ABSTRACT
Murid herpesvirus 4 (MuHV-4) is a B cell-tropic gammaherpesvirus that can be studied in vivo. Despite viral evasion, type I interferons (IFN-I) limit its spread. After MuHV-4 inoculation into footpads, IFN-I protect lymph node subcapsular sinus macrophages (SSM) against productive infection; after peritoneal inoculation, they protect splenic marginal zone macrophages, and they limit MuHV-4 replication in the lungs. While invasive infections can be used to test specific aspects of host colonization, it is also important to understand natural infection. MuHV-4 taken up spontaneously by alert mice enters them via olfactory neurons. We determined how IFN-I act in this context. Blocking IFN-I signaling did not increase neuronal infection but allowed the virus to spread to the adjacent respiratory epithelium. In lymph nodes, a complete IFN-I signaling block increased MuHV-4 lytic infection in SSM and increased the number of dendritic cells (DC) expressing viral green fluorescent protein (GFP) independently of lytic infection. A CD11c+ cell-directed signaling block increased infection of DC only. However, this was sufficient to increase downstream infection, consistent with DC providing the main viral route to B cells. The capacity of IFN-I to limit DC infection indicated that viral IFN-I evasion was only partly effective. Therefore, DC are a possible target for IFN-I-based interventions to reduce host colonization.
IMPORTANCE Human gammaherpesviruses infect B cells and cause B cell cancers. Interventions to block virus binding to B cells have not stopped their infection. Therefore, we must identify other control points that are relevant to natural infection. Human infections are difficult to analyze. However, gammaherpesviruses colonize all mammals. A related gammaherpesvirus of mice reaches B cells not directly but via infected dendritic cells. We show that type I interferons, an important general antiviral defense, limit gammaherpesvirus B cell infection by acting on dendritic cells. Therefore, dendritic cell infection is a potential point of interferon-based therapeutic intervention.
KEYWORDS: dendritic cell, gammaherpesvirus, interferons, respiratory tract
INTRODUCTION
Type I interferons (IFN-I) are a potent defense against many viruses and restrict host colonization even when there is viral IFN-I evasion (1). Herpesviruses have multiple IFN-I evasion genes. Murid herpesvirus 4 (MuHV-4), a relative of Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), inhibits interferon regulatory factor 3 (IRF3) signaling via open reading frame 36 (ORF36) (2) and TANK binding kinase 1 via ORF11 (3). It also downregulates STAT-1 and STAT-2 via M2 (4), degrades the IFN-I receptor (IFNAR) via ORF54 (5), and inhibits downstream responses via ORF37 (6). Nonetheless, MuHV-4 given in the lungs of IFNAR-deficient mice shows increased lung and spleen colonization (7), implying that IFN-I still affords some protection at these sites. Understanding how such protection works could provide a basis for better infection control.
MuHV-4, like EBV and KSHV, persists as a mainly latent infection of B cells (8). Therefore, reducing B cell infection is a key therapeutic goal. MuHV-4 can pass through B cells even when they are exposed to IFN-I (9). Viral latency in B cells may help to achieve this, as a major action of IFN-I is to inhibit protein synthesis, which would have the greatest impact on lytic infection. Notably, enforcing lytic infection leads to early, IFN-I-dependent MuHV-4 attenuation (9). Transcriptional suppression of MuHV-4 M2 by IFN-I further promotes latency in B cells, possibly directly (10) and possibly via the reduced STAT-1-dependent suppression of ORF50 (11). The barriers to IFN-I acting directly on B cell infection mean that it may have to act indirectly, by targeting upstream events.
How EBV and KSHV reach B cells is unknown. MuHV-4 does so by using olfactory neurons and sustentacular cells for host entry (12) and then infecting dendritic cells (DC) and being transferred from them to B cells in the draining superficial cervical lymph nodes (SCLN) (13). Infection subsequently spreads to the spleen. MuHV-4 inoculated into footpads (intrafootpad [i.f.] inoculation) reaches lymph nodes (LN) directly, where it infects subcapsular sinus macrophages (SSM) before reaching B cells (14). MuHV-4 inoculated intraperitoneally (i.p.) infects the spleen directly (as well as other peritoneal organs) and specifically targets marginal zone (MZ) macrophages (MZM) (15, 16). From MZM, infection spreads sequentially to MZ B cells, follicular DC, and then follicular B cells. MuHV-4 delivered to the lungs by a large-volume (30 μl) intranasal (i.n.) inoculation under anesthesia (17) infects alveolar macrophages (AM), and then type 1 alveolar epithelial cells (AEC1) (18), before reaching the mediastinal LN (MLN) to infect B cells. Thus, regardless of the inoculation route, MuHV-4 reaches B cells via myeloid cells (19). Gammaherpesviruses presumably colonized B cells long before the primate/rodent divergence (for example, there are gammaherpesviruses of marsupials [20]), so KSHV and possibly also EBV are likely to follow similar routes. Close parallels between the spread of MuHV-4 and normal immune cell communications (15), plus the similar exploitation of myeloid cells for access to lymphocytes by other viruses (21, 22), suggest that this a general gateway and thus that targeting myeloid cell infection provides a possible general way to reduce lymphocyte infections (23).
The importance of IFN-I for host defense against MuHV-4 makes it a potentially important regulator of myeloid cell infection. Consistent with this idea, IFNAR blockade increases SSM infection by i.f. inoculation of MuHV-4 (24) and MZM infection by i.p. inoculation of MuHV-4 (9). However, mucosal infection spreads mainly via DC, and clearly, this operates in immunocompetent mice despite the presence of IFN-I. The increased splenic infection of IFNAR−/− mice given MuHV-4 in their lungs (7) implies greater spread to lymphoid tissue, but the cell types involved are unknown, and increased lung infection makes knock-on effects in other organs difficult to exclude. Here we analyzed the impact of IFN-I blockade on MuHV-4 spread after olfactory infection and used myeloid cell subset-specific IFNAR disruption to understand how IFN-I signaling to different cell types regulates host colonization.
RESULTS
In the lungs, IFN-I regulates epithelial and myeloid cell infections.
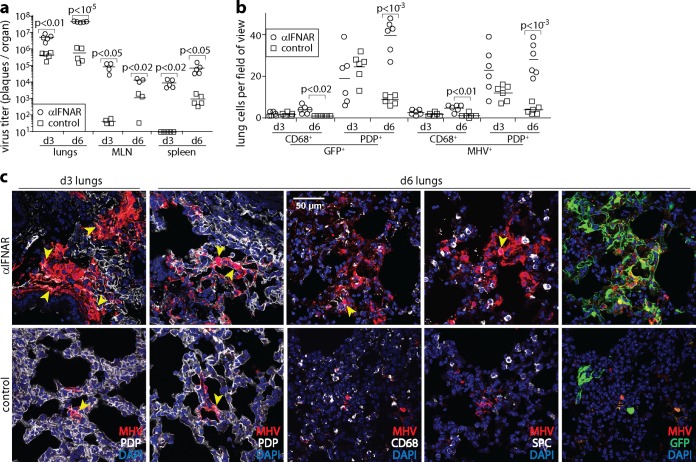
Lung infection by MuHV-4 has an unknown physiological significance, as it has been demonstrated only in anesthetized mice. However, it provides a well-characterized experimental reference point. Mice lacking IFNAR show increased lung infection (7). To identify the cell types involved, C57BL/6 mice were given an IFNAR-blocking antibody (anti-IFNAR) or not and then given green fluorescent protein (GFP)-expressing MuHV-4 i.n. (in 30 μl under anesthesia) (Fig. 1). Virus titers 3 and 6 days later showed significantly increased lung, MLN, and spleen infections in anti-IFNAR-treated mice. Usually, by day 6, most MuHV-4 lung infection is located in AEC1 (podoplanin-positive [PDP+]cells) (18). AM are also infected (CD68+ cells), but AEC2 (surfactant protein C-positive [SPC+] type 2 alveolar epithelial cells) are spared. With anti-IFNAR treatment, MuHV-4 lytic antigen staining with polyclonal immune serum showed that most of the extra productive infection was located in AEC1. However, the number of infected AM showed an equivalent fold increase. Lytic cycle-independent viral GFP expression (25) showed a similar distribution of infection. AEC2 remained uninfected. Thus, when IFN-I signaling was blocked, lung infection increased without noticeably altering the cell types involved.
FIG 1.
IFNAR blockade increases MuHV-4 lytic infection of AM and AEC1 in the lungs. (a) C57BL/6 mice were given an IFNAR-blocking antibody i.p. (αIFNAR) or not (control) and then given MHV-GFP i.n. (104 PFU in 30 μl under anesthesia). At days 3 and 6, the titers of infectious virus in lungs were determined by a plaque assay, and the titers of total recoverable virus (lytic plus latent) in MLN and spleens were determined by an infectious-center assay. Symbols show data for individual mice. Bars show means. Significant differences are indicated. (b) After infection as described above for panel a, day 3 and day 6 lung sections were stained for virus-expressed GFP and lytic antigens (MHV). Lung myeloid cells, predominantly AM, were identified by staining for CD68. AEC1 were identified by staining for podoplanin (PDP). We counted infected cells in at least 4 fields of view per section, across 2 sections from each of 3 mice per group. Symbols show mean counts per section. Bars show group means. Significant differences between groups are shown. (c) In representative images of MHV staining quantitated as described above for panel b, yellow arrows show example dual-positive cells. Colocalization with surfactant protein C-positive (SPC+) AEC2 was rare (<2% of infected cells). Dual staining is also shown for GFP and MHV (colocalization appears in yellow). Viral GFP expression was generally more extensive than MHV staining, implying that not all infection was lytic, but it had a similar distribution.
IFN-I limits nasal MuHV-4 spread.
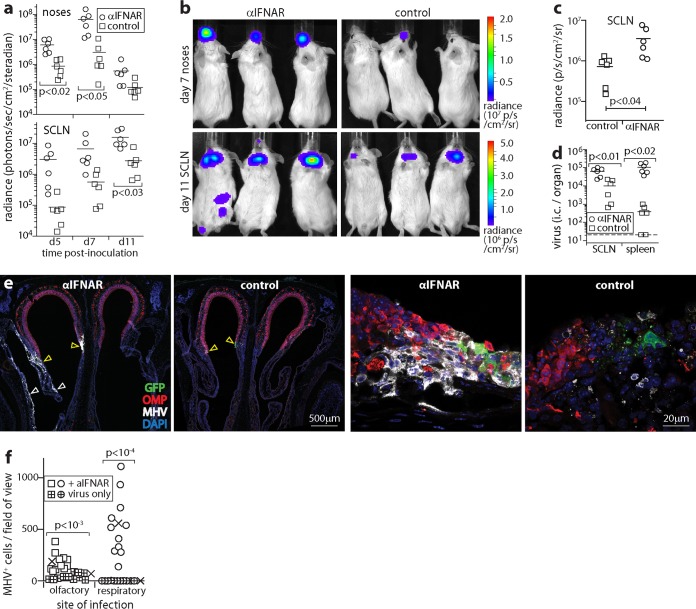
We next tested upper respiratory tract infection. For an overview, BALB/c mice were given anti-IFNAR treatment or not and then given luciferase-positive MuHV-4 (5 μl without anesthesia) i.n., and infection was tracked by live imaging of light emission (Fig. 2). Anti-IFNAR treatment increased nose signals at days 5 and 7 (Fig. 2a and b). SCLN (anterior cervical) signals were also increased at days 5 and 7. However, because the kinetics of the spread of infection from noses to SCLN varied between individual mice, this increase did not reach statistical significance until day 11.
FIG 2.
IFNAR blockade increases MuHV-4 spread in the upper respiratory tract. (a) BALB/c mice were given anti-IFNAR treatment or not (control) and then given MHV-LUC i.n. (105 PFU in 5 μl without anesthesia). Infection was monitored by luciferin injection and live imaging of light emission (radiance). Symbols show nose and neck (SCLN) signals of individual mice. Bars show means. Significant differences are shown. (b) Example images from panel a showing nose signals at day 7 and SCLN signals at day 11. (c) Ex vivo luciferase imaging of dissected SCLN at day 11 confirms greater colonization in anti-IFNAR-treated mice. Symbols show counts for individual mice. Bars show means. (d) Infectious-center (i.c.) assays at day 11 confirm greater infection of SCLN and spleens in anti-IFNAR-treated mice. Symbols show data for individual mice. Bars show means. The dashed line indicates the assay sensitivity limit. (e) C57BL/6 mice were given anti-IFNAR treatment or not (control) and then given MHV-GFP i.n. as described above for panel a. At day 7, coronal nose sections were stained for viral GFP and lytic antigens (MHV) and for olfactory marker protein (OMP) to identify olfactory neurons. The left-hand low-magnification images show example anti-IFNAR-treated and control mice, each with infection at the olfactory/respiratory epithelial junction (yellow arrows). In the anti-IFNAR-treated mouse, infection had spread to the respiratory epithelium (white arrows). The right-hand, high-magnification images show areas of olfactory infection, with sparing of olfactory neurons in the sample from a mouse given anti-IFNAR treatment despite extensive infection. Images are representative of results for 3 mice per group. (f) MHV+ olfactory and respiratory epithelial cells were counted across sections from 3 infected mice per group, with or without anti-IFNAR treatment, as described above for panel e. Crosses show means, and other symbols show counts per field of view. Anti-IFNAR treatment significantly increased both olfactory and respiratory epithelial infections, with a larger effect on respiratory epithelial infection.
Dissection and ex vivo luciferase imaging of organs at day 11 confirmed that cervical signals came from the SCLN (Fig. 2c). Spleen signals were also evident in some anti-IFNAR-treated mice, whereas they were not evident in controls. Neither live imaging nor ex vivo imaging showed infection spreading to the brain or lungs of anti-IFNAR-treated mice. Infectious-center assays at day 11 (Fig. 2d) confirmed significantly greater SCLN and spleen infections in anti-IFNAR-treated mice than in controls. Thus, anti-IFNAR treatment increased MuHV-4 infection in normally colonized sites, the nose, SCLN, and spleen, but did not allow nasal infection to spread to new organs such as the brain (via olfactory neurons) or the lungs (via the respiratory tract).
IFN-I protects the nasal respiratory epithelium.
To visualize infected cells in the nose, C57BL/6 mice were given anti-IFNAR treatment or not and infected i.n. with MHV-GFP, and nose sections were stained for MuHV-4 lytic antigens and GFP at day 6. We identified olfactory neurons by staining for olfactory marker protein (OMP) (Fig. 2e). Again, anti-IFNAR treatment increased infection. MuHV-4 infects olfactory neurons, but most lytic infection occurs in the adjacent (OMP-negative [OMP−]) sustentacular cells (13). Anti-IFNAR treatment did not change this outcome: lytic infection increased in OMP− but not OMP+ olfactory cells, and there was no sign of spread to the olfactory bulbs (data not shown). Instead, infection spread to the respiratory epithelium. Infection often occurs where the olfactory epithelium merges with the respiratory epithelium, presumably because this anterocaudal olfactory region is particularly exposed to inhaled inocula. The respiratory epithelium is normally spared. After anti-IFNAR treatment, it was extensively involved, with infection being evident in 3/3 mice versus 0/3 controls (Fig. 2f). Therefore, IFN-I limited MuHV-4 spread to the respiratory epithelium. IFNAR-treated mice also showed more subepithelial virus spread, but neuronal infection evidently had other restraints.
IFNAR blockade increases SSM and DC infections in LN.
Myeloid cells play a central role in IFN-I responses, both producing large amounts of IFN-I and being prominent sites of its action. Plasmacytoid DC are “prearmed” to produce large amounts of IFN-α and IFN-β, while conventional DC and other myeloid cells produce IFN-α in response to IFN-β (26). After i.f. MuHV-4 inoculation, plasmacytoid DC depletion increases the spread of infection much less than does anti-IFNAR treatment (24), so conventional myeloid cells such as SSM (27) probably account for most IFN-I production. Anti-IFNAR treatment greatly increases infection of SSM by i.f. inoculation of MuHV-4 (14), so they are also a prominent site of IFN-I action. Relatively little B cell infection comes from SSM; instead, it comes from DC (13, 14, 24), so they may respond less well than SSM to IFN-I.
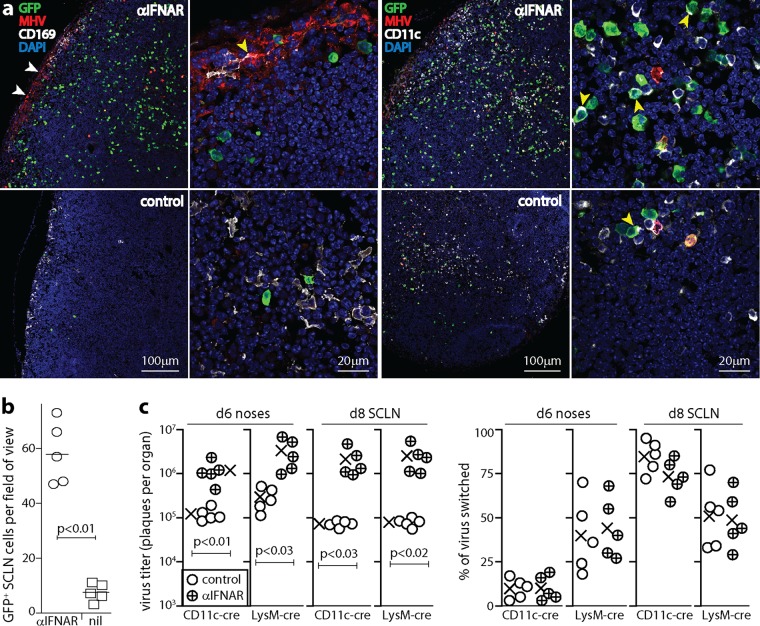
To identify where IFN-I act in LN after olfactory infection, C57BL/6 mice were given anti-IFNAR treatment or not and then given MHV-GFP i.n. (5 μl), and SCLN sections were examined at day 6 (Fig. 3a and b). Anti-IFNAR treatment increased viral GFP and lytic antigen expression levels but in different distributions: lytic antigens were abundant around the subcapsular sinus, while lytic cycle-independent viral GFP+ cells were abundant in the LN parenchyma. Most GFP+ cells were CD11c+. CD11c is not exclusive to DC (28), but immunostaining of sections generally detects only CD11chi cells; for example, in the low-magnification view in Fig. 3a, SSM are not detectably CD11c+, their lower level expression level is evident only at a high magnification (Fig. 4a) (29), and the CD11chi cells visible in LN at a low magnification are predominantly DC. Both anti-IFNAR-treated and control LN contained CD11c+ GFP+ cells, but the former had significantly more of these cells. Therefore, IFN-I regulated a strongly lytic infection of SSM and a less strongly lytic infection of DC.
FIG 3.
IFNAR blockade increases MuHV-4 LN infection. (a) C57BL/6 mice were given anti-IFNAR treatment or not (control) and then given MHV-GFP i.n. (105 PFU in 5 μl). At day 6, SCLN sections were stained for viral GFP and lytic antigens (MHV), for CD169 to identify SSM, and for CD11c to identify DC. Nuclei were stained with DAPI. Low-magnification images (left panels of each antibody set) show marked increases of MHV staining around the subcapsular sinus (white arrows) and GFP staining throughout the LN with anti-IFNAR treatment. GFP is expressed from an EF1α promoter in the viral genome, independently of lytic infection (55). CD11c staining of SSM is weak and thus is difficult to see at a low magnification. High-magnification images (right panels of each antibody set) show the subcapsular sinus for CD169 (the yellow arrow shows an MHV+ SSM) and example GFP+ DC in the LN substance for CD11c (yellow arrows). (b) Quantitation of GFP+ cells across at least 2 SCLN sections for each of 3 mice per group shows a significant increase after anti-IFNAR treatment. Symbols show mean counts for each section (at least 5 fields of view per section). Bars show group means. (c) CD11c-cre and lysM-cre mice were treated or not with anti-IFNAR and then given MuHV-4 with a floxed color-switching cassette (MHV-RG) (105 PFU i.n. in 5 μl). Noses were assayed for infectious virus at day 6 by a plaque assay, and SCLN were explanted to recover both lytic and latent virus at day 8. Recovered plaques were also typed as being red (mCherry+, native) or green (GFP+, switched) fluorescent. Percent switching was calculated as follows: 100 × green plaques/(green plaques + red plaques). Circles show data for individual mice. Crosses show means. Significant differences are shown. Regardless of anti-IFNAR treatment, lysM-cre mice had more virus switching in noses, and CD11c-cre mice had more virus switching in LN (P < 0.02).
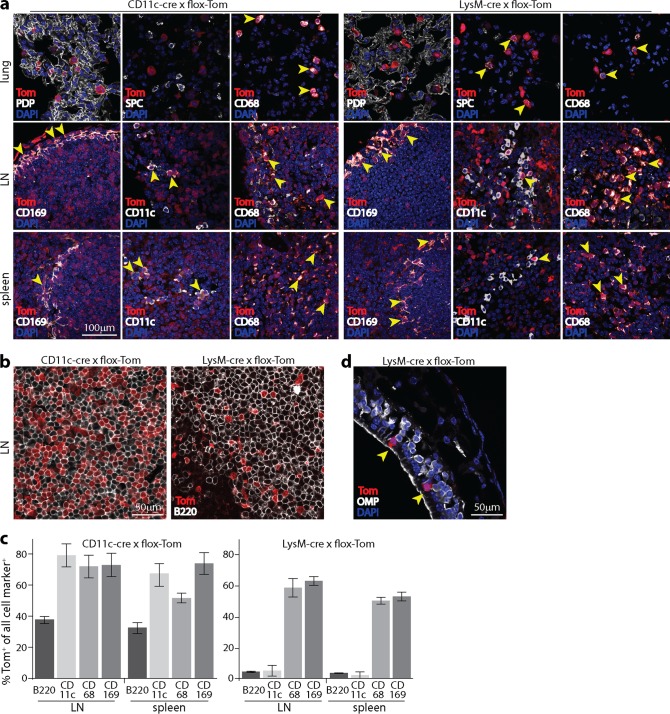
FIG 4.
Distribution of lymphoid cre expression in CD11c-cre and lysM-cre mice. (a) Naive CD11c-cre × flox-Tom and lysM-cre × flox-Tom mice were analyzed for tdTomato (Tom) expression in tissue sections. In lungs, CD11c-cre expression gave Tom+ AM (CD68+) but not AEC1 (PDP+) or AEC2 (SPC+). lysM-cre expression gave Tom+ AEC2 and AM. In LN, the expression of both CD11c-cre and lysM-cre gave Tom+ SSM (CD169+) and other Tom+ myeloid cells (CD68+). CD11c-cre labeled most DC (CD11chi). LysM-cre labeled <10% of DC. In spleens, both CD11c-cre and lysM-cre labeled myeloid cells in the MZ. CD11c-cre labeled most (although not all) CD11c+ cells, while lysM-cre labeled <5% of CD11c+ cells. (b) Example Tom+ fluorescence in B220+ cells of CD11c-cre+ and lysM-cre+ LN. (c) Summary of Tom expression in LN and spleens of lysM-cre+ and CD11c-cre+ mice. Bars show the mean percentages of Tom+ cells ± standard errors of the means for each cell type marker. Cells counts are for at least 3 sections per organ. (d) The olfactory epithelium of lysM-cre × flox-Tom mice contains occasional Tom+ cells, but they are OMP− and thus are not olfactory neurons. They also lack the characteristic epithelium-spanning morphology of sustentacular cells and thus seem likely to be intraepithelial myeloid cells.
IFNAR blockade does not change the predominant route of i.n. MuHV-4 inoculation through LN.
To test the significance of increased SSM and DC infections for viral spread, we gave i.n. to cre+ mice MuHV-4 with a floxed color-switching cassette (MHV-RG) (Fig. 3c). MHV-RG makes mCherry (red) until cre switches it to GFP (green) (19). Thus, counting of red and green plaques reveals the extent of viral passage through cre+ cells. SSM express lysM (14) and CD11c (29), and DC express CD11c but rarely express lysM (30). MHV-RG passing from noses to SCLN switches more in CD11c-cre than in lysM-cre mice (19), consistent with DC providing the main route to B cells (13). Anti-IFNAR treatment increased virus titers in both cre+ mouse strains. However, the percentage of virus switching showed no significant change: regardless of anti-IFNAR treatment, it was higher in lysM-cre than in CD11c-cre mice for noses and was higher in CD11c-cre than in lysM-cre mice for SCLN (Fig. 3c). Therefore, IFN-I limited the spread of MuHV-4 but did not alter the predominant route of spread, via DC.
Cell subset-specific IFNAR disruption.
Because anti-IFNAR treatment increased epithelial infection, increased SSM and DC infections could have been secondary effects. To test more specifically how IFN-I signaling to SSM and DC contributes to MuHV-4 control, we analyzed CD11c-cre × flox-IFNAR (CD11c+ IFNARflox/flox) and lysM-cre × flox-IFNAR mice. These mice should lack IFN-I signaling to CD11c+ and LysM+ cells. Viral fluorochrome switching depends on cre being present at the time of infection; host loxP sites can be recombined earlier. To check the cumulative distribution of cre expression in CD11c-cre and lysM-cre mice, we crossed them with the flox-Tom reporter strain (Fig. 4). Here cre uses the same recognition elements to excise an upstream stop cassette, so the distribution of Tom+ cells should match IFNAR disruption in flox-IFNAR crosses. (The relatively normal leukocyte subsets of IFNAR−/− mice [31] argue against a large effect of IFNAR on cell survival.)
In CD11c-cre × flox-Tom mouse lungs, Tom+ cells were CD68+ CD11c+ SPC− PDP−, consistent with cre expression in DC and AM (both CD68+ CD11c+) and not in AEC1 (PDP+) or AEC2 (SPC+). In LN, Tom expression was seen in CD11c+ cells in the LN substance (presumably DC) and in CD11c+ CD169+ cells lining the subcapsular sinus (SSM) (Fig. 4a). However, not all CD11c+ cells were Tom+, and 38.9% ± 4.4% of B220+ cells were also Tom+ (mean counts ± standard deviations [SD] for >2,000 cells across 10 sections) (Fig. 4b and c). Similarly, in spleens, Tom was expressed in most but not all CD11c+ cells and in 35.3% ± 8.0% of B220+ cells. Some Tom+ B220+ cells may have been plasmacytoid DC (CD11clo B220hi), but they account for <1% of spleen cells. Therefore, CD11c-cre expression was found mainly in CD11c+ cells but less strictly so than originally reported (32) and more in line with what was recently reported for another CD11c-cre transgenic line (33).
LysM-cre is described as being expressed in almost all granulocytes, >80% of macrophages and <20% of DC (30). Recombination has also been noted for AEC2 (18), neurons (34), and some B cells (35). Lungs of lysM-cre × flox-Tom mice contained both CD68+ CD11c+ and SPC+ Tom+ cells, consistent with AM and AEC2 expressing lysM. In LN, CD169+ SSM and other CD68+ macrophages were Tom+, as were CD169+ splenic MZM. Relatively few (<10%) CD11c+ and B220+ cells were Tom+. Olfactory neurons were not Tom+ (Fig. 4d). Thus, comparison of lysM-cre × flox-IFNAR mice with CD11c-cre × flox-IFNAR mice distinguished SSM (CD11c+ LysM+) from DC (CD11c+ lysM−) IFN-I signaling. To control for possible effects of CD11c-cre expression in B cells, we also crossed flox-IFNAR mice with B cell-specific CD19-cre mice (36–38).
IFNAR signaling to CD11c− lysM+ cells limits MuHV-4 spleen infection.
We gave MHV-GFP i.n. to CD11c-cre × flox-IFNAR and lysM-cre × flox-IFNAR mice, either in 30 μl with anesthesia (lung infection) or in 5 μl without anesthesia (upper respiratory tract infection). We compared these mice with cre-negative flox-IFNAR mice, with or without anti-IFNAR treatment (Fig. 5). By day 6 of lung infection, the peak of acute viral lytic replication, anti-IFNAR treatment increased virus titers in lungs, MLN, and spleens. Neither cre+ strain had significantly increased lung titers. Therefore, the IFNAR-dependent restriction of lung infection did not depend on signaling to AM or DC. Surprisingly, MLN virus titers were also not increased in cre+ mice. Therefore, complete IFNAR blockade increased MLN infection mainly by increasing lung infection rather than by acting on myeloid cells in the MLN. Splenic virus titers were increased in lysM-cre × flox-IFNAR but not CD11c-cre × flox-IFNAR mice, consistent with IFN-I defending MZM (9), which were more consistently lysM+ than CD11c+ (Fig. 4a).
FIG 5.
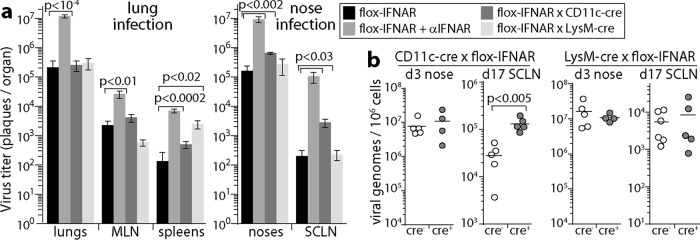
IFNAR loss from CD11c+ cells impairs MuHV-4 control in lymphoid rather than epithelial sites. (a) CD11c-cre × flox-IFNAR, lysM-cre × flox-IFNAR, and cre− flox-IFNAR mice were given MuHV-4 i.n. at either 104 PFU in 30 μl under anesthesia (lung infection) or 105 PFU in 5 μl without anesthesia (nose infection). For complete IFNAR blockade, cre− flox-IFNAR mice were given anti-IFNAR treatment. At day 6, organs were assayed for infectious virus by a plaque assay (lungs and noses) or by an infectious-center assay to also detect reactivatable latent virus (SCLN, MLN, and spleens). Bars show mean titers ± standard errors of the means for 5 mice per group. Significant differences between groups are shown. (b) Cre+ mice or cre− littermate controls were infected i.n. with MuHV-4 (105 PFU in 5 μl without anesthesia). Noses at day 3 and SCLN at day 17 were assayed for infection by quantitative PCR of viral DNA. Signals were normalized by the cellular DNA load, assayed in parallel for each sample. Circles show data for individual mice. Horizontal bars show means. Significant differences are shown.
IFNAR signaling to DC limits SCLN infection.
Nose infection, like lung infection, was increased by anti-IFNAR treatment and not by lysM-cre-dependent or CD11c-cre-dependent IFNAR disruption (Fig. 5a). This result was consistent with most of the infectious virus in noses coming from epithelial cells rather than myeloid cells. However, SCLN infection was significantly increased in CD11c-cre × flox-IFNAR mice. Viral genome measurements (Fig. 5b) similarly showed no difference between loads in noses of cre+ and cre− mice at day 3 (early lytic infection) and increased loads in CD11c-cre × flox-IFNAR SCLN at day 17 (latency). The normal SCLN infection of lysM-cre × flox-IFNAR mice argued that IFNAR signaling to DC (CD11c+ lysM−) was more restrictive than that to SSM (CD11c+ lysM+).
Visualization of LN infection in CD11c-cre × flox-IFNAR mice.
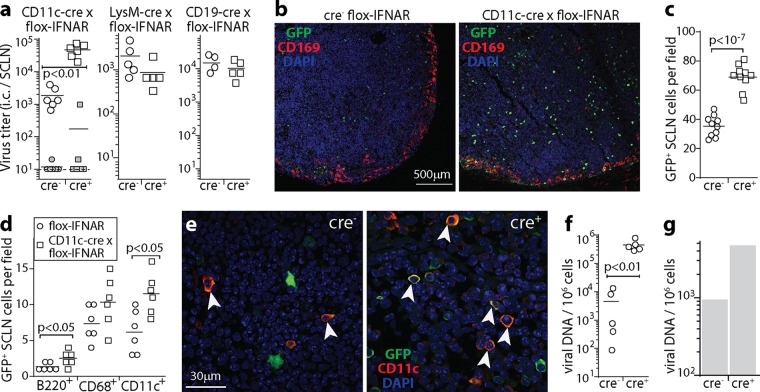
To understand better how IFNAR signaling to CD11c+ cells limited LN infection, we gave mice MHV-GFP i.n. (5 μl) and examined SCLN at day 9, when DC are transferring infectious virus to B cells (Fig. 6). CD11c-cre × flox-IFNAR mice had higher virus titers than did cre− controls, while lysM-cre × flox-IFNAR mice had titers that were the same as those in the controls (Fig. 6a). The titers in CD19-cre × flox-IFNAR mice were also the same as those in the controls, so IFNAR signaling to B cells did not restrict infection. In CD11c-cre × flox-IFNAR LN, titers determined by an infectious-center assay increased, while those determined by a plaque assay did not, so increased infection was latent rather than lytic.
FIG 6.
Tracking lymphoid infection in CD11c-cre × flox-IFNAR mice. (a) CD11c-cre × flox-IFNAR, lysM-cre × flox-IFNAR, CD19-cre × flox-IFNAR, and cre− littermate control mice were given MHV-GFP i.n. (105 PFU in 5 μl). At day 9, infectious-center (i.c.) assays recovered significantly more virus from SCLN of CD11c-cre × flox-IFNAR mice than from controls (open symbols). The closed symbols show plaque titers of preformed infectious virus (freeze-thawed samples), for which CD11c-cre × flox-IFNAR mice did not differ significantly from controls, nor did the i.c. titers in LysM-cre × flox-IFNAR and CD19-cre × flox-IFNAR mice differ from those in controls. Symbols show data for SCLN of individual mice, and bars show group means. Data in the 3 graphs are from experiments done at different times, so comparisons can be made between cre+ and cre− groups within each graph but not between graphs. (b) Mice were infected as described above for panel a. At day 9, SCLN sections were stained for viral GFP plus CD169 to identify SSM. Nuclei were stained with DAPI. (c) Total GFP+ cell counts for SCLN sections from infected CD11c-cre × flox-IFNAR and cre− controls, as described above for panel b, for 10 sections from 3 mice per group, counting at least 5 fields of view per section. Symbols show mean counts for each section, and bars show group means. (d) At day 9, SCLN sections from mice, as described above for panel b, were stained for viral GFP and for B220 (B cells), CD68 (myeloid cells), CD169 (SSM), or CD11c (DC) to identify types of infected cells. Staining was quantitated for 6 sections from 3 mice per group, counting at least 5 fields of view per section. Symbols show mean counts for each section. Bars show group means. Significant differences between groups are shown. Few CD169+ cells were GFP+, so these cells were not counted. (e) Example CD11c staining quantitated as described above for panel d. Colocalization appears in yellow. Arrows show example CD11c +GFP+ cells. (f) CD11c-cre × flox-IFNAR mice and cre− littermate controls were infected as described above for panel a. At day 17, spleens were analyzed for infection by quantitative PCR of viral genomes, normalized by the number cellular genome copies in each sample. Circles show data for individual mice. Bars show means. (g) Mice were infected as described above for panel a, day 17 spleens were pooled from 3 mice per group, and B220hi B cells were purified by flow cytometric sorting. Viral loads were then measured by quantitative PCR as described above for panel f. B cell colonization was greater in CD11c-cre × flox-IFNAR mice than in cre− controls, so when CD11c+ cells lacked IFNAR signaling, more infection passed to B cells.
LN sections showed increased GFP+ cell numbers in CD11c-cre × flox-IFNAR mice (Fig. 6b and c). As with anti-IFNAR treatment, these cells were distributed throughout the LN substance rather than around the subcapsular sinus. Many of these cells were CD11c+ and thus were likely to be DC (Fig. 6d and e). Numbers of B220+ GFP+ cells also increased, consistent with virus transfer from DC to B cells, as did splenic viral genome loads at day 17 (Fig. 6f). Flow cytometric sorting of B220+ spleen cells at day 17 of i.n. infection (Fig. 6g) confirmed higher B cell viral genome loads in CD11c-cre × flox-IFNAR mice than in cre− flox-IFNAR controls. Thus, IFN-I signaling to DC limited their infection and consequently limited the spread of infection to B cells.
IFN-I signaling to myeloid cells also regulates splenic infection.
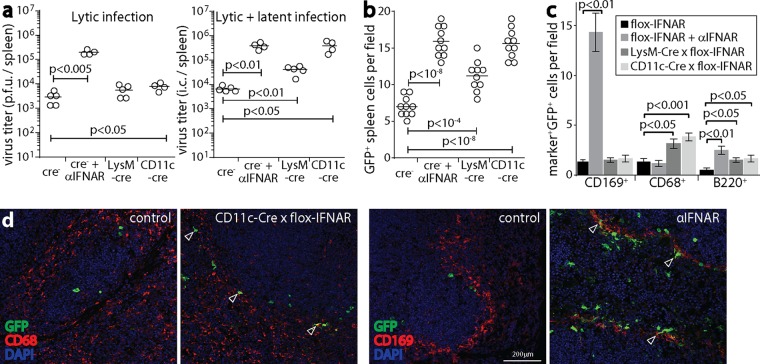
The normal LN and increased spleen infections of i.n. infected LysM-cre × flox-IFNAR mice (Fig. 5a) suggested that IFN-I also restricts MuHV-4 spread by signaling to MZM. To track spleen infection without complicating upstream changes in SCLN infection, we used i.p. inoculation to deliver MHV-GFP directly (15) to the spleens of cre+ and cre− flox-IFNAR mice (Fig. 7). We analyzed spleens at day 4, when infectious virus is passing through myeloid cells (16). Anti-IFNAR treatment increased the titers of infectious virus, consistent with IFN-I restricting lytic MZM infection (9). In CD11c-cre × flox-IFNAR mice, the amount of infectious virus increased only marginally, but the number of infectious centers (lytic virus plus latent virus) increased comparably to those in anti-IFNAR-treated mice. LysM-cre × flox-IFNAR mice similarly showed more latent rather than lytic infection (Fig. 7a). Thus, as in LN, IFN-I acted on myeloid cells to limit their infection upstream of the initiation of the lytic cycle.
FIG 7.
IFNAR signaling to myeloid cells restricts splenic infection by i.p. inoculation of MuHV-4. (a) CD11c-cre × flox-IFNAR, lysM-cre × flox-IFNAR, and cre− flox-IFNAR mice were given MHV-GFP i.p. (105 PFU). As a positive control, cre− mice were also given anti-IFNAR treatment. At day 4, spleens were assayed for lytic infection by a plaque assay and assayed for total recoverable virus (lytic plus latent infection) by an infectious-center (i.c.) assay. Circles show titers in individual mice. Bars show means. Significant differences are shown. (b) Mice were infected as described above for panel a. At day 4, spleen sections were stained for viral GFP. GFP+ cell counts across 10 sections from 3 mice for each group were increased in all groups compared to those for the cre− negative controls. Symbols show mean counts per section from at least 5 fields of view per section. Bars show group means. (c) Spleen sections, as described above for panel b, were stained for viral GFP plus markers of MZM (CD169), myeloid cells in general (CD68), and B cells (B220). Anti-IFNAR treatment increased MZM infection, whereas CD11c-cre and lysM-cre expression increased myeloid cell infection without a focus on MZM. All groups showed significantly more B cell infection than that in cre− negative controls. Bars show mean counts ± standard errors of the means for 10 sections from 3 mice, counting at least 5 fields of view per section. (d) Examples of myeloid cell staining from panel c show increased CD169+ cell infection with anti-IFNAR treatment and increased CD68+ cell infection with CD11c-cre expression. CD11c-cre+ and lysM-cre+ mice had infection around the MZ, but little infection colocalized with CD169, and GFP+ cells were more evenly distributed across the MZ and white pulp. Arrows show example positive cells.
Immunostaining of spleen sections showed viral GFP+ cell numbers consistent with the infectious-center assay results: anti-IFNAR treatment and CD11c-cre expression led to equivalent increases, and lysM-cre expression led to a smaller but still significant increase (Fig. 7b). However, anti-IFNAR treatment led to a markedly different GFP+ cell distribution (Fig. 7c and d). It increased mainly MZ infection, and many GFP+ cells were CD169+ (MZM). In CD11c-cre × flox-IFNAR and lysM-cre × flox-IFNAR mice, GFP+ cells were less concentrated in the MZ, and infection was increased in CD68+ (myeloid cells generally) rather than in CD169+ cells. As many MZM express lysM+ or CD11c+ (Fig. 4a) cells, the greater effect of anti-IFNAR treatment on their infection suggested that after i.p. inoculation of MuHV-4, CD11c− lysM− cells, presumably in other peritoneal organs (39), are a significant, IFN-I-regulated source of infectious virus seeding in MZM.
Numbers of infected B cells were low, as expected at this early time point (16), but cre+ and anti-IFNAR-treated mice all showed more B cell infection (B220+ GFP+) than did cre− flox-IFNAR controls, so again, IFN-I limited viral spread to B cells.
DISCUSSION
Most viruses reproduce as fast as they can. Herpesviruses limit their lytic replication to evade immune defenses. Gammaherpesviruses also do so to drive the proliferation of infected lymphocytes. MuHV-4 establishes a largely latent infection of B cells. Epithelial and myeloid cells have a greater propensity for lytic infection and provide transport between hosts (epithelial) and into the B cell reservoir (myeloid) (40). MuHV-4 has several lytic-cycle IFN-I evasion genes. However, they must act before IFN-I signaling, as this suppresses their expression. Thus, preemptive IFN-I signaling to uninfected cells still limits lytic viral spread. In contrast, transcriptional quiescence makes latent viral genomes inherently IFN-I resistant. Therefore, MuHV-4 should be most vulnerable to IFN-I in epithelial and myeloid cells. Myeloid cell infection, as the immediate gateway to B cells, is of particular interest. We tested how IFN-I signaling to myeloid cells contributes to defense against natural mucosal infection. In peripheral sites, where most new virus comes from epithelial cells, it had little impact; in lymphoid tissue, where myeloid cells transmit infection to B cells, it was important and limited mainly the colonization of DC.
To understand complex infections, we must distinguish direct from knock-on effects. Blocking all IFN-I signaling increased lytic SSM infection. Blocking IFN-I signaling to just CD11c+ cells did not, even though SSM are CD11c+. Therefore, increased SSM infection reflected increased peripheral infection, which then seeded to SSM via lymphatics. This was associated with olfactory infection spreading to subepithelial and respiratory epithelial cells. Increased SSM infection after IFNAR blockade is recapitulated by i.f. inoculation of MuHV-4, which directly delivers cell-free virions to LN (24). Normally, olfactory infection bypassed SSM by reaching LN in migrating DC.
Blocking of IFNAR after i.n. infection increased the number of viral GFP+ DC in LN, and this effect was reproduced by a CD11c+ cell-specific IFNAR blockade without increasing peripheral infection. Therefore, here IFN-I directly protected DC. IFN-I production by conventional DC depends on positive feedback through IFNAR (41), so disrupting IFNAR in CD11c+ cells might have reduced the protection of CD11c− cells by DC-derived IFN-I. However, peripheral infection was not increased, and most of the additional GFP+ cells were CD11c+. Therefore, IFN-I appeared to suppress DC infection directly. In spleens, where MuHV-4 spreads via lysM+ MZM and CD11c+ DC (15), IFN-I signaling to lysM+ and CD11c+ cells restricted infection primarily in lysM+ and CD11c+ cells. Thus, IFN-I signaling to myeloid cells consistently limited normal virus flux through lymphoid tissue. After invasive inoculations, IFN-I more prominently blocks atypical pathways of MuHV-4 spread. For example, after i.f. inoculation, it protects SSM (24), and after i.p. inoculation, it prevents widespread splenic macrophage infection (9). IFNAR blockade also increased SSM and splenic macrophage infections after olfactory MuHV-4 inoculation, but the effect was less dramatic than that after invasive inoculations because virus delivery to the relevant cells was less dramatic.
In both LN and spleens, increased DC infection was associated with higher latent rather than lytic virus titers and with more CD11c+ cells expressing lytic-cycle-independent viral GFP but not viral lytic antigens. Some DC infection must be lytic, as it was passed to B cells, but the extent of lytic infection evidently had limits other than IFN-I. Instead, IFNAR blockade seemed to rescue viral genomes from silencing or destruction. A relevant IFN-I action is its upregulation of antiviral ND10 domains (42). The viral ORF75c tegument protein normally disrupts these domains (43, 44) but presumably does so less well when they are primed by IFN-I. When IFNAR blockade increases vesicular stomatitis virus replication, there is greater immune priming (45), so MuHV-4 may restrict unnecessary lytic infection to reach B cells with minimal immunostimulation.
Because DC infection plays a key role in the MuHV-4 life cycle, it is protected by viral evasion, for example, through major histocompatibility complex (MHC) class I downregulation (46). However, viral compromises may be necessary to allow the exploitation of host immune functions. For example, CD4+ T cells may be difficult for MuHV-4 to evade because it needs CD4+ T cell engagement to drive infected B cell proliferation in germinal centers (47). The need of MuHV-4 for normal DC functions such as migration and B cell engagement may limit its capacity to evade DC-directed IFN-I. CD4+ T cells engage DC and produce IFN-γ, which synergizes with IFN-I (48) and shows myeloid cell-specific MuHV-4 suppression (49). Normally, MuHV-4 reaches DC before a strong CD4+ T cell response is mounted (50). However, immune priming allows earlier defense, and CD4+ T cells contribute to protection against MuHV-4 in a live-attenuated vaccine (51). Thus, DC-directed IFN-I signaling might be a way to reduce gammaherpesvirus colonization of B cells.
MATERIALS AND METHODS
Mice.
BALB/c, C57BL/6J, flox-IFNAR (52), ROSA26-loxP-stop-loxP-tdTomato (flox-Tom) (53), CD11c-cre (32), lysM-cre (30), and CD19-cre (36) mice were maintained at University of Queensland animal units. The flox-IFNAR allele is functional until recombination by cre deletes exon 10 to truncate it upstream of the transmembrane domain and thus prevent signaling. For cell type-specific IFNAR disruption, the CD11c-cre, lysM-cre, and CD19-cre strains were backcrossed onto the homozygous flox-IFNAR strain. The floxed allele was detected by PCR using primers 5′-CTTTTTGGATCGATCCATAACTTCG (loxP specific), 5′-GACAACTTTTTGGATATCAAGAAAGC (wild-type specific), and 5′-CTGGATTATAACTGGATTAGGTGTTG (common). The cre transgenes were maintained as heterozygous transgenes. To identify transgenic CD11c-cre and lysM-cre expression, each strain was crossed with the homozygous flox-Tom strain, and 6-week-old cre+ F1 progeny were analyzed. Mice were given MuHV-4 when they were 6 to 8 weeks old, either i.n. in 30 μl under isoflurane anesthesia to inoculate the lungs (3 × 104 PFU), i.n. in 5 μl without anesthesia to inoculate the upper respiratory tract (105 PFU), or i.p. (105 PFU). We used a higher dose for upper than for lower respiratory tract inoculations because experimental fluid delivery to the upper respiratory tract is less efficient: most of a 5-μl inoculum is swallowed (17). Thirty microliters of i.n. inocula also reaches the upper respiratory tract. However, little fluid is retained here, as most goes to the lungs (17), and MuHV-4 also replicates more extensively in the lungs than in the upper respiratory tract, so lung infection dominates the outcome. Luciferase-positive MuHV-4 infection (MHV-LUC) (39) was imaged by i.p. injection of d-luciferin (2 mg; Pure Science) and scanning with a charge-coupled-device camera (IVIS Spectrum) and analyzed with Living Image software (PerkinElmer). IFN-αβR signaling was blocked by i.p. injection of monoclonal antibody (MAb) MAR-5A3 (100 μg/mouse every 2 days) (Bio X Cell). Animal experiments were approved by the University of Queensland Animal Ethics Committees in accordance with Australian National Health and Medical Research Council (NHMRC) guidelines. Statistical comparison was performed by using Student's 2-tailed unpaired t test unless otherwise stated.
Cells and viruses.
Bovine hamster kidney (BHK-21) fibroblasts (ATCC CCL-10) were grown in Dulbecco's modified Eagle's medium with 2 mM glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (complete medium). All viruses were derived from bacterial artificial chromosome (BAC)-cloned (54) MuHV-4 clone g2.4 (55). MHV-LUC makes luciferase under the control of an ectopic viral M3 lytic promoter (39). MHV-GFP expresses GFP from an EF1α promoter, inserted between the 3′ ends of ORFs 57 and 58. This operates independently of viral lytic gene expression (25). MHV-RG (19) has an M3 promoter driving loxP-flanked mCherry plus a stop cassette, upstream of GFP, at the same site. Thus, it forms mCherry+ plaques until loxP recombination by cre deletes mCherry plus the stop cassette, when it forms GFP+ plaques. The loxP sites of the color-switching cassette have an altered spacer region to make them incompatible with those flanking the BAC cassette. MuHV-4 was grown and titers were determined on BHK-21 cells. Virions were harvested from an infected cell culture by ultracentrifugation (30,000 × g for 120 min), and cell debris was removed by low-speed centrifugation (500 × g for 10 min).
Virus assays.
To determine the titer of infectious MuHV-4, culture-grown stocks or freeze-thawed organ homogenates were plated onto BHK-21 cells (56). To determine the titer of total reactivatable MuHV-4, organs were disrupted into single-cell suspensions and then plated onto BHK-21 cells. The cells were cultured in complete medium for 3 h, overlaid with complete medium plus 0.3% carboxymethyl cellulose, cultured for 4 days, and then fixed with 1% formaldehyde and stained with 0.1% toluidine blue for plaque counting.
Immunostaining.
Organs were fixed in 1% formaldehyde–10 mM sodium periodate–75 mM l-lysine (18 h at 4°C), equilibrated in 30% sucrose (24 h at 4°C), and then frozen in OCT. Sections (6 μm) were air dried (1 h, 23°C), washed three times in phosphate-buffered saline (PBS), blocked with 0.3% Triton X-100–5% normal donkey serum (1 h at 23°C), and then incubated (18 h at 4°C) with combinations of antibodies to GFP (rabbit or goat polyclonal antibody [PAb]), CD11c (hamster MAb HL-3; BD Pharmingen), surfactant C precursor (goat PAb; Santa Cruz Biotechnology), olfactory marker protein (goat PAb; Wako Chemicals), CD68 (rat MAb FA-11; Abcam), B220 (rat MAb RA3-6B2), CD169 (rat MAb 3D6.112) (Serotec), podoplanin (goat PAb; R&D Systems), and MuHV-4 (polyclonal rabbit serum). The latter was raised by 2 subcutaneous inoculations of MuHV-4 virions (109 PFU). Like previously described immune sera (57), it recognizes a range of virion proteins by Western blotting and strongly recognizes the products of ORF4 (gp70), M7 (gp150), and ORF65 (p20). After incubation with the primary antibodies, sections were washed three times in PBS; incubated (for 1 h at 23°C) with combinations of Alexa 568-conjugated donkey anti-rat IgG PAb, Alexa 488- or Alexa 647-conjugated donkey anti-rabbit IgG PAb, Alexa 647-conjugated donkey anti-mouse IgM PAb, and Alexa 488-conjugatged donkey anti-goat PAb (Life Technologies); and then washed three times in PBS, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in Prolong Gold (Life Technologies). tdTomato fluorescence was visualized directly. Images were captured with a Zeiss LCM510 confocal microscope or a Nikon epifluorescence microscope and analyzed with ImageJ.
Viral genome quantitation.
MuHV-4 genomic positions 24832 to 25071 were amplified by PCR (Rotor Gene 3000; Corbett Research) from 10 ng DNA (NucleoSpin tissue kit; Macherey-Nagel). PCR products quantified with Sybr green (Invitrogen) were compared to a standard curve of a cloned template amplified in parallel and distinguished from paired primers by melting-curve analysis. Correct sizing was confirmed by electrophoresis and ethidium bromide staining. The amount of cellular DNA in the same samples was quantified by amplifying a β-actin gene fragment.
Flow cytometric sorting.
Splenocytes of infected mice were separated from erythrocytes by centrifuging freshly isolated spleen cell suspensions on a Ficoll gradient. B cells were stained with phycoerythrin-conjugated anti-B220 MAb (BD Biosciences), washed twice, and sorted into B220+ and B220− fractions on a BD FACSAria II instrument. DNA was extracted from each sorted population for viral genome quantitation.
ACKNOWLEDGMENTS
We thank Ian Fraser for flox-Tom and lysM-cre mice, Gabrielle Belz for CD11c-cre mice, and Chris Engwerda for flox-IFNAR mice. We thank Michael Nefedov for help with flow cytometric sorting.
The work was supported by grants from the Australian Research Council (FT130100138) and the NHMRC (project grants 1064015, 1079180, and 1122070).
REFERENCES
- 1.Schulz KS, Mossman KL. 2016. Viral evasion strategies in type I IFN signaling—a summary of recent developments. Front Immunol 7:498. doi: 10.3389/fimmu.2016.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang S, Kim KS, Flano E, Wu TT, Tong LM, Park AN, Song MJ, Sanchez DJ, O'Connell RM, Cheng G, Sun R. 2009. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5:166–178. doi: 10.1016/j.chom.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang HR, Cheong WC, Park JE, Ryu S, Cho HJ, Youn H, Ahn JH, Song MJ. 2014. Murine gammaherpesvirus 68 encoding open reading frame 11 targets TANK binding kinase 1 to negatively regulate the host type I interferon response. J Virol 88:6832–6846. doi: 10.1128/JVI.03460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang X, Shin YC, Means RE, Jung JU. 2004. Inhibition of interferon-mediated antiviral activity by murine gammaherpesvirus 68 latency-associated M2 protein. J Virol 78:12416–12427. doi: 10.1128/JVI.78.22.12416-12427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leang RS, Wu TT, Hwang S, Liang LT, Tong L, Truong JT, Sun R. 2011. The anti-interferon activity of conserved viral dUTPase ORF54 is essential for an effective MHV-68 infection. PLoS Pathog 7:e1002292. doi: 10.1371/journal.ppat.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheridan V, Polychronopoulos L, Dutia BM, Ebrahimi B. 2014. A shutoff and exonuclease mutant of murine gammaherpesvirus-68 yields infectious virus and causes RNA loss in type I interferon receptor knockout cells. J Gen Virol 95:1135–1143. doi: 10.1099/vir.0.059329-0. [DOI] [PubMed] [Google Scholar]
- 7.Dutia BM, Allen DJ, Dyson H, Nash AA. 1999. Type I interferons and IRF-1 play a critical role in the control of a gammaherpesvirus infection. Virology 261:173–179. doi: 10.1006/viro.1999.9834. [DOI] [PubMed] [Google Scholar]
- 8.Sunil-Chandra NP, Efstathiou S, Nash AA. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol 73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 9.Tan CS, Lawler C, May JS, Belz GT, Stevenson PG. 2016. Type I interferons direct gammaherpesvirus host colonization. PLoS Pathog 12:e1005654. doi: 10.1371/journal.ppat.1005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal P, Krueger BE, Oldenburg D, Andry KA, Beard RS, White DW, Barton ES. 2011. A gammaherpesvirus cooperates with interferon-alpha/beta-induced IRF2 to halt viral replication, control reactivation, and minimize host lethality. PLoS Pathog 7:e1002371. doi: 10.1371/journal.ppat.1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin MM, Canny S, Steed A, Virgin HW. 2010. Murine gammaherpesvirus 68 has evolved gamma interferon and stat1-repressible promoters for the lytic switch gene 50. J Virol 84:3711–3717. doi: 10.1128/JVI.02099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milho R, Frederico B, Efstathiou S, Stevenson PG. 2012. A heparan-dependent herpesvirus targets the olfactory neuroepithelium for host entry. PLoS Pathog 8:e1002986. doi: 10.1371/journal.ppat.1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaspar M, May JS, Sukla S, Frederico B, Gill MB, Smith CM, Belz GT, Stevenson PG. 2011. Murid herpesvirus-4 exploits dendritic cells to infect B cells. PLoS Pathog 7:e1002346. doi: 10.1371/journal.ppat.1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frederico B, Chao B, Lawler C, May JS, Stevenson PG. 2015. Subcapsular sinus macrophages limit acute gammaherpesvirus dissemination. J Gen Virol 96:2314–2327. doi: 10.1099/vir.0.000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frederico B, Chao B, May JS, Belz GT, Stevenson PG. 2014. A murid gamma-herpesviruses exploits normal splenic immune communication routes for systemic spread. Cell Host Microbe 15:457–470. doi: 10.1016/j.chom.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Chao B, Frederico B, Stevenson PG. 2015. B-cell-independent lymphoid tissue infection by a B-cell-tropic rhadinovirus. J Gen Virol 96:2788–2793. doi: 10.1099/vir.0.000188. [DOI] [PubMed] [Google Scholar]
- 17.Tan CS, Frederico B, Stevenson PG. 2014. Herpesvirus delivery to the murine respiratory tract. J Virol Methods 206:105–114. doi: 10.1016/j.jviromet.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Lawler C, Milho R, May JS, Stevenson PG. 2015. Rhadinovirus host entry by co-operative infection. PLoS Pathog 11:e1004761. doi: 10.1371/journal.ppat.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frederico B, Milho R, May JS, Gillet L, Stevenson PG. 2012. Myeloid infection links epithelial and B cell tropisms of murid herpesvirus-4. PLoS Pathog 8:e1002935. doi: 10.1371/journal.ppat.1002935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaz P, Whiteley PL, Wilks CR, Browning GF, Gilkerson JR, Ficorilli N, Devlin JM. 2012. Detection of a second novel gammaherpesvirus in a free-ranging koala (Phascolarctos cinereus). J Wildl Dis 48:226–229. doi: 10.7589/0090-3558-48.1.226. [DOI] [PubMed] [Google Scholar]
- 21.De Witte L, de Vries RD, van der Vlist M, Yuksel S, Litjens M, de Swart RL, Geijtenbeek TB. 2008. DC-SIGN and CD150 have distinct roles in transmission of measles virus from dendritic cells to T-lymphocytes. PLoS Pathog 4:e1000049. doi: 10.1371/journal.ppat.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piguet V, Steinman RM. 2007. The interaction of HIV with dendritic cells: outcomes and pathways. Trends Immunol 28:503–510. doi: 10.1016/j.it.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillet L, Frederico B, Stevenson PG. 2015. Host entry by gamma-herpesviruses—lessons from animal viruses? Curr Opin Virol 15:34–40. doi: 10.1016/j.coviro.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Lawler C, Tan CS, Simas JP, Stevenson PG. 2016. Type I interferons and NK cells restrict gammaherpesvirus lymph node infection. J Virol 90:9046–9057. doi: 10.1128/JVI.01108-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.May JS, Stevenson PG. 2010. Vaccination with murid herpesvirus-4 glycoprotein B reduces viral lytic replication but does not induce detectable virion neutralization. J Gen Virol 91:2542–2552. doi: 10.1099/vir.0.023085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swiecki M, Colonna M. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iannacone M, Moseman EA, Tonti E, Bosurgi L, Junt T, Henrickson SE, Whelan SP, Guidotti LG, von Andrian UH. 2010. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature 465:1079–1083. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hume DA. 2011. Applications of myeloid-specific promoters in transgenic mice support in vivo imaging and functional genomics but do not support the concept of distinct macrophage and dendritic cell lineages or roles in immunity. J Leukoc Biol 89:525–538. doi: 10.1189/jlb.0810472. [DOI] [PubMed] [Google Scholar]
- 29.Farrell HE, Bruce K, Lawler C, Cardin RD, Davis-Poynter NJ, Stevenson PG. 2016. Type 1 interferons and NK cells limit murine cytomegalovirus escape from the lymph node subcapsular sinus. PLoS Pathog 12:e1006069. doi: 10.1371/journal.ppat.1006069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 31.Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. 1995. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci U S A 92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caton ML, Smith-Raska MR, Reizis B. 2007. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J Exp Med 204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones MB, Ryan SO, Johnson JL, Cobb BA. 2016. Dendritic cell-specific Mgat2 knockout mice show antigen presentation defects but reveal an unexpected CD11c expression pattern. Glycobiology 26:1007–1013. doi: 10.1093/glycob/cww056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orthgiess J, Gericke M, Immig K, Schulz A, Hirrlinger J, Bechmann I, Eilers J. 2016. Neurons exhibit Lyz2 promoter activity in vivo: implications for using LysM-Cre mice in myeloid cell research. Eur J Immunol 46:1529–1532. doi: 10.1002/eji.201546108. [DOI] [PubMed] [Google Scholar]
- 35.Ye M, Iwasaki H, Laiosa CV, Stadtfeld M, Xie H, Heck S, Clausen B, Akashi K, Graf T. 2003. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19:689–699. doi: 10.1016/S1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- 36.Rickert RC, Roes J, Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoda M, Li T, Pihkala JP, Koni PA. 2006. Role of MHC class II on memory B cells in post-germinal center B cell homeostasis and memory response. J Immunol 176:2122–2133. doi: 10.4049/jimmunol.176.4.2122. [DOI] [PubMed] [Google Scholar]
- 38.Turner VM, Gardam S, Brink R. 2010. Lineage-specific transgene expression in hematopoietic cells using a Cre-regulated retroviral vector. J Immunol Methods 360:162–166. doi: 10.1016/j.jim.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Milho R, Smith CM, Marques S, Alenquer M, May JS, Gillet L, Gaspar M, Efstathiou S, Simas JP, Stevenson PG. 2009. In vivo imaging of murid herpesvirus-4 infection. J Gen Virol 90:21–32. doi: 10.1099/vir.0.006569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marques S, Efstathiou S, Smith KG, Haury M, Simas JP. 2003. Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. J Virol 77:7308–7318. doi: 10.1128/JVI.77.13.7308-7318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Weerd NA, Samarajiwa SA, Hertzog PJ. 2007. Type I interferon receptors: biochemistry and biological functions. J Biol Chem 282:20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 42.Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Gaspar M, Gill MB, Lösing JB, May JS, Stevenson PG. 2008. Multiple functions for ORF75c in murid herpesvirus-4 infection. PLoS One 3:e2781. doi: 10.1371/journal.pone.0002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling PD, Tan J, Sewatanon J, Peng R. 2008. Murine gammaherpesvirus 68 open reading frame 75c tegument protein induces the degradation of PML and is essential for production of infectious virus. J Virol 82:8000–8012. doi: 10.1128/JVI.02752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honke N, Shaabani N, Cadeddu G, Sorg UR, Zhang DE, Trilling M, Klingel K, Sauter M, Kandolf R, Gailus N, van Rooijen N, Burkart C, Baldus SE, Grusdat M, Löhning M, Hengel H, Pfeffer K, Tanaka M, Häussinger D, Recher M, Lang PA, Lang KS. 2011. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol 13:51–57. doi: 10.1038/ni.2169. [DOI] [PubMed] [Google Scholar]
- 46.Smith CM, Gill MB, May JS, Stevenson PG. 2007. Murine gammaherpesvirus-68 inhibits antigen presentation by dendritic cells. PLoS One 2:e1048. doi: 10.1371/journal.pone.0001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Usherwood EJ, Ross AJ, Allen DJ, Nash AA. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J Gen Virol 77:627–630. doi: 10.1099/0022-1317-77-4-627. [DOI] [PubMed] [Google Scholar]
- 48.Luker GD, Prior JL, Song J, Pica CM, Leib DA. 2003. Bioluminescence imaging reveals systemic dissemination of herpes simplex virus type 1 in the absence of interferon receptors. J Virol 77:11082–11093. doi: 10.1128/JVI.77.20.11082-11093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steed A, Buch T, Waisman A, Virgin HW. 2007. Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner. J Virol 81:6134–6140. doi: 10.1128/JVI.00108-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson PG, Doherty PC. 1998. Kinetic analysis of the specific host response to a murine gammaherpesvirus. J Virol 72:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClellan JS, Tibbetts SA, Gangappa S, Brett KA, Virgin HW. 2004. Critical role of CD4 T cells in an antibody-independent mechanism of vaccination against gammaherpesvirus latency. J Virol 78:6836–6845. doi: 10.1128/JVI.78.13.6836-6845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. 2006. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood 108:3253–3261. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- 53.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74:6964–6974. doi: 10.1128/JVI.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efstathiou S, Ho YM, Minson AC. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol 71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 56.de Lima BD, May JS, Stevenson PG. 2004. Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J Virol 78:5103–5112. doi: 10.1128/JVI.78.10.5103-5112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gillet L, Adler H, Stevenson PG. 2007. Glycosaminoglycan interactions in murine gammaherpesvirus-68 infection. PLoS One 2:e347. doi: 10.1371/journal.pone.0000347. [DOI] [PMC free article] [PubMed] [Google Scholar]