–E1904. [DOI] [PubMed] [Google Scholar]
- 11.Takagi M, Shinohara H, Nagashima Y, Hasegawa Y, Narumi S, Hasegawa T. A novel dominant negative mutation in the intracellular domain of GHR is associated with growth hormone insensitivity. Clin Endocrinol (Oxf). 2016;85(4):669–671. [DOI] [PubMed] [Google Scholar]
- 12.Iida K, Takahashi Y, Kaji H, Takahashi MO, Okimura Y, Nose O, Abe H, Chihara K. Functional characterization of truncated growth hormone (GH) receptor-(1-277) causing partial GH insensitivity syndrome with high GH-binding protein. J Clin Endocrinol Metab. 1999;84(3):1011–1016. [DOI] [PubMed] [Google Scholar]
- 13.Iida K, Takahashi Y, Kaji H, Nose O, Okimura Y, Abe H, Chihara K. Growth hormone (GH) insensitivity syndrome with high serum GH-binding protein levels caused by a heterozygous splice site mutation of the GH receptor gene producing a lack of intracellular domain. J Clin Endocrinol Metab. 1998;83(2):531–537. [DOI] [PubMed] [Google Scholar]
- 14.Ayling RM, Ross R, Towner P, Von Laue S, Finidori J, Moutoussamy S, Buchanan CR, Clayton PE, Norman MR. A dominant-negative mutation of the growth hormone receptor causes familial short stature. Nat Genet. 1997;16(1):13–14. [DOI] [PubMed] [Google Scholar]
- 15.Aisenberg J, Auyeung V, Pedro HF, Sugalski R, Chartoff A, Rothenberg R, Derr MA, Hwa V, Rosenfeld RG. Atypical GH insensitivity syndrome and severe insulin-like growth factor-I deficiency resulting from compound heterozygous mutations of the GH receptor, including a novel frameshift mutation affecting the intracellular domain. Horm Res Paediatr. 2010;74(6):406–411. [DOI] [PubMed] [Google Scholar]
- 16.de Bruin C, Mericq V, Andrew SF, van Duyvenvoorde HA, Verkaik NS, Losekoot M, Porollo A, Garcia H, Kuang Y, Hanson D, Clayton P, van Gent DC, Wit JM, Hwa V, Dauber A. An XRCC4 splice mutation associated with severe short stature, gonadal failure, and early-onset metabolic syndrome. J Clin Endocrinol Metab. 2015;100(5):E789–E798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backeljauw PF, Miller BS, Dutailly P, Houchard A, Lawson E, Hale DE, Reiner B, Sperling MA; MS316 Study Group . Recombinant human growth hormone plus recombinant human insulin-like growth factor-1 coadministration therapy in short children with low insulin-like growth factor-1 and growth hormone sufficiency: results from a randomized, multicenter, open-label, parallel-group, active treatment-controlled trial. Horm Res Paediatr. 2015;83(4):268–279. [DOI] [PubMed] [Google Scholar]
- 18.Aalbers AM, Chin D, Pratt KL, Little BM, Frank SJ, Hwa V, Rosenfeld RG. Extreme elevation of serum growth hormone-binding protein concentrations resulting from a novel heterozygous splice site mutation of the growth hormone receptor gene. Horm Res. 2009;71(5):276–284. [DOI] [PubMed] [Google Scholar]
- 19.Metherell LA, Akker SA, Munroe PB, Rose SJ, Caulfield M, Savage MO, Chew SL, Clark AJ. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. Am J Hum Genet. 2001;69(3):641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David A, Camacho-Hübner C, Bhangoo A, Rose SJ, Miraki-Moud F, Akker SA, Butler GE, Ten S, Clayton PE, Clark AJ, Savage MO, Metherell LA. An intronic growth hormone receptor mutation causing activation of a pseudoexon is associated with a broad spectrum of growth hormone insensitivity phenotypes. J Clin Endocrinol Metab. 2007;92(2):655–659. [DOI] [PubMed] [Google Scholar]
- 21.Derr MA, Fang P, Sinha SK, Ten S, Hwa V, Rosenfeld RG. A novel Y332C missense mutation in the intracellular domain of the human growth hormone receptor does not alter STAT5b signaling: redundancy of GHR intracellular tyrosines involved in STAT5b signaling. Horm Res Paediatr. 2011;75(3):187–199. [DOI] [PubMed] [Google Scholar]