ABSTRACT
RNA viruses accumulate mutations to rapidly adapt to environmental changes. Enterovirus A71 (EV-A71) causes various clinical manifestations with occasional severe neurological complications. However, the mechanism by which EV-A71 evolves within the human body is unclear. Utilizing deep sequencing and haplotype analyses of viruses from various tissues of an autopsy patient, we sought to define the evolutionary pathway by which enterovirus A71 evolves fitness for invading the central nervous system in humans. Broad mutant spectra with divergent mutations were observed at the initial infection sites in the respiratory and digestive systems. After viral invasion, we identified a haplotype switch and dominant haplotype, with glycine at VP1 residue 31 (VP1-31G) in viral particles disseminated into the integumentary and central nervous systems. In vitro viral growth and fitness analyses indicated that VP1-31G conferred growth and a fitness advantage in human neuronal cells, whereas VP1-31D conferred enhanced replication in human colorectal cells. A higher proportion of VP1-31G was also found among fatal cases, suggesting that it may facilitate central nervous system infection in humans. Our data provide the first glimpse of EV-A71 quasispecies from oral tissues to the central nervous system within humans, showing broad implications for the surveillance and pathogenesis of this reemerging viral pathogen.
IMPORTANCE EV-A71 continues to be a worldwide burden to public health. Although EV-A71 is the major etiological agent of hand, foot, and mouth disease, it can also cause neurological pulmonary edema, encephalitis, and even death, especially in children. Understanding selection processes enabling dissemination and accurately estimating EV-A71 diversity during invasion in humans are critical for applications in viral pathogenesis and vaccine studies. Here, we define a selection bottleneck appearing in respiratory and digestive tissues. Glycine substitution at VP1 residue 31 helps viruses break through the bottleneck and invade the central nervous system. This substitution is also advantageous for replication in neuronal cells in vitro. Considering that fatal cases contain enhanced glycine substitution at VP1-31, we suggest that the increased prevalence of VP1-31G may alter viral tropism and aid central nervous system invasion. Our findings provide new insights into a dynamic mutant spectral switch active during acute viral infection with emerging viral pathogens.
KEYWORDS: enterovirus A71, quasispecies, capsid protein VP1, pathogenesis
INTRODUCTION
Human enterovirus A71 (EV-A71) is a major causative agent of hand, foot, and mouth disease (HFMD). EV-A71 belongs to the Enterovirus genus in the Picornaviridae family, which includes the Enterovirus A to H and J species. EV-A71 is classified as an Enterovirus A species based on its genome sequence (1, 2). Several HFMD outbreaks associated with cocirculation of EV-A71 have been reported in the Asia-Pacific region, including Taiwan, China, Japan, Malaysia, Singapore, Vietnam, Cambodia, Australia, and Thailand (3–7).
HFMD manifests as a common viral rash of infants or children with fever, which usually causes low-grade vesicles on the buccal mucosa and tongue and small, tender papulovesicular lesions on the hands and feet, as well as occasionally on the buttocks and genitals (8, 9). Most EV-A71 infections are asymptomatic or self-limited; however, severe neurological complications such as encephalomyelitis and sudden death occasionally occur, especially in children (8). EV-A71 transmission between hosts occurs through oral-fecal or oral-oral routes (10). Tissues in the respiratory and digestive tracts, such as the tonsillar crypt epithelium, are suspected of being initial infection sites that support active viral replication (11). Following the initial infection, the virus invades the host and disseminates from the skeletal muscle to the motor neuron junctions, entering the motor nuclei via peripheral motor nerves and the central nervous system (CNS) in mouse models (12, 13). EV-A71 encephalomyelitis from autopsy studies exhibited intense inflammation in the spinal cord, brainstem, and cerebellar dentate nucleus (14). The inflammation was typical of viral encephalitis, and anterior horn cells at all levels of the cord showed neurotropism with viral antigens or RNA in neuronal bodies. Virus antigens and RNA could also be detected in other inflammation sites, including the medulla, fourth ventricle, midbrain, hypothalamus, and subthalamic nucleus (14). Viral antigens or RNA is detected mostly in neurons with neuronophagia or degeneration, suggesting that viral cytolysis is an important mechanism for tissue injury. In addition, other mechanisms of tissue injury, such as immune-mediated or bystander effects, contribute to pathogenesis, as viral antigens and RNA are more focal than expected from the extent and severity of inflammation (15). Based on human autopsy results supported by findings in mouse models, EV-A71 enters the CNS through peripheral motor nerves. Because peripheral motor nerves are the first neurons encountered, motor and adjacent neurons in the spinal cord, medulla, pons, and midbrain experience the most severe inflammation. Therefore, it has been suggested that the virus spreads from the skeletal muscle to motor neuron junctions, up to the peripheral motor nerves and motor nuclei in the CNS (12).
EV-A71 is a nonenveloped, positive-sense, single-stranded RNA virus that consists of 60 copies each of four capsid proteins (VP1, VP2, VP3, and VP4) that form a symmetrical icosahedral structure. Consequently, the external capsid proteins contain antigenic determinants and play important roles in binding receptors on the surfaces of susceptible cells. Several receptors or attachment factors have been identified for EV-A71 binding or entry, including human scavenger receptor class B, member 2 (SCARB2) (16), human P-selectin glycoprotein ligand-1 (PSGL1) (17), sialylated glycans (18), annexin II (19), vimentin (20), and nucleolin (21). Receptor expression may not explain neuron tropism, as the receptor molecules or proteins identified to date are ubiquitously expressed in various tissues instead of being specifically expressed in the CNS, although EV-A71 may use unidentified receptors to enter the CNS.
As an RNA virus, EV-A71 exhibits dynamic evolution. An error-prone viral RdRp in EV-A71 leads to high mutation rates and increased genetic diversity (22), facilitating mutational robustness in quasispecies (reviewed by Lauring et al. [23]). To date, many investigations have clarified that RNA viruses generate 10−4 to 10−6 mutations per nucleotide, averaging 1 mutation per genome in each replication cycle (24). Such large population diversity can be observed in both experimental and natural infections with RNA viruses, and numerous combinations of mutations are generated, resulting in new phenotypic constraints during each cycle of replication. During the virus life cycle, viruses are constantly challenged by selective pressure, such as when infecting a new host with a distinct immune system (25), when infecting different types of cells, or during exposure to different chemical agents or antiviral drugs (26). Previously, it was proposed that mutational robustness provides advantages for viruses to adapt to new environments under various selective pressures (27). For example, reducing the mutation rate of poliovirus without changing the replication rate causes decreased virulence in mice (28, 29). Similarly, a high-fidelity EV-A71 variant with an L123F mutation in protein 3D (3D-L123F) and a G64S mutation in protein 3D (3D-G64S) attenuates virulence and pathogenesis in vivo (30), indicating that the low fidelity of RdRp facilitates viral replication within hosts. Conversely, extremely low fidelity deleteriously affects viral fitness, leading to viral extinction (31). Most random single mutations in vesicular stomatitis virus reduce virus replication, and 40% are lethal mutations (32). A similar scenario was observed with other viruses, including tobacco etch virus and the phages (31). These findings suggest that the mutation rates of RNA viruses approach the maximum tolerable error rate to increase population maintenance while avoiding extinction. In complex environments such as those with virus spreading in host populations, the ability to generate highly diverse mutant spectra enables viruses to survive in various intrahost environments, including selective pressures generated by tissue adaptation and immune responses (28, 29, 33). During the course of viral infection and spreading among hosts, viral quasispecies frequently face selection events whereupon a preferred “haplotype” becomes dominant in the viral population. Because high genome diversity can confer a fitness advantage for viruses in adapting to a new environment or escaping immune responses, the question arises whether the haplotype diversity of quasispecies or potential virulence factors hidden in quasispecies contribute to EV-A71 virulence and pathogenesis. Thus, knowledge of the evolutionary dynamics under selective pressure facilitates an understanding of EV-A71 pathogenesis within hosts, which is directly relevant for improving clinical therapy and disease management (34).
Next-generation sequencing has become a powerful tool for dissecting genomic diversity in viral quasispecies (35), offering deep-coverage sequence data from up to millions of reads and providing exceptional resolution for investigating the mutant spectra of complex virus populations (36). To define the role of mutant spectra in EV-A71 pathogenesis, we analyzed EV-A71 quasispecies of isolates from discrete tissues of an individual with a fatal outcome and from patients with different disease severities, using deep sequencing to investigate the dynamics of EV-A71 mutant spectra. Our data were analyzed to identify novel virulence variants in the CNS that were associated with fatal disease, which may be used to more precisely explore viral transmission across tissues and define viral factors contributing to disease outcomes, leading to a better understanding of EV-A71 pathogenesis in humans.
RESULTS
Dynamic VP1 residue 31 (VP1-31) haplotype changes among diverse tissues.
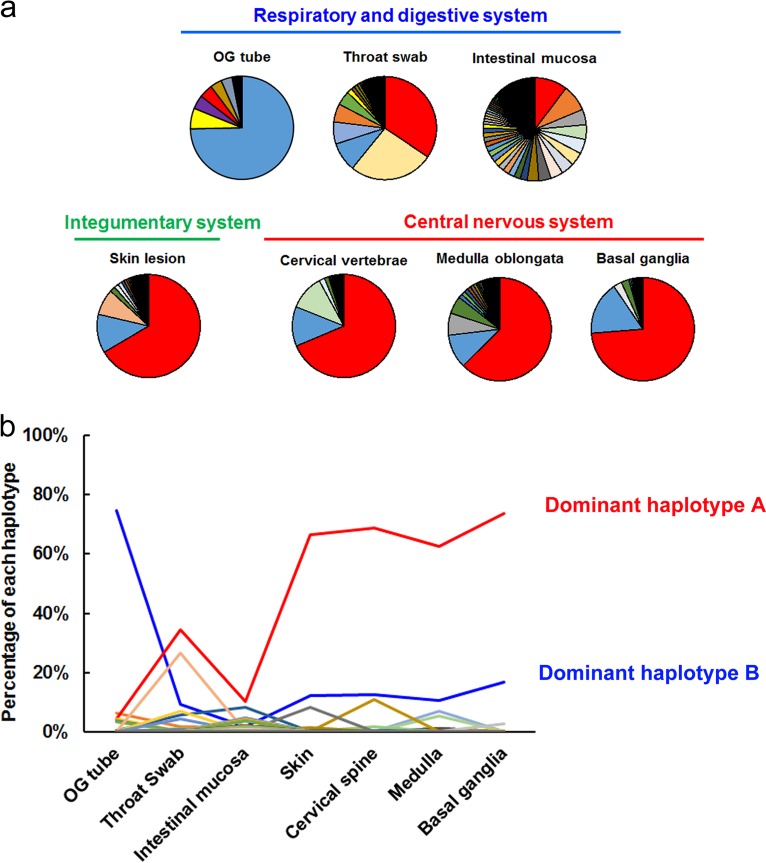
We first investigated viral quasispecies and EV-A71 dissemination across diverse specimens and tissues obtained in 1998 at National Cheng Kung University Hospital from a child with fatal disease. EV-A71 isolates were collected from seven autopsy specimens or tissues from the (i) respiratory and digestive systems (orogastric tube swab, throat swab, and intestinal mucosa), (ii) integumentary system (skin lesion), and (iii) CNS (cervical vertebrae, medulla oblongata, and basal ganglia) (Fig. 1a). We amplified the whole genomes of isolates and analyzed their variants in quasispecies via deep sequencing.
FIG 1.
Divergent EV-A71 mutant spectra among various tissues in humans. (a) Pie charts displaying the proportion of each haplotype among the indicated specimens or tissues according to haplotype prediction results by using the QuasiRecomb program. Among all seven pie plots, each haplotype is represented by a unique color, which represents the distribution of each haplotype across each specimen or tissue. (b) Plot showing the frequency of distribution of each haplotype in different tissues. Each line with a unique color indicates the proportion of changes of a haplotype among specimens or tissues in the respiratory and digestive systems, integumentary system, and central nervous system. OG, orogastric.
Diverse traits contributing to virus infectivity might also be conferred by combinations of various mutations present in individual virus genomes, referred to as “haplotypes.” Haplotype reconstruction is used to identify each variant sequence and its frequency of distribution among mutant spectra, which can reveal dynamic changes of haplotypes within hosts. To precisely quantify the genetic diversity of EV-A71 quasispecies in various tissues, we reconstructed the haplotypes of collected isolates, using QuasiRecomb software (37). To determine dynamic changes of the haplotypes within the human, we first analyzed their proportions in all specimens along the EV-A71 infection route. A major B-to-A haplotype switch from peripheral to CNS specimens was clearly apparent (Fig. 1b). Dominant haplotype A increased in prevalence from the orogastric tube (4.3%) to the basal ganglia (73.7%), whereas the dominant haplotype B was reduced (74.7% to 16.7%). A comparison of the haplotype frequencies revealed that the two dominant haplotypes (A and B) switched in the specimens from the initial infection sites to the CNS. This haplotype switch demonstrated an evolutionary force acting on EV-A71.
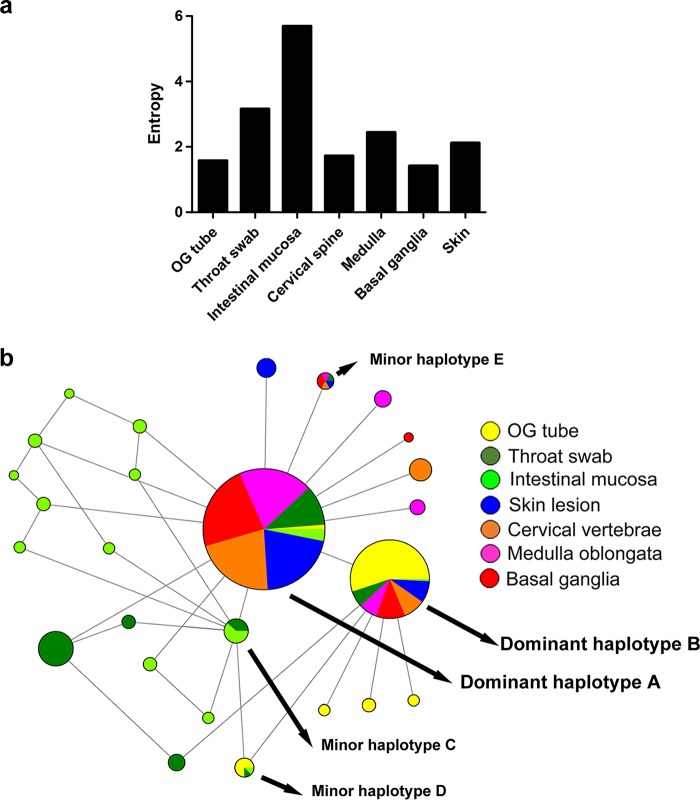
According to the estimated haplotype distribution, we next defined intrahost virus diversity by measuring Shannon entropy, H. Consistent with the single-nucleotide variation (SNV) results, the highest diversity was found in samples from the throat swab (H = 3.15) and intestinal mucosa (H = 5.69) (Fig. 2a). As EV-A71 transmits through the oral-fecal or oral-oral routes between human hosts, the nasopharyngeal and gastric-intestinal tissues are thought to be primary infection sites (11). Together, these results indicated that EV-A71 exhibited broad mutant spectra in throat swabs and intestinal mucosa, which are suggested to be primary infection sites in human dissemination routes. This phenomenon may facilitate viral adaptation to different hosts, tissues, or environments. However, as viruses invaded the integumentary system and CNS, the genetic diversity decreased. The reduction of mutant spectra implied that selective pressure potentially arose during viral dissemination within the host, forming a bottleneck with the tissues around the external lumens to specific sites in the integumentary system and CNS.
FIG 2.
Dynamic haplotype switching between the respiratory tract and the CNS. (a) The Shannon entropy value for each viral quasispecies isolated from the indicated tissues was calculated based on the frequency of distribution of all haplotypes detected in the indicated specimens or tissues. (b) Phylogeny networks and frequency of appearance of each haplotype from different tissues. The sizes of the solid circles indicate the total abundance of each haplotype among all specimens or tissues. Different colors displayed in each solid-color circle show the abundance contributed by the indicated specimens or tissues.
We further verified the genetic association of haplotypes in various tissues. To combine the frequency and genetic evolution results, we performed Network analysis (38) to cluster haplotypes from all tissues based on their frequency in mutant spectra by the size of the associated circles. Clearly, dominant haplotypes A and B accounted for a rather high proportion of haplotypes among the mutant spectra (Fig. 2b). Network results also revealed sporadic haplotypes in the respiratory and digestive systems. Taken together, these data showed that dominant haplotypes A and B, respectively, appeared among various tissues with high frequency. We suggest that the diverse mutant spectra of viral variants containing dominant haplotypes A and B evolved from the respiratory and digestive tracts to the CNS in the host. Those haplotypes colonizing the intestinal mucosa showed broadened, evolving mutant spectra, which might be advantageous for adapting in the gut or result from natural viral evolution under less selective pressure from the gut.
To identify which region of the EV-A71 genome that the evolutionary force acted upon, we compared the sequences of the two dominant haplotypes (A and B). Unexpectedly, only one nonsynonymous mutation was identified: an aspartic acid (D)-to-glycine (G) mutation at VP1 residue 31. Thus, we speculated that VP1 residue 31 was altered by evolutionary pressure and that the VP1-31 D-to-G mutation may increase viral adaption and fitness as the virus spreads from the respiratory and digestive systems to the CNS in humans. We considered that a selective bottleneck shaped the evolutionary pathway and drove a haplotype switch after the virus invaded the respiratory and digestive systems. The VP1-31 substitution might act under selective pressure to facilitate EV-A71 dissemination to specific sites, including the integumentary system and CNS, which potentially contributed to EV-A71 infectivity and dissemination in the human body.
The selection bottleneck appearing in viral quasispecies from different tissues involved VP1 residue 31.
To identify the selective pressure that acted on a specific viral gene(s) to help the virus break through the tissue tropism bottleneck, we quantified the frequency of synonymous (πS) and nonsynonymous (πN) mutations in each protein. We used the π value, representing the diversity of viral quasispecies, to quantify the average number of pairwise differences per nucleotide site among virus populations. Compared with the normal dN and dS values among individual strains, πN and πS values emphasized the nonsynonymous and synonymous substitution rates in viral quasispecies existing in an individual strain. In general, πN > πS or πS > πN indicates that positive (diversifying) or negative (purifying) selection is acting on the viral genome to introduce new mutations or to remove deleterious mutations, respectively. Most tissues except for the orogastric tube showed πN > πS in the VP1 and 3A proteins (Table 1). Considering that the VP1 and 3A proteins, respectively, determine viral entry (39) and endoplasmic reticulum-to-Golgi traffic (40–43) during enterovirus replication, our data suggested that these two proteins might acquire various nonsynonymous mutations to help the virus colonize these tissues.
TABLE 1.
Nonsynonymous and synonymous nucleotide diversity of EV-A71 genes in various tissues of the autopsy case
| Tissue or specimen | Diversity value | Result for indicated proteina |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP4 | VP2 | VP3 | VP1 | 2A | 2B | 2C | 3A | 3B | 3C | 3D | ||
| OG tube | πN | 0.000000 | 0.000181 | 0.000072 | 0.000359 | 0.000340 | 0.000174* | 0.000090 | 0.000275 | 0.000146 | 0.000545 | 0.000097 |
| πS | 0.000625 | 0.000351 | 0.000307 | 0.001611 | 0.003251 | 0.000111 | 0.000421 | 0.000460 | 0.009626 | 0.005091 | 0.000557 | |
| Throat swab | πN | 0.000304 | 0.000958 | 0.000180 | 0.000803 | 0.000215 | 0.000000 | 0.000195 | 0.001629 | 0.000000 | 0.000150 | 0.000350 |
| πS | 0.001374 | 0.001389 | 0.001825 | 0.000098 | 0.003374 | 0.000000 | 0.001239 | 0.001489 | 0.015991 | 0.003906 | 0.000360 | |
| Intestinal mucosa | πN | 0.000000 | 0.000065 | 0.000275 | 0.000405 | 0.000063 | 0.000097 | 0.000123 | 0.001912 | 0.000000 | 0.000219 | 0.000956 |
| πS | 0.000000 | 0.000195 | 0.003767 | 0.000385 | 0.004618 | 0.000795 | 0.002090 | 0.000535 | 0.000000 | 0.000483 | 0.003844 | |
| Skin lesion | πN | 0.000638 | 0.000028 | 0.000097 | 0.000553 | 0.000078 | 0.000071 | 0.000025 | 0.000987 | 0.000000 | 0.000097 | 0.000379 |
| πS | 0.000000 | 0.000233 | 0.000148 | 0.000226 | 0.000317 | 0.000000 | 0.000485 | 0.000000 | 0.015170 | 0.000411 | 0.000069 | |
| Cervical vertebrae | πN | 0.000252 | 0.000143 | 0.000066 | 0.000686 | 0.000086 | 0.000276 | 0.000047 | 0.003739 | 0.000000 | 0.000054 | 0.000510 |
| πS | 0.000969 | 0.000000 | 0.000194 | 0.000412 | 0.000459 | 0.000278 | 0.000779 | 0.000000 | 0.000000 | 0.000230 | 0.000302 | |
| Medulla oblongata | πN | 0.000197 | 0.000528 | 0.000112 | 0.000779 | 0.000328 | 0.000203 | 0.000410 | 0.002101 | 0.000000 | 0.000138 | 0.000229 |
| πS | 0.000618 | 0.000351 | 0.000406 | 0.000345 | 0.000920 | 0.000251 | 0.000592 | 0.000000 | 0.000000 | 0.001727 | 0.000368 | |
| Basal ganglia | πN | 0.000000 | 0.000174 | 0.000245 | 0.000721 | 0.000000 | 0.000000 | 0.000099 | 0.001578 | 0.000000 | 0.000051 | 0.000705 |
| πS | 0.000000 | 0.000345 | 0.000202 | 0.000628 | 0.000347 | 0.000247 | 0.000081 | 0.000954 | 0.000000 | 0.000729 | 0.000889 | |
Bold values represent results for genes for which πN > πS.
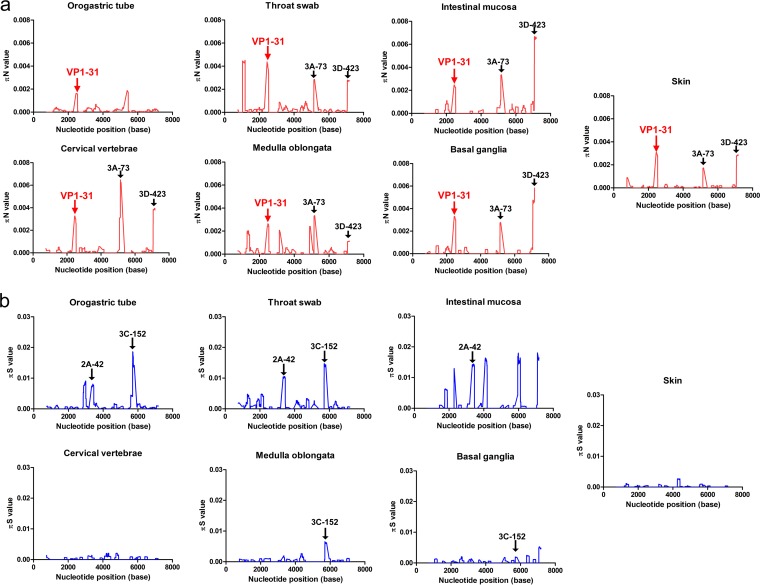
To further define the genetic sites where selection occurred, we estimated the πN and πS values with a sliding window of 50 codons and a step of 1 codon. Divergent πN and πS peak distributions indicated that various amino acid residues were positively or negatively selected in different tissues (Fig. 3; see Table S1 in the supplemental material). Among the examined tissues, only the nonsynonymous VP1-31 substitution (nucleotide position 2532) between aspartic acid (D) and glycine (G) appeared in all πN plots with low πS values from different tissues (Fig. 3a). Thus, the VP1-31 SNV was the only residue subjected to consistent and positive selection pressure in all tissues through which the virus spread, suggesting that EV-A71 might maintain a dynamic, balanced distribution of VP1-31 variants from the initial host infection and that positive selection pressure might fine-tune the relative proportions of VP1-31 polymorphisms in viral quasispecies among various tissues. Two other mutations with high πN but low πS values were frequently identified among these mutant spectra: the S73C mutation in protein 3A (3A-S73C) (nucleotide position 5283) and the W423R mutation in protein 3D (3D-W423R) (nucleotide position 7205) were commonly found in all isolates, except for those in orogastric tube samples (Table S1). The results indicated that these residues broadly accumulated nonsynonymous mutations during viral dissemination in humans.
FIG 3.
Genetic-diversity contributions of nonsynonymous and synonymous mutations of viruses isolated from various tissues. The πN (a) and πS (b) values were calculated throughout each viral genome using a 50-codon sliding window and a 1-codon step. High πN/πS values indicate cases where the SNVs evolved under positive selection, whereas low πN/πS values indicate SNVs that evolved under purifying selection. Amino acid residues for the major peaks of πN and πS are noted in the plots. The arrows indicate the πN peaks of the VP1-31, 3A-73, and 3D-423 residues and πS peaks of 2A-42 and 3C-152 residues among different specimens or tissues, respectively.
In addition to the πN peaks for amino acid residues under positive selection pressure, many πS peaks (Fig. 3b; Table S1), located at potential purifying-selection sites, occurred in the mutant spectra. Among various πS peaks in various tissues, we identified two common purifying-selection sites (protein 2A, residue 42 [2A-42], and protein 3C, residue 152 [3C-152]). The πS peak at 2A-42 was found in all respiratory and digestive system samples (orogastric tube, throat swab, and intestinal mucosa), whereas the πS peak for 3C-152 was found in orogastric tube, throat swab, medulla oblongata, and basal ganglia samples. Other sporadic πS peaks (Table S1) included VP1-186, VP1-194, 2A-40, 2A-49, 2A-97, 3C-115, 3D-48, 3D-164, and 3D-182 in the orogastric tube, VP2-158, VP3-83, VP3-166, 2A-47, and 2C-247 in throat swabs, VP2-61, VP2-240, 2C-38, 3D-17, and 3D-55 in the intestinal mucosa, 2C-109 in skin lesions, VP1-281 in cervical vertebrae, 3D-25 in the medulla oblongata, and VP1-67, 3D-187, and 3D-421 in the basal ganglia. These data suggested that purification, but not diversification, may be favored at these sites, although the functions of these residues are unclear. Among these positively and negatively selected SNVs, the VP1-31 residue was the only consistently selected site in all tissues during viral dissemination.
The proportion of VP1-31G correlated with disease severity in patients with EV-A71 infection.
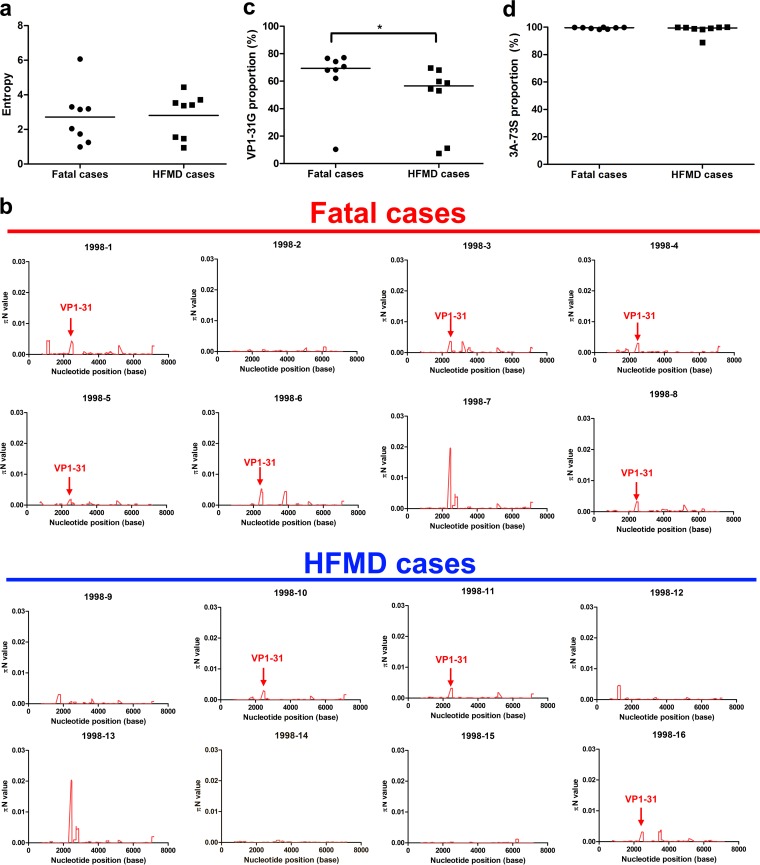
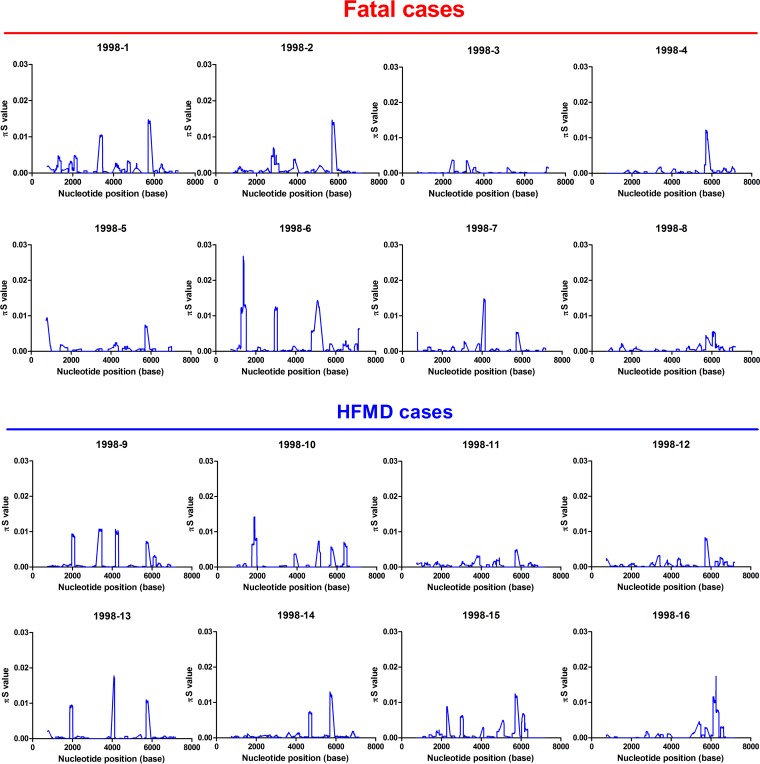
To determine whether evolutionary selection commonly acts on the mutant spectra of other EV-A71 strains while infecting human hosts, we collected 15 additional EV-A71 strains, 7 of which were isolated from fatal cases and 8 of which were from HFMD cases. To rule out genetic diversity resulting from different genotypes or specimen types, the collected strains were consistently isolated from throat swabs and belonged to genotype C2, which is the same as that of the strain from the first fatal autopsy case. We first analyzed the mutant spectra of these strains by determining the frequency of the haplotypes involved. Analysis of the reconstructed haplotypes in the mutant spectra showed that the haplotype entropy did not correlate with patient disease severity (Fig. 4a). We further analyzed the selection pressure that acted on a specific viral protein(s) in these strains by quantifying πN and πS. Similar to the findings with the autopsy, 68.75% (11/16) and 62.5% (10/16) of the strains evolved under positive selection at the VP1 and 3A proteins, respectively (Table S2). Thus, adaptation of the VP1 and 3A proteins during viral evolution might play important roles in EV-A71 invasion into the CNS in infected patients. Further cross-matching of strain mutations with πN > πS values indicated that two SNVs were found in the autopsy cases: the VP1-31 SNV was found in six of eight viruses from fatal cases and in four of eight HFMD cases; the 3A-S73C SNV was found in five of eight viruses from either fatal or HFMD cases (Fig. 4b and 5; Table S2). To determine whether the VP1-31G or 3A-73C mutations correlated with EV-A71 disease severity, we compared the frequencies of the two SNV mutations in viruses from fatal cases and HFMD cases according to the deep-sequencing result. Analysis of virus isolates from patients with differing disease severities indicated that the frequency of VP1-31G, but not 3A-73C, was significantly higher (Mann-Whitney U test, P < 0.05) among patients with fatal disease than among those with HFMD (Fig. 4c and d). We speculate that VP1-31G might confer higher replication and a fitness advantage for adapting to CNS tissues. As more haplotypes with VP1-31G appear at the initial infection sites, this increases the potential for EV-A71 to invade the CNS and cause severe neurological disease. Thus, VP1-31G may be a potential viral determinant that contributes to intrahost EV-A71 spread, as well as disease severity in different hosts.
FIG 4.
Higher proportion of VP1-31G viruses from fatal cases than HFMD cases. (a) The Shannon entropies of quasispecies isolated from patients with fatal disease or HFMD were compared based on the frequency of distribution of all haplotypes. The 1998-1 virus was isolated from a throat swab of an autopsy case. The center values represent the Shannon entropies among the selected fatal and HFMD cases. The proportions of VP1-31G (b) and 3A-73S (c) among virus isolates from patients with fatal disease or HFMD were determined based on the SNV calling results from deep-sequencing reads. The values represented by the horizontal lines are mean proportions among the selected fatal and HFMD cases. The Mann-Whitney U test was used to calculate P values. *, P < 0.05. (d) The πN values for EV-A71 from fatal cases and HFMD cases were calculated for each viral genome using a 50-codon sliding window and a 1-codon step.
FIG 5.
Genetic diversity contributions of synonymous mutations of viruses isolated from patients with different disease severities. The values of πS of EV-A71 from fatal cases and HFMD cases were calculated throughout each viral genome using a 50-codon sliding window and 1-codon step. The 1998-1 virus is the virus isolated from the throat swab of the autopsy case.
VP1-31 determined thermostability, viral replication, and fitness in vitro.
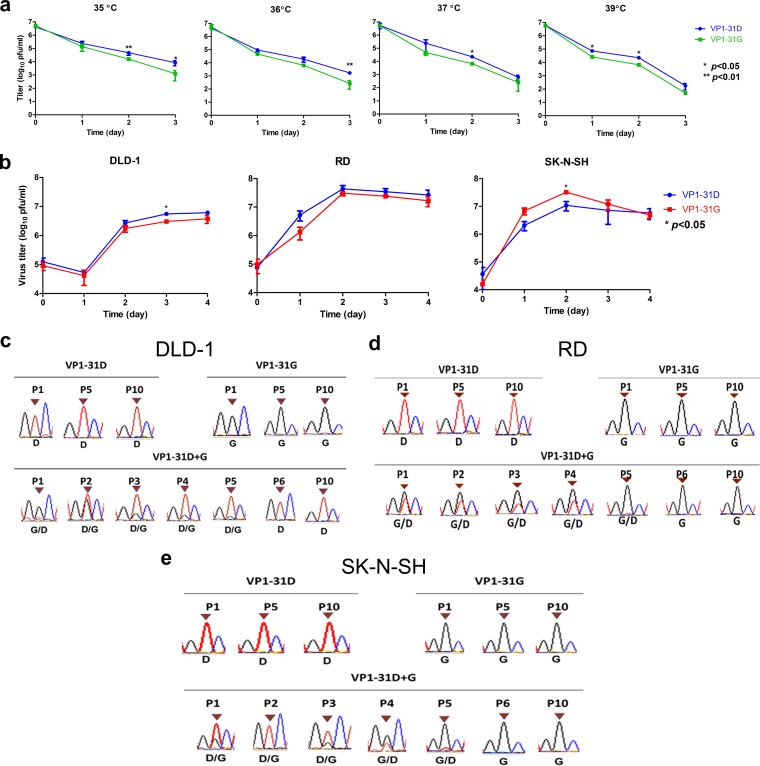
The VP1 protein determines virus binding and uncoating by interacting with cellular receptors and serves as the major target protein of neutralizing antibodies. As the VP1-D31G substitution may constitute a potential viral mutation to facilitate EV-A71 bottleneck breakthrough and invasion into CNS tissues, we next examined the effect of the VP1-D31G mutation on viral characteristics and replication in vitro. We generated recombinant viruses according to the haplotype sequences of dominant haplotype A (VP1-31G) and B (VP1-31D). As capsid protein VP1 is critical for virion structure stability and virus entry, we first examined virus thermostability. The thermostability of infectious VP1-31G virus particles decreased faster than VP1-31D particles at all tested temperatures (Fig. 6a), implying a reduction in virion stability in VP1-31G virus.
FIG 6.
Effects of VP1-31 substitution on thermostability, viral replication, and viral fitness. Panels a and b display, respectively, the thermostability and viral replication of a virus with VP1-31 substitutions. (a) A total of 107 PFU of the VP1-31G (green) or VP1-31D (blue) virus was incubated at different temperatures. Viruses were harvested after the indicated number of days and titrated in plaque assays (n = 6). The values and error bars are the mean and ±1 standard deviation, respectively. A paired Student's t test was used to calculate P values. (b) DLD-1, RD, and SK-N-SH cells were infected with VP1-31G (red) or VP1-31D (blue) at an MOI of 0.1, as indicated. Viral titers were determined at the indicated days postinfection. The values and error bars are the mean ±1 standard deviation, respectively. A 2-tailed paired Student's t test was used to calculated P values. (c) DLD-1, RD, and SK-N-SH cells were individually inoculated or coinoculated with the VP1-31G and VP1-31D viruses, using the same amount of each virus. Viruses were harvested once obvious cytopathic effects were observed. Harvested viruses were sequentially passaged in the same cell line. The dominant haplotype appearing in various passages of viruses were defined by Sanger sequencing of the VP1-31 residue.
We next examined the effect of the VP1-31 substitution on viral infection. Because the VP1-31G haplotype (dominant haplotype A) frequency increased from the respiratory and digestive systems to the CNS in infected individuals, we utilized human colorectal adenocarcinoma DLD-1 cells (digestive system), human rhabdomyosarcoma RD cells (muscle), and human neuroblastoma SK-N-SH cells (CNS) to simulate VP1-31G/D viral infection in various tissues. The VP1-31G virus exhibited lower titers in DLD-1 cells than did the VP1-31D virus (Fig. 6b). In the RD cells, no significant difference in viral replication was observed between strains; however, the VP1-31G virus exhibited significantly increased growth compared with the VP1-31D virus in SK-N-SH cells.
To assess viral fitness, we performed coinfections with both VP1-31 haplotype viruses in the different cell lines. Virus mixtures were continuously passaged in the respective cells, and the dominant virus in the viral population was determined by cDNA sequencing, targeting the VP1-31 residue. Similar to the viral replication results, virus competition demonstrated that the VP1-31G virus exhibited low viral fitness in colorectal cells but high viral fitness in muscle and neuronal cells (Fig. 6c to e). In the colorectal cells (Fig. 6c), both viruses were mixed in the population in the first five passages but VP1-31D emerged as the only haplotype in the sixth passage. This change was not due to sequential passaging, as 10 passages of each virus individually did not alter the VP1-31 residue, but rather reflected the higher viral fitness of the VP1-31D virus in cells originating from the digestive system. Conversely, VP1-31G viruses showed high viral fitness in RD muscle cells (Fig. 6d) and SK-N-SH neuronal cells (Fig. 6e). In these cell types, only VP1-31G could be detected after six passages, suggesting that the VP1-31G virus exhibited higher viral fitness in cells derived from the muscular system and CNS. Together, the viral growth and fitness results were consistent with the observed haplotype distribution in the mutant spectra (Fig. 1b and 2b), implying that VP1-31G conferred the most fitness for CNS infection but that VP1-31D conferred more fitness for respiratory and digestive tract tissue infection.
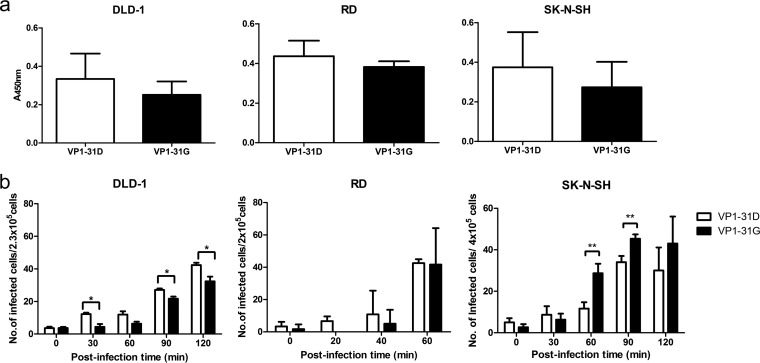
To further define the viral replication step affected by the VP1-D31G mutation, we examined the contribution to virus binding and uncoating. No significant difference in virus binding was observed between virus variants in enzyme-linked immunosorbent assays (ELISAs) (Fig. 7a), consistent with the unlikelihood of changes in the inaccessible internal VP1 protein surface impacting the ability of virions to bind cellular receptors. Next, neutral red-labeled viruses were utilized to determine the uncoating rates of the VP1-31D and VP1-31G viruses. Similar to the viral replication results, the VP1-31D and VP1-31G viruses released their RNA faster in DLD-1 and SK-N-SH cells, respectively (Fig. 7b), with no obvious difference observed in RD cells. Even though the RNA release results (Fig. 7b) displayed approximately 2-fold changes in various tissues, these effects would be obviously amplified after multiple replication cycles. Our viral fitness results (Fig. 6b to d) also demonstrated that the VP1-31 mutation contributed to viral fitness, which was obvious after three to five passages in vitro. Taken together, our data suggest that the VP1-31D and VP1-31G viruses exhibited different viral replication and fitness in vitro. The accordance with the relative variant frequencies across tissues indicates that EV-A71 adapted to different tissues by changing its RNA release ability through the VP1-D31G substitution.
FIG 7.
Effects of VP1-31 substitution on virus binding and RNA release. (a) VP1-31G and VP1-31D viruses were absorbed to DLD-1, RD, and SK-N-SH cells, as indicated. Bound viruses were quantified by ELISA. Values and error bars are the mean ±1 standard deviation, respectively. (b) DLD-1, RD, and SK-N-SH cells were infected with neutral red-labeled VP1-31G and VP1-31D viruses, as indicated. Based on the characteristics of a neutral red-labeled virus, which can be inactivated under light exposure unless the virus releases its RNA, the numbers of viruses that released their RNA genome into the cells were determined using the infectious center assay. Values and error bars are the mean ±1 standard deviation, as indicated. A paired Student's t test was used to calculate P values. The error bars represent 1 standard deviation from triplicate experimental results.
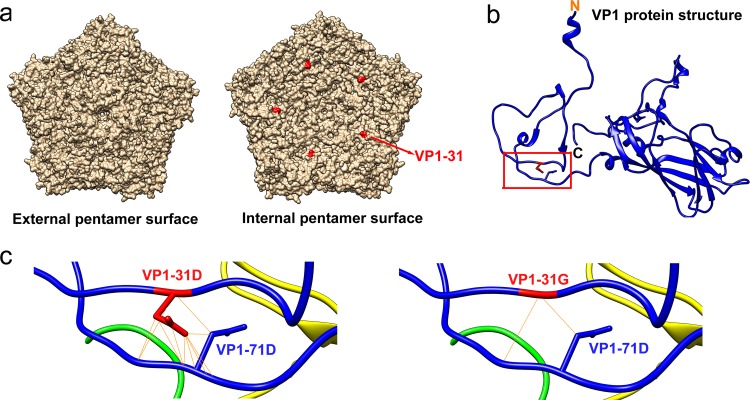
To define the potential structural effects of this substitution on the virion, we utilized the EV-A71 crystal structure (PDB code 4AED) to localize VP1-31 on the capsid protein pentamer and to predict the possible changes by protein modeling. According to the pentamer structure (Fig. 8a), the VP1-31 residue is exposed on the virion interior rather than at the exterior surface. Thus, this residue cannot be accessed by cellular receptors or neutralizing antibodies, indicating that the VP1-D31G substitution might not change receptor binding ability to cells or antibody recognition by humoral immunity in the host.
FIG 8.
Localization of the VP1-31 residue and potential protein structure changes at the VP1-31 substitution site. (a) The external and internal surfaces of the EV-A71 capsid pentamer are shown, as generated using the UCSF Chimera program, version 1.9. The red residue indicates the location of the VP1-31 residue in the outer (left panel) or internal (right panel) surface of the pentamer structure (PDB code 4AED). (b) The VP1-31 residue (red) is displayed on the VP1 protein ribbon structure. N and C indicate the N- and C-terminal ends of the VP1 protein. The red box region is enlarged in panel c of this figure. (c) Diagram displaying protein modeling of the structural consequences of the VP1-31 D-to-G mutation. Blue and red sticks expanding from the backbone structure represent the side chains of VP1-71D (in both panels), VP1-31D (left panel), and VP1-31G (right panel) residues. Solid yellow lines indicate potential interactions between atoms of VP1-71D and VP1-31D (left panel) or VP1-31G (right panel).
The VP1 N terminus forms a loop-like structure to stabilize the mature virion (44). Upon virus uncoating, the proximal end of the VP1 N-terminal extension rearranges to exit through the capsid at the quasi-3-fold axis near the base of the canyon, thereby loosening the capsid structure on the virion. The N-terminal VP1 region then migrates across the top of the VP2 protein and binds the membrane. This connection may be involved in viral RNA transfer into cells. Our protein-modeling results indicated that the VP1-31 residue was predicted to interact with asparagine at VP1-71, both of which are within the N-terminal region of VP1 (Fig. 8b); however, VP1-D31G exhibited reduced salt bridge formation (Fig. 8c). Since N-terminal extension is essential for virus binding and RNA release, we believe that the potential decrease in the VP1-31 and VP1-71 interaction strength upon VP1-D31G substitution might reduce thermostability and conversely increase the release of viral RNA.
DISCUSSION
RNA viruses generally contain highly diverse quasispecies due to the low fidelity of their RNA polymerases. When facing selection pressures with various strengths, such as tissue tropism, environment stresses, or host immune responses, the viruses displayed changing haplotypes as an evolutionary response. A broad mutant spectrum may increase the chance of successfully adapting to the new selective pressures, such as when establishing infection and colonization at the initial infection sites. Our results defined a selection bottleneck that exists when EV-A71 invades from the respiratory and digestive systems into other tissues, including the CNS, as illustrated in Fig. 9. When EV-A71 infects human hosts through fecal-oral or oral-oral routes, virus initially enters the body through the lumen of the respiratory and digestive tracts and colonizes the epithelial tissues of either the tonsillar crypt (11) or intestinal mucosa in the respiratory and digestive systems, followed by spreading to other tissues. Virus collected from the orogastric tube that spans from the mouth to the stomach represents the mutant spectrum in the lumen of the alimentary canal. This mutant spectrum comprises the dominant haplotype B with the VP1-31D substitution (Fig. 2c and d). Our in vitro experiment results indicated that the VP1-31D variant increases virion stability (Fig. 6a), which potentially facilitates the maintenance of virus infectivity in cell-free environments, such as the lumen of the alimentary canal. This substitution also increased viral infectivity and fitness (Fig. 6b and c) in the cells originating from the digestive system. These advantages may aid viral adaptation to environmental changes and resistance to high body temperatures due to a fever following viral transmission to other tissues. These advantages may also explain why the VP1-31D haplotype was still present at a low proportion among tissues from the respiratory and digestive tracts to the brain after selection. The VP1-31D variant also helps viruses infect epithelial cells and then colonize the alimentary canal. Following virus colonization of initial infection sites in the alimentary canal, we observed that the dominant haplotype switched from VP1-31D to VP1-31G, as EV-A71 disseminated to tissues of the integumentary system and CNS. In contrast to the replication advantage of VP1-31D in cells of the digestive system, VP1-31G exhibits higher growth and fitness in neuronal cells of the CNS in vitro (Fig. 6b and c). We speculate that the differing tissue tropism of EV-A71 between the respiratory and digestive systems versus the integumentary system or CNS forms a selection bottleneck for viral populations with mutant spectra, resulting in the adaptive VP1-31G haplotype becoming dominant in neuronal tissues. Once EV-A71 establishes infection in the respiratory and intestinal tracts, the VP1-31G mutation may facilitate bottleneck breakthrough and invasion into the skin and CNS. Together, these data indicate that EV-A71 quasispecies utilize the dynamic balance of the diverse haplotype population, especially the VP1-31D and VP1-31G haplotypes, to cooperatively maintain population plasticity and facilitate viral adaptation and spread among various tissues.
FIG 9.
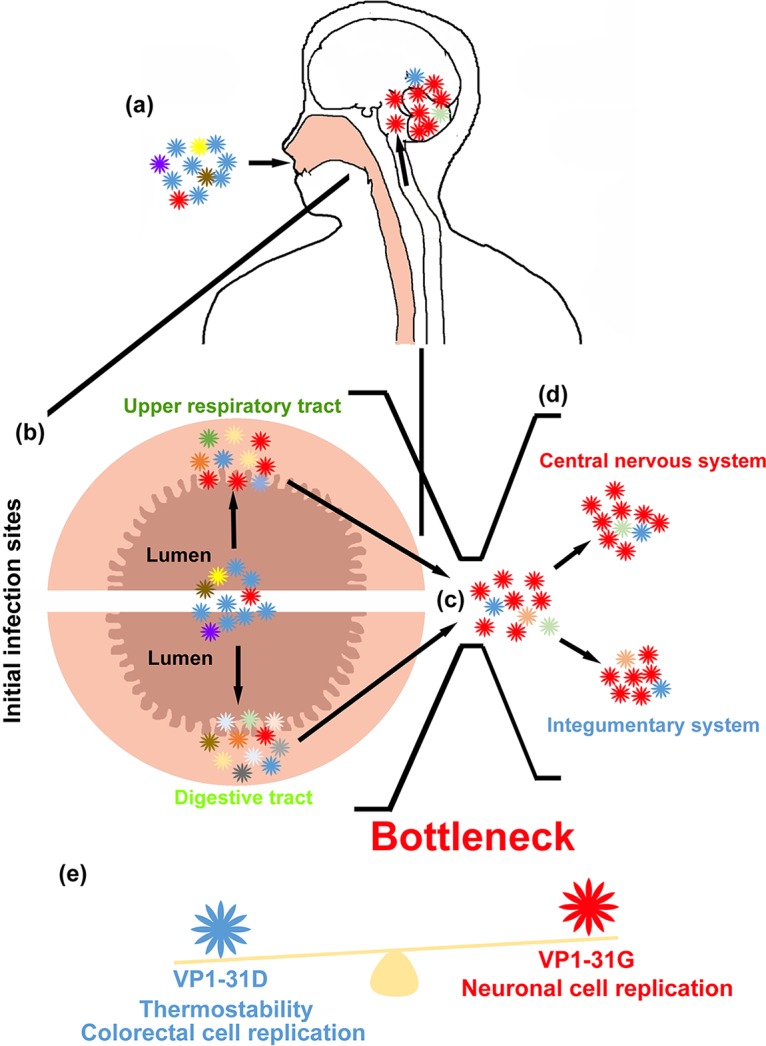
Model of EV-A71 haplotype evolution under bottleneck selection in humans. (a) A broad mutant spectrum of EV-A71, which has major VP1-31D (blue) and minor VP1-31G (red) haplotypes and can infect humans through oral-oral or oral-fecal routes through the respiratory and digestive tract. (b) After entry into the human body, these haplotypes infect epithelial cells in the pharynges (upper respiratory tract) and intestines (digestive tract) and colonize at the initial infection sites. (c) Once the virus colonizes, it next invades other tissues, including the integumentary system and CNS. The mutant spectrum is shaped by a selection bottleneck such as tissue tropism, which changes the haplotype composition in these tissues. (d) The haplotype with VP1-31G becomes dominant in skin, spinal, and brain tissues, which causes HFMD and severe neurological diseases. (e) These evolutionary changes may depend on the effects of the VP1-31 substitution on virus property. The VP1-31D virus showed high thermostability, viral growth, and fitness in colorectal cells, whereas the VP1-31G virus showed better replication fitness in neuronal cells.
Despite the two dominant haplotypes, the minor haplotypes in the intestinal mucosa displayed a series of clustering variants, compared with other haplotypes (Fig. 2b). To survive in a complex environment involving mucosal immunity and microbes, complex haplotypes can occur in the intestines to facilitate virus adaption. According to network analysis results, we observed three minor haplotypes (C to E) coexisting in various tissues. Two haplotypes coexisted in collected isolates from the respiratory and digestive systems (intestinal mucosa, throat swab, and orogastric tube): minor haplotype C was identified in both the intestinal mucosa and throat swab specimens, and minor haplotype D was identified in all three respiratory and digestive system specimens. Since determining the genetic sequences of viral haplotypes, like the footprints of viral evolution, can help us identify variants to trace their transmission between different tissues, the observed coexistence of minor haplotypes C and D among the respiratory and digestive systems indicated that viruses can be transmitted between the orogastric tube, throat swab, and intestinal mucosa. Interestingly, we also identified the minor haplotype E, which appeared in the throat swab, skin lesion, cervical vertebrae, medulla oblongata, and basal ganglia but not in the intestinal mucosa, which is suspected as being the initial replication site of enteroviruses. Because this haplotype was present only in the throat swab, and not in the intestinal mucosa, this haplotype potentially reflects another dissemination route, whereby EV-A71 directly invades the host after replication in the upper respiratory tract and spreads to other tissues, including the CNS.
In addition, we examined whether selective pressure commonly existed at VP1 residue 31 in EV-A71. We analyzed the mutant spectra of an additional 15 EV-A71 strains from patients with various disease severities. All these circulating strains were isolated from throat swabs and belonged to the same genotype (C2) circulating in the 1998 outbreak in Taiwan. The πN and πS values provided an indication that approximately 60% of circulating EV-A71 strains face bottleneck selection at VP1 residue 31 (Fig. 3), and more viruses showed high πN values in fatal cases. Our data showed that VP1 residue 31 is common at positive-selection sites among circulating EV-A71. Studying the VP1-31G proportion among these strains further indicated that a higher proportion of VP1-31G appeared among the mutant spectra of EV-A71 in throat swabs in fatal cases (Fig. 4c). Since the EV-A71 VP1-31G haplotype increased in the mutant, this raised the possibility that EV-A71 could break through the bottleneck and invade CNS tissues, promoting fatality in patients with EV-A71 infection. To further determine if VP1-31 selection occurred across genotypes, we also examined the properties of VP1-31G virus isolates belonging to genotypes B4 and B5, but no VP1-31 variation was identified in these mutant spectra (data not shown). Thus, we suggest that our identified selection bottleneck (and even unidentified bottlenecks for other genotypes) might shape unique evolutionary pathways and act at a different genetic site(s) for each genotype. Although we may not have defined all virulence determinants causing severe neurological diseases among various EV-A71 strains or genotypes in this study, our novel findings demonstrated that the viral quasispecies utilized the dynamic balance of the diverse population to cooperatively maintain population plasticity and facilitate viral adaptation and spread when facing a selection bottleneck between the respiratory and digestive systems and the CNS, which may be associated with the viral pathogenesis of EV-A71.
Several animal models, including neonatal mice, transgenic mice, and monkeys, have been used to investigate EV-A71 pathogenesis. Although EV-A71 caused neurological symptoms in these models, the routes of infection were mostly intraspinal, intracerebral, intratracheal, intraperitoneal, intramuscular, and subcutaneous routes. Only rare models successfully and consistently established infection in animals through natural fecal-oral and oral-oral routes. In addition, in the limited animal models that employed the natural oral routes, the virus was not detected in the intestinal mucosa, which differs from our observations in humans. We suggested that the different tissue tropisms observed between humans and other available animal models will result in distinct selection pressure for virus quasispecies during dissemination in humans and animals. To investigate enterovirus dissemination in humans, we directly analyzed EV-A71 quasispecies among various tissues in autopsy patient specimens. To further evaluate the important role of the VP1-31 substitution identified in the autopsy patient, we analyzed additional EV-A71 isolates from other patients. Both the results from the autopsy case and various isolates from patients indicated that the VP1-31 substitution was associated with the ability of the virus to invade the CNS to cause severe neurological infection, resulting in fatalities. Thus, continuously analyzing the mutant spectra of circulating strains is necessary to survey virus evolution and to understand viral pathogenesis.
In addition to VP1-31G, other unidentified virulence determinants in viral quasispecies may also contribute to disease severity (13, 45–53), although few studies have been conducted to examine the correlation between viral variation and disease outcome. Data from a genetic-variation study focused on human EV-A71 strains by comparing sequences available in the GenBank database suggested that the substitution of VP1-N31D in genotype C, as well as VP1-E145G/Q, VP1-E164D/K, and VP1-T292N/K in genotype B, was associated with neurological symptoms (54). Data from another study, using viral strains from various specimens and collection dates, also revealed that VP1-L97R contributes to neuronal cell tropism (55). All of these virus substitutions represent useful tools for predicting the disease severity of EV-A71.
In conclusion, we identified a novel selection bottleneck appearing between the respiratory and digestive systems and the CNS during EV-A71 dissemination in humans. This bottleneck shaped the mutant spectra of EV-A71 in these tissues and drove a dominant haplotype switch from VP1-31D to VP1-31G. The proportion of VP1-31G virus also increased among fatal cases of EV-A71 infection. The VP1-31G virus, with the dominant haplotype in the CNS, exhibited high viral growth and fitness in neuronal cells, indicating that VP1-31G might contribute to CNS invasion and cause neurological disease in patients. Together, the process of haplotype selection might not only associate with viral pathogenesis in EV-A71-infected patients but also correlate with disease severity among different EV-A71-infected individuals with various disease severities, especially for those haplotypes isolated from fatal cases. VP1-31G represents a potential viral factor that resides among quasispecies of EV-A71 isolates and contributes to viral dissemination and disease severity within humans.
MATERIALS AND METHODS
Clinical EV-A71 isolates.
A retrospective study of 22 clinical isolates from inpatients and outpatients from 1998 were obtained from collections of the NCKU Virology Laboratory, as approved by the Institutional Review Board of National Cheng Kung University Hospital (A-ER-104-365). Fifteen virus strains isolated from throat swab specimens were randomly selected for quasispecies analysis: seven from fatal cases and eight from HFMD cases. In addition, seven viral strains isolated from various tissues or specimens from one autopsy case were analyzed. RD (rhabdomyosarcoma; Bioresource Collection and Research Center, Taiwan), DLD-1 (human colorectal adenocarcinoma; Bioresource Collection and Research Center, Taiwan), and SK-N-SH (human neuroblastoma; American Type Culture Collection, USA) cells were maintained, respectively, in Dulbecco's modified Eagle's medium (DMEM) (RD cells), RPMI 1640 medium (DLD-1 cells), or minimum essential medium (SK-N-SH cells) containing 10% fetal bovine serum plus 2 mM l-glutamine, 100 IU penicillin, and 100 μg streptomycin per ml at 37°C. The cells were tested and found to be free of mycoplasma contamination. To prepare EV-A71 stocks, viruses were propagated for one passage and titrated by performing plaque assays in RD cells.
Virus cDNA amplification and next-generation sequencing.
Our sequences were derived from cDNA of EV-A71 isolates that had been isolated from Vero cells in 1998. In that year, we performed virus isolation for all strains by following the same procedure in our clinical virology laboratory. Since 1998, the isolates have been stored at −80°C to retain all viral haplotypes as best as possible. We recently selected isolates with limited passages (less than two passages, only in Vero cells) to reduce potential selective effects on the virus sequencing results. Without any further passaging, we directly extracted total RNA from these isolates and performed reverse transcriptase PCR (RT-PCR) experiments to amplify complete viral genomes as we studied the viral population of each strain. The cDNA amplicons of EV-A71 isolates from various patients were prepared as previously described (52). RNA purification from EV-A71 isolates was performed using a QIAamp viral RNA kit (Qiagen). After extraction of virus RNA, cDNA was synthesized using EV-A71-specific primers and SuperScript III (Life Technologies), as recommended by the manufacturer. RNA was added to the SuperScript III RT reaction mix containing 400 nM reverse primer. Reverse transcription was performed at 55°C for 60 min. Synthesized cDNA was treated with RNase H (Life Technologies) and RNase T1 and stored at −20°C. To amplify cDNA amplicons from different EV-A71 isolates, High-Fidelity KOD Plus DNA polymerase (Toyobo) was activated at 95°C for 5 min, and PCR was performed for 25 cycles (denaturation at 95°C for 1 min and annealing-extension at 68°C for 8 min). PCR products were analyzed and extracted from 1% agarose gels for complete genome sequencing with the cDNA amplicons by shotgun sequencing with the 454 pyrosequencing system (Roche), using a commercial sequencing service (Genomics). After retrieving the shotgun reads at various depths, we used Burrows-Wheeler Aligner (BWA version 0.7.6) (56) or CLC Bio Genomic Workbench software (Qiagen) to assemble the combined reads into contigs/scaffolds and generate full-genome alignments between the assembled and target genomes. Sequencing reads were first trimmed based on a quality value of >30. Duplicate reads were then filtered by the samtools program. Only limited-duplicate reads (approximately 1 to 5% of the total reads) were identified for each read set. Both mutations and indels were counted as variants (mismatches) by CLC Bio Genomic Workbench. Genetic variations of viruses were determined by calculating the ratio of mismatch mutations to total reads at each genomic site. The genetic diversity of mutant spectra was quantified with SNPgenie software (57) and displayed as the π value for each SNV. The value of π indicated the genetic diversity of the viral mutant spectra, showing the average number of mutations per nucleotide site among sequence variants. The π value results illustrated the numbers of SNVs associated with each specimen or tissue. Alignment results were utilized to predict the haplotype composition of each isolate with QuasiRecomb (37). Based on the frequency of distribution results of the haplotype predictions, we calculated Shannon entropies and haplotype distributions for the various viruses (58). Haplotype frequencies and sequences were also used to define phylogenic networks among haplotypes between various tissues, using Network software (59).
Virion and protein structure of EV-A71.
The EV-A71 capsid pentamer and VP1 capsid protein structures (PDB code 4AED) (60) were viewed using the UCSF Chimera program, version 1.9 (61). Potential interactions between the indicated amino acid residues were identified among neighboring atoms of the residues.
Construction of infectious cDNA clones.
After extraction of viral RNA, cDNA was synthesized using EV-A71-specific primers and SuperScript III (Life Technologies) as recommended by the manufacturer. The synthesized cDNA was treated with RNase H and RNase T1 and stored at −20°C. To construct infectious cDNA clones of EV-A71 isolates, synthesized cDNA was used as the template for PCR using KOD Plus polymerase (Toyobo). PCR amplicons were purified by phenol-chloroform extraction and cloned using a TOPO XL PCR cloning kit (Invitrogen). Specific primers designed based on the published EV-A71 sequences were used for complete genome sequencing of these cDNA constructs.
In vitro transcription and transfection.
All cDNA constructs were digested with MluI to produce DNA templates, purified by phenol extraction and ethanol precipitation, and then dissolved in RNase-free water. For in vitro RNA transcription reactions, 5 μg linear DNA template was added to a buffer containing 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol, 3.125 mM each nucleoside triphosphate (NTP), 50 U RNasin RNase inhibitor, and 30 U T7 polymerase (Promega) in a total volume of 50 μl. Transcription reactions were terminated by the addition of 6 U RNase-free DNase (Promega) and incubation at 37°C for 30 min. The RNA concentration was determined by measurement of the optical density at 260 nm and checked by denaturing agarose gel electrophoresis. Then, 2 μg RNA was transfected into RD cells to produce recombinant viruses, as previously described (52).
Thermostability, virus growth, and fitness assays.
To assess thermostability, 107 PFU of EV-A71 VP1-31D and VP1-31G viruses were incubated at the temperatures indicated on the figure. After a 1- to 3-day incubation, the numbers of infectious viruses were titrated by performing plaque assays. For viral growth, SK-N-SH, RD, and DLD-1 cell monolayers were infected with various EV-A71 recombinants at a multiplicity of infection (MOI) of 0.1, as described above. Infected cells were harvested at different intervals, and whole-cell lysates were prepared for plaque assays to determine virus titers after a single freeze-thaw. For viral fitness, equal quantities of wild-type and mutant EV-A71 (106 PFU) virus were used to coinfect the indicated cells, and viruses were harvested after >75% of the cells appeared to exhibit cytopathic effects. Virus mixtures were continuously passaged in cells, and the cDNAs of viruses after the indicated passages were sequenced to determine the dominant virus in the viral mixtures that defined the viral fitness of the compared viruses.
Virus-binding assay.
To examine virus-binding ability by ELISA, 2 × 104 cells/well were seeded in 96-well plates and incubated for 2 days. The cells were washed with phosphate-buffered saline (PBS) and various recombinant EV-A71 strains with mutations were added to cells at an MOI of 20 for a 1-h incubation at 4°C. Unbound virus was washed away from the cells with ice-cold PBS. Cells were fixed using 80% acetone and stained with a monoclonal antibody against EV-A71 (MAb 979; Millipore) at a 1:2,000 dilution in 1% bovine serum albumin for 1 h. This was followed by washing with PBS-T (0.05% Tween 20 in PBS) and staining with a peroxidase-conjugated, goat-anti-mouse IgG polyclonal antibody (Cell Signaling Technology) at a 1:2,000 dilution in 1% bovine serum albumin for 1 h. After washing with PBS-T, freshly prepared substrate, 3,3′,5,5′-tetramethylbenzidine (100 μl; Invitrogen), was added to each well and the absorbance was measured at 450 nm.
Virus RNA release assay.
Neutral red (NR)-labeled viruses were prepared by infecting RD cells with the mutant viruses indicated on the figure in the presence of 10 μg ml−1 neutral red. At 3 days postinfection in the dark, cell lysates were harvested and subjected to three rounds of freeze-thawing. Cell debris was removed by low-speed centrifugation, and virus titers were determined by performing plaque assays. To analyze viral RNA release of mutant viruses, confluent cells were infected with the indicated mutant viruses at an MOI of 1. Following virus absorption to the indicated cells at 37°C for 2 h, unbound viruses were washed away using DMEM. After the postinfection time indicated on the figure, NR-labeled viruses that did not uncoat their capsids were inactivated by light exposure for 30 min. To determine the numbers of viruses that had released their RNA, infected cells were suspended using trypsin, and 104 cells were added to plates with confluent RD cells for plaque counts. After 4 days, the plaques in 104 infected cells were counted to obtain the numbers of viruses that had released RNA into the cells.
Ethics statement.
A retrospective study of clinical isolates and leftover specimens from the 1998 outbreak were obtained. All subjects for this study were approved by the Institutional Review Board of National Cheng Kung University Hospital (A-ER-104-365). Obtaining informed consent was not feasible because (i) the risk associated with the investigation is the lowest, (ii) the potential risk of participants is not higher than that of nonparticipants, and (iii) the exception of informed consent did not change the benefit to participants; this was approved by the Institutional Review Board. All methods were performed in accordance with the relevant guidelines and regulations.
Accession number(s).
The accession number of the high-throughput sequencing data at the NCBI Sequence Read Archive database is SRP104040.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Center of Infectious Disease and Signaling Research, National Cheng Kung University, Aim for the Top University Project, Ministry of Education; a Centers of Disease Control, Ministry of Health and Welfare, grant; a National Health Research Institute grant; and Ministry of Science and Technology, Taiwan, grant 105-2320-B-006-032-MY3. Computational analyses and data mining were performed using the system provided by the Bioinformatics Core at the National Cheng Kung University, supported by the Ministry of Science and Technology, Taiwan.
S.-W.H., Y.-H.H., and J.-R.W. designed the experiments. S.-W.H., Y.-H.H., H.-P.T., and P.-H.K. performed the experiments. S.-W.H., Y.-H.H., S.-M.W., C.-C.L., and J.-R.W. analyzed the data. S.-W.H. and J.-R.W. wrote the manuscript.
We declare that no conflicts of interest exist.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01062-17.
REFERENCES
- 1.Pulli T, Koskimies P, Hyypia T. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30–38. doi: 10.1006/viro.1995.1450. [DOI] [PubMed] [Google Scholar]
- 2.Brown BA, Pallansch MA. 1995. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res 39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 3.Tu PV, Thao NT, Perera D, Huu TK, Tien NT, Thuong TC, How OM, Cardosa MJ, McMinn PC. 2007. Epidemiologic and virologic investigation of hand, foot, and mouth disease, southern Vietnam, 2005. Emerg Infect Dis 13:1733–1741. doi: 10.3201/eid1311.070632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwai M, Masaki A, Hasegawa S, Obara M, Horimoto E, Nakamura K, Tanaka Y, Endo K, Tanaka K, Ueda J, Shiraki K, Kurata T, Takizawa T. 2009. Genetic changes of coxsackievirus A16 and enterovirus 71 isolated from hand, foot, and mouth disease patients in Toyama, Japan between 1981 and 2007. Jpn J Infect Dis 62:254–259. [PubMed] [Google Scholar]
- 5.Zhang Y, Wang D, Yan D, Zhu S, Liu J, Wang H, Zhao S, Yu D, Nan L, An J, Chen L, An H, Xu A, Xu W. 2010. Molecular evidence of persistent epidemic and evolution of subgenotype B1 coxsackievirus A16-associated hand, foot, and mouth disease in China. J Clin Microbiol 48:619–622. doi: 10.1128/JCM.02338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puenpa J, Theamboonlers A, Korkong S, Linsuwanon P, Thongmee C, Chatproedprai S, Poovorawan Y. 2011. Molecular characterization and complete genome analysis of human enterovirus 71 and coxsackievirus A16 from children with hand, foot and mouth disease in Thailand during 2008-2011. Arch Virol 156:2007–2013. doi: 10.1007/s00705-011-1098-5. [DOI] [PubMed] [Google Scholar]
- 7.Zong W, He Y, Yu S, Yang H, Xian H, Liao Y, Hu G. 2011. Molecular phylogeny of coxsackievirus A16 in Shenzhen, China, from 2005 to 2009. J Clin Microbiol 49:1659–1661. doi: 10.1128/JCM.00010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. 2010. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol 9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 9.Gantt S, Yao L, Kollmann TR, Casper C, Zhang J, Self SG. 2013. Implications of age-dependent immune responses to enterovirus 71 infection for disease pathogenesis and vaccine design. J Pediatric Infect Dis Soc 2:162–170. doi: 10.1093/jpids/pit017. [DOI] [PubMed] [Google Scholar]
- 10.Pallansch MA, Roos RP. 2001. Enteroviruses: poliovirus, coxsackieviruses, echoviruses, and newer enteroviruses, p 723–775. In Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 11.He Y, Ong KC, Gao Z, Zhao X, Anderson VM, McNutt MA, Wong KT, Lu M. 2014. Tonsillar crypt epithelium is an important extra-central nervous system site for viral replication in EV71 encephalomyelitis. Am J Pathol 184:714–720. doi: 10.1016/j.ajpath.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Tan SH, Ong KC, Wong KT. 2014. Enterovirus 71 can directly infect the brainstem via cranial nerves and infection can be ameliorated by passive immunization. J Neuropathol Exp Neurol 73:999–1008. doi: 10.1097/NEN.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 13.Wang YF, Chou CT, Lei HY, Liu CC, Wang SM, Yan JJ, Su IJ, Wang JR, Yeh TM, Chen SH, Yu CK. 2004. A mouse-adapted enterovirus 71 strain causes neurological disease in mice after oral infection. J Virol 78:7916–7924. doi: 10.1128/JVI.78.15.7916-7924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong KC, Wong KT. 2015. Understanding enterovirus 71 neuropathogenesis and its impact on other neurotropic enteroviruses. Brain Pathol 25:614–624. doi: 10.1111/bpa.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong KT, Munisamy B, Ong KC, Kojima H, Noriyo N, Chua KB, Ong BB, Nagashima K. 2008. The distribution of inflammation and virus in human enterovirus 71 encephalomyelitis suggests possible viral spread by neural pathways. J Neuropathol Exp Neurol 67:162–169. doi: 10.1097/nen.0b013e318163a990. [DOI] [PubMed] [Google Scholar]
- 16.Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med 15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med 15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- 18.Yang B, Chuang H, Yang KD. 2009. Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol J 6:141. doi: 10.1186/1743-422X-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang SL, Chou YT, Wu CN, Ho MS. 2011. Annexin II binds to capsid protein VP1 of enterovirus 71 and enhances viral infectivity. J Virol 85:11809–11820. doi: 10.1128/JVI.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du N, Cong H, Tian H, Zhang H, Zhang W, Song L, Tien P. 2014. Cell surface vimentin is an attachment receptor for enterovirus 71. J Virol 88:5816–5833. doi: 10.1128/JVI.03826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su PY, Wang YF, Huang SW, Lo YC, Wang YH, Wu SR, Shieh DB, Chen SH, Wang JR, Lai MD, Chang CF. 2015. Cell surface nucleolin facilitates enterovirus 71 binding and infection. J Virol 89:4527–4538. doi: 10.1128/JVI.03498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 23.Lauring AS, Frydman J, Andino R. 2013. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS, Lu L, Newton PN, Klenerman P. 2009. Genetic history of hepatitis C virus in East Asia. J Virol 83:1071–1082. doi: 10.1128/JVI.01501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojosnegros S, Beerenwinkel N, Antal T, Nowak MA, Escarmis C, Domingo E. 2010. Competition-colonization dynamics in an RNA virus. Proc Natl Acad Sci U S A 107:2108–2112. doi: 10.1073/pnas.0909787107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasa Y, Michor F, Nowak MA. 2003. Evolutionary dynamics of escape from biomedical intervention. Proc Biol Sci 270:2573–2578. doi: 10.1098/rspb.2003.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeiffer JK, Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog 1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng T, Kwang J. 2014. Attenuation of human enterovirus 71 high-replication-fidelity variants in AG129 mice. J Virol 88:5803–5815. doi: 10.1128/JVI.00289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanjuan R, Moya A, Elena SF. 2004. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci U S A 101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanjuan R, Moya A, Elena SF. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A 101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domingo E, Martin V, Perales C, Grande-Perez A, Garcia-Arriaza J, Arias A. 2006. Viruses as quasispecies: biological implications. Curr Top Microbiol Immunol 299:51–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beerenwinkel N, Gunthard HF, Roth V, Metzner KJ. 2012. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol 3:329. doi: 10.3389/fmicb.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 37.Topfer A, Zagordi O, Prabhakaran S, Roth V, Halperin E, Beerenwinkel N. 2013. Probabilistic inference of viral quasispecies subject to recombination. J Comput Biol 20:113–123. doi: 10.1089/cmb.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandelt HJ, Dress AW. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol 1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 39.Chen P, Song Z, Qi Y, Feng X, Xu N, Sun Y, Wu X, Yao X, Mao Q, Li X, Dong W, Wan X, Huang N, Shen X, Liang Z, Li W. 2012. Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2. J Biol Chem 287:6406–6420. doi: 10.1074/jbc.M111.301622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arita M, Wakita T, Shimizu H. 2012. Valosin-containing protein (VCP/p97) is required for poliovirus replication and is involved in cellular protein secretion pathway in poliovirus infection. J Virol 86:5541–5553. doi: 10.1128/JVI.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teterina NL, Pinto Y, Weaver JD, Jensen KS, Ehrenfeld E. 2011. Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J Virol 85:4284–4296. doi: 10.1128/JVI.02398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci U S A 97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doedens JR, Giddings TH Jr, Kirkegaard K. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol 71:9054–9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaini Z, Phuektes P, McMinn P. 2012. Mouse adaptation of a sub-genogroup B5 strain of human enterovirus 71 is associated with a novel lysine to glutamic acid substitution at position 244 in protein VP1. Virus Res 167:86–96. doi: 10.1016/j.virusres.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Zaini Z, McMinn P. 2012. A single mutation in capsid protein VP1 (Q145E) of a genogroup C4 strain of human enterovirus 71 generates a mouse-virulent phenotype. J Gen Virol 93:1935–1940. doi: 10.1099/vir.0.043893-0. [DOI] [PubMed] [Google Scholar]
- 47.Yeh MT, Wang SW, Yu CK, Lin KH, Lei HY, Su IJ, Wang JR. 2011. A single nucleotide in stem loop II of 5′-untranslated region contributes to virulence of enterovirus 71 in mice. PLoS One 6:e27082. doi: 10.1371/journal.pone.0027082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Duo J, Liu J, Ma C, Zhang L, Wei Q, Qin C. 2011. A mouse muscle-adapted enterovirus 71 strain with increased virulence in mice. Microbes Infect 13:862–870. doi: 10.1016/j.micinf.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Li R, Zou Q, Chen L, Zhang H, Wang Y. 2011. Molecular analysis of virulent determinants of enterovirus 71. PLoS One 6:e26237. doi: 10.1371/journal.pone.0026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kung YH, Huang SW, Kuo PH, Kiang D, Ho MS, Liu CC, Yu CK, Su IJ, Wang JR. 2010. Introduction of a strong temperature-sensitive phenotype into enterovirus 71 by altering an amino acid of virus 3D polymerase. Virology 396:1–9. doi: 10.1016/j.virol.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 51.Arita M, Shimizu H, Nagata N, Ami Y, Suzaki Y, Sata T, Iwasaki T, Miyamura T. 2005. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J Gen Virol 86:1391–1401. doi: 10.1099/vir.0.80784-0. [DOI] [PubMed] [Google Scholar]
- 52.Huang SW, Wang YF, Yu CK, Su IJ, Wang JR. 2012. Mutations in VP2 and VP1 capsid proteins increase infectivity and mouse lethality of enterovirus 71 by virus binding and RNA accumulation enhancement. Virology 422:132–143. doi: 10.1016/j.virol.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 53.Arita M, Ami Y, Wakita T, Shimizu H. 2008. Cooperative effect of the attenuation determinants derived from poliovirus Sabin 1 strain is essential for attenuation of enterovirus 71 in the NOD/SCID mouse infection model. J Virol 82:1787–1797. doi: 10.1128/JVI.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, Wu X, Huang K, Li L, Zheng L, Wan C, He ML, Zhao W. 2014. The variations of VP1 protein might be associated with nervous system symptoms caused by enterovirus 71 infection. BMC Infect Dis 14:243. doi: 10.1186/1471-2334-14-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cordey S, Petty TJ, Schibler M, Martinez Y, Gerlach D, van Belle S, Turin L, Zdobnov E, Kaiser L, Tapparel C. 2012. Identification of site-specific adaptations conferring increased neural cell tropism during human enterovirus 71 infection. PLoS Pathog 8:e1002826. doi: 10.1371/journal.ppat.1002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson CW, Moncla LH, Hughes AL. 2015. SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31:3709–3711. doi: 10.1093/bioinformatics/btv449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers MB, Song T, Sebra R, Greenbaum BD, Hamelin ME, Fitch A, Twaddle A, Cui L, Holmes EC, Boivin G, Ghedin E. 2015. Intrahost dynamics of antiviral resistance in influenza A virus reflect complex patterns of segment linkage, reassortment, and natural selection. mBio 6:e02464-14. doi: 10.1128/mBio.02464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 60.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.