Abstract
Computational fluid dynamics (CFD) is a promising tool to aid in clinical diagnoses of cardiovascular diseases. However, it uses assumptions that simplify the complexities of the real cardiovascular flow. Due to high-stakes in the clinical setting, it is critical to calculate the effect of these assumptions in the CFD simulation results. However, existing CFD validation approaches do not quantify error in the simulation results due to the CFD solver’s modeling assumptions. Instead, they directly compare CFD simulation results against validation data. Thus, to quantify the accuracy of a CFD solver, we developed a validation methodology that calculates the CFD model error (arising from modeling assumptions). Our methodology identifies independent error sources in CFD and validation experiments, and calculates the model error by parsing out other sources of error inherent in simulation and experiments. To demonstrate the method, we simulated the flow field of a patient-specific intracranial aneurysm (IA) in the commercial CFD software star-ccm+. Particle image velocimetry (PIV) provided validation datasets for the flow field on two orthogonal planes. The average model error in the star-ccm+ solver was 5.63 ± 5.49% along the intersecting validation line of the orthogonal planes. Furthermore, we demonstrated that our validation method is superior to existing validation approaches by applying three representative existing validation techniques to our CFD and experimental dataset, and comparing the validation results. Our validation methodology offers a streamlined workflow to extract the “true” accuracy of a CFD solver.
Keywords: computational fluid dynamics, particle image velocimetry, cardiovascular flow, biomedical industry, hemodynamics, ASME V&V 20, validation protocol, patient-specific, CFD validation, PIV
Introduction
Image-based computational fluid dynamics (CFD) has been extensively used in research to simulate blood flow in cardiovascular systems [1–4]. Recently, there has been an increasing interest in using CFD for clinical management of cardiovascular diseases [5–9]. However, because of the potential direct impact on patients’ lives, the margin for error in the results of a cardiovascular CFD simulation is undoubtedly small. Thus, the accuracy of these CFD-based tools must be quantified before their translation into clinic. Unfortunately, there are no guidelines specifically tailored to validate CFD solvers in the context of cardiovascular flows.
Because cardiovascular flows are multiphysics and complex by nature, CFD models of these flows generally use simplifying modeling assumptions (e.g., rigid vessel walls and Newtonian blood properties) [2,3] to reduce the complexity of the computational model. These assumptions affect the performance of the CFD solver and can potentially affect the simulated flow field in the intended cardiovascular system. Evaluating the error in simulation results due to these CFD modeling assumptions is crucial for assessing the validity of a CFD solver. However, previous CFD validation studies in cardiovascular flows only performed either qualitative or quantitative comparisons of simulation results with validation data (generally in vitro experimental measurements) [10–13]. Directly comparing the simulation and experimental data in this way does not provide any information about the CFD solver’s modeling assumptions, and thus is not sufficient to validate the accuracy of a CFD solver. Thus, a validation methodology that quantifies the performance of a CFD solver by evaluating the error due to the CFD modeling assumptions is needed.
To this end, we developed a validation methodology for CFD that can quantify the error due to its modeling assumptions. We adopted and streamlined the concepts of the American Society of Mechanical Engineers Verification & Validation 20 Standard (ASME V&V 20) [14] and calculated CFD model error, by parsing out other sources of error in simulation and validation datasets. The CFD model error is an estimate of “how good” the assumptions of the candidate CFD solver are at simulating the physics of a given flow.
We applied this validation methodology in a clinically relevant context by simulating blood flow in a patient-specific intracranial aneurysm (IA) using the commercial CFD software package star-ccm+ (v10.02, CD-adapco, Melville, NY). We validated the simulated IA flow field against experimental velocity field measurements from two-dimensional particle image velocimetry (PIV) in an identical silicone aneurysm model. From the simulation and experimental results, we calculated the validation error and uncertainties to quantify the model error using our validation framework. To demonstrate the advantage of using our validation methodology over existing validation approaches [10–13], we applied three existing validation techniques to our CFD and experimental results, and compared the results from each validation technique with our validation results.
Methods
CFD Validation Methodology.
Existing CFD validation studies [11–13,15] quantify the difference between CFD results and validation data as the error in the CFD simulation. However, this difference does not reflect the accuracy of the CFD solver. This difference is also affected by other error sources (in both simulation and experimental data), and does not capture the error due to CFD solver’s modeling assumptions. Let us assume that CFD is used to predict a physical quantity, (where is the true value). The simulation provides a numerical result, , as a prediction of . The error in simulation result () is
| (1) |
As shown in Fig. 1, the CFD error () has three sources
Fig. 1.
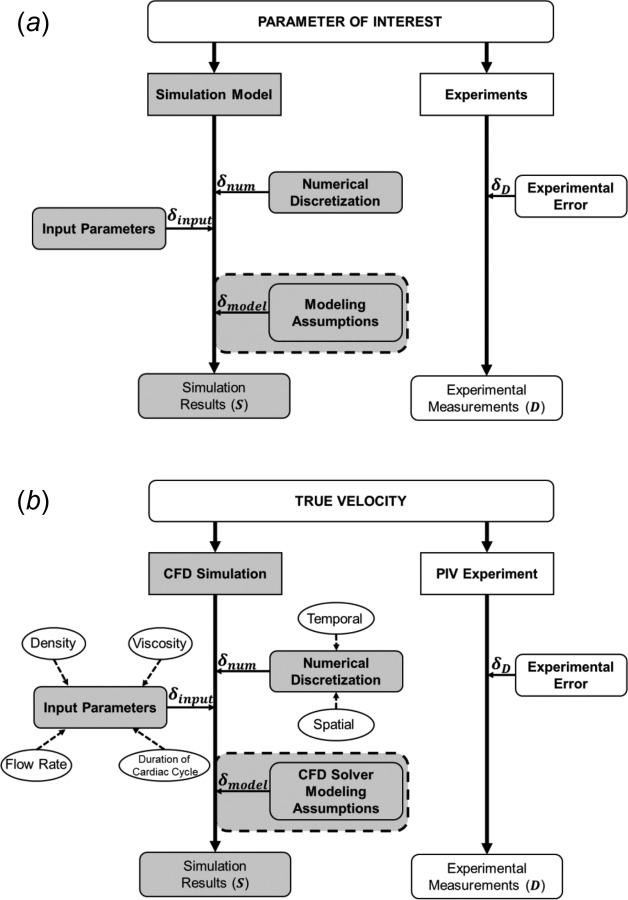
Overview of the validation concept with sources of error in round-edged rectangular boxes. (shaded box, error due to modeling assumptions) represents the true accuracy of the simulation model. (a) Generalized sources of errors in a computational model and experimental measurements. (b) Sources of errors considered while calculating based on our example CFD simulation and PIV experiments on the IA model.
-
(1)
numerical error (), arising from the numerical discretization;
-
(2)
input parameter error (), error in the CFD result () due to the errors in the input parameters into the simulation;
-
(3)
model error (), arising from the CFD modeling assumptions.
Therefore, the overall simulation error can be expressed as
| (2) |
Combining Eqs. (1) and (2) gives
| (3) |
There are two possible scenarios when calculating , given the numerical prediction . First, the true value of the physical quantity is known. In this case, we need to estimate and , and use Eq. (3) to calculate . Second, the true value is unknown. In this case, an in vitro or in vivo validation experiment is required to predict the measured value, , for the physical quantity. Consequently, the error in experimental measurement () is
| (4) |
Substituting the value of the true value, , from Eq. (4) into Eq. (3) gives
which can be rewritten as
| (5) |
where (), the difference between CFD results and experimental measurements, is referred to as the validation error
| (6) |
Thus, Eq. (5) can be written in terms of the validation error () as
| (7) |
The terms in parenthesis in RHS can be lumped together and represented by a validation uncertainty () term. Thus, the model error can be written in terms of validation error and uncertainty as
| (8) |
where
| (9) |
Since all three terms in RHS in Eq. (9) are systematic error and independent of each other [16,17], can expressed as the root mean square of their uncertainties as follows:
| (10) |
Where
-
(1)
Numerical Uncertainty () is the uncertainty due to numerical discretization in the CFD solver.
-
(2)
Input Parameter Uncertainty () is the uncertainty due to input parameters in the CFD simulation.
-
(3)
Experimental Uncertainty () is the uncertainty in the experimental measurements.
The model error can then be expressed as the combination of and as
| (11) |
Determination of , and , and eventually as well as determining becomes essential in calculating as shown by Eq. (11).
Numerical Uncertainty ().
The uncertainty in CFD results due to the spatial and temporal discretization of the flow domain is called the numerical uncertainty (). Based on Richardson extrapolation or h-extrapolation [18,19], is calculated by analyzing the differences in CFD results for systematically refined discretization. CFD results for at least two discretization (the so-called coarse and fine discretization) are required to evaluate . Based on these two discretization, the grid refinement factor is defined as the ratio of coarse to fine discretization
| (12) |
where is the representative cell, mesh or grid size for spatial discretization, and time step for temporal discretization.
The difference in in the two discretization is defined as the estimated error (), which is calculated as
| (13) |
where and are the results of the output parameter of interest in the coarse and fine discretization, respectively.
Then, grid convergence index (GCI) and are calculated from , the factor of safety (), and the expansion factor (). The recommended values of and are 1.25 and 1.15, respectively [14]
| (14) |
| (15) |
where is defined as the observed order of the method, and is equal to 1 for systematically refined discretization. Equations (12)–(15) are used to calculate the uncertainties due to spatial and temporal discretizations [14].
Progressive refinement of the spatial discretization of the flow domain should result in convergence of , and the converged value of should be used to calculate . Therefore, to demonstrate that the value of converges, we recommend to use at least four consecutively refined meshes, and then calculate on the three consecutive mesh pairs in the sequence. It is possible that four meshes are not enough to observe the convergence of . In these cases, further grid refinement should be performed to ascertain proper convergence of [14,20].
Input Parameter Uncertainty ().
Input parameter uncertainty () is defined as the uncertainty in CFD results due to the error in input parameters and is defined as
| (16) |
where is the associated standard uncertainty of the input parameter, is the nominal parameter value, and is the number of uncorrelated input parameters. The term is called the scaled sensitivity coefficient, which can be calculated by using a second-order central finite difference approximation for the term as follows:
| (17) |
Experimental Uncertainty ().
The experimental uncertainty () is the uncertainty in the experimental results, a consequence of variabilities in the experimental setup. The general methodology used to calculate requires evaluating the uncertainties of the individual measured variables during an experiment, and then calculating the overall uncertainty in the experimental results due to variations in the measured variables [14].
Example: Validation of CFD Simulation in an Intracranial Aneurysm
Intracranial Aneurysm Model Selection and Phantom Fabrication.
As a test-bed for application of our validation methodology in a cardiovascular blood flow model, we chose a representative patient-specific saccular internal carotid artery aneurysm with 9 mm diameter. Under institutional review board approval from University at Buffalo, we obtained the three-dimensional (3D) angiographic image of this patient IA (Fig. 2(a)) recorded at the Gates Vascular Institute. We segmented the image using virtual modeling tool kit [21] (vmtk) to obtain a triangulated surface stereolithographic (stl) geometry of the IA model (Fig. 2(b)). Subsequently, using a 3D printer (Objet Eden 500V, Stratasys Ltd., Eden Prarie, MN), we printed a physical model of the IA geometry. Employing the lost wax technique [22] on the 3D printed geometry, we fabricated an optically clear silicone (Sylgard 184 elastomer, Dow Corning, Midland, MI) cube containing hollow aneurysm-vessel geometry (Fig. 2(c)).
Fig. 2.
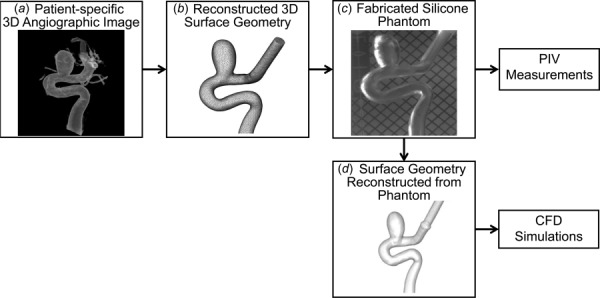
The patient-specific IA model used for validation of star-ccm+ CFD solver: (a) 3D angiographic image of the IA, (b) surface geometry reconstructed from the angiographic image, (c) clear silicone aneurysm phantom fabricated for PIV experiment, and (d) reconstructed surface geometry from reimaging the silicone phantom
PIV Setup.
Particle image velocimetry experiments were performed on the IA phantom to obtain planar velocity measurements on two orthogonal planes in the IA sac (Fig. 3). A blood-mimicking fluid (47.4% water, 36.9% glycerol, and 15.7% NaI) [23] was prepared and fine-tuned [24] to match the refractive index of the silicone phantom (1.42) to eliminate the refraction of the light sheet. Table 1 shows properties of the fine-tuned fluid. A flow loop was then setup (Fig. 3(a)) and seeded with 3.2 μm neutrally buoyant fluorescent tracer particles. A peristaltic pump (Harvard Bioscience, Inc., Holliston, MA) generated physiological waveforms with the duration of a cardiac cycle as 0.85 s (heart rate = 70 beats/min). An inline flow probe and a flowmeter (AD Instruments, Inc., Colorado Springs, CO) were connected upstream to the phantom to record the mean flow rate in the loop [25]. The recorded data were transferred to a computer using an in-house labview code (National Instruments, Austin, TX). A state-of-the-art PIV system (LaVision, Inc., Ypsilanti, MI) was arranged next to the flow loop that included a neodymium-doped yttrium aluminum garnet (Nd:YAG) laser (LaVision, Goettingern, Germany), with a wavelength of 532 nm, and a high-speed 16 bit CMOS camera at 5.5 megapixels. The PIV system acquired image pairs at 40 time-points in one cycle, with 100 image pairs acquired for each time-point. To obtain the velocity vectors, a multipass cross-correlation analysis was used on each image pair with the interrogation window size of 64 × 64 pixels in the first pass and 32 × 32 pixels in the second pass. The vectors were then averaged to obtain the time-averaged in-plane velocity vectors for the IA model. The PIV experiment was performed on two orthogonal planes: sagittal and transverse planes (Fig. 3(b)) to obtain in-plane time-averaged velocity vectors.
Fig. 3.
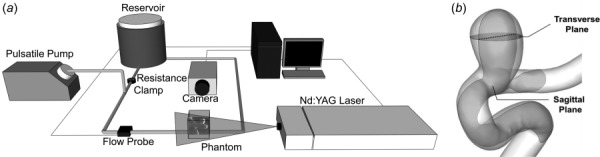
PIV setup and orthogonal planes in the IA model for PIV data acquisition: (a) schematic diagram of the PIV experimental setup and (b) IA geometry with orthogonal sagittal and transverse planes used for PIV flow measurements. The dotted line of intersection of the planes was used as the domain of validation (validation line) for the star-ccm+ CFD solver.
Table 1.
Fluid properties and flow conditions. Properties of the blood-mimicking fluid and inlet boundary conditions for both the flow loop in the experimental setup and CFD simulations.
| Fluid | |
|---|---|
| Refractive index | 1.42 |
| Viscosity (cP) | 4.15 |
| Density (kg/m3) | 1238 |
| Inlet flow conditions | |
| Flow rate (ml/min) | 256 |
| Reynolds number | 358 |
| Duration of a cardiac cycle (s) | 0.85 |
CFD Methods.
The IA phantom was re-imaged using a Toshiba Infinix C-arm machine (Toshiba Medical Systems Corp., Tokyo, Japan) and accurate surface imaging of the IA model was obtained [11]. The re-imaged file was segmented into a surface stl file (Fig. 2(d)) and was used to perform CFD simulations. The 3D computational domain of the IA model was discretized into polyhedral cells, with four prism layers near the wall. A finite volume solver in star-ccm+ was used to solve the flow-governing Navier–Stokes equations under pulsatile, incompressible, and laminar flow conditions. The aneurysm wall was assumed rigid with no-slip boundary condition, and blood was assumed as Newtonian. Traction-free boundary condition was implemented at the outlet of the IA model. We matched the inlet flow rate, flow waveform, density, and viscosity values with the experiment (Table 1). Each simulation was run for three flow waveforms to allow for convergence of the solution, and results from the final waveform were used as the solution of the simulation.
CFD Solver Parameter Optimization.
To obtain the optimal parameter settings for the star-ccm+ solver, we tested the sensitivity of the simulation results to the following CFD parameters: (1) spatial discretization, (2) temporal resolution, and (3) order of accuracy of the solver discretization scheme. We first performed a grid convergence analysis where the IA flow domain was discretized into ten different mesh sizes, with number of elements ranging from 69,920 to 12.8 million. Then, we performed a temporal resolution analysis on the spatially optimized flow domain, by varying the number of time steps from 170 to 2833 per flow waveform. Finally, the spatially and temporally optimized computational flow domain was tested for different orders of accuracy of the spatial and temporal discretization of the star-ccm+ CFD solver. Six simulations including combinations of following spatial and temporal discretization schemes were performed: first and second-order implicit unsteady discretization schemes for temporal discretization (first and second order temporal), and first and second-order accurate upwind schemes and third-order accurate central differencing schemes (first, second, and third order spatial). In total, 21 simulations were performed on the IA model. In each of these simulations, we monitored average velocity magnitude () and validation error percentage () along the line of intersection of the sagittal and transverse planes, matching the planes of PIV imaging.
Validation Analysis.
We applied the validation methodology (Eqs. (6), (10), and (11)) on the simulated flow field by star-ccm+ CFD solver at the optimized settings. The validation analysis was performed on the line of intersection of the sagittal and transverse planes (see Fig. 3(b)) which shall be referred to as the validation line. The time-averaged magnitude of velocity in the sagittal plane along the validation line was used as the physical quantity to be validated. The validation line was divided into 100 equidistant points (with normalized distance from = 0 to = 1), on which velocity magnitude () from both CFD () and experiment () were obtained to calculate validation error ().
The numerical uncertainty () was calculated using Eqs. (12)–(15) on the simulations results for both spatial discretization and temporal resolution. To check for convergence of spatial numerical uncertainty, (spatial component of ) was calculated for consecutive mesh pairs in the grid convergence analysis up until the converged mesh. We then normalized by the average experimental velocity magnitude () to represent it as the percentage of the average flow velocity magnitude for each consecutive mesh pair in the sequence. The uncertainty due to temporal resolution was calculated using the same Eqs. (12)–(15) based on the optimal time resolution (fine temporal discretization), and the preceding temporal resolution (coarse temporal discretization) from the temporal resolution analysis.
To calculate , we examined the effect of four input parameters on the CFD results: density (), viscosity (), flow rate (), and duration of a cardiac cycle () also shown in oval boxes in Fig. 1(b). The systematic uncertainties associated with , , and were based on ten separate experimental measurements, while the uncertainty in was obtained from the specifications of the flowmeter (Table 2). Based on the nominal values and systematic uncertainties of each input parameter, two simulations were performed by perturbing the input to the simulations (). The results of these simulations were used to calculate scaled sensitivity coefficient () using Eq. (17). Equation (16) was then used to calculate overall that includes contributions from all four input parameters. Eight simulations were performed, two each for density, viscosity, flow rate, and duration of a cardiac cycle to calculate .
Table 2.
Input parameter uncertainty analysis. Results of the average input parameter uncertainty () analysis. represents the corresponding input parameters to the CFD simulations, with representing their systematic uncertainties obtained from the experiments. The nominal values of the parameters were taken from Table 1. The scaled sensitivity coefficients () calculated for each input parameter is also shows, which was fed into Eq. (16) to calculate .
| Input parameter () | Nominal parameter value () | Systematic uncertainty () | Scaled sensitivity coefficients () |
|---|---|---|---|
| Flow rate, Q (ml/min) | 256 | ±4% | 5.271 |
| Viscosity, μ (cP) | 4.15 | 0.029 | −0.004 |
| Density, ρ (kg/m3) | 1238 | 0.001 | −0.104 |
| Duration of a cardiac cycle, T (s) | 0.85 | 0.004 | 0.045 |
The experimental uncertainty () was calculated using the correlation statistics algorithm [26] in the lavision davis 8.1 software. The term included uncertainty in velocity measurements due to factors like background noise, out-of-plane particle motion, inaccurate focusing of the camera, and nonhomogeneous seeding density of particles during the PIV experiment [27].
Comparison With Existing Validation Techniques.
To demonstrate the advantage of our validation methodology over existing validation approaches, we applied three existing validation techniques to our CFD and experimental results (Table 3). For qualitative comparison of the flow field, velocity vectors in the sagittal and transverse planes were plotted for both CFD results and experimental measurements. For line comparison and validation error quantification [11], velocity magnitudes in both CFD results and experimental measurements along the validation line were plotted, and was quantified. For the angular and magnitude similarity quantification [13], the average angular similarity index (ASI) and magnitude similarity index (MSI) were quantified and averaged along the validation line. Equations used to calculate ASI and MSI are provided in Table 3. Finally, for our validation methodology, we plotted (by pointwise plotting E and ) along the validation line.
Table 3.
List of representative validation techniques in literature that were applied to our CFD and experimental results. Also listed are the validation parameter of interest and equations to quantify that parameter for each validation technique.
| Validation approach | Study | Quantity for comparison | Equation |
|---|---|---|---|
| Qualitative comparison | Ford et al. [12] | None | None |
| Line comparison and validation error quantification | Hoi et al. [11] | Error (E) | |
| Angular and magnitude similarity | Raschi et al. [13] | ASI, MSI | |
| Sheng et al. [10] | |||
| CFD solver model error () | Current | Model error () |
Results
CFD Solver Parameter Optimization Results.
Figure 4 shows the result of the parameter optimization on velocity magnitude and validation error percentage averaged on the validation line ( and , respectively) for each solver parameter, including the number of elements in mesh discretization, the number of time steps in temporal resolution, and the order of accuracy of solver discretization scheme. As we progressively refined the mesh, time step, or solver accuracy, convergence was reached when the difference in between successive simulations was less than 1%. Figure 4(a) shows the grid convergence analysis between 69,920 and 12.8 million elements in the discretization of IA model. After an initial dip, increased steadily with successive mesh refinement until its asymptotic value of 9.8 cm/s. The successive difference in between 9.6 million to 12.8 million elements was 0.7%, meeting the convergence criterion. Grid size of 9.6 million elements ( = 6.28%) was used for subsequent analyses.
Fig. 4.
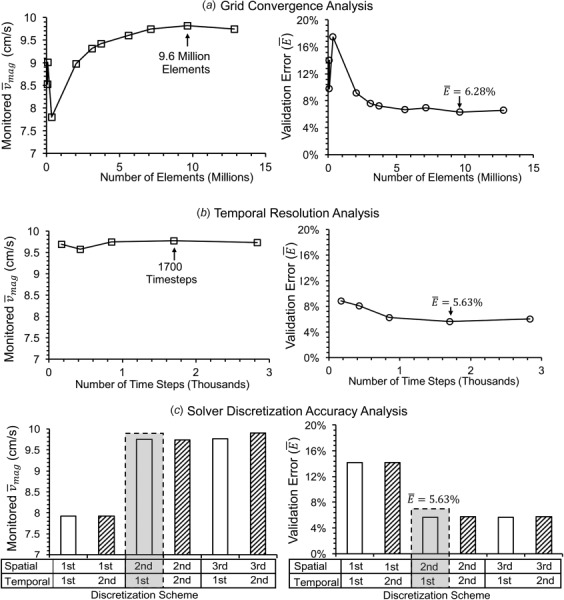
Velocity magnitude (, left) and validation error percentage (, right) monitored for different solver parameters in star-ccm+. (a) Grid convergence analysis. (b) Temporal resolution analysis. (c) Solver discretization accuracy analysis (spatial: first, second, and third; temporal: first (unfilled) and second (hatched)). The optimal solver parameters are indicated by an arrow in the plots (left) for grid sensitivity and temporal resolution analysis, and by the shaded gray rectangle in the solver discretization analysis. The corresponding minimum for the optimum parameters are also indicated in the plots on the right.
Figure 4(b) plots the temporal resolution analysis, where the cardiac cycle was discretized between 170 and 2833 time steps. We found that for all the tested temporal resolutions, the differences between successive were less than 1%, meeting the convergence criterion. However, decreased from an initial 8.85% to a minimum of 5.63% at 1700 time steps. Therefore, this temporal resolution was adopted for subsequent analyses.
Figure 4(c) shows the solver discretization accuracy analysis for different combinations of spatial (first, second, and third) and temporal orders of accuracy (first—unfilled, second—hatched). When the spatial accuracy was fixed, the order of temporal accuracy did not make any significant difference in , and produced similar . However, increasing spatial accuracy from first to second order resulted in 20% increase in the monitored and a reduction of from 14.13% to 5.63%. Furthermore, increasing the spatial discretization scheme to third order did not produce significant difference in and . Since star-ccm+solver treats second order spatial and first order temporal schemes as default, this combination was used in subsequent analyses.
Based on the above mentioned optimization analysis, we chose the following CFD solver settings to simulate flow in our IA model: (1) unstructured grids consisting of polyhedral cells and prism layers at the wall with a total of 9.6 million elements; (2) cardiac cycle resolution of 1700 time steps in the physiologically pulsatile waveform; and (3) second order spatial and first order temporal discretization accuracy.
Validation Results.
Figure 5 shows results of the validation analysis on star-ccm+ solver along the validation line in the IA model. Figure 5(a) lists the equations for calculating the model error (), the validation error (), and the validation uncertainty (). Figure 5(b) shows the percentage of the spatial numerical uncertainty (, normalized by the experimental average velocity magnitude, ) plotted for each consecutive mesh pair in the sequence used in the grid convergence analysis (in Fig. 4(a)). As shown in Fig. 5(b), as the mesh pairs in the sequence are changed from coarse grid pair (1-2) to fine grid pair (8-9), the value of progressively decreases resulting in a converging value of 3.30% at the 8-9 mesh pair, where the number of mesh elements were 7.1 million and 9.6 million, respectively. Figure 5(c) plots pointwise along the validation line as the combination of (circles) and (bars). The value of ranged from −5.42 cm/s to 3.57 cm/s with dissimilar contributions from and along the validation line. varied between −4.60 cm/s and 0.90 cm/s, with highest magnitude of at = 0.89. The value of ranged between ±0.08 cm/s and ±2.60 cm/s, with maximum value occurring around = 0.4. The highest magnitude of occurred at = 0.89 with a large negative (−4.63 cm/s) and small (0.79 cm/s), which means that at this location, CFD grossly under-predicted the velocity values as compared to the experimental measurements.
Fig. 5.
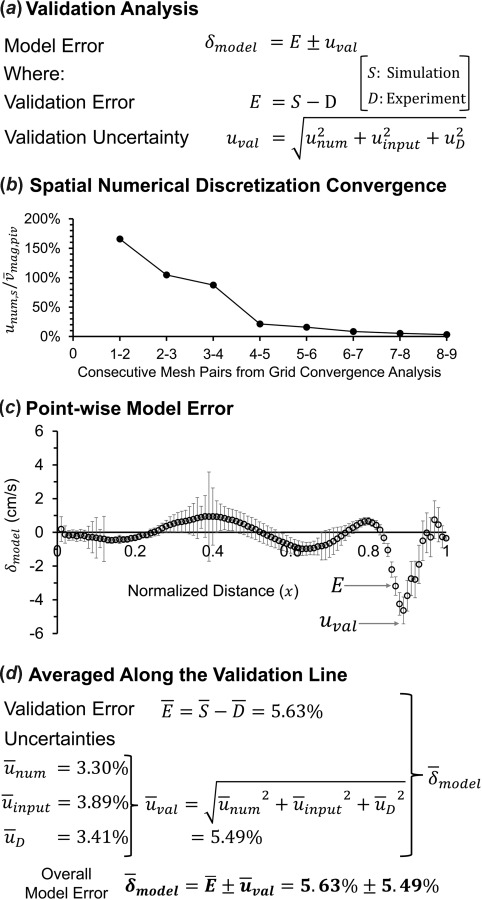
Result of the validation analysis on star-ccm+ CFD solver along the validation line. (a) Equations used to quantify the model error (). (b) Spatial numerical uncertainty () percentage normalized by the average experimental velocity magnitude () for each combination of mesh pairs in the sequence from the grid convergence analysis in Fig. 4(a). (c) Pointwise plot of along the validation line, hollow circles are the validation error () values, and error bars are the validation uncertainty () at each point. (d) The average values of the validation error () and the uncertainties () along the validation line, resulting in the overall average model error ().
As shown in Fig. 5(d), we also calculated , , , and (percentage based on PIV measurements) averaged over the validation line, which were 5.63%, 3.30%, 3.89%, and 3.41%, respectively. The combined uncertainty, i.e., was 5.49%. Consequently, the overall averaged on the validation line was 5.63 ± 5.49%.
Comparison With Existing Validation Techniques.
Figure 6 shows results of application of four validation approaches to our CFD and experimental results. In the Qualitative Comparison of Velocity Flow Field section, we describe each validation result and insight gained about the CFD solver’s accuracy from each validation analysis.
Fig. 6.
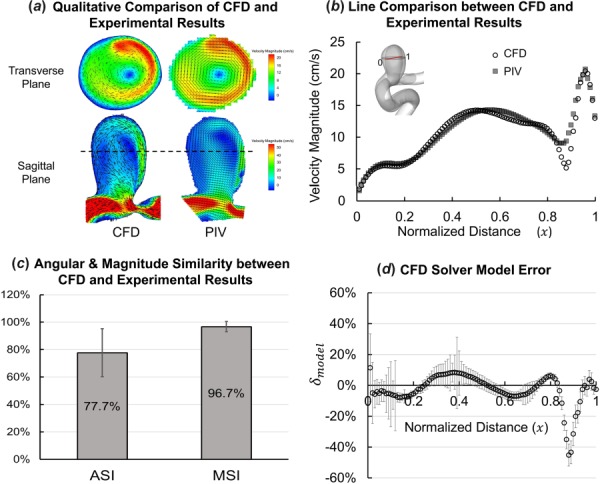
Results of application of different validation approaches to the CFD results and experimental measurements. (a)–(c) Previous validation techniques and (d) proposed validation methodology. (a) Qualitative comparison showing velocity magnitude contours with in-plane velocity vectors of constant length plotted on top (in black) to illustrate the flowdirection in the sagittal plane in CFD and PIV. (b) Pointwise velocity magnitudes from star-ccm+ simulation (CFD, hollow circles) and experimental measurement (PIV, shaded squares) plotted along the validation line normalized from 0 to 1. (c) Average ASI and MSI calculated over the validation line. (d) Pointwise mode error () plotted along the validation line showing the error and uncertainty in star-ccm+ simulation results.
Qualitative Comparison of Velocity Flow Field.
Figure 6(a) shows visual comparison of time-averaged in-plane velocity field on the transverse and sagittal planes. Velocity vectors and magnitude contours show good agreement in the flow patterns between simulation (CFD) and experimental results (PIV) in both the planes. A 3D vortex can be seen, rotating clockwise in the transverse plane and counter-clockwise in the sagittal plane. The location of the vortex generally matches between CFD and PIV. Although the general flow patterns match between the CFD and experimental results, qualitative comparison does not provide any information about the CFD solver’s accuracy.
Line Comparison and Validation Error Quantification.
Figure 6(b) shows the velocity magnitude in CFD results and experimental measurements plotted along the validation line. Simulation results agree well with PIV measurement data along the validation line, except for 0.85 ≤ x ≤ 0.91, where CFD shows a deeper valley than PIV. The average E along the validation line was 5.63%. This validation approach quantifies E, but E alone does not represent the accuracy of the CFD solver itself.
Angular and Magnitude Similarity Between CFD and Experimental Results.
As shown in Fig. 6(c), the average ASI was 77.7% (standard deviation = 17.5%) and MSI was 97.2% (standard deviation = 3.7%) along the validation line. This indicates that there was high similarity in the magnitude and good similarity in the angles of the velocity vectors between CFD results and experimental measurements. Since ASI and MSI are normalized indices, they can be misleading as absolute differences in the CFD and experimental results can be diminished by normalizing the values.
CFD Solver Model Error.
Figure 6(d) shows the result of our validation methodology on the validation line, where is plotted as a combination of (circles) and (bars). E and together represent the variations in CFD-predicted results, which include the contributions from different error sources in the results (refer to Fig. 1(b) for sources of errors). Mathematically, , the combination of E and , represents the error in star-ccm+ due to its modeling assumptions along the validation line. Thus, provides the true accuracy of star-ccm+ along the validation line against the given PIV experimental measurements.
Discussion
Importance of Calculating the Model Error ().
In this study, we present a systematic validation methodology, where is the validation quantity of interest, representing the error due to CFD modeling assumptions. For our example case, we used PIV experiments as surrogate for in vivo flow in the IA. However, since the star-ccm+ simulation matched the PIV experimental fluid and flow conditions, resulting did not capture the effect of common CFD assumptions like Newtonian fluid, generalized inlet boundary conditions, and rigid wall. To capture the effect of these assumptions on the CFD solver, more accurate in vivo flow velocity data (e.g., 4D pc-MRI) is required as the validation data.
Our results show that when compared to existing validation techniques in literature, arising from our validation approach provides most precise information about the accuracy of star-ccm+ on the validation line. While the existing validation techniques mentioned in Table 3 and Figs. 6(a)–6(c) may be acceptable in a research setting, they may not be sufficient for validating CFD solvers for clinical application in cardiovascular systems. By quantifying , our validation framework provides a sense of “credibility” of a CFD solver in its intended context of use.
Optimization of CFD Solver Parameters.
As our results suggest, identifying the optimal CFD solver parameters prior to assessing the solver’s accuracy is critical, since not doing so could result in wide-ranging simulation results and validation error ( = 5.63–17.47% in this study). Thus, the true accuracy of a CFD solver cannot be assessed if optimal solver parameters are not used. This can be exemplified by a recent study organized by the FDA to assess the interlaboratory differences in CFD solvers from different research groups [15,28,29]. In that study, members of the research community were invited to perform steady-state CFD simulations of flow in an ideal axisymmetric nozzle, which was chosen to represent a generic biomedical device. The results showed that the standard deviation of simulated velocity among the participants was greater than 60% of the mean velocity value, with only 4 out of 28 simulation results falling in the confidence intervals of the experimental PIV measurements. We suspect that the choice of CFD solver settings was partially the cause of the large discrepancies in CFD results between the participating groups. Since participants were free to choose CFD solver settings and were not required to optimize them, the considerable variations between CFD results and modest agreement in global and local flow behaviors between CFD and experiment may have been due to suboptimal solver parameters. We believe that if a validation framework (like the one presented in this study) was used, error sources in each participant’s CFD results could have been identified, thus elucidating reasons for the large discrepancies among CFD solvers.
Flow-Specific Considerations in Validation Analysis.
Not only can suboptimal CFD solver settings cause larger model error: inherent complexities in the simulated flow field could also do so. Our validation results displayed significant spatial variation, especially in the region > 0.8 (Fig. 7), which coincided with the largest variation of . To understand what might have caused this variation, we examined the simulated aneurysmal flow field in further detail. Figure 7 shows the presence of a strong impingement jet entering the IA sac, which coincides with higher velocity magnitude and gradient. In this impingement zone (IZ) (0.8 1), was substantially higher (13.92%) than in the nonimpingement zone (4.45%). Furthermore, was almost double in the impingement zone, while was much lower (see Supplementary Material, which is available under the “Supplemental Data” tab for this paper on the ASME Digital Collection). We believe that large where the flow is impinging arises from high velocity gradients near the aneurysm wall. This high sensitivity of velocity due to input boundary conditions can lead to inaccurate velocity predictions, and have potentially contributed to higher . Furthermore, lower in the impingement zone could be due to faster convergence of velocity during grid convergence analysis, since there are higher velocity values in the impingement zone compared to the nonimpingement zone. This analysis shows that complex flow phenomenon, like an impingement jet, can result in high model error. Thus, complexities in the underlying flow field should be closely considered when performing a validation analysis.
Fig. 7.
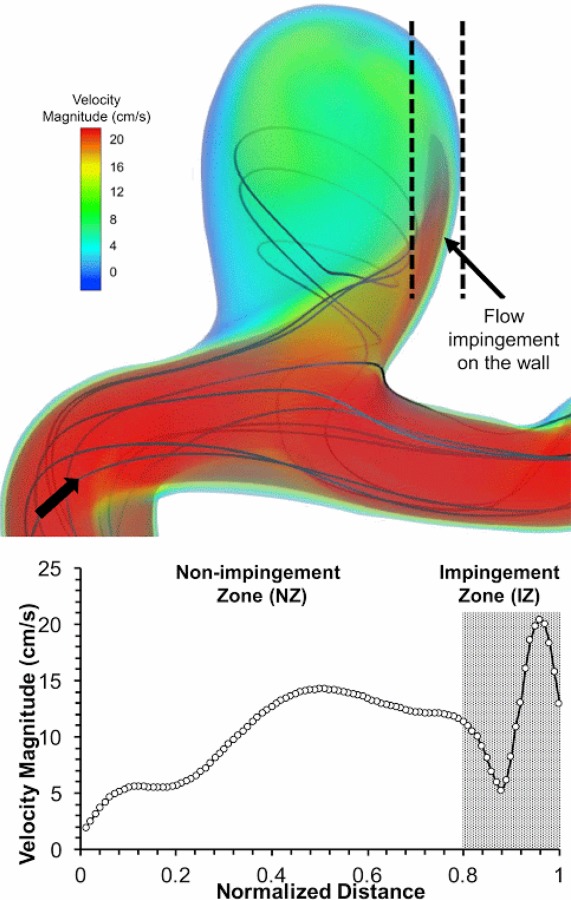
Visualization of 3D flow dynamics using velocity streamlines and volume rendering of velocity magnitude showing the complex 3D flow in the IA model (arrow: flow direction). A strong flow jet impinges on the aneurysm wall, resulting in high velocity and gradients in the region 0.8 0.1, marked as the IZ.
Limitations.
Our study has some limitations. First, velocity measurements from two-dimensional PIV experiments did not accurately capture the complex 3D flow field inside the IA (Fig. 7). Measurements from 4D pc-MRI could provide in vivo validation data for the IA model, which can be used to measure the accuracy of star-ccm+ in simulating the flow in this IA model. Second, we did not consider the uncertainties due to geometrical errors like imaging and segmentation of the IA phantom as a part of input parameters. Geometry is an important input parameter to the CFD solver, and should be considered as a part of input parameter uncertainty in future studies. Third, we used a line as the domain of interest in the IA model to demonstrate the application of our validation methodology. We did not quantify the accuracy of star-ccm+ solver throughout the 3D computational flow domain. Our results should not be considered as the accuracy of star-ccm+ in this IA model.
Supplementary Material
Acknowledgment
We would like to thank Dr. Matthew Ringuette, Dr. Jianping Xiang and Christopher Martensen for their assistance in the PIV experimental setup, and Hamidreza Rajabzadeh-Oghaz, Dr. Yiemeng Hoi and Dr. Kristian Debus for helpful discussions. We also acknowledge the Center for Computational Research at the University at Buffalo for providing the computational support for CFD simulations.
Contributor Information
Nikhil Paliwal, Department of Mechanical and Aerospace Engineering, , University at Buffalo, , Buffalo, NY 14260;; Toshiba Stroke and Vascular Research Center, , University at Buffalo, , Buffalo, NY 14203
Robert J. Damiano, Department of Mechanical and Aerospace Engineering, , University at Buffalo, , Buffalo, NY 14260; Toshiba Stroke and Vascular Research Center, , University at Buffalo, , Buffalo, NY 14203
Nicole A. Varble, Department of Mechanical and Aerospace Engineering, , University at Buffalo, , Buffalo, NY 14260; Toshiba Stroke and Vascular Research Center, , University at Buffalo, , Buffalo, NY 14203
Vincent M. Tutino, Toshiba Stroke and Vascular Research Center, , University at Buffalo, , Buffalo, NY 14203; Department of Biomedical Engineering, , University at Buffalo, , Buffalo, NY 14260
Zhongwang Dou, Department of Mechanical and Aerospace Engineering, , University at Buffalo, , Buffalo, NY 14260.
Adnan H. Siddiqui, Toshiba Stroke and Vascular Research Center, , University at Buffalo, , Buffalo, NY 14260; Department of Neurosurgery, , University at Buffalo, , Buffalo, NY 14226
Hui Meng, Department of Mechanical and Aerospace Engineering, , University at Buffalo, , 324 Jarvis Hall, , Buffalo, NY 14260;; Toshiba Stroke and Vascular Research Center, , University at Buffalo, , Buffalo, NY 14203; Department of Biomedical Engineering, , University at Buffalo, , Buffalo, NY 14260; Department of Neurosurgery, , University at Buffalo, , Buffalo, NY 14226 , e-mail: huimeng@buffalo.edu
Funding Data
National Institutes of Health (No. R01 NS091075).
Toshiba Medical System Corporation (NIH Grant No. R03 NS090193).
References
- [1]. Meng, H. , Wang, Z. , Hoi, Y. , Gao, L. , Metaxa, E. , Swartz, D. D. , and Kolega, J. , 2007, “ Complex Hemodynamics at the Apex of an Arterial Bifurcation Induces Vascular Remodeling Resembling Cerebral Aneurysm Initiation,” Stroke, 38(6), pp. 1924–1931. 10.1161/STROKEAHA.106.481234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Steinman, D. A. , Milner, J. S. , Norley, C. J. , Lownie, S. P. , and Holdsworth, D. W. , 2003, “ Image-Based Computational Simulation of Flow Dynamics in a Giant Intracranial Aneurysm,” Am. J. Neuroradiology, 24(4), pp. 559–566.http://www.ajnr.org/content/24/4/559 [PMC free article] [PubMed] [Google Scholar]
- [3]. Cebral, J. R. , Castro, M. A. , Burgess, J. E. , Pergolizzi, R. S. , Sheridan, M. J. , and Putman, C. M. , 2005, “ Characterization of Cerebral Aneurysms for Assessing Risk of Rupture by Using Patient-Specific Computational Hemodynamics Models,” Am. J. Neuroradiology, 26(10), pp. 2550–2559.http://www.ajnr.org/content/26/10/2550 [PMC free article] [PubMed] [Google Scholar]
- [4]. Longest, P. W. , and Vinchurkar, S. , 2007, “ Validating CFD Predictions of Respiratory Aerosol Deposition: Effects of Upstream Transition and Turbulence,” J. Biomech., 40(2), pp. 305–316. 10.1016/j.jbiomech.2006.01.006 [DOI] [PubMed] [Google Scholar]
- [5]. Min, J. K. , Leipsic, J. , Pencina, M. J. , Berman, D. S. , Koo, B.-K. , van Mieghem, C. , Erglis, A. , Lin, F. Y. , Dunning, A. M. , and Apruzzese, P. , 2012, “ Diagnostic Accuracy of Fractional Flow Reserve From Anatomic CT Angiography,” JAMA, 308(12), pp. 1237–1245. 10.1001/2012.jama.11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Taylor, C. A. , Fonte, T. A. , and Min, J. K. , 2013, “ Computational Fluid Dynamics Applied to Cardiac Computed Tomography for Noninvasive Quantification of Fractional Flow Reserve: Scientific Basis,” J. Am. Coll. Cardiol., 61(22), pp. 2233–2241. 10.1016/j.jacc.2012.11.083 [DOI] [PubMed] [Google Scholar]
- [7]. Larrabide, I. , Villa-Uriol, M. C. , Cardenes, R. , Barbarito, V. , Carotenuto, L. , Geers, A. J. , Morales, H. G. , Pozo, J. M. , Mazzeo, M. D. , Bogunovic, H. , Omedas, P. , Riccobene, C. , Macho, J. M. , and Frangi, A. F. , 2012, “ AngioLab—A Software Tool for Morphological Analysis and Endovascular Treatment Planning of Intracranial Aneurysms,” Comput. Methods Programs Biomed., 108(2), pp. 806–819. 10.1016/j.cmpb.2012.05.006 [DOI] [PubMed] [Google Scholar]
- [8]. Xiang, J. , Antiga, L. , Varble, N. , Snyder, K. V. , Levy, E. I. , Siddiqui, A. H. , and Meng, H. , 2016, “ AView: An Image-Based Clinical Computational Tool for Intracranial Aneurysm Flow Visualization and Clinical Management,” Ann. Biomed. Eng., 44(4), pp. 1085–1096. 10.1007/s10439-015-1363-y [DOI] [PubMed] [Google Scholar]
- [9]. Villa-Uriol, M. C. , Berti, G. , Hose, D. R. , Marzo, A. , Chiarini, A. , Penrose, J. , Pozo, J. , Schmidt, J. G. , Singh, P. , Lycett, R. , Larrabide, I. , and Frangi, A. F. , 2011, “ @neurIST Complex Information Processing Toolchain for the Integrated Management of Cerebral Aneurysms,” Interface Focus, 1(3), pp. 308–319. 10.1098/rsfs.2010.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Sheng, J. , Meng, H. , and Fox, R. O. , 1998, “ Validation of CFD Simulations of a Stirred Tank Using Particle Image Velocimetry Data,” Can. J. Chem. Eng., 76(3), pp. 611–625. 10.1002/cjce.5450760333 [DOI] [Google Scholar]
- [11]. Hoi, Y. , Woodward, S. H. , Kim, M. , Taulbee, D. B. , and Meng, H. , 2006, “ Validation of CFD Simulations of Cerebral Aneurysms With Implication of Geometric Variations,” ASME J. Biomech. Eng., 128(6), pp. 844–851. 10.1115/1.2354209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Ford, M. D. , Nikolov, H. N. , Milner, J. S. , Lownie, S. P. , DeMont, E. M. , Kalata, W. , Loth, F. , Holdsworth, D. W. , and Steinman, D. A. , 2008, “ PIV-Measured Versus CFD-Predicted Flow Dynamics in Anatomically Realistic Cerebral Aneurysm Models,” ASME J. Biomech. Eng., 130(2), p. 021015. 10.1115/1.2900724 [DOI] [PubMed] [Google Scholar]
- [13]. Raschi, M. , Mut, F. , Byrne, G. , Putman, C. M. , Tateshima, S. , Viñuela, F. , Tanoue, T. , Tanishita, K. , and Cebral, J. R. , 2012, “ CFD and PIV Analysis of Hemodynamics in a Growing Intracranial Aneurysm,” Int. J. Numer. Methods Biomed. Eng., 28(2), pp. 214–228. 10.1002/cnm.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].ASME, 2009, “ Standard for Verification and Validation in Computational Fluid Dynamics and Heat Transfer,” American Society of Mechanical Engineers, New York, Standard No. ASME-V&V-20-2009.https://www.asme.org/products/codes-standards/v-v-20-2009-standard-verification-validation
- [15]. Hariharan, P. , Giarra, M. , Reddy, V. , Day, S. W. , Manning, K. B. , Deutsch, S. , Stewart, S. F. C. , Myers, M. R. , Berman, M. R. , Burgreen, G. W. , Paterson, E. G. , and Malinauskas, R. A. , 2011, “ Multilaboratory Particle Image Velocimetry Analysis of the FDA Benchmark Nozzle Model to Support Validation of Computational Fluid Dynamics Simulations,” ASME J. Biomech. Eng., 133(4), p. 041002. 10.1115/1.4003440 [DOI] [PubMed] [Google Scholar]
- [16]. Coleman, H. , and Stern, F. , 1997, “ Uncertainties and CFD Code Validation,” ASME J. Fluids Eng., 119(4), pp. 795–803. 10.1115/1.2819500 [DOI] [Google Scholar]
- [17].BIPM, 1995, “ Guide to the Expression of Uncertainty in Measurement,” Bureau International des Poids et Mesures, Paris, France.
- [18]. Richardson, L. F. , 1911, “ The Approximate Arithmetical Solution by Finite Differences of Physical Problems Involving Differential Equations, With an Application to the Stresses In a Masonry Dam,” Trans. R. Soc. London, 210(459–470), pp. 307–357. 10.1098/rsta.1911.0009 [DOI] [Google Scholar]
- [19]. Richardson, L. F. , 1927, “ The Deferred Approach to the Limit,” Philos. Trans. R. Soc. London, 226(636–646), pp. 299–361. 10.1098/rsta.1927.0008 [DOI] [Google Scholar]
- [20]. Eça, L. , and Hoekstra, M. , 2003, “ An Evaluation of Verification Procedures for CFD Applications,” 24th Symposium on Naval Hydrodynamics, Fukuoka, Japan, July 8–13, pp. 8–13.https://www.nap.edu/read/10834/chapter/39
- [21]. Antiga, L. , Piccinelli, M. , Botti, L. , Ene-Iordache, B. , Remuzzi, A. , and Steinman, D. A. , 2008, “ An Image-Based Modeling Framework for Patient-Specific Computational Hemodynamics,” Med. Biol. Eng. Comput., 46(11), pp. 1097–1112. 10.1007/s11517-008-0420-1 [DOI] [PubMed] [Google Scholar]
- [22]. Kerber, C. W. , and Heilman, C. B. , 1992, “ Flow Dynamics in the Human Carotid Artery—I: Preliminary Observations Using a Transparent Elastic Model,” Am. J. Neuroradiology, 13(1), pp. 173–180. [PMC free article] [PubMed] [Google Scholar]
- [23]. Yousif, M. Y. , Holdsworth, D. W. , and Poepping, T. L. , 2011, “ A Blood-Mimicking Fluid for Particle Image Velocimetry With Silicone Vascular Models,” Exp. Fluids, 50(3), pp. 769–774. 10.1007/s00348-010-0958-1 [DOI] [PubMed] [Google Scholar]
- [24]. Hopkins, L. , Kelly, J. , Wexler, A. , and Prasad, A. , 2000, “ Particle Image Velocimetry Measurements in Complex Geometries,” Exp. Fluids, 29(1), pp. 91–95. 10.1007/s003480050430 [DOI] [Google Scholar]
- [25]. Zhao, M. , Amin-Hanjani, S. , Ruland, S. , Curcio, A. , Ostergren, L. , and Charbel, F. , 2007, “ Regional Cerebral Blood Flow Using Quantitative MR Angiography,” Am. J. Neuroradiology, 28(8), pp. 1470–1473. 10.3174/ajnr.A0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Wieneke, B. , 2015, “ PIV Uncertainty Quantification From Correlation Statistics,” Meas. Sci. Technol., 26(7), p. 074002. 10.1088/0957-0233/26/7/074002 [DOI] [Google Scholar]
- [27]. Adrian, R. J. , and Westerweel, J. , 2011, Particle Image Velocimetry, Cambridge University Press, New York. [Google Scholar]
- [28]. Stewart, S. F. , Hariharan, P. , Paterson, E. G. , Burgreen, G. W. , Reddy, V. , Day, S. W. , Giarra, M. , Manning, K. B. , Deutsch, S. , and Berman, M. R. , 2013, “ Results of FDA’s First Interlaboratory Computational Study of a Nozzle With a Sudden Contraction and Conical Diffuser,” Cardiovasc. Eng. Technol., 4(4), pp. 374–391. 10.1007/s13239-013-0166-2 [DOI] [Google Scholar]
- [29]. Stewart, S. F. , Paterson, E. G. , Burgreen, G. W. , Hariharan, P. , Giarra, M. , Reddy, V. , Day, S. W. , Manning, K. B. , Deutsch, S. , and Berman, M. R. , 2012, “ Assessment of CFD Performance in Simulations of an Idealized Medical Device: Results of FDA’s First Computational Interlaboratory Study,” Cardiovasc. Eng. Technol., 3(2), pp. 139–160. 10.1007/s13239-012-0087-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


