ABSTRACT
The cocirculation of zoonotic highly pathogenic avian influenza virus (HPAIV) of subtype H5N1 and avian influenza virus (AIV) of subtype H9N2 among poultry in Egypt for at least 6 years should render that country a hypothetical hot spot for the emergence of reassortant, phenotypically altered viruses, yet no reassortants have been detected in Egypt. The present investigations proved that reassortants of the Egyptian H5N1 clade 2.2.1.2 virus and H9N2 virus of the G1-B lineage can be generated by coamplification in embryonated chicken eggs. Reassortants were restricted to the H5N1 subtype and acquired between two and all six of the internal segments of the H9N2 virus. Five selected plaque-purified reassortant clones expressed a broad phenotypic spectrum both in vitro and in vivo. Two groups of reassortants were characterized to have retarded growth characteristics in vitro compared to the H5N1 parent virus. One clone provoked reduced mortality in inoculated chickens, although the characteristics of a highly pathogenic phenotype were retained. Enhanced zoonotic properties were not predicted for any of these clones, and this prediction was confirmed by ferret inoculation experiments: neither the H5N1 parent virus nor two selected clones induced severe clinical symptoms or were transmitted to sentinel ferrets by contact. While the emergence of reassortants of Egyptian HPAIV of subtype H5N1 with internal gene segments of cocirculating H9N2 viruses is possible in principle, the spread of such viruses is expected to be governed by their fitness to outcompete the parental viruses in the field. The eventual spread of attenuated phenotypes, however, would negatively impact syndrome surveillance on poultry farms and might foster enzootic virus circulation.
IMPORTANCE Despite almost 6 years of the continuous cocirculation of highly pathogenic avian influenza virus H5N1 and avian influenza virus H9N2 in poultry in Egypt, no reassortants of the two subtypes have been reported. Here, the principal compatibility of the two subtypes is shown by forcing the reassortment between copassaged H5N1 und H9N2 viruses in embryonated chicken eggs. The resulting reassortant viruses displayed a wide range of pathogenicity including attenuated phenotypes in chickens, but did not show enhanced zoonotic propensities in the ferret model.
KEYWORDS: Egypt, highly pathogenic avian influenza, reassortment, viral fitness, zoonosis
INTRODUCTION
Influenza pandemics are associated with the emergence of novel zoonotic influenza A viruses which efficiently transmit between humans (1, 2). In the past, reassortment between influenza A viruses of animal and human origin preceded the emergence of such viruses in at least the last three pandemics (H2N2 viruses in 1957, H3N2 viruses in 1968, H1N1 viruses in 2009) (3). Transspecies transmission of fully avian or porcine influenza A viruses and adaptation to the human host are considered an alternative mechanism for the generation of human pandemic viruses (4, 5).
Several avian influenza viruses (AIV) with enhanced zoonotic potential have been described over the past decade. These include highly pathogenic AIV (HPAIV) H5N1 of the goose/Guangdong (gs/GD) lineage (6) and further AIV subtypes that are of low pathogenicity in avian hosts: H7N9 (7), H9N2 (8), and H10N8 (9). Since their reemergence in 2003, gs/GD-like H5 HPAIV have evolved rapidly, extended their geographic distribution from Southeast Asia into Africa, Europe, and North America, and caused devastating outbreaks in wild birds and poultry. In several countries, gs/GD-like viruses have established an endemic status in poultry populations. It is such a status that is associated with a high risk of exposure and spillover transmission to humans (6, 10). By April of 2017, according to the World Health Organization (WHO), HPAIV H5N1 was associated with more than 858 reported human cases with a fatality rate of approximately 60% (11). Among the affected countries, Egypt has reported the highest number of human cases of HPAIV H5N1 infections (11). The currently circulating lineages of gs/GD HPAIV H5N1 express different degrees of zoonotic propensity, but to date, none of them has undergone an adaptation toward efficient human-to-human transmission (6, 10). While humans are still considered dead-end hosts for gs/GD HPAIV H5 strains, previous studies showed that limited genetic changes suffice to adapt these viruses for efficient airborne transmission in ferrets, an animal model system in which influenza mimics that in humans (12).
For the past 5 years, low-pathogenic avian influenza viruses (LPAIV) of the H7N9 subtype have emerged in eastern China (7). While LPAIV are still strictly avian-adapted viruses that do not induce significant clinical signs in gallinaceous poultry, human hosts infected with these viruses are nevertheless prone to develop severe and lethal disease (13). The emergence of these viruses was heavily influenced by reassortment events with LPAIV of subtype H9N2 of the G1 lineage, which contributed the whole cassette of the six so-called internal (i.e., nonhemagglutinin [non-HA], nonneuraminidase [non-NA]) genome segments (7). A similar upsurge in zoonotic potential was also observed in H10 viruses in southern China following reassortment between H9N2 G1-like viruses (9). G1-like H9N2 viruses arose in southern China in the mid-1990s and have spread through all of southern and central Asia to the Middle East and North Africa, causing frequent and economically tangible outbreaks among poultry and establishing an endemic status in many of the affected countries (8). A hallmark of the G1 lineage of H9N2 viruses is the substitutional mutation Q224L in the hemagglutinin (HA) protein, which broadens the receptor binding specificity toward human-like alpha-2-6-linked sialic acids (8). Consequently, 32 human cases of infection caused by the H9N2 G1 lineage but not by the other lineages have been reported, with cases being reported from China (n = 25), Bangladesh (n = 3), and Egypt (n = 4) (14). Recent serology data suggested that the incidence of exposure to and possible infection with H9N2 G1-like viruses in humans may be considerably underestimated (15, 16). Unlike infection with gs/GD H5 or H7N9 viruses, most of the reported cases of human H9N2 infection developed mild signs of disease, and no evidence of human-to-human transmission has been recorded (17, 18).
In Egypt, the continuous cocirculation of gs/GD HPAIV H5N1 of clade 2.2.1.2 and LPAIV H9N2 G1 has been observed in poultry populations since 2012 (19). Although ample opportunities for the exchange of genome segments between these two subtypes have been expected, no reassortants have been detected in Egypt so far, and both subtypes have maintained unchanged genotypes since their introduction in 2006 and 2010, respectively (19). This is surprising, since reassortment events between HPAIV H5N1, although of a clade different from that of the Egyptian viruses, and LPAIV H9N2 were reported from Dubai (20) and Bangladesh (21). A recent study using a reverse genetics-based system indicated at least partial genetic incompatibility of some genome segments of HPAIV H5N1 in the Egyptian LPAI H9N2 backbone (22).
The current study was conducted to clarify whether incompatibilities between HPAIV H5N1 clade 2.2.1.2 and H9N2 G1 strains that prevent further reassortment under natural conditions might exist. Forced coamplification of the two subtypes in embryonated chicken eggs (ECE) finally produced five H5N1 reassortants carrying different genome segments of H9N2; their phenotypes were analyzed in vitro and in vivo.
RESULTS
The compatibility for natural reassortment between the Egyptian HPAIV H5N1 and LPAIV H9N2 strains was investigated by coinfecting embryonated chicken eggs with a representative (parental) strain of each virus. Putative and actual reassortants were investigated in silico and in vitro as well as in vivo for replication kinetics, virulence, and zoonotic properties.
In silico analysis of zoonotic potential of reassortants of HPAIV H5N1 and H9N2.
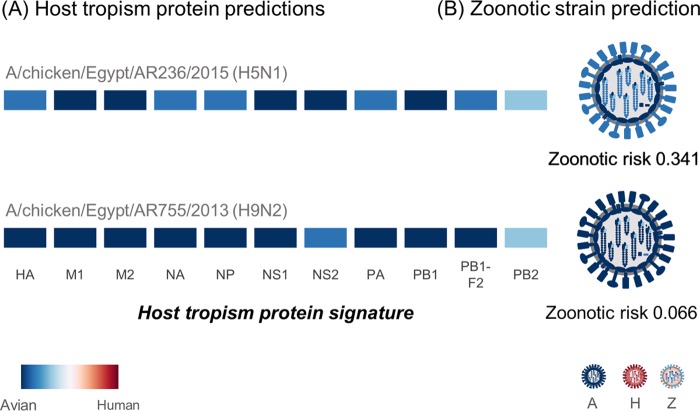
The Egyptian HPAIV H5N1 and LPAIV H9N2 parent strains and putative reassortants were first analyzed for their zoonotic potential using the FluLeap influenza zoonotic prediction system, a machine learning approach based on the protein sequences (23). Both the H5N1 and H9N2 parent strains were predicted to be of low zoonotic risk. The host tropism protein signatures of both strains exhibited a classic avian signature (24), with all 11 proteins individually predicted to have avian tropisms (Fig. 1). Indeed, both strains were predicted to be avian, with very low zoonotic probability estimates of 0.341 for the H5N1 strain and 0.066 for the H9N2 strain being found on the basis of the host tropism protein signatures. This translates to low to almost no zoonotic risk for both parent strains, indicating that they are fully adapted avian strains and do not share similar signatures with zoonotic strains elsewhere.
FIG 1.
Host tropism and zoonotic prediction between the Egyptian HPAIV H5N1 and LPAIV H9N2 strains. (A) The host tropism signatures of proteins from both parent strains depict the individual host tropism predictions of 11 viral proteins (HA, M1, M2, NA, NP, NS1, NS2, PA, PB1, PB1-F2, and PB2), determined using deduced protein sequences. The intensity of the color represents the confidence in the avian or human tropism prediction by the individual protein prediction models. Both parent strains carry a typical avian tropism signature with 11 avian protein tropisms. (B) The signatures were next used for a second layer of machine learning prediction for classification of avian (A), human (H), or zoonotic (Z) strains, where both strains were predicted to show low zoonotic risks.
In silico simulation of reassortment between the H5N1 and H9N2 parent strains using their host tropism protein signatures generated a list of 128 possible unique reassortant strains. This simulation was performed by sequentially swapping each viral segment represented by the individual host tropism protein prediction. A zoonotic risk analysis of the reassortant progeny again revealed that none of the 128 reassortant strains would carry a high zoonotic risk. Only one reassortant strain with a zoonotic risk of 0.354, which was slightly higher than that for the H5N1 parent strain, was predicted, and it was generated by the H5N1 parent strain acquiring the NS segment from the H9N2 strain. This suggests that neither the parent strains nor the progeny that can potentially be generated through reassortment have the zoonotic potential to cause widespread infection in humans.
Generation and detection of reassortants.
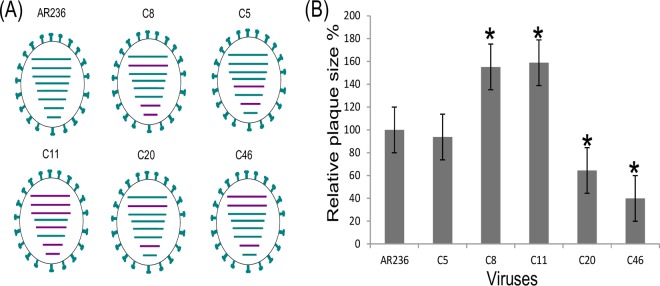
An HPAIV H5N1 strain and an LPAIV H9N2 strain isolated from Egypt were coamplified in ECE. Plaque-purified clones selected after two rounds of virus neutralization in the presence of H5-specific antiserum (n = 50 clones) and H9-specific antiserum (n = 50 clones) and subsequent reamplification in ECE were screened for their internal gene segment composition using a set of 12 SYBR green-based real-time reverse transcriptase quantitative PCRs (RT-qPCRs) that distinguished between the internal gene segments of H5N1 and H9N2. No reassortants were observed among the H9-positive clones selected in the presence of the H5-specific antiserum. In contrast, different genotypes were detected among the H5N1-positive clones. Two sequential plaque purification passages were sufficient to obtain in total five monogenotypic purified reassortant clones. These clones were designated C5, C8, C11, C20, and C46. Figure 2A depicts the genotypes of the selected reassortants, which were determined on the basis of SYBR green-based RT-qPCRs. C5 contained H9 NP and M, C8 contained H9 PB1, M, and NS, C11 contained all six internal H9 segments, C20 contained H9 PB1 and M, and, finally, C46 contained PB1, PB2, and M of the H9N2 virus. No HA/NA reassortants (H5N2 or H9N1) were detected among the total of 100 clones examined.
FIG 2.
Genotype (A) and plaque morphology (B) of five reassortant clones of HPAIV H5N1 and H9N2 strains from Egypt generated by coinfection of embryonated chicken eggs. (A) The eight viral genome segments are indicated by dashes, which indicate, from top to bottom, PB2, PB1, PA, HA, NP, NA, M, and NS segments. Gene segments of HPAIV H5N1 parent strain A/chicken/Egypt/AR236/2015 (AR236) are colored in green, and those of the H9N2 subtype are shown in purple. The genotypes of five reassortant clones (C5, C8, C11, C20, and C46, all of the H5N1 subtype) are shown. (B) The size of 25 randomly selected plaques per clone in infected MDCK cells was measured using ImageJ software. The plaque size of parent strain AR236 was considered 100%. *, statistically significant differences in relation to the parental virus (AR236) (P < 0.001).
Sequence and structural analysis of five H5 reassortant clones.
The genotypes determined on the basis of the SYBR green-based RT-qPCRs were confirmed by Sanger sequencing of the whole genome of the selected clones (C5, C8, C11, C20, and C46). Further sequence analyses focused on detection of mutations between the reassortant clone and the parental viruses. Substitution mutations I18V in PB1 and F612V in PA of clone C5 were identified. Compared to the original sequence deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database, an A156T substitutional mutation was observed in the HA of all five reassortant clones as well as in the HPAIV parent strain A/chicken/Egypt/AR236/2015 (AR236). Additionally, the T4I and T11M mutations were found in PB2 of the H9N2 reassortant clones harboring this gene segment as well as the H9N2 parent virus. No further amino acid mutations were detected in other gene segments of any of the clones compared to the sequences of those gene segments of the parental viruses.
Characterization of H5 reassortant clones in vitro.
The in vitro growth properties of the five selected reassortants, as well as the HPAIV H5N1 parent strain, comprised a plaque morphology in MDCK II cells. The C8 and C11 viruses formed significantly (P < 0.0001) larger plaques (4.32 and 4.39 mm, respectively) than the H5N1 parent virus AR236 (2.76 mm), while C20 and C46 revealed significantly (P < 0.0001) smaller plaques (1.85 and 1.45 nm, respectively). The results of a percentage-based comparison are shown in Fig. 2B (where the average size of the H5N1 parent virus was considered 100%).
Replication kinetics were likewise studied in MDCK II cells employing a multistep growth curve approach with a multiplicity of infection (MOI) of 0.001. Reassortant clones C5, C8, and C11 replicated faster than the parent virus (AR236), reaching a peak titer of 106.5 50% tissue culture infective doses (TCID50)/ml within 24 h (Fig. 3). The replication characteristics of clone C20 were indistinguishable from those of the HPAIV H5N1 parent strain. The replication kinetics of clone C46 were retarded, achieving a titer of only 105.2 TCID50/ml within 24 h and a maximum titer of not more than 105.6 TCID50/ml after 48 h.
FIG 3.
Replication kinetics (multistep growth curve) of different reassortant avian influenza viruses (clones C5, C8, C11, C20, and C46) and the HPAIV H5N1 parent strain (A/chicken/Egypt/AR236/2015) in MDCK cells. Subconfluent monolayers of MDCK cells were infected with each of the reassortant clones (C5, C8, C11, C20, and C46) or the parental virus (HPAIV H5N1 A/chicken/Egypt/AR236/2015) at a multiplicity of infection of 0.001. Virus titers in the supernatants were measured in triplicate at 1, 8, 24, 48, and 72 h. Statistically significant differences from the titers of the parental virus (AR236) were observed for C5, C8, and C11 at 8 h after infection and for C46 at 24 h and at 48 h after infection (P < 0.01).
Antigenicity was not investigated, since no amino acid substitutions in either the HA or the NA of the clones were detected.
Characterization of H5 reassortant clones in vivo. (i) Chickens.
The results of the intravenous pathogenicity index (IVPI) study are presented in Fig. 4A. Three patterns of virus-induced mortality were distinguishable: infection with clones C8 and C11 and parent strain AR236 rapidly led to the death of all inoculated birds within 48 to 72 h postinfection (p.i.). Clones C5 and C20 induced lethal disease and mortality that was significantly retarded compared to the disease and mortality induced by C8, C11, and AR236 (P < 0.001 in all cases); all animals were dead at 5 days postinfection (dpi). C46 induced mortality in only 3 out of 10 chickens. However, all inoculated birds developed severe disease within 2 dpi, consisting of depression, anorexia, diarrhea, ruffled feathers, cyanosis of the comb, and subcutaneous hemorrhage on the feet and shank, but recovered by 10 dpi (Table 1).
FIG 4.
Survival of chickens after intravenous (A) or oculonasal (B) inoculation with reassortant clones C5, C8, C11, C20, and C46 or HPAIV H5N1 parent strain A/chicken/Egypt/AR236/2015. Statistically significant differences compared to the survival of chickens inoculated with the parental virus (AR236) were observed for C5, C20, and C46 for both routes of inoculation (P < 0.01).
TABLE 1.
Pathogenicity indices and percent mortality after intravenous or oculonasal inoculation of chickens with reassortant clone C5, C8, C11, C20, or C46 or parent strain AR236
| Virus or clone | Intravenous inoculation |
Oculonasal inoculation |
||||
|---|---|---|---|---|---|---|
| Index | % mortality | P valuea | Index | % mortality | P value | |
| AR236 | 2.91 | 100 | 2.71 | 100 | ||
| C5 | 2.35 | 100 | <0.001 | 2.38 | 100 | 0.001 |
| C8 | 2.96 | 100 | 1 | 2.78 | 100 | 0.114 |
| C11 | 2.87 | 100 | 0.331 | 2.59 | 100 | 1 |
| C20 | 2.47 | 100 | <0.001 | 2.45 | 100 | 0.005 |
| C46 | 1.49 | 30 | <0.001 | 1.45 | 50 | 0.001 |
P values were calculated by log-rank tests on survival curves in comparison to the survival of the HPAIV H5N1 parent strain A/chicken/Egypt/AR236/2015 (AR236) and adjusted by use of the Bonferroni comparison.
Oculonasal inoculation of chickens was performed to evaluate the pathogenicity of the clones following infection via a natural route. The results strictly paralleled the data obtained by the IVPI assay (Fig. 4B): all birds in the C8-, C11-, and AR236-infected groups died within 72 h p.i. C20 and C5 infection provoked death by 4 and 5 dpi, respectively, with a significant delay in comparison to the time to death provoked by infection with C8, C11, and AR236, similar to the findings of the IVPI assay (P < 0.01 in all cases). In contrast, 5 out of 10 birds infected with C46 survived. The birds which succumbed to C46 infection died by 8 dpi. The surviving birds showed severe symptoms, as described for the IVPI assay with C46, but recovered by 14 dpi. The surviving birds in the C46-infected group had seroconverted by enzyme-linked immunosorbent assay (ELISA) and an H5 hemagglutination inhibition (HI) test by day 14 (average HI titer range, 1:32 to 1:128; data not shown). Sentinel birds in all groups (including the C46-infected group) developed clinical signs of disease by 3 to 4 dpi. All sentinel birds died by 8 dpi.
Excretion of the inoculated viruses was detected by RT-qPCR of oropharyngeal (OP) and cloacal (CL) swab samples collected at 2 dpi from all infected birds of the respective groups (Fig. 5). The infectivity equivalent titer in the oropharyngeal and cloacal swab specimens from the C8- and AR236-infected groups was significantly (P < 0.05) higher than that in the specimens from the other groups. The C46- and C20-infected groups revealed a significantly (P < 0.01) reduced shedding compared to the AR236-infected group. The surviving birds in the C46-infected group continued to shed virus up to 10 dpi (OP swabs, 5/5 birds; CL swabs, 1/5 birds). Effective transmission of all reassortants and the parental virus to two sentinel chickens per group was ascertained at 4 dpi by detection of RNA in OP and CL swab samples. OP virus excretion was significantly more pronounced than CL shedding for all clones and the parental virus (P < 0.05).
FIG 5.
Oropharyngeal and cloacal shedding of different reassortant clones (C5, C8, C11, C20, and C46) and of the HPAIV H5N1 parent strain (A/chicken/Egypt/AR236/2015) in 8-week-old White Leghorn specific-pathogen-free chickens at day 2 postinfection. Six groups of 8 week-old SPF chickens were inoculated oculonasally with 105 TCID50 of five reassortant clones (C5, C8, C11, C20, and C46) or with the H5N1 parent strain (AR236). Viral shedding in oropharyngeal and cloacal swab specimens at 2 dpi was determined. The y axis represents the calculated TCID50 milliliter−1 equivalents. The x axis indicates the different reassortant clones. Within each box plot, the center line represents the median, while the top and bottom borders mark the 75th and 25th percentiles, respectively. Whiskers represent the minimum and maximum values. Extreme outliers are indicated by circles.
Virus tropism was assessed using immunohistochemistry for influenza A virus nucleoprotein antigen in brain, heart, lung, liver, kidney, spleen, pancreas, duodenum, and cecum, including cecal tonsil, tissues from one chicken (in the group infected with C8) or two chickens (in the groups infected with C5, C11, C20, C46, and AR236) in each group sacrificed at day 2. The median scores for endothelial cells across all organs within all groups were greater than or equal to those for parenchymal cells across all organs, which demonstrated the endotheliotropism and systemic (highly pathogenic) disease course induced by the wild-type virus and all reassortants (Table 2). Notably, all virus strains (AR236, C8, C11) which induced the death of the chickens within 72 dpi exhibited a coalescing to diffuse endothelial influenza A virus nucleoprotein immunoreactivity within all organs. The reassortants which induced a moderately prolonged disease course (C5, C20) revealed a multifocal endothelial influenza A virus nucleoprotein immunoreactivity within all organs, and the chicken infected with the less virulent C46 strain exhibited only focal to oligofocal influenza A virus nucleoprotein-immunoreactive endothelia.
TABLE 2.
Influenza A virus tissue topography in experimentally inoculated chickens sacrificed on day 2 p.i. as revealed by immunohistochemistry
| Tissue type | Median scorea for animals infected with: |
|||||
|---|---|---|---|---|---|---|
| AR236 | C8 | C11 | C20 | C5 | C46 | |
| Endothelial cells (median for all organs) | 3.0 | 3.0 | 3.0 | 2.0 | 2.0 | 1.0 |
| Parenchymal cells (median for all organs) | 1.5 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Parenchymal cells | ||||||
| Lungs | 3.0 | 3.0 | 3.0 | 2.0 | 2.0 | 2.0 |
| Heart | 2.5 | 2.0 | 2.0 | 1.0 | 1.0 | 1.0 |
| Liver | 1.0 | 1.0 | 2.0 | 1.5 | 1.5 | 2.0 |
| Spleen | 2.0 | 1.0 | 1.5 | 1.0 | 1.0 | 1.0 |
| Brain | 2.0 | 1.0 | 0.5 | 1.5 | 1.0 | 1.0 |
| Kidneys | 1.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.5 |
| Pancreas | 1.0 | 0.0 | 0.5 | 0.5 | 0.0 | 0.5 |
| Cecum with cecal tonsils | 1.0 | 1.0 | 0.0 | 0.5 | 0.0 | 0.0 |
| Duodenum | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
The values represent the median score for all animals within each group. Scores are as follows: 0, no influenza A virus nucleoprotein immunoreactivity; 1, focal to oligofocal influenza A virus nucleoprotein immunoreactivity; 2, multifocal influenza A virus nucleoprotein immunoreactivity; 3, coalescing to diffuse influenza A virus nucleoprotein immunoreactivity.
The semiquantitative scores for the involvement of parenchymal cells differed between the different organs, but the general trend was comparable between the strains. Multifocal to diffuse parenchymal influenza A virus nucleoprotein immunoreactivity of all strains was observed in the lungs. On the basis of their morphology and distribution, the infected cells were most likely alveolar epithelial cells and/or alveolar macrophages. Notably, the parabronchial and bronchial epithelia were not affected. Other organs with oligofocal to multifocal influenza A virus nucleoprotein immunoreactivity included the heart, liver, spleen, and brain. Focal to multifocal parenchymal influenza A virus nucleoprotein immunoreactivity in the kidneys was also shown for the wild-type AR236 strain. Only focal to oligofocal parenchymal influenza A virus nucleoprotein immunoreactivity in the kidneys, pancreas, and cecal tonsils was seen for some of the reassortants and the wild type. Notably, no influenza A virus nucleoprotein immunoreactivity was observed in the mucosa of the duodenum or cecum in any of the groups.
A marked difference between histopathology findings and nucleoprotein immunoreactivity was found, as the influenza A virus nucleoprotein was abundant within all organs but histopathological lesions were only infrequent and mild. In particular, the high viral load observed within the lungs had no histopathological counterpart. The only obvious histopathological change within the lungs was mild to moderate congestive hyperemia, a variable degree of alveolar edema, and pyknosis and karyorrhexis of lymphocytes within the bronchus-associated lymphoid tissues in the AR236-infected chicken. Lymphocytic pyknosis, karyorrhexis, and loss were also present in the spleens and cecal tonsils of AR236- and C11-infected chickens. The most obvious histopathological lesion was mild, acute, multifocal necrotizing encephalitis, observed in both chickens infected with AR236 and one out of two chickens infected with C20, C11, and C46.
(ii) Ferrets.
The inoculated ferrets developed very mild symptoms of disease, restricted to sneezing at 2 dpi and an increase in body temperature of 1.8 to 2.6°C, which was observed among the inoculated ferrets of all groups from 1 to 7 dpi. Moreover, a slight decrease in body weight (a 2 to 4% decrease compared to their body weight at 0 dpi) was also recorded in the infected animals but not in the sentinel animals. The infected ferrets continued to shed virus in nasal washes by 5 dpi, and only one ferret from the AR236-infected group excreted virus at a low titer in the nasal wash for more than 5 days (up to 9 dpi) (Fig. 6). All infected ferrets were found to be seropositive for influenza A virus antibodies by ELISA and HI assay (average titer range, 1:16 to 1:64) at the end of the observation period at 11 dpi. Sentinel animals did not develop any signs of disease, showed a constant body temperature, and had no episode of weight loss. Virus was not detected in any of the nasal washes obtained by the end of the observation period. In addition, sentinel ferrets stayed seronegative by an NP-specific ELISA.
FIG 6.
Viral shedding in nasal wash fluids of ferrets infected with reassortant clone C11 or C46 or with the HPAIV H5N1 parent strain (A/chicken/Egypt/AR236/2015). The y axis represents the calculated TCID50 milliliter−1 equivalents extrapolated from the RT-qPCR Cq values obtained from a titration of viral infectivity of the parental virus (AR236) in MDCK cells. The x axis indicates the results for the different reassortant clones on 5 different days. Each bar represents a single ferret.
Influenza A virus nucleoprotein was restricted to the cells of respiratory organs of the three AR236-, C11-, and C46-infected ferrets assessed, whereas heart, thymus, tongue, tonsilla palatina, lymphonodus tracheobronchialis medius, spleen, liver, pancreas, duodenum, colon, kidney, adrenal gland, skeletal musculature, brain, ganglion trigeminale, bulbus olfactorius, nervus opticus, and retina revealed only negative results by immunohistochemistry. Concerning the respiratory tract, influenza A virus nucleoprotein was detected in the nasal mucosa of the C46-infected ferret, the tonsilla pharyngealis of the AR236-infected ferret (the tonsilla pharyngealis was assessed in this ferret only), a single submucosal gland of the trachea of the AR236-infected ferret, and a mild and variable proportion of the lungs of all three ferrets. In detail, small amounts of focally to multifocally distributed, variable combinations of influenza A virus nucleoprotein-immunoreactive bronchiolar epithelia, pneumocytes, and/or alveolar macrophages were detected in two out of six lung lobes in the AR236-infected ferret, five out of six lung lobes in the C11-infected ferret, and two out of six lung lobes in the C46-infected ferret (Fig. 7).
FIG 7.
(A, B) C11-infected ferret lung at 4 dpi. (A) Mild, acute, catarrhal bronchiolitis with protein-rich fluid, desquamated and degenerating epithelia (arrow), and scant neutrophils within the lumen. (B) Focal spot of influenza A virus nucleoprotein-immunoreactive cellular debris, epithelial cells, and alveolar macrophages interpreted as remnants of a necrotic bronchiolus. (C, D) AR236-infected ferret lung at 4 dpi. (C) Moderate, acute, catarrhal and suppurative (broncho-)pneumonia with intra-alveolar neutrophils (arrow), macrophages (arrowhead), and protein-rich edema. (D) Multifocal accumulation of influenza A virus nucleoprotein-immunoreactive cellular debris, alveolar macrophages, and/or type II pneumocytes. (E, F) C46-infected ferret nasal conchae at 4 dpi. (E) Mild, acute, focal degeneration of epithelial cells (arrow) within the respiratory mucosa. (F) Multiple influenza A virus nucleoprotein-immunoreactive epithelial cells within superficial and deeper layers of the respiratory mucosa. (G, H) C46-infected ferret lung at 4 dpi. (G) Moderate, subacute, coalescing, proliferative pneumonia with hyperplastic type II pneumocytes (arrow) and alveolar histiocytosis. (H) A discrete, large round cell with an influenza A virus nucleoprotein-immunoreactive nucleus and cytoplasm interpreted to be an alveolar macrophage or desquamated type II pneumocyte. (A, C, E, G) Hematoxylin and eosin stain. (B, D, F, G) Influenza A virus nucleoprotein immunohistochemistry by the avidin-biotin-peroxidase complex method with 3-amino-9-ethylcarbazol as the chromogen and hematoxylin counterstain. Bars = 20 μm.
In agreement with the immunohistochemical results, histopathology revealed a mild, acute, multifocal catarrhal and necrotizing bronchiolitis in five out of six lung lobes in the C11-infected ferret. A slightly more pronounced and locally variable mild to moderate, acute to subacute, multifocal, necrotizing bronchiolitis and catarrhal and suppurative pneumonia with prominent protein-rich alveolar edema, alveolar histiocytosis, mild and variable type II pneumocyte proliferation, and lymphohistiocytic infiltration were observed in all lung lobes of the AR236-infected ferret. Four out of six lung lobes in the C46-infected ferret displayed a locally variable mild to moderate, subacute, oligofocal to coalescing, proliferative pneumonia with prominent type II pneumocyte proliferation, alveolar histiocytosis, catarrhal bronchiolitis, and bronchiolar and alveolar collapse. In contrast, all other organs revealed no histopathological alterations.
DISCUSSION
In the present study, we assessed the natural ability of Egyptian HPAIV H5N1 and LPAIV H9N2 strains to reassort with each other. Our findings suggest that reassortment is possible in principal and that reassortment may affect the phenotype in vitro and in vivo, leading, in one case, to remarkable growth retardation and attenuation of the H5N1 reassortant virus.
Following the establishment of an endemic status of zoonotic HPAIV H5N1 of clade 2.2.1.X in poultry in Egypt after 2007 (25), the epidemiological situation was further complicated after the introduction and endemic circulation in poultry of potential zoonotic G1-like H9N2 viruses (14). Spillover infection of HPAIV H5N1 caused 858 cases of human infection with a case fatality ratio of 60%, while only 4 clinically mild cases of human H9N2 infection were reported from Egypt (14). The two different subtypes carry distinct but homogeneous sets of internal genome segments. This situation should have fostered reassortment, but despite the intensive cocirculation of the two subtypes in poultry for at least 5 years, no reassortant viruses have been detected (19, 26) in Egypt so far.
The lack of detection of reassortant H5N1 and/or H9N2 viruses in Egypt may have several reasons. (i) Surveillance for circulating viruses may have been insufficient to detect reassortants since the monitoring focused on HA (and NA) genes because of their relevance for vaccine selection. This is emphasized when comparing sequence database entries: among all entries for Egyptian H5N1 viruses in GISAID's EpiFlu database (n = 1,299), 1,122 HA and 572 NA sequences but only 172 full genome sequences (13.2%) are represented. Moreover, among the 219 published Egyptian H9N2 viruses among the viruses in GISAID's EpiFlu database, 210 HA and 142 NA sequences but only 50 full genome sequences (22.8%) are found. Thus, there is a lack of full genome sequencing that could have detected the circulation of reassortants. Alternatively, we have previously established (19) and also used here a set of 12 SYBR green-based RT-qPCRs that can be used instead of sequencing to identify each of the internal gene segments of the two subtypes.
(ii) HPAIV H5N1 and H9N2 may have circulated in mutually exclusive host populations or may have been separated by geographical or temporal barriers. The likelihood of the importance of these scenarios is low, as the cocirculation of both viruses on the same poultry farms at the same time, the coinfection of chickens with both viruses, and the presence of both viruses in the same poultry field samples have been repeatedly reported (27–29), However, the emergence and perpetuation of the now extinct HPAIV H5N1 2.2.1.1 clade of vaccine escape mutants from 2008 to 2011 were highly restricted to industrial poultry farms. These viruses did not spill over to backyard holdings, where viruses of another clade (2.2.1.2) prevailed (25). Thus, divergent evolution was fostered even by the sectoral separation of poultry populations in Egypt.
(iii) The genetic incompatibility for reassortment between these subtypes may have played a role. Several mechanisms that directly restrict the generation of reassortants during coinfection of different influenza A virus subtypes have been described: (i) an inability of the RNA-dependent RNA polymerase to transcribe and/or replicate heterotypic viral genome segments, (ii) incompatible protein-protein interactions, (iii) the inhibitory effects of viral proteins of one subtype on the other, or (iv) the incompatibility of virus-specific packaging signals required for effective segment incorporation into budding virions (30–32). Incompatibility between different influenza virus subtypes, that is, between equine H7N7 and human H3N2 (33) and between bat influenza virus and other avian and mammalian influenza A virus subtypes (34), was described previously. Incompatibility between the two subtypes investigated here seems unlikely, because H9N2 G1 viruses have been notoriously implicated as versatile donors of internal segments, creating a bunch of highly zoonotic reassortant viruses in Southeast Asia (21), including the gs/GD lineage of H5N1, as well H7N9 and H10N8 viruses in China (7, 9). However, a recent study using a reverse genetics-based system to create reassortants indicated the incompatibility of the Egyptian NA and NP genome segments of HPAIV H5N1 in the Egyptian LPAI H9N2 backbone but did not report any reassortants of subtype H5N1 (22).
The natural reassortment compatibility between Egyptian HPAIV H5N1 and LPAIV H9N2 was investigated here for the first time. The findings presented here clearly show that reassortants with various genotypes could be generated, at least by coinfection in embryonated chicken eggs. The reassortants analyzed here originated from a single coinfection experiment in eggs; they were selected from a group of 100 plaque-picked viruses on the basis of the diversity of their genotypes. Thus, the possibility that more reassortants with different genotypes might have been present in the cloud of virus progeny cannot be excluded. In the limited set of clones analyzed here for virulence and replication kinetics in vitro and in vivo, the unidirectional transfer of internal segments from H9N2 to H5N1 was evident. However, the possibility that H9N2 viruses could have harbored internal segments of H5N1 origin cannot be excluded, and analysis of a substantial further number of H9N2 clones may have shown that H9N2 viruses harboring internal segments of H5N1 origin were generated as well.
The small plaque size measured for clones 46 and 20 correlated with reduced replication in MDCK II cells and also with attenuated virulence in chickens. Acquisition of H9N2-derived PB1 and M segments contributed to attenuation (C20), which was enhanced by the acquisition of the PB2 segment (C46), while the H9N2 NS segment counteracted attenuation (C8). The emergence of a highly attenuated reassortant phenotype like that of C46 would have epidemiological implications. (i) Infected chickens showed prolonged survival, and the surviving chickens excreted virus through the full observation period. This could serve as a stealth mechanism facilitating the spread and establishment of endemicity of such viruses among poultry populations. (ii) The surviving chickens displayed substantial HI titers against H5N1 and by serology were indistinguishable from vaccinated chickens. The spread of such viruses may lead, as a beneficial effect, to an increased level of population immunity but, at the same time, increase selection pressure on H5N1 viruses, which in turn would promote antigenic drift.
A large plaque size (C8, C11) correlated with enhanced replication in MDCK cells, especially during the first 12 h. Evidence for the further increased virulence of these clones could not be measured because of the already very high virulence of the HPAIV H5N1 parent strain in chickens at the chosen inoculation dose. C11 harbors the whole set of six internal gene segments of the H9N2 virus and in this respect resembled the highly zoonotic H7N9 and H10N8 viruses described in China (7, 9). However, no changes in the virulence or transmissibility of C11 were observed in the ferret model. In contrast, the H7N9 virus displayed a high degree of virulence in ferrets and was transmitted between ferrets by contact and respiratory droplets (35, 36). This study was limited to an examination of the effect of reassortment between these viruses. Consecutive passages of the reassortants selected here in chickens and ferrets might have led to adaptive mutations that could have resulted in increased virulence, particularly in ferrets. Zoonotic potential is one of the most important characteristics that needs to be assessed for emerging animal influenza viruses, and the reliable prediction of zoonotic potential from nucleotide sequence data would significantly improve the current necessity to carry out animal infection studies. Here the machine learning prediction results indicating that the viruses were fully avian adapted and presented low to no zoonotic risks were confirmed in the ferret model, as neither severe clinical signs nor the transmission of two selected clones and the HPAIV H5N1 parent strain by contact was detected. A similar result has previously been reported for another HPAIV H5N1 clade 2.2.1.2 strain from Egypt (37).
In summary, natural reassortants of G1-like H9N2 and HPAIV H5N1 strains can be generated following coinfection in ECE. Failure to detect such reassortants in the field in Egypt is unlikely due to genetic incompatibilities between these subtypes. In the limited set of clones analyzed here, such reassortants still represented a minority population among the HPAIV H5N1 parent isolates after two rounds of amplification in ECE, yet among these reassortants, a clone (C46) with grossly retarded growth characteristics in vitro and an attenuated phenotype in vivo was also rescued. Competition among reassortants in the field in Egypt is expected to favor the fittest clones. It can be argued that so far no genotype that would outcompete the optimally fit H9N2 and HPAIV H5N1 parent strains has evolved. The wide spectrum of phenotypic variation in terms of virulence seen here in this limited set of reassortants was unexpected. In particular, the attenuation of one of the clones (C46), leading to reduced mortality and prolonged virus excretion by infected birds, might foster the spread of such viruses in the field. Therefore, enhanced surveillance and continuous examination of the whole genome of both subtypes are required to improve the early detection of reassortant viruses. Moreover, further reverse genetics-based studies are recommended to explore the specific impact of each gene segment on the virulence, transmissibility, and fitness of reassortant viruses.
MATERIALS AND METHODS
Ethics statement.
All procedures involving virus isolation, propagation, and the generation of reassortants were handled in biosafety level 3 (BSL3) facilities at the Friedrich Loeffler Institute (FLI) after permission was obtained from the FLI biosafety council. All animal experiments were carried out in strict accordance with a protocol legally approved by the Ethics Commission of the Ministry of Agriculture of the federal state of Mecklenburg-Vorpommern, Germany (LALLF MV 7221.3-1-033/15). All animal experiments were conducted in BSL3 animal facilities at FLI.
Origin of viruses and virus propagation.
The viruses used in the current study were obtained from the FLI strain repository and had previously been provided by the National Laboratory for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Giza, Egypt: LPAIV H9N2 (A/chicken/Egypt/AR755/2013; sequence data are available at the Global Initiative on Sharing All Influenza Data [GISAID] platform under accession numbers EPI557458 to EPI557465) and HPAIV H5N1 (A/chicken/Egypt/AR236/2015; GISAID accession numbers EPI573253 to EPI573260). The viruses were propagated in specific-pathogen-free (SPF) embryonated chicken eggs (ECE) according to standard procedures.
Plaque assays.
Plaque assays were conducted to obtain purified virus clones. In addition, plaque size was used as an in vitro characteristic of individual clones. Briefly, 10-fold serial dilutions of each virus were incubated on confluent MDCK II cell monolayers in six-well plates for 1 h at 37°C. The inoculum was removed, and the cell layers were carefully washed twice with phosphate-buffered saline (PBS) before being overlaid with Eagle's modified essential medium (EMEM) containing 1.8% (wt/vol) Bacto agar (Becton, Dickinson and Company, NJ, USA), 1.2% (wt/vol) bovine serum albumin fraction V (Roth, Karlsruhe, Germany), and 100 U/ml of penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA). Cells infected with LPAIV were grown in the presence of 1 μg/ml N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin in the semisolid overlay (Sigma, St. Louis, MO, USA). After 48 h of incubation at 37°C, plaques were visualized by crystal violet staining, and the areas of 25 plaques for each clone were measured using ImageJ software (38).
Sequencing.
Whole-genome sequences of the selected clones were amplified using the primers previously described by Höper et al. (39) and a SuperScript III one-step reverse transcriptase PCR (RT-PCR) system (Invitrogen). The gene-specific RT-PCR amplicons were size separated by agarose gel electrophoresis, excised, and purified from the gels using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Further, purified PCR products were used directly for cycle sequencing reactions using a BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) as described previously by Naguib et al. (19). The reaction products were purified using a NucleoSeq kit (Macherey-Nagel, Düren, Germany) and sequenced on an ABI Prism 3100 genetic analyzer (Life Technologies, CA, USA). Assembly and editing of the sequences were carried out with Geneious software (version 9.0.5) (40).
Generation of reassortant viruses.
Natural reassortants, i.e., reassortants not produced by reverse genetics techniques, were generated by the simultaneous inoculation of HPAIV H5N1 and LPAIV H9N2 isolates at an equal ratio of infectivity into SPF ECE. Allantoic fluid obtained from the initial amplification round after 36 h of incubation was diluted 1:100, and 100 μl polyclonal antiserum against the H5N1 strain and 100 μl polyclonal antiserum against the H9N2 strain, raised in chickens, were incubated at 37°C for 2 h. Following incubation with antibodies, a new batch of ECE was inoculated for virus amplification. This cycle was repeated once. Following the second round of virus neutralization/amplification, the allantoic fluids were harvested and plated to allow plaque formation. Initially, 50 clones were picked from each neutralization/amplification lot. Plugs of semisolid agar medium representing a single, isolated plaque were resuspended in EMEM, and RNA was extracted from part of the clarified resuspension fluid. Eluates were tested for the presence of RNA specific for H5, H9, N1, and N2 by RT-qPCRs as described by Hoffmann et al. (41). Clones that harbored only a single HA and/or NA subtype were further analyzed by SYBR green-based RT-qPCRs specific for each of the internal gene segments of the H5N1 and the H9N2 subtypes as previously described (19). Pure clones were obtained and finally amplified in ECE after three consecutive cycles of the picking of plaques for each of the selected reassortant viruses.
Replication kinetics of plaque-purified reassortant clones and parental viruses.
To determine viral multistep growth curves, monolayers of MDCK II cells in six-well plates were infected at a multiplicity of infection (MOI) of 0.001 (42) at 37°C for 1 h. The titer of infectivity of the virus inoculum was determined by the plaque formation assay. The cell numbers in a representative well of a six-well plate showing about 80 to 90% confluence were counted to calculate the MOI. After 1 h of incubation, the virus inoculum was removed and the cells were washed once with sterile PBS. Cells were overlaid with serum-free medium and incubated at 37°C in 5% CO2 in the absence of trypsin. After 1, 8, 24, 48, and 72 h, the supernatant was harvested and stored at −80°C. Removal of the cell debris from the thawed suspension was performed by centrifugation at 800 × g for 5 min. The titers of the virus yield were determined by measuring the 50% tissue culture infective dose (TCID50) in MDCK II cells. In brief, monolayers of MDCK II cells at 80 to 90% confluence were incubated with 10-fold serial dilutions of the collected viruses for 1 h at 37°C. Then, the medium was carefully removed from the cells, which were then washed gently with PBS, and new serum-free medium was added. Finally, after 72 h, viral cytopathic effects were recorded as the readout and the titers were calculated according to the method of Reed and Muench (43). Each experiment was performed in triplicate, and the average and standard deviations from the experiments were determined and plotted.
Pathogenicity and transmissibility in chickens.
White Leghorn chickens hatched at FLI from SPF ECE purchased from Lohmann Animal Health, Cuxhaven, Germany, were raised until they were 8 weeks old. A total of 132 chickens were separated into two sections. In the first experiment, the intravenous pathogenicity index (IVPI) of selected reassortant clones was determined (IVPI study). Sixty chickens were divided into six groups of 10 birds each. The second experiment examined the pathobiology and transmissibility of the viruses after inoculation via a natural infection route; this experiment included 72 chickens separated into six groups (each with 10 inoculated chickens and 2 sentinel chickens).
IVPI study.
To assess the pathogenicity of the viruses according to a legally prescribed method, chickens were inoculated intravenously with 0.1 ml of 1:10-diluted allantoic fluid containing 128 to 256 hemagglutinating units of five selected reassortants as well as the HPAIV H5N1 parent strain. The birds were monitored daily for 10 days for clinical signs and mortality. Indices were calculated according to World Organization for Animal Health (OIE) regulations (44).
Pathobiology and transmissibility.
Virus-infected allantoic fluid was diluted using PBS in order to obtain an inoculum with a titer of 105.0 TCID50 per 0.5 ml. Ten birds in each group were inoculated conjunctivally (CV) and intranasally (i.n.) with a total of 105.0 TCID50 of one of the five reassortant viruses or the HPAIV H5N1 parent strain; two birds per group were kept as sentinels and added to the infected birds at the first day postinoculation (dpi). In order to compare virus shedding patterns via the respiratory or digestive tract, oropharyngeal (OP) and cloacal (CL) swab specimens were collected at 2, 4, 7, and 10 dpi. Clinical signs were scored daily through the following 11 days as follows: 0, healthy (no abnormal signs); 1, sick (showing one of the following symptoms: ruffled feathers, respiratory distress, depression, facial edema, cyanosis of the comb and wattles, or diarrhea); 2, severely sick (showing two or more signs); and 3, dead. Moreover, two infected birds from each inoculated group were sacrificed (except for the group inoculated with C8, in which only one bird was sacrificed) and sampled at 2 dpi to study the virus topography and histopathological lesions.
Pathogenicity and transmissibility in ferrets.
Two reassortants (C11 and C46) were selected, in addition to the H5N1 HPAIV parent strain, to assess their pathogenicity and transmissibility in the ferret model. The two viruses were selected on the basis of the genotype and the pathogenicity results in chickens. A total of 12 ferrets (Mustela putorius furo) were obtained from V. von Messling, Paul-Ehrlich-Institut, Langen, Germany. All ferrets tested negative for influenza A virus antibodies prior to infection. Ferrets were grouped into three groups of four individuals each and housed in cages enriched with tubes and various bedding materials to serve species-specific behavorial patterns of the animals. The cages were separated by double layers of stainless steel to prevent direct contact between the groups. For intranasal (i.n.) infection with 105 TCID50 of virus as the inoculum, three ferrets in each virus-infected group were anesthetized by inhalation of 5% isoflurane in O2 prior to infection. The ferrets were monitored daily for clinical signs (nasal discharge; labored breathing; reduced activity; body temperature, determined by use of a subcutaneous implantable temperature-logging device [Plexx, Elst, The Netherlands]; body weight; and neurological symptoms). To assess transmission by direct contact, an additional ferret was housed with each group from 1 dpi onwards. Nasal wash samples were collected prior to infection and at 1, 3, 5, 7, and 9 dpi from all of the ferrets while they were under light inhalation anesthesia by applying 1 ml of PBS to each nostril and then recollecting the PBS. One ferret per virus-infected group was sacrificed at 4 dpi, which was performed by exsanguination while the ferret was under deep anesthesia. Ferrets underwent a complete necropsy under biosafety level 3 conditions. All remaining ferrets were euthanized as described above at 11 dpi (the end of the experiment).
Real-time RT-PCR.
Viral RNA was extracted from the OP and CL swab specimens (obtained from chickens) or nasal wash fluid (obtained from ferrets) using a QIAamp viral RNA kit (Qiagen). Quantification of the titer (TCID50) of virus from the collected swab specimens was based on the quantification cycle (Cq) values obtained by real-time reverse transcriptase quantitative PCR (RT-qPCR). Correlation of the Cq values and the infectivity titers was done as outlined by Pantin-Jackwood et al. (45). Briefly, a standard Cq curve for virus quantification was generated using viral RNA extracted from serially diluted (log10) HPAIV H5N1 suspensions with a known infectivity titer. An RT-qPCR targeting the matrix (M) gene (46) and an AgPath-ID one-step RT-qPCR kit (Ambion, CA USA) were employed. To relate M-specific Cq values to viral infectivity in the examined swab specimens, the Cq values from these extracts were plotted on the generated standard curve linking infectivity with Cq values. RT-qPCR was carried out on a Bio-Rad CFX1000 real-time PCR system (Bio-Rad, CA, USA) using software supplied with the machine. The resulting values were labeled the “infectivity equivalent titer.”
Histopathology and immunohistochemistry.
Tissues were fixed in 10% neutral buffered formalin for 21 days, processed, and embedded in paraffin wax using a Leica ASP 300S fully enclosed tissue processor (Leica Biosystems, Nussloch, Germany), sectioned to a thickness of 2 to 4 μm using a Hyrax M55 electronic rotary microtome (Carl Zeiss Microimaging GmbH, Jena, Germany), mounted on Superfrost Plus glass slides (Menzel Gläser, Braunschweig, Germany), stained with hematoxylin and eosin using a Medite TST 44.000C automatic tissue stainer (Medite, Burgdorf, Germany), and screened for histopathological changes using an Axio Imager M2 microscope equipped with 10×, 20×, and 40× Plan Neofluar objectives and an AxioCam ICc3 3.3-megapixel digital camera (Carl Zeiss Microscopy GmbH, Jena, Germany). Immunohistochemistry was performed on serial sections to detect influenza A virus antigen using the avidin-biotin-peroxidase complex method (Vectastain PK 6100; Vector Laboratories, Burlingame, CA, USA) with citric buffer (10 mM, pH 6.0) pretreatment, a polyclonal rabbit anti-influenza A virus FPV/Rostock/34 nucleoprotein antiserum (diluted 1:750) (47), 3-amino-9-ethylcarbazol as the chromogen (Agilent Technologies, Santa Clara, CA, USA), and hematoxylin counterstain. The specificity of the immunohistochemical reaction was confirmed by the use of negative tissues from noninfected chicken and ferret archival diagnostic cases, as well as validated positive avian tissues from the recent H5N8 outbreak (48). Serial sections of the negative controls were treated with rabbit serum instead of the primary polyclonal antiserum. The distribution of influenza A virus nucleoprotein was semiquantitatively assessed for each organ, discriminating endothelial and parenchymal immunoreactivity by scoring on a severity scale of 0 to 3, where 0 was negativity for immunoreactive nuclei, 1 was focal or oligofocal immunoreactive nuclei, 2 was multifocal immunoreactive nuclei, and 3 was coalescing to diffuse immunoreactive nuclei.
Serology.
Blood samples were collected from the chickens or ferrets prior to the animal experiment and from the surviving animals at the end of the experiment. Serum samples were examined for AIV-specific antibodies using an influenza A virus nucleoprotein (NP) antibody competition ELISA kit (Idexx, Hoofddorp, The Netherlands) according to the producer's recommendations. Positive samples were further examined by a hemagglutination inhibition (HI) assay using the homologous H9N2 and H5N1 viruses, applying standard protocols described by the World Organization for Animal Health (OIE) (44).
In silico analysis of different reassortant probabilities.
The two parent strains of H5N1 and H9N2 were analyzed for their zoonotic risks and reassortant possibilities using the FluLeap influenza zoonotic prediction system (http://fluleap.bic.nus.edu.sg), based on machine learning approaches (23). The sequences of 11 viral proteins translated from the genomic sequences of the two strains (HA, M1, M2, NA, NP, NS1, NEP, PA, PB1, PB1-F2, and PB2) were first subjected to individual host tropism predictions. The protein sequences, represented by amino acid compositions and physicochemical properties, were individually predicted to have either avian or human protein tropism using the random forest machine learning classifier (49). The host tropism protein prediction results for the 11 proteins of each strain were then combined as a host tropism protein signature, which illustrates the underlying avian or human tropism of the strains (24).
A second layer of machine learning prediction was next applied to the entire host tropism protein signatures of both parent strains for prediction of the zoonotic potential of each strain. Using the host tropism prediction results for the 11 proteins as features, the random forest strain prediction model classifies the strain as avian, human, or zoonotic (23), with the accompanying zoonotic probability estimate denoting the zoonotic propensity of the strain.
Lastly, a reassortment simulation was performed for the two parent strains using their host tropism protein signatures. The eight RNA segments, characterized by the individual host tropism protein predictions, were sequentially recombined for segments with different predictions, generating a list of possible unique reassortant strains. The computationally simulated reassortant progeny were then analyzed for their zoonotic risks using the zoonotic strain prediction model as described above.
Statistical analysis.
Variations in plaque sizes and in virus shedding patterns were evaluated by the Kruskal-Wallis test and the post hoc Mann-Whitney-Wilcoxon test with the Bonferroni correction. In order to compare replication kinetics, measurements were log10 transformed and analyzed using a linear mixed-effects model including random effects for repeated measures for the same sample. Statistically significant differences in replication kinetics were first confirmed by a two-way analysis of variance (ANOVA) and further assessed by pairwise comparisons of the adjusted means per virus and per time point using z-tests and the Benjamini-Hochberg procedure for adjusting for multiple testing. Differences in survival probability were evaluated by means of log-rank tests (50) with the Bonferroni correction. P values of less than 0.05 were considered significant. All analyses were conducted using R (version 3.3.1) software (R Foundation for Statistical Computing, http://www.r-project.org) with the packages survival, nlme, and multcomp.
ACKNOWLEDGMENTS
We thank Diana Wessler, Aline Maksimov, and Silvia Schuparis, FLI, Germany, for excellent technical support. We are grateful to animal keepers F. Klipp and B. Berger.
M. M. Naguib is the recipient of a doctoral scholarship from the German Academic Exchange Service (DAAD).
We declare no conflict of interest for any of the authors.
REFERENCES
- 1.Yen HL, Webster RG. 2009. Pandemic influenza as a current threat. Curr Top Microbiol Immunol 333:3–24. doi: 10.1007/978-3-540-92165-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Cao B. 2017. Pandemic and avian influenza A viruses in humans: epidemiology, virology, clinical characteristics, and treatment strategy. Clin Chest Med 38:59–70. doi: 10.1016/j.ccm.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Bui CM, Chughtai AA, Adam DC, MacIntyre CR. 2017. An overview of the epidemiology and emergence of influenza A infection in humans over time. Arch Public Health 75:15. doi: 10.1186/s13690-017-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katz JM, Veguilla V, Belser JA, Maines TR, Van Hoeven N, Pappas C, Hancock K, Tumpey TM. 2009. The public health impact of avian influenza viruses. Poult Sci 88:872–879. doi: 10.3382/ps.2008-00465. [DOI] [PubMed] [Google Scholar]
- 5.Horimoto T, Kawaoka Y. 2001. Pandemic threat posed by avian influenza A viruses. Clin Microbiol Rev 14:129–149. doi: 10.1128/CMR.14.1.129-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harfoot R, Webby RJ. 2017. H5 influenza, a global update. J Microbiol 55:196–203. doi: 10.1007/s12275-017-7062-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 8.Fusaro A, Monne I, Salviato A, Valastro V, Schivo A, Amarin NM, Gonzalez C, Ismail MM, Al-Ankari A-R, Al-Blowi MH, Khan OA, Maken Ali AS, Hedayati A, Garcia Garcia J, Ziay GM, Shoushtari A, Al Qahtani KN, Capua I, Holmes EC, Cattoli G. 2011. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol 85:8413–8421. doi: 10.1128/JVI.00219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M, Li X, Yuan H, Zhou J, Wu J, Bo H, Xia W, Xiong Y, Yang L, Gao R, Guo J, Huang W, Zhang Y, Zhao X, Zou X, Chen T, Wang D, Li Q, Wang S, Chen S, Hu M, Ni X, Gong T, Shi Y, Li J, Zhou J, Cai J, Xiao Z, Zhang W, Sun J, Li D, Wu G, Feng Z, Wang Y, Chen H, Shu Y. 2015. Genetic diversity of avian influenza A (H10N8) virus in live poultry markets and its association with human infections in China. Sci Rep 5:7632. doi: 10.1038/srep07632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai S, Qin Y, Cowling BJ, Ren X, Wardrop NA, Gilbert M, Tsang TK, Wu P, Feng L, Jiang H, Peng Z, Zheng J, Liao Q, Li S, Horby PW, Farrar JJ, Gao GF, Tatem AJ, Yu H. 2016. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997-2015: a systematic review of individual case data. Lancet Infect Dis 16:e108–e118. doi: 10.1016/S1473-3099(16)00153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. 2016. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2016. WHO, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/ Accessed 1 January 2017. [Google Scholar]
- 12.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, Meng L, Hong Z, Tu W, Cao Y, Li L, Ding F, Liu B, Wang M, Xie R, Gao R, Li X, Bai T, Zou S, He J, Hu J, Xu Y, Chai C, Wang S, Gao Y, Jin L, Zhang Y, Luo H, Yu H, He J, Li Q, Wang X, Gao L, Pang X, Liu G, Yan Y, Yuan H, Shu Y, Yang W, Wang Y, Wu F, Uyeki TM, Feng Z. 2014. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 370:520–532. doi: 10.1056/NEJMoa1304617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FluTrackers. 2017. Global cumulative H9N2 partial case list 1998-2017. FluTrackers.com Inc, Winter Park, FL: https://flutrackers.com/forum/forum/flutrackers-high-pathogenic-h5n1-h1n08-h5n8-h5n6-h5n3-tracking-outbreaks-spread/745429-flutrackers-global-cumulative-h9n2-partial-case-list-1998-2017 Accessed 6 February 2017. [Google Scholar]
- 15.Khan SU, Anderson BD, Heil GL, Liang S, Gray GC. 2015. A systematic review and meta-analysis of the seroprevalence of influenza A(H9N2) infection among humans. J Infect Dis 212:562–569. doi: 10.1093/infdis/jiv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruin E, Zhang X, Ke C, Sikkema R, Koopmans M. 20 February 2017. Serological evidence for exposure to avian influenza viruses within poultry workers in southern China. Zoonoses Public Health. doi: 10.1111/zph.12346. [DOI] [PubMed] [Google Scholar]
- 17.WHO. 2016. Influenza at the human-animal interface. WHO, Geneva, Switzerland: www.who.int/entity/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_10_03_2016.pdf Accessed 16 January 2017. [Google Scholar]
- 18.Alexander DJ, Brown IH. 2009. History of highly pathogenic avian influenza. Rev Sci Tech 28:19–38. doi: 10.20506/rst.28.1.1856. [DOI] [PubMed] [Google Scholar]
- 19.Naguib MM, Arafa AS, El-Kady MF, Selim AA, Gunalan V, Maurer-Stroh S, Goller KV, Hassan MK, Beer M, Abdelwhab EM, Harder TC. 2015. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co-circulating in Egypt. Infect Genet Evol 34:278–291. doi: 10.1016/j.meegid.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Naguib MM, Kinne J, Chen H, Chan KH, Joseph S, Wong PC, Woo PC, Wernery R, Beer M, Wernery U, Harder TC. 2015. Outbreaks of highly pathogenic avian influenza H5N1 clade 2.3.2.1c in hunting falcons and kept wild birds in Dubai implicate intercontinental virus spread. J Gen Virol 96:3212–3222. doi: 10.1099/jgv.0.000274. [DOI] [PubMed] [Google Scholar]
- 21.Marinova-Petkova A, Shanmuganatham K, Feeroz MM, Jones-Engel L, Hasan MK, Akhtar S, Turner J, Walker D, Seiler P, Franks J, McKenzie P, Krauss S, Webby RJ, Webster RG. 2016. The continuing evolution of H5N1 and H9N2 influenza viruses in Bangladesh between 2013 and 2014. Avian Dis 60:108–117. doi: 10.1637/11136-050815-Reg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moatasim Y, Kandeil A, Mostafa A, Elghaffar SKA, El Shesheny R, Elwahy AHM, Ali MA. 15 June 2017. Single gene reassortment of highly pathogenic avian influenza A H5N1 in the low pathogenic H9N2 backbone and its impact on pathogenicity and infectivity of novel reassortant viruses. Arch Virol. doi: 10.1007/s00705-017-3434-x. [DOI] [PubMed] [Google Scholar]
- 23.Eng CLP, Tong JC, Tan TW. 2017. Predicting zoonotic risk of influenza A viruses from host tropism protein signature using random forest. Int J Mol Sci 18:E1135. doi: 10.3390/ijms18061135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eng CL, Tong JC, Tan TW. 2016. Distinct host tropism protein signatures to identify possible zoonotic influenza A viruses. PLoS One 11:e0150173. doi: 10.1371/journal.pone.0150173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelwhab EM, Hassan MK, Abdel-Moneim AS, Naguib MM, Mostafa A, Hussein IT, Arafa A, Erfan AM, Kilany WH, Agour MG, El-Kanawati Z, Hussein HA, Selim AA, Kholousy S, El-Naggar H, El-Zoghby EF, Samy A, Iqbal M, Eid A, Ibraheem EM, Pleschka S, Veits J, Nasef SA, Beer M, Mettenleiter TC, Grund C, Ali MM, Harder TC, Hafez HM. 2016. Introduction and enzootic of A/H5N1 in Egypt: virus evolution, pathogenicity and vaccine efficacy ten years on. Infect Genet Evol 40:80–90. doi: 10.1016/j.meegid.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Arafa AS, Naguib MM, Luttermann C, Selim AA, Kilany WH, Hagag N, Samy A, Abdelhalim A, Hassan MK, Abdelwhab EM, Makonnen Y, Dauphin G, Lubroth J, Mettenleiter TC, Beer M, Grund C, Harder TC. 2015. Emergence of a novel cluster of influenza A(H5N1) virus clade 2.2.1.2 with putative human health impact in Egypt, 2014/15. Euro Surveill 20(13):2–8. doi: 10.2807/1560-7917.ES2015.20.13.21085. [DOI] [PubMed] [Google Scholar]
- 27.Naguib MM, El-Kady MF, Luschow D, Hassan KE, Arafa AS, El-Zanaty A, Hassan MK, Hafez HM, Grund C, Harder TC. 2017. New real time and conventional RT-PCRs for updated molecular diagnosis of infectious bronchitis virus infection (IBV) in chickens in Egypt associated with frequent co-infections with avian influenza and Newcastle disease viruses. J Virol Methods 245:19–27. doi: 10.1016/j.jviromet.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arafa AS, Hagag NM, Yehia N, Zanaty AM, Naguib MM, Nasef SA. 2012. Effect of cocirculation of highly pathogenic avian influenza H5N1 subtype with low pathogenic H9N2 subtype on the spread of infections. Avian Dis 56:849–857. doi: 10.1637/10152-040812-Reg.1. [DOI] [PubMed] [Google Scholar]
- 29.Hassan KE, Shany SA, Ali A, Dahshan AH, El-Sawah AA, El-Kady MF. 2016. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult Sci 95:1271–1280. doi: 10.3382/ps/pew068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald SM, Nelson MI, Turner PE, Patton JT. 2016. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat Rev Microbiol 14:448–460. doi: 10.1038/nrmicro.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC. 2013. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog 9:e1003421. doi: 10.1371/journal.ppat.1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerber M, Isel C, Moules V, Marquet R. 2014. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol 22:446–455. doi: 10.1016/j.tim.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Hatta M, Watanabe S, Neumann G, Kawaoka Y. 2008. Compatibility among polymerase subunit proteins is a restricting factor in reassortment between equine H7N7 and human H3N2 influenza viruses. J Virol 82:11880–11888. doi: 10.1128/JVI.01445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou B, Ma J, Liu Q, Bawa B, Wang W, Shabman RS, Duff M, Lee J, Lang Y, Cao N, Nagy A, Lin X, Stockwell TB, Richt JA, Wentworth DE, Ma W. 2014. Characterization of uncultivable bat influenza virus using a replicative synthetic virus. PLoS Pathog 10:e1004420. doi: 10.1371/journal.ppat.1004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yum J, Ku KB, Kim HS, Seo SH. 2015. H7N9 influenza virus is more virulent in ferrets than 2009 pandemic H1N1 influenza virus. Viral Immunol 28:590–599. doi: 10.1089/vim.2015.0052. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 37.Arafa AS, Yamada S, Imai M, Watanabe T, Yamayoshi S, Iwatsuki-Horimoto K, Kiso M, Sakai-Tagawa Y, Ito M, Imamura T, Nakajima N, Takahashi K, Zhao D, Oishi K, Yasuhara A, Macken CA, Zhong G, Hanson AP, Fan S, Ping J, Hatta M, Lopes TJ, Suzuki Y, El-Husseiny M, Selim A, Hagag N, Soliman M, Neumann G, Hasegawa H, Kawaoka Y. 2016. Risk assessment of recent Egyptian H5N1 influenza viruses. Sci Rep 6:38388. doi: 10.1038/srep38388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. 2015. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Höper D, Hoffmann B, Beer M. 2009. Simple, sensitive, and swift sequencing of complete H5N1 avian influenza virus genomes. J Clin Microbiol 47:674–679. doi: 10.1128/JCM.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann B, Hoffmann D, Henritzi D, Beer M, Harder TC. 2016. Riems influenza A typing array (RITA): an RT-qPCR-based low density array for subtyping avian and mammalian influenza A viruses. Sci Rep 6:27211. doi: 10.1038/srep27211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ping J, Lopes TJ, Nidom CA, Ghedin E, Macken CA, Fitch A, Imai M, Maher EA, Neumann G, Kawaoka Y. 2015. Development of high-yield influenza A virus vaccine viruses. Nat Commun 6:8148. doi: 10.1038/ncomms9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 44.World Organization for Animal Health. 2015. Avian influenza. World Organization for Animal Health, Paris, France. [Google Scholar]
- 45.Pantin-Jackwood M, Swayne DE, Smith D, Shepherd E. 2013. Effect of species, breed and route of virus inoculation on the pathogenicity of H5N1 highly pathogenic influenza (HPAI) viruses in domestic ducks. Vet Res 44:62. doi: 10.1186/1297-9716-44-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann B, Harder T, Lange E, Kalthoff D, Reimann I, Grund C, Oehme R, Vahlenkamp TW, Beer M. 2010. New real-time reverse transcriptase polymerase chain reactions facilitate detection and differentiation of novel A/H1N1 influenza virus in porcine and human samples. Berl Munch Tierarztl Wochenschr 123:286–292. [DOI] [PubMed] [Google Scholar]
- 47.Klopfleisch R, Werner O, Mundt E, Harder T, Teifke JP. 2006. Neurotropism of highly pathogenic avian influenza virus A/chicken/Indonesia/2003 (H5N1) in experimentally infected pigeons (Columbia livia f. domestica). Vet Pathol 43:463–470. doi: 10.1354/vp.43-4-463. [DOI] [PubMed] [Google Scholar]
- 48.Pohlmann A, Starick E, Harder T, Grund C, Hoper D, Globig A, Staubach C, Dietze K, Strebelow G, Ulrich RG, Schinkothe J, Teifke JP, Conraths FJ, Mettenleiter TC, Beer M. 2017. Outbreaks among wild birds and domestic poultry caused by reassorted influenza A(H5N8) clade 2.3.4.4 viruses, Germany, 2016. Emerg Infect Dis 23:633–636. doi: 10.3201/eid2304.161949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eng CL, Tong JC, Tan TW. 2014. Predicting host tropism of influenza A virus proteins using random forest. BMC Med Genomics 7(Suppl 3):S1. doi: 10.1186/1755-8794-7-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bland JM, Altman DG. 2004. The logrank test. BMJ 328:1073. doi: 10.1136/bmj.328.7447.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]