ABSTRACT
Our genomes are dominated by repetitive elements. The majority of these elements derive from retrotransposons, which expand throughout the genome through a process of reverse transcription and integration. Short interspersed nuclear elements, or SINEs, are an abundant class of retrotransposons that are transcribed by RNA polymerase III, thus generating exclusively noncoding RNA (ncRNA) that must hijack the machinery required for their transposition. SINE loci are generally transcriptionally repressed in somatic cells but can be robustly induced upon infection with multiple DNA viruses. Recent research has focused on the gene expression and signaling events that are modulated by SINE ncRNAs, particularly during gammaherpesvirus infection. Here, we review the biology of these SINE ncRNAs, explore how DNA virus infection may lead to their induction, and describe how novel gene regulatory and immune-related functions of these ncRNAs may impact the viral life cycle.
KEYWORDS: B2, DNA virus, MHV68, SINE, noncoding RNA, retrotransposon
INTRODUCTION
Over one million retrotransposons called short interspersed nuclear elements, or SINEs, reside in our genome (1–3). Their impressive copy number is a direct result of their “copy and paste” mechanism of replication, which occurs by reverse transcription of expressed SINE RNAs, followed by integration of newly copied SINE cDNA. SINEs are unique in that they are transcribed by RNA polymerase III (pol III), a polymerase responsible for the expression of essential housekeeping genes such as transfer RNAs (tRNAs) and the 5S rRNA. Like other pol III genes, SINEs produce short noncoding RNAs (ncRNAs) (typically <500 bp) and therefore do not encode the protein machinery to assist with their retrotransposition (4). Proteins with reverse transcriptase and integrase activities required for replication of SINEs are provided by pol II-dependent retrotransposons called long interspersed nuclear elements (LINEs) (5, 6). Thus, SINEs are not self-sufficient; they parasitize not only our genomes but other retrotransposons as well.
There is incredible diversity regarding the abundance and types of SINEs in animal genomes, with the latter defined in part by the pol III gene from which they are ancestrally derived. The most abundant SINE in the human genome is the Alu SINE, named for an AluI restriction site present in these repeats (1). The copy number of Alu is over 1 million, with each element approximately 300 bases long organized as a dimeric structure, with each monomer derived from the pol III-transcribed 7SL RNA gene (7). The human genome also contains a much less abundant SINE called SVA, whose copy number reaches only approximately 3,000 and likely relies on RNA pol II for its transcription (8). In mice, four different classes of SINEs are present: B1 (∼550,000 copies), B2 (∼350,000), ID (∼80,000), and B4 (∼400,000) elements (3). B1 and B2 are the best studied murine SINEs—they are known to be active in the mouse genome (6). The B1 elements, like Alu elements, are derived from 7SL, while B2 elements most closely resemble a tRNA gene (9, 10). B1 and B2 are both monomeric in structure and are ∼135 bases long and ∼200 bases long, respectively (4).
Like their ancestral and modern pol III gene counterparts, the internal promoter of a SINE element comprises two stretches of conserved sequences called the A and B boxes. These sequences, which are classified as type II pol III promoters, bind a transcription factor complex called TFIIIC, which then recruits the TFIIIB complex and pol III to the promoter for transcription. Although pol III genes produce the most abundant RNAs in the cell, SINEs are expressed at extremely low levels in healthy somatic cells. This has been attributed to rampant DNA methylation marks found at repetitive sequences in the genome (11–13), but a recent study has also highlighted the importance of histone methylation in control of SINE expression (14). It is thought that the silencing of SINEs is a preventive measure against uncontrolled amplification that could be detrimental to the integrity of the host genome.
SINE loci are normally silent, but they can be robustly induced upon infection with a number of DNA viruses, including members of the Adenoviridae, Polyomaviridae, Parvoviridae, and Herpesviridae families, as well as in certain developmental and stress-related contexts (15–22). Simian virus 40 (SV40), minute virus of mice (MVM), and murine gammaherpesvirus 68 (MHV68) lead to induction of murine B1 and B2 SINEs (18, 22, 23), and Alu induction has been described for both adenovirus and herpes simplex virus infection (15–17). While SINE expression might correlate with increased retrotransposition frequency, SINE retrotransposition is a rare event, and it is not known whether DNA virus infection leads to the amplification of genomic SINE copies. However, SINE ncRNAs have demonstrated functions separate from those occurring during retrotransposition, and there is now evidence that SINE ncRNA transcripts influence viral replication, particularly in the context of immune signaling.
DNA VIRUSES UPREGULATE RNA POL III ACTIVITY
The transcriptional activation of SINEs during infection is likely linked, in part, to the increased pol III activity that is often observed upon infection with DNA tumor viruses. Enhanced expression of pol III genes such as tRNAs and 5S rRNA is associated with transformation of cells (24) and may contribute to the oncogenic environment during DNA tumor virus infection. Furthermore, many DNA viruses require the activity of pol III for the expression of select viral ncRNAs, including adenoviral viral-associated (VA) RNAs, MHV68 tRNA-micro RNA (miRNA) encoding RNAs (TMERs), and Epstein-Barr virus (EBV)-encoded small RNAs (EBERs) (25–27). Pol III activation during DNA tumor virus infection is frequently linked to functions associated with viral oncogenes. During EBV infection, the viral EBNA1 protein boosts production of transcription factor complexes (TFIIIC, TFIIIB, c-Myc) required for the expression of pol III genes (28, 29). Similarly, adenovirus and SV40 promote TFIIIC expression and/or activity through the action of the oncogenic E1A and the large T antigen proteins, respectively (30–34). The large T antigens of polyomavirus and SV40 also elevate pol III activity by sequestering the retinoblastoma protein (Rb), thereby preventing it from binding and inhibiting Brf1-TFIIIB association with pol III promoters (30, 31, 35).
One key question is the extent to which increased pol III transcription factor availability drives the induction of SINEs (or other pol III genes). TFIIIC induction correlates well with phenotypes observed during EBV infection, as there is increased transcription of TFIIIC-dependent 5S rRNA, tRNA, and 7SL RNA genes (type I and II promoters), but not with TFIIIC-independent U6 or 7SK genes (type III promoters) (29). In contrast, while the gammaherpesvirus MHV68 induces B1 and B2 SINEs (type II promoters), levels of other endogenous type I/II pol III promoters containing genes such as 5S, tRNA, and 7SL RNAs remain stable (23). We have not observed changes in TFIIIC levels during MHV68 infection (unpublished observations), suggesting that additional mechanisms drive selective pol III promoter activation in this context.
Given the vast overrepresentation of SINE promoters relative to those of other pol III genes and the observation that SINE loci are epigenetically silenced (11–14), other signaling events presumably help direct virus-induced retrotransposon activation. In this regard, sequencing of independently expressed B2 SINE RNAs (SINE-seq) during MHV68 infection reveals that only a subset of B2 SINE loci in the genome are induced, suggesting that additional layers of transcriptional regulation are operational (36). This is supported by the observation that intact promoter elements do not predict B2 SINE expression levels or strictly define which loci are active during infection (36).
CONSEQUENCES OF SINE EXPRESSION
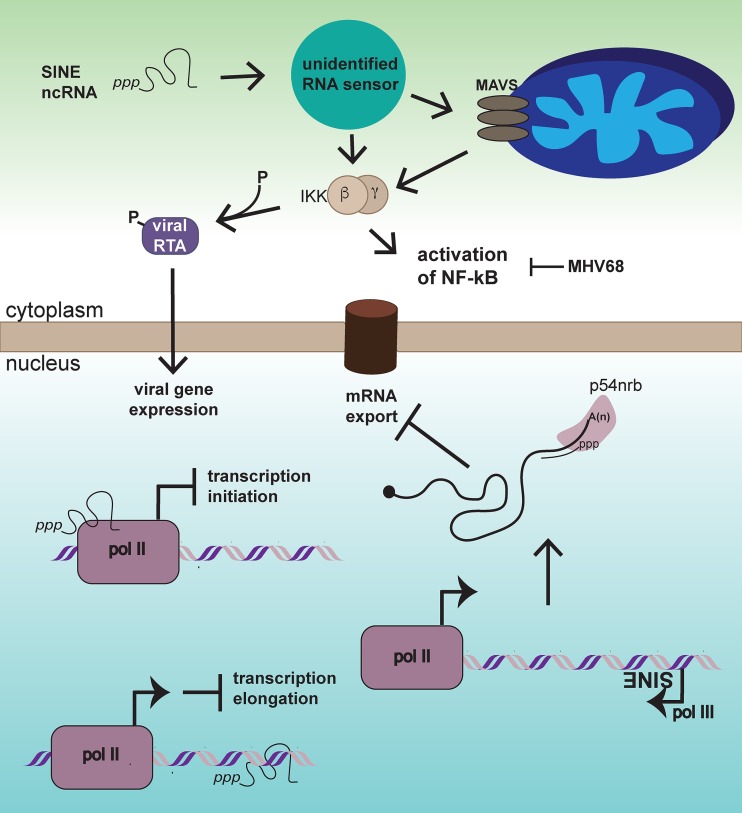
When induced, SINE ncRNAs are localized to both the nucleus and cytoplasm, affecting distinct cellular process in each cellular compartment (Fig. 1). In the nucleus, SINE ncRNAs induced upon heat shock downregulate a subset of pol II genes by binding directly to pol II and preventing transcription initiation (37, 38). A recent CHART-seq study designed to identify B2 ncRNA-associated chromatin further suggested that nuclear B2 ncRNA may block transcriptional elongation by acting as a molecular “speed bump” (29). Induction of specific SINE loci could also impact gene expression by altering the chromatin context of neighboring genes or by directly interacting with nascent transcripts, as many SINEs are embedded within mRNAs. In the case of MHV68 infection of 3T3 cells, SINE-seq analyses revealed that the majority of induced B2 SINE loci are located within or proximal to RNA pol II transcribed genes, including in the antisense orientation within 3′ untranscribed regions (UTRs) of mRNAs (36). As proof-of-concept, B2 ncRNAs expressed from the SGOL2 locus specifically during MHV68 infection were shown to base pair with SGOL2 mRNA, leading to nuclear retention of this transcript through a mechanism involving binding by the p54nrb protein. Additionally, RNA pol II genes located proximal to MHV68-induced SINEs were ontologically enriched in the regulation of gene expression (36). Although the functional consequences of these observations toward the viral life cycle have yet to be explored, they suggest that identifying which SINE loci become activated is relevant from a gene regulatory perspective.
FIG 1.
SINE ncRNAs can impact cellular function by distinct mechanisms. Once expression is induced by viral infection or other contexts, nuclear SINE ncRNA can disrupt transcription by binding and inhibiting RNA pol II at the step of initiation, or by binding to genomic loci and inhibiting pol II elongation. Independently expressed SINEs embedded in an antisense orientation to mRNAs can base pair with their cognate mRNA, causing recognition by p54nrb and blocking of nuclear export. In the cytoplasm, SINE ncRNAs trigger NF-κB, likely due to a resemblance to pathogen-associated molecular patterns (PAMPs), i.e., 5′ triphosphate end and double-stranded regions. In the context of MHV68 infection, SINE ncRNAs activate NF-κB through both MAVS-dependent and -independent mechanisms. MHV68 co-opts IKKβ to phosphorylate the major viral transcriptional regulator RTA to enhance viral gene expression, while simultaneously blocking the downstream effects of NF-κB activation. The factor(s) responsible for sensing SINE ncRNAs remains unidentified. See the text for references.
Recent findings indicate that SINE ncRNAs exported to the cytoplasm can impact the course of viral infection by stimulating immune signaling pathways (23). SINE ncRNA contains double-stranded regions and usually possesses 5′ triphosphate ends (39–42), and several lines of evidence lend support to the idea that these RNAs could be immune stimulatory. First, introduction of Alu or B2 SINE plasmids into various cell types is sufficient to activate the NF-κB signaling pathway (23, 43). Furthermore, transfection of in vitro transcribed Alu or B2 ncRNA can lead to cytokine release or cytotoxicity, respectively (although the sensing of transfected SINE RNA is likely distinct in some aspects to endogenous transcription) (44, 45). There is also evidence of a role for SINE ncRNA in human disease, suggesting that control of SINE expression is biologically relevant beyond preventing the spread of these repetitive elements. Alu and B2 ncRNAs are implicated in the degeneration of the retinal pigmented epithelium during geographic atrophy, a type of age-related macular degeneration (46). An abundance of SINE ncRNAs in this tissue leads to activation of NF-κB and the inflammasome, ultimately leading to apoptosis of these cells (43). Additionally, Alu expression is elevated in association with altered immune-related gene expression patterns in the absence of the Alu-binding protein Ro60, a protein recognized by autoantibodies in patients with systemic lupus erythematosus (SLE) or Sjögren's syndrome (44).
Given the above observations, it seems logical that virus-induced SINEs might play a role in the innate immune response to infection. Indeed, induced B2 SINE ncRNA in the cytoplasm was recently shown to contribute to the rapid activation of NF-κB that occurs during lytic MHV68 infection (Fig. 1) (23). NF-κB activation by B2 SINE ncRNA is partially dependent on the presence of the mitochondrial antiviral-signaling protein (MAVS) adaptor protein, which is known to be stimulated by RIG-I-like receptors following binding of specific RNA structures (23, 47). However, the full extent of NF-κB stimulation cannot be explained by MAVS signaling, and the identity of the participating RNA sensor(s) during MHV68 infection remains unknown (23). Surprisingly, suppressing B2 ncRNA accumulation reduced viral gene expression and replication, indicating that B2 SINEs are proviral for MHV68 (23). This occurs on account of a clever twist: MHV68 coopts a kinase within the NF-κB pathway (IKKß), redirecting it to phosphorylate and thereby boost the activity of the major viral transcriptional regulator RTA (23, 48, 49). Downstream NF-κB signaling is subsequently inhibited by the virus, enabling MHV68 to avoid NF-κB-induced antiviral cytokine production (23, 49). Whether other viruses similarly hijack SINE-induced signaling or are instead restricted by the immune stimulatory capacity of these ncRNAs remains an exciting avenue for future studies.
FINAL REMARKS
While it is clear that SINE activation occurs following infection with a wide array of DNA viruses, much remains to be done to define the mechanism of SINE induction, especially with regard to the specificity observed at pol III loci, as well as whether similar induction occurs upon RNA virus infection. Additionally, it will be important to decipher how these abundant ncRNAs function during virus replication, either through regulation of RNA pol II gene expression in the nucleus or stimulation of cytoplasmic immune signaling. Exploring how cells sense and respond to virus-induced SINE ncRNAs should provide key insight into novel retrotransposon-linked components of cell signaling during infection.
ACKNOWLEDGMENTS
We apologize to authors whose work could not be cited due to length restrictions.
B.A.G. is a Howard Hughes Investigator. Funding includes NIH grants CA136367 and CA160556.
There are no conflicts of interest regarding the publication of this article.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, et al. 2001. The sequence of the human genome. Science 291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Kramerov DA, Vassetzky NS. 2011. SINEs. Wiley Interdiscip Rev RNA 2:772–786. doi: 10.1002/wrna.91. [DOI] [PubMed] [Google Scholar]
- 5.Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 6.Dewannieux M, Heidmann T. 2005. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol 349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 7.Ullu E, Tschudi C. 1984. Alu sequences are processed 7SL RNA genes. Nature 312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- 8.Hancks DC, Kazazian HH Jr. 2010. SVA retrotransposons: evolution and genetic instability. Semin Cancer Biol 20:234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels GR, Deininger PL. 1985. Repeat sequence families derived from mammalian tRNA genes. Nature 317:819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- 10.Krayev AS, Kramerov DA, Skryabin KG, Ryskov AP, Bayev AA, Georgiev GP. 1980. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res 8:1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid CW. 1991. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res 19:5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochanek S, Renz D, Doerfler W. 1993. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J 12:1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. 2015. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun 6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang KL, Latchman DS. 1989. HSV infection induces increased transcription of Alu repeated sequences by RNA polymerase III. FEBS Lett 258:255–258. doi: 10.1016/0014-5793(89)81667-9. [DOI] [PubMed] [Google Scholar]
- 16.Panning B, Smiley JR. 1993. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol Cell Biol 13:3231–3244. doi: 10.1128/MCB.13.6.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panning B, Smiley JR. 1994. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology 202:408–417. doi: 10.1006/viro.1994.1357. [DOI] [PubMed] [Google Scholar]
- 18.Singh K, Carey M, Saragosti S, Botchan M. 1985. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature 314:553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- 19.Fornace AJ Jr, Mitchell JB. 1986. Induction of B2 RNA polymerase III transcription by heat shock: enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res 14:5793–5811. doi: 10.1093/nar/14.14.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu WM, Chu WM, Choudary PV, Schmid CW. 1995. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res 23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klawitter S, Fuchs NV, Upton KR, Munoz-Lopez M, Shukla R, Wang J, Garcia-Canadas M, Lopez-Ruiz C, Gerhardt DJ, Sebe A, Grabundzija I, Merkert S, Gerdes P, Pulgarin JA, Bock A, Held U, Witthuhn A, Haase A, Sarkadi B, Lower J, Wolvetang EJ, Martin U, Ivics Z, Izsvak Z, Garcia-Perez JL, Faulkner GJ, Schumann GG. 2016. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun 7:10286. doi: 10.1038/ncomms10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams WP, Tamburic L, Astell CR. 2004. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology 327:233–241. doi: 10.1016/j.virol.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 23.Karijolich J, Abernathy E, Glaunsinger BA. 2015. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-κB pathway. PLoS Pathog 11:e1005260. doi: 10.1371/journal.ppat.1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White RJ. 2004. RNA polymerase III transcription and cancer. Oncogene 23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 25.Vachon VK, Conn GL. 2016. Adenovirus VA RNA: an essential pro-viral non-coding RNA. Virus Res 212:39–52. doi: 10.1016/j.virusres.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Feldman ER, Kara M, Oko LM, Grau KR, Krueger BJ, Zhang J, Feng P, van Dyk LF, Renne R, Tibbetts SA. 2016. A gammaherpesvirus noncoding RNA is essential for hematogenous dissemination and establishment of peripheral latency. mSphere 1: pii:e00105-15. doi: 10.1128/mSphere.00105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baglio SR, van Eijndhoven MA, Koppers-Lalic D, Berenguer J, Lougheed SM, Gibbs S, Leveille N, Rinkel RN, Hopmans ES, Swaminathan S, Verkuijlen SA, Scheffer GL, van Kuppeveld FJ, de Gruijl TD, Bultink IE, Jordanova ES, Hackenberg M, Piersma SR, Knol JC, Voskuyl AE, Wurdinger T, Jimenez CR, Middeldorp JM, Pegtel DM. 2016. Sensing of latent EBV infection through exosomal transfer of 5′pppRNA. Proc Natl Acad Sci U S A 113:E587–E596. doi: 10.1073/pnas.1518130113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen TJ, O'Neil JD, Dawson CW, Hu C, Chen X, Yao Y, Wood VH, Mitchell LE, White RJ, Young LS, Arrand JR. 2010. Epstein-Barr virus-encoded EBNA1 enhances RNA polymerase III-dependent EBER expression through induction of EBER-associated cellular transcription factors. Mol Cancer 9:241. doi: 10.1186/1476-4598-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felton-Edkins ZA, Kondrashov A, Karali D, Fairley JA, Dawson CW, Arrand JR, Young LS, White RJ. 2006. Epstein-Barr virus induces cellular transcription factors to allow active expression of EBER genes by RNA polymerase III. J Biol Chem 281:33871–33880. doi: 10.1074/jbc.M600468200. [DOI] [PubMed] [Google Scholar]
- 30.Larminie CG, Sutcliffe JE, Tosh K, Winter AG, Felton-Edkins ZA, White RJ. 1999. Activation of RNA polymerase III transcription in cells transformed by simian virus 40. Mol Cell Biol 19:4927–4934. doi: 10.1128/MCB.19.7.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felton-Edkins ZA, White RJ. 2002. Multiple mechanisms contribute to the activation of RNA polymerase III transcription in cells transformed by papovaviruses. J Biol Chem 277:48182–48191. doi: 10.1074/jbc.M201333200. [DOI] [PubMed] [Google Scholar]
- 32.Hoeffler WK, Kovelman R, Roeder RG. 1988. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell 53:907–920. doi: 10.1016/S0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 33.Hoeffler WK, Roeder RG. 1985. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell 41:955–963. doi: 10.1016/S0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- 34.Gaynor RB, Feldman LT, Berk AJ. 1985. Transcription of class III genes activated by viral immediate early proteins. Science 230:447–450. doi: 10.1126/science.2996135. [DOI] [PubMed] [Google Scholar]
- 35.Gjidoda A, Henry RW. 2013. RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim Biophys Acta 1829:385–392. doi: 10.1016/j.bbagrm.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karijolich J, Zhao Y, Alla R, Glaunsinger B. 2017. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res 45:6194–6208. doi: 10.1093/nar/gkx180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol 11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 38.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Espinoza CA, Goodrich JA, Kugel JF. 2007. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA 13:583–596. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labuda D, Sinnett D, Richer C, Deragon JM, Striker G. 1991. Evolution of mouse B1 repeats: 7SL RNA folding pattern conserved. J Mol Evol 32:405–414. doi: 10.1007/BF02101280. [DOI] [PubMed] [Google Scholar]
- 41.Maraia RJ. 1991. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res 19:5695–5702. doi: 10.1093/nar/19.20.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinnett D, Richer C, Deragon JM, Labuda D. 1991. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J Biol Chem 266:8675–8678. [PubMed] [Google Scholar]
- 43.Kerur N, Hirano Y, Tarallo V, Fowler BJ, Bastos-Carvalho A, Yasuma T, Yasuma R, Kim Y, Hinton DR, Kirschning CJ, Gelfand BD, Ambati J. 2013. TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Invest Ophthalmol Vis Sci 54:7395–7401. doi: 10.1167/iovs.13-12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung T, Pratt GA, Sundararaman B, Townsend MJ, Chaivorapol C, Bhangale T, Graham RR, Ortmann W, Criswell LA, Yeo GW, Behrens TW. 2015. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 350:455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zovoilis A, Cifuentes-Rojas C, Chu HP, Hernandez AJ, Lee JT. 2016. Destabilization of B2 RNA by EZH2 activates the stress response. Cell 167:1788–1802 e1713. doi: 10.1016/j.cell.2016.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vazquez C, Horner SM. 2015. MAVS coordination of antiviral innate immunity. J Virol 89:6974–6977. doi: 10.1128/JVI.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong X, Feng H, Sun Q, Li H, Wu TT, Sun R, Tibbetts SA, Chen ZJ, Feng P. 2010. Murine gamma-herpesvirus 68 hijacks MAVS and IKKbeta to initiate lytic replication. PLoS Pathog 6:e1001001. doi: 10.1371/journal.ppat.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong X, Feng P. 2011. Murine gamma herpesvirus 68 hijacks MAVS and IKKbeta to abrogate NFκB activation and antiviral cytokine production. PLoS Pathog 7:e1002336. doi: 10.1371/journal.ppat.1002336. [DOI] [PMC free article] [PubMed] [Google Scholar]