Abstract
Polybrominated diphenyl ethers (PBDEs) were used as flame-retardant additives in a wide range of polymers. The generations born when environmental concentrations of PBDEs reached their maximum account in the United States for one-fifth of the total population. We hypothesized that exposure to PBDEs during sensitive developmental windows might result in long-lasting changes in liver metabolism. The present study was based on experiments with CD-1 mice and human hepatocellular carcinoma cells (human hepatoma cell line, HepG2). Pregnant mice were exposed to 0.2 mg/kg 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) from gestation day 8 until postnatal day 21. The metabolic health-related outcomes were analyzed on postnatal day 21 and postnatal week 20 in male offspring. Several groups of metabolic genes, including ribosomal and mitochondrial genes, were significantly upregulated in the liver at both points. Genes regulated via mechanistic target of rapamycin (mTOR) pathway, the gatekeeper of metabolic homeostasis, were whether up- or downregulated at both measurement points. On postnatal day 21, but not week 20, both mTOR complexes in the liver were activated, as measured by phosphorylation of their targets. mTOR complexes were also activated by BDE-47 in HepG2 cells in vitro. The following changes were observed at week 20: a decreased number of polyploid hepatocytes, suppressed cytoplasmic S6K1, twofold greater blood insulin-like growth factor-1 and triglycerides, and 2.5-fold lower expression of fatty acid uptake membrane receptor CD36 in liver tissue. Thus, perinatal exposure to environmentally relevant doses of BDE-47 in laboratory mice results in long-lasting changes in liver physiology. Our evidence suggests involvement of the mTOR pathway in the observed metabolic programming of the liver.
Keywords: Freeform/Key Words: polybrominated diphenyl ether, mTOR, metabolism, rodent, ribosome, hyperlipidemia
Developmental exposure to BDE-47 activates mTOR in mouse liver and in HepG2 cells and programs altered expression of metabolic genes in the liver and increased concentrations of triglycerides in blood.
Polybrominated diphenyl ethers (PBDEs) were used as flame-retardant additives in a wide range of polymers; thus, the concentrations increased exponentially for 30 years in human blood, milk, and tissues [1]. Recently, PBDEs were withdrawn from commerce in North America and Europe because of their environmental persistence and bioaccumulative properties [2]. Waste and recycling sites, the indoor use of PBDE-containing products [3], the global circulation of PBDEs toward the northern hemisphere, and the high potency of PBDEs for bioconcentration in food chains [4] have all contributed to the long-term persistence of human PBDE exposure all over the world. The results from human studies and animal experiments suggest that accumulated PBDEs mobilize during pregnancy, causing high-dose exposure of the developing organism via cord blood and breast milk [5–7]. PBDEs easily cross the placenta [8] and have been found in most fetal samples in North America [9, 10]. Toddlers are exposed to higher PBDEs doses than adults because of their greater rates of dust ingestion [11] and greater food intake per kilogram of body weight [6]. Manufacturing of commercial products containing PBDEs began in 1965 [12], and the concentrations of PBDE in human tissues reached a plateau in the 2000s [13]. These numbers started to decrease only the most recent years [14, 15]. Thus, the generations that were exposed in utero and during early postnatal life to highest doses of PBDEs have now reached 0 to 15 years of age. According to demographic data, approximately 60 million Americans are <15 years of age [16], comprising one-fifth of the US population.
An emerging hypothesis links the metabolic diseases epidemic with exposures to endocrine disruptors during vulnerable windows of development [17–19]. The prevalence of obesity and type 2 diabetes has increased dramatically in children in the United States during the past few decades. More than 60% of children aged ≥10 years either are or will become obese later in life, and one in four overweight children has impaired glucose tolerance [18]. According to the Centers for Disease Control and Prevention, approximately two of every five Americans will develop type 2 diabetes at some point during their adult life [20]. Hyperlipidemia, prevalent throughout the developed world, is the primary risk factor for myocardial infarction, which is the most common cause of mortality in the developed world, with >700,000 deaths attributed to the disease in the United States annually [21]. Thus, an urgent need exists to identify environmental factors that have the potential to reprogram metabolic health and to understand the molecular events triggered by environmental exposures.
PBDEs disrupt hormonal signaling through the thyroid and steroid axes [22] and interfere with the insulin-like growth factor (IGF) axis [23]. Cord blood levels of IGF-1, the major regulator of anabolism, correlated with breast milk levels of PBDEs in a Taiwan birth cohort [24], and placental IGF1 expression correlated with cord blood PBDEs in a China population exposed to electronic waste [25]. Several studies have reported a positive association between the body mass index in nursing women and PBDE concentrations in their blood, milk, or placenta for the following congeners: BDE-209 [7, 26]; BDE-47, -99, and -100 [27, 28]; and BDE-28, and -153 [27]. The body mass index was inversely associated with serum PBDE levels in the 7-year-old Mexican-American children from the CHAMACOS (Center for the Health Assessment of Mothers and Children of Salinas) cohort [29]. In several animal studies, including our low-dose rodent model, developmental exposures to BDE-47 resulted in increased body weights [23, 30]. PBDE-induced growth changes might be linked to changes in fatty acid and carbohydrate metabolism. However, it is not yet clear whether these exposures could induce long-lasting changes in metabolic health. In our experiments, rats perinatally exposed to BDE-47 had increased cholesterol levels [31], increased glucose uptake by tissues [23], and systemic activation of pathways responsible for the metabolism of lipids and carbohydrates [31]. Thus, emerging evidence has linked PBDE with perturbed liver metabolic health, in particular.
In the present study, we found that perinatal exposure to the most prevalent PBDE congener in human samples, 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47), results in long-lasting changes in the expression of metabolic genes in the liver, accompanied by increased blood triglycerides. We also discuss a possibility that a mechanistic target of rapamycin (mTOR)-centered molecular pathway, a central pathway of metabolic homeostasis maintenance [32], is involved in liver metabolism programming by BDE-47.
1. Materials and Methods
A. Animals and Treatment
CD-1 mice—an outbred strain that is the most commonly used in toxicology studies in the United States—were used in the present study [33]. We obtained 8-week-old male (weight, 30 to 35 g) and female (weight, 27 to 30 g) mice from Charles River Laboratories (Kingston, NY), which were housed in a temperature-controlled (23°C ± 2°C) and humidity-controlled (40% ± 10%) environment, with a 12-hour light/dark cycle and food and water available ad libitum. After 3 days of acclimation, the mice were bred, and the day of vaginal plug detection was considered pregnancy day 1. The dams were assigned to one of two treatment groups (n = 5 per group) according to weight and were exposed to tocopherol-stripped corn oil (MP Biomedicals, Solon, OH) or 0.2 mg/mL solution of BDE-47 (AccuStandard, Inc., New Haven, CT; 100% purity) in tocopherol-stripped corn oil daily from pregnancy day 8 through postpartum day 21. The females were fed 1 µL/g body weight from the tip of a pipette, resulting in exposure of 0.2 mg/kg body weight daily. This method of exposure is routinely used in our laboratory as a substitution for oral gavage, because the latter method induces a substantial stress response by the endocrine system, which could interfere with analyzed health outcomes [34]. The dams were allowed to deliver naturally, and the litters were not culled to maintain the consistency of nutrient distribution among the same number of fetuses and pups during the pre- and postnatal periods and to avoid catch-up growth [35]. The dams and pups were kept together until weaning on postnatal day (PND) 21, when the male and female pups were separated. Using cervical dislocation, the dams and one randomly selected male pup per litter were euthanized on PND21, and three to five randomly selected male pups per litter were euthanized on postnatal week (PNW) 20. Only male pups were used for further analysis to avoid any interaction of the measured health outcomes with hormonal fluctuations due to estrus cycle. All the mice were euthanized between 9 am and 11 am after 2 hours of fasting. Tissue samples were collected immediately on euthanasia. The blood samples were then centrifuged at 3000g for 10 minutes, and serum was collected and stored at −80°C. The liver, right inguinal subcutaneous fat pad, and gonadal fat from the right side of each mouse were collected, weighed, snap-frozen in liquid nitrogen, and stored at −80°C. The left lateral lobe was excised from each liver before freezing and fixed in 4% paraformaldehyde in 0.1 M phosphate-buffered saline for 24 hours at room temperature, washed in 0.1 M phosphate-buffered saline, and stored in 70% ethanol at 4°C. The design of the experiment with the mice is shown in Fig. 1(a). All procedures met the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the institutional animal care and use committee of the University of Massachusetts, Amherst approved the present study.
Figure 1.
Schemes of experimental design of gene expression studies used in the present report: (a) present study and (b) Suvorov and Takser in 2010 [31] and Suvorov and Takser in 2011 [44]. BW, body weight.
B. RNA Extraction and Sequencing
Total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific Life Sciences, Waltham, MA) and quantified using a NanoDrop 1000 instrument (Thermo Fisher Scientific Life Sciences, Wilmington, DE). RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples of RNA with integrity values >8 were used for library preparation in accordance with the recommendations of library preparation and sequencing kit manuals. The NEBNext Poly(A) messenger RNA (mRNA) Magnetic Isolation Module (E7490; New England BioLabs Inc., Ipswich, MA) was then used to isolate intact poly(A)+ RNA from 3 µg total RNA. Libraries were constructed using NEBNext mRNA Library Prep Reagent Set for Illumina (Illumina, San Diego, CA) with multiplexing indexes from NEBNext Multiplex Oligos for Illumina (E6100 and E7335, respectively; New England BioLabs Inc.). The quality and purity of the libraries was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany), and the concentration of the libraries was measured using real-time polymerase chain reaction with primers for P5 and P7 flow cell oligo sequences using the KAPA Library Quantification Kit (KR0405; Kapa Biosystems, Boston, MA) in a 384-well plate on a CFX384 Touch Real-Time Polymerase Chain Reaction Detection System (Bio-Rad, Hercules, CA). High-throughput sequencing was performed using a NextSeq500 sequencing system (Illumina) in the Genomic Resource Laboratory of the University of Massachusetts, Amherst. Complementary DNA libraries were single-end sequenced in 75 cycles using the NextSEquation 500 High Output Kit (FC-404-1005; Illumina) in a multiplex run of 10 samples. All sequencing data were uploaded to the Gene Expression Omnibus (GEO) public repository and assigned GEO series accession number GSE85221.
C. Bioinformatic Analysis of Mouse RNA-Sequencing Data
To analyze the data from our mRNA sequencing experiment, read filtering, trimming, and demultiplexing were performed via the BaseSpace cloud computing service supported by Illumina (available at: https://basespace.illumina.com/home/index). Furthermore, the preprocessed reads were mapped to the reference mouse genome (MM10) using the TopHat2 aligner [36]. BAM files were uploaded to Qlucore Omics Explorer software (Qlucore AB, Lund, Sweden) to generate heat maps of differentially expressed transcripts [37]. Aligned reads were then used for assembly of transcripts using Cufflinks, version 2.1.1, and differential expression of reference transcripts using Cuffdiff, version 2.1.1 [38]. Differential expression data sets were further used for gene set enrichment analysis (GSEA, Broad Institute, Boston, MA; available at: www.broadinstitute.org/gsea). This approach is particularly effective for the identification of biologically important changes in gene expression that are associated with relatively small effects across multiple members of a gene set [39]. The details of the method and statistical approaches used by GSEA have been previously described [40, 41]. We used GSEA against the hallmark collection of data sets and data sets from Reactome and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Additionally, we created 2 gene sets from the published data to specifically interrogate changes in the expression of genes that are transcriptionally regulated downstream the mTOR pathway in rodent liver. The gene set of mTOR complex 2 (mTORC2)-regulated genes was extracted from a study [42] in which gene expression was analyzed in the livers of control and Rictor (component of mTORC2) liver-specific knockout mice that were fasted overnight and then given ad libitum access to food for 3 hours. The gene set of mTOR complex 1 (mTORC1)-regulated genes was extracted from another study from the same group [39] in which two groups of adult Sprague-Dawley rats were fasted for 48 hours. One group was then subjected to a dimethyl sulfoxide (DMSO) vehicle and the other group to selective mTORC1 inhibitor rapamycin (2.5 mg/kg body weight). Both groups were then refed and euthanized 3 hours after refeeding. According to the results of the GSEA, the genes regulated downstream of the mTOR pathway were either up- or downregulated by BDE-47 (see the Results section). To further explore which biological functions controlled by the mTOR pathway were activated and suppressed, we built two lists of genes, the first of which was created by merging together positively regulated genes (GSEA ranking list metrics >0) from three gene sets of mTOR-regulated genes: one from the hallmark collection and two created by us, as described previously. Downregulated genes (GSEA ranking list metrics <0) from the same three gene sets were merged into another gene list. These two gene lists were than analyzed using the DAVID Functional Annotation Clustering tool with the default settings (available at: david.ncifcrf.gov) [43].
D. Blood Circulating Factors
IGF-1 was analyzed in the serum of the dams and male offspring euthanized on PND21 and PNW20, respectively, using the RayBio mouse IGF-1 enzyme-linked immunosorbent assay kit (ELM-IGF1; RayBiotech, Inc., Norcross, GA). The insulin and triglyceride levels were measured in the blood of the offspring euthanized on PNW20 using the insulin enzyme-linked immunosorbent assay kit (catalog no. ab100578; Abcam, Cambridge, MA) and triglyceride quantification kit (catalog no. ab65336; Abcam), respectively. High-density lipoproteins (HDLs) and low-density lipoproteins (LDLs) and very-low-density lipoproteins (VLDLs) were analyzed in the same blood samples using an HDL and LDL+VLDL cholesterol assay kit (STA-391; Cell Biolabs, Inc., San Diego, CA). The glucose levels were measured using an Accu-Check Compact Plus blood glucose monitoring system (Roche Diagnostics GmbH, Berlin, Germany) as previously described [23]. For all serum measurements at PNW20, the analysis was performed for three to five individual mice per litter. These values were then averaged and used in a t test, with a subsequent Dunnett’s test to compare five exposed litters against five control litters.
E. Liver Histologic Examination
To prepare the paraffin sections, paraformaldehyde-fixed lateral lobes of livers collected on PNW20 were dehydrated through a series of alcohols and xylene and embedded with Paraplast X-tra paraffin (Leica Biosystems, Wetzlar, Germany) using the Excelsior ES Tissue Processor (Thermo Fisher Scientific Life Sciences, Runcorn, Cheshire, UK). Five-micrometer sections were cut on a Microm HM 355S microtome (Thermo Fisher Scientific Life Sciences, Walldorf, Germany) and mounted on Colorfrost Plus slides (Thermo Fisher Scientific Life Sciences). The sections were processed through xylene to remove the paraffin, hydrated by a series of alcohols, and stained with Harris' hematoxylin and eosin. The sections were viewed through a Zeiss Axio Observer Z1 inverted light microscope with ZEN imaging software (Carl Zeiss AG, Oberkochen, Germany), at ×10 and ×40. Images were captured at 88,000 dpi (dots per inch) using the Axiocam 506 color digital camera (Carl Zeiss AG). The slides were first examined for morphological differences between exposure groups, and the difference in nucleus size in hepatocytes and frequency of binuclear hepatocytes were noted. To quantify these differences, three images were taken from every slide from nonoverlapping areas, and three boxes 1000 × 1000 µm were randomly placed on each image. The number of hepatocytes with single and double nuclei was calculated for each box, and the size of all nuclei in single-nuclear hepatocytes was measured using ZEN software by an operator unaware of the exposure group. All values for each slide (nine boxes) were averaged for the final statistical analysis.
F. Cell Culture and Treatment
The human hepatoma cell line HepG2 were purchased from ATCC (ATCC no. HB-8065; lot no. 62591368; Manassas, VA). The cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY), 2 mM l-glutamine, 110 mg/L sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin and incubated at 37°C in a humidified atmosphere containing 5% carbon dioxide. BDE-47 was prepared in DMSO as a 50-mM stock solution. When the cells had grown to 80% confluence in 100-mm culture plates, the media were discarded, and fresh medium containing 1 µM BDE-47 was supplemented. The cells were divided into three groups: control, 3-hour treated group, and 24-hour treated group. The control group was treated with DMSO for 24 hours. Each group had three replicates.
G. Protein Phosphorylation Analysis
Phosphorylation of protein targets of mTORC1 and mTORC2 was analyzed by Western blotting in liver samples of male offspring euthanized on PND21 and PNW20 and in HepG2 cells stimulated by BDE-47. The treated and control HepG2 cells were harvested with 0.25% trypsin and washed twice in ice-cold phosphate-buffered saline. The liver and HepG2 cells samples were lysed in T-PER tissue protein extraction buffer (catalog no. 78510; Thermo Fisher Scientific Life Sciences, Rockford, IL) and M-PER mammalian protein extraction buffer (catalog no. 78503; Thermo Fisher Scientific Life Sciences), respectively. In both cases, buffers contained a protease and phosphatase inhibitor cocktail (catalog no. 78442; Thermo Fisher Scientific Life Sciences). A microplate-based BCA Protein Assay Kit (catalog no. 23227; Thermo Fisher Scientific Life Sciences) was then used to determine the protein concentrations. Western blot analyses were performed after separating the proteins on 4% to 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (catalog no. 456-1094; Bio-Rad, Hercules, CA) and transferring them to a polyvinylidene difluoride membrane (0.2 μm) under wet conditions using a Bio-Rad Mini Trans-Blot Cell (catalog no. 1703935, Hercules, CA). The following primary antibodies were used at the indicated dilutions: anti-phospho-Akt (ser473; 1:2000; catalog no. 4060), anti-AKT (1:2000; catalog no. 4691), anti-actin (1:2000; catalog no. 4970), anti-phospho-p70S6 kinase (Thr389; 1:1000; catalog no. 9205), and anti-p70S6 kinase (1:1000; catalog no. 9202; all from Cell Signaling Technology, Danvers, MA). Proteins were visualized using secondary antibody conjugated with horseradish peroxidase (1:5000, catalog no. ab6721; Abcam), and Pierce ECL enhanced chemiluminescence reagent (catalog no. 32106; Thermo Fisher Scientific Life Sciences). Western blot densitometry was quantified using Image Studio Lite (LI-COR Biosciences, Lincoln, NE), version 5.2, software.
H. Reanalysis of Published Transcriptional Data Sets
To test whether changes in gene expression in response to BDE-47 exposure in previously published studies matched the profile of altered mTOR signaling, we reanalyzed transcriptomic data sets previously produced by our research group [31, 44]. To the best of our knowledge, no other studies focusing on the transcriptomic effects of developmental BDE-47 have been reported. In our previous studies, changes in gene expression in the liver [31] and brain frontal lobes [44] of Wistar rats were analyzed in response to exposure to BDE-47. Dams were exposed to 0.002 and 0.2 mg/kg body weight BDE-47 by intravenous injections on the 15th day of pregnancy and at 1, 5, 10, 15, and 20 days after delivery [Fig. 1(b)]. The pups were weaned at PND21 and were not exposed directly to BDE-47. Liver samples were collected from three female and three male pups per exposure group at PND27, and brain frontal lobe samples were collected from three female pups per exposure group at PND41. Gene expression analysis was performed using Illumina BeadChips RatRef‐12. The raw data were preprocessed and normalized with the Bioconductor lumi package [45]. Gene expression data were submitted to GEO (GEO accession nos. GSE19868 for liver samples and GSE19867 for brain samples).
To test for the signature of altered mTOR signaling in these data sets, we analyzed, using GSEA, the changes in expression of five gene sets regulated via mTOR pathway from a curated C2 collection of Molecular Signatures Database (available at: http://www.broadinstitute.org/gsea/msigdb/collections.jsp). In response to nutrients and growth factors, mTORC1 positively regulates cell growth and proliferation by inducing ribosome biogenesis [46, 47], mitochondrial biogenesis [48, 49], and adipogenesis [47, 50]. In addition, activated mTORC1 suppresses autophagy [51] and blocks hepatic ketogenesis. To address changes in ribosome biogenesis, the “KEGG ribosome” gene set was used. The data set includes 88 human genes of ribosomal proteins and RNA and is curated by KEGG (available at: http://www.genome.jp/kegg/). To study the alteration in mitochondria biogenesis, we used the “Mootha mitochondria” gene set, which includes 447 human mitochondrial genes [41]. To analyze the coordinated changes in genes participating in autophagy, we used the gene set “Reactome ER [endoplasmic reticulum] phagosome pathway.” The gene set consists of 61 human genes and is curated by a pathway database Reactome (available at: http://www.reactome.org/). The gene set “Reactome PPARα activates gene expression” from the same database was used to interrogate changes in peroxisome proliferator-activated receptor-α (PPARα) targets, which include metabolic genes responsible for ketone body production. The gene set contains 104 human genes known to be activated by PPARα from a diversity of studies. Finally, changes in expression of PPARγ targets were analyzed using the gene set “Wang classic adipogenic targets of PPARγ,” which includes 26 adipogenic genes induced by PPARγ during adipogenesis in mouse 3T3-L1 preadipocytes [52].
2. Results
We found no important relationship between the litter size and exposure to BDE-47, with the number of pups varying from 11 to 15 per litter. No weight differences were observed between the control and exposed dams and pups throughout the experiment. On PNW20, the body weight of the mice was 50.8 ± 0.3 g in the control mice and 49.4 ± 1.9 g in the exposed mice. No substantial changes were observed in the weight of the livers or adipose tissues. On PNW20, the weight of the livers, inguinal subcutaneous adipose tissue from the right side of body, and gonadal adipose tissue from the right side of body in the control mice was 2.16 ± 0.15 g, 0.32 ± 0.05 g, and 0.79 ± 0.11 g, respectively. In the exposed mice, the corresponding respective weights were 2.13 ± 0.17 g, 0.34 ± 0.03 g, and 0.83 ± 0.10 g. All data are shown as the mean ± standard error (SE).
A. Transcriptional Changes in Murine Liver After Perinatal Exposure to BDE-47
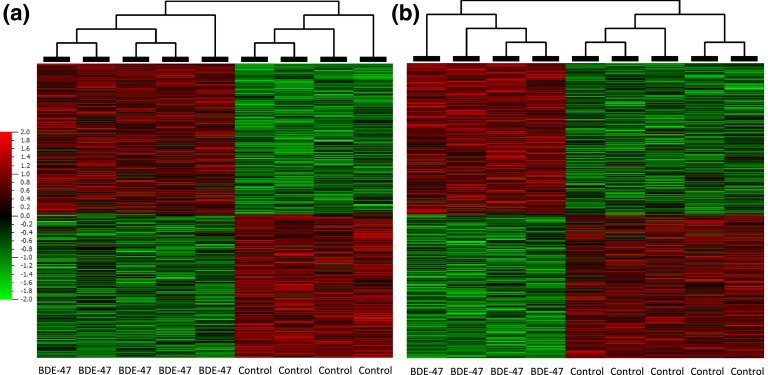
We analyzed the transcriptional changes in genes in male mouse livers at the last day of perinatal exposure (PND21) and long after the end of exposure (PNW20) using an RNA-sequencing approach and subsequent GSEA. Sequencing was completed with a minimum 30 and 20 million reads per sample for RNA samples extracted from the livers on PND21 and PNW20, respectively. A total of 1857 genes were regulated on PND21 and 1850 genes on PNW20, with false discovery rate (FDR) of q ≤ 0.05 (Fig. 2). The lists of 22,923 (PND21 data set) and 22,929 (PNW20 data set) assembled unique transcripts with known identifiers (genes, noncoding RNAs) were used for GSEA. Gene sets enriched with a nominal P ≤ 0.05 and FDR q ≤ 0.2 were selected for further analysis using more stringent criteria than those recommended by the GSEA developers (FDR q ≤ 0.25; available at: http://software.broadinstitute.org/gsea/doc/) to focus on the most significantly altered biological processes. The gene sets that satisfied these criteria at least one time point (PND21 and PNW20) are listed in Table 1. The most important metabolic gene sets included “oxidative phosphorylation,” which was positively enriched at both time points [Fig. 3(a) and 3(b)]; “bile acid metabolism,” which was negatively enriched at both time points, although the difference was substantial on PND21 only; “peptide chain elongation” [Fig. 3(c) and 3(d)] and “ribosome” [Fig. 3(e) and 3(f)], which were positively enriched at both time points, although with a small difference on PND21; and “steroid hormones” and “metabolism of steroid hormones and vitamins A and D,” which were negatively enriched at both time points, although the difference was substantial on PND21 only. The “mTORC1 signaling” data set was positively enriched on PND21 and negatively on PNW20. However, at both time points, most genes of this data set were either up- or downregulated [Fig. 4(a) and 4(b)]. The same binary distribution was observed for mTOR pathway genes from the C2 collection [Fig. 4(c) and 4(d)] and for the gene sets of mTORC1- and mTORC2-regulated genes in rodent livers created from studies by Bolyen et al. [39] and Lamming et al. [42] [Fig. 4(e-h)]. To analyze which biological functions controlled by the mTOR pathway were activated and which were suppressed, we uploaded merged gene lists of the mTOR-controlled genes that had been up- or downregulated by BDE-47 to DAVID Functional Annotation Clustering. The top enriched clusters of positively regulated biological functions were associated with ribosome, mitochondria, proteasome, and oxidative stress at both time points (Table 2). The top enriched clusters of inhibited biological functions included cholesterol/steroid/lipid metabolism and hexose metabolism at both time points and, in addition, organic acid/amino acid biosynthesis on PNW20 (Table 2).
Figure 2.
Heat maps of genes significantly regulated (P ≤ 0.05) in mouse livers by perinatal exposure to BDE-47: (a) 1857 genes regulated on PND21 and (b) 1850 genes regulated on PNW20. Heat bar shows Log2 values of fold change of differentially expressed genes.
Table 1.
Top GSEA-Enriched Gene Sets on PND21 and/or PNW20 in Livers of Mice Exposed Perinatally to BDE-47
| Gene Set | PND21 |
PNW20 |
||||
|---|---|---|---|---|---|---|
| NES | NOM P | FDR q | NES | NOM P | FDR q | |
| Hallmark collection of gene sets | ||||||
| MYC targets (version 1) | 1.72a | < 0.0005 | 0.01 | 0.99 | 0.497 | 1.00 |
| Unfolded protein response | 1.65a | 0.002 | 0.02 | −0.56 | 1.000 | 1.00 |
| G2M checkpoint | 1.65a | < 0.0005 | 0.01 | −0.84 | 0.859 | 1.00 |
| E2F targets | 1.58a | < 0.0005 | 0.02 | −0.61 | 1.000 | 1.00 |
| Oxidative phosphorylation | 1.52a | 0.005 | 0.04 | 1.52a | < 0.0005 | 0.15 |
| mTORC1 signaling | 1.41a | 0.014 | 0.11 | −0.88 | 0.762 | 1.00 |
| DNA repair | 1.33a | 0.049 | 0.20 | 1.41a | 0.004 | 0.20 |
| Bile acid metabolism | 1.34a | 0.045 | 0.19 | −0.53 | 1.000 | 1.00 |
| Peroxisome | −1.40a | 0.023 | 0.15 | 0.71 | 0.989 | 1.00 |
| Ultraviolet response (downregulated by ultraviolet) | −1.47a | 0.009 | 0.10 | −0.81 | 0.892 | 1.00 |
| KRAS signaling (downregulated by KRAS) | −1.56a | 0.002 | 0.05 | −0.88 | 0.734 | 1.00 |
| Myogenesis | −1.66a | < 0.0005 | 0.02 | 1.01 | 0.388 | 1.00 |
| KEGG and reactome collections of gene sets | ||||||
| Reactome: peptide chain elongation | 1.56 | 0.015 | 0.35 | 1.82a | < 0.0005 | 0.13 |
| KEGG: ribosome | 1.59 | < 0.0005 | 0.33 | 1.77a | < 0.0005 | 0.20 |
| Reactome: influenza viral RNA transcription and replication | 1.63 | 0.002 | 0.33 | 1.75a | 0.003 | 0.20 |
| Reactome: 3′ UTR mediated translation regulation | 1.55 | < 0.0005 | 0.34 | 1.75a | < 0.0005 | 0.16 |
| Reactome: voltage-gated potassium channels | −1.68a | 0.006 | 0.10 | 0.95 | 0.52 | 1.00 |
| Reactome: platelet aggregation plug formation | −1.69a | 0.002 | 0.10 | −0.74 | 0.86 | 1.00 |
| Reactome: steroid hormones | −1.70a | < 0.0005 | 0.09 | −1.37 | 0.06 | 1.00 |
| Reactome: Grb2:SOS provides linkage to MAPK signaling for integrins | −1.70a | < 0.0005 | 0.09 | −0.63 | 0.92 | 1.00 |
| Reactome: potassium channels | −1.70a | 0.002 | 0.10 | −0.97 | 0.54 | 1.00 |
| Reactome: P130Cas linkage to MAPK signaling for integrins | −1.71a | < 0.0005 | 0.09 | −0.40 | 1.00 | 1.00 |
| KEGG: complement and coagulation cascade | −1.71a | < 0.0005 | 0.09 | −0.88 | 0.68 | 1.00 |
| Reactome: metabolism of steroid hormones and vitamins A and D | −1.72a | 0.002 | 0.08 | −1.11 | 0.31 | 1.00 |
| Reactome: integrin alpha IIB beta3 signaling | −1.76a | 0.002 | 0.06 | −0.64 | 0.95 | 1.00 |
| Reactome: formation of fibrin clot, clotting cascade | −1.80a | < 0.0005 | 0.04 | −0.73 | 0.84 | 1.00 |
Abbreviations: FRD q, false discovery rate q value; MAPK, mitogen-activated protein kinase; NES, normalized enrichment score; NOM P, nominal P value; UTR, untranslated region.
Gene sets enriched with nominal P ≤ 0.05 and FDR q ≤ 0.2.
Figure 3.
GSEA enrichment of three gene sets in mouse liver RNA-sequencing data on PND21 (a, c, and e) and PNW20 (b, d, and f) in response to perinatal exposure to BDE-47: (a and b) oxidative phosphorylation; (c and d) peptide chain elongation; and (e and f) ribosomal genes. GSEA plot legend: 1, running enrichment score for the gene set; 2, vertical lines show where the members of the gene set appear in the ranked list of genes; and 3 and 4, ranked list of differentially expressed genes from the most upregulated (left of each plot) to the most downregulated (right of each plot). Regulation is shown in the heat bar (3) and the bar plot (4).
Figure 4.
GSEA enrichment of four gene sets in mouse liver RNA-sequencing data on PND21 (a, c, e, and g) and PNW20 (b, d, f, and h) in response to perinatal exposure to BDE-47: (a and b) genes activated by mTORC1 (Hallmark collection); (c and d) genes of mTOR signaling pathway (C2 collection); (e and f) genes regulated in rodent liver via mTORC1 [39]; and (g and h) gene regulated in rodent liver via mTORC2 [42]. The GSEA plot legend is as provided for Fig. 3.
Table 2.
Top Enriched DAVID Clusters of Biological Functions Controlling Downstream mTOR Signaling Pathway
| Cluster of Enriched Terms | PND21 |
PNW20 |
||
|---|---|---|---|---|
| ES | Benjamini P Value | ES | Benjamini P Value | |
| Positively regulated function | ||||
| Ribosome | 13.2 | 1.5E-3–2.3E-17 | 14.9 | 1.9E-2–8.4E-21 |
| Mitochondrion | 9.2 | 5.9E-3–5.6E-13 | 8.8 | 3.3E-3–6.2E-13 |
| Proteasome | 6.7 | 1.9E-4–1.4E-6 | 2.5 | 9.6E-1–1.6E-4 |
| Response to oxidative stress | 2.7 | 7.0E-1–4.9E-3 | NS | NS |
| Glutathione metabolism | 2.6 | 3.0E-1–1.7E-4 | 2.5 | 8.1E-1–7.7E-3 |
| Negatively regulated function | ||||
| Cholesterol/steroid/lipid metabolism | 2.9 | 1.5E-1–4.1E-2 | 3.3 | 7.2E-1–3.8E-4 |
| Hexose metabolism | 2.4 | 7.6E-1 – 2.9E-2 | 3.2 | 8.4E-2–3.4E-2 |
| Lipid catabolism | 2.3 | 7.1E-1–2.5E-2 | NS | NS |
| Organic acid/amino acid metabolism | NS | NS | 4.1 | 6.8E-1–6.4E-4 |
Abbreviations: ES, enrichment score; NS, not significant.
Other data sets enriched in GSEA covered a broad range of biological functions. Some among these several enriched gene sets indicated activation of stress-response mechanisms at one or both time points, including positive enrichment of “unfolded protein response” and “DNA repair” gene sets and negative enrichment of an “ultraviolet response” gene set composed of genes downregulated by ultraviolet exposure. Other gene sets indicated inhibition of the complement and coagulation cascade (“Reactome: platelet aggregation plug formation,” “Reactome: Grb2:SOS provides linkage to MAPK [mitogen-activated protein kinase] signaling for integrins,” “Reactome: P130Cas linkage to MAPK [mitogen-activated protein kinase] signaling for integrins,” “KEGG: complement and coagulation cascade,” “Reactome: integrin alpha IIB beta3 signaling,” and “Reactome: formation of fibrin clot, clotting cascade”) and suppression of potassium channels (“Reactome: voltage-gated potassium channels” and “Reactome: potassium channels”).
B. Perinatal Exposure to BDE-47 Decreases Percentage of Polyploid Hepatocytes in Murine Liver on PNW20
Visual inspection of hematoxylin and eosin-stained sections of liver lateral lobes from exposed and control animals revealed a lower percentage of binuclear hepatocytes and hepatocytes with large nuclei, considered an indication of polyploidy [53] [Fig. 5(a) and 5(b)]. To quantify these differences, the greatest width of each nucleus was measured, and the number of binuclear hepatocytes was counted in nine 1000 × 1000-μm boxes per slide. The proportion of binuclear hepatocytes was 31.9% ± 3.4% in the control mice and 23.0% ± 2.5% in the livers of the exposed mice [mean ± SE; P = 0.06; Fig. 5(c)]. The average nuclear diameter in the single-nuclear hepatocytes was significantly smaller in the exposed mice (8.96 ± 0.11 µm) compared with that in the controls [9.36 ± 0.12 µm; mean ± SE; P = 0.05; Fig. 5(d)]. We assumed that given that the increased nuclear size is due to polyploidy, several size classes might exist of nuclei different in ploidy in mouse livers. To test this hypothesis, we explored the distribution of nuclear sizes by combining the measurements from all the mice and plotting the quantities of nuclei in bins of 0.2 μm [Fig. 5(e)]. The appearance of the plot showed that the nucleus size has a multimodal distribution, likely corresponding to the existence of hepatocytes with several levels of ploidy. The well-pronounced gap between two distribution peaks that separates the visually large nuclei and visually “normal” nuclei corresponded to approximately 10.1 µm. To test whether a difference was present in the number of hepatocytes with large nuclei in the exposed and control mice, we calculated the proportion of nuclei >10.1 µm in every mouse. The control mice had a proportion of 32.0% ± 3.5% of large nuclei, and the BDE-47–exposed mice have a proportion of 21.9% ± 2.3% of large nuclei [mean ± SE; P = 0.04; Fig. 5(f)].
Figure 5.
Perinatal exposure to BDE-47 resulted in reduced number of polyploid hepatocytes on PNW20: (a) representative image of liver section from control mouse (1, example of large polyploid nucleus; 2, example of binuclear hepatocyte); (b) representative image of liver section from exposed mouse; (c) changes in percentage of binuclear hepatocytes; (d) changes in nucleus width; (e) nucleus size distribution across all mice from both exposure groups (red line indicates gap between 2 distribution peaks used in the present study to delineate normal and large nuclei); and (f) changes in percentage of large nuclei. All data presented as mean ± SE; n = 5/exposure group; P value calculated using t test.
C. Changes in mTOR Activity in Murine Liver After Perinatal Exposure to BDE-47
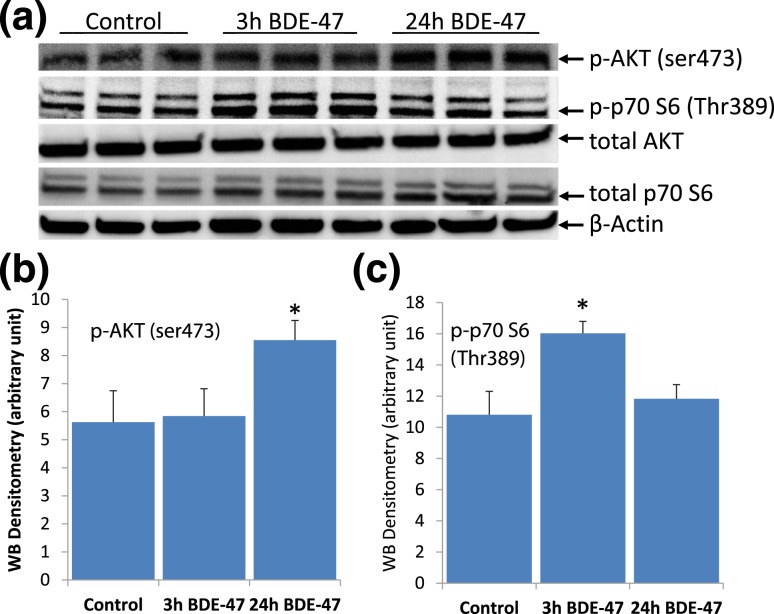
Increased transcription of ribosomal and mitochondrial proteins and changes in the expression of genes of the mTORC1 pathway and the genes regulated downstream of mTORC1 and mTORC2 indicated a possibility of mTOR pathway involvement in the response to BDE-47 in the livers of the exposed mice [46, 47]. To test directly whether BDE-47 changes the activity of mTOR complexes in the liver, we measured the phosphorylation of well-characterized mTORC1 and mTORC2 targets: phospho-p70S6 kinase (Thr389) and phospho-Akt (ser473), respectively (Fig. 6). The activity of both mTOR complexes in the livers of the exposed mice was significantly increased on PND21 and had returned to a normal level by PNW20. To control for changes in total p70S6, we used anti-p70S6 antibody (catalog no. 9202; Cell Signaling Technology), which also binds to p85S6, an alternative translation variant of S6K1 [54]. Expression of p85S6 was significantly decreased in the 20-week-old mice perinatally exposed to BDE-47 [Fig. 5(d) and 5(e)].
Figure 6.
Activity of mTORC1 and mTORC2 was increased (a) on PND21 but not (d) on PNW20 in livers of mice exposed to BDE-47 perinatally, as measured by phosphorylation of p70S6 at Thr389 and Akt at Ser473, respective targets of mTORC1 and mTORC2. Expression of p85S6 (cytoplasmic S6K1) was suppressed in exposed mice on PNW20. (b and c) Quantification of phospho (p)-Akt and phospho-p70S6, respectively, on PND21. (e) Quantification of total p85S6 on PNW20. Data presented as mean ± SE; n = 3/exposure group; P value calculated using the t test.
D. Changes in Circulating IGF-1, Insulin, and Glucose After Perinatal Exposure to BDE-47
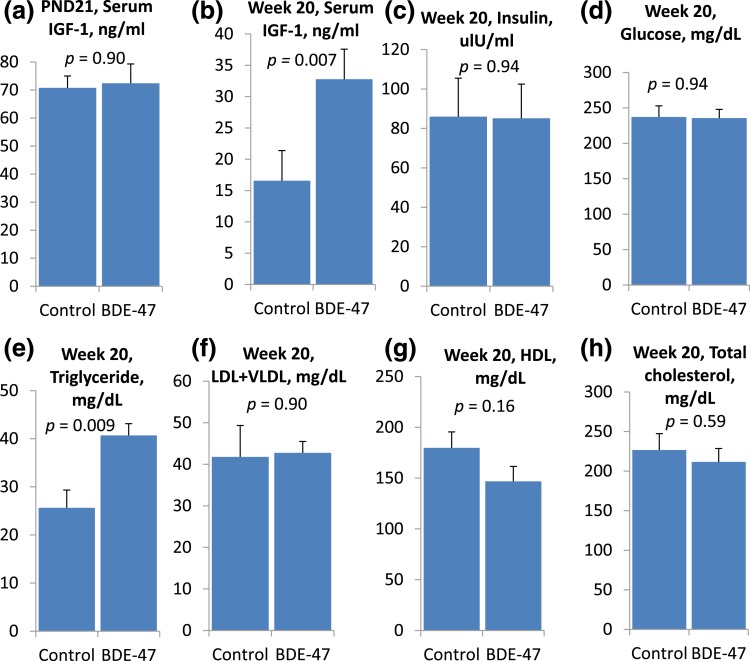
PBDEs are known to affect the IGF axis [23–25, 31, 44, 55, 56]. Thus, we hypothesized that changes in the expression of metabolic genes, including the effects of BDE-47 on the mTOR pathway, are mediated by the availability of circulating IGF-1. To test this hypothesis, we measured IGF-1 in the blood serum of the dams and male offspring euthanized on PND21 and PNW20. No substantial differences were found in the serum IGF-1 levels of the dams and male pups on PND21 [Fig. 7(a)]. By week 20, the concentrations of circulating IGF-1 had decreased more than four times in the control mice compared with the PND21 levels [Fig. 7(b)]. Additionally, by week 20, the concentrations of IGF-1 had increased twofold in the exposed mice compared with the age-matched controls. No changes in fasting insulin and glucose levels were found in male mice on PNW20 [Fig. 7(c) and 7(d)].
Figure 7.
Effect of perinatal exposure to BDE-47 on serum IGF-1 on (a) PND21 and (b) PNW20 and (c) insulin, (d) glucose, (e) triglycerides, (f) LDL+VLDL, (g) HDL, and (h) total cholesterol on PNW20. All data presented as mean ± SE; n = 5 (averaged litter values)/exposure group; P value calculated using Dunnett’s test.
E. Changes in Circulating Lipoproteins and Triglycerides and in Expression of Triglyceride Metabolism Genes in Liver After Perinatal Exposure to BDE-47
The most well-characterized metabolic complication in patients taking the mTORC1 inhibitor rapamycin is hyperlipidemia [57–59]. Rapamycin-associated dyslipidemia has been reported in 45% of liver transplant patients [60] and in ~40% of renal transplant patients [61]. To test whether BDE-47 is able to induce long-lasting changes in circulating lipids in an animal model, we measured the circulating triglycerides, HDL, and LDL+VLDL levels in the serum of mouse offspring on PNW20 after perinatal exposure to BDE-47. Total cholesterol was calculated according to the equation: LDL + VLDL + HDL + triglycerides/5. Blood triglyceride levels were significantly increased twofold in exposed mice [Fig. 7(e)]. The concentrations of circulating lipoproteins were unaltered in the exposed mice [Fig. 7(f–H)]. To check whether any associated changes were present in liver triglyceride transport and metabolism at the level of gene expression, we manually checked expression of the enzymes responsible for triglyceride uptake (Slc27 fatty acid transporters and CD36), synthesis (fatty acid synthesis and acetyl-coenzyme A carboxylases), and export (microsomal triglyceride transfer protein and apolipoprotein B) [62, 63]. Expression of only one gene was significantly altered: CD36 was downregulated 2.5-fold, with a FDR of q = 0.02.
F. Changes in mTOR Activity in HepG2 Cells After Exposure to BDE-47
The effects of BDE-47 on mTOR signaling in liver cells might be direct or might be mediated through changes in the levels of circulating hormones induced by exposure. To test whether BDE-47 changes the activity of mTOR complexes in vitro, we measured phosphorylation of mTORC1 and mTORC2 targets in HepG2 cells after exposure to BDE-47 for 3 and 24 hours (Fig. 8). The activity of mTORC1 was significantly induced by BDE-47 at 3 hours after the start of exposure and had returned to normal levels after 24 hours of exposure. In contrast, the activity of mTORC2 was not changed at the 3-hour point but was significantly induced after 24 hours of exposure.
Figure 8.
In HepG2 cells exposed to BDE-47, mTORC1 activity increased within 3 hours and mTORC2 activity increased within 24 hours: (a) representative Western blots showing phosphorylation status of p70S6 at Thr389 and Akt at Ser473 in control cells and cells exposed to 1 µM for 3 and 24 hours; (b) quantification of phospho (p)-Akt; (c) quantification of phospho-p70S6. Data presented as mean ± SD; n = 3/exposure group; *P < 0.01, t test).
G. Coordinated Regulation of Genes in Rat Tissues: Revising Published Transcriptomic Studies
We hypothesized that the metabolic response to PBDE exposure in mammalian organisms is mediated by the mTOR-centered pathway. If this hypothesis were true, groups of functionally related molecules, targets of the pathway, would be coregulated. Thus, we predicted that ribosomal, mitochondrial, and adipogenesis genes would be regulated in one direction and phagosomal genes and ketone body production genes would be regulated in the opposite direction in exposed animals. To test this prediction, we performed GSEA using previously published transcriptomic experiments. The results of the GSEA corresponded to our prediction for all five genomic data sets used in the present study (Table 3), although statistical significance was low for the enrichment of some gene sets. The enrichment scores of all five data sets after developmental exposure to BDE-47 were very similar to those for female brain frontal lobes on PND41 and livers on PND27.
Table 3.
GSEA Enrichment of Functional Gene Groups Coregulated by mTOR-Centered Pathway in Rats Exposed to PBDE
| Variable | Suvorov and Takser |
|
|---|---|---|
| [31] | [44] | |
| Species | Rat | Rat |
| Sex | Male | Female |
| Tissue | Liver | Brain frontal lobes |
| PND | 27 | 41 |
| Interval after dosing (days) | 7 | 21 |
| Normalized ES/Nom P value | ||
| KEGG ribosome | 1.39/0.034a | 1.40/0.033a |
| Mootha mitochondria | 1.22/0.056 | 1.22/0.039a |
| Reactome ER phagosome pathway | −1.73/0.002a | −1.72/0.000a |
| Reactome PPARα activates gene expression | −1.00/0.438 | −1.00/0.434 |
| Wang classic adipogenic targets of PPARγ | 0.98/0.502 | 0.99/0.491 |
Abbreviations: ER, endoplasmic reticulum; ES, enrichment score; NOM P value, nominal P value; NS, not significant.
Nominal P < 0.05.
3. Discussion
We found that perinatal exposure to 0.2 mg/kg body weight of BDE-47 results in long-lasting changes in the expression of several groups of metabolic genes in the mouse liver. In particular, genes of oxidative phosphorylation and ribosomal proteins were increased in expression 17 weeks after exposure had stopped. Changes in gene expression were associated with a long-lasting twofold increase in circulating triglycerides and IGF-1, a decrease in the percentage of polyploid and binuclear hepatocytes, and substantial suppression of expression of p85S6 kinase in the liver. In a previous study [64], intravenous exposure of pregnant rats to 0.2 mg/kg body weight of BDE-47 resulted in 234.3 ng BDE-47/g lipid in the adipose tissue of the dams and 1054.7 ng BDE-47/g lipid in the adipose tissue of the pups. These concentrations are comparable with those of the North American human population (mean concentration in adipose tissue of adult subjects, 399 ng/g lipids) [65]. Given that the rate of absorption after oral administration of BDE-47 is 75% to 85% in rodents [66–68] and that the rate of BDE-47 elimination is ~10 times greater in mice than in rats [68, 69], we suggest that the exposure used in the present study is also relevant for the North American human population. To further increase the relevance of our exposure paradigm to human exposures, we dosed the mice during the perinatal period to match that of the human PBDE exposure peaks during the perinatal period of life resulting from the active transport of PBDE via cord blood and breast milk [5–7] and the greater rates of dust ingestion [11] and greater food intake per kilogram of body weight in toddlers [6].
Several lines of evidence suggest that the metabolic effects of BDE-47 might be mediated by changes in the mTOR pathway. mTOR is an atypical serine/threonine kinase that is present in two distinct complexes: mTORC1 and mTORC2. Both of these complexes were activated on PND21 but not on PNW20 in mouse livers. Additionally, we observed changes in the expression of genes participating in mTORC1 signaling pathways and genes regulated downstream mTORC1 and mTORC2. In particular, mTORC1 regulates ribosomal biogenesis [46, 47] and mitochondrial biogenesis [48, 49]. Ribosomal genes and genes of oxidative phosphorylation, which are mostly mitochondrial genes, were among the most highly enriched at both PND21 and PNW20.
One unexpected finding of the present study was the substantial and long-lasting suppression of the p85S6 kinase in the livers of mice perinatally exposed to BDE-47. This kinase, along with p70S6 and p31S6, is a product of the RPS6KB1 gene. One alternative splicing variant of the gene encodes the p31 kinase and another produces two alternative translation variants: p70 and p85 [70, 71]. Little is known about the regulation of translation that results in one S6K1 protein or another. All three S6K1 are targets of mTOR-mediated phosphorylation [72] but have different cellular localization; p31 is exclusively nuclear, p85 cytoplasmic, and p70 is subject to mTORC1-controlled shuttling between the cytoplasm and the nucleus [72]. Although many substrates of S6K1 have been reported [70, 71], the main target of S6K1 is ribosomal protein S6, a component of the 40S ribosome subunit. Phosphorylation of S6 induces protein synthesis at the ribosome, suggesting that S6K1 regulates cell growth by affecting the translation process [73]. Thus, long-lasting molecular changes in the livers of mice perinatally exposed to BDE-47 include somewhat controversial decreases in cytoplasmic S6K1 responsible for translation activation and increases in expression of ribosomal genes. This controversy suggests a compensatory nature of some of the observed changes at the molecular level.
Given that PBDEs are known to affect the IGF axis [23–25, 31, 44, 55, 56] and given that IGF-1 activates mTOR signaling [74–76], we hypothesized initially that changes in the liver metabolic profile in response to BDE-47 exposure might be IGF-1-dependent. However, no substantial differences were found in the serum IGF-1 of the male pups on PND21, the point by which persisting changes in the altered expression of liver metabolic genes has already been established. Thus, it is unlikely that changes in circulating IGF-1 after BDE-47 exposure is a causative factor for changes in liver metabolism. Additionally, activation of mTORC1 and mTORC2 was observed in HepG2 cells after 3 and 24 hours of treatment by BDE-47, respectively. This fact suggests that BDE-47 directly affects mTOR signaling in liver cells rather than acting via changes in circulating hormones. IGF-1 is one of the major activators of the mTOR pathway [47, 74]. It was surprising, therefore, that the twofold increase in circulating IGF-1 on PNW20 was not associated with increased activity of mTOR complexes in mouse livers. We hypothesized that changes in IGF-1 secretion might also result from some compensatory mechanism that keeps essential cellular processes at physiological levels instead of developmentally reprogrammed pathways responsible for metabolism homeostasis maintenance.
In our experiments, perinatal exposure to BDE-47 resulted in a long-lasting, almost twofold, increase in circulating triglycerides. This increase is likely explained by the decreased expression of CD36 in the liver of the exposed mice. CD36 is a multifunctional membrane receptor involved in fatty acid uptake [77, 78]. CD36 is regulated at the translational level downstream of mTOR [79, 80]. Recent studies have demonstrated that reduced expression of CD36 improves insulin sensitivity [78, 81] but increases circulating lipids [81]. Improved glucose uptake in rats perinatally exposed to BDE-47 was reported in our previous study [23], and these animals had increased blood cholesterol levels [31]. Thus, it is likely that the long-lasting metabolic effects of perinatal BDE-47 exposure in rodent models consist, among others, in suppression of CD36, associated with increased blood lipids and increased insulin sensitivity. Increased insulin sensitivity is usually considered a desirable shift in metabolism; however, other changes in liver metabolic reactions induced by perinatal BDE-47 exposure in our studies and studies reported by other groups [56, 82–84] produced, as yet poorly understood, shifts in metabolic homeostasis maintenance that might result in multiple disadvantages.
Perinatal exposure to BDE-47 resulted in a substantial decrease in the percentage of binuclear and polyploid hepatocytes in the mouse livers on PNW20. Polyploidy is a common characteristic of the mammalian liver [85]. In rodents, hepatocyte polyploidization starts at the time of weaning and continues throughout development and aging [86]. After the end of the proliferative stage, the liver undergoes gradual polyploidization, during which several ploidy classes of hepatocytes emerge, including tetraploid and octoploid cell classes with one or two nuclei [87]. In adult rodents, the degree of polyploidy reaches 85% in C57Bl mice [88] and Wistar rats [89]. Liver polyploidization is associated with terminal differentiation and aging [90–92]. It was shown recently that generation of binuclear tetraploid hepatocytes is dependent on Akt activity [86, 93]. The investigators demonstrated that selective inhibition of phosphatidylinositol 3-kinase (PI3K) in rat livers or in cultured hepatocytes results in substantially reduced Akt and S6 ribosomal phosphorylation and a drastic decrease in cytokinesis failure events. Cytokinesis failure was mTORC1-independent, revealing that Akt controls the cytokinesis program and consequent generation of binuclear tetraploid hepatocytes directly or through mTORC2 [86]. This second possibility is supported in that mTORC2 has an important role in regulating the actin cytoskeleton and cytokinesis [94].
The results of the present study have demonstrated that BDE-47 at low doses can activate both mTOR complexes in mouse livers and in human hepatocellular carcinoma cells, suggesting common mechanisms of activation. One such mechanism consists of PI3K activation. PI3K is a member of the classic PI3K/Akt/mTORC1 cascade [74]. PI3K-dependent binding of mTORC2 to ribosomes was also shown recently as a mechanism of mTORC2 activation [95]. Another mechanism involves post-translational modifications of the mTORC2 complex member Rictor by mTORC1-dependent S6K1 [96]. It was also reported that Rac1 regulates both mTORC1 and mTORC2 by binding directly to mTOR and mediating mTORC1 and mTORC2 localization at specific membranes in response to stimulation by growth factors [97]. Additional studies are needed to dissect the molecular mechanisms upstream of mTOR that are affected by BDE-47.
Given that exposure in our mouse experiment ceased on PND21 and many effects linked to the mTOR pathway were found 17 weeks later, we hypothesize that modulation of mTOR signaling in the liver during a sensitive developmental window might have a permanent programming effect on liver physiology. The initial half-life for BDE-47 is 1 to 3 days in mice, and the terminal half-life was calculated to be 23 days [69], suggesting that the observed differences between control and exposed mice cannot be attributed to the direct effect of BDE-47 retained in the animals’ tissues. The mechanisms of programming by BDE-47 are not clear; however, we speculate that opposing forces might be involved in these changes, including abnormal setting of mTOR pathway activity in response to developmental exposure and compensatory response aimed at restoring normal cellular physiology. For example, the expression of mTORC1- and mTORC2-regulated genes in our study was altered in both directions (up and down), implicating opposing forces involved in their regulation. Substantial suppression of major translation regulator cytoplasmic S6K1 was associated with substantial upregulation of ribosomal genes. A twofold elevated blood IGF-1 level was not accompanied by increased activity of mTOR complexes. Given that the mTOR pathway has numerous feedback loops [32, 42, 47, 74, 98, 99], it is not surprising that dissecting a long-lasting compensatory network induced by developmental modulation of mTOR activity will be challenging. Additional studies are needed to better characterize the molecular mechanisms affected by developmental exposure to PBDEs and the resulting metabolic phenotype in laboratory animals and humans.
4. Conclusions
The results of the present study have shown that perinatal exposure to environmentally relevant doses of BDE-47 in laboratory mice results in long-lasting changes in liver metabolism associated with a twofold increase in circulating triglycerides. Given that one-fifth of the North American population was exposed to high environmental doses of PBDEs during perinatal development, the possibility of long-term programming of metabolic health by developmental exposures to this group of chemicals raises important concerns and requires mechanistic studies to provide a background for the development of preventive and protective interventions against environmentally induced metabolic disorders. The present study has also provided experimental evidence for a hypothesis linking PBDE exposure with long-lasting metabolic changes via an mTOR-centered molecular mechanism—a major pathway of regulation of growth and metabolism, linked to aging and to the development of cancer, obesity, type 2 diabetes, male infertility, and neurodevelopmental and neurodegenerative diseases. The potential ability of environmental exposures to affect the mTOR pathway, the pathway involved in the pathogenesis of conditions representing major public health threats, calls for further mechanistic research.
Acknowledgments
Acknowledgments
The present report was produced with funding from a University of Massachusetts start-up fund to A.S.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BDE-47
- 2,2′,4,4′-tetrabromodiphenyl ether
- DMSO
- dimethyl sulfoxide
- GEO
- Gene Expression Omnibus
- GSEA
- gene set enrichment analysis
- HDL
- high-density lipoprotein
- IGF
- insulin-like growth factor
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LDL
- low-density lipoprotein
- mRNA
- messenger RNA
- mTOR
- mechanistic target of rapamycin
- mTORC1
- mTOR complex 1
- mTORC2
- mTOR complex 2
- PBDE
- polybrominated diphenyl ether
- PI3K
- phosphatidylinositol 3-kinase
- PND
- postnatal day
- PNW
- postnatal week
- PPAR
- peroxisome proliferator-activated receptor
- SE
- standard error
- VLDL
- very-low-density lipoprotein.
References and Notes
- 1.Hites RA. Polybrominated diphenyl ethers in the environment and in people: a meta-analysis of concentrations. Environ Sci Technol. 2004;38(4):945–956. [DOI] [PubMed] [Google Scholar]
- 2.Dodson RE, Perovich LJ, Covaci A, Van den Eede N, Ionas AC, Dirtu AC, Brody JG, Rudel RA. After the PBDE phase-out: a broad suite of flame retardants in repeat house dust samples from California. Environ Sci Technol. 2012;46(24):13056–13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradman A, Castorina R, Gaspar F, Nishioka M, Colón M, Weathers W, Egeghy PP, Maddalena R, Williams J, Jenkins PL, McKone TE. Flame retardant exposures in California early childhood education environments. Chemosphere. 2014;116:61–66. [DOI] [PubMed] [Google Scholar]
- 4.Waszak I, Dabrowska H, Góra A. Bioaccumulation of polybrominated diphenyl ethers (PBDEs) in flounder (Platichthys flesus) in the southern Baltic Sea. Mar Environ Res. 2012;79:132–141. [DOI] [PubMed] [Google Scholar]
- 5.Antignac JP, Cariou R, Maume D, Marchand P, Monteau F, Zalko D, Berrebi A, Cravedi JP, Andre F, Le Bizec B. Exposure assessment of fetus and newborn to brominated flame retardants in France: preliminary data. Mol Nutr Food Res. 2008;52(2):258–265. [DOI] [PubMed] [Google Scholar]
- 6.Schecter A, Päpke O, Harris TR, Tung KC, Musumba A, Olson J, Birnbaum L. Polybrominated diphenyl ether (PBDE) levels in an expanded market basket survey of U.S. food and estimated PBDE dietary intake by age and sex. Environ Health Perspect. 2006;114(10):1515–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Z, Jiao Y, Hu Y, Sun Z, Zhou X, Feng J, Li J, Wu Y. Levels of tetrabromobisphenol A, hexabromocyclododecanes and polybrominated diphenyl ethers in human milk from the general population in Beijing, China. Sci Total Environ. 2013;452-453:10–18. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Ruan X, Li Y, Yan M, Qin Z. Polybrominated diphenyl ethers (PBDEs) in aborted human fetuses and placental transfer during the first trimester of pregnancy. Environ Sci Technol. 2013;47(11):5939–5946. [DOI] [PubMed] [Google Scholar]
- 9.Herbstman JB, Sjödin A, Apelberg BJ, Witter FR, Patterson DG, Halden RU, Jones RS, Park A, Zhang Y, Heidler J, Needham LL, Goldman LR. Determinants of prenatal exposure to polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs) in an urban population. Environ Health Perspect. 2007;115(12):1794–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucet J, Tague B, Arnold DL, Cooke GM, Hayward S, Goodyer CG. Persistent organic pollutant residues in human fetal liver and placenta from Greater Montreal, Quebec: a longitudinal study from 1998 through 2006. Environ Health Perspect. 2009;117(4):605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilford BH, Shoeib M, Harner T, Zhu J, Jones KC. Polybrominated diphenyl ethers in indoor dust in Ottawa, Canada: implications for sources and exposure. Environ Sci Technol. 2005;39(18):7027–7035. [DOI] [PubMed] [Google Scholar]
- 12.Suvorov A, Takser L. Facing the challenge of data transfer from animal models to humans: the case of persistent organohalogens. Environ Health. 2008;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan JJ, Rawn DF. The brominated flame retardants, PBDEs and HBCD, in Canadian human milk samples collected from 1992 to 2005; concentrations and trends. Environ Int. 2014;70:1–8. [DOI] [PubMed] [Google Scholar]
- 14.Zota AR, Linderholm L, Park JS, Petreas M, Guo T, Privalsky ML, Zoeller RT, Woodruff TJ. Temporal comparison of PBDEs, OH-PBDEs, PCBs, and OH-PCBs in the serum of second trimester pregnant women recruited from San Francisco General Hospital, California. Environ Sci Technol. 2013;47(20):11776–11784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law RJ, Covaci A, Harrad S, Herzke D, Abdallah MA, Fernie K, Toms LM, Takigami H. Levels and trends of PBDEs and HBCDs in the global environment: status at the end of 2012. Environ Int. 2014;65:147–158. [DOI] [PubMed] [Google Scholar]
- 16.United States Census Bureau. Census data. Available at: http://www.census.gov/data.html. Updated 2016. Accessed 15 February 2017.
- 17.Newbold RR, Jefferson WN, Padilla-Banks E, Haseman J. Developmental exposure to diethylstilbestrol (DES) alters uterine response to estrogens in prepubescent mice: low versus high dose effects. Reprod Toxicol. 2004;18(3):399–406. [DOI] [PubMed] [Google Scholar]
- 18. National Institutes of Health. Funding opportunity announcement: role of environmental chemical exposures in the development of obesity, type 2 diabetes and metabolic syndrome (R21). Posted date, May 11, 2012; Open date, September 5, 2012. Available at: https://grants.nih.gov/grants/guide/pa-files/PA-12-185.html. Accessed 15 February 2017.
- 19. doi: 10.1186/s12940-015-0042-7. Heindel JJ, vom Saal FS, Blumberg B, et al. Parma consensus statement on metabolic disruptors. Environ Health. 2015;14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregg EW, Zhuo X, Cheng YJ, Albright AL, Narayan KM, Thompson TJ. Trends in lifetime risk and years of life lost due to diabetes in the USA, 1985-2011: a modelling study. Lancet Diabetes Endocrinol. 2014;2(11):867–874. [DOI] [PubMed] [Google Scholar]
- 21.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER III, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB; American Heart Association Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 2015;131(4):e29–e322. [DOI] [PubMed] [Google Scholar]
- 22.Ren XM, Guo LH. Molecular toxicology of polybrominated diphenyl ethers: nuclear hormone receptor mediated pathways. Environ Sci Process Impacts. 2013;15(4):702–708. [DOI] [PubMed] [Google Scholar]
- 23.Suvorov A, Battista MC, Takser L. Perinatal exposure to low-dose 2,2′,4,4′-tetrabromodiphenyl ether affects growth in rat offspring: what is the role of IGF-1? Toxicology. 2009;260(1-3):126–131. [DOI] [PubMed] [Google Scholar]
- 24.Shy CG, Huang HL, Chao HR, Chang-Chien GP. Cord blood levels of thyroid hormones and IGF-1 weakly correlate with breast milk levels of PBDEs in Taiwan. Int J Hyg Environ Health. 2012;215(3):345–351. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Yekeen TA, Xiao Q, Wang Y, Lu F, Huo X. Placental IGF-1 and IGFBP-3 expression correlate with umbilical cord blood PAH and PBDE levels from prenatal exposure to electronic waste. Environ Pollut. 2013;182:63–69. [DOI] [PubMed] [Google Scholar]
- 26.Frederiksen M, Thomsen M, Vorkamp K, Knudsen LE. Patterns and concentration levels of polybrominated diphenyl ethers (PBDEs) in placental tissue of women in Denmark. Chemosphere. 2009;76(11):1464–1469. [DOI] [PubMed] [Google Scholar]
- 27.Daniels JL, Pan IJ, Jones R, Anderson S, Patterson DG Jr, Needham LL, Sjödin A. Individual characteristics associated with PBDE levels in U.S. human milk samples. Environ Health Perspect. 2010;118(1):155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croes K, Colles A, Koppen G, Govarts E, Bruckers L, Van de Mieroop E, Nelen V, Covaci A, Dirtu AC, Thomsen C, Haug LS, Becher G, Mampaey M, Schoeters G, Van Larebeke N, Baeyens W. Persistent organic pollutants (POPs) in human milk: a biomonitoring study in rural areas of Flanders (Belgium). Chemosphere. 2012;89(8):988–994. [DOI] [PubMed] [Google Scholar]
- 29.Bradman A, Castorina R, Sjödin A, Fenster L, Jones RS, Harley KG, Chevrier J, Holland NT, Eskenazi B. Factors associated with serum polybrominated diphenyl ether (PBDE) levels among school-age children in the CHAMACOS cohort. Environ Sci Technol. 2012;46(13):7373–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gee JR, Moser VC. Acute postnatal exposure to brominated diphenylether 47 delays neuromotor ontogeny and alters motor activity in mice. Neurotoxicol Teratol. 2008;30(2):79–87. [DOI] [PubMed] [Google Scholar]
- 31.Suvorov A, Takser L. Global gene expression analysis in the livers of rat offspring perinatally exposed to low doses of 2,2′,4,4′-tetrabromodiphenyl ether. Environ Health Perspect. 2010;118(1):97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15(6):555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shayne CG, ed. Animal Models in Toxicology, 3rd ed. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- 34. doi: 10.1186/1476-069X-13-46. Vandenberg LN, Welshons WV, Vom Saal FS, Toutain PL, Myers JP. Should oral gavage be abandoned in toxicity testing of endocrine disruptors? Environ Health. 2014;13(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suvorov A, Vandenberg LN. To cull or not to cull? Considerations for studies of endocrine-disrupting chemicals. Endocrinology. 2016;157(7):2586–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. doi: 10.1186/gb-2013-14-4-r36. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jimbo R, Fernandez-Rodriguez J, Sul YT, Johansson CB. Principal component analysis: a novel analysis to evaluate the characteristics of osseointegration of different implant surfaces. Implant Dent. 2011;20(5):364–368. [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boylan JM, Sanders JA, Neretti N, Gruppuso PA. Profiling of the fetal and adult rat liver transcriptome and translatome reveals discordant regulation by the mechanistic target of rapamycin (mTOR). Am J Physiol Regul Integr Comp Physiol. 2015;309(1):R22–R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. [DOI] [PubMed] [Google Scholar]
- 42.Lamming DW, Demirkan G, Boylan JM, Mihaylova MM, Peng T, Ferreira J, Neretti N, Salomon A, Sabatini DM, Gruppuso PA. Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2). FASEB J. 2014;28(1):300–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 44.Suvorov A, Takser L. Delayed response in the rat frontal lobe transcriptome to perinatal exposure to the flame retardant BDE-47. J Appl Toxicol. 2011;31(5):477–483. [DOI] [PubMed] [Google Scholar]
- 45.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. [DOI] [PubMed] [Google Scholar]
- 46.Conn CS, Qian SB. mTOR signaling in protein homeostasis: less is more? Cell Cycle. 2011;10(12):1940–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126(Pt 8):1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koyanagi M, Asahara S, Matsuda T, Hashimoto N, Shigeyama Y, Shibutani Y, Kanno A, Fuchita M, Mikami T, Hosooka T, Inoue H, Matsumoto M, Koike M, Uchiyama Y, Noda T, Seino S, Kasuga M, Kido Y. Ablation of TSC2 enhances insulin secretion by increasing the number of mitochondria through activation of mTORC1. PLoS One. 2011;6(8):e23238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shende P, Plaisance I, Morandi C, Pellieux C, Berthonneche C, Zorzato F, Krishnan J, Lerch R, Hall MN, Rüegg MA, Pedrazzini T, Brink M. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123(10):1073–1082. [DOI] [PubMed] [Google Scholar]
- 50.Zhang HH, Huang J, Düvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4(7):e6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, Qiang L, Farmer SR. Identification of a domain within peroxisome proliferator-activated receptor gamma regulating expression of a group of genes containing fibroblast growth factor 21 that are selectively repressed by SIRT1 in adipocytes. Mol Cell Biol. 2008;28(1):188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Enesco HE, Samborsky J. Liver polyploidy: influence of age and of dietary restriction. Exp Gerontol. 1983;18(1):79–87. [DOI] [PubMed] [Google Scholar]
- 54.Rosner M, Schipany K, Hengstschläger M. p70 S6K1 nuclear localization depends on its mTOR-mediated phosphorylation at T389, but not on its kinase activity towards S6. Amino Acids. 2012;42(6):2251–2256. [DOI] [PubMed] [Google Scholar]
- 55.Haave M, Folven KI, Carroll T, Glover C, Heegaard E, Brattelid T, Hogstrand C, Lundebye AK. Cerebral gene expression and neurobehavioural development after perinatal exposure to an environmentally relevant polybrominated diphenylether (BDE47). Cell Biol Toxicol. 2011;27(5):343–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunnick JK, Brix A, Cunny H, Vallant M, Shockley KR. Characterization of polybrominated diphenyl ether toxicity in Wistar Han rats and use of liver microarray data for predicting disease susceptibilities. Toxicol Pathol. 2012;40(1):93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutcher JP. Mammalian target of rapamycin (mTOR) Inhibitors. Curr Oncol Rep. 2004;6(2):111–115. [DOI] [PubMed] [Google Scholar]
- 58.Murgia MG, Jordan S, Kahan BD. The side effect profile of sirolimus: a phase I study in quiescent cyclosporine-prednisone-treated renal transplant patients. Kidney Int. 1996;49(1):209–216. [DOI] [PubMed] [Google Scholar]
- 59.Blum CB. Effects of sirolimus on lipids in renal allograft recipients: an analysis using the Framingham risk model. Am J Transplant. 2002;2(6):551–559. [DOI] [PubMed] [Google Scholar]
- 60.Mathe D, Adam R, Malmendier C, Gigou M, Lontie JF, Dubois D, Martin C, Bismuth H, Jacotot B. Prevalence of dyslipidemia in liver transplant recipients. Transplantation. 1992;54(1):167–170. [PubMed] [Google Scholar]
- 61.Kahan BD, Yakupoglu YK, Schoenberg L, Knight RJ, Katz SM, Lai D, Van Buren CT. Low incidence of malignancy among sirolimus/cyclosporine-treated renal transplant recipients. Transplantation. 2005;80(6):749–758. [DOI] [PubMed] [Google Scholar]
- 62.Larsson SL, Skogsberg J, Björkegren J. The low density lipoprotein receptor prevents secretion of dense apoB100-containing lipoproteins from the liver. J Biol Chem. 2004;279(2):831–836. [DOI] [PubMed] [Google Scholar]
- 63.Mashek DG. Hepatic fatty acid trafficking: multiple forks in the road. Adv Nutr. 2013;4(6):697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suvorov A, Girard S, Lachapelle S, Abdelouahab N, Sebire G, Takser L. Perinatal exposure to low-dose BDE-47, an emergent environmental contaminant, causes hyperactivity in rat offspring. Neonatology. 2009;95(3):203–209. [DOI] [PubMed] [Google Scholar]
- 65.Johnson-Restrepo B, Kannan K, Rapaport DP, Rodan BD. Polybrominated diphenyl ethers and polychlorinated biphenyls in human adipose tissue from New York. Environ Sci Technol. 2005;39(14):5177–5182. [DOI] [PubMed] [Google Scholar]
- 66.Orn U, Klasson-Wehler E. Metabolism of 2,2′,4,4′-tetrabromodiphenyl ether in rat and mouse. Xenobiotica. 1998;28(2):199–211. [PubMed] [Google Scholar]
- 67.Huwe J, Hakk H, Lorentzsen M. Bioavailability and mass balance studies of a commercial pentabromodiphenyl ether mixture in male Sprague-Dawley rats. Chemosphere. 2007;66(2):259–266. [DOI] [PubMed] [Google Scholar]
- 68.Sanders JM, Chen LJ, Lebetkin EH, Burka LT. Metabolism and disposition of 2,2′,4,4′-tetrabromodiphenyl ether following administration of single or multiple doses to rats and mice. Xenobiotica. 2006;36(1):103–117. [DOI] [PubMed] [Google Scholar]
- 69.Staskal DF, Diliberto JJ, DeVito MJ, Birnbaum LS. Toxicokinetics of BDE 47 in female mice: effect of dose, route of exposure, and time. Toxicol Sci. 2005;83(2):215–223. [DOI] [PubMed] [Google Scholar]
- 70.Meyuhas O, Dreazen A. Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog Mol Biol Transl Sci. 2009;90:109–153. [DOI] [PubMed] [Google Scholar]
- 71.Tavares MR, Pavan IC, Amaral CL, Meneguello L, Luchessi AD, Simabuco FM. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. [DOI] [PubMed] [Google Scholar]
- 72.Rosner M, Hengstschläger M. Nucleocytoplasmic localization of p70 S6K1, but not of its isoforms p85 and p31, is regulated by TSC2/mTOR. Oncogene. 2011;30(44):4509–4522. [DOI] [PubMed] [Google Scholar]
- 73.Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, Pende M. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene. 2014;33(4):474–483. [DOI] [PubMed] [Google Scholar]
- 74.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garbacz WG, Lu P, Miller TM, Poloyac SM, Eyre NS, Mayrhofer G, Xu M, Ren S, Xie W. Hepatic overexpression of CD36 improves glycogen homeostasis and attenuates high-fat diet-induced hepatic steatosis and insulin resistance. Mol Cell Biol. 2016;36(21):2715–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson CG, Tran JL, Erion DM, Vera NB, Febbraio M, Weiss EJ. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology. 2016;157(2):570–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang C, Yan Y, Hu L, Zhao L, Yang P, Moorhead JF, Varghese Z, Chen Y, Ruan XZ. Rapamycin-mediated CD36 translational suppression contributes to alleviation of hepatic steatosis. Biochem Biophys Res Commun. 2014;447(1):57–63. [DOI] [PubMed] [Google Scholar]
- 80.Wang C, Hu L, Zhao L, Yang P, Moorhead JF, Varghese Z, Chen Y, Ruan XZ. Inflammatory stress increases hepatic CD36 translational efficiency via activation of the mTOR signalling pathway. PLoS One. 2014;9(7):e103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knøsgaard L, Kazankov K, Birkebæk NH, Holland-Fischer P, Lange A, Solvig J, Hørlyck A, Kristensen K, Rittig S, Vilstrup H, Grønbæk H, Handberg A. Reduced sCD36 following weight loss corresponds to improved insulin sensitivity, dyslipidemia and liver fat in obese children. Eur J Clin Nutr. 2016;70(9):1073–1077. [DOI] [PubMed] [Google Scholar]
- 82.Dunnick JK, Nyska A. Characterization of liver toxicity in F344/N rats and B6C3F1 mice after exposure to a flame retardant containing lower molecular weight polybrominated diphenyl ethers. Exp Toxicol Pathol. 2009;61(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cowens KR, Simpson S, Thomas WK, Carey GB. Polybrominated diphenyl ether (PBDE)-induced suppression of phosphoenolpyruvate carboxykinase (PEPCK) decreases hepatic glyceroneogenesis and disrupts hepatic lipid homeostasis. J Toxicol Environ Health A. 2015;78(23-24):1437–1449. [DOI] [PubMed] [Google Scholar]
- 84.Nash JT, Szabo DT, Carey GB. Polybrominated diphenyl ethers alter hepatic phosphoenolpyruvate carboxykinase enzyme kinetics in male Wistar rats: implications for lipid and glucose metabolism. J Toxicol Environ Health A. 2013;76(2):142–156. [DOI] [PubMed] [Google Scholar]
- 85.Gentric G, Desdouets C. Polyploidization in liver tissue. Am J Pathol. 2014;184(2):322–331. [DOI] [PubMed] [Google Scholar]
- 86.Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest. 2009;119(7):1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Celton-Morizur S, Desdouets C. Liver physiological polyploidization: microRNA-122 a key regulator. Clin Res Hepatol Gastroenterol. 2017;41:123–125. [DOI] [PubMed] [Google Scholar]
- 88.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467(7316):707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saeter G, Schwarze PE, Nesland JM, Juul N, Pettersen EO, Seglen PO. The polyploidizing growth pattern of normal rat liver is replaced by divisional, diploid growth in hepatocellular nodules and carcinomas. Carcinogenesis. 1988;9(6):939–945. [DOI] [PubMed] [Google Scholar]
- 90.Kudryavtsev BN, Kudryavtseva MV, Sakuta GA, Stein GI. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64(6):387–393. [DOI] [PubMed] [Google Scholar]
- 91.Gupta S. Hepatic polyploidy and liver growth control. Semin Cancer Biol. 2000;10(3):161–171. [DOI] [PubMed] [Google Scholar]
- 92.Schmucker DL. Hepatocyte fine structure during maturation and senescence. J Electron Microsc Tech. 1990;14(2):106–125. [DOI] [PubMed] [Google Scholar]
- 93.Celton-Morizur S, Merlen G, Couton D, Desdouets C. Polyploidy and liver proliferation: central role of insulin signaling. Cell Cycle. 2010;9(3):460–466. [DOI] [PubMed] [Google Scholar]
- 94.Baker K, Kirkham S, Halova L, Atkin J, Franz-Wachtel M, Cobley D, Krug K, Maček B, Mulvihill DP, Petersen J. TOR complex 2 localises to the cytokinetic actomyosin ring and controls the fidelity of cytokinesis. J Cell Sci. 2016;129(13):2613–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144(5):757–768. [DOI] [PubMed] [Google Scholar]
- 96.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29(21):5657–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42(1):50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang G, Murashige DS, Humphrey SJ, James DE. A positive feedback loop between Akt and mTORC2 via SIN1 phosphorylation. Cell Reports. 2015;12(6):937–943. [DOI] [PubMed] [Google Scholar]
- 99.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]