ABSTRACT
Since 1999, Caenorhabditis elegans has been extensively used to study microbe-host interactions due to its simple culture, genetic tractability, and susceptibility to numerous bacterial and fungal pathogens. In contrast, virus studies have been hampered by a lack of convenient virus infection models in nematodes. The recent discovery of a natural viral pathogen of C. elegans and development of diverse artificial infection models are providing new opportunities to explore virus-host interplay in this powerful model organism.
KEYWORDS: Caenorhabditis elegans, Flock House virus, innate immunity, Orsay virus, RNA interference, virus-host interactions, vesicular stomatitis virus
INTRODUCTION
In the past 40 years, the free-living nematode Caenorhabditis elegans has been used in diverse fields such as neurobiology, RNA interference (RNAi), and pathogen-host interactions. As a model, C. elegans offers many advantages, including genetic tractability, small size, and inexpensive culture. Its sexually dimorphic nature and short reproductive cycle make genetic crosses and rearing large numbers of animals simple. As C. elegans naturally feeds on bacteria (1), one can often initiate intestinal infection by using the microbe of interest as a food source. Furthermore, the transparent body of C. elegans permits visualization of fluorescently tagged pathogens and tissue pathologies. Well-defined techniques for genetically modifying the laboratory strain (N2) make identifying nematode factors influencing pathogen susceptibilities relatively straightforward (2, 3). RNAi is also easily performed by feeding nematodes bacteria expressing double-stranded RNAs (dsRNAs) targeting the gene of interest. This “feeding” RNAi response spreads to most tissues and can be inherited, allowing inhibition of gene expression in adults and progeny.
Although lacking the classic adaptive immunity found in vertebrates that relies on specialized immune cells, C. elegans employs a variety of innate and nonclassic adaptive immune responses that are shared with other metazoans (i.e., antiviral RNAi) (4, 5). An estimated 38% of the ∼20,250 protein-encoding genes in C. elegans have human orthologs (6), and several antimicrobial pathways found in mammals are also found in worms (4, 5, 7–9). Therefore, C. elegans can be used to explore conserved immunological mechanisms.
Since the establishment of a Pseudomonas aeruginosa infection model in 1999 (10, 11), dozens of bacterial and fungal pathogens (including several that infect humans) have been shown to cause disease in C. elegans (reviewed in [1, 9]). Studies of these predominantly extracellular pathogens have largely shaped our understanding of C. elegans immunity. However, with the recent discovery of natural intracellular pathogens of C. elegans (12, 13) and the development of nonnatural virus infection models (14–18), this is beginning to change.
Here virus-C. elegans interaction models (Table 1) are discussed in terms of their features, advantages/disadvantages, and contributions to our understanding of C. elegans antiviral immunity (Fig. 1). For brevity, the most prominent RNA virus models are discussed, although it should be noted that a DNA virus model using the poxvirus vaccinia virus was also reported in C. elegans (18).
TABLE 1.
RNA virus models in Caenorhabditis elegans
| Virus | Family | Viral genomeb | Infection strategy | Pathology | Horizontal/vertical transmission | References |
|---|---|---|---|---|---|---|
| Flock House virus | Nodaviridae | (+)ssRNA, bipartite | Transgenic animals expressing viral genome | None reported | No | 15 |
| Vesicular stomatitis virus | Rhabdoviridae | (−)ssRNA | Primary cell culture and microinjection of animals | N2 infections limited primarily to muscle; involvement of multiple tissues in RNAi-deficient animals; lethal infection | Vertical transmission | 14,16,17 |
| Orsay virus | Nodaviridaea | (+)ssRNA, bipartite | Natural/experimental infections in animals | N2 infections largely asymptomatic; infection of intestinal cells; frequent symptoms in RNAi-deficient animals: disappearance of intestinal cell nuclei and storage granules and cell fusion, delayed egg laying; nonlethal infection | Horizontal transmission | 13,35 |
Based on phylogenetic analyses, but formal classification has not been given by the International Committee for the Taxonomy of Viruses.
Positive and negative polarity ssRNA virus genomes are indicated by (+) and (−), respectively.
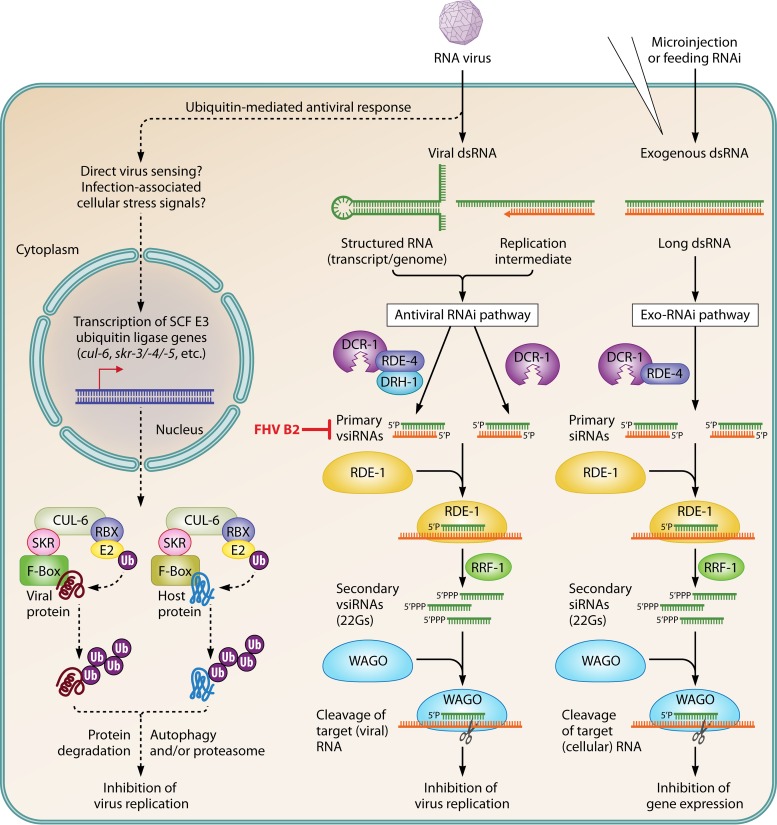
FIG 1.
RNAi- and ubiquitin-mediated restriction of RNA virus infection in C. elegans (adapted from [17, 28, 43]). Virus infection and replication leads to the production of viral dsRNA that triggers the antiviral RNAi pathway. Viral dsRNAs are recognized and cleaved by a DCR-1/RDE-4/DRH-1 complex into 23 nucleotide (nt)-long primary vsiRNAs. DCR-1 may also produce primary vsiRNAs in a parallel, RDE-4/DRH-1-independent pathway (28). Duplex vsiRNAs complex with the argonaute RDE-1, and one strand of the duplex is lost. These complexes recruit the RdRP RFF-1 to viral RNA to synthesize 22 nt-long secondary vsiRNAs with 5′guanines (22Gs) that complex with worm-specific argonautes (WAGOs). WAGO-22G complexes direct cleavage of complementary viral RNAs, thereby inhibiting virus replication. FHV B2 blocks the antiviral RNAi pathway by inhibiting the function of primary vsiRNAs (29). Also shown is the Exo-RNAi pathway, which is triggered by exogenous, long dsRNAs and uses similar machinery to produce siRNAs but does not require DRH-1. Virus infection also elicits, through direct viral sensing or cellular stress signals associated with infection, the upregulation of host genes encoding Skp1-related (SKR)-Cullin (CUL)-6-F-box (SCF) E3 ubiquitin (Ub) ligase complex components. These associate with other SCF components (RBX, E2 ubiquitin-conjugating enzymes, etc.) to form antiviral ubiquitin ligase complexes. Different F-box proteins may provide target specificity, allowing ubiquitination and degradation of proviral host factors and/or viral proteins (and possibly virions [not shown]) through proteasome- and autophagy-dependent mechanisms (43).
RNA VIRUS-C. Elegans MODELS
Nonnatural virus models.
(i) Flock House virus.
The first in vivo system for studying virus replication in C. elegans used transgenic nematodes encoding a Flock House virus (FHV) replicon (15). FHV is an alphanodavirus that can replicate in a broad range of plant and animal cells (15). The positive-sense single-stranded RNA (ssRNA) FHV genome is bipartite: segment 1 (RNA1) encodes the viral RNA-dependent RNA polymerase (RdRP), and segment 2 (RNA2) encodes the capsid protein precursor (19). The FHV RdRP replicates both RNA1 and RNA2 and transcribes a subgenomic RNA (RNA3) from the replicative intermediate of RNA1 (19). RNA3 encodes B1, an uncharacterized protein, and B2, a dsRNA-binding protein with RNAi suppressor activity (15).
In the FHV model, transgenic nematodes encode RNA1 and RNA2 under a heat-inducible promoter that drives the initial production of RNA1 and RNA2 (15). Newly made viral RdRP replicates these genome segments and transcribes RNA3. Although all three FHV RNAs are produced in C. elegans, virions do not appear to form (R. Lui, personal communication), suggesting that one or more host factors required for virus assembly may be missing. Nevertheless, FHV genome replication triggers a potent, RNAi-mediated antiviral response (15). Consequently, B2-deficient FHV replicons cannot detectably replicate in wild-type (N2) worms but can in mutant animals with deficient feeding RNAi responses (15, 20). These observations provided some of the first evidence for a C. elegans antiviral RNAi pathway that shared components with the “exo-RNAi” pathway (21, 22), which is trigged by exogenous dsRNAs as in feeding RNAi. These shared RNAi components include the single nematode Dicer, DCR-1, its binding partner and dsRNA-binding protein, RDE-4, the Argonaute protein, RDE-1, and the RdRP, RRF-1 (Fig. 1).
Later work by Lu et al. (20) identified additional host antiviral RNAi factors using a RNAi screen that was based on the rescue of a B2-deficient FHV replicon that encoded green fluorescent protein (GFP) in place of B2. This screen identified ∼35 worm factors that when knocked down relieved FHV restriction (indicated by GFP expression). One of the identified factors, Dicer-related helicase-1 (DRH-1), has recently garnered much attention.
DRH-1 physically interacts with DCR-1/RDE-4 complexes (23) and is required for fulminate antiviral RNAi responses but is dispensable for exo-RNAi (20, 24). Interestingly, DRH-1 shares homology with mammalian retinoic acid-inducible gene-I (RIG-I)-like helicases (20, 24), which act as cytosolic sensors of viral RNA that activate the vertebrate interferon response (25). Remarkably, human RIG-I domains can substitute for the corresponding DRH-1 domains to promote antiviral RNAi in C. elegans (24). These observations suggest that DRH-1 might function as a sensor of viral RNA, analogous to RIG-I, but instead trigger an RNAi response (26). Others have argued that DRH-1 acts downstream of viral RNA sensing because cleavage of viral dsRNA by DCR-1 to produce primary virus-derived small interfering RNAs (vsiRNAs) is still observed in drh-1 mutants (20, 27). Interestingly, DCR-1 can also initiate primary vsiRNA production in the absence of RDE-4, albeit less efficiently (28). Therefore, the role of DRH-1 in antiviral RNAi remains enigmatic.
The FHV model is an elegant genetic system to identify and characterize antiviral RNAi machinery and can also be used to identify viral suppressors of RNAi (29). However, given the apparent lack of virion formation in worms, the FHV system cannot be used to study virus entry, assembly, exit, or transmission, all of which require a bona fide viral pathogen.
(ii) Vesicular stomatitis virus.
In 2005, two reports demonstrated that vesicular stomatitis virus (VSV), an arbovirus with a broad invertebrate and vertebrate host range, could replicate in primary cells derived from C. elegans embryos (14, 16). VSV is a Rhabdoviridae family member and encodes five proteins from its negative-sense ssRNA genome, although additional open reading frames (ORFs) can be inserted into the VSV genome using reverse genetics (30, 31). Using recombinant VSV strains encoding GFP to track infection, and reverse transcriptase PCR (RT-PCR) and plaque assays, these initial studies showed that VSV replication was enhanced in cells derived from RNAi-deficient nematodes (14, 16). These studies provided the first fully infectious virus model system in C. elegans and, along with the FHV model, established RNAi as an antiviral defense mechanism in the worm. However, there are major drawbacks to using C. elegans cell cultures to explore virus-host interactions. For one, it is necessary to isolate embryonic cells from embryos to establish cultures because an immortalized C. elegans cell line has not been developed. In addition, isolation of embryonic cells is both labor intensive and technically challenging. These challenges may explain why there has been such a limited number of virological studies using C. elegans cell cultures.
To overcome these limitations and create an in vivo model, we developed techniques to microinject VSV into nematodes (17). Microinjection of VSV particles leads to a dose-dependent, lethal infection that can be scored and tracked in real time using recombinant VSV strains expressing fluorescent proteins. VSV is predominantly restricted to muscle tissue in N2 nematodes and is not efficiently transmitted horizontally. VSV restriction to muscle appears to be partially mediated by the antiviral RNAi response because drh-1 mutants experience infection in multiple tissues, enhanced VSV replication, and earlier mortality. The reduced vsiRNA production in drh-1 mutants likely explains this hypersensitivity. Unexpectedly, we also observed infection of oocytes and vertical transmission in drh-1 mutants after somatic injection of VSV. However, vertical transmission was observed only when mutants were cultured on medium containing the DNA synthesis inhibitor fluorodeoxyuridine. Previous studies have suggested that this compound can modulate stress responses in C. elegans (32). We suspect that this may inadvertently impede additional antiviral pathways that guard against vertical transmission. Finally, we found progeny derived from adults infected with VSV to be more resistant to VSV challenge compared to progeny from naive mothers, and this phenomenon was dependent upon specific antiviral RNAi factors. Heritable RNAi responses were previously reported in the FHV replicon system (33), suggesting the fascinating possibility that nematodes use transgenerational RNAi responses as a form of immunological memory to prevent infection.
While the need to microinject VSV to establish infection may limit use of the VSV model in large-scale screens, it is the only model in which infection can be directly observed in living nematodes. Moreover, the lethal nature of VSV infection provides a convenient readout for identifying host backgrounds with differential virus susceptibilities. Finally, the VSV model can be used to study antiviral responses in nonintestinal tissue and vertical transmission, providing unique opportunities not afforded by other models.
Natural virus models: Orsay virus.
Two recent studies of wild-caught nematodes identified the first natural viral pathogens of Caenorhabditis worms: Orsay virus (OV), Santeuil virus, and Le Blanc virus (13, 34). OV infects C. elegans but not the related nematode Caenorhabditis briggsae, whereas both Santeuil and Le Blanc viruses can infect C. briggsae but not C. elegans (13, 34). Experimental infection by these viruses is obtained by adding filtered lysates of infected animals to cultures of naive nematodes. This leads to horizontal transmission among animals, likely through a fecal-oral route. These viruses primarily infect intestinal cells (35) and cause intestinal abnormalities but do not affect brood sizes or longevity and are not vertically transmitted (13).
Each nematode virus encodes a bipartite, positive-sense ssRNA genome, and based on genome sequence, the three viruses are most closely related to one another, although they are also related to alphanodaviruses (13, 34, 36). Several unique characteristics of these nematode viruses have suggested that they represent a new and distinct group of nodavirus-like pathogens. For one, the OV capsid structure is more similar to that of piscine betanodaviruses than that of alphanodaviruses (36, 37). Furthermore, although these nematode viruses encode an RdRP and capsid protein (CP) on RNA1 and RNA2 segments, respectively, they also encode an additional ORF downstream of the CP on RNA2 called δ that is unrelated to any known protein, and these viruses also do not encode the subgenomic (RNA3) segment typically found in nodaviruses (13, 38).
Recent studies suggest that OV uses ribosomal frame shifting to produce δ protein predominantly as a CP-δ fusion (38) that forms a virion-associated fiber resembling those used by reoviruses and adenoviruses for entry (36, 39). Using an OV reverse genetic system (39), OV strains were created that were either unable to produce free δ protein (but still produced CP-δ) or encoded mutations predicted to disrupt the CP-δ fiber structure (36). OV mutants unable to produce free δ replicated normally, but strains with structure-disrupting substitutions in CP-δ were either unviable or dramatically reduced in their infectivity (36). These studies have implicated the CP-δ protein in mediating host cell attachment and entry. Using CP-δ as probe to identify cell receptors may help us understand the intestine-specific tropism of OV.
OV replication, like that of FHV and VSV, is restricted by RNAi, but whether this response is heritable remains controversial (40, 41). OV infections in N2 animals are typically asymptomatic, whereas RNAi-deficient mutants often exhibit significant intestinal pathologies (13). Recently, a 159-bp deletion in the drh-1 gene was found in the wild isolate (JU1580) that OV was originally identified in, possibly explaining why it exhibited severe intestinal abnormalities (26). Intriguingly, Felix et al. (13) reported variation in OV susceptibility in a collection of diverse wild nematode strains that did not always correlate with each strain's RNAi proficiency, suggesting that other virus resistance mechanisms exist (42).
A major advance in C. elegans antiviral immunity came from studies of another group of obligate intracellular pathogens, the microsporidia. These fungal-related pathogens have been recovered from wild-caught nematodes, and one species, Nematocida parisii, causes a lethal intestinal infection in C. elegans (12). Examination of transcriptional responses to N. parisii infection identified a set of differentially regulated genes (DEGs) that shared a large degree of overlap with DEGs found after OV infection but little overlap with DEGs observed after infections with extracellular pathogens (43, 44). This suggested that there might be a unique host response to intracellular pathogens. Many of the DEGs that were upregulated encode ubiquitylation-related machinery, in particular members of Skp1-Cull-F-box (SCF) class of multisubunit E3 ubiquitin ligases (43). Furthermore, RNAi knockdown of core SCF ligase components such as CUL-6 promoted N. parisii and OV infection (43). These studies suggest that C. elegans responds to intracellular infection by upregulation of core SCF ligase components such as CUL-6 as well as F-box proteins that modulate SCF ligase complex target specificity. Interestingly, F-box proteins are greatly expanded in worms (>500) compared to humans (∼69), possibly reflecting an evolutionary pathogen-host arms race (43, 45). Use of different F-box adaptors may allow SCF ligases to ubiquitinate pathogen and/or host proteins required for pathogen replication (43). This might target these proteins for destruction through autophagy and/or proteasome-mediated pathways, thereby inhibiting infection (Fig. 1) (43).
Because OV is restricted to intestinal cells (even in RNAi-deficient animals [35]) and not vertically transmitted, the OV model may not be amenable to studies of antiviral defenses in nonintestinal tissues. However, this model is well suited for studies of virus-host coevolution; horizontal transmission; virus entry, assembly, and exit; and intestinal antiviral defenses.
CONCLUSIONS AND FUTURE DIRECTIONS
Efforts by many investigators to develop new virus models in C. elegans have yielded several complementary systems for investigating virus-host interplay. The FHV replicon system is well suited for identifying host factors that restrict virus replication postentry, including components of the antiviral RNAi machinery. The OV model allows study of aspects of the viral life cycle not afforded by the FHV system, such as virus entry and exit and horizontal transmission. Growing evidence suggests that interactions among enteric viruses, intestinal flora, and host can influence the course of viral disease in other metazoans (46). With the discovery of OV as a natural enteric pathogen of C. elegans and the worm's susceptibility to many other intestinal microbes (1), the nematode may prove useful in exploring how such “transkingdom” interactions modulate pathogenesis and horizontal transmission.
Recent OV and N. parisii studies have provided initial insights into nematode transcriptional responses invoked by intracellular pathogens (43, 44, 47). These investigations have revealed a role for ubiquitin-mediated defenses in the control of intracellular enteric pathogens (43). How these responses are regulated and whether they are active in nonintestinal tissues remain open questions. The capacity of VSV to infect muscle and spread to other somatic and germ line tissues (17) may provide avenues to identify common and tissue-specific antiviral defenses, including those that protect against vertical transmission. The growing threat of arboviruses and other pathogenic viruses to public health emphasizes the need to develop new means to understand viral disease. Perhaps many big mysteries surrounding virus-host interactions can be solved in this tiny worm.
ACKNOWLEDGMENTS
I apologize to colleagues whose work was not directly cited in this article due to space limitations.
This work was supported by the Endowed Scholars Program at the University of Texas Southwestern Medical Center.
REFERENCES
- 1.Schulenburg H, Felix MA. 2017. The natural biotic environment of Caenorhabditis elegans. Genetics 206:55–86. doi: 10.1534/genetics.116.195511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Feng X, Guang S. 2016. Targeted genome engineering in Caenorhabditis elegans. Cell Biosci 6:60. doi: 10.1186/s13578-016-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutscher LM, Shaham S. 2014. Forward and reverse mutagenesis in C. elegans. WormBook 1–26. doi: 10.1895/wormbook.1.167.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ermolaeva MA, Schumacher B. 2014. Insights from the worm: the C. elegans model for innate immunity. Semin Immunol 26:303–309. doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewbank JJ, Pujol N. 2016. Local and long-range activation of innate immunity by infection and damage in C. elegans. Curr Opin Immunol 38:1–7. doi: 10.1016/j.coi.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Shaye DD, Greenwald I. 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One 6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abnave P, Conti F, Torre C, Ghigo E. 2014. What RNAi screens in model organisms revealed about microbicidal response in mammals? Front Cell Infect Microbiol 4:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan MW, Shapira M. 2011. Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol 13:497–507. doi: 10.1111/j.1462-5822.2011.01570.x. [DOI] [PubMed] [Google Scholar]
- 9.Gravato-Nobre MJ, Hodgkin J. 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol 7:741–751. doi: 10.1111/j.1462-5822.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 10.Tan MW, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troemel ER, Felix MA, Whiteman NK, Barriere A, Ausubel FM. 2008. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol 6:2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D. 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. 2005. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schott DH, Cureton DK, Whelan SP, Hunter CP. 2005. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A 102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gammon DB, Ishidate T, Li L, Gu W, Silverman N, Mello CC. 2017. The antiviral RNA interference response provides resistance to lethal arbovirus infection and vertical transmission in Caenorhabditis elegans. Curr Biol 27:795–806. doi: 10.1016/j.cub.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu WH, Lin YL, Wang JP, Liou W, Hou RF, Wu YC, Liao CL. 2006. Restriction of vaccinia virus replication by a ced-3 and ced-4-dependent pathway in Caenorhabditis elegans. Proc Natl Acad Sci U S A 103:4174–4179. doi: 10.1073/pnas.0506442103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball LA, Johnson KL. 1999. Reverse genetics of nodaviruses. Adv Virus Res 53:229–244. doi: 10.1016/S0065-3527(08)60350-4. [DOI] [PubMed] [Google Scholar]
- 20.Lu R, Yigit E, Li WX, Ding SW. 2009. An RIG-I-like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog 5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishok A. 2013. Biology and mechanisms of short RNAs in Caenorhabditis elegans. Adv Genet 83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- 22.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 23.Tabara H, Yigit E, Siomi H, Mello CC. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109:861–871. doi: 10.1016/S0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 24.Guo X, Zhang R, Wang J, Ding SW, Lu R. 2013. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci U S A 110:16085–16090. doi: 10.1073/pnas.1307453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Errett JS, Gale M. 2015. Emerging complexity and new roles for the RIG-I-like receptors in innate antiviral immunity. Virol Sin 30:163–173. doi: 10.1007/s12250-015-3604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashe A, Belicard T, Le Pen J, Sarkies P, Frezal L, Lehrbach NJ, Felix MA, Miska EA. 2013. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife 2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffman SR, Lu J, Guo X, Zhong J, Jiang H, Broitman-Maduro G, Li WX, Lu R, Maduro M, Ding SW. 2017. Caenorhabditis elegans RIG-I homolog mediates antiviral RNA interference downstream of Dicer-dependent biogenesis of viral small interfering RNAs. MBio 8:pii:e00264-17. doi: 10.1128/mBio.00264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, Zhang R, Wang J, Lu R. 2013. Antiviral RNA silencing initiated in the absence of RDE-4, a double-stranded RNA binding protein, in Caenorhabditis elegans. J Virol 87:10721–10729. doi: 10.1128/JVI.01305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Lu R. 2013. Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J Virol 87:5414–5423. doi: 10.1128/JVI.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson ND, Stillman EA, Whitt MA, Rose JK. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A 92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whelan SP, Ball LA, Barr JN, Wertz GT. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A 92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson EN, Corkins ME, Li JC, Singh K, Parsons S, Tucey TM, Sorkac A, Huang H, Dimitriadi M, Sinclair DA, Hart AC. 2016. C. elegans lifespan extension by osmotic stress requires FUdR, base excision repair, FOXO, and sirtuins. Mech Ageing Dev 154:30–42. doi: 10.1016/j.mad.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rechavi O, Minevich G, Hobert O. 2011. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franz CJ, Zhao G, Felix MA, Wang D. 2012. Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J Virol 86:11940. doi: 10.1128/JVI.02025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franz CJ, Renshaw H, Frezal L, Jiang Y, Felix MA, Wang D. 2014. Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology 448:255–264. doi: 10.1016/j.virol.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Fan Y, Guo YR, Yuan W, Zhou Y, Holt MV, Wang T, Demeler B, Young NL, Zhong W, Tao YJ. 2017. Structure of a pentameric virion-associated fiber with a potential role in Orsay virus entry to host cells. PLoS Pathog 13:e1006231. doi: 10.1371/journal.ppat.1006231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo YR, Hryc CF, Jakana J, Jiang H, Wang D, Chiu W, Zhong W, Tao YJ. 2014. Crystal structure of a nematode-infecting virus. Proc Natl Acad Sci U S A 111:12781–12786. doi: 10.1073/pnas.1407122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H, Franz CJ, Wu G, Renshaw H, Zhao G, Firth AE, Wang D. 2014. Orsay virus utilizes ribosomal frameshifting to express a novel protein that is incorporated into virions. Virology 450-451:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang H, Franz CJ, Wang D. 2014. Engineering recombinant Orsay virus directly in the metazoan host Caenorhabditis elegans. J Virol 88:11774–11781. doi: 10.1128/JVI.01630-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sterken MG, Snoek LB, Bosman KJ, Daamen J, Riksen JA, Bakker J, Pijlman GP, Kammenga JE. 2014. A heritable antiviral RNAi response limits Orsay virus infection in Caenorhabditis elegans N2. PLoS One 9:e89760. doi: 10.1371/journal.pone.0089760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashe A, Sarkies P, Le Pen J, Tanguy M, Miska EA. 2015. Antiviral RNA interference against Orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans. J Virol 89:12035–12046. doi: 10.1128/JVI.03664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balla KM, Troemel ER. 2013. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell Microbiol 15:1313–1322. doi: 10.1111/cmi.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TL, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. 2014. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog 10:e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen K, Franz CJ, Jiang H, Jiang Y, Wang D. 2017. An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes. BMC Genomics 18:303. doi: 10.1186/s12864-017-3689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas JH. 2006. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res 16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeiffer JK, Virgin HW. 2016. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351:pii:aad5872. doi: 10.1126/science.aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarkies P, Ashe A, Le Pen J, McKie MA, Miska EA. 2013. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res 23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]