Abstract
Achillea millefolium and Achillea ptarmica are both plants belonging to the Asteracea family and are traditionally used for their medicinal properties. It has already been shown that some N-alkylamides (NAAs) are responsible for these pharmacological actions. Therefore, in the present study, the NAA content of the two plants was analytically characterised. Different extracts were prepared from the roots, the leaves, the stems and the flowers. The structures of NAAs have been assigned in ethanolic extracts of Achillea millefolium and Achillea ptarmica using high performance liquid chromatography – electrospray ionisation – mass spectrometry (HPLC–ESI–MS) and gas chromatography – electron impact – mass spectrometry (GC–EI–MS). Using both analytical techniques, the structures of 14 and 15 NAAs have been assigned in Achillea ptarmica and Achillea millefolium, respectively. Structures of two new NAAs, previously never observed in Achillea ptarmica, were assigned: deca-2E,6Z,8E-trienoic acid 2-methylbutylamide (homospilanthol) or a related isomeric compound and deca-2E,4E-dienoic acid N-methyl isobutylamide. The structure of homospilanthol or a related isomeric compound was also assigned in Achillea millefolium for the first time.
Keywords: N-alkylamides, Achillea millefolium, Achillea ptarmica, HPLC–ESI–MS, GC–EI–MS
1. Introduction
The genus Achillea, mainly distributed in the Northern Hemisphere, consists of more than 120 species worldwide. Achillea species have been used in traditional folk medicine for many years to treat various diseases and are especially known to cure slow-healing wounds, which explains the name of the genus Achillea [1], [2], [3]. Two species of the genus Achillea (millefolium and ptarmica) will be discussed in detail, i.e., Achillea millefolium L. (A. millefolium) and Achillea ptarmica L. (A. ptarmica), both belonging to the Anthemideae tribe and Asteraceae plant family. A. millefolium and A. ptarmica plants are both ethnopharmacologically used to treat stomach disorders [4], [5].
A. millefolium, also known as yarrow, consists of several closely related species, named a species complex or aggregate. A diversity of pharmacological properties is ascribed to this plant, such as spasmolytic, anti-inflammatory, analgesic, haemostatic, antidiabetic, cholagogue, antitumor, antioxidant, antifungal, antiseptic and liver protective effects. Furthermore, tea from A. millefolium is used to treat diseases of the gastrointestinal tract, like dyspepsia, flatulence, abdominal pain, diarrhea, stomachache and digestive complaints. In a double-blind randomized clinical trial, it has been shown that tea prepared from powder of the flowers of A. millefolium relieved the severity of pain in primary dysmenorrhea. A. millefolium can be consumed as essential oil, infusion or alcohol extract, decoction, hydroalcoholic, methanolic or aqueous extract [1], [2], [6], [7], [8], [9], [10], [11], [12].
An aqueous extract of the aerial parts of the plant protected the gastric mucosa in Wistar rats against ethanol- and indomethacin- induced gastric lesions. It also healed acetic acid induced chronic gastric lesions [6]. Potrich et al. [13] reported that the antioxidant properties are at least partly responsible for the gastroprotective effects of the extract. Furthermore, an A. millefolium extract of the aerial parts (hexane: ether:methanol (1:1:1, v/v/v)) showed antimicrobial activity against Staphylococcus aureus, Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa and Salmonella enteritidis [9]. Safety studies in Wistar rats showed that there were no signs of relevant toxicity after daily treatment with the extract in a concentration of 0.3–1.2 g/kg (p.o.) for 28 or 90 days [6].
A. millefolium is used in cosmetics as it has been proven in vivo that a 2% A. millefolium extract has a rejuvenating effect on the appearance and feeling of the skin surface [14]. The extract is a biological ingredient in 65 cosmetic product formulations and creams used to accelerate the wound healing rate consisting of 2%, 5% or 10% A. millefolium extract [15], [16]. Furthermore, there exist European national pharmacopoeia monographs about A. millefolium, such as the Hungarian, German, Austrian, Czech, French, and Romanian pharmacopoeias, extra pharmacopoeia Martindale, British herbal pharmacopoeia and the Polish herbal compendium [17].
Another plant belonging to the Asteracea family is Achillea ptarmica L. (sneezewort yarrow). Althaus et al. [18] reported that a dichloromethane extract of flowering aerial parts of A. ptarmica was found to possess antiprotozoal activity in vitro. The extract showed anti-Trypanosoma brucei rhodesiense activity (IC50 of 0.67 µg/mL) as well as anti-Plasmodium falciparum activity (IC50 of 6.6 µg/mL) [18].
Of special pharmacological interest, the genus Achillea produces several N-alkylamides (NAAs), which are secondary metabolites in plants and because of their wide structural diversity, they are classified according to a structural classification system, indicated as FxMy. The F and M stand for the fatty acid chain and the amine part of the NAA, respectively. X and y represent numbers (1−13), indicative for the structure of the chains. Both chains are linked to each other through an amide bond [19]. Achillea NAAs consist of the more widespread isobutylamides and phenethyl amides, and especially saturated and unsaturated 5- and 6-ring alkylamides (piperidides, pyrrolidides, piperideides, and pyrrolideides). C10-, C11- and C14-olefinic and acetylenic alkylamides are characteristic for the genus Achillea and are mainly found in the roots of the plant [2], [3], [20], [21], [22].
Typical NAAs present in A. millefolium are NAAs consisting of a C10 fatty acid chain linked to a piperideide function. 2E,4E,6Z-decatrienoic acid piperideide is the main compound, while the corresponding all trans-isomer only occurs in small amounts. N-isobutyl-2,4-decadiene amide (pellitorine), 2,4-decadienoic acid piperidide, 2,4-decadienoic acid piperideide and 2,4,6-decatrienoic acid piperideide are NAAs identified using gas chromatography–mass spectrometry (GC–MS) in the roots of A. distans Willd. subsp. distans; however, the correct stereoisomer could not be determined. A. distans Willd. subsp. distans belongs to the A. millefolium aggregate [2]. Greger and Hofer [23] identified 17 NAAs in A. millefolium, while Greger and Werner [24] identified two additional NAAs, namely undeca-2E,4E-diene-8,10-diynoic acid piperidide and tetradeca-2E,4E,12Z-triene-8,10-diynoic acid isobutylamide. Besides NAAs, other compounds present in A. millefolium L. are volatile oils, sesquiterpene lactones, flavonoids, amino acids, polyacetylenes, polysaccharides, phenolic acids, fatty acids, vitamins, alkanes, alkaloids and bases, saponins, sterols, sugars, coumarins, and tannins [17].
The main components of A. ptarmica are flavonoids, some essential oils and NAAs (carboxamides of olefinic and polyynic carboxylic acids with various amines) [18]. Kuropka et al. [25] and Althaus et al. [18] identified five and six NAAs in A. ptarmica, respectively.
In this study, a thorough NAA profiling of ethanolic extracts from the roots, flowers, leaves and stems of A. millefolium and A. ptarmica was performed using high performance liquid chromatography–electrospray ionisation – mass spectrometry (HPLC–ESI–MS) and gas chromatography – electron impact – mass spectrometry (GC–EI–MS). This study led to the structural assignment of NAAs, previously never reported in either plant.
2. Materials and methods
2.1. Chemicals and reagents
Ultrapure water of 18.2 MΩ.cm quality was produced by an Arium 611 purification system (Sartorius, Göttingen, Germany). Acetic acid was purchased from Sigma-Aldrich (Diegem, Belgium), while denaturated ethanol (95% ethanol denaturated with 5% diethylether) was obtained from Chem-Lab (Zedelgem, Belgium). Absolute ethanol and HPLC gradient grade acetonitrile (ACN) were purchased from Fisher Scientific (Erembodegem, Belgium). Pellitorine was purchased from Adipogen Life Sciences (99.8% purity determined by HPLC).
2.2. Plant material and extraction
A. millefolium (type: Wesersandstein) and A. ptarmica (type: The Pearl) were bought at the tree nursery ‘De Bock’ in Belgium (Oudenaarde). Plants were harvested in September 2013. The fresh stems, the flowers, the roots and the leaves were collected from the plants and washed with ultrapure water. The plant parts were dried at room temperature for approximately six weeks. The dried plant parts were cut into smaller pieces (parts of approximately 1 cm) with a scissor. Extraction was performed at a ratio of plant part: solvent (m/v), ranging from 1/12 to 1/64 for A. millefolium and from 1/12 to 1/34 for A. ptarmica. As extraction solvent, denaturated ethanol: H2O (90:10, v/v) was used. After maceration in darkness for approximately 48 h at room temperature (20–27 °C, 230 rpm), the plant parts were removed by filtration (Whatman, UK). Thereafter, the extraction solvent was removed using a rotavapor (Heidolph and Büchi), and protected from light. The extraction yield of A. millefolium and A. ptarmica was between 3%–8% (m/m) and 3%–11% (m/m), respectively. The extract was kept in the dark at 4 °C until analysis.
2.3. HPLC-UV/ESI-MS analysis
The HPLC-MS analyses were done on an HPLC system which consisted of a Spectra System SN4000 interface, a Spectra System SCM1000 degasser, a Spectra System P1000XR pump, a Spectra System AS3000 autosampler, and a Finnigan LCQ Classic ion trap mass spectrometer in positive ion mode (all Thermo, San José, CA, USA) equipped with Xcalibur 2.0 software (Thermo) for data acquisition. The plant extracts (roots, stems, leaves, flowers) were dissolved in ACN:H2O (50:50, v/v) (final extract ranging between 5 and 155 mg/mL) and filtered over a 0.45 µm PVDF membrane HPLC filter (Whatman) before analysis. 10 µL of this solution was injected on a Grace Prevail C18 column (250mm × 4.6 mm, 5 µm) using a Waters HPLC equipped with a Waters 2487 Dual Absorbance detector set at 260 nm. A linear gradient with a flow rate of 1.0 mL/min was applied as follows: t=0 min: A: B (80:20, v/v), t=0–150 min: A: B (10:90, v/v), t=150–151 min: A: B (80:20, v/v), and t=151–166 min: A: B (80:20, v/v) (with A =1% acetic acid in H2O and B = ACN). The needle was rinsed with methanol. ESI was conducted with a capillary voltage of 3 kV. Nitrogen was used as sheath and auxiliary gas. The temperature of the heated capillary was kept at 275 °C. MS-MS spectra were obtained by collision induced dissociation (CID) of the parent m/z, with the relative collision energy set to 35%. Structural assignment was based on the parent m/z values and fragmentation ions. Assuming all NAA peaks have a response factor of 1 relative to pellitorine, the total amount of NAAs was estimated as 0.6% (m/m) in A. ptarmica and 0.2% (m/m) in A. millefolium.
2.4. GC-EI-MS analysis
The GC–MS analyses were performed on an Agilent 6890 instrument consisting of an automatic injector 7683 and coupled to a Mass Selective Detector 5973 (Agilent). The mass detecter was operated in EI mode (70 eV). The output signal was recorded and processed using Instrument Analysis MSD Chemstation (Agilent). The root plant extracts were dissolved in absolute ethanol (final extract ranging between 31 and 35 mg/mL) and samples were injected by the instrument’s autosampler with an injection volume of 1 µL. Ethanol was used to rinse the syringe between injections (3 x wash post injection). An HP-5MS column (30 m × 0.25 mm, 0.25 µm) (Agilent, Belgium) was used for separation. The column oven was programmed with an initial oven temperature of 100 °C, and increased to 180 °C at a rate of 10 °C/min, ramped to 200 °C at a rate of 1 °C/min, followed by increasing the temperature to 320 °C at a rate of 10 °C/min and held at 320 °C for 1 min. The total run time was 41 min. The split ratio was set at 10:1 (v/v). The injector and MS transfer line temperature were kept at 210 °C and 250 °C, respectively. Helium (Air Products and Chemicals, Allentown, PA, USA) was used as a carrier gas with a head pressure of 71.3 kPa resulting in an average velocity of 37 cm/s. The ion source and quadrupole temperature were 150 °C and 230 °C, respectively. The MS detection scan range was between m/z 40 and 550.
3. Results and discussion
The structures of N-alkylamides in the ethanolic A. millefolium and A. ptarmica extracts were assigned based on their MS spectra. NAAs have characteristic CID fragmentation patterns, related to the amide part of the compound and typical fragment losses are presented in Table 1 [26], [27], [28]. The typical fragment losses including the loss of the alkyl group directly attached to the amine; the loss of the entire amine functional group, resulting in an acylium ion; the loss of the amide portion of the molecule and saturation of one of the double bonds on the alkyl chain; and the loss of the amide portion, resulted in the loss of characteristic m/z values. From the corresponding alkyl chain of the two last losses and from the m/z value of the acylium ion, the number of carbons present in the alkyl chain can be determined [29]. Furthermore, also with electron impact, characteristic product ions of spilanthol, as a prototypical NAA, are formed [30], [31], [32].
Table 1.
Characteristic fragment ions of NAAs after CID fragmentation.
| Amine alkyl group | Loss of alkyl group directly attached to the amine | Loss of entire amine functional group | Loss of the amide portion and saturation of one of the double bonds on the alkyl chain | Loss of the amide portion | |
|---|---|---|---|---|---|
| Isobutylamide | Ion | [(M+H)-C4H8]+ | [(M+H)-C4H11N]+ | [(M+H)-C5H9NO]+ | [(M+H)-C5H11NO]+ |
| -m/z | 56 | 73 | 99 | 101 | |
| Phenylethylamide | Ion | [(M+H)-C8H8]+ | [(M+H)-C8H11N]+ | [(M+H)-C9H9NO]+ | [(M+H)-C9H11NO]+ |
| -m/z | 104 | 121 | 147 | 149 | |
| 2-methyl isobutylamide | Ion | [(M+H)-C5H10]+ | [(M+H)-C5H13N]+ | [(M+H)-C6H11NO]+ | [(M+H)-C6H13NO]+ |
| -m/z | 70 | 87 | 113 | 115 | |
| N-methyl isobutylamide | Ion | [(M+H)-C4H8]+ | [(M+H)-C5H13N]+ | [(M+H)-C6H11NO]+ | [(M+H)-C6H13NO]+ |
| -m/z | 56 | 87 | 113 | 115 | |
| 4-hydroxy phenylethylamide | Ion | [(M+H)-C8H8O]+ | [(M+H)-C8H11NO]+ | [(M+H)-C9H9NO2]+ | [(M+H)-C9H11NO2]+ |
| -m/z | 120 | 137 | 163 | 165 | |
| 4-methoxy phenylethylamide | Ion | [(M+H)-C9H10O]+ | [(M+H)-C9H13NO]+ | [(M+H)-C10H11NO2]+ | [(M+H)-C10H13NO2]+ |
| -m/z | 134 | 151 | 177 | 179 | |
3.1. A. ptarmica
3.1.1. N-alkylamide profiling using HPLC–ESI–MS
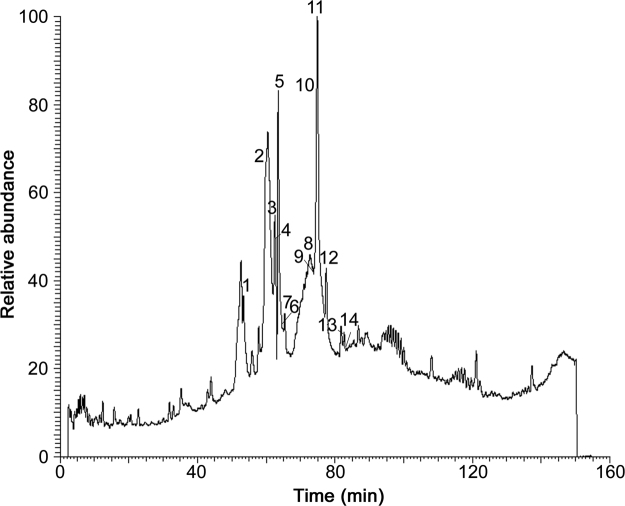
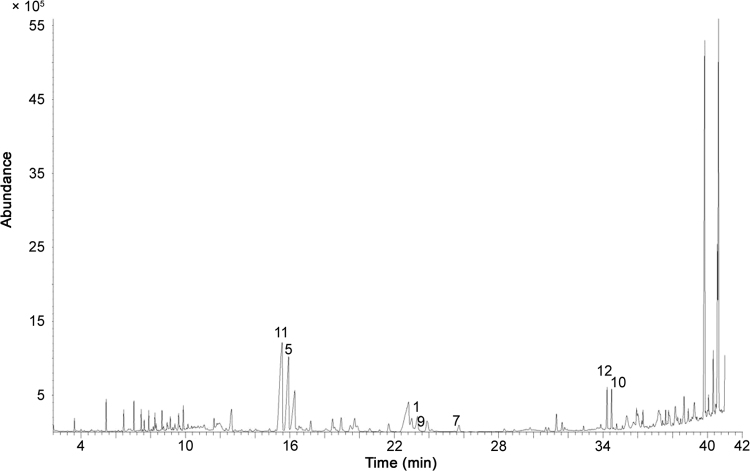
The total ion chromatogram (TIC) of the root extract of A. ptarmica is given in Fig. 1. Peak labels correspond to NAA designations.
Fig. 1.
TIC of Achillea ptarmica obtained using HPLC–ESI–MS.
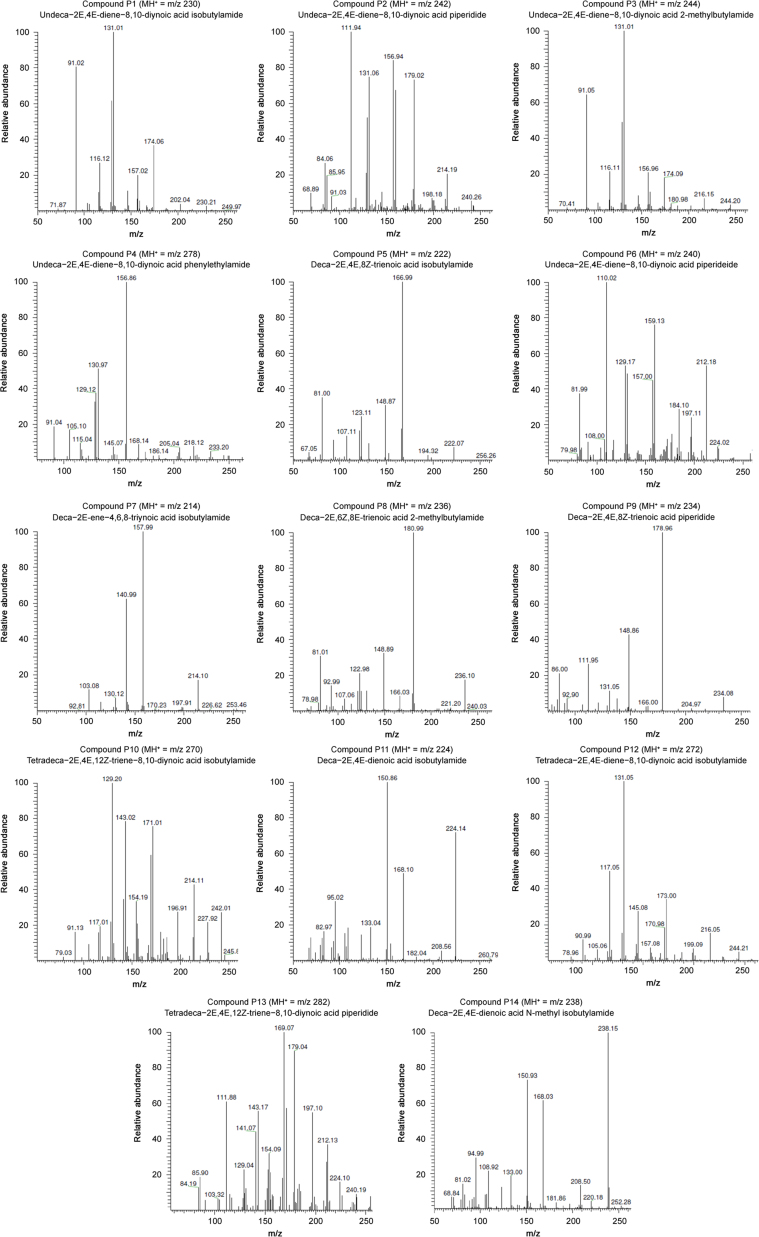
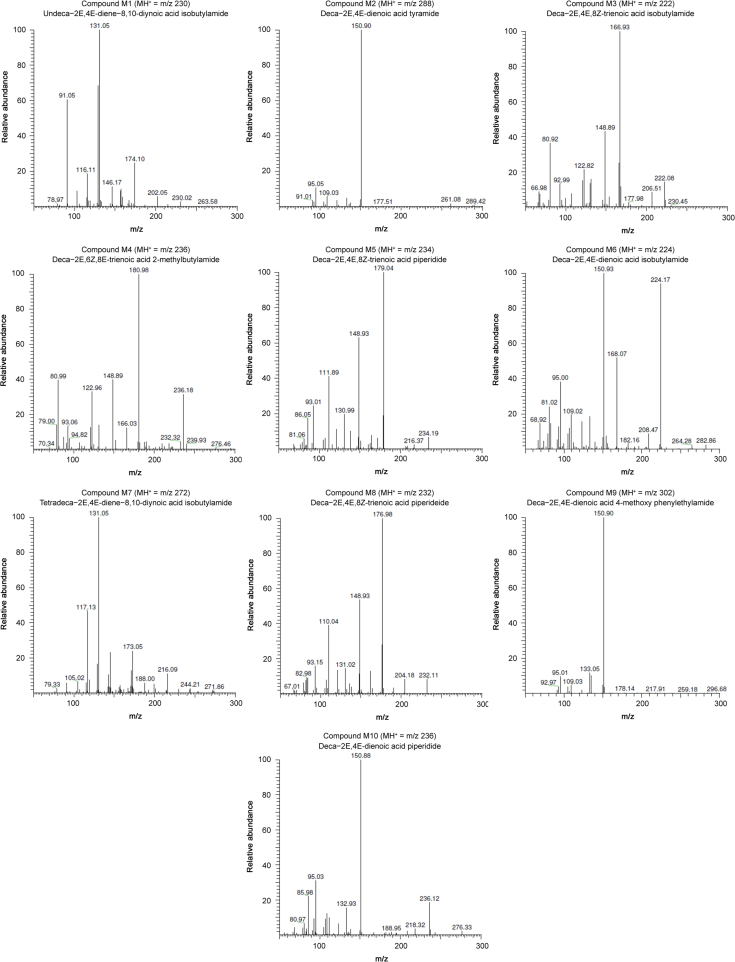
The major ions observed in the MS1 spectra correspond to the protonated forms of NAAs. The MS2 spectra are shown in Fig. 2, while an overview of all the NAAs assigned in A. ptarmica is given in Table 2 with their corresponding retention time (Rt), structure, chemical name, molecular weight (MW, average mass) and classification. Structures of fourteen NAAs were assigned with different types of amides in the A. ptarmica extract: 6 NAAs having an isobutylamide function (compounds P1, P5, P7, P10, P11 and P12), 3 NAAs with a piperidide function (saturated 6-ring C5H10N, compounds P2, P9, and P13), 1 NAA with a piperideide function (unsaturated 6-ring C5H8N, compound P6), 2 NAAs with a 2-methylbutylamide function (compounds P3 and P8), 1 NAA with a phenylethylamide function (compound P4) and 1 NAA having a N-methyl isobutylamide function (compound P14). Furthermore, the fatty acid chain of NAAs varies in length (C10, C11, and C14) and consists of many sites of unsaturated bonds (double and triple bonds) (Table 2). Complex MS-MS spectra are due to fragmentation of this chain [26]. As can be seen in Table 2, NAAs with terminal alkynes elute early with reversed phase HPLC [29].
Fig. 2.
MS2 fragmentation spectra (CID) of N-alkylamides in the A. ptarmica extract.
Table 2.
N-alkylamides in the ethanolic A. ptarmica extract using HPLC–ESI–MS and/or GC–EI–MS.
| Compound | Rt (HPLC) (min)a | Rt (GC) (min) | Structure | Chemical name | MW (g/mol) | Classification |
|---|---|---|---|---|---|---|
| P1 (=M1) | 53.4 [17.7%] | 23.3 |  |
Undeca-2E,4E-diene-8,10-diynoic acid isobutylamide | 229.32 | F3M1 |
| P2 | 59.7 [14.3%] | – | 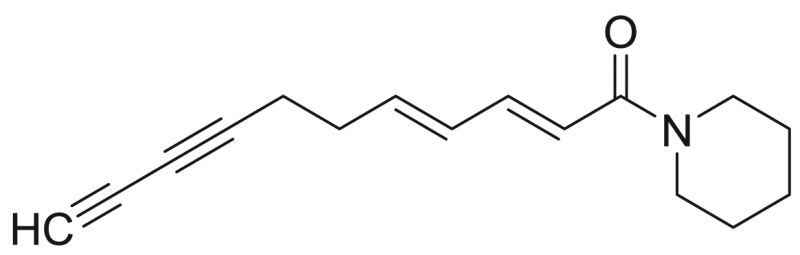 |
Undeca-2E,4E-diene-8,10-diynoic acid piperidide | 241.33 | F3M5 |
| P3 | 62.4 [3.4%] | – |  |
Undeca-2E,4E-diene-8,10-diynoic acid 2-methylbutylamide | 243.35 | F3M1 |
| P4 | 62.6 [0.1%] | – | 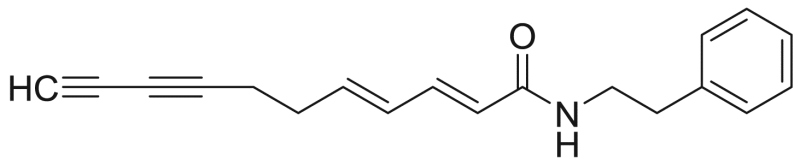 |
Undeca-2E,4E-diene-8,10-diynoic acid phenylethylamide | 277.37 | F3M11 |
| P5 (=M3) | 63.4 [73.6%] | 15.9 | 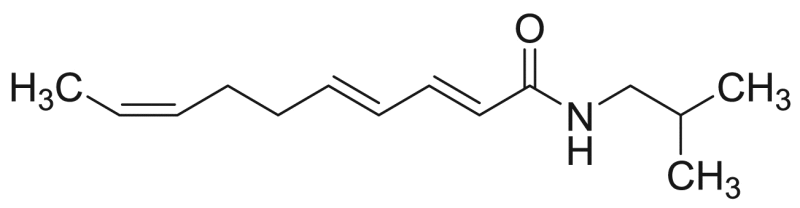 |
Deca-2E,4E,8Z-trienoic acid isobutylamide (8,9-dehydropellitorine) | 221.34 | F3M1 |
| P6 | 65.0 [1.5%] | – | 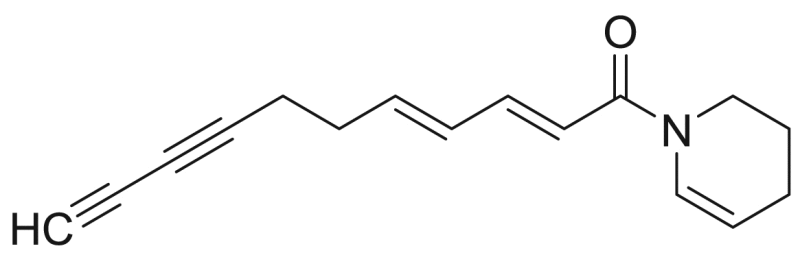 |
Undeca-2E,4E-diene-8,10-diynoic acid piperideide | 239.32 | F3M5 |
| P7 | 65.4 [7.0%] | 25.6 |  |
Deca-2E-ene-4,6,8-triynoic acid isobutylamide | 213.28 | F3M1 |
| P8 (=M4) | 72.5 [3.4%] | – | 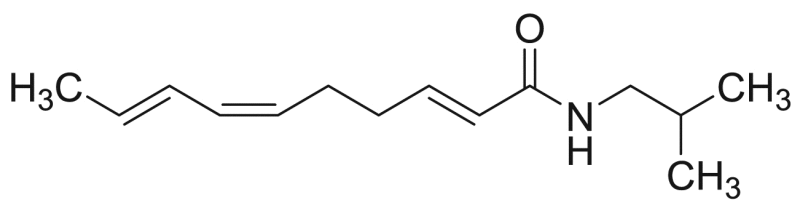 |
Deca-2E,6Z,8E-trienoic acid 2-methylbutylamide (homospilanthol) | 235.37 | F3M1 |
| P9 (=M5) | 73.6 [1.3%] | 23.4 | 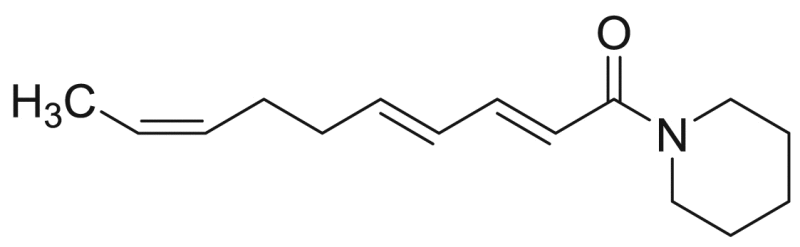 |
Deca-2E,4E,8Z-trienoic acid piperidide | 233.35 | F3M5 |
| P10 (=M17) | 74.5 [11.0%] | 34.5 | 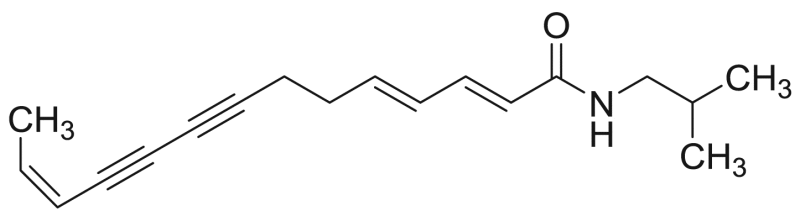 |
Tetradeca-2E,4E,12Z-triene-8,10-diynoic acid isobutylamide | 269.39 | F3M1 |
| P11 (=M6) | 74.9 [100.0%] | 15.5 | 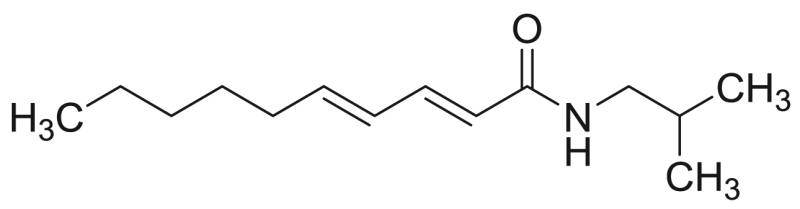 |
Deca-2E,4E-dienoic acid isobutylamide (pellitorine) | 223.36 | F3M1 |
| P12 (=M7) | 77.5 [13.6%] | 34.2 |  |
Tetradeca-2E,4E-diene-8,10-diynoic acid isobutylamide (anacycline) | 271.40 | F3M1 |
| P13 | 82.4 [1.5%] | – | 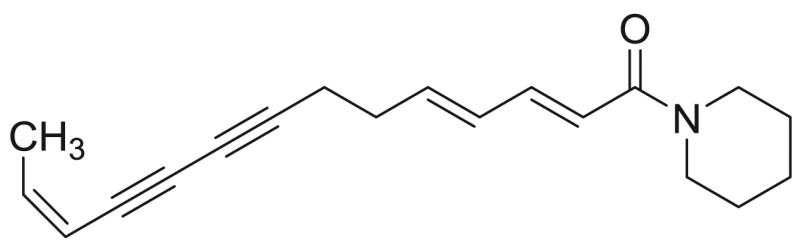 |
Tetradeca-2E,4E,12Z-triene-8,10-diynoic acid piperidide | 281.39 | F3M5 |
| P14 | 83.3 [4.6%] | – | 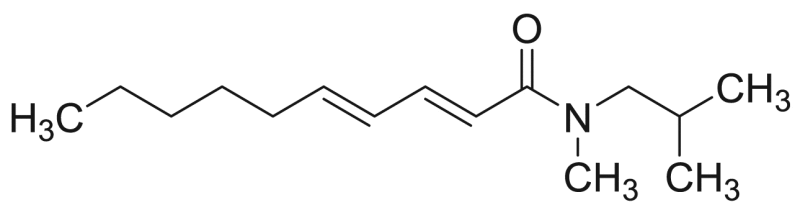 |
Deca-2E,4E-dienoic acid N-methyl isobutylamide | 237.38 | F3M1 |
-: not applicable
Between brackets: estimated relative quantity to pellitorine from total ion chromatogram.
Characteristic fragment ions of NAAs with an isobutylamide group are formed by CID (Table 1) and these typical m/z values are indicated in bold in Table 3 for compounds P1, P5, P7, P10, P11 and P12. In case of compound P1, there were cleavages in the fatty acid chain between C1-C2 (m/z 129) and between C2-C3 (m/z 116). The product ions with m/z 174, 157, 131, 129, 116 and 91 have been reported previously for undeca-2E,4E-diene-8,10-diynoic acid isobutylamide [26], [28], [33]. For compound P11 (deca-2E,4E-dienoic acid isobutylamide or pellitorine), product ions with m/z 182, 168, 154, 151, 133, 123, 109, 95, 83 and 69 were consistent with values reported in literature [28], [34]. In addition, a cleavage between C4-C5 (m/z 140) and C9-C10 (m/z 209) of the fatty acid chain was observed. For compound P12 (tetradeca-2E,4E-diene-8,10-diynoic acid isobutylamide or anacycline), there was a cleavage in the fatty acid chain between C1-C2 (m/z 171), C3-C4 (m/z 145) and C6-C7 (m/z 167). Moreover, the assignment of compounds P1, P11 and P12 was also based on comparison of the retention time [28]. For compound P5 (deca-2E,4E,8Z-trienoic acid isobutylamide), cleavages occurred in the fatty acid between C1-C2 (m/z 121) and C8-C9 (m/z 194). Furthermore, as the fatty acid chain contained doubly allylic carbon atoms, there was the formation of a distonic radical cation (C6-C7, m/z 167) and cationic species with the loss of H, due to cleavage between C5-C6 (m/z 152) [34]. There were cleavages between C1-C2 (m/z 169), C3-C4 (m/z 143), C5-C6 (m/z 117) and C12-C13 (m/z 242) for compound P10 (tetradeca-2E,4E,12Z-triene-8,10-diynoic acid isobutylamide).
Table 3.
MS1 and MS2 information of N-alkylamides in the ethanolic A. ptarmica extract using HPLC–ESI–MS.
| Compound | [M+H]+ | Product ions (m/z) | Losses (m/z) |
|---|---|---|---|
| P1 | 230 | 215; 202; 174; 159; 157; 146; 133; 131; 129; 128; 123; 121; 117; 116; 115; 110; 105; 98; 93; 91; 79; 72 | −15; −28; -56; −71; −73; −84; −97; -99; −101; −102; −107; −109; −113; −114; −115; −120; −125; −132; −137; −139; −151; −158 |
| P2 | 242 | 222; 214; 198; 179; 176; 159; 157; 145; 131; 129; 112; 91; 86; 84; 69 | −20; −28; 44, −63; −66; −83; −85; −97; −111; −113; −130; −151; −156; −158; −173 |
| P3 | 244 | 216; 181; 174; 157; 146; 131; 129; 116; 103; 91; 70 | −28; −63; -70; -87; −98; -113; -115; −128; −141; −153; −174 |
| P4 | 278 | 245; 218; 205; 186; 174; 168; 157; 145; 131; 129; 128; 115; 105; 91 | −33; −60; −73; −92; -104; −110; −121; −133; −147; −149; −150; −163; −173; −187 |
| P5 | 222 | 194; 168; 167; 152; 149; 132; 131; 123; 121; 110; 107; 100; 93; 91; 81; 67 | −28; −54; −55; −70; -73; −90; −91; −99; −101; −112; −115; −122; −129; −131; −141; −155 |
| P6 | 240 | 224; 212; 184; 177; 159; 157; 155; 129; 110; 108; 82; 80 | −16; −28; −56; −63; −81; −83; −85; −111; −130; −132; −158; −160 |
| P7 | 214 | 198; 172; 159; 158; 141; 130; 115; 103; 89; 72 | −16; −42; −55; -56; -73; −84; −99; −111; −125; −142 |
| P8 | 236 | 221; 182; 181; 180; 166; 151; 149; 138; 125; 123; 121; 107; 95; 93; 81; 79 | −15; −54; −55; −56; −70; −85; −87; −98; −111; −113; −115; −129; −141; −143; −155; −157 |
| P9 | 234 | 205; 179; 166; 149; 131; 112; 93; 86 | −29; −55; −68; −85; −103; −122; −141; −148 |
| P10 | 270 | 246; 242; 228; 214; 197; 179; 171; 169; 154; 143; 129; 117; 105; 91; 79 | −24; −28; −42; -56; -73; −91; -99; -101; −116; −127; −141; −153; −165; −179; −191 |
| P11 | 224 | 209; 204; 182; 168; 154; 151; 140; 133; 123; 109; 105; 95; 83; 69 | −15; −20; −42; −56; −70; −73; −84; −91; -101; −115; −119; −129; −141; −155 |
| P12 | 272 | 244; 216; 199; 173; 171; 167; 145; 131; 117; 91; 81 | −28; −56; −73; −99; −101; −105; −127; −141; −155;−181; −191 |
| P13 | 282 | 254; 240; 224; 212; 197; 179; 169; 165; 141; 143; 129; 112; 103; 86; 84 | 28; −42; −58; −70; −85; −103; −113; −128; −141; −139; 153; −170; −179; −196; −198 |
| P14 | 238 | 220; 210; 209; 197; 182; 168; 151; 133; 109; 95; 81; 69 | −18; −28; −29; −41; −56; −70; −87; −105; −129; −143; −157; 169 |
In bold: characteristic product ions or losses.
NAAs having a phenylethylamide alkyl group showed similarly formed fragments as the isobutylamide NAAs, i.e., the typical losses, which are presented in Table 1. The typical m/z values of these losses for compound P4 (undeca-2E,4E-diene-8,10-diynoic acid phenylethylamide) are indicated in bold in Table 3. Furthermore, a tropylium ion was formed (benzene and α C) (m/z 91). A cleavage was observed in the fatty acid between C1-C2 (m/z 129). Product ions of compound P4 with m/z 168, 157, 131, 105 and 91 were consistent with values reported earlier [28]. Moreover, assignment of the NAA was also based upon retention time comparison [28].
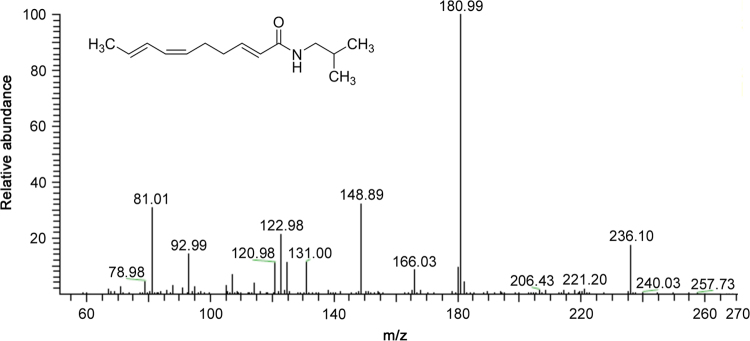
Characteristic losses of the NAAs with a 2-methyl isobutylamide function are presented in Table 1 and indicated for compounds P3 and P8 in Table 3. For compound P3, a cleavage occurred in the fatty acid chain between C1-C2 (m/z 129), C2-C3 (m/z 116) and C6-C7 (m/z 181). The MS spectrum of compound P8 corroborates well with the structure of deca-2E,6Z,8E-trienoic acid 2-methylbutylamide (homospilanthol) or a related isomeric compound and has never previously been reported in this plant (Fig. 3). A cleavage was observed in the fatty acid between C6-C7 (m/z 182) and C9-C10 (m/z 221). Furthermore, structural assignment of compound P8 was done based on comparison of retention time and product ions. Product ions with m/z 166, 149, 123, 121 and 81 were already reported for this NAA [27].
Fig. 3.
MS2 spectra of deca-2E,6Z,8E-trienoic acid 2-methylbutylamide or homospilanthol (compound P8) with [M+H]+= m/z 236.
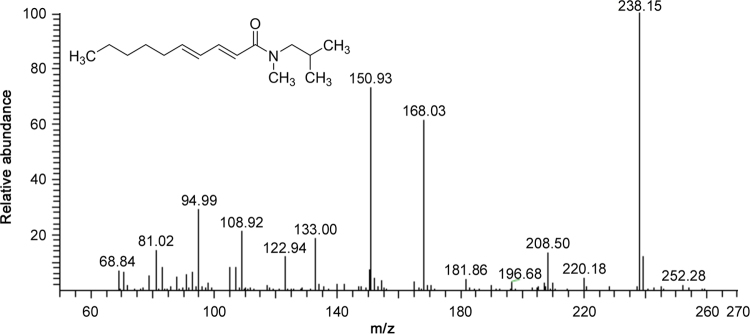
The characteristic losses for NAAs having a N-methyl isobutylamide function are shown in Table 1 and indicated in Table 3 for compound P14. Interestingly, deca-2E,4E-dienoic acid N-methyl isobutylamide is the second NAA, never previously reported in this plant. The MS2 spectrum of compound 14 is shown in Fig. 4. A cleavage occurred in the fatty acid part between C8-C9 (m/z 209). Moreover, the structural assignment of compound P14 was also based on comparison of the retention time and the product ions m/z 182, 168, 151 and 109, which were previously reported [28].
Fig. 4.
MS2 spectra of deca-2E,4E-dienoic acid N-methyl isobutylamide (compound P14) with [M+H]+= m/z 238.
For compounds P2 (undeca-2E,4E-diene-8,10-diynoic acid piperidide), P9 (deca-2E,4E,8Z-trienoic acid piperidide) and P13 (tetradeca-2E,4E,12Z-triene-8,10-diynoic acid piperidide), having a piperidide function, acylium ions were formed with m/z values of 157, 149 and 197, respectively, corresponding with a loss of m/z 85. There was a cleavage in the fatty acid chain of these compounds between C6-C7 (m/z 179) (Table 3). Furthermore, cleavages were found in the fatty acid chain between C3-C4 (m/z 143) and C12-C13 (m/z 254) of compound P13. Compound P9 has doubly allylic carbon atoms in the fatty acid part and formed a distonic radical cation due to cleavage between C6-C7 (m/z 179) [34]. An acylium ion was also formed for compound P6, having a piperideide function, with a m/z value of 157 (loss of m/z 83) and as compound P6 contains doubly allylic carbon atoms, a distonic radical cation was formed (C6-C7, m/z 177) (Table 3) [34].
All the reported NAAs were found in the roots of A. ptarmica, except for compound P4, which was only found in the leaves. Furthermore, compounds P1, P2, P3, P5, P6 and P7 were also observed in the leaves, while compounds P1, P2, P3, P5 and P6 were found in the stem as well (data not shown). No NAAs could be found in the flowers, as the concentration of NAAs was probably too low. In conclusion, the highest amount of NAAs was found in the roots, which is consistent with the literature [35].
3.1.2. N-alkylamide profiling using GC–EI–MS
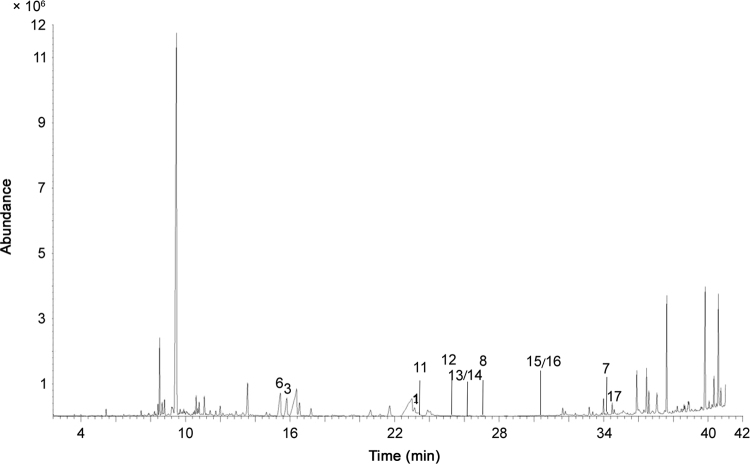
Compounds P1, P5, P7, P9, P10, P11 and P12, observed using HPLC–ESI–MS, were also observed with GC–EI–MS. The TIC is shown in Fig. 5, using the same numbers as indicated in HPLC-MS.
Fig. 5.
TIC of Achillea ptarmica obtained using GC–EI–MS.
The molecular ions were detected for all compounds. Product ions characteristic for isobutylamide NAAs were found for compounds P1, P5, P7, P10, P11 and P12. These product ions are indicated in bold in Table 4. This was also done for compound P9, an NAA having a piperidide function. Typical product ions for the amide part are also indicated in bold in Table 4. Furthermore, σ-cleavages were observed in the fatty acid chain of all the NAAs. These product ions were for compound P1: m/z 63/166 (C6-C7), m/z 77 (C5-C6), m/z 103 (C3-C4); for compound P5: m/z 206 (C9-C10), m/z 41 (C7-C8), m/z 55/166 (C6-C7), m/z 69 (C5-C6), m/z 95 (C3-C4), m/z 100/121 (C1-C2); for compound P7: m/z 198 (C9-C10), m/z 63 (C5-C6), m/z 87 (C3-C4), m/z 113 (C1-C2); for compound P10: m/z 254 (C13-C14), m/z 41 (C11-C12), m/z 103 (C6-C7), m/z 117 (C5-C6), m/z 143 (C3-C4), m/z 169 (C1-C2); for compound P11: m/z 208 (C9-C10), m/z 194 (C8-C9), m/z 180 (C7-C8), m/z 166 (C6-C7), m/z 152 (C5-C6), m/z 126 (C3-C4); for compound P12: m/z 256 (C13-C14), m/z 242 (C12-C13), m/z 43/228 (C11-C12), m/z 67 (C9-C10), m/z 91 (C7-C8), m/z 105/166 (C6-C7), m/z 119/152 (C5-C6), m/z 100/171 (C1-C2) and for compound P9: m/z 41 (C7-C8), m/z 55 (C6-C7), m/z 69 (C5-C6), m/z 95/138 (C3-C4), m/z 121/112 (C1-C2). In addition, product ions were also detected consistent with a (H-)rearrangement between C4-C5 in the fatty acid chain, for compound P5: m/z 81, m/z 140; for compound P10: m/z 129; for compound P11: m/z 83; for compound P12: m/z 131; and for compound P9: m/z 81, m/z 152. The product ions of compound P11 with m/z 223, 208, 180, 166, 151, 113, 110, 96, 81, 66, 57, 55, 53 and 41 were also described by Lazarevic et al. [2]. Compound P1 was also recognized by the Nist library.
Table 4.
MS information of N-alkylamides in the ethanolic A. ptarmica extract using GC–EI–MS.
| Compound | [M]+ | Product ions (m/z) (% intensity relative to base peak) |
|---|---|---|
| P1 | 229 | 41 (10%), 43, 51, 55, 57 (9%), 63 (10%), 66 (23%), 67 (15%), 72, 77, 81 (8%), 94, 102, 103, 110 (10%), 115, 123, 127 (27%), 128 (64%), 129 (19%), 157 (100%), 166 (14%), 172 (9%), 178, 186, 214 (9%), 228 (17%), 229 (13%), 233 |
| P5 | 221 | 41 (18%), 43, 53 (10%), 55 (51%), 57 (39%), 65 (10%), 66 (28%), 67 (31%), 68 (12%), 69, 72, 77 (8%), 79(16%), 81 (21%), 82 (9%), 91 (9%), 96 (11%), 95 (13%), 94 (15%), 93 (19%), 100, 107 (10%), 110 (26%), 121 (8%), 122 (9%), 131 (8%), 139, 140, 149 (100%), 150 (16%), 152 (18%), 157, 164, 166 (50%), 167 (17%), 178, 192, 206 (11%), 221 (41%), 222 |
| P7 | 213 | 41 (8%), 43, 57, 63 (9%), 69, 73, 77, 81, 85, 86 (11%), 87 (20%), 91, 95, 102, 108, 113 (18%), 114 (6%), 128 (6%), 133, 141 (100%), 142 (15%), 152, 156, 157 (26%), 170, 171 (16%), 198, 213 (43%), 214 (8%) |
| P9 | 233 | 41 (65%), 42 (12%), 43 (41%), 45 (10%), 53 (28%), 54 (28%), 55 (92%), 56 (15%), 57 (33%), 58 (14%), 60 (14%), 63 (9%), 65 (23%), 66 (45%), 67 (81%), 68 (25%), 69 (62%), 70 (14%), 71 (12%), 73 (20%), 77 (26%), 78 (14%), 79 (53%), 80 (25%), 81 (84%), 82 (40%), 83 (49%), 84 (70%), 85 (20%), 91 (29%), 93 (38%), 94 (28%), 95 (54%), 96 (31%), 97 (18%), 98 (16%), 101 (11%), 105 (15%), 106 (9%), 107 (21%), 108 (12%), 109 (24%), 110 (23%), 111 (15%), 112 (38%), 113 (10%), 115 (11%), 117 (9%), 119 (11%), 120 (10%), 121 (17%), 122 (13%), 123 (17%), 124 (12%), 125 (9%), 126 (10%), 127 (24%), 128 (49%), 129 (22%), 131 (14%), 133 (12%), 134 (8%), 135 (12%), 136 (11%), 137 (27%), 138 (46%), 139 (14%), 143 (12%), 144 (10%), 148 (10%), 149 (17%), 150 (29%), 151 (28%), 152 (9%), 157 (75%), 158 (15%), 162 (10%), 164 (29%), 165 (15%), 166 (13%), 167 (8%), 172 (10%), 177 (10%), 178 (100%), 179 (13%), 228 (16%), 229 (16%), 232 (9%), 233 (50%), 234 (11%), 280 (10%) |
| P10 | 269 | 40 (8%), 41 (36%), 43 (24%), 44 (8%), 51 (10%), 53 (11%), 54 (9%), 55 (31%), 56 (12%), 57 (70%), 58 (14%), 63 (11%), 65 (16%), 66 (30%), 67 (40%), 68 (18%), 69 (20%), 71 (11%), 72 (9%), 73 (16%), 74 (8%), 75 (16%), 76 (9%), 77 (80%), 78 (11%), 79 (16%), 81 (19%), 82 (19%), 83 (13%), 84 (8%), 85 (9%), 91 (21%), 93 (15%), 94 (11%), 95 (27%), 96 (12%), 97 (12%), 98 (14%), 102 (15%), 103 (62%), 105 (9%), 107 (11%), 108 (9%), 109 (15%), 110 (16%), 111 (8%), 115 (25%), 117 (13%), 128 (19%), 129 (34%), 131 (10%), 135 (10%), 141 (51%), 142 (17%), 143, 147 (9%), 149 (12%), 152 (25%), 153 (29%), 154 (38%), 155 (25%), 157 (8%), 165 (10%), 166 (20%), 167 (54%), 168 (12%), 169 (24%), 171 (10%), 178, 185 (10%), 187 (10%), 197 (22%), 198 (11%), 207 (15%), 212 (8%), 223, 239 (9%), 254, 268 (20%), 269 (100%), 270 (23%), 281 (13%), 355 (8%), 410 |
| P11 | 223 | 41 (12%), 43, 53 (8%), 55 (8%), 57, 60, 66 (11%), 67 (13%), 69 (10%), 72, 71, 73, 77, 79 (8%), 81 (40%), 83, 89, 94, 95 (12%), 96 (40%), 97, 98 (8%), 103, 109, 110 (12%), 113 (9%), 120, 123, 126, 138, 145, 152 (35%), 151 (100%), 166 (8%), 167, 180 (8%), 194, 208 (10%), 223 (35%), 224 |
| P12 | 271 | 41 (34%), 43 (17%), 51 (10%), 53 (10%), 55 (29%), 57 (80%), 58 (8%), 63 (17%), 65 (26%), 66 (52%), 67 (45%), 68 (15%), 69 (11%), 73 (9%), 77 (58%), 78 (11%), 79 (38%), 81 (17%), 82 (12%), 91 (26%), 93 (9%), 94 (20%), 95 (17%), 96 (9%), 98 (9%), 100 (8%), 103 (18%), 105 (17%), 107 (8%), 110 (29%), 111 (11%), 115 (31%), 117 (18%), 119, 127 (14%), 128 (55%), 129 (100%), 130 (23%), 131 (13%), 134, 141 (35%), 142 (22%), 143 (68%), 144 (23%), 145 (9%), 152 (17%), 153 (10%), 155 (18%), 156 (12%), 157 (27%), 158 (10%), 166 (36%), 167 (55%), 168 (10%), 169 (15%), 170 (11%), 171 (36%), 172 (15%), 173, 186 (10%), 199 (75%), 200 (22%), 214 (13%), 223, 228 (8%), 242 (13%), 243, 256 (33%), 257 (9%), 270 (23%), 271 (57%,), 281 (8%), 297, 355 (8%) |
Note: if no % is indicated between brackets that means intensity <7% of base peak.
Product ions indicated in bold: characteristic product ions for the amide functional group of the NAA.
3.2. A. millefolium
3.2.1. N-alkylamide profiling using HPLC–ESI–MS
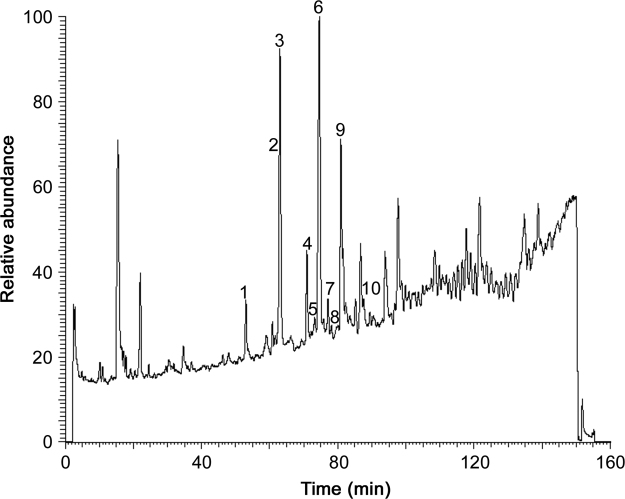
The TIC of the root extract of A. millefolium is shown in Fig. 6 in which peak labels correspond to NAA designations.
Fig. 6.
TIC of Achillea millefolium obtained using HPLC–ESI–MS.
In the MS1 spectra, the major ions are the protonated forms of the NAAs. MS2 spectra are presented in Fig. 7. All the NAAs assigned in A. millefolium are summarised in Table 5 with their corresponding retention time (Rt), structure, chemical name, molecular weight (MW, average mass) and classification. Structures of ten NAAs were assigned in the A. millefolium extract with different types of amides: 4 NAAs having an isobutylamide function (compounds M1, M3, M6, M7), 2 NAAs with a piperidide function (compounds M5, M10), 1 NAA with a piperideide function (compound M8), 1 NAA with a 2-methylbutylamide function (compound M4), 1 NAA with a 4-hydroxyphenylethylamide function (compound M2) and 1 NAA having a 4-methoxy phenylethylamide function (compound M9).
Fig. 7.
MS2 fragmentation spectra (CID) of N-alkylamides in the A. millefolium extract.
Table 5.
N-alkylamides in the ethanolic A. millefolium extract using HPLC-ESI-MS and/or GC-EI-MS.
| Compound | Rt (HPLC) (min)a | Rt (GC) (min) | Structure | Chemical name | MW (g/mol) | Classification |
|---|---|---|---|---|---|---|
| M1 (=P1) | 53.1 [13.7%] | 23.3 |  |
Undeca−2E,4E-diene−8,10-diynoic acid isobutylamide | 229.32 | F3M1 |
| M2 | 62.9 [21.0%] | – | 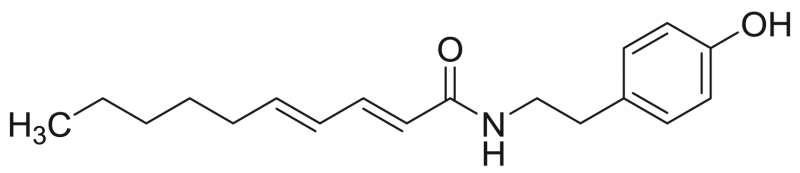 |
Deca−2E,4E-dienoic acid tyramide | 287.40 | F3M12 |
| M3 (=P1) | 63.1 [86.6%] | 15.8 |  |
Deca−2E,4E,8Z-trienoic acid isobutylamide | 221.34 | F3M1 |
| M4 (=P8) | 71.6 [3.7%] | – |  |
Deca−2E,6Z,8E-trienoic acid 2-methylbutylamide (homospilanthol) | 235.37 | F3M1 |
| M5 (=P9) | 73.4 [5.9%] | – | 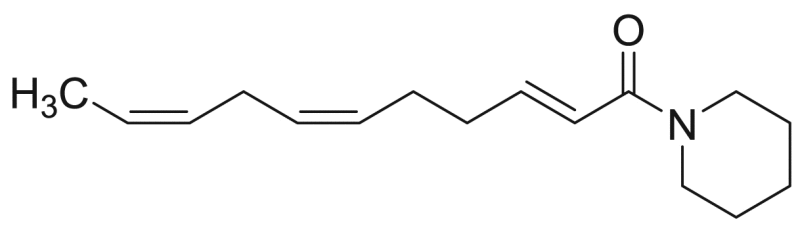 |
Deca−2E,4E,8Z-trienoic acid piperidide | 233.35 | F3M5 |
| M6 (=P11) | 74.6 [100.0%] | 15.4 |  |
Deca−2E,4E-dienoic acid isobutylamide (pellitorine) | 223.36 | F3M1 |
| M7 (=P12) | 77.2 [5.8%] | 34.2 | 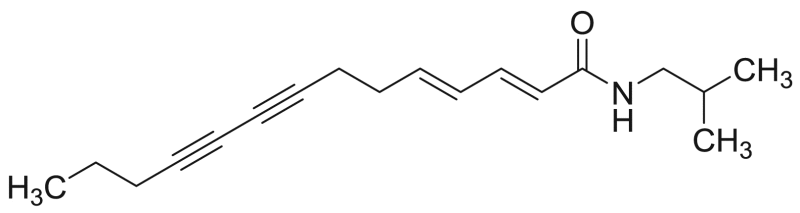 |
Tetradeca−2E,4E-diene−8,10-diynoic acid isobutylamide (anacycline) | 271.40 | F3M1 |
| M8 | 78.3 [2.3%] | 27.1 |  |
Deca−2E,4E,8Z-trienoic acid piperideide | 231.34 | F3M5 |
| M9 | 80.9 [30.3%] | – |  |
Deca−2E,4E-dienoic acid 4-methoxy phenylethylamide | 301.43 | F3M12 |
| M10 | 87.6 [5.4%] | – | 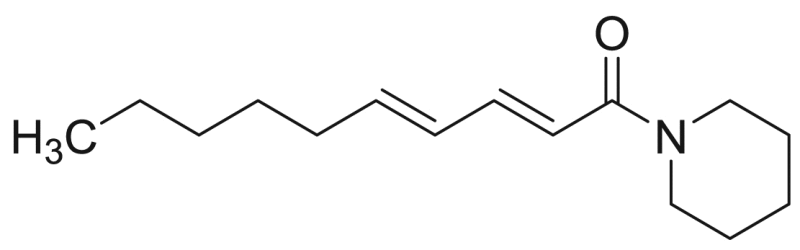 |
Deca−2E,4E-dienoic acid piperidide | 235.37 | F3M5 |
| M11 | – | 23.4 | 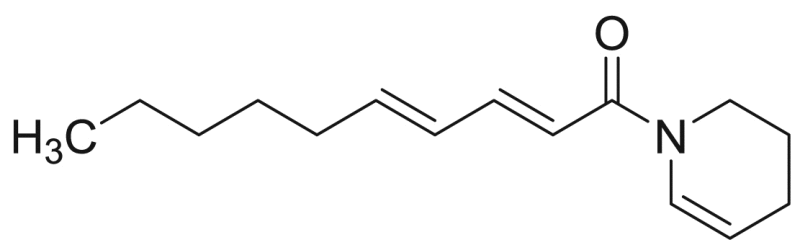 |
Deca−2E,4E-dienoic acid piperideide | 233.35 | F3M5 |
| M12 | – | 25.3 | 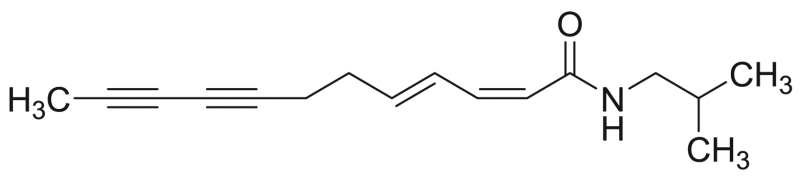 |
Dodeca−2Z,4E-diene−8,10-diynoic acid isobutylamide | 243.35 | F3M1 |
| M13 | – | 26.3 |  |
Deca−2E,4E,6Z-trienoic acid piperideide | 231.34 | F3M5 |
| M14 | – | 26.3 | 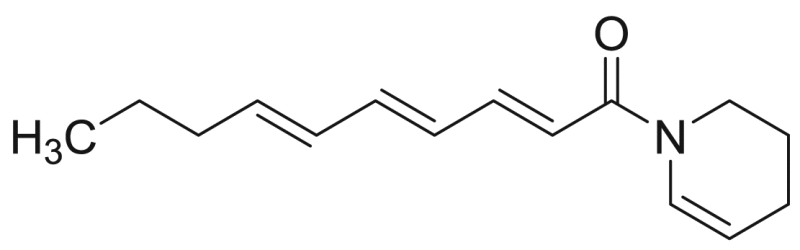 |
Deca−2E,4E,6E-trienoic acid piperideide | 231.34 | F3M5 |
| M15 | – | 30.3 |  |
Deca−2E,4E,6Z,8Z-tetraenoic acid piperideide | 229.32 | F3M5 |
| M16 | – | 30.3 | 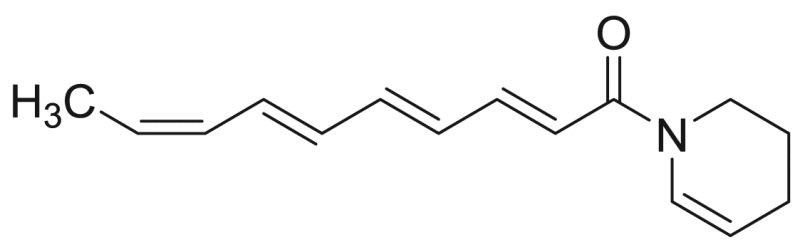 |
Deca−2E,4E,6E,8Z-tetraenoic acid piperideide | 229.32 | F3M5 |
| M17 (=P10) | – | 34. 5 |  |
Tetradeca−2E,4E,12Z-triene−8,10-diynoic acid isobutylamide | 269.39 | F3M1 |
-: not applicable
Between brackets: estimated relative quantity to pellitorine from total ion chromatogram.
Table 1 contains characteristic fragment ions formed by CID for NAAs with an isobutylamide, a 2-methylbutylamide, a 4-hydroxyphenylethylamide and 4-methoxy phenylethylamide function. Typical m/z values of fragment losses of NAAs with an isobutylamide function are indicated in bold in Table 6 for compounds M1 (undeca-2E,4E-diene-8,10-diynoic acid isobutylamide), M3 (deca-2E,4E,8Z-trienoic acid isobutylamide), M6 (deca-2E,4E-dienoic acid isobutylamide or pellitorine) and M7 (tetradeca-2E,4E-diene-8,10-diynoic acid isobutylamide or anacycline). For compound M1, there was a cleavage in the fatty acid chain between C1-C2 (m/z 129), C2-C3 (m/z 116), C3-C4 (m/z 103) and C4-C5 (m/z 90). Moreover, for compounds M1 and M6 of A. millefolium and compounds P1 and P11 of A. ptarmica, identical product ions were previously reported [26], [28], [33], [34]. Furthermore, cleavages between C1-C2 (m/z 123) and C4-C5 (m/z 140) of the fatty acid chain in compound M6 were observed. For compound M7, there was a cleavage in the fatty acid chain between C1-C2 (m/z 171), C3-C4 (m/z 145) and C8-C9 (m/z 79). Moreover, the assignment of compounds M1, M6 and M7 was also based on comparison of the retention time [28]. For compound M3, cleavages occurred in the fatty acid chain between C1-C2 (m/z 121) and C8-C9 (m/z 194). As compound M3 contains doubly allylic carbon atoms, there is the formation of a distonic radical cation due to cleavage between C6-C7 (m/z 167). This distonic radical cation undergoes a hydrogen rearrangement to form an acylium ion and the subsequent loss of CO results in a C5 cation (m/z 67) [34].
Table 6.
MS1 and MS2 information of N-alkylamides in the ethanolic A. millefolium extract using HPLC–ESI–MS.
| Compound | [M+H]+ | Product ions (m/z) | Losses (m/z) |
|---|---|---|---|
| M1 | 230 | 215; 202; 188; 174; 157; 156; 146; 131; 129; 116; 115; 103; 91; 90; 79 | −15; −28; −42; -56; -73; −74; −84; −99; −101; −114; −115; −127; −139; −140; −151 |
| M2 | 288 | 261; 178; 164; 151; 138; 133; 121; 109; 95; 91; 90 | −27; −110; −124; −137; −150; −155; −167; −179; −193; −197; −198 |
| M3 | 222 | 206; 194; 178; 168; 167; 149; 131; 123; 121; 107; 93; 81; 67 | −15; −28; −44; −54; −55; −73; −91; −99; −101; −115; −129; −141; −155 |
| M4 | 236 | 232; 190; 181; 166; 149; 131; 123; 95; 93; 81; 79; 70 | −4; −46; −55; −70; -87; −105; −113; −141; −143; −155; −157; −166 |
| M5 | 234 | 216; 206; 193; 179; 164; 149; 131; 112; 93; 86; 81 | −18; −28; −41; −55; −70; −85; −103; −122; −141; −148; −153 |
| M6 | 224 | 208; 182; 168; 151; 140; 133; 123; 109; 95; 81; 69 | −16; −42; -56; -73; −84; −91; −101; −115; −129; −143; −155 |
| M7 | 272 | 244; 216; 199; 188; 173; 171; 157; 145; 131; 117; 105; 91; 81; 79 | −28; −56; −73; −84; -99; −101; −115; −127; −141; −155; −167; −181; −191; −193 |
| M8 | 232 | 204; 177; 162; 149; 131; 110; 93; 83; 67 | −28; −55; −70; −83; −101; −122; −139; −149; −165 |
| M9 | 302 | 297; 259; 218; 152; 151; 133; 109; 95; 93 | −5; −43; −84; −150; −151; −169; −193; −207; −209 |
| M10 | 236 | 218; 189; 167; 151; 133; 123; 109; 95; 86; 81 | −18; −47; −69; −85; −103; −113; −127; −141; −150; −155 |
In bold: characteristic product ions or losses.
NAAs having a 4-hydroxyphenylethylamide also showed typical fragment ions and are described in Table 1 and indicated in bold in Table 6 for compound M2 (deca-2E,4E-dienoic acid tyramide). A cleavage occurred in the fatty acid chain between C2-C3 (m/z 178) and a tropylium ion was formed (benzene and α-C) (m/z 91). Moreover, the assignment of compound M2 was also based on comparison of the retention time and product ions: m/z 178, 151, 133, 121 and 95 were previously reported [28].
Characteristic fragment ions for NAAs with a 4-methoxy phenylethylamide function are described in Table 1 and indicated in Table 6 for compound M9 (deca-2E,4E-dienoic acid 4-methoxy phenylethylamide). A cleavage occurred in the fatty acid chain between C4-C5 (m/z 218) and C7-C8 (m/z 259).
For NAAs having a 2-methyl isobutylamide function, the characteristic fragmentation ions are summarised in Table 1. Typical fragment losses of compound M4 are marked in Table 6. The obtained MS spectra corroborate well with the structure of deca-2E,6Z,8E-trienoic acid 2-methylbutylamide (homospilanthol) or a related isomeric compound. This NAA has never been reported before in A. millefolium. A cleavage occurred in the fatty acid part between C3-C4 (m/z 95) and C4-C5 (m/z 81). Compound M4 (A. millefolium) was also reported for the first time in A. ptarmica (compound P8) and is shown in Fig. 3 together with its MS2 spectrum.
In case of NAAs having a piperidide function, namely compounds M5 (deca-2E,4E,8Z-trienoic acid piperidide) and M10 (deca-2E,4E-dienoic acid piperidide), there was the formation of acylium ions with m/z values of 149 and 151, respectively, corresponding with a loss of m/z 85. There was a cleavage in the fatty acid chain of compound M5 between C6-C7 (m/z 179), C7-C8 (m/z 193) and C8-C9 (m/z 206). Furthermore, compound M5, containing doubly allylic carbon atoms in the fatty acid part, formed a distonic radical cation due to cleavage between C6-C7 (m/z 179) and a cationic species as a result of the cleavage between C5-C6 and loss of a hydrogen atom (m/z 164) [34]. For compound M8, possessing a piperideide function, there were cleavages in the fatty acid chain between C6-C7 (m/z 177) and C8-C9 (m/z 204) and the formation of a distonic radical cation (C6-C7, m/z 177). This cation undergoes a hydrogen rearrangement to form an acylium ion and the subsequent loss of CO results in a C5 cation (m/z 67). There is also the formation of a cationic species (C5-C6+ loss of H, m/z 162) [34]. An acylium ion was also formed in case of compound M8, with a m/z value of 149 (loss of m/z 83).
The previously mentioned NAAs in A. millefolium were all observed in the roots, while in the stems only compounds M1, M3, M5, M6 and M10 were found. Due to too low NAA concentrations in the flowers and leaves, no NAAs could be observed.
3.2.2. N-alkylamide profiling using GC–EI–MS
Using GC-EI-MS, compounds M1, M3, M6 and M7, observed with HPLC-ESI-MS, were also found. The TIC is shown in Fig. 8, with the same numbers as indicated in HPLC-MS. Additional compounds assigned using GC–MS were indicated starting numbering from 11.
Fig. 8.
TIC of Achillea millefolium obtained using GC–EI–MS.
Other NAAs were structurally assigned using GC–MS and were not observed with HPLC–MS: compounds M11 (deca-2E,4E-dienoic acid piperideide), M12 (dodeca-2Z,4E-diene-8,10-diynoic acid isobutylamide), M13 (deca-2E,4E,6Z-trienoic acid piperideide) and M17 (tetradeca-2E,4E,12Z-triene-8,10-diynoic acid isobutylamide). With the current MS information, no distinction can be made between isomeric compounds M8 (deca-2E,4E,8Z-trienoic acid piperideide), M13 (deca-2E,4E,6Z-trienoic acid piperideide) and M14 (deca-2E,4E,6E-trienoic acid piperideide) and between compound M15 (deca-2E,4E,6Z,8Z-tetraenoic acid piperideide) and its isomer M16 (deca-2E,4E,6E,8Z-tetraenoic acid piperideide). For all compounds, the molecular ions were detected, except for compound M12. Characteristic product ions were found for compounds M1, M3, M6, M7, M12 and M17, having an isobutylamide function and are indicated in bold in Table 7. This was also done for compounds M8, M11, M13/14 and M15/16, which are NAAs having a piperideide function. Furthermore, in all NAAs, σ-cleavages were observed in the fatty acid chain. These product ions were for compound M1: m/z 204 (C9-C10), m/z 63/166 (C6-C7), m/z 77 (C5-C6), m/z 103 (C3-C4), m/z 129 (C1-C2); for compound M3: m/z 206 (C9-C10), m/z 41/180 (C7-C8), m/z 55/166 (C6-C7), m/z 152 (C5-C6), m/z 95 (C3-C4), m/z 121 (C1-C2); for compound M6: m/z 208 (C9-C10), m/z 194 (C8-C9), m/z 43/180 (C7-C8), m/z 57 (C6-C7), m/z 152 (C5-C6), m/z 97 (C3-C4), m/z 123 (C1-C2); for compound M7: m/z 242 (C12-C13), m/z 43/228 (C11-C12), m/z 67 (C9-C10), m/z 91 (C7-C8), m/z 105 (C6-C7), m/z 171 (C1-C2); for compound M12: m/z 77 (C6-C7), m/z 143 (C1-C2); for compound M17: m/z 254 (C13-C14), m/z 41 (C11-C12), m/z 103 (C6-C7); for compound M8: m/z 41 (C7-C8), m/z 55 (C6-C7), m/z 69 (C5-C6), m/z 95 (C3-C4), m/z 121/110 (C1-C2); for compound M11: m/z 218 (C9-C10), m/z 204 (C8-C9), m/z 190 (C7-C8), m/z 57 (C6- C7), m/z 162 (C5-C6), m/z 97 (C3-C4), m/z 123 (C1-C2); for compound M13/14: m/z 202 (C8-C9), m/z 43 (C7-C8), m/z 69 (C5-C6), m/z 95 (C3-C4); and for compound M15/16: m/z 41 (C7-C8), m/z 67 (C5-C6), m/z 93 (C3-C4), m/z 119 (C1-C2). In addition, product ions were also detected consistent with a (H-)rearrangement between C4-C5 in the fatty acid chain, i.e., for compound M3: m/z 81; for compound M6: m/z 83; for compound M17: m/z 129; for compound M8: m/z 81; for compound M11: m/z 83; and for compound M13/14: m/z 81. Compound M6 in the A. millefolium extract corresponds to compound P11 in the A. ptarmica extract and product ions were already described in the literature for pellitorine [2]. Moreover, the following product ions of compound M11 were reported as well: m/z 233, 162, 151, 95, 81, 81, 69, 67, 66, and 55 [2]. Compound M1 was also recognized by the Nist library.
Table 7.
MS information of N-alkylamides in the ethanolic A. millefolium extract using GC-EI-MS.
| Compound | [M]+ | Product ions (m/z) (% intensity relative to base peak) |
|---|---|---|
| M1 | 229 | 41 (27%), 43 (12%), 53 (8%), 54 (8%) 55 (32%), 56, 58 (17%), 63 (11%), 65 (11%), 66 (27%), 67 (39%), 68 (10%), 69 (20%), 77 (11%), 79 (22%), 81 (26%), 82 (26%), 83 (23%), 84 (23%), 91 (14%), 93 (13%), 94 (13%), 95 (23%), 96 (12%), 97 (12%), 103, 107 (8%), 108 (9%), 109 (9%), 110 (14%), 115 (15%), 123, 127 (25%), 128 (69%), 129 (22%,) 133, 138 (12%), 143 (8%), 151, 157 (100%), 158 (15%), 166 (14%), 172 (8%), 178 (18%), 186, 204, 214 (9%), 222, 228 (16%), 229 (14%), 233, 264, 280 |
| M3 | 221 | 41 (18%), 53 (9%), 55 (52%), 57 (37%), 65 (10%), 66 (27%), 67 (30%), 68 (11%), 77 (9%), 79 (16%), 81 (20%), 82 (10%), 91 (8%), 93 (19%), 94 (15%), 95 (12%), 96 (11%), 98, 100, 107 (8%), 110 (26%), 121 (8%), 122 (9%), 131, 149 (100%), 150 (15%), 152 (19%), 164, 166 (50%), 167 (16%), 178, 180, 192, 206 (11%), 221 (40%), 222 |
| M6 | 223 | 41 (11%), 43, 53, 55 (8%), 57, 66 (10%), 67 (12%), 69 (10%), 72, 77, 79 (8%), 81 (38%), 83, 95 (12%), 96 (38%), 97, 98, 109, 110 (11%), 113 (10%), 123, 124, 151 (100%), 152 (33%), 166, 180, 194, 208 (10%), 223 (33%) |
| M7 | 271 | 41 (4%), 43 (28%), 51 (11%), 53 (13%), 55 (37%), 57 (77%), 58 (8%), 63 (15%), 65 (23%), 66 (49%), 67 (51%), 68 (30%), 69 (22%), 71 (12%), 72 (12%), 73 (20%), 75 (8%), 76 (8%), 77 (48%), 78 (11%), 79 (42%), 80 (11%), 81 (24%), 82 (22%), 83 (16%), 84 (10%), 85 (9%), 91 (31%), 92 (9%), 93 (16%), 94 (20%), 95 (32%), 96 (13%), 97 (13%), 98 (15%), 103 (17%), 105 (17%), 107 (13%), 109 (15%), 110 (25%), 111 (11%), 115 (31%), 116 (10%), 117 (22%), 118 (9%), 119 (10%), 120, 121 (11%), 123 (9%), 127 (16%), 128 (48%), 129 (100%), 130 (17%), 131 (11%), 141 (35%), 142 (19%), 143 (58%), 144 (25%), 145 (11%), 147 (11%), 151 (9%), 152 (15%), 153 (9%), 155 (16%), 156 (11%), 157 (20%), 158 (11%), 159 (11%), 166 (52%), 167 (52%), 169 (18%), 171 (38%), 172 (13%), 175, 186 (15%), 187 (12%), 189 (13%), 199 (69%), 200 (17%), 207 (12%), 214 (12%), 228 (9%), 242 (16%), 256 (40%), 270 (26%), 271 (50%), 272 (12%), 281 (12%), 294, 341, 355, 408 |
| M8 | 231 | 41 (54%), 43 (60%), 54 (30%), 55 (88%), 56 (19%), 57 (42%), 60 (24%), 67 (43%), 68 (24%), 69 (56%), 71 (30%), 73 (67%), 77 (23%), 79 (48%), 80 (23%), 81 (79%), 82 (36%), 83 (40%), 85 (23%), 91 (51%), 93 (34%), 95 (59%), 96 (24%), 97 (21%), 107 (100%), 108 (28%), 109 (39%), 110 (23%), 115 (22%), 121 (22%), 122 (20%), 129 (27%), 133 (21%), 135 (20%), 147 (30%), 149 (64%), 185 (18%), 189 (23%), 231 (73%) |
| M11 | 233 | 41 (49%), 42 (10%), 43 (30%), 53 (21%), 54 (20%), 55 (75%), 56 (18%), 57 (18%), 60 (9%), 65 (13%), 66 (24%), 67 (67%), 68 (26%), 69 (53%), 70 (10%), 71 (8%), 73 (11%), 77 (23%), 78 (9%), 79 (53%), 80 (22%), 81 (100%), 82 (42%), 83 (80%), 84 (43%), 85 (12%), 91 (24%), 93 (31%), 94 (20%), 95 (57%), 96 (25%), 97 (22%), 98 (8%), 105 (11%), 107 (20%), 108 (12%), 109 (23%), 110 (16%), 111 (8%), 112 (16%), 115 (11%), 121 (13%), 122 (8%), 123 (13%), 127 (9%), 131 (9%), 133 (14%), 135 (12%), 136 (9%), 137 (18%), 138 (34%), 139 (10%), 145, 149 (10%), 150 (21%), 151 (73%), 152 (13%), 157 (16%), 162 (28%), 164 (17%), 165 (9%), 167, 176 (9%), 177 (10%), 178 (59%), 179 (14%), 180 (9%), 190, 204 (9%), 209, 218, 233 (82%), 234 (15%), 264 (8%), 280 (10%) |
| M12 | 243 | 41 (17%), 42, 43 (91%), 45 (8%), 53, 55 (21%), 56 (8%), 57 (15%), 60 (11%), 65, 67 (18%), 68 (8%), 69 (19%), 73 (16%), 77 (11%), 79 (14%), 80 (8%), 81 (20%), 82 (12%), 87 (12%), 88 (10%), 89 (11%), 91 (8%), 93 (12%), 95 (16%), 96 (8%), 97 (13%), 98 (8%), 99 (10%), 101 (9%), 105 (9%), 109 (10%), 110, 111 (11%), 114 (10%), 115 (43%), 116 (8%), 121 (8%), 127 (11%), 129 (15%), 135, 139 (9%), 143 (100%), 144 (15%), 151 (10%), 152 (11%), 153 (18%), 157 (15%), 165 (8%), 171 (10%), 182 (25%), 185 (50%), 186 (51%), 187 (8%), 199 (8%), 200, 207, 213 (8%), 218, 224, 241 (8%), 284 (10%) |
| M13/14 | 231 | 41 (23%), 43 (23%), 53 (8%), 55 (60%), 57, 60, 67 (15%), 68 (12%), 69 (14%), 73 (10%), 77 (28%), 79 (32%), 80 (10%), 81 (14%), 82 (19%), 83 (29%), 85, 91 (30%), 93 (10%), 95 (15%), 96, 105 (10%), 107 (87%), 108 (10%), 109 (11%), 119 (11%), 129 (9%), 135 (10%), 149 (100%), 150 (10%), 163, 174, 189, 202, 207, 231 (77%), 232 (18%), 280 |
| M15/16 | 229 | 40 (19%), 41 (78%), 43 (90%), 44 (17%), 45 (17%), 53 (21%), 54 (15%), 55 (90%), 56 (25%), 57 (80%), 60 (25%), 65 (18%), 67 (85%), 68 (61%), 69 (58%), 70 (18%), 71 (31%), 73 (30%), 77 (45%), 79 (63%), 80 (32%), 81 (87%), 82 (53%), 83 (43%), 85 (48%), 91 (90%), 92 (20%), 93 (58%), 94 (70%), 95 (93%), 96 (40%), 97 (27%), 103 (21%), 105 (29%), 107 (51%), 108 (26%), 109 (100%), 110 (36%), 111 (20%), 115 (16%), 117 (16%), 119 (61%), 121 (42%), 122 (51%), 123 (30%), 124 (18%), 125 (26%), 129 (28%), 133 (23%), 135 (66%), 136 (18%), 137 (61%), 138 (15%), 145 (16%), 147 (85%), 149 (23%), 151 (34%), 152 (34%), 153 (59%), 159 (17%), 161 (20%), 163 (19%), 164 (19%), 165 (24%), 177 (30%), 189 (28%), 191 (18%), 203 (27%), 205 (19%), 218 (25%), 229 (52%), 259 (17%), 274 (23%), 280 (15%), 284 (16%) |
| M17 | 269 | 41 (17%), 55 (9%), 57 (38%), 63 (10%), 65 (10%), 66 (24%), 67 (20%), 68 (8%), 75 (8%), 77 (58%), 91 (10%), 94 (8%), 95 (9%), 102 (12%), 103 (47%), 110 (12%), 115 (13%), 128 (17%), 129 (20%), 141 (37%), 142 (14%), 152 (18%), 153 (28%), 154 (31%), 155 (24%), 166 (14%), 167 (48%), 168 (11%), 169 (19%), 175, 184, 197 (17%), 212 (9%), 226, 240, 254, 268 (16%), 269 (100%), 270 (21%) |
Note: if no % is indicated between brackets , that means intensity <7% of base peak.
Product ions indicated in bold: characteristic product ions for the amide functional group of the NAA.
4. Conclusion
In this research, the N-alkylamide profiling in two ethanolic plant extracts of the Achillea genus, namely Achillea ptarmica and Achillea millefolium, was performed using two different analytical techniques, HPLC–ESI–MS and GC–EI–MS, allowing tentative structural assignments. Our obtained MS spectra corroborate well with the structures of these NAAs, although full confirmation of the identity can be obtained by nuclear magnetic resonance (NMR) and synthetic standards. In the A. ptarmica extract, a total of 14 NAAs were assigned: six NAAs having an isobutylamide function, three NAAs with a piperidide function, two NAAs with a 2-methylbutylamide function, one NAA with a phenylethylamide function, one NAA having a piperideide function and one NAA having a N-methyl isobutylamide function. Using both analytical methods, compounds P1, P5, P7, P9, P10, P11 and P12 were reported. Compounds P2, P3, P4, P6, P8, P13 and P14 were assigned with HPLC–ESI–MS, but not with GC–EI–MS . Interestingly, it is the first time that compounds P8 and P14 are reported in A. ptarmica. The MS spectra corroborate well with the structures of deca-2E,6Z,8E-trienoic acid 2-methylbutylamide (homospilanthol) or a related isomeric compound and deca-2E,4E-dienoic acid N-methyl isobutylamide, respectively. In the A. millefolium extract, 15 NAAs were assigned: six NAAs having a isobutylamide function, one NAA with a 4-hydroxyphenylethylamide function, one NAA with a 2-methylbutylamide function, two NAAs having a piperidide function, four NAAs with a piperideide function and one NAA with a 4-methoxyphenylethylamide function. Using HPLC–MS and GC–MS, five NAAs were assigned using both analytical techniques: compounds M1, M3, M6, M7 and M8, whereas compounds M2, M4, M5, M9 and M10 were only assigned using HPLC–ESI–MS. Furthermore, five additional NAAs were reported using GC–EI–MS: compounds M11, M12, M13/14, M15/16 and M17. Like in the A. ptarmica extract, the MS spectra of compound M4 in A. millefolium extract corroborate well with the structure of homospilanthol or a related isomeric compound. This is the first time that homospilanthol or a related isomeric compound has been assigned in A. ptarmica and A. millefolium.
Acknowledgments
The authors would like to thank the Special Research Fund of Ghent University (BOF 01D23812 to Lien Taevernier).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Si X.T., Zhang M.L., Shi Q.W. Chemical constituents of the plants in the genus Achillea. Chem. Biodivers. 2006;3:1163–1180. doi: 10.1002/cbdv.200690119. [DOI] [PubMed] [Google Scholar]
- 2.Lazarevic J., Radulovic N., Zlatkovic B. Composition of Achillea distans Willd. subsp. distans root essential oil. Nat. Prod. Res. 2010;24:718–731. doi: 10.1080/14786410802617292. [DOI] [PubMed] [Google Scholar]
- 3.Nemeth E., Bernath J. Biological activities of yarrow species (Achillea spp.) Curr. Pharm. Des. 2008;14:3151–3167. doi: 10.2174/138161208786404281. [DOI] [PubMed] [Google Scholar]
- 4.Rigat M., Bonet M.A., Garcia S. Studies on pharmaceutical ethnobotany in the high river Ter valley (Pyrenees, Catalonia, Iberian Peninsula) J. Ethnopharmacol. 2007;113:267–277. doi: 10.1016/j.jep.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Savikin K., Zdunic G., Menkovic N. Ethnobotanical study on traditional use of medicinal plants in South-Western Serbia, Zlatibor district. J. Ethnopharmacol. 2013;146:803–810. doi: 10.1016/j.jep.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Cavalcanti A.M., Baggio C.H., Freitas C.S. Safety and antiulcer efficacy studies of Achillea millefolium L. after chronic treatment in Wistar rats. J. Ethnopharmacol. 2006;107:277–284. doi: 10.1016/j.jep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Karamenderes C., Apaydin S. Antispasmodic effect of Achillea nobilis L. subsp. sipylea (O. Schwarz) Bassler on the rat isolated duodenum. J. Ethnopharmacol. 2003;84:175–179. doi: 10.1016/s0378-8741(02)00296-9. [DOI] [PubMed] [Google Scholar]
- 8.Tajik H., Jalali F.S.S., Sobhani A. In vitro Assessment of Antimicrobial Efficacy of Alcoholic Extract of Achillea millefolium in Comparison with Penicillin Derivatives. J. Anim. Vet. Adv. 2008;7:508–511. [Google Scholar]
- 9.Stojanovic G., Radulovic N., Hashimoto T. In vitro antimicrobial activity of extracts of four Achillea species: the composition of Achillea clavennae L. (Asteraceae) extract. J. Ethnopharmacol. 2005;101:185–190. doi: 10.1016/j.jep.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Dias M.I., Barros L., Duenas M. Chemical composition of wild and commercial Achillea millefolium L. and bioactivity of the methanolic extract, infusion and decoction. Food Chem. 2013;141:4152–4160. doi: 10.1016/j.foodchem.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Fierascu I., Ungureanu C., Avramescu S.M. In Vitro Antioxidant and Antifungal Properties of Achillea millefolium L. Rom. Biotech. Lett. 2015;20:10626–10636. [Google Scholar]
- 12.Jenabi E., Fereidoony B. Effect of Achillea millefolium on Relief of Primary Dysmenorrhea: a Double-Blind Randomized Clinical Trial. J. Pediatr. Adolesc. Gynecol. 2015;28:402–404. doi: 10.1016/j.jpag.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Potrich F.B., Allemand A., da Silva L.M. Antiulcerogenic activity of hydroalcoholic extract of Achillea millefolium L.: involvement of the antioxidant system. J. Ethnopharmacol. 2010;130:85–92. doi: 10.1016/j.jep.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Pain S., Altobelli C., Boher A. Surface rejuvenating effect of Achillea millefolium extract. Int. J. Cosmet. Sci. 2011;33:535–542. doi: 10.1111/j.1468-2494.2011.00667.x. [DOI] [PubMed] [Google Scholar]
- 15.Hemmati A., Arzi A., Amin M. Effect of Achillea millefolium extract in wound healing of rabbit. J. Nat. Remedies. 2/2. 2002:164–167. [Google Scholar]
- 16.Final report on the safety assessment of Yarrow (Achillea millefolium) Extract IJT. 2001;20(Suppl. 2):79–84. doi: 10.1080/10915810160233785. [DOI] [PubMed] [Google Scholar]
- 17.EMA/HMPC/290309/2009, Assessment report on Achillea millefolium L., herba, 2010
- 18.Althaus J.B., Kaiser M., Brun R. Antiprotozoal activity of Achillea ptarmica (Asteraceae) and its main alkamide constituents. Molecules. 2014;19:6428–6438. doi: 10.3390/molecules19056428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boonen J., Bronselaer A., Nielandt J. Alkamid database: chemistry, occurrence and functionality of plant N-alkylamides. J. Ethnopharmacol. 2012;142:563–590. doi: 10.1016/j.jep.2012.05.038. [DOI] [PubMed] [Google Scholar]
- 20.Greger H., Zdero C., Bohlmann F. Pyrrole amides from achillea-Ageratifolia. Phytochemistry. 1987;26:2289–2291. [Google Scholar]
- 21.Greger H., Hofer O., Werner A. Biosynthetically simple C 18-alkamides from achillea species. Phytochemistry. 1987;26:2235–2242. [Google Scholar]
- 22.Christensen L.P. Acetylenes and Related-compounds in anthemideae. Phytochemistry. 1992;31:7–49. [Google Scholar]
- 23.Greger H., Hofer O. Polyenoic acid piperideides and other alkamides from achillea-millefolium. Phytochemistry. 1989;28:2363–2368. [Google Scholar]
- 24.Greger H., Werner A. Comparative HPLC analyses of alkamides within the achillea millefolium group. Planta Med. 1990;56:482–486. doi: 10.1055/s-2006-961017. [DOI] [PubMed] [Google Scholar]
- 25.Kuropka G., Koch M., Glombitza K.W. Acid amides from achillea ptarmica. Planta Med. 1986;41:244–245. [Google Scholar]
- 26.Cech N.B., Eleazer M.S., Shoffner L.T. High performance liquid chromatography/electrospray ionization mass spectrometry for simultaneous analysis of alkamides and caffeic acid derivatives from Echinacea purpurea extracts. J. Chromatogr. A. 2006;1103:219–228. doi: 10.1016/j.chroma.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Boonen J., Baert B., Burvenich C. LC-MS profiling of N-alkylamides in Spilanthes acmella extract and the transmucosal behaviour of its main bio-active spilanthol. J. Pharm. Biomed. Anal. 2010;53:243–249. doi: 10.1016/j.jpba.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Boonen J., Sharma V., Dixit V.K. LC-MS N-alkylamide profiling of an ethanolic anacyclus pyrethrum root extract. Planta Med. 2012;78:1787–1795. doi: 10.1055/s-0032-1315371. [DOI] [PubMed] [Google Scholar]
- 29.Spelman K., Wetschler M.H., Cech N.B. Comparison of alkylamide yield in ethanolic extracts prepared from fresh versus dry Echinacea purpurea utilizing HPLC-ESI-MS. J. Pharm. Biomed. Anal. 2009;49:1141–1149. doi: 10.1016/j.jpba.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leng T.C., Ping N.S., Lim B.P. Detection of bioactive compounds from Spilanthes acmella (L.) plants and its various in vitro culture products. J. Med. Plants Res. 2011;5:371–378. [Google Scholar]
- 31.Nakatani N., Nagashima M. Pungent alkamides from spilanthes acmella L. var. oleracea Clarke. Biosci. Biotech. Bioch. 1992;56:759–762. doi: 10.1271/bbb.56.759. [DOI] [PubMed] [Google Scholar]
- 32.Costa S.S., Arumugam D., Gariepy Y. Spilanthol extraction using microwave: calibration curve for gas chromatography. Chem. Eng. Trans. 2013;32:1783–1788. [Google Scholar]
- 33.Bae S.S., Ehrmann B.M., Ettefagh K.A. A Validated liquid chromatography-electrospray Ionization-mass spectrometry method for quantification of spilanthol in Spilanthes acmella (L.) Murr. Phytochem. Anal. 2010;21:438–443. doi: 10.1002/pca.1215. [DOI] [PubMed] [Google Scholar]
- 34.Hiserodt R.D., Pope B.M., Cossette M. Proposed mechanisms for the fragmentation of doubly allylic alkenamides (tingle compounds) by low energy collisional activation in a triple quadrupole mass spectrometer. J. Am. Soc. Mass Spectr. 2004;15:1462–1470. doi: 10.1016/j.jasms.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Stuart D.L., Wills R.B.H. Effect of drying temperature on alkylamide and cichoric acid concentrations of Echinacea purpurea. J. Agr. Food Chem. 2003;51:1608–1610. doi: 10.1021/jf026213k. [DOI] [PubMed] [Google Scholar]