Abstract
Context:
Kisspeptin receptor (KISS1R) is expressed in hypothalamic gonadotropin-releasing hormone neurons and responsible for pubertal onset and reproductive functions. KISS1R mutations remain a rare cause of congenital hypogonadotropic hypogonadism (CHH).
Objective:
The aim of this study was to identify the genetic cause of CHH in a patient and to functionally characterize a KISS1R mutation.
Design:
The patient was a 47-year-old Japanese man whose parents were first cousins. He lacked secondary sexual characteristics owing to normosmic CHH. Exon segments for the KISS1R gene in this patient were screened for mutations. Functional analyses were performed using HEK293 cells expressing KISS1R mutants. Molecular dynamics simulations were performed to compare the ligand-KISS1R mutant complex with those of wild-type KISS1R variants.
Results:
A homozygous mutation (c.440C>T, p.P147L) in KISS1R was identified. The P147L mutation did not affect either receptor expression level or subcellular localization in the recombinant expression system. Intracellular calcium measurements and cellular dielectric spectroscopy demonstrated that the P147L mutation impaired receptor function to an extent more severe than that of a previously reported L148S mutation. A receptor-ligand binding assay showed the P147L mutation causes a substantial loss of ligand-binding affinity. Molecular dynamics simulations revealed the P147L mutation decreases the contact surface area of the ligand-receptor complex in an expanded ligand-binding pocket.
Conclusion:
We identified a loss-of-function mutation in KISS1R associated with CHH. Our results demonstrated that the P147L mutation causes a severe phenotype and functional impairment resulting from the loss of ligand-binding affinity due to an expanded ligand-binding pocket.
Keywords: congenital hypogonadotropic hypogonadism, KISS1R, kisspeptin receptor, ligand binding, missense mutation
We identified a novel KISS1R mutation (P147L) and demonstrated that a conformational change in the ligand-binding pocket of the mutant receptor leads to congenital hypogonadotropic hypogonadism.
Congenital hypogonadotropic hypogonadism (CHH) is characterized by deficient or absent pubertal development caused by hyposecretion of gonadotropin. The prevalence of CHH is approximately one case in 8000 individuals [1]. CHH is divided primarily in two clinical categories: CHH with anosmia or hyposmia (Kallmann syndrome) and CHH with normal smell (normosmic CHH). Normosmic CHH has diverse genetic causes, including mutations of KISS1/KISS1R, TAC3/TACR3, GNRH1/GNRHR, LEP/LEPR, HESX1, FSHB, and LHB [2]. KISS1R mutations are found in 2% of patients with normosmic CHH [1].
A 54-amino acid peptide kisspeptin (KP; encoded by KISS1) and its receptor, KISS1R, in the hypothalamus regulate the secretion of gonadotropin-releasing hormone (GnRH) and are responsible for pubertal onset and reproductive functions [3]. The human KISS1R gene is located on chromosome 19, consists of five exons, and encodes 398 amino acids that form a seven-transmembrane G-protein–coupled receptor (GPCR) [4]. In 2003, two research groups independently identified different KISS1R mutations in patients with CHH [5, 6]. These reports showed that loss-of-function mutations in KISS1R cause CHH in mice and humans.
The frequency of mutations in KISS1R-associated CHH is relatively low. At least 24 KISS1R mutations have been reported, including heterozygous mutations [1, 7]. Moreover, few studies have been conducted to evaluate the functions of these mutations. Therefore, the structure-function relationship between wild-type and mutant receptors remains unknown. Here, we identified a loss-of-function mutation in KISS1R found in a Japanese patient with normosmic CHH, elucidating the role of this KISS1R mutation in the pathogenesis of CHH by functional analysis in vitro.
1. Case Report
The subject was a 47-year-old, single Japanese man whose parents were first cousins. His lack of secondary sexual characteristics was noted by a dermatologist when he was admitted to our hospital for treatment of a refractory left leg ulcer. His height, body weight, and arm span were 163.4 cm, 47 kg, and 172.0 cm, respectively. Testicular atrophy, micropenis, and lack of pubic hair (Tanner stage I) were observed, but no hyposmia was detected from an olfaction test using thiamine propyldisulfide. Endocrine test results are shown in Table 1. Luteinizing hormone (LH) and free testosterone were below detectable levels, and follicle-stimulating hormone (FSH) levels were below normal range, whereas levels of other anterior pituitary hormones were normal. A continuous-loading test for GnRH indicated normal gonadotropin response, and magnetic resonance imaging showed no abnormal morphology of the hypothalamus, the pituitary, or the olfactory bulbs. Therefore, we diagnosed CHH. Additionally, we diagnosed osteoporosis owing to decreased bone density of the lumbar vertebrae (L2–4) to 51% of the average found in young adults. Because the patient did not desire fertility, he was continuously given testosterone by intramuscular injection. The leg ulcer was cured after testosterone supplementation; however, it recurred due to the patient self-terminating the treatment. His stretched penile length was 2.5 cm, even after testosterone supplementation for a decade. Written informed consent for DNA-sequencing analysis and permission for publication were obtained from the patient and his brothers. This study was approved by the ethics committee of the University of Miyazaki.
Table 1.
Endocrinological Characteristics of the Patient
| Variable | Laboratory Value | Normal Range |
|---|---|---|
| ACTH, pg/mL | 16.3 | 7.4–55.7 |
| Cortisol, µg/dL | 11.9 | 4.5–21.1 |
| DHEA-S, µg/dL | 133 | 70–495 |
| TSH, μIU/mL | 1.32 | 0.35–4.94 |
| Free T4, ng/dL | 1.14 | 0.70–1.48 |
| Free T3, pg/mL | 2.58 | 1.71–3.71 |
| GH, ng/mL | 1.14 | <2.47 |
| IGF-1, ng/mL | 122 | 90–250 |
| LH, IU/L | <0.1 | 1.2–7.1 |
| FSH, IU/L | 0.8 | 2.0–8.3 |
| Total testosterone, ng/mL | 0.33 | 2.07–7.61 |
| Free testosterone, pg/mL | <0.6 | 4.7–21.6 |
| 17β-estradiol, pg/mL | <10 | 10–40 |
| PRL, ng/mL | 7.4 | 3.5–19.4 |
| GnRH loading test, IU/La | ||
| Basal LH | <0.1 | |
| Peak LH (90 minutes) | 0.6 | |
| Basal FSH | 0.8 | |
| Peak FSH (120 minutes) | 3.7 | |
| GnRH continuous loading test, IU/Lb | ||
| Basal LH | 0.3 | |
| Peak LH (30 minutes) | 5.4 | |
| Basal FSH | 2.1 | |
| Peak FSH (120 minutes) | 5.6 | |
| hCG loading test, day 4 free testosterone, pg/mLc | <0.6 | |
| 75-g oral glucose tolerance test | ||
| Fasting plasma glucose, mg/dL | 91 | 70–110 |
| Peak plasma glucose, mg/dL (15 minutes) | 131 | |
| 30-minute plasma glucose, mg/dL | 113 | |
| Fasting plasma insulin, µU/mL | 5.6 | 1.7–10.4 |
| Peak plasma insulin, µU/mL (15 minutes) | 46.4 | |
| 30-minute plasma insulin, µU/mL | 28.1 |
Abbreviations: ACTH, adrenocorticotropic hormone; DHEA-S, dehydroepiandrosterone sulfate; FSH, follicle-stimulating hormone; GH, growth hormone; hCG, human chorionic gonadotropin; IGF-1, insulin-like growth factor-1; LH, luteinizing hormone; PRL, prolactin; TSH, thyroid-stimulating hormone.
Blood sampling with intravenous administration of GnRH (0.1 mg).
Blood sampling with intravenous administration of GnRH (0.1 mg) at day 4 after 3 days of intravenous administration of GnRH (0.5 mg).
Blood sampling at day 4 the morning after 3 days of intramuscular administration of hCG (5000 U).
2. Material and Methods
A. DNA-Sequencing Analysis
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and amplified by polymerase chain reaction (PCR) using KOD FX polymerase (Toyobo, Osaka, Japan). The primers for KISS1R and TACR3 are shown in Supplemental Table 1 (32.9KB, docx) . The PCR products were sequenced using a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
B. Construction of Expression Vectors
Mutations were introduced by PCR into the wild-type KISS1R sequence cloned in the pcDNA3.1(−) expression vector. To study cell surface expression of the mutant receptors, FLAG tags were added at the N-terminal end of KISS1R. The details are provided in the Supplemental Materials and Methods (32.9KB, docx) .
C. Cell Culture and Transfection
Human embryonic kidney (HEK) 293 cells were cultured in Dulbecco’s modified Eagle medium with 10% fetal bovine serum, 50 IU/mL penicillin, and 50 µg/mL streptomycin at 37°C and 5% CO2. Cells were transiently transfected with expression vectors using ViaFect (Promega, Madison, WI) or FuGENE6 (Promega) according to the manufacturer’s instructions.
D. Intracellular Calcium Measurements
At 5-hours posttransfection, cells were plated on 96-well black plates (Corning, Corning, NY) at 3 × 104 cells/well. At 24-hours posttransfection, the changes in intracellular calcium caused by agonist stimulation were detected using a FLIPR Tetra fluorescence-imaging plate reader (Molecular Devices, Sunnyvale, CA) as described previously [8]. We used human kisspeptin-10 (KP-10) and kisspeptin-54 (KP-54; Peptide Institute, Osaka, Japan) as KISS1R agonists.
E. Cellular Dielectric Spectroscopy
At 5-hours posttransfection, KISS1R-HEK293 cells were reseeded at 4.5 × 104 cells/well on CellKey Standard 96-well plates (Molecular Devices). At 24-hours posttransfection, the medium was replaced with 20 mM HEPES-buffered Hanks balanced salt solution (pH 7.4) containing 0.1% bovine serum albumin (BSA). Changes in cellular impedance induced by agonist stimulation were measured by cellular dielectric spectroscopy using a CellKey System (Molecular Devices) [9].
F. Quantitative Real-Time PCR
Total RNA was extracted from KISS1R-HEK293 cells by using an RNeasy Mini Kit (Qiagen) and reverse transcribed into complementary DNA (cDNA) using a High-Capacity RNA-to-cDNA Kit (Applied Biosystems). Quantitative real-time PCR reactions were conducted with a Thermal Cycler Dice Real Time System II (TaKaRa, Otsu, Japan) using a TaqMan gene expression assay containing primers for human KISS1R (assay ID: Hs00261399_m1; Applied Biosystems) or a housekeeping gene (human GAPDH; assay ID: Hs02758991_g1), TaqMan Fast Advanced Master Mix (Applied Biosystems), and 1 µg of cDNA.
G. Immunocytochemistry
FLAG-tagged-KISS1R-expressing HEK293 (FLAG-KISS1R-HEK293) cells cultured in Nunc Laboratory-Tek II CC2 Chamber Slide System four-well chambers (Thermo Fisher Scientific, Waltham, MA) were fixed with 4% paraformaldehyde, blocked, and permeabilized with 5% normal goat serum, 0.1% BSA, and 0.3% Triton X-100. Cells were incubated with the anti-FLAG M2 antibody [Research Resource Identifier (RRID): AB_262044; 1:500 dilution; Sigma-Aldrich, St. Louis, MO] for 2 hours at 37°C, followed by incubation with Alexa Fluor 488 goat anti-mouse immunoglobulin G (RRID: AB_2534069; 1:1000 dilution; Life Technologies) for 1 hour at room temperature. Nuclear staining was performed using Cellstain 4′,6-diamidino-2-phenylindole solution (1:1000 dilution; Dojindo, Kumamoto, Japan). Cells were imaged using a C2 confocal laser-scanning microscope system (Nikon, Tokyo, Japan).
H. Western Blot
FLAG-KISS1R-HEK293 cells were scraped into HEPES buffer (25 mM HEPES, pH 7.4, 10 mM MgCl2, and 0.25 M sucrose) and homogenized on ice. The homogenate was centrifuged at 800g at 4°C for 20 minutes and the supernatant was centrifuged twice at 100,000g at 4°C for 1 hour. The cell pellet was solubilized with Cell-LyEX MP (TOYO B-Net, Tokyo, Japan). A 1-μg aliquot of this fraction with urea (final concentration, 5 M) was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis [10] and transferred to a polyvinylidene fluoride membrane. The transfer membrane was incubated with anti-FLAG M2 antibody (1:1000 dilution) overnight at 4°C, followed by incubation with anti-mouse immunoglobulin G horseradish-peroxidase-linked antibody (RRID: AB_330924; 1:10,000 dilution; Cell Signaling Technology, Danvers, MA) for 1 hour at room temperature. The immunoreactive band was visualized with ImmunoStar LD (Wako Pure Chemicals, Osaka, Japan) and analyzed using a Fusion FX7 Chemiluminescence Imaging System (Vilber-Lourmat, Eberhardzell, Germany). To calculate relative KISS1R expression, Na,K-ATPase (RRID: AB_2060983; 1:1000 dilution; Cell Signaling Technology) was used as a loading control.
I. Receptor-Ligand Binding Assay
The cell membrane fraction was extracted as described, except for pellet resuspension in HEPES buffer. The cell membrane fraction (2 µg of protein) was incubated with 0.1 to 1.6 nM 125I-labeled KP-10 (125I-KP-10; PerkinElmer, Yokohama, Japan) in the presence (nonspecific binding) or absence (total binding) of 1 µM unlabeled KP-10 in a total of 200 µL of HEPES buffer containing 0.05% BSA for 2 hours at 25°C. After incubation, bound 125I-KP-10 was isolated by vacuum filtration through glass-fiber filter paper (GF/B; Whatman, Maidstone, United Kingdom), placed on a 1225 Sampling Manifold (Millipore, Billerica, MA), and the filter was thoroughly washed with HEPES buffer and dried for 1 hour at 80°C. The radioactivity was measured using a Tri-Carb 2810TR liquid-scintillation analyzer (PerkinElmer) after adding 5 mL of Ultima Gold F scintillation cocktail (PerkinElmer). Specific binding was calculated by subtracting nonspecific binding from total binding.
J. Protein Modeling and Molecular Dynamics Simulations
Protein structure modeling and molecular dynamics simulations were performed using the Molecular Operating Environment (version 2015.1001; Chemical Computing Group, Montreal, PQ, Canada). Homology models of wild-type KISS1R were generated based on the nociceptin receptor (Protein Data Bank code: 4EA3). The details of this method are provided in Supplemental Materials and Methods (32.9KB, docx) .
K. Statistical Analysis
Data are shown as the mean ± standard error of the mean. GraphPad Prism 7 software (GraphPad Software, La Jolla, CA) was used for statistical analysis and curve fitting, and one-way ANOVA was used for comparisons among the three groups. Two-way analysis of variance and repeated measures combined with the Tukey multiple comparison test were applied for experiments with two factors, genotyping, and amounts of agonist. P < 0.05 was considered statistically significant.
2. Results
A. Identification of a KISS1R Mutation Associated With CHH
We investigated genetic defects in the KISS1R and TACR3 [11] genes in this patient with CHH. A homozygous mutation (c.440C>T, p.P147L) in exon 3 of the KISS1R gene was identified in the patient by sequence analysis [Fig. 1(A)]. No mutation was found in the TACR3 gene. Although his older and younger brothers were heterozygous for the mutation [Fig. 1(B)], their pubertal development (pubic hair: Tanner stage V) and gonadotropin and testosterone levels were intact (Supplemental Table 3 (32.9KB, docx) ).
Figure 1.
Family pedigree corresponding to a KISS1R mutation. (A) Electropherograms of DNA sequences in the patient showing homozygous mutations in the KISS1R gene. (B) Pedigree of the reported family with a homozygous KISS1R c.440C>T, p.P147L mutation. The patient is indicated by an arrow. Squares represent men, circles represent women, and slashes indicate deceased members. There was no family history of hypogonadism. KISS1R sequencing was performed only in the patient and his brothers.
B. The P147L Mutation is an Almost Complete Loss-of-Function Mutation
To evaluate the functional properties of the KISS1R P147L mutant, we constructed HEK293 cells transiently expressing KISS1R wild-type or mutant variants (P147L, identified in this study; L148S, reported by Seminara et al. [6]) and measured changes in intracellular calcium concentration induced by KISS1R agonists. KP-10 and KP-54 induced concentration-dependent, robust increases in intracellular calcium concentration in wild-type KISS1R-expressing cells [half maximal effective concentration, 2.54 ×10−10 and 1.85 ×10−9 M, respectively; Fig. 2(A) and 2(B)]. In contrast, no response was observed, even at 1 × 10−6 M of KP-10 in KISS1R P147L-expressing cells. As shown by Wacker et al. [12], the potency and efficacy of these peptides in KISS1R L148S-expressing cells was much less than those observed for wild-type–expressing cells.
Figure 2.
Dose-response relationships of the changes in intracellular calcium concentration for (A) KP-10 and (B) KP-54 in HEK293 cells transiently expressing KISS1R wild-type (filled circle), P147L (filled triangle), L148S (open triangle), or transfected with empty vector (Mock, open circle). Dose-response relationships of changes in cellular impedance for (C) KP-10 and (D) KP-54 in HEK293 cells transiently expressing wild-type KISS1R and variants (n = 3 for each genotype).
We then performed cellular dielectric spectroscopy, which can measure complex impedance changes in cell morphology associated with ligand binding. KP-10 and KP-54 induced concentration-dependent increases in impedance in wild-type KISS1R-expressing cells [half maximal effective concentration, 4.14 ×10−10 and 7.69 ×10−10 M, respectively; Fig. (2)C and 2(D)]. The potency and efficacy of these peptides for KISS1R L148S-expressing cells were less than those for wild-type–expressing cells, whereas no response was observed, even at 1 ×10−6 M KP-10 in KISS1R P147L-expressing cells. These results showed that both the P147L and L148S mutations impaired KISS1R function, and that the P147L mutation caused a more severe loss of function relative to that observed in the L148S mutation.
C. The P147L Mutation Does Not Affect KISS1R Expression or Subcellular Localization
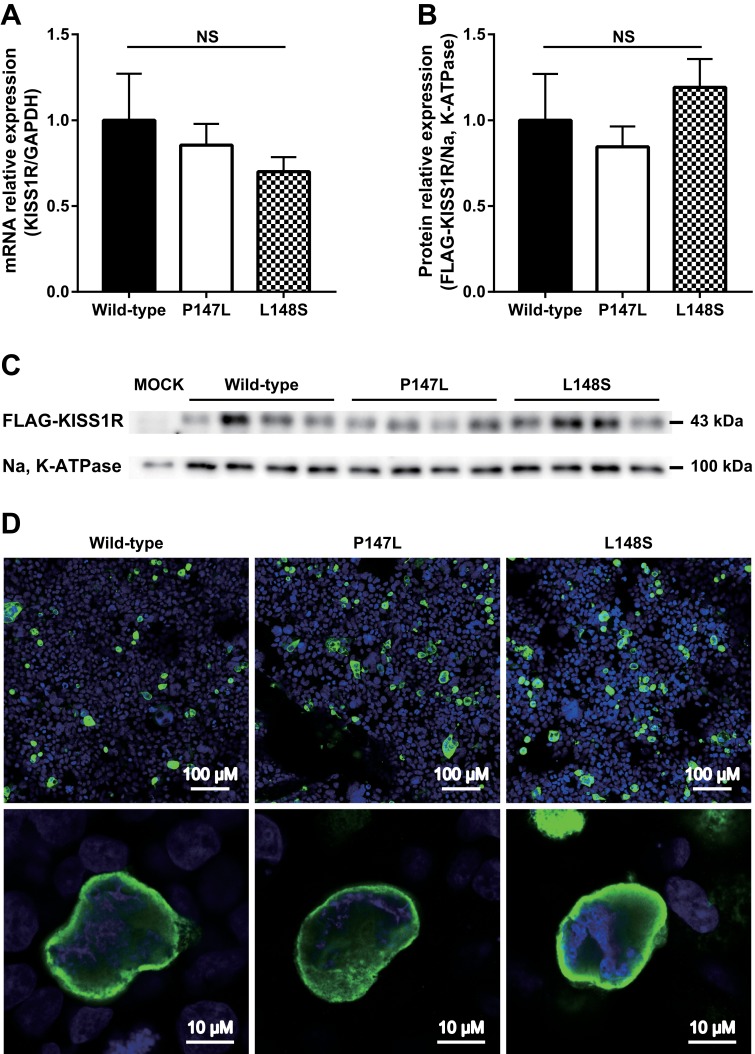
Results from quantitative real-time PCR analysis showed no significant differences in relative mRNA expression among the KISS1R wild-type, P147L, or L148S variants and validated similar levels of transfection efficiency by the KISS1R-expression vectors [Fig. 3(A)]. To observe the effect of the P147L mutation on transport of the receptor to the cell surface membrane, we constructed HEK293 cells transiently expressing KISS1R wild-type or mutant variants with N-terminal FLAG tags. Western blot analysis revealed no significant differences in relative FLAG-KISS1R expression in the cell membrane fraction among the wild-type, P147L, and L148S variants [Fig. 3(B) and 3(C)]. We then performed immunocytochemistry to analyze the effect of the P147L mutation on KISS1R translation and subcellular localization. Immunocytochemistry results showed no differences in subcellular localization patterns of FLAG-KISS1R among the wild-type, P147L, and L148S variants [Fig. 3(D)]. These findings indicated that neither the P147L nor the L148S mutation affected KISS1R transcription, translation efficiency, or its transport to the cell surface membrane.
Figure 3.
The KISS1R P147L mutation does not affect the expression or subcellular localization of KISS1R. (A) Quantitative real-time PCR analysis of KISS1R mRNA expression in HEK293 cells transfected with KISS1R wild-type, P147L, or L148S variants (n = 4 for each genotype). (B, C) Western blot analysis of FLAG-tagged-KISS1R protein in the cell membrane fraction of HEK293 cells expressing FLAG-tagged-KISS1R wild-type, P147L, L148S, or transfected with empty vector (Mock) (n = 4 for each genotype). (D) Reproducible subcellular localization of FLAG-tagged-KISS1R in HEK293 cells expressing FLAG-tagged-KISS1R wild-type, P147L, or L148S variants. FLAG is green; nuclei are blue. NS, not significant.
D. The P147L Mutation Causes a Significant Loss of Ligand-Binding Affinity
We performed a receptor-ligand binding assay to analyze the effect of the P147L mutation on the ligand-binding affinity of the receptor. The assay revealed that the ligand-binding affinities of the KISS1R P147L and L148S mutants were significantly lower than that observed in the wild-type variant (Fig. 4). Specifically, the P147L mutant showed no significant binding at from 0.1 to 1.6 nM 125I-KP-10. These results indicated that the reduction in ligand-binding affinity was a consequence of the P147L and L148S mutations, with increased severity of this impairment detected in the P147L mutation.
Figure 4.
The KISS1R P147L mutation causes a significant loss of ligand-binding affinity. Radioactivity of bound 125I-KP-10 in the cell membrane fraction of HEK293 cells expressing KISS1R wild-type (filled circle), P147L (filled triangle), or L148S (open triangle) variants (n = 4 for each genotype).
E. The P147L Mutation Dramatically Changes the Conformation of the Ligand-Binding Pocket
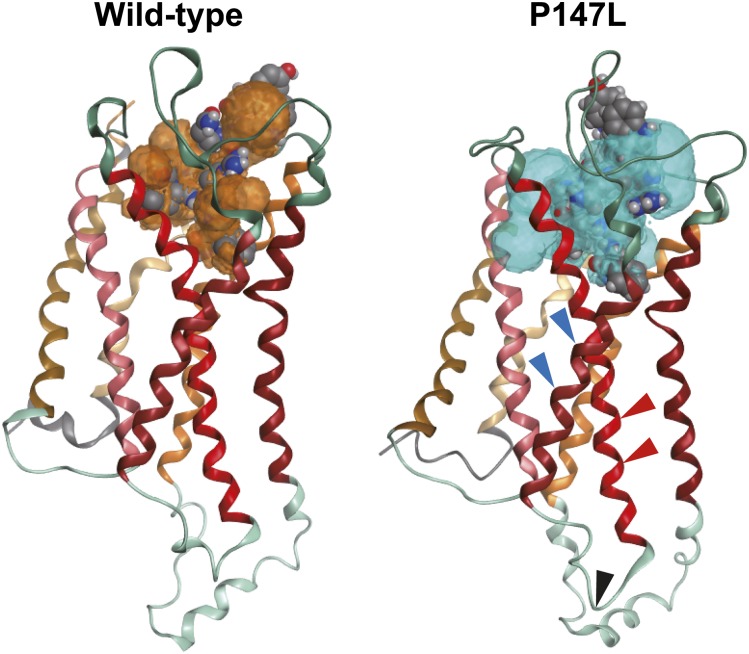
To analyze the effect of the P147L mutation on receptor conformation and receptor-ligand interactions, we performed in silico analysis of protein-structure modeling and molecular dynamics simulations using Molecular Operating Environment (Chemical Computing Group). Molecular dynamics simulations revealed that the binding free energies of the KP-10-KISS1R complexes were higher in those of the P147L mutant compared with those observed in the wild-type variant (Table 2). Consequently, the equilibrium constants (Ka = [ligand-receptor]/[ligand][receptor]) were lower in the P147L mutant, consistent with results from the receptor-ligand binding assay. Additionally, the contact surface areas of the KP-10-KISS1R complexes decreased in the P147L mutant. Furthermore, comparison of the lowest energy conformations revealed that the unoccupied cavity volume of the ligand-binding pocket was greater in the KISS1R P147L mutant as compared with that observed in the wild-type variant (Fig. 5). These results suggested that the loss of ligand-binding affinity observed in the P147L mutant was caused by the expansion of the ligand-binding pocket.
Table 2.
Molecular Dynamics Simulations of the KP-10-KISS1R Complexes
| Variable | Wild Type | P147L Mutant |
|---|---|---|
| Binding free energy, kcal/mol | ||
| GBVI/WSA dG | −14.37 ± 0.01 | −12.81 ± 0.01 |
| London dG | −22.29 ± 0.01 | −18.53 ± 0.02 |
| Ka calculated from binding free energy | ||
| GBVI/WSA dG | 3.465 × 1010 | 2.486 × 109 |
| London dG | 2.231 × 1016 | 3.898 × 1013 |
| Contact surface area, Ǻ2 | 1275.2 ± 0.6 | 1189.9 ± 0.7 |
| Unoccupied cavity volume of the ligand-binding pocket, Ǻ3 | 1069 | 1644 |
GBVI/WSA dG and London dG are scoring functions in the Molecular Operating Environment (Chemical Computing Group) to estimate the binding free energy [13].
Figure 5.
The P147L mutation dramatically changes the conformation of the ligand-binding pocket. Molecular dynamics simulations showing comparisons of unoccupied cavity volume in the ligand-binding pocket of wild-type KISS1R and the P147L mutant at the lowest energy conformations. Black triangles indicate the position of the P147L mutation, red triangles indicate transmembrane domain 3, and blue triangles indicate transmembrane domain 4 of KISS1R. Gray bubbles represent KP-10, and orange (wild-type) and light blue (P147L) bubbles represent the ligand-binding pockets. Red and blue parts of KP-10 represent oxygen and nitrogen atoms, respectively.
3. Discussion
In this study, we identified a KISS1R mutation in a Japanese patient with CHH. This P147L mutation appears to be rare, because it was not found in searches of the Exome Aggregation Consortium database [14], which covers exome sequences of 60,706 European, African, Asian, and American individuals, and was also not found in searches of the Integrative Japanese Genome Variation database [15], which covers whole-genome sequences of 2049 Japanese individuals. Functional analyses revealed that the P147L variant constitutes an almost complete loss-of-function mutation, resulting in a severe phenotype corresponding to lack of pubertal development. Moreover, we showed that the mutation in the second intracellular loop of the KISS1R caused the loss of ligand-binding affinity, due to the expanded ligand-binding pocket. These results emphasized that the KISS1/KISS1R system plays an important role in GnRH neurons, which are responsible for pubertal onset and reproductive functions.
The phenotype of the patient in this study included lack of pubertal development due to severe hypogonadism (pubic hair: Tanner stage I; LH level <0.1 IU/L; and FSH level of 0.8 IU/L). In contrast, the phenotype of the male patients who were homozygous for the L148S mutation constituted delayed pubertal development (pubic hair: Tanner stage II–III; LH level, 0.5 to 4.1 IU/L; and FSH level, 0.7 to 3.7 IU/L) [16, 17]. Fertility was restored in these four patients by gonadotropin administration. The phenotype of the patients appeared more severe in those harboring the P147L mutation as compared with the L148S mutation, in accordance with the results of our functional analyses. The stretched penile length of the patient was −2.2 standard deviations of the mean penile length of Japanese full-term newborn infants [18]. It could thus be deduced that loss-of-function mutations of KISS1R cause prenatal testosterone deficiency [19]. Although the brothers of the patient were heterozygous for the P147L mutation, their associated phenotype was normal. This disease is considered to be inherited in an autosomal recessive manner. The roles for the KISS1/KISS1R system in glucose-stimulated insulin secretion in pancreatic β cells remain a matter of controversy [20, 21]. In our patient, a 75-g oral glucose-tolerance test indicated favorable insulin secretion based on a homeostasis-model assessment of β-cell function at 72% and an insulinogenic index of 1.0 (Table 1). These values are almost equivalent to the averages observed in normal Japanese glucose-tolerant subjects [22]. Therefore, the KISS1/KISS1R system was not considered crucial for glucose-stimulated insulin secretion.
KISS1R is classified as a rhodopsin-like, class A GPCR, and the Pro147 and Leu148 residues located in the second intracellular loop are highly conserved in class A GPCRs. These two amino acids form a hydrophobic interface adjacent to the GTPase region of Gαq. A previous study showed that the L148S mutation inhibits ligand-induced catalytic activation of Gαq, whereas it does not affect KISS1R expression, localization, or its ligand-binding affinity [12]. We thus surmised that the P147L mutation also impaired G-protein coupling with the receptor. However, our study demonstrated that the loss of ligand-binding affinity was the most fundamental cause of impaired function in the P147L mutation, which was distant from the ligand-binding site. There are various mechanisms by which KISS1R mutations cause impaired function, including impaired transcription [6] and intracellular transport to the cell membrane [1, 23, 24]. As in the case of the KISS1R A287E mutation, which impaired transport to the cell membrane [1] despite its location in the third extracellular loop, which is not a typical region associated with receptor misfolding, the relationship between the site of the mutation in the GPCR with the molecular mechanism of functional impairment is not straightforward. In general, the original amino acid at the site of the P147L mutation, proline, acts as a structural hinge or swivel that significantly affects protein conformation [25]. Mutation of proline residues in the transmembrane domain causes various impairments to receptor function, including loss of ligand-binding affinity [26–31]. Moreover, mutations of several amino acids in the second intracellular loop of the class A GPCR melanocortin-3 receptor caused not only impaired G-protein coupling with the receptor but also a loss of or decrease in ligand-binding affinity [32]. Computer analyses in the current study suggested that the P147L mutation increases binding free energy owing to decreased contact surface area in the KP-10-KISS1R complex along with the significantly expanded ligand-binding pocket. A previous study showed the size of the contact surface area associated with the protein-protein complex was positively correlated with binding affinity [33]. Taken together, these reports support our data that the P147L mutation causes the loss of ligand-binding affinity, owing to an expanded ligand-binding pocket.
In conclusion, we identified a loss-of-function mutation in the KISS1R associated with CHH. To our knowledge, this is the first KISS1R mutation found in a Japanese patient, and the severe phenotypic features associated with hypogonadotropic hypogonadism demonstrated the deleterious effects of this mutation. This study confirmed the role of the KISS1/KISS1R system in CHH pathogenesis and contributes to a better knowledge of the structure-function relationship of GPCR. However, little is known about hypogonadotropic hypogonadism-associated symptoms in patients harboring KISS1R mutations. Further identification of KISS1 and KISS1R mutations are needed to define the precise genotype-phenotype relationship.
Acknowledgments
We thank Dr. Nagako Horikawa (Department of Dermatology, Faculty of Medicine, University of Miyazaki, Japan) for referring the patient and performing a laboratory test; Ms. Miki Oshikawa for technical support involving DNA-sequencing analysis and site-directed mutagenesis; and Dr. Yoshiro Kimura (Science and Technology System Division, Ryoka Systems, Japan) for conducting protein modeling and molecular dynamics simulations.
Author Contributions: K.S. and M.N. designed the experiments. T.Y. identified the P147L mutation in the proband. M.Y. and M.M. performed intracellular calcium measurements and cellular dielectric spectroscopy. K.S. performed the other experiments and analyzed data. A.M. and H.S. supervised the experimental procedures. K.S., M.Y., H.Y., and M.N. wrote the paper, and all authors revised the manuscript.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CHH
- congenital hypogonadotropic hypogonadism
- FSH
- follicle-stimulating hormone
- GnRH
- gonadotropin-releasing hormone
- GPCR
- G-protein–coupled receptor
- HEK
- human embryonic kidney
- KISS1R
- kisspeptin receptor
- KP
- kisspeptin
- LH
- luteinizing hormone
- RRID
- Research Resource Identifier.
References and Notes
- 1.Francou B, Paul C, Amazit L, Cartes A, Bouvattier C, Albarel F, Maiter D, Chanson P, Trabado S, Brailly-Tabard S, Brue T, Guiochon-Mantel A, Young J, Bouligand J. Prevalence of KISS1 Receptor mutations in a series of 603 patients with normosmic congenital hypogonadotrophic hypogonadism and characterization of novel mutations: a single-centre study. Hum Reprod. 2016;31(6):1363–1374. [DOI] [PubMed] [Google Scholar]
- 2.Valdes-Socin H, Rubio Almanza M, Tomé Fernández-Ladreda M, Debray FG, Bours V, Beckers A. Reproduction, smell, and neurodevelopmental disorders: genetic defects in different hypogonadotropic hypogonadal syndromes. Front Endocrinol (Lausanne). 2014;5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skorupskaite K, George JT, Anderson RA. The kisspeptin-GnRH pathway in human reproductive health and disease. Hum Reprod Update. 2014;20(4):485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM, Chambers JK, Murdock P, Steplewski K, Shabon U, Miller JE, Middleton SE, Darker JG, Larminie CG, Wilson S, Bergsma DJ, Emson P, Faull R, Philpott KL, Harrison DC. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276(31):28969–28975. [DOI] [PubMed] [Google Scholar]
- 5.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100(19):10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349(17):1614–1627. [DOI] [PubMed] [Google Scholar]
- 7.Demirbilek H, Ozbek MN, Demir K, Kotan LD, Cesur Y, Dogan M, Temiz F, Mengen E, Gurbuz F, Yuksel B, Topaloglu AK. Normosmic idiopathic hypogonadotropic hypogonadism due to a novel homozygous nonsense c.C969A (p.Y323X) mutation in the KISS1R gene in three unrelated families. Clin Endocrinol (Oxf). 2015;82(3):429–438. [DOI] [PubMed] [Google Scholar]
- 8.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402(6762):656–660. [DOI] [PubMed] [Google Scholar]
- 9.Verdonk E, Johnson K, McGuinness R, Leung G, Chen YW, Tang HR, Michelotti JM, Liu VF. Cellular dielectric spectroscopy: a label-free comprehensive platform for functional evaluation of endogenous receptors. Assay Drug Dev Technol. 2006;4(5):609–619. [DOI] [PubMed] [Google Scholar]
- 10.Soulié S, Møller JV, Falson P, le Maire M. Urea reduces the aggregation of membrane proteins on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1996;236(2):363–364. [DOI] [PubMed] [Google Scholar]
- 11.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Imamoglu S, Akalin NS, Yuksel B, O’Rahilly S, Semple RK. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41(3):354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wacker JL, Feller DB, Tang XB, Defino MC, Namkung Y, Lyssand JS, Mhyre AJ, Tan X, Jensen JB, Hague C. Disease-causing mutation in GPR54 reveals the importance of the second intracellular loop for class A G-protein-coupled receptor function. J Biol Chem. 2008;283(45):31068–31078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbeil CR, Williams CI, Labute P. Variability in docking success rates due to dataset preparation. J Comput Aided Mol Des. 2012;26:775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukunaga H, Yokoya A, Taki Y, Prise KM. Radiobiological implications of Fukushima nuclear accident for personalized medical approach. Tohoku J Exp Med. 2017;242(1):77–81. [DOI] [PubMed] [Google Scholar]
- 16.Bo-Abbas Y, Acierno JS Jr, Shagoury JK, Crowley WF Jr, Seminara SB. Autosomal recessive idiopathic hypogonadotropic hypogonadism: genetic analysis excludes mutations in the gonadotropin-releasing hormone (GnRH) and GnRH receptor genes. J Clin Endocrinol Metab. 2003;88(6):2730–2737. [DOI] [PubMed] [Google Scholar]
- 17.Pallais JC, Bo-Abbas Y, Pitteloud N, Crowley WF Jr, Seminara SB. Neuroendocrine, gonadal, placental, and obstetric phenotypes in patients with IHH and mutations in the G-protein coupled receptor, GPR54. Mol Cell Endocrinol. 2006;254-255:70–77. [DOI] [PubMed] [Google Scholar]
- 18.Matsuo N, Ishii T, Takayama JI, Miwa M, Hasegawa T. Reference standard of penile size and prevalence of buried penis in Japanese newborn male infants. Endocr J. 2014;61(9):849–853. [DOI] [PubMed] [Google Scholar]
- 19.Wiygul J, Palmer LS. Micropenis. Sci World J. 2011;11:1462–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song WJ, Mondal P, Wolfe A, Alonso LC, Stamateris R, Ong BW, Lim OC, Yang KS, Radovick S, Novaira HJ, Farber EA, Farber CR, Turner SD, Hussain MA. Glucagon regulates hepatic kisspeptin to impair insulin secretion. Cell Metab. 2014;19(4):667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowe JE, Foot VL, Amiel SA, Huang GC, Lamb MW, Lakey J, Jones PM, Persaud SJ. GPR54 peptide agonists stimulate insulin secretion from murine, porcine and human islets. Islets. 2012;4(1):20–23. [DOI] [PubMed] [Google Scholar]
- 22.Iwao T, Sakai K, Ando E. Relative contribution of insulin secretion and sensitivity at different stages of glucose tolerance: non-obese versus obese Japanese subjects. Intern Med. 2014;53(5):383–390. [DOI] [PubMed] [Google Scholar]
- 23.Nimri R, Lebenthal Y, Lazar L, Chevrier L, Phillip M, Bar M, Hernandez-Mora E, de Roux N, Gat-Yablonski G. A novel loss-of-function mutation in GPR54/KISS1R leads to hypogonadotropic hypogonadism in a highly consanguineous family. J Clin Endocrinol Metab. 2011;96(3):E536–E545. [DOI] [PubMed] [Google Scholar]
- 24.Chevrier L, de Brevern A, Hernandez E, Leprince J, Vaudry H, Guedj AM, de Roux N. PRR repeats in the intracellular domain of KISS1R are important for its export to cell membrane. Mol Endocrinol. 2013;27(6):1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sansom MS, Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol Sci. 2000;21(11):445–451. [DOI] [PubMed] [Google Scholar]
- 26.Conner AC, Hay DL, Simms J, Howitt SG, Schindler M, Smith DM, Wheatley M, Poyner DR. A key role for transmembrane prolines in calcitonin receptor-like receptor agonist binding and signalling: implications for family B G-protein-coupled receptors. Mol Pharmacol. 2005;67(1):20–31. [PubMed] [Google Scholar]
- 27.Hong S, Ryu KS, Oh MS, Ji I, Ji TH. Roles of transmembrane prolines and proline-induced kinks of the lutropin/choriogonadotropin receptor. J Biol Chem. 1997;272(7):4166–4171. [DOI] [PubMed] [Google Scholar]
- 28.Kolakowski LF Jr, Lu B, Gerard C, Gerard NP. Probing the “message:address” sites for chemoattractant binding to the C5a receptor. Mutagenesis of hydrophilic and proline residues within the transmembrane segments. J Biol Chem. 1995;270(30):18077–18082. [DOI] [PubMed] [Google Scholar]
- 29.Mazna P, Grycova L, Balik A, Zemkova H, Friedlova E, Obsilova V, Obsil T, Teisinger J. The role of proline residues in the structure and function of human MT2 melatonin receptor. J Pineal Res. 2008;45(4):361–372. [DOI] [PubMed] [Google Scholar]
- 30.Stitham J, Martin KA, Hwa J. The critical role of transmembrane prolines in human prostacyclin receptor activation. Mol Pharmacol. 2002;61(5):1202–1210. [DOI] [PubMed] [Google Scholar]
- 31.Wess J, Nanavati S, Vogel Z, Maggio R. Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. EMBO J. 1993;12(1):331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, Tao YX. Functions of the DRY motif and intracellular loop 2 of human melanocortin 3 receptor. J Mol Endocrinol. 2014;53(3):319–330. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Sawyer N, Regan L. Protein-protein interactions: general trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22(4):510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]