ABSTRACT
The size of lentiviral DNA reservoirs reflects the effectiveness of immune responses against lentiviruses. So far, abundant information has been gathered on the control of HIV-1 replication. Understanding the innate mechanisms contributing to containment of the HIV DNA reservoir, however, are only partly clarified and are relevant to guiding interventions for reservoir containment or eradication. We studied the contribution of natural killer (NK) cell functional features in HIV patients controlling replication either spontaneously (HIV controllers [HIC]) or after progression and antiretroviral treatment (progressor patients [PP]). An inverse correlation between HIV DNA copy numbers (either total or integrated) in circulating CD4+ cells and NK cell function was observed. Induced interferon gamma (IFN-γ) production and NKp46/NKp30 activating receptor-induced expression correlated inversely with reservoir size. The correlation was present not only for a homogeneous cohort of HIC patients but also when PP were included in the analysis. Adaptive (NKG2C+ CD57+) NK cell features were not associated with reservoir size. However, a distinct set of 370 differentially expressed transcripts was found to underlie functional differences in NK cells controlling HIV DNA reservoir size. In proof-of-principle in vitro experiments of CD4+ cell infection with HIV-1, purified NK cells with the above-mentioned functional/transcriptional features displayed 10- and 30-fold higher abilities to control HIV replication and DNA burdens in vitro, respectively, than those of other NK cells. Thus, NK cells with a specific functional and transcriptional signature contribute to control of the HIV reservoir in CD4+ cells. Their selection, expansion, and/or adoptive transfer may support strategies to eradicate HIV-1 infection or to safely deescalate antiretroviral treatment.
IMPORTANCE The most relevant feature of HIV-1 infection is represented by its DNA reservoir size in the body, which guarantees lifelong infection and resumption of virus replication after antiretroviral treatment interruption. So far, there has been little success in the identification of factors contributing to HIV-1 reservoir containment. In this study, by studying quantitative total and integrated HIV-1 DNA levels and NK cells in HIV-1 patients with either progressive or nonprogressive disease, we observed that inducible IFN-γ and natural cytotoxicity receptor (NCR) expression in a specific subset of NK cells with a characteristic transcriptional signature represents a correlate for HIV-1 reservoir control. This represents an advance in our understanding of the mechanism(s) that controls the lentivirus reservoir. Monitoring, selection, expansion, and adoptive transfer of these NK cells may allow monitoring of treatment efficacy and the likelihood of reservoir control and may support protocols for HIV-1 eradication.
KEYWORDS: DNA, HIV-1, reservoir, interferons, lentiviruses, natural killer cells
INTRODUCTION
Since their original description as cells endowed with cytotoxic antitumor activity and CD16 and CD56 antigen expression (1–4), our view of natural killer (NK) cells has been upgraded with a number of additional functions, including a finely tuned balance between activating natural cytotoxicity receptor (NCR; NKp46, NKp30, and NKp44) function and inhibitory killer Ig-like inhibitory receptor (KIR) function, cytokine production and responsiveness, Toll-like receptor (TLR) and chemokine receptor expression, and identification of NK cell developmental stages from CD34+ cell precursors (5–12). In addition, a quite remarkable heterogeneity of circulating NK cell phenotypes has been revealed by mass cytometry (13) and computer-assisted flow cytometric analysis (14).
Similarly, the original view of prevalent NK cell antitumor activity has evolved to encompass the fundamental role of these cells in the control of virus infections. Indeed, NK cell absence in humans, or their removal in mice, leads to disseminated herpesvirus, influenza virus, and Ebola virus infections (15–18). Their relevance in control of virus infections is reflected by the quantity of mechanisms that have been developed to evade NK cell activity, including skewed cytokine production (19, 20), reduced NCR expression with an activated NK cell phenotype (21–23), and NCR ligand shedding or downmodulation on target cells (22, 24–27). A distinct pattern of NCR expression and induction associates with a benign course of HIV infection in a minority of HIV-infected patients with low or undetectable viremia (HIV controllers [HIC], long-term nonprogressors [LTNP], and elite controllers [EC]) (28–30) and in chronically hepatitis C virus (HCV)-infected patients who respond to treatment (31, 32). In addition, epidemiological DNA carriage studies showed an association between specific KIR:HLA carriage and control of HIV-1 replication (33–35). The impact of coordinate KIR:HLA genotype carriage on disease progression so far partly eludes understanding due to the variability of molecule transcription and surface expression on T and NK cells. Accordingly, the in vitro effect of HLA:KIR carriage on NK cell control of HIV replication (HIVp24) in patients with delayed disease progression remains controversial (36, 37). During infections with different viruses, including human cytomegalovirus (HCMV) (38, 39), murine CMV (MCMV) (40), and possibly chikungunya virus, dengue virus, and hantavirus (41, 42), persistent or transient expansions of a specific NK cell subset bearing NKG2C in humans and its homolog in mice have been described. These cell expansions are interpreted as possible memory-like features of NK cells with resemblance to adaptive immune T cell responses (11, 12).
With regard to NK cells in HIV-1 infection, scientific focus has so far concentrated on their control of HIV-1 replication and of plasma viral load (RNA), leading, among other achievements, to the identification of particular KIR:HLA haplotypes or NCR expression regulation that may control virus replication (30, 33, 37) and to their cooperation with adaptive CD8+ cytotoxic T lymphocyte (CTL) responses. The hallmark of lentivirus infection is, however, represented by the persistence of integrated or partly episomal DNA in long-lived cells—referred to as reservoir—which guarantees lifelong infection (43).
Focuses on different reservoir sites and cells, including CXCR5+ PD1+ Tfh cells (44, 45), CD32 CD4+ T cells (46), and tissue monocytes/macrophages, are currently being actively pursued. Peripheral blood HIV-1 DNA in circulating CD4+ T cells represents an acknowledged site for estimating the total HIV reservoir in HIV-infected patients (43, 47–49). The reservoir is still detected even after 5 to 14 years of successful (i.e., with undetectable plasma viral RNA) antiretroviral therapy (ART) (50, 51). Persistence of HIV DNA is maintained in a relatively constant nonreplicating pool of central memory CD4+ T cells in peripheral blood, lymph nodes, and gut-associated lymphoid tissue (GALT) (49) and in monocytes and follicular dendritic CD4+ T cells (43). Quantitative determination of HIV DNA in peripheral blood mononuclear cells (PBMC) contributes to defining the risk of disease progression in infected patients (52), in whom low levels of HIV DNA in CD4+ PBMC are associated with nonprogressive disease (HIC) (53) with posttreatment control of viremia (54). Accordingly, one of the major therapeutic objectives for lentiviruses in general and HIV-1 in particular is represented by the containment of the HIV DNA reservoir size and its targeting with novel therapeutic strategies (55).
Limited information is available so far on the mechanism(s) that contributes to the containment of the HIV-1 reservoir in vivo. To address this issue with a specific focus on innate immune cells, we studied the relationship between NK cell function and total and integrated HIV DNA copy numbers in circulating CD4+ PBMC in two widely diverging clinical cohorts (HIC and progressor patients). We report here that the degrees of inducibility of interferon gamma (IFN-γ) production and NCRs have a linear relationship to quantitative control of the HIV-1 reservoir in infected patients over a wide spectrum of disease activity and are associated with a specific NK cell transcriptional signature.
RESULTS
IFN-γ production by NK cells is associated with control of HIV DNA.
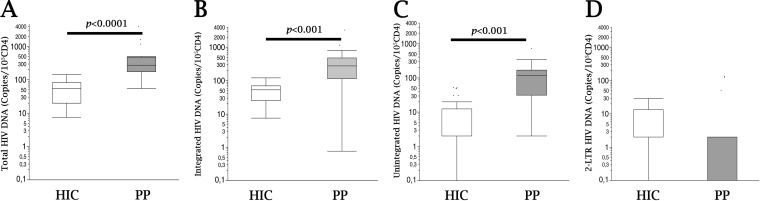
We first studied the quantitative HIV-1 reservoir in circulating PBMC of progressor patients (PP) who had been treated successfully and were persistently suppressed for >18 months on ART and in a cohort of HIV-1 controller patients (HIC) with spontaneously stable CD4+ counts of >500/μl and low or undetectable plasma viremia. As expected, a higher viral reservoir was detected in PP than in HIC (Fig. 1A to C) in terms of total, integrated, and unintegrated (i.e., linear DNA and 1- and 2-long-terminal-repeat [LTR] circles) viral DNAs (P < 0.001). In contrast, the evaluation of 2-LTR DNA was not different between the two patient groups (Fig. 1D). A relatively wide range of numbers of HIV-1 DNA copies/105 CD4+ PBMC was detected in PP but also in HIC, suggesting that ample variations possibly exist in the mechanism(s) underlying control of the HIV reservoir in vivo both across patient groups (i.e., HIV versus PP) and within relatively select groups, such as HIC. We therefore next sought to study possible parameters that could be associated with and explain the wide dispersion of the HIV-1 reservoir observed in HIC. In view of the reported association of innate NK cell control with virus replication for HIV (30, 37) and the absence of an association of adaptive CD8 CTL function with posttreatment control or HIC (56), we next studied whether circulating HIV DNA levels could be associated with specific differences in NK cell function.
FIG 1.
HIV DNA in PBMC from patients with different disease courses. Comparisons of total (A), integrated (B), unintegrated (C), and 2-LTR (D) HIV DNAs (copies/105 CD4+ cells) in HIC (EC and LTNP) (white boxes) and PP (gray boxes) are shown. For box plot analysis, bars represent ranges, lines represent median values, and the box tops and bottoms represent the 75th and 25th percentiles, respectively (P < 0.001 or P < 0.0001 by the Mann-Whitney U test). HIC, HIV controllers (n = 20); PP, progressor patients (previously progressing patients who are successfully treated and have persistently undetectable HIV RNA [<50 copies/ml]) (n = 10).
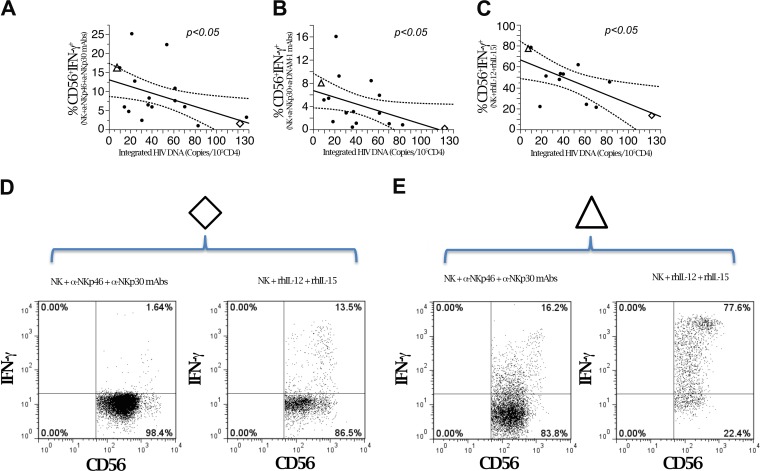
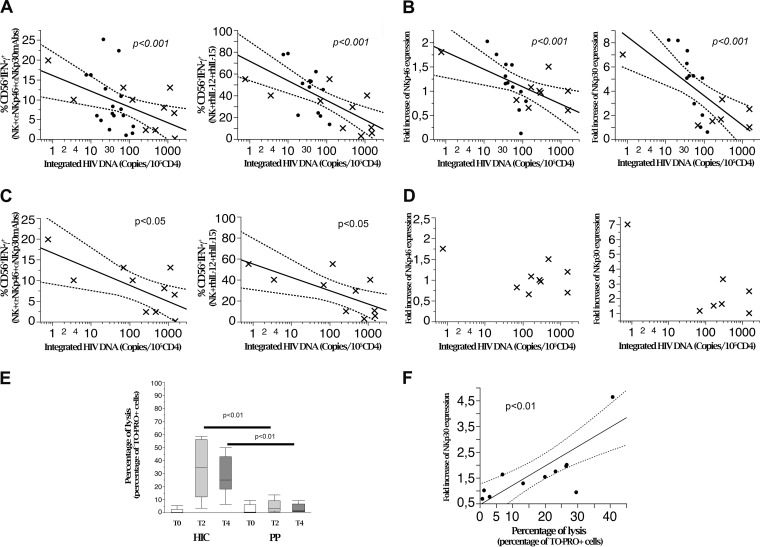
To this end, HIV DNA copy numbers in PBMC (expressed as numbers/CD4+ PBMC) were first compared to NK cell IFN-γ levels by Spearman's rho. NK cells were gated as shown in Fig. 2. Lower HIV DNA loads were detected in patients with larger proportions of IFN-γ-producing NK cells. This significant inverse association was detected using all three NK cell-triggering strategies and was not restricted to a single activating stimulus (Table 1; Fig. 3A to C). Representative examples of two HIC patients with different proportions of NK cells producing IFN-γ and highly diverging HIV DNA reservoirs (8 versus 122 copies/105 CD4 cells) are shown in Fig. 3D and E. Data relevant to these two divergent patients are outlined in Fig. 3A to C by open diamonds and open triangles, respectively. Nonparametric correlation analysis showed a significant correlation for total and integrated DNAs but not for unintegrated DNA (Table 1).
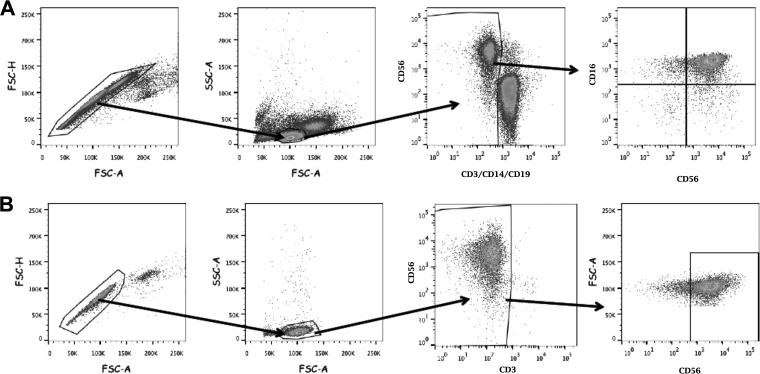
FIG 2.
Gating strategy for selection of NK cells. (A) Gating strategy for NK cell identification, analysis, and sorting among PBMC. (B) Gating strategy for identification and characterization of PBMC-derived, purified NK cells in vitro after activation in vitro.
TABLE 1.
Nonparametric analysis of correlation of quantitated HIV DNA (total, integrated, or unintegrated) with IFN-γ production and with activating receptor-induced expression on NK cells after activation in vitroa
| Graphic representation, data set | Variable 1 | Variable 2 | Spearman ρ | P > |ρ| |
|---|---|---|---|---|
| Fig. 2 | IFN-γ + (a) | I HIV DNA | −0.4858 | 0.0088* |
| IFN-γ + (b) | I HIV DNA | −0.5101 | 0.0013* | |
| IFN-γ + (c) | I HIV DNA | −0.5831 | 0.0002* | |
| Fig. 2, NS | IFN-γ + (a) | T HIV DNA | −0.5976 | 0.0013* |
| IFN-γ + (b) | T HIV DNA | −0.4329 | 0.0190* | |
| IFN-γ + (c) | T HIV DNA | −0.4148 | 0.0486* | |
| IFN-γ + (a) | U HIV DNA | −0.2274 | 0.2851 | |
| IFN-γ + (b) | U HIV DNA | 0.0446 | 0.8286 | |
| IFN-γ + (c) | U HIV DNA | −0.0887 | 0.7098 | |
| Fig. 3A | FI % NKp46 expression | I HIV DNA | −0.5536 | 0.0075* |
| FI % NKp30 expression | I HIV DNA | −0.6873 | 0.0033* | |
| Fig. 3B | IFN-γ + (a) | FI % NKp46 expression | 0.7169 | 0.0130* |
| FI % NKp30 expression | IFN-γ + (a) | 0.7945 | 0.0035* | |
| Fig. 3C | I HIV DNA | FI MFI for NKp46 expression | −0.6561 | 0.0205* |
| I HIV DNA | FI MFI for NKp30 expression | −0.6333 | 0.0365* | |
| IFN-γ + (a) | FI MFI for NKp46 expression | 0.8389 | 0.0024* | |
| IFN-γ + (a) | FI MFI for NKp30 expression | 0.7939 | 0.0061* | |
| Fig. 3, NS | FI % NKp46 expression | T HIV DNA | −0.6423 | 0.0032* |
| FI % NKp30 expression | T HIV DNA | −0.6527 | 0.0039* | |
| FI % NKp46 expression | U HIV DNA | 0.056 | 0.7117 | |
| FI % NKp30 expression | U HIV DNA | −0.1989 | 0.1956 | |
| T HIV DNA | FI MFI for NKp46 expression | −0.6555 | 0.0207* | |
| U HIV DNA | FI MFI for NKp46 expression | −0.487 | 0.0657 | |
| T HIV DNA | FI MFI for NKp30 expression | −0.6872 | 0.0195* | |
| U HIV DNA | FI MFI for NKp30 expression | −0.5612 | 0.0724 | |
| Fig. 4A | IFN-γ + (a) | I HIV DNA | −0.6048 | 0.0003* |
| IFN-γ + (b) | I HIV DNA | −0.4375 | 0.0053* | |
| Fig. 4B | FI % NKp46 expression | I HIV DNA | −0.5954 | 0.0056* |
| FI % NKp30 expression | I HIV DNA | −0.831 | <.0001* | |
| Fig. 4C | IFN-γ + (a) | I HIV DNA | −0.7185 | 0.0127* |
| IFN-γ + (b) | I HIV DNA | −0.7219 | 0.0121* | |
| Fig. 4D | FI % NKp46 expression | I HIV DNA | −0.0251 | 0.9489 |
| FI % NKp30 expression | I HIV DNA | −0.2523 | 0.5852 | |
| Fig. 4F | FI % NKp30 expression | % cytolysis | 0.7107 | 0.0142* |
| Fig. 4, NS | IFN-γ + (a) | T HIV DNA | −0.5976 | 0.0013* |
| IFN-γ + (b) | T HIV DNA | −0.4329 | 0.0190* | |
| FI % NKp46 expression | T HIV DNA | −0.5051 | 0.0231* | |
| FI % NKp30 expression | T HIV DNA | −0.5258 | 0.0250* | |
| IFN-γ + (a) | T HIV DNA (only PP) | −0.7165 | 0.0059* | |
| IFN-γ + (b) | T HIV DNA) (only PP) | −0.8988 | <0.0001* | |
| FI % NKp46 expression | T HIV DNA (only PP) | 0.0418 | 0.9149 | |
| FI % NKp30 expression | T HIV DNA (only PP) | −0.1441 | 0.7578 | |
| IFN-γ + (a) | U HIV DNA | −0.2274 | 0.2851 | |
| IFN-γ + (b) | U HIV DNA | 0.0446 | 0.8286 | |
| FI % NKp46 expression | U HIV DNA | −0.2325 | 0.324 | |
| FI % NKp30 expression | U HIV DNA | −0.3976 | 0.114 | |
| IFN-γ + (a) | U HIV DNA (only PP) | 0.1215 | 0.7219 | |
| IFN-γ + (b) | U HIV DNA (only PP) | 0.4014 | 0.221 | |
| FI % NKp46 expression | U HIV DNA (only PP) | 0.1187 | 7611 | |
| FI % NKp30 expression | U HIV DNA (only PP) | 0.3964 | 0.3786 | |
| NKG2D expression on resting NK cells | I HIV DNA | −0.1258 | 0.4518 | |
| NKG2D expression on resting NK cells | T HIV DNA | −0.1832 | 0.2516 | |
| NKG2D expression on resting NK cells | U HIV DNA | 0.0519 | 0.7603 |
NS, not shown; I, integrated DNA; T, total DNA; U, unintegrated DNA; IFN-γ + NK, % CD56+ IFN-γ+ NK cells; FI, fold increase; (a), after rhIL-12-plus-rhIL-15 activation; (b), NK cells after anti-NKp46 and anti-NKp30 MAb activation; (c), after anti-NKG2D and DNAM-1 activation. HIV DNA was reported as the number of copies/10e4 CD4 cells. Group composition refers to the group comparison in the respective figure (indicated in column 1), unless otherwise specified. Asterisks in the final column indicate ••••.
FIG 3.
HIV DNA is inversely correlated with induced IFN-γ production in NK cells from HIC. (A to C) Linear correlation analysis of quantitated integrated HIV DNA (copies/105 CD4+ cells) and purified NK cell IFN-γ production upon triggering with activating receptors or cytokines. (A) Anti-NKp46 and anti-NKp30. Data are representative for 17 patients. (B) Anti-NKp30 and anti-DNAM-1. Data are representative for 16 patients. (C) rhIL-12 plus rhIL-15. Data are representative for 12 patients. (D and E) Flow cytometric analysis of peripheral NK cells producing IFN-γ from two patients with widely diverging HIV DNA copy numbers (marked with open diamonds and open triangles in panels A to C), i.e., high (D) and low (E) HIV DNA copy numbers. CD3− cells were gated by flow cytometric analysis and analyzed for CD56 and CD16 expression and for intracellular IFN-γ production.
Taken together, these data indicate that the size of the HIV DNA reservoir is closely associated with the ability of NK cells to produce IFN-γ in response to multiple triggering signals mimicking signaling of NCR-ligand recognition on target cells (e.g., B7-H6 through NKp30 or PVR/Nectin-2 via DNAM-1) or signaling by mature dendritic cells (DCs) (e.g., DNAM-NKp30 or recombinant human interleukin-12 [rhIL-12]/rhIL-15). The overall ability to produce IFN-γ contributes to up to 10-fold differences (as indicated by the HIV DNA range) (Fig. 1A and B) in the control of the lentivirus reservoir even among a homogeneous cohort of HIC patients with excellent control of viral replication.
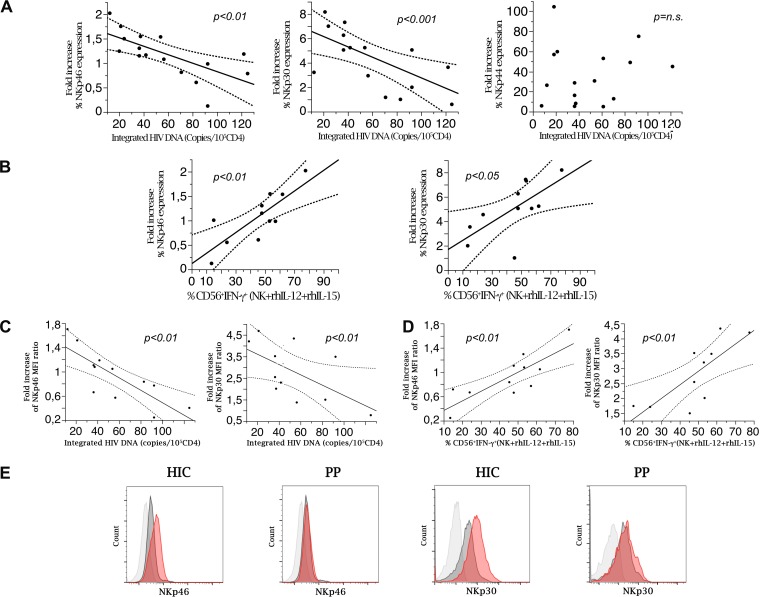
Inducible expression of NKp46 and NKp30 upon NK cell activation correlates with HIV DNA reservoir and with IFN-γ production.
We next studied whether inducible NCR expression by NK cells, which has been associated with benign disease courses in different chronic infections (tuberculosis [TB] [57], HCV infection [31], and HIV infection [30, 58]), can also contribute to control of the size of the virus reservoir. To this end, flow cytometric analysis of NCR expression was performed on highly purified NK cells before and after rhIL-2 activation (see Fig. 2 for the gating strategy). The inducibility of NCRs (Fig. 4E), expressed as the fold increase over baseline (see Materials and Methods), was then correlated with the number of HIV DNA copies. As shown in Fig. 4A and C and Table 1, the inducibility of both NKp46 and NKp30, expressed as proportions of expressing cells and also as molecule density (mean fluorescence intensity [MFI]) on NK cells, was inversely correlated with the amount of HIV DNA (number of copies/105 CD4+ PBMC) detected in PBMC from HIC patients (P < 0.01 and P < 0.001, respectively). Similar correlations were detected for total HIV DNA but not for unintegrated DNA (Table 1). No correlation was observed between NKp44 inducibility and reservoir size (Fig. 4A).
FIG 4.
Induced NCR expression by NK cells is inversely associated with the HIV DNA reservoir and correlates with IFN-γ production. (A) Integrated HIV DNA copy numbers are inversely correlated with the fold increase in NKp46 or NKp30 expression on purified NK cells after in vitro activation with rhIL-2 in HIV controller patients. (B) Peripheral NK cell IFN-γ production is directly correlated with the fold increase in NCR (NKp46 and NKp30) expression on purified activated NK cells in HIV controller patients. The graphs show correlation analysis of NK cells producing IFN-γ in response to cytokine stimulation (rhIL12+rh-IL-15) and fold increases of NKp46 and NKp30 expression on purified NK cells after in vitro activation with rh-IL-2 (n = 12). (C) The HIV DNA reservoir in HIC (n = 12) is inversely correlated with fold increase of NCR molecule density, expressed as MFI, on purified NK cells after in vitro activation with rh-IL-2. Molecule densities are expressed as ratios of sample MFI to control MFI to account for intersample variability (P < 0.01). (D) Peripheral NK cell IFN-γ production in response to cytokine stimulation (rhIL-12/rhIL-15) is directly correlated with fold increases of NKp46 and NKp30 molecule density, expressed as MFI, on purified NK cells after in vitro activation with rhIL-2 (P < 0.01). Data are representative for 12 different patients. (E) Representative shifts in NKp46 and in NKp30 molecule expression and density after stimulation with rIL-2 in vitro for highly purified NK cells in former progressor patients after 24 month of successful cART (PP) and in HIV controller patients (HIC). The graphs show the results of flow cytometric analysis and overlay histogram representations. Light gray histogram, negative control; dark gray histogram, resting cell NCR (NKp46 and NKp30) expression; red histogram, NCR expression after 48 h of culture in the presence of rIL-2 200 IU/ml.
Analysis of IFN-γ production and NCR induction upon NK cell activation revealed that these two NK cell functions are directly correlated. Indeed, by inducing in vitro activation with rhIL-12 plus rhIL-15—i.e., a condition independent of NCR triggering and a proxy for NK cell activation by mature DCs—we detected a direct correlation between IFN-γ secretion and de novo expression of NKp46 and NKp30 (P < 0.01 and P < 0.05, respectively) (Fig. 4B and D).
Thus, these findings indicate that the size of the lentivirus reservoir in a subset of patients with efficient control of virus replication (HIC) correlates inversely with the functional efficiency of NK cells in terms of not only IFN-γ production but also inducible expression of NKp46 and NKp30.
NK cell function is associated with control of HIV DNA in PBMC with divergent disease courses.
The inverse association between two distinct NK cell functional capabilities and the size of the HIV DNA reservoir in HIC raised the question of whether this could be applied only to the minority (<0.5 to 2%) of patients with exceptionally conserved NK cell function (HIC) or if it could represent a general mechanism also applying to patients who do not control HIV-1 replication and have progressive disease, CD4+ T cell loss, and the need for ART (PP).
To address this question, we analyzed NK cell function (IFN-γ production and induction of NCR expression) in a group of stably virologically suppressed PP on combination ART (cART). HIV DNA levels in PBMC from these patients were significantly higher than HIV DNA levels in the PBMC of HIC patients (17 versus 10 copies/105 CD4+ PBMC; P < 0.0001 by the U test) (Fig. 1).
Analyses of pooled (PP plus HIC) patient samples confirmed the inverse relationship between NK cell function and HIV DNA copies (Table 1). This correlation was detected for both IFN-γ production (Fig. 5A) and induction of NKp46 and NKp30 expression versus HIV DNA copies in PBMC (Fig. 5B). Following pooled analysis, PP samples were also evaluated alone. As with HIC (Fig. 4), an inverse relationship was observed between virus reservoir size and IFN-γ production (Fig. 5C), while this was not the case for NCR inducibility (Fig. 5D). This was not unexpected, as a lack of NKp46 and NKp30 NCR inducibility (but not NKp44 inducibility) is a hallmark of previously progressive disease even after successful cART (30).
FIG 5.
NK cell function inversely correlates with HIV DNA reservoir size across widely divergent disease courses. (A) Linear correlation analysis of integrated HIV DNA with IFN-γ production by NK cells triggered by activating receptors (NKp46 and NKp30) (n = 27) or by cytokines (rhIL-12 plus rhIL-15) (n = 27) in HIC (dots) and PP. (B) Correlation analysis of integrated HIV DNA and fold increases of NCR (NKp46 [n = 21] and NKp30 [n = 19]) expression on purified NK cells after in vitro activation with rhIL-2 in HIC (dots) and PP (x). Data are representative of 21 experiments. (C) Correlation analysis for PP alone (x) of integrated HIV DNA and IFN-γ production by NK cells triggered by activating receptors (NKp46 and NKp30) (n = 10) or by cytokines (rhIL-12 plus rhIL-15) (n = 10). (D) Correlation analysis for PP alone (x) of integrated HIV DNA and fold increases of NCR (NKp46 [n = 9] and NKp30 [n = 7]) expression on purified NK cells after in vitro activation with rh-IL-2 (n.s., not significant). (E) Purified NK cell cyototoxicity before and after 2 and 4 days of in vitro activation in the presence of rhIL-2 (200 U/ml). A PKH-26/TO-PRO3 flow cytometric cytotoxicity assay was performed. Box plots indicate the percentages of target cell lysis in the presence of NKp30-specific MAbs in HIC (n = 14) and PP (n = 7). In the box plots, bars represent range, lines represent median values, and box tops and bottoms represent 75th and 25th percentiles, respectively (P < 0.01 by the Mann–Whitney U test). (F) Correlation analysis of NK cell cytotoxicity and NKp30 induction upon in vitro rhIL-2 activation in HIC (n = 6) and PP (n = 5). HIC, HIV-controller patients; PP, progressor patients who were aviremic and needed cART to control viral replication.
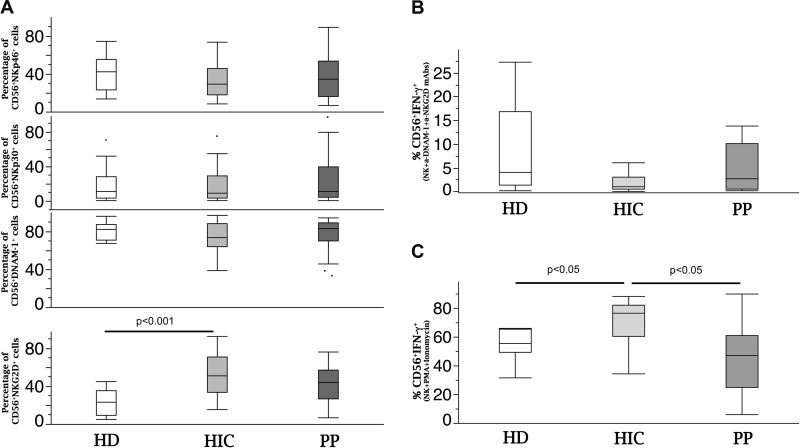
Since expression of NCRs is clearly inducible only in HIC, not in PP, we verified whether differences in cytotoxic activity would actually reflect differences in activating surface receptor expression after in vitro activation of purified NK cells. Indeed, levels of NCR-mediated redirected killing of target cells were similar for HIC and PP at baseline (freshly drawn purified NK cells; T0), while they were significantly increased in HIC versus PP after 2 and 4 days of in vitro activation in the presence of IL-2 (P < 0.01), with 2- to 10-fold increases over baseline values for different patients (Fig. 5E). Interestingly, NK cell cytotoxicity after activation in vitro directly correlated with the fold increase in NCR expression (Fig. 5F). It should be noted that the differences observed in functional induction of NCRs did not correspond to differences in baseline expression of the NKp30, NKp46, or DNAM-1 activating receptor on NK cells (Fig. 6A; see Fig. 2 for the gating strategy), and the frequencies of NKp30+ and NKp46+ NK cells did not correlate with IFN-y production. In addition, IFN-y production inversely correlated with the reservoir size not only under conditions with NK cell triggering via NKp46/30 but also via IL-15/IL-12 and NKp30/DNAM-1 triggering. These findings indicate that the key functional feature with regard to IFN-y production is not represented by basal receptor expression but rather by intrinsic production upon polyfunctional triggering.
FIG 6.
Baseline expression of activating receptors on NK cells and IFN-γ-producing potential under NKG2D and DNAM-1 triggering and under maximal stimulation. (A) Flow cytometric analysis of peripheral NK cells in HD (white), HIC (light gray), and PP (dark gray). (B and C) IFN-γ production by NK cells upon NKG2D- and DNAM-1 triggering (B) or via PMA and ionomycin (C). For box plot analysis, bars represent ranges, lines represent median values, and box tops and bottoms represent 75th and 25th percentiles, respectively (P < 0.05 or P < 0.001 by the Mann–Whitney U test). HD, healthy donors (n = 10); HIC, HIV controller patients (n = 20); PP, progressor patients (n = 10).
With regard to NKG2D, its expression was increased in HIC compared to that in healthy donors (HD) (Fig. 5A), with comparable IFN-γ production upon its triggering in combination with DNAM-1 to reproduce the interaction with DCs in vitro (Fig. 6B). No defect emerged in the general potential of NK cells from PP to produce IFN-γ compared to that of cells from HD, as determined following maximal stimulation with phorbol myristate acetate (PMA) plus ionomycin (Fig. 6C), and HIC had increased IFN-γ-producing potential compared to both PP and HD (Fig. 6C).
Thus, these results show that inducibility of NCR expression parallels acknowledged important NK cell functions, such as IFN-γ production, and contributes to editing downstream NCR-mediated NK cell cytotoxicity. The relationship of these functional activities with quantitative HIV DNA assessment independent of the ability to control HIV-1 replication and CD4+ cell loss suggests that different set points for the virus reservoir (HIV DNA) are associated with a continuum of quantitative NK cell functions. The satisfactory control of HIV DNA copies observed not only in HIC but also in some PP after successful ART suggests a preferential role for in vivo NK cell function in the control of the lentivirus burden (DNA) even when failing to control the viral load/replication (RNA).
Distinct NK cell transcriptional signatures underlie the control of reservoir size.
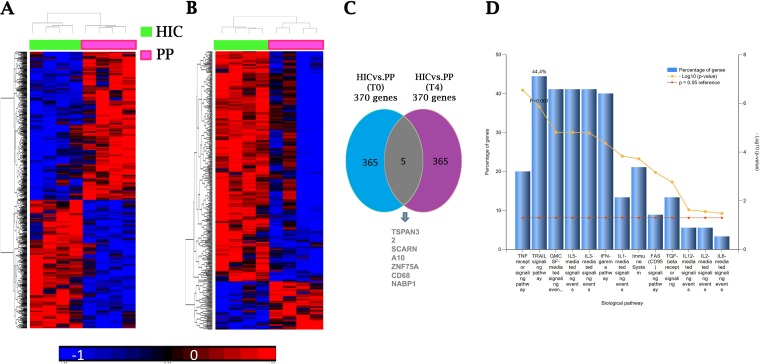
Upon the identification of IFN-γ production and the extent of NCR (NKp46 and NKp30) modulation as inverse correlates of HIV reservoir size in CD4+ PBMC, we decided to address some open questions, including whether the strong correlations observed could indeed be based on substantial differences in transcriptional programs. We wanted to test the possibility that increased transcripts for NKp46 (NCR1) and NKp30 (NCR3) could be detected, or rather whether the increased de novo expression of NCR in NK cells from HIC patients would depend predominantly on posttranscriptional regulation. In addition, we also wanted to explore additional differences in transcriptional signatures that could help to identify specific surface markers for future selection strategies.
To this end, we performed a full transcriptional characterization of purified peripheral NK cells. Multiple samples of purified NK cells, either resting or activated in vitro with rhIL-2, were obtained from different patients (4 HIC and 4 PP) and evaluated by microarray analysis. Comparative transcript analysis (Student's t test cutoff, P < 0.05) identified 370 genes which were differentially expressed by purified resting NK cells derived from HIC and PP (Fig. 7A). Interestingly, a set of 370 transcripts were also found to be differentially expressed in NK cells of HIC versus PP following rhIL-2 stimulation (Student's t test cutoff, P < 0.05) (Fig. 7B). By assessing the identities of the transcripts that were differentially expressed under resting and activated conditions, however, only 5 genes were found to be overlapping between the two sets of 370 transcripts (Fig. 7C). In NK cells derived from HIC patients, of the 370 genes differentially expressed upon rhIL-2 stimulation, 306 (see Table S1 in the supplemental material) and 64 (Table S2) were upregulated and downregulated, respectively. Interestingly, genes encoding activating receptors of NK cells, such as NCR3 (NKp30) and NCR1 (NKp46), were found to be upregulated in NK cells derived from HIC patients upon rhIL-2 stimulation (Table S1), thus indicating that in NK cells from these patients, NCR induction is transcriptionally regulated and differs from that in PP. Under the same conditions, other transcripts coding for activating NK cell receptors were also upregulated, including NKG2D (KLRK1), NKG2F (KLRC4), and NKp80 (KLRF1), while transcripts coding for an inhibitory receptor (CTLA4) were downregulated. Transcripts for NKG2C (KLRC2) showed only a trend toward increased transcription (P = 0.17; fold increase = 2.5). In addition, significantly increased expression of the inhibitory receptor transcripts KIR2DL4 and KIR3DL3 was detected at baseline for HIC compared to that for PP. At the same time, the induction of KIR3DS1 confirmed previous reports (37) and may contribute to HIV DNA control only in the fraction of HIC carrying the appropriate HLA class I molecules.
FIG 7.
Whole-genome microarray analysis demonstrates the existence of differential gene expression patterns of NK cells between HIC and PP. (A) Heat map based on genes differentially expressed HIC versus PP (P < 0.05 by the Student t test and fold change of >1.5) (370 genes for time T0). (B) Heat map based on genes differentially expressed in HIC versus PP (P < 0.05 by the Student t test and fold change of >1.5) (370 genes for time T4). (C) Venn diagram showing genes in common between T0 and T4. (D) Gene Ontology enrichment analysis of purified NK cells by FunRich. Biological pathways overrepresented in genes differentially expressed in HIC versus PP are shown. T0, resting NK cells; T4, NK cells after 4 days of IL-2 culture; HIC, HIV controller patients (n = 4); PP, progressor patients (n = 4).
We then explored the functional pathways among the differentially expressed transcripts by FunRich analysis. The entire cohort of 370 transcripts that were differentially expressed between HIC and PP following rhIL-2 stimulation showed an involvement of IFN-γ, rhIL-2/rhIL-12, and tumor necrosis factor (TNF) receptor signaling pathways (Fig. 7D). Furthermore, functional analysis performed by use of the David tool showed that genes that were upregulated in HIC upon rhIL-2 stimulation (n = 306) were mostly involved in the NF-κB pathway, in lysosome signaling, and in mechanisms shared with lymphocyte cytotoxicity (Table S3).
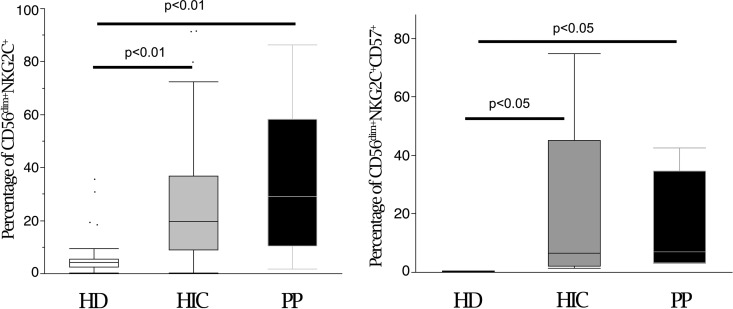
The different transcriptional signatures of purified NK cells in HIC may reflect an inherent, and possibly genetically encoded, regulation of NK cell function, or rather may derive from a memory-like expansion of one or a few NK cell subsets following persistent lentiviral infection. Both the NKG2C activating receptor and CD57 are expressed on CD56dim memory-like NK cells that undergo expansion after HCMV and other virus infections (39, 40). Therefore, we studied the expression of these markers on NK cells from our patient cohorts. In line with the KLRC2 findings by microarray analysis, flow cytometric analysis of peripheral NK cells from HIC and PP showed similar proportions of memory-like NK cells (HIC versus PP) in terms of both CD56dim NKG2C+ and CD56dim CD57+ NKG2C+ NK cells, while these were virtually absent in HD, as expected, due to a lower CMV seroprevalence (Fig. 8). Taken together, these results show that differences in major NK cell functional activity in patients who control the lentiviral reservoir (HIV DNA) rely on inherent NK cell transcriptional regulation, with a distinctive signature involving primarily several activating NK cell receptors. Thus, an NK cell enrichment that is sufficiently high to be detected clearly by macroarray analysis is present. This was unrelated to differential involvement in HIV-1 reservoir control of the whole subset of memory-like (NKG2C+ CD57+) NK cells. However, the possibility cannot be discarded that a particular subset of cells in this memory-like population may be involved and may encompass adaptive NK cells with inducible NCR/NKG2D/NKp80, as indicated by in vitro and microarray analyses.
FIG 8.
NK cells from HIC and PP show similar proportions of memory-like NK cells. CD3− CD14− CD19− cells were gated by flow cytometric analysis and analyzed for CD56 and NKG2C/CD57 expression in HD (white), HIC (gray), and PP (black). In the box plots, bars represent ranges, lines represent median values, and box tops and bottoms represent 75th and 25th percentiles, respectively (P < 0.05 or P < 0.01 by the Mann-Whitney U test). Data are representative of 30 experiments. HD, healthy donors (n = 10); HIC, HIV controller patients (n = 10); PP, progressor patients (n = 10).
Transcriptionally unique NK cells from HIC patients control HIV integration in CD4+ T cells in vitro.
In order to directly verify that differences in NK cell function and significant differences in transcriptional signature indeed determine different DNA integration statuses and contribute to the lentivirus reservoir size, we then studied the effect of coculturing NK cells in an in vitro infection experimental model.
Purified activated HD CD4+ blasts were cultured in vitro and infected with HIV-1Bal at a high multiplicity of infection (MOI = 0.1). After removal of excess virus, highly purified (>98%) activated NK cells from patients were added to the cultures at a 0.5:1 (NK:CD4) cell ratio. This experimental design aimed at studying in vitro control of the DNA reservoir by NK cells while monitoring successful infection as represented by HIV RNA/p24. To explore the effects of predominant IFN-γ production and NCR expression on infected CD4+ cells and to minimize the physiological inhibitory effect of KIR:HLA engagement, preactivation of purified NK cells was performed before coculture in an allogeneic system.
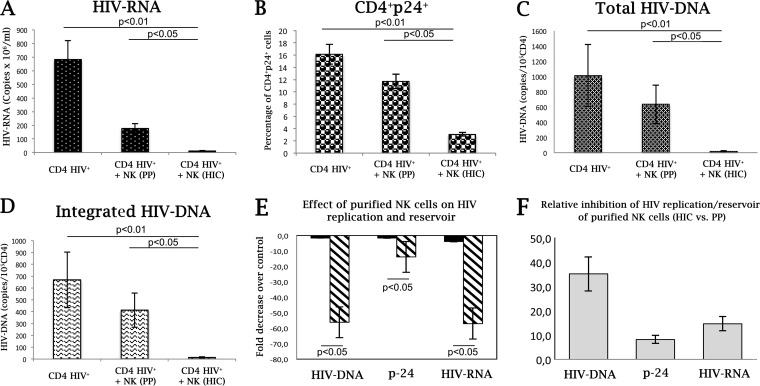
Virus replication, as measured by numbers of supernatant RNA copies and by HIV p24 expression by infected CD4+ T cells, peaked at day 7 after infection. NK cells from HIC patients efficiently controlled virus replication compared to those from PP (Fig. 9A and B). An 88% reduction of p24+ CD4+ T cells was detected in culture by flow cytometry, and a 93% reduction in HIV RNA was detected by quantitative reverse transcription-PCR (qRT-PCR). Quantitative evaluation of HIV DNA was performed by RT-PCR and showed that HIC NK cells sharply controlled total and integrated HIV-1 DNA accumulation during peak virus replication, with 98% and 96% reductions in total and integrated HIV DNAs, respectively, compared to the levels in cultures supplemented with purified NK cells from PP (Fig. 9C and D). In comparing the effects of NK cells from different donors (HIC versus PP) on HIV DNA accumulation (Fig. 9E), purified HIC NK cells with specific transcriptional signatures had a 35-fold larger effect than that of NK cells from PP (Fig. 9F). A more contained comparative potency in containing HIV-1 replication was observed (14- and 8-fold for HIV RNA and HIV p24, respectively) (Fig. 9F). Compared to NK cells from PP, lacking NCR and IFN-γ inducibility, the presently characterized NK cells, with a characteristic function and transcriptional signature, had an approximately 3-fold higher efficacy in containing HIV DNA accumulation over HIV replication.
FIG 9.
Purified NK cells from HIC patients control HIV integration in vitro. IL-2/PHA CD4+ T lymphocytes were infected with HIV-1Bal at an MOI of 0.01 in the presence or absence of activated NK cells from PP or HIC. After 7 days of culture, virus replication was monitored by measuring HIV RNA (copies per milliliter) in the supernatant (A), p24 levels via intracellular staining of CD3+56− T cells by flow cytometry (B), and viral reservoir accumulation by quantitative assay of total HIV DNA (copies/105 CD4+ cells) (C) and of integrated HIV DNA (D). (E) Fold decreases in virus accumulation in the presence of the given purified NK cells over that in control cultures without NK cells. (F) Relative potencies of purified NK cells from HIC versus NK cells from PP in inhibiting virus replication or DNA integration/accumulation. HIC, HIV controller patients (n = 6); PP, progressor patients (n = 6).
Taken together, these experiments show that NK cells with a specific transcriptional signature and function provide a remarkable contribution to limiting HIV DNA in vitro, thus confirming the in vivo observations.
DISCUSSION
Control of the DNA reservoir, rather than only RNA, represents the main goal for the treatment of lentiviruses, and ultimately for retrovirus eradication and clearance (43, 58–60). However, the immune mechanism(s) contributing to lentivirus DNA reservoir size shaping and control still eludes our full understanding. NK cells are involved in the control of RNA and DNA viruses belonging to different virus families, including the control of retrovirus replication (11, 16, 32, 38, 61).
In the present work, we explored the impact of NK cell function and NK cell characterization on the containment of the lentivirus reservoir in vivo and in vitro by using HIV-1 as a model. The results provide evidence that transcriptionally characterized NK cells substantially contribute to HIV reservoir containment. The induction of IFN-γ and NCR (NKp46 and NKp30) expression represents a quantitative functional correlate of lentiviral reservoir size containment. Previous work showed that NK cells with specific HLA:KIR carriage may control viral replication (RNA production) without information on the HIV reservoir (37) and also that nonprogressing disease may be unrelated to carriage of KIR3DS1 (36). In the present in vivo and in vitro settings, no HLA:KIR carriage was associated with HIC status, and the in vitro experimental strategy was chosen to minimize the interference of HLA:KIR interaction with NCR expression/inducibility and IFN-γ production on HIV DNA accumulation. A doubtless need has emerged for future work to include combined analyses of activating and inhibitory NK cell receptors and of the actual expression of KIR3DL1/3DS1 and of HLA-Bw4 and the role of other soluble factors produced by NK cells in addition to IFN-γ.
The present finding of higher levels of HIV DNA in CD4+ PBMC from cART-treated PP is in line with previous reports (53) and with the association of more rapid disease progression in patients with higher CD4+ T cell-associated circulating HIV DNA (52). In fact, the reservoir size decays with shallower slopes after the first year(s) of cART, remains constant and fails to decrease below detectability in most patients (50, 51), and is independent of CD8+ CTL activity (51). Virus replication invariably resumes upon cART interruption (47, 48, 50), with a remarkable exception to this concept being a very small group of early cART-treated patients (posttreatment controller patients [PTCP]) (4, 5). The present findings on NK cell-associated control of HIV reservoir size provide a novel reading frame for the above observations, suggesting that NK cell functional potential in terms of NCR and IFN-γ inducibility represents two pivotal elements of proviral DNA containment. Indeed, cART has a remarkable impact on HIV replication in CD4+ cells; however, it has limited effects on the recovery of NK cell function (62, 63) and on DNA size reduction (51). An adaptive CTL-mediated control of DNA reservoir size is unlikely, as suggested by the heterogeneity of CD8+ CTL function in PTCP (64) and by the lack of correlation between CD8+ CTL activation and the DNA reservoir (51). Thus, according to the present data, the failure to fully reconstitute NK cell function upon cART in PP is likely to contribute to the limited decrease in HIV reservoir size over time (50, 51), and ultimately to the renewed HIV replication observed upon treatment interruption in most patients (47, 48). In view of the reported lack of CD8+ CTL control in PTCP, it may be hypothesized that, in these patients, NK cells may represent an important immune mechanism leading to HIV reservoir and spontaneous posttreatment control.
cART has been shown to reconstitute CD4+ cell numbers, to leave NK cell phenotype and function largely unaffected, and to provide a minimal contribution to HIV reservoir decrease. For this reason, it appears unlikely that the effects on HIV reservoir size and NK cell function in treated patients are due to cART. It remains to be determined whether some patients with active replication and progression to low CD4 numbers may still limit the reservoir size in the presence of robust innate immune defenses (e.g., NK cell function) or whether, in some PP, cART may also be effective for NK cell function and contribute to the HIV reservoir. Further focus is needed to understand whether the NK cell function/transcriptional profile is preset and innate or may be affected by virus replication or by cART and to what extent poor control of the CD4+ cell HIV reservoir by poor IFN-γ producers may be improved.
If confirmed, the present characterization of transcriptionally and functionally distinctive NK cells may be exploited for the prospective identification of HIV patients with good chances of success for undergoing cART simplification to monotherapy (65) or cART interruption (e.g., PTCP) (58, 66, 67).
In the present analysis, NK cells controlling HIV-1 reservoir size displayed a set of 370 differentially expressed NK cell genes/transcripts that included, among others, the principal functional NK cell pathways (i.e., cytokine production, cytokine response, and NCR induction) that were assessed by phenotypic and functional assays. This indicates that an inherent individual regulation of NK cell gene expression underlies functional differences and affects the control of the HIV-1 reservoir set point and disease course (52). This finding is in line with previous observations showing that the NK cell transcriptional signature underlies specific disease courses in chronically HCV-infected patients who respond to treatment (58, 68). In view of the considerable size of the NK cell phenotype spectrum, with up to 30,000 to 100,000 phenotypes (13), the detection of a specialized NK cell population of sufficient size to identify a distinct transcriptional signature may reflect enrichment of circulating NK cells, thus raising the question of their origin. NK cell-mediated memory-like reactions with large expansions of NKG2C+ CD57+ NK cells have been detected during infections with different viruses in humans (38, 42, 69) and in animal models (40). Enrichment of memory-like NK cells of known phenotype (NKG2C+ CD57+) was not involved here, since both patient groups (HIC and PP) had similarly large circulating pools of putative memory-like (NKG2C+ CD57+) NK cells, while functional and transcriptional differences were detected only for patients with the smaller lentiviral reservoir size (HIC). Support of the view that control of the lentiviral reservoir cannot be ascribed to imbalances of the memory-like NK cell subset as a whole is further provided by the efficient IFN-γ production by NK cells upon rhIL-2 or rhIL-12/rhIL-15 stimulation. This is in contrast to the diminished responses to rhIL-12 and rhIL-18 in NKG2C+ NK cells from HCMV donors (42) and to reduced IFN-γ production upon H1N1 influenza virus antigen or pertussis toxin stimulation (70). Moreover, memory-like NK cell expansion in HIV patients is due to HCMV rather than to persistent HIV-1 infection (71). Therefore, two possible hypotheses may be proposed to explain our present findings. First, the enrichment in NK cells with such a distinct transcriptional signature in patients with a small HIV-1 reservoir size may represent a novel type of “trained” or adaptive NK cells, either independent of or representing a specialized subset of “memory-like” NKG2C+ NK cells that is expanded and functional in HIC but not in PP. These cells may be identified by their ability to induce transcription of NKp46, NKp30, NKG2D, and NKp80 while retaining CD57 and NKG2C expression. Alternatively, in agreement with the high environmental influence of NK cell activating receptor expression (13), they may represent an inherent rather than an acquired inborn feature, therefore predating HIV infection and contributing to lentiviral containment following virus entry. In this case, their expansion may be detected in the healthy population at a low frequency (about 1 to 2%) superimposable to HIC prevalence among HIV-infected patients. Further work is needed to fully identify the origin and the presence at low frequency of these phenotypes in the overall population.
Finally, current programs and protocols for HIV eradication, including histone deacetylase inhibitor (HDACi) administration and immunotherapy, reportedly need improved immunotherapeutic strategies to clear virus in reactivated cells and to limit new virus integration (43, 55, 59, 72, 73). Selection and expansion of NK cells with high NCR and IFN-γ inducibility may represent a useful tool to efficiently clear HIV-1 when combined within the frame of virus eradication strategies exploiting latency exit induction and immuno-/antiretroviral therapy (72–74).
In conclusion, HIV reservoir size is dependent on the presence of a defined NK cell population with a specific transcriptional signature: high NCR (NKp46 and NKp30) and IFN-γ inducibility upon NCR and cytokine receptor engagement. These cells may represent a novel tool offering relevant promise for translational applications, including patient monitoring, treatment interruption, and virus eradication.
MATERIALS AND METHODS
Patients.
Five infectious disease units caring for HIV patients enrolled 20 HIV-1-infected controller patients (10 EC and 10 LTNP) and 10 HIV-1-infected patients with progressive disease (Table 2). Patients were being monitored within a program for surveillance and treatment of HIV-1 infection. EC patients had positive HIV-1 serology, were ART naive, and had CD4+ counts of ≥450 cells/μl at all visits for ≥7 years of follow-up, with no clinical evidence of disease progression. LTNP met the same definitions as those for EC, with the exception of HIV RNA being detectable during the years of observation, at <2,000 copies/ml. A group of HIV-infected progressor patients on ART with HIV RNA levels of <50 copies/ml for at least 24 months and CD4+ T cell levels of >350/μl were considered aviremic progressor controls (progressor patients [PP]). For these patients, evidence from clinical records at cART initiation showed that they had viral replication and low CD4+ cell numbers. HD (n = 10) were recruited locally among blood bank donors. Carriage of KIR:HLA alleles by HIC and PP was analyzed as described previously (30). No significant association for HLA-Bw4:KR3DL1/S1 was detected in HIC versus PP.
TABLE 2.
Demographic and clinical patient dataa
| Patient group | Age (yr) | No. of males | No. of females | % CD4+ cells at baseline | No. of CD4+ cells at baseline | Last % CD4+ cells | Last CD4+ count | Length of HIV+ status (yr) |
|---|---|---|---|---|---|---|---|---|
| EC | 52 ± 4.41 | 3 | 7 | 35 ± 6.59 | 1,033 ± 479.8 | 35.8 ± 5.07 | 739.5 ± 347.6 | 20 ± 4.4 |
| LTNP | 49 ± 12.2 | 4 | 6 | 36 ± 7.83 | 876 ± 378 | 34.6 ± 8.33 | 776 ± 362 | 18 ± 8.3 |
| PP | 51 ± 5.9 | 4 | 6 | 9.9 ± 5.56 | 85 ± 75.5 | 24.6 ± 8.33 | 325 ± 182 | 17 ± 4.9 |
With the exception of gender data, numbers represent means ± SD.
Exclusion criteria were current or previous ART, age of <18 years, pregnancy, cancer, and treatment for HCV infection during the previous 6 months.
The study was authorized by the local ethical committee. Patients were enrolled after providing informed consent. Peripheral blood mononuclear cells (PBMC) were obtained by density gradient centrifugation (Ficoll-Hypaque) and processed or cryopreserved at −80°C.
Abs.
The following panel of mouse anti-human monoclonal antibodies (MAbs) was used: PC7-conjugated anti-CD56 (clone N901; Immunotech-Coulter), fluorescein isothiocyanate (FITC)- and allophycocyanin (APC)-conjugated anti-CD3 (clone HIT3A; BioLegend), APC-conjugated anti-CD19 and anti-CD14 (clones HIB19 and M5E2, respectively; BioLegend), phycoerythrin (PE)- and FITC-conjugated anti-CD16 (clone 3G8; BD Pharmingen), 7-aminoactinomycin D (7AAD) (BD Pharmingen), anti-NKp46 (BAB281 and IgG1), anti-NKp30 (7A6 and IgG1), anti-NKp44 (ZIN231 and IgG1), anti-NKG2D (BAT221 and IgG1), and anti-DNAM-1 (F22 and IgG1). FITC-conjugated (Southern Biotechnology) and PE-conjugated goat anti-mouse anti-isotype Abs (BD Pharmingen) were used as controls.
Immunofluorescence analysis.
Cells were analyzed by eight-color flow cytometry. Briefly, cells were incubated with primary MAbs followed by PE- or FITC-conjugated anti-isotype-specific goat anti-mouse secondary reagents. Direct staining was performed by use of fluorochrome-conjugated MAbs as the third step. For cytofluorimetric analysis, cells were gated using forward and side light scatter parameters (FACSCantoII; BD), and 10,000 events were always acquired on CD3/14/19− CD56+ gating. MFI was used to quantify molecule density. To reduce interassay variability, the MFI ratio (MFIsample/MFIcontrol) was used to compare samples/groups. Data were analyzed using FlowJo (Tree Star, Inc.). Gating strategies are shown in Fig. 2.
NK cell activating receptor induction assay.
NK cells were isolated from PBMC by negative immunomagnetic separation using an NK cell isolation kit II (Miltenyi Biotec) as previously described (75). NK cell purity was ≥95%.
NKp46, NKp30, and NKp44 expression on highly purified NK cells before and 2 and 4 days after in vitro culture with rhIL-2 (200 U/ml) (Proleukin; Chiron Corp.) was analyzed by flow cytometry. Receptor expression changes were determined by comparing the MFI ratio to the fresh NK cell MFI ratio, as follows: (MFIratio ACT − MFIratio baseline)/MFIratio baseline × 100 (ACT stands for “activated”).
Cytotoxicity assay.
NK cell cytotoxic activity was determined using a PKH26 and TO-PRO3 (Sigma-Aldrich and Invitrogen, respectively) assay as previously described (76). FcγR+ P815 mouse mastocytoma cell lines were used as target cells and were labeled with PKH26 (76). P815-PKH26+ cells were incubated with rhIL-2-activated NK cells at a 1:1 effector:target (E:T) ratio. Cultures were incubated for 4 h at 37°C in 5% CO2 in RPMI 1620 medium (BioWhittaker, Lonza) supplemented with 10% fetal calf serum (FCS) (complete medium [CM]), in the presence or absence of MAbs, and then placed on ice until flow cytometric analysis. Spontaneous and maximal target cell deaths were determined by PKH26 labeling of cells cultured alone and permeabilized with BD Cytofix/Cytoperm reagent (BD Pharmingen), respectively. To identify dead cells, 5 μl of a 10 μM stock solution of TO-PRO3 was added to each tube immediately before analysis. Cells were analyzed by FACSCanto II flow cytometry (BD), and 10,000 events were collected. Specific lysis was calculated by use of the following formula for dye-labeled cells: (sample − spontaneous)/(total − spontaneous) × 100.
IFN-γ production assay.
PBMC were stimulated using FcγR+ P815 target cells at a 10:1 E:T ratio in complete medium in the presence or absence of an anti-NKp30, anti-NKp46, anti-NKp30, and/or anti-DNAM-1 MAb mixture (0.1 μg ml−1), rhIL-12 (20 ng ml−1), and rhIL-15 (40 ng ml−1) (PeproTech), as previously described (75).
HIV infection.
PBMC from healthy HIV-1-seronegative donors were activated with purified phytohemagglutinin (PHA) (0.25% [vol/vol]) and rhIL-2 (50 U/ml) for 5 days in RPMI 1620 complete medium (BioWhittaker, Lonza) supplemented with 10% FCS, l-glutamine (2 mM), and a 1% antibiotic mixture (penicillin-streptomycin [5 mg/ml]). Activated CD4+ cells were positively purified by use of antibody-coated magnetic beads (Miltenyi Biotec) and then cultured with rhIL-2 (50 U/ml) for 1 week. The cells were infected with HIV-1 (HIVBal; a kind gift from G. Poli, Milan, Italy) at an MOI of 0.01 for 2 h at 37°C in a humidified 5% CO2 atmosphere, washed twice to remove excess virus, and cultured (5 × 105 cells/ml) in CM with rhIL-2 (50 U/ml) alone or with purified/activated NK cells in triplicate at a 1:2 ratio. Supernatants and cells were collected on day 7 and cryopreserved.
Total, unintegrated, and integrated HIV DNA quantification.
DNAs were isolated from 1.0 × 106 to 2.0 × 106 PBMC from HIC and PP and from 3.0 × 105 in vitro-infected CD4+ blasts. A constant HIV-1-negative donor background of 1 × 107 uninfected and unstimulated PBMC was mixed with the cell pellets to ensure high DNA extraction efficiencies. Briefly, total and unintegrated HIV DNAs (the ensemble of linear and 1- and 2-LTR circular DNAs) were selectively separated by an optimized chromatographic procedure (a method utilized to separate the high-molecular-weight DNA containing the proviral HIV DNA from the low-molecular-weight DNA, consisting of unintegrated HIV DNA) (77) and analyzed by quantitative SYBR green I real-time PCR (qPCR), using a single set of specific primers selected in the highly conserved LTR-Gag region of the HIV-1 genome (78) and able to detect all HIV-1 subtypes in the M group. The qPCR measurements of 2-LTR circles were performed using primers flanking the dual-repeat cassette within the circular form. PCRs were carried out with a final volume of 50 μl in a model 7500 real-time PCR system (Applied Biosystems, Thermo Fisher Scientific Inc.), using an HIV-1 DNA qPCR kit (Diatheva s.r.l., Fano, Italy) according to the above-developed assay. Each sample was analyzed in triplicate in the first PCR (1st qPCR), which consisted of two wells containing 0.5 μg each plus one well containing 1.0 μg of DNA (to ensure the detection of the target even at a low copy number, i.e., near the quantification limit [QL] [2 copies/PCR]), or the equivalent quantity of elution fraction from chromatographic separation to quantify unintegrated forms. For samples with HIV DNA values of <30 copies (coefficient of variation of 21%), a 2nd PCR experiment was performed. Six 0.5-μg replicates were assayed for samples which in the 1st qPCR had been quantified in the range of 2 to 30 copies; three 0.5-μg replicates and three 1.0-μg replicates were tested for samples which in the 1st qPCR had been quantified near or detected below the QL. In case of negative amplification, a PCR spike test was performed by adding 2 or 10 copies of our plasmid (pPBS) standard to the samples to exclude the presence of inhibitors. The HIV-1 DNA copy number was estimated by interpolation of the experimentally determined threshold cycle (CT) based on standard curves generated using half-log serial dilutions from 103 to 2 copies. The amount of integrated HIV DNA was obtained by subtracting the amount of unintegrated DNA from the amount of total HIV DNA. The agreement between the indirect and direct measurements of integrated HIV DNA has been demonstrated before (A. Casabianca, unpublished data). Total, unintegrated, integrated, and 2-LTR DNA copy numbers were determined by adding up the copy numbers from the 0.5/1.0-μg replicates tested and were expressed relative to 1 μg of DNA and then normalized to numbers of copies/105 CD4+ T cells. Values of <2 copies/μg were considered to have a value of 1 for statistical analyses.
Serum and supernatant HIV RNA levels were quantified by use of a commercial kit according to the manufacturer's instructions (Nuclisens EasyQ HIV-1 2.0; bioMérieux SA).
Microarray analysis.
Total RNA was purified from freshly isolated, purified (>98%) NK cells after flow cytometric sorting for 4 HIC patients and 4 PP by use of miRNeasy minikits (Qiagen, Germantown, MD, USA) according to the manufacturer's protocol. RNA quality and quantity were estimated using Nanodrop spectrophotometry (Thermo Scientific, Waltham, MA, USA) and an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). First- and second-strand cDNAs were synthesized from 10 ng of total RNA by use of a Nugen Ovation Pico WTA system V2 (Nugen Technologies, San Carlos, CA, USA) following the manufacturer's instructions. cDNAs were fragmented and biotinylated by use of the Nugen Encore biotin module (Nugen Technologies, San Carlos, CA, USA) and hybridized to GeneChip Human Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA, USA). The arrays were washed and stained on a GeneChip 450 fluidics station and scanned by a GeneChip 3000 scanner (Affymetrix, Santa Clara, CA, USA). Global gene expression profiling of NK cells was analyzed using Partek Genomics Suite (St. Louis, MO). Functional analysis was performed using David analysis (https://david.ncifcrf.gov/) and FunRich analysis (functional enrichment analysis tool).
Analysis of genetic carriage of HLA and KIR was performed on PBMC as previously described (30) and failed to show significant gene carriage enrichments.
Statistical analysis.
Statistical analysis was performed using the Mann-Whitney U test for comparisons between unpaired data sets. Correlation analysis was performed by Spearman's test and by two-way direct correlation for graphical representation. Analysis was performed using JMP 10.0 (SAS).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help of our patients and of all the colleagues in the infectious disease units who cared for them over the years in Sanremo, Savona, Genova, La Spezia, and Alessandria, Italy (G. Ferrea, P. De Leo, G. Mazzarello, G. Cassola, G. Cenderello, M. Guerra, S. Boni, S. Artioli, F. Bisio, E. Nicco, and E. Mantia), as well as the help with viral load determination from Bianca Bruzzone, whose contribution and dedication were fundamental to this study.
F.M. planned and performed experiments, analyzed data, discussed data, and wrote the manuscript. F.B., A.C., and C.O. performed experiments, analyzed data, discussed results, and contributed to the writing of the manuscript. M.L.A. performed experiments, analyzed and interpreted data, and contributed to manuscript preparation. A.D.B., E.P., C.D., L.N., L.T., and G.O. cared for patients, contributed samples, verified the accuracy of required clinical conditions, discussed data and results, and participated in manuscript revision. M.M., E.W., and F.M.M. supervised experiments and data analysis and participated in discussions of the draft manuscript. L.M. discussed and interpreted the data and wrote the manuscript. A.D.M. supervised patient inclusion and sample collection, planned experiments, analyzed and discussed data, verified data analysis, and wrote the manuscript.
This work was supported by grants awarded by the Associazione Italiana Ricerca sul Cancro (AIRC) (IG 2010 project 10225 and “Special Program Molecular Clinical Oncology 5×1000” project 9962 to L.M.); the Istituto Superiore di Sanità (ISS) (Programma nazionale di ricerca sull’AIDS, Accordi di collaborazione scientifica 45G.11 and 40H69 to A.D.M.); and the Ministero della Salute (RF-2010-2316197 to A.D.M.).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00647-17.
REFERENCES
- 1.Rosenberg EB, Herberman RB, Levine PH, Halterman RH, McCoy JL, Wunderlich JR. 1972. Lymphocyte cytotoxicity reactions to leukemia-associated antigens in identical twins. Int J Cancer 9:648–658. doi: 10.1002/ijc.2910090323. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL, Le AM, Phillips JH, Warner NL, Babcock GF. 1983. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J Immunol 131:1789–1796. [PubMed] [Google Scholar]
- 3.Perussia B, Starr S, Abraham S, Fanning V, Trinchieri G. 1983. Human natural killer cells analyzed by B73.1, a monoclonal antibody blocking Fc receptor functions. I. Characterization of the lymphocyte subset reactive with B73.1. J Immunol 130:2133–2141. [PubMed] [Google Scholar]
- 4.Lanier LL, Chang C, Azuma M, Ruitenberg JJ, Hemperly JJ, Phillips JH. 1991. Molecular and functional analysis of human natural killer cell-associated neural cell adhesion molecule (N-CAM/CD56). J Immunol 146:4421–4426. [PubMed] [Google Scholar]
- 5.Sivori S, Falco M, Marcenaro E, Parolini S, Biassoni R, Bottino C, Moretta L, Moretta A. 2002. Early expression of triggering receptors and regulatory role of 2B4 in human natural killer cell precursors undergoing in vitro differentiation. Proc Natl Acad Sci U S A 99:4526–4531. doi: 10.1073/pnas.072065999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. 1998. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med 187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, Marcenaro E, Accame L, Malaspina A, Biassoni R, Bottino C, Moretta L, Moretta A. 1999. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med 190:1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, Moretta L, Moretta A. 1999. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur J Immunol 29:1656–1666. [DOI] [PubMed] [Google Scholar]
- 9.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. 2001. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol 19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 10.Montaldo E, Del Zotto G, Della Chiesa M, Mingari MC, Moretta A, De Maria A, Moretta L. 2013. Human NK cell receptors/markers: a tool to analyze NK cell development, subsets and function. Cytometry A 83:702–713. doi: 10.1002/cyto.a.22302. [DOI] [PubMed] [Google Scholar]
- 11.Cerwenka A, Lanier LL. 2016. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol 16:112–123. doi: 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 12.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA. 2013. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5: 208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gondois-Rey F, Cheret A, Mallet F, Bidaut G, Granjeaud S, Lecuroux C, Ploquin M, Muller-Trutwin M, Rouzioux C, Avettand-Fenoel V, De Maria A, Pialoux G, Goujard C, Meyer L, Olive D. 2017. A mature NK profile at the time of HIV primary infection is associated with an early response to cART. Front Immunol 8:54. doi: 10.3389/fimmu.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etzioni A, Eidenschenk C, Katz R, Beck R, Casanova JL, Pollack S. 2005. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr 146:423–425. doi: 10.1016/j.jpeds.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Warfield KL, Perkins JG, Swenson DL, Deal EM, Bosio CM, Aman MJ, Yokoyama WM, Young HA, Bavari S. 2004. Role of natural killer cells in innate protection against lethal Ebola virus infection. J Exp Med 200:169–179. doi: 10.1084/jem.20032141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, Zakay-Rones Z, Porgador A, Mandelboim O. 2006. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol 7:517–523. [DOI] [PubMed] [Google Scholar]
- 18.Biron CA, Byron KS, Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 19.Crotta S, Brazzoli M, Piccioli D, Valiante NM, Wack A. 2010. Hepatitis C virions subvert natural killer cell activation to generate a cytokine environment permissive for infection. J Hepatol 52:183–190. doi: 10.1016/j.jhep.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 20.De Maria A, Fogli M, Mazza S, Basso M, Picciotto A, Costa P, Congia S, Mingari M, Moretta L. 2007. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol 37:445–455. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- 21.Amadei B, Urbani S, Cazaly A, Fisicaro P, Zerbini A, Ahmed P, Missale G, Ferrari C, Khakoo SI. 2010. Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology 138:1536–1545. doi: 10.1053/j.gastro.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Maria A, Fogli M, Costa P, Murdaca G, Puppo F, Mavilio D, Moretta A, Moretta L. 2003. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44). Eur J Immunol 33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 23.Fogli M, Costa P, Murdaca G, Setti M, Mingari MC, Moretta L, Moretta A, De Maria A. 2004. Significant NK cell activation associated with decreased cytolytic function in peripheral blood of HIV-1-infected patients. Eur J Immunol 34:2313–2321. doi: 10.1002/eji.200425251. [DOI] [PubMed] [Google Scholar]
- 24.Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, Barker E. 2007. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood 110:1207–1214. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grauwet K, Cantoni C, Parodi M, De Maria A, Devriendt B, Pende D, Moretta L, Vitale M, Favoreel HW. 2014. Modulation of CD112 by the alphaherpesvirus gD protein suppresses DNAM-1-dependent NK cell-mediated lysis of infected cells. Proc Natl Acad Sci U S A 111:16118–16123. doi: 10.1073/pnas.1409485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerboni C, Neri F, Casartelli N, Zingoni A, Cosman D, Rossi P, Santoni A, Doria M. 2007. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol 88:242–250. doi: 10.1099/vir.0.82125-0. [DOI] [PubMed] [Google Scholar]
- 27.Matusali G, Tchidjou HK, Pontrelli G, Bernardi S, D'Ettorre G, Vullo V, Buonomini AR, Andreoni M, Santoni A, Cerboni C, Doria M. 2013. Soluble ligands for the NKG2D receptor are released during HIV-1 infection and impair NKG2D expression and cytotoxicity of NK cells. FASEB J 27:2440–2450. doi: 10.1096/fj.12-223057. [DOI] [PubMed] [Google Scholar]
- 28.Cao Y, Qin L, Zhang L, Safrit J, Ho DD. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med 332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 29.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen OJ, Demarest JF, Montefiori D, Orenstein JM, Fox C, Schrager LK, Margolick JB, Buchbinder S, Giorgi JV, Fauci AS. 1995. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N Engl J Med 332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 30.Marras F, Nicco E, Bozzano F, Di Biagio A, Dentone C, Pontali E, Boni S, Setti M, Orofino G, Mantia E, Bartolacci V, Bisio F, Riva A, Biassoni R, Moretta L, De Maria A. 2013. Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci U S A 110:11970–11975. doi: 10.1073/pnas.1302090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bozzano F, Picciotto A, Costa P, Marras F, Fazio V, Hirsch I, Olive D, Moretta L, De Maria A. 2011. Activating NK cell receptor expression/function (NKp30, NKp46, DNAM-1) during chronic viraemic HCV infection is associated with the outcome of combined treatment. Eur J Immunol 41:2905–2914. doi: 10.1002/eji.201041361. [DOI] [PubMed] [Google Scholar]
- 32.Marras F, Bozzano F, Ascierto ML, De Maria A. 2014. Baseline and dynamic expression of activating NK cell receptors in the control of chronic viral infections: the paradigm of HIV-1 and HCV. Front Immunol 5:305. doi: 10.3389/fimmu.2014.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malnati MS, Ugolotti E, Monti MC, Battista D, Vanni I, Bordo D, Sironi F, Larghero P, Marco ED, Biswas P, Poli G, Vicenzi E, Riva A, Tarkowski M, Tambussi G, Nozza S, Tripodi G, Marras F, De Maria A, Pistorio A, Biassoni R. 2017. Activating killer immunoglobulin receptors and HLA-C: a successful combination providing HIV-1 control. Sci Rep 7:42470. doi: 10.1038/srep42470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31:429–434. [DOI] [PubMed] [Google Scholar]
- 35.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connell KA, Han Y, Williams TM, Siliciano RF, Blankson JN. 2009. Role of natural killer cells in a cohort of elite suppressors: low frequency of the protective KIR3DS1 allele and limited inhibition of human immunodeficiency virus type 1 replication in vitro. J Virol 83:5028–5034. doi: 10.1128/JVI.02551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. 2007. Differential natural killer cell mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med 204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 39.Della Chiesa M, Falco M, Podestà M, Locatelli F, Moretta L, Frassoni F, Moretta A. 2012. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood 119:399–410. doi: 10.1182/blood-2011-08-372003. [DOI] [PubMed] [Google Scholar]
- 40.Sun JC, Beilke JN, Lanier LL. 2009. Adaptive immune features of natural killer cells. Nature 457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petitdemange C, Wauquier N, Devilliers H, Yssel H, Mombo I, Caron M, Nkoghè D, Debrè P, Leroy E, Vieillard V. 2016. Longitudinal analysis of natural killer cells in dengue virus-infected patients in comparison to chikungunya and chikungunya/dengue virus-infected patients. PLoS Negl Trop Dis 10:e0004499. doi: 10.1371/journal.pntd.0004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjorkstrom NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michalsson J, Malmberg K-J, Klingstrom J, Ahlm C, Ljunggren H-G. 2011. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med 208:13–21. doi: 10.1084/jem.20100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maldarelli F. 2016. The role of HIV integration in viral persistence: no more whistling past the proviral graveyard. J Clin Invest 126:438–447. doi: 10.1172/JCI80564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pallikkuth S, Sharkey M, Babic DZ, Gupta S, Stone GW, Fischl MA, Stevenson M, Pahwa S. 2015. Peripheral T follicular helper cells are the major HIV reservoir within central memory CD4 T cells in peripheral blood from chronically HIV-infected individuals on combination antiretroviral therapy. J Virol 90:2718–2728. doi: 10.1128/JVI.02883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Descours B, Petitjean G, Lopez-Zaragoza JL, Bruel T, Raffel R, Psomas C, Reynes J, Lacabaratz C, Levy Y, Schwartz O, Lelievre JD, Benkirane M. 2017. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 543:564–567. doi: 10.1038/nature21710. [DOI] [PubMed] [Google Scholar]
- 47.Chun T-W, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 49.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, Faria NR, Lemey P, Albert J, Hunt P, Loeb L, Pilcher C, Poole L, Hatano H, Somsouk M, Douek D, Boritz E, Deeks SG, Hecht FM, Palmer S. 2015. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis 212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viard J-P, Burgard M, Hubert J-B, Aaron L, Rabian C, Pertuiset N, Lourenço M, Rothschild C, Rouzioux C. 2004. Impact of 5 years of maximally successful highly active antiretroviral therapy on CD4 cell count and HIV-1 DNA level. AIDS 18:45–49. doi: 10.1097/00002030-200401020-00005. [DOI] [PubMed] [Google Scholar]
- 51.Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, Riddler SA, McMahon DK, Hong F, Mellors JW. 2014. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouzioux C, Hubert J-B, Burgard M, Deveau C, Goujard C, Bary M, Séréni D, Viard J-P, Delfraissy J-F, Meyer L. 2005. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis 192:46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- 53.Sáez-Cirión A, Hamimi C, Bergamaschi A, David A, Versmisse P, Mélard A, Boufassa F, Barré-Sinoussi F, Lambotte O, Rouzioux C, Pancino G. 2011. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood 118:955–964. doi: 10.1182/blood-2010-12-327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Assoumou L, Weiss L, Piketty C, Burgard M, Melard A, Girard P-M, Rouzioux C, Costagliola D, ANRS 116 SALTO Study Group. 2015. A low HIV-DNA level in peripheral blood mononuclear cells at antiretroviral treatment interruption predicts a higher probability of maintaining viral control. AIDS 29:2003–2007. doi: 10.1097/QAD.0000000000000734. [DOI] [PubMed] [Google Scholar]
- 55.Olesen R, Vigano S, Rasmussen TA, Søgaard OS, Ouyang Z, Buzon M, Bashirova A, Carrington M, Palmer S, Brinkmann CR, Yu XG, Østergaard L, Tolstrup M, Lichterfeld M. 2015. Innate immune activity correlates with CD4 T cell-associated HIV-1 DNA decline during latency-reversing treatment with panobinostat. J Virol 89:10176–10189. doi: 10.1128/JVI.01484-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sáez-Cirión A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard J-P, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, ANRS VISCONTI Study Group. 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 9:e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bozzano F, Costa P, Passalacqua G, Dodi F, Ravera S, Pagano G, Canonica GW, Moretta L, De Maria A. 2009. Functionally relevant decreases in activatory receptor expression on NK cells are associated with pulmonary tuberculosis in vivo and persist after successful treatment. Int Immunol 21:779–791. doi: 10.1093/intimm/dxp046. [DOI] [PubMed] [Google Scholar]
- 58.Bozzano F, Nasi M, Bertoncelli L, Nemes E, Prati F, Marras F, Mussini C, Moretta L, Cossarizza A, De Maria A. 2011. NK-cell phenotype at interruption underlies widely divergent duration of CD4+-guided antiretroviral treatment interruption. Int Immunol 23:109–118. doi: 10.1093/intimm/dxq462. [DOI] [PubMed] [Google Scholar]
- 59.Rasmussen TA, Tolstrup M, Søgaard OS. 2016. Reversal of latency as part of a cure for HIV-1. Trends Microbiol 24:90–97. doi: 10.1016/j.tim.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Cillo AR, Mellors JW. 2016. Which therapeutic strategy will achieve a cure for HIV-1? Curr Opin Virol 18:14–19. doi: 10.1016/j.coviro.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. 2001. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 62.Bisio F, Bozzano F, Marras F, Di Biagio A, Moretta L, De Maria A. 2013. Successfully treated HIV-infected patients have differential expression of NK cell receptors (NKp46 and NKp30) according to AIDS status at presentation. Immunol Lett 152:16–24. doi: 10.1016/j.imlet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 63.Lichtfuss GF, Cheng W-J, Farsakoglu Y, Paukovics G, Rajasuriar R, Velayudham P, Kramski M, Hearps AC, Cameron PU, Lewin SR, Crowe SM, Jaworowski A. 2012. Virologically suppressed HIV patients show activation of NK cells and persistent innate immune activation. J Immunol 189:1491–1499. doi: 10.4049/jimmunol.1200458. [DOI] [PubMed] [Google Scholar]
- 64.Sáez-Cirión A, Sinet M, Shin SY, Urrutia A, Versmisse P, Lacabaratz C, Boufassa F, Avettand-Fènoël V, Rouzioux C, Delfraissy J-F, Barré-Sinoussi F, Lambotte O, Venet A, Pancino G, ANRS EP36 HIV Controllers Study Group. 2009. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol 182:7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 65.Mondi A, Fabbiani M, Ciccarelli N, Colafigli M, D'Avino A, Borghetti A, Gagliardini R, Cauda R, De Luca A, Di Giambenedetto S. 2015. Efficacy and safety of treatment simplification to atazanavir/ritonavir + lamivudine in HIV-infected patients with virological suppression: 144 week follow-up of the AtLaS pilot study. J Antimicrob Chemother 70:1843–1849. doi: 10.1093/jac/dkv037. [DOI] [PubMed] [Google Scholar]
- 66.Hocqueloux L, Prazuck T, Avettand-Fenoel V, Lafeuillade A, Cardon B, Viard J-P, Rouzioux C. 2010. Long-term immunovirologic control following antiretroviral therapy interruption in patients treated at the time of primary HIV-1 infection. AIDS 24:1598–1601. doi: 10.1097/QAD.0b013e32833b61ba. [DOI] [PubMed] [Google Scholar]
- 67.Lodi S, Meyer L, Kelleher AD, Rosinska S, Ghosn J, Sannes M, Porter K. 2012. Immunovirologic control 24 months after interruption of antiretroviral therapy initiated close to HIV seroconversion. Arch Intern Med 172:1252–1255. doi: 10.1001/archinternmed.2012.2719. [DOI] [PubMed] [Google Scholar]
- 68.Ascierto ML, Bozzano F, Bedognetti D, Marras F, Schechterly C, Matsuura K, Picciotto A, Marenco S, Zhao Y, DeGiorgi V, Sommariva M, Moretta L, Wang E, Alter HJ, Marincola FM, De Maria A. 2015. Inherent transcriptional signatures of NK cells are associated with response to IFNalpha + rivabirin therapy in patients with hepatitis C virus. J Transl Med 13:77. doi: 10.1186/s12967-015-0428-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muccio L, Bertaina A, Falco M, Pende D, Meazza R, Lopez-Botet M, Moretta L, Locatelli F, Moretta A, Chiesa MD. 2016. Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing αβ+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica 101:371–381. doi: 10.3324/haematol.2015.134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen CM, White MJ, Bottomley C, Lusa C, Rodriguez-Galan A, Turner SE, Goodier MR, Riley EM. 2015. Impaired NK cell responses to pertussis and H1N1 influenza vaccine antigens in human cytomegalovirus-infected individuals. J Immunol 194:4657–4667. doi: 10.4049/jimmunol.1403080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, Lopez-Botet M. 2006. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis 194:38–41. doi: 10.1086/504719. [DOI] [PubMed] [Google Scholar]
- 72.Liu B, Zou F, Lu L, Chen C, He D, Zhang X, Tang X, Liu C, Li L, Zhang H.. 2016. Chimeric antigen receptor T cells guided by the single-chain Fv of a broadly neutralizing antibody specifically and effectively eradicate virus reactivated from latency in CD4+ T-lymphocytes isolated from HIV-1-infected individuals receiving suppressive combined antiretroviral therapy. J Virol 90:9712–9724. doi: 10.1128/JVI.00852-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang H, Buisson S, Bossi G, Wallace Z, Hancock G, So C, Ashfield R, Vuidepot A, Mahon T, Molloy P, Oates J, Paston SJ, Aleksic M, Hassan NJ, Jakobsen BK, Dorrell L. 2016. Elimination of latently HIV-infected cells from antiretroviral therapy-suppressed subjects by engineered immune-mobilizing T-cell receptors. Mol Ther 24:1913–1925. doi: 10.1038/mt.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. 2016. Latency reversal and viral clearance to cure HIV-1. Science 353:aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Maria A, Bozzano F, Cantoni C, Moretta L. 2011. Revisiting human natural killer cell subset function revealed cytolytic CD56(dim)CD16+ NK cells as rapid producers of abundant IFN-gamma on activation. Proc Natl Acad Sci U S A 108:728–732. doi: 10.1073/pnas.1012356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee-MacAry AE, Ross EL, Davies D, Laylor R, Honeychurch J, Glennie MJ, Snary D, Wilkinson RW. 2001. Development of a novel flow cytometric cell-mediated cytotoxicity assay using the fluorophores PKH-26 and TO-PRO-3 iodide. J Immunol Methods 252:83–92. doi: 10.1016/S0022-1759(01)00336-2. [DOI] [PubMed] [Google Scholar]
- 77.Casabianca A, Orlandi C, Canovari B, Scotti M, Acetoso M, Valentini M, Petrelli E, Magnani M. 2014. A real time PCR platform for the simultaneous quantification of total and extrachromosomal HIV DNA forms in blood of HIV-1 infected patients. PLoS One 9:e111919. doi: 10.1371/journal.pone.0111919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casabianca A, Gori C, Orlandi C, Forbici F, Federico Perno C, Magnani M. 2007. Fast and sensitive quantitative detection of HIV DNA in whole blood leucocytes by SYBR green I real-time PCR assay. Mol Cell Probes 21:368–378. doi: 10.1016/j.mcp.2007.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.