Mechanosensing depicts the ability of a cell to sense mechanical cues, which under some circumstances is mediated by the surface receptors. In this review, a four-step model is described for receptor-mediated mechanosensing. Platelet GPIb, T-cell receptor, and integrins are used as examples to illustrate the key concepts and players in this process.
Abstract
Mechanosensing describes the ability of a cell to sense mechanical cues of its microenvironment, including not only all components of force, stress, and strain but also substrate rigidity, topology, and adhesiveness. This ability is crucial for the cell to respond to the surrounding mechanical cues and adapt to the changing environment. Examples of responses and adaptation include (de)activation, proliferation/apoptosis, and (de)differentiation. Receptor-mediated cell mechanosensing is a multistep process that is initiated by binding of cell surface receptors to their ligands on the extracellular matrix or the surface of adjacent cells. Mechanical cues are presented by the ligand and received by the receptor at the binding interface; but their transmission over space and time and their conversion into biochemical signals may involve other domains and additional molecules. In this review, a four-step model is described for the receptor-mediated cell mechanosensing process. Platelet glycoprotein Ib, T-cell receptor, and integrins are used as examples to illustrate the key concepts and players in this process.
INTRODUCTION
In all forms of life, survival is based upon the ability to adapt to environmental pressures, including diverse sets of mechanical forces. Force therefore plays an important role in the shaping, development, and maintenance of tissues and organs. Virtually all organisms have evolved structures from the macroscale (organs, tissues) to the microscale (cells) and nanoscale (molecular assemblies, single proteins) that are sensitive and responsive to myriad forces, including compressive, tensile, shear stress, and hydrostatic pressure. At the cellular level, mechanobiology is concerned with how the cell detects, interprets, responds, and adapts to the mechanical environment. At the molecular level, mechanobiology includes not only enlisting the molecular players and elucidating their interconnections, but also understanding the design and working principles of various mechanosensing machineries so as to re-engineer them for specific applications.
Mechanobiology includes the long history of investigations on mechanosensation, referred to as an organism’s active response to environmental mechanical stimuli, such as the functioning of the auditory and haptic system (Gillespie and Walker, 2001; Ingber, 2006). The received signals travel across multicellular tissues/organs to the central nervous system (along the route of a reflex arc), so as to trigger the awareness of the organism and its response. The initial reception of the mechanical stimulations, although presented in a macroscopic scale, is via somatic cells. Certain membrane proteins are found to convert extracellularly applied mechanical stimuli into intracellular chemical signals by opening/closing channels formed by their transmembrane domains (TMDs) to enable/disable movement of substances across the cell membrane (Ingber, 2006).
Mechanobiology is much broader than mechanosensation that can be initiated only by limited types of neurological cells using “professional” components for reception of highly specific types of mechanical signals. By comparison, a wide variety of other cells in all tissues and organs are endowed with machineries that allow them to sense and respond to mechanical cues in their microenvironment, which are also subjects of mechanobiology research. In these cases, the reception and processing of, and the response to the mechanical signals are all accomplished in a single cell. Receptor–ligand engagement is absent in the initiation of mechanosensation but is required in such important type of mechanosensing—the receptor-mediated cell mechanosensing. In this review, we will focus on receptor-mediated mechanosensing by cells, discuss its requirements and steps, and study how a cell can use such an elegant process to sense and respond to the mechanical environment.
Cells can support mechanical loads via specific or nonspecific structures. As an example of the latter, pressure is borne by the entire cell surface. By comparison, targeted mechanical stimulations are usually applied to specific receptors on cells in direct physical contact with the extracellular matrix (ECM) or adjacent cells through ligand engagement, resulting in receptor-mediated cell mechanosensing. Receptor-mediated cell mechanosensing is of physiological importance, because it plays a crucial role in cell (de)activation, (de)differentiation, proliferation/apoptosis, and many other cellular processes (Orr et al., 2006; Vogel and Sheetz, 2006). For example, focal adhesion assembly enables reinforcement of cells’ attachment to the ECM, which requires initial adhesion to transduce force signals to the cytoskeleton, which then regulates actin (de)polymerization and integrin convergence (Wozniak et al., 2004; Roca-Cusachs et al., 2012). Cell spreading (Qiu et al., 2014), contraction (Lam et al., 2011; Sheehy et al., 2012; Garcia and Garcia, 2014; Iskratsch et al., 2014; Myers et al., 2016), migration (Pathak and Kumar, 2012; Schaefer and Hordijk, 2015; Paluch et al., 2016), and differentiation (Engler et al., 2006; Moore and Sheetz, 2011; Wen et al., 2014) also depend on adhesion receptor–mediated mechanosensing, as evidenced by their sensitivity to substrate stiffness (Elosegui-Artola et al., 2016), which in population can even fulfill sophisticated tissue- and organ-level tasks like ECM remodeling (Cox and Erler, 2011).
In these examples of receptor-mediated mechanosensing, adhesion molecules (e.g., cadherins and integrins) have been suggested as, at least parts of, the mechanosensing machineries, for their perturbation alters whether and how a cell responds to the changing mechanical environment. Using the same strategy, many intracellular proteins have also been identified as candidate molecules that play a role in mechanosensing, including adaptor and scaffolding proteins (e.g., talin and vinculin), kinases and phosphatases (e.g., focal adhesion kinase, FAK), and even cytoskeletal components (i.e., actin filament and microtubule). Unlike cell surface receptors that are at the first-line of mechanosensing, these molecules are further downstream and play different roles, for example, transmitting the mechanical signals into the cell, converting them into biochemical signals, integrating and interpreting these signals for decision making, and dispatching the decisions to the appropriate cellular organelles for actions. There have been a number of excellent reviews for these aspects of mechanobiology (Dahl et al., 2008; Kuo, 2013; Romet-Lemonne and Jegou, 2013; Seong et al., 2013; Haining et al., 2016), and they will not be the focus here.
In addition to responding to externally applied forces, cells can internally generate and exert forces on cell surface receptors. For example, forces from actin polymerization, retrograde flow of the actin cytoskeleton, and myosin II–dependent contraction may be transmitted to T- or B-cell receptors (TCR or BCR) bound to peptide–major histocompatibility complex (pMHC) molecules or antibodies anchored to a surface during signaling activation (Hu and Butte, 2016; Murugesan et al., 2016; Hong et al., 2017), cell motility (Mempel et al., 2004; Kim et al., 2009), and formation of immunological synapses and kinapses (Mossman et al., 2005; Sims et al., 2007; Dustin, 2008; Ilani et al., 2009; Hammer and Burkhardt, 2013). Indeed, internal forces generated by T- and B-cells have been observed being exerted via respectively engaged pMHCs and antigens on the TCR (Bashour et al., 2014a, b; Liu et al., 2016; Ma et al., 2016) and BCR (Wan et al., 2015), respectively. External force applied to the TCR has been shown to induce intracellular calcium flux (Kim et al., 2009; Li et al., 2010; Liu et al., 2014a; Pryshchep et al., 2014; Feng et al., 2017), regulate pMHC dissociation kinetics from the TCR (Liu et al., 2014a; Das et al., 2015) and pre-TCR (Mallis et al., 2015; Das et al., 2016), and potentiate cytotoxic T-cell killing of target cells (Basu et al., 2016). Both T-cells (Axmann et al., 2012; Liu et al., 2014a; Das et al., 2015, 2016; Hong et al., 2015) and B-cells (Natkanski et al., 2013) are able to use mechanical forces to amplify antigen discrimination. In these examples of the emerging field of mechanoimmunology, TCR and BCR represent signal receptors. Unlike receptors for soluble agonists, these receptors bind immobilized ligands and, as such, are subject to mechanical forces. Thus, signaling via these receptors can be modulated or even triggered by force (Upadhyaya, 2017).
Receptor-mediated mechanosensing of a cell is hardly a onetime deal in physiology. Rather, it is usually a complex process composed of many concurrent, sequential, and coordinated signaling events triggered by the same and/or different types of receptors throughout the cell surface (e.g., in focal adhesions, clusters of integrins receive signals simultaneously) over a period of time. Moreover, the ability of cells to provide feedback to the sensed mechanical stimulations and to adjust the force and adhesion strength accordingly (via deformation and modifying cytoskeletal support) enables constant cross-talk between the cell and the environment (Roca-Cusachs et al., 2017). Nonetheless, focusing on the cell sensing of a single wave of unidirectional mechanical signal may simplify the problem and provide a useful angle to begin the mechanistic investigation. In this review, a four-step model is described for the receptor-mediated cell mechanosensing process, covering the presentation, reception, transmission, and transduction of a single wave of mechanical signal. Kinetics of receptor–ligand interaction and mechanics of the molecular players are discussed. Biophysical nanotools commonly used to study receptor-medicated mechanosensing on cells are summarized. Platelet glycoprotein Ib-IX, T-cell receptor, and integrins are used as examples to illustrate the key players in this process.
FOUR-STEP MODEL FOR RECEPTOR-MEDIATED CELL MECHANOSENSING
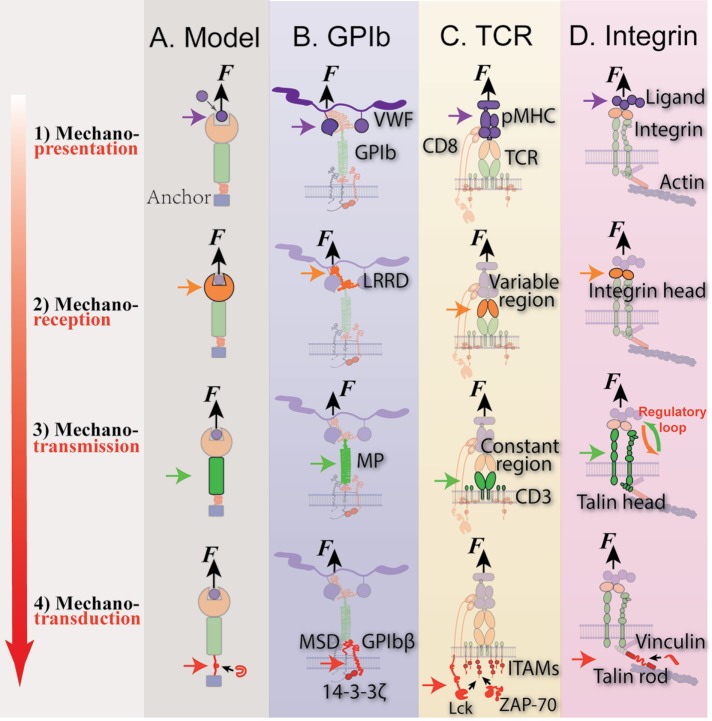
Upon ligand engagement, cell surface receptors can transduce signals across the membrane. When force is exerted upon receptor–ligand bond formation, this can initiate cell mechanosensing. As demonstrated in Figure 1A, in many cases, such a mechanosensing process may be broken down into four steps carried out by distinct structural components of the receptor–ligand axis:
Step 1. Mechanopresentation: In this step, mechanical cues are presented for the cell to sense. If the force to be sensed is exerted to a receptor, mechanopresentation requires a ligand that is anchored on a surface (mechanopresenter) to support the force upon its exertion (Figure 1A, step 1, purple). In contrast, soluble ligands do not sustain force, and therefore cannot present mechanical cues.
Step 2. Mechanoreception: This is the step during which the mechanopresenting ligand is engaged with a cell surface receptor onto which force is exerted (Figure 1A, step 2, orange). The receptor is termed a mechanoreceptor, because it is the molecule that receives the mechanical signal, which may induce conformational changes in the binding site of the receptor or the ligand to alter the bond properties.
Step 3. Mechanotransmission: This is the step performed by the mechanotransmitter, whereby the mechanical signal propagates away from the ligand binding site (the business end of the mechanoreceptor) toward the cell interior (Figure 1A, step 3, green). Note that the propagating mechanical signal is not limited to the mechanical force only. As we will show later in the examples of TCR- and integrin-mediated mechanosensing, propagation of force-induced molecular conformational changes should also be counted as part of mechanotransmission, notwithstanding that it may also facilitate mechanotransduction.
Step 4. Mechanotransduction: This is the step when the mechanical cue is translated into a biochemical signal. Note that this definition is different from that used in some other articles, wherein “mechanotransduction” is referred to as the entire “cell mechanosensing” process. Usually, a certain region of the receptor or its linked subunit(s) undergo(es) conformational changes in response to the force waveform (cf. Figure 2), enabling a biochemical event to occur in the cytoplasm. The molecule that contains the structure(s) undergoing mechanical changes, together with other participants of the chemical event, is termed the mechanotransducer (Figure 1A, step 4, red).
FIGURE 1:
Model of mechanosensing with three exemplary systems (GPIb, TCR, and integrin), which is broken into four steps: 1) mechanopresentation; 2) mechanoreception; 3) mechanotransmission; and 4) mechanotransduction. Purple, orange, green, and red arrows indicate the location of each step carried out by a molecule or molecular assembly: the mechanopresenter, mechanoreceptor, mechanotransmitter, and mechanotransducer, respectively. Black arrows indicates external force, F. Steps of mechanosensing of a generic model (A) and three model systems, GPIb (B), TCR (C), and integrin (D), were depicted in correspondence to the proposed model. Due to the existence of multiple mechanosensing mechanisms in integrins, the talin unfolding mechanism is selected as representative in D.
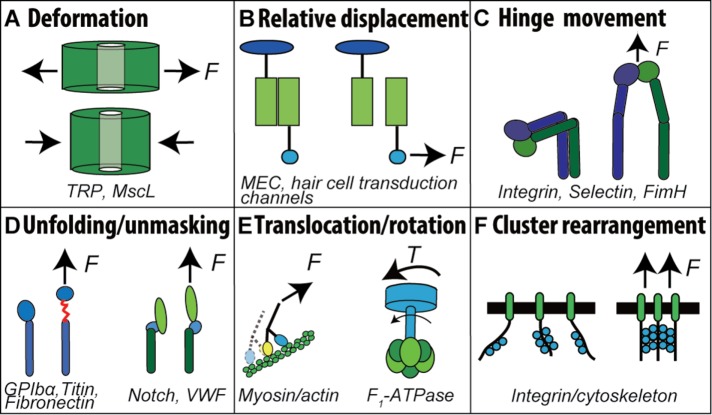
FIGURE 2:
Mechanisms of protein mechanosensitivity. Mechanosensitive proteins contain a motif or motifs that can change structure in response to mechanical forces, giving rise to (A) deformation, (B) relative displacement, (C) hinge movement, (D) unfolding and unmasking, (E) translocation and rotation, and (F) cluster rearrangement.
Importantly, a key property of mechanotransduction is the capability to distinguish different mechanical stimulations via either a digital or analogous mechanism. In other words, it may react to certain stimulations only, remaining inert to others (digital), or the intensity of the triggered biochemical signal may correlate with the strength of the mechanical stimulation (analogous) (Ju et al., 2016). Mechanotransmission and mechanotransduction are similar in some aspects. However, the central distinction between these two steps is that the latter translates mechanical signals into chemical signals, but the former does not. It is also important to note that a molecule may play more than one role in the mechanosensing process. For example, platelet glycoprotein (GP) Ibα acts as the mechanoreceptor, mechanotransmitter, and also part of mechanotransducer in GPIb-mediated cell mechanosensing, which will be demonstrated below in the section that discusses Platelet mechanosensing via single GPIb.
KINETIC AND MECHANICAL ASPECTS
Kinetic constraints of mechanopresentation and mechanoreception
The first two steps of mechanosensing require binding of a mechanoreceptor to an immobilized ligand to allow force to be exerted on the receptor–ligand bond. The receptor–ligand binding kinetics that governs how fast the bond associates and dissociates, quantified by an on-rate and a force-dependent off-rate, are critical to mechanosensing, because they determine the magnitude, duration, and frequency of force application. In other words, kinetics places a constraint on mechanopresentation and mechanoreception, because it limits the frequency, magnitude, and duration of the force that can be presented by a mechanopresenter to a mechanoreceptor. For example, the large volumetric fraction of red blood cells in the bloodstream creates a margination effect that pushes platelets toward the vascular wall (Vahidkhah et al., 2014). This may decrease the gap distance between the von Willebrand factor (VWF)-bearing vascular bed surface and the GPIb-bearing platelet surface, thereby increasing the apparent on-rate of their association (Ju et al., 2015c) and in turn enabling more frequent mechanopresentation and mechanoreception. Even in cases of signal receptors that can be triggered in the absence of force, force may still modulate the signal initiation by impacting the durability of ligand engagement.
At the single-bond level, bond lifetime, that is, the duration of a bond (which is inversely related to off-rate) can be prolonged, unaffected, or shortened by force, defining the catch, ideal, or slip bond behavior, respectively (Dembo et al., 1988). The physics of the catch bond has been an intriguing problem for biophysicists ever since it was first demonstrated in biological experiments (Marshall et al., 2003). Interested readers are referred to reviews that summarized current models of the catch bond and the underlying mechanisms (Rakshit and Sivasankar, 2014; Liu et al., 2015a). Catch and slip bond characteristics can be important for cell mechanosensing via regulation of bond strength or durability. As demonstrated in the TCR (Liu et al., 2014a) and GPIb (Ju et al., 2016) systems, the levels of intracellular calcium triggered by the receptor–ligand bonds display a similar force dependency to their corresponding lifetimes. Interactions of TCR with agonist pMHC and of GPIb with wild-type (WT) VWF form catch-to-slip bonds, and the levels of Ca2+ induced by exerting force on the two receptors show the same pattern, first increasing and then decreasing with increasing force (Liu et al., 2014a; Ju et al., 2016). Furthermore, the force at which the bond lifetime becomes the longest coincides with the force at which the Ca2+ level become the highest for both interactions. Moreover, changing the respective ligands to antagonist pMHC and type 2B von Willebrand disease (VWD) mutant VWF converts their respective interactions with TCR and GPIb to slip-only bonds, and the induced Ca2+ level also changes its pattern, monotonically decreasing with force in both cases (Liu et al., 2014a; Ju et al., 2016).
Mechanical changes in mechanosensing molecules
Like any materials, biomolecules undergo mechanical changes in response to mechanical loads. Molecules of mechanosensing machineries usually possess certain motifs that are sensitive to mechanical perturbations (termed mechanosensitive domains, or MSDs). Mechanical changes in a MSD may carry out certain mechanosensing function by coupling directly or allosterically to a biochemical event, such as binding to, proteolysis by, or enzymatic modification by another molecule (Ingber, 2006). In this process, the mechanical signals are converted into biochemical signals to propagate further downstream inside the cell, fulfilling mechanotransduction. To date, the most-studied mechanisms of mechanical changes can be classified into six categories:
Deformation (Figure 2A): Changes in the environment impose mechanical forces and/or hydrophobic effects onto the mechanosensitive proteins and cause deformation of their global shape. By adopting this mechanism, pore-forming membrane ion channels like transient receptor potential channels and bacterial large-conductance mechanosensitive channels can deform in response to membrane tension change and control the channel opening/closing (Sukharev et al., 1999; Orr et al., 2006).
Relative displacement (Figure 2B): Two subunits of a transmembrane channel are respectively linked with two external structures. Deflections of these structures cause their relative displacement, which mediates the opening/closing of the channel. This mechanism is mostly studied under mechanosensation, such as in deformation of the skin, vibration of a fly’s bristle, and oscillation of a hair cell’s hair bundle (Gillespie and Walker, 2001).
Hinge movement (Figure 2C): For proteins or their subunits that consist of two or more distinct globular domains connected by a hinge region, such as integrin (Luo and Springer, 2006), selectin (Somers et al., 2000), and FimH (Le Trong et al., 2010), force can relieve the conformational constraints and allosterically promote hinge opening. Take integrin as an example: many studies have demonstrated that force applied through ligand binding facilitates the conformational switch from a bent to an extended conformation (Chen et al., 2012, 2016; Springer and Dustin, 2012).
Unfolding and unmasking (Figure 2D): Force can unfold and unmask a specific protein domain to expose cryptic cleaving, binding, or enzymatic sites. For example, several recent studies have demonstrated that force can unfold distinct domains of GPIbα to mediate signal transduction (Zhang et al., 2015; Deng et al., 2016; Ju et al., 2016). Force-induced exposure of a binding site has been shown for intracellular proteins such as talin and vinculin, as well as extracellular proteins such as fibronectin and VWF (Smith et al., 2007; Sing and Alexander-Katz, 2010; Yao et al., 2014; Elosegui-Artola et al., 2016). Force-induced exposure of an enzymatic cleavage site has also been shown for Notch activation (Stephenson and Avis, 2012) and VWF proteolysis (Zhang et al., 2009; Wu et al., 2010).
Translocation and rotation (Figure 2E): External force applied to a protein noncovalently complexed with a subunit or subunits of a polymeric filament can drag the protein to translocate along the filament, repeatedly breaking old bonds with the subunit(s) from one side and forming new bonds with the subunit(s) from the other side. This mode of relative movement has been observed in the traveling of linear motors (myosins, kinesins, and dyneins) on their respective filamentous tracks (F-actin and microtubules) (Cross, 2006; Gebhardt et al., 2006; Kodera et al., 2010; Cross and McAinsh, 2014). Myosins are important in adhesion-mediated cell cytoskeletal rearrangement and deformation, and have been postulated to play a critical role in substrate rigidity sensing of cells (Holle and Engler, 2011). Kinesins mediate chromosome segregation during anaphase, a key step in cell mitosis. Application of an external force can cause backward stepping of a myosin-V on actin, opposite to the direction of ATP-consuming walking (Gebhardt et al., 2006). Rotational movement, on the other hand, occurs in rotary motors, for example, ATPases, where a rod-shaped subunit inserted into the center of a circular complex of subunits can rotate relative to the circular complex (Noji et al., 1997; Nakanishi-Matsui et al., 2010). For example, the F1 portion of ATP synthase, F1-ATPase, hydrolyzes ATP to create a torque that rotates its γ subunit relative to α3β3 subunits; reversely, an externally applied torque can rotate the γ subunit to result in ATP synthesis (Itoh et al., 2004; Rondelez et al., 2005).
Cluster rearrangement (Figure 2F): The external force is dispersed on a group of clustered proteins, which, through their mutual interactions, act on the adjacent structures and change their arrangement. In focal adhesion, a large number of high-affinity integrins are recruited at the dorsal side of the cell, which affects the dynamic structural rearrangement of the cytoskeleton via linkage between the integrin tail and actin network (Schwartz and DeSimone, 2008). This and the “translocation/rotation” model both achieve mechanosensitivity via cooperation within a group of molecules, in contrast to the other “single-molecule” models.
Depending on the specific physiological function of the mechanotransducer, the type of conformational change (Figure 2, A–F) that it adopts may vary. A specific type of conformational change induces a biochemical signal and generates a functional output, yet it also puts forth limitations. For example, the cluster rearrangement mechanism (Figure 2F) found in mechanosensing of large clusters of integrins in focal adhesions is able to facilitate the rearrangement of the actin skeleton. By comparison, on a platelet, the unfolding of the MSD on a single GPIb is apparently not sufficient to mediate the actin network rearrangement. Instead, its intraplatelet calcium flux triggering has to be via a pathway that can be fulfilled by a single GPIb (Ju et al., 2016).
Besides performing mechanotransduction on cell surface receptors and intracellular molecules, mechanical changes can also occur and be important to other steps of mechanosensing. Taking GPIb mechanosensing as an example, in normal blood circulation, VWF multimer, a mechanopresenter in GPIb mechanosensing, adopts a folded, globular conformation that shields the GPIbα binding sites in its A1 domain and the ADAMTS-13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) cleavage site in its A2 domain (Mendolicchio and Ruggeri, 2005). Only when it experiences sufficient fluid shear will its globular structure be straightened. The exposure of the A1 functional site makes it available for GPIbα binding, whereas A2 unfolding allows its cleavage by ADAMTS-13 to reduce its size, which becomes less adhesive. Therefore, the mechanosensitivity of VWF allows its mechanopresentation to be modulated by force before engaging the GPIbα (Fu et al., 2017).
While the two previous subsections discuss kinetics and mechanics separately, the two can be coupled at the level of single receptor–ligand pair. For example, conformational changes can be induced in the receptor or ligand by the bonding force, which in turn modulates their bond strength. Indeed, force has been suggested to expose the cryptic sites of fibronectin, which modifies its binding preference against different integrin species (Vogel, 2006). The leucine-rich repeat domain (LRRD) in GPIbα is unfolded by the force on the VWF–GPIbα bond, which in turn strengthens this interaction. The prolonged bond lifetime in turn increases the unfolding likelihood of the MSD in GPIbα (Ju et al., 2015b), thus enhancing calcium triggering in platelets (Ju et al., 2016). Furthermore, the waveform of the receptor–ligand binding can also affect the occurrence of mechanical events. For example, only abruptly increasing, but not constant, forces are able to unfold the LRRD of GPIbα, whereas both modes of force application are able to unfold the MSD of GPIbα (Ju et al., 2016).
NANOTOOLS FOR STUDYING RECEPTOR-MEDIATED CELL MECHANOSENSING
Dynamic force spectroscopy
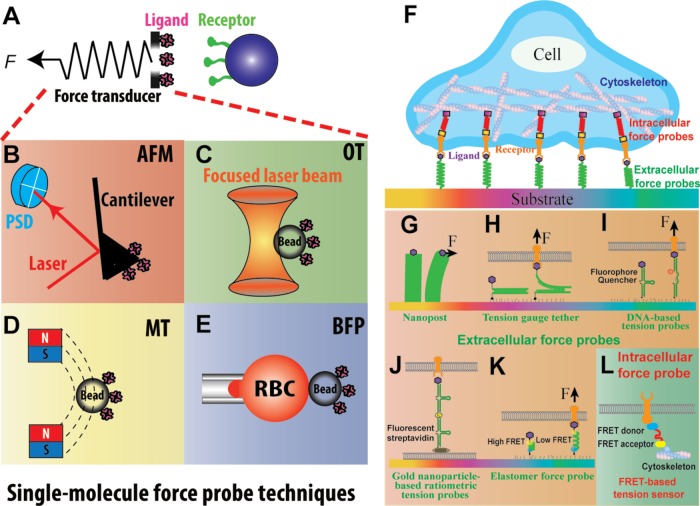
Electron microscopy, crystallography, and antibody mapping are the most commonly used approaches to visualize conformations and characterize behaviors of purified proteins (Springer and Dustin, 2012). However, these approaches only take snapshots of proteins’ stable states and lack real-time details of transient processes. With the exception of antibody mapping, these methods cannot be used to investigate the coupling of protein conformational changes with subsequent signaling events on live cells. Over the past two decades, dynamic force spectroscopy (DFS) (Neuman and Nagy, 2008; Dulin et al., 2013; Liu et al., 2015a) has provided various biomechanical approaches for manipulation, characterization, and visualization of single receptor–ligand interaction and conformational change with tunable force (Liu et al., 2015a). DFS analyses are usually performed using ultrasensitive force probes (Figure 3A) such as atomic force microscopy (AFM; Figure 3B), optical tweezers (Figure 3C), magnetic tweezers (Figure 3D), and biomembrane force probes (BFPs; Figure 3E), but recently they have been conducted using a novel centrifuge force microscope that enables high-throughput measurements (Halvorsen and Wong, 2010; Yang et al., 2016). With nanometer spatial, submillisecond temporal, and piconewton force resolutions, these techniques can be used to induce, follow, and analyze single-molecule mechanical events in real time, thus revealing individual molecular details inaccessible by conventional methods.
FIGURE 3:
Single-molecule force probe techniques (A–E) and microscopic probes for cell tractions and intracellular forces (F–L) techniques. (A) A generic force probe that applies forces (F) to the receptor–ligand bond spanning a surface and a force transducer. (B) Atomic force microscopy (AFM): force is applied to individual molecules tethered between a functionalized cantilever and a surface. (C) Optical tweezers (OT): a protein-coated bead is held by a laser beam. (D) Magnetic tweezers (MT): permanent/electrical magnets are used to manipulate a protein-coated magnetic bead. (E) Biomembrane force probe (BFP): the protein-coated bead is attached to the apex of a micropipette-aspirated red blood cell (RBC). (B–E) Force is determined respectively by cantilever deflection (B), bead displacement (C), gradient of the magnetic field (D), and RBC deformation (E). (F) Extracellular and intracellular tension sensors allow microscopic observation of endogenous forces experienced by different proteins. (G) Nanopost: the deflections of the polydimethylsiloxane posts reflect the lateral components of tractions exerted by adhered cells. (H) Tension gauge tethers (TGTs): DNA strands with defined tension tolerances are repurposed to test the tension required to activate cell adhesion. (I) Molecular tension-based fluorescence microscopic probe. The fluorophore and quencher are coupled to report the force-induced unfolding of the DNA hairpin, thereby unquenching the fluorescence to report the molecular forces. (J) Gold nanoparticle-based ratiometric tension probes on supported lipid bilayer monitor hairpin opening due to force while controlling for clustering of mobile ligands using a secondary fluorescent readout. (K) Elastomer force probes report drop in FRET signal due to the elongation of the nanospring domain upon force application. (L) Intracellular force probes genetically inserted into domains of intracellular proteins allow force measurement of key domains involved in mechanotransduction inside the cell.
In a DFS experiment, automated precise movement brings together ligands and receptors on the respective force probe (i.e., an AFM cantilever in Figure 3B or a bead in Figure 3, C–E) and target (i.e., a bead in Figure 3A, right, a cell, or a molecule-functionalized surface) with controlled contact time, area, and force. The following separation of the two surfaces applies a piconewton-level force to the receptor–ligand bond to induce mechanical changes in the molecules and modulate bond dissociation. Usually, a DFS experiment is set to perform in either the force-ramp or force-clamp mode. For force-ramp DFS, the force is continuously ramped until the bond ruptures (Figure 4C). For force-clamp DFS, the force is ramped to and clamped at a preset level until the bond ruptures. The bond lifetime is the clamp period (Figure 4E). The rupture forces measured in force-ramp DFS and bond lifetimes measured in force-clamp DFS are used to characterize the force-dependent dissociation (off-rate) of receptor–ligand bonds (Liu et al., 2015a). In addition, multiplexed modes of operation can also be used to exert different force waveforms on the molecular bond, for example, jump-and-ramp DFS (Evans et al., 2004) and cyclic-force DFS (Kong et al., 2013; Li et al., 2016a). In general, DFS experiments have been done to analyze force-dependent receptor–ligand (un)binding and protein domain unfolding/refolding. In the context of receptor-mediated cell mechanosensing, the force-dependent receptor–ligand (un)binding may be involved in mechanopresentation and mechanoreception, whereas protein domain unfolding/refolding may be involved in mechanotransmission and/or mechanotransduction. Thus, DFS analysis provides a tool to dissect the steps of a receptor-mediated cell mechanosensing process and examine them one at a time.
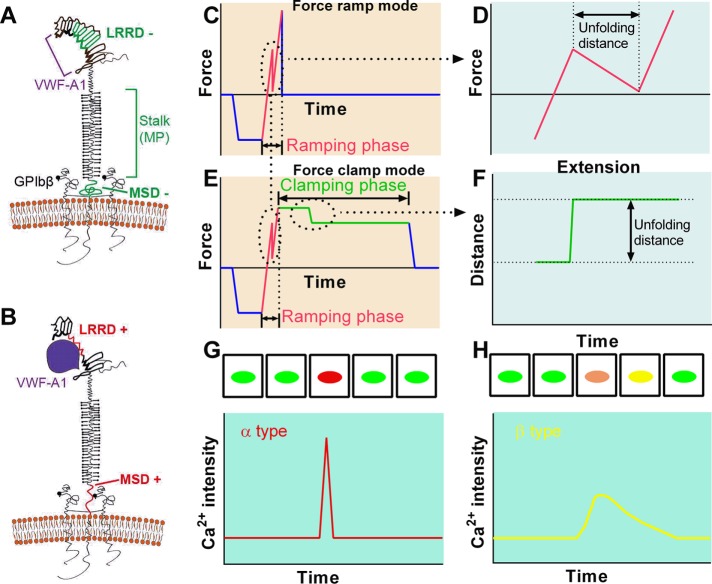
FIGURE 4:
Identification and characterization of GPIb mechanosensing mechanism. (A, B) Schematics of GPIb on the platelet membrane, highlighting the folded (−) and unfolded (+) LRRD and MSD. Ligand binding domain for VWF-A1 and other regions are indicated. (C–F) Illustrative BFP force traces showing zoom-in views of unfolding signatures in both ramping and clamping phases. (G, H) Illustrative analysis of GPIb-mediated single-platelet Ca2+ flux. Top, pseudo-colored images of intracellular Ca2+ in platelets in time sequences. Bottom, time courses of normalized intracellular Ca2+ intensity of the α (G) and β (H) types.
However, DFS alone is not sufficient to fully characterize cell mechanosensing without readout of the intracellular signaling induced by the mechanical cues. To directly observe single receptor-mediated cell mechanosensing events on a living cell requires performing DFS with concurrent imaging of intracellular signaling (e.g., calcium flux), which was recently achieved by us with fluorescence BFP (Liu et al., 2014a; Chen et al., 2015; Ju et al., 2016) and others using fluorescence optical tweezers (Kim et al., 2009; Feng et al., 2017). This allows one to apply a controlled force waveform and concurrently analyze the resulting receptor–ligand unbinding, receptor mechanical events, and intracellular signaling. In the context of investigating receptor-mediated cell mechanosensing, different force waveforms created by the different DFS modes can respectively mimic the abrupt, sustained, and even more complex mechanical cues a cell may receive physiologically. As demonstrated in recent studies, only durable forces applied via force-clamp, but not transient forces applied by force-ramp, on the respective TCR–pMHC and GPIbα–VWF bonds are able to effectively trigger calcium signaling in T-cells and platelets, respectively (Liu et al., 2014a; Ju et al., 2016). This indicates that cells can distinguish different force waveforms and respond by eliciting different calcium signals. More recently, the dual biomembrane force probe (dBFP) has been developed, which allows examination of the signal reception, initiation, and transduction from one receptor to another on a single cell step by step in space and time. It has demonstrated that the mechano-signals induced by TCR–pMHC and GPIbα–VWF lead to the up-regulation of integrin functions on a T-cell and a platelet, respectively (Ju et al., 2017).
Magnetic twisting cytometry
Another powerful technique that has been extensively used in studies of mechanobiology is magnetic twisting cytometry (MTC) (Wang et al., 1993; Kasza et al., 2011). Instead of applying force, as in DFS experiments, MTC applies a twisting torque to the cell surface via an adherent ligand-functionalized magnetic bead (Andresen Eguiluz et al., 2017). It is a high-throughput method, as more than one bead is often placed on the surface of a cell, and the responses of many cells are measured spontaneously. This method often involves the engagement of multiple receptors to the bead surface ligands, because a single receptor–ligand bond cannot bear a torque about long axis of the bond. Yet, from the perspective of each single bond, the load still takes the form of mechanical force. A direct readout of an MTC experiment is the rotational angle of the bead in response to the applied torque. Apparent cell stiffness can be calculated as the ratio of the twisting torque to the rotational angle of the bead. MTC experiments generated the first evidence of integrin-mediated mechanosensing by observing stiffening of the cell body in response to a torque (Wang et al., 1993). Moreover, when MTC was combined with fluorescence microscopy (Zhang et al., 2017), various cell signaling events following the application of torque on cadherin-expressing cells (le Duc et al., 2010; Barry et al., 2014; Kim et al., 2015; Muhamed et al., 2016) and up-regulation of dihydrofolate reductase transcription induced by force-regulated integrin ligation (Tajik et al., 2016) were observed.
Microscopic probes for cell traction and internal force
Mechanosensing includes two aspects: the cell senses mechanical forces exerted upon it and the cell actively reaches out to extract mechanical information embedded in its environment (e.g., stiffness, texture, and microtopology). The latter is important, because probing the mechanical properties of the surroundings can influence a cell’s behavior, for example, leading cancer cells to become metastatic and mesenchymal stem cells to differentiate into other specialized cell types (Vogel and Sheetz, 2006; DuFort et al., 2011). A major limitation of the ultrasensitive force techniques depicted in Figure 3, A–E, is that they externally apply forces to the cell but provide no information on whether, and if so, how, the cell actively exerts force, and what cytoplasmic structures support such force intracellularly. To overcome such limitations, a variety of microscopic probes have been developed in recent years to visualize cell tractions and internal forces (Figure 3F). The limitation, however, is that, unlike DFS, which can control the amount of mechanical stimulation fed to the cell, experiments using microscopic probes are incapable of observing cell mechanosensing of a single wave of mechanical signal, but rather provide a summed result of all mechanosensing events that occurred over the whole cell–substrate interface. Moreover, they cannot dissect the individual steps of mechanosensing independently, again unlike DFS.
The first class of the microscopic probes, traction force microscopy, measures bulk traction forces generated by adherent cells. A cell is seeded onto a ligand-coated surface of an elastic substrate, the deformation of which is measured microscopically. The substrate is either an elastic hydrogel containing fluorescent fiducial beads (Plotnikov et al., 2014; Muhamed et al., 2016) or arrays of flexible micro/nanoposts (Figure 3G) (Tan et al., 2003; Fu et al., 2010). The substrate stiffness in both cases is measurable, so that deconvolution of the fiducial beads’ displacements or deflections of the micro/nanoposts can generate a map of traction force (Sabass et al., 2008; Jin et al., 2017). A three-dimensional version of this technique has also been developed by seeding cells within a cubical hydrogel/biopolymer matrix. Fluorescently tagged beads, again, were used as indicators of local deformation (Legant et al., 2010; Toyjanova et al., 2014; Steinwachs et al., 2016). In the context of cell mechanosensing, traction force microscopy has been instrumental in understanding focal adhesion-related cell mechanosensing (Legant et al., 2010) and the adhesion activity of T-cells (Basu and Huse, 2016). Working with MTC in parallel, traction force microscopy has been used to study how force priming on cadherin cooperated with cell rigidity sensing to induce intracellular signaling (Andresen Eguiluz et al., 2017). Combining with other live-cell imaging methods, traction force microscopy allows one to concurrently visualize the distribution and dynamics of traction forces and relevant molecular players during cellular processes (Feghhi et al., 2016). Furthermore, using the basic setup of traction force microscopy, a technique called monolayer stress microscopy has been developed, in which a cluster or monolayer of cells instead of a single cell is seeded. The tension on the junction of adjacent cells and the internal pressure of the cells are calculated using Newton’s laws, given that force balance is achieved on each individual cell (Trepat and Fredberg, 2011). This technique is capable of capturing the mechanical interaction between the cells; however, it cannot provide accurate absolute values for the force parameters, because the mechanical properties of cells are still unknown in such settings (Roca-Cusachs et al., 2017).
The second class of microscopic force probes inserts a polymer (e.g., a double-stranded DNA or a DNA hairpin) between the ligand and the substrate anchor, which extends when the tension applied via engaged cell receptors exceeds a threshold. Two types of DNA-based force probes have been developed, one to limit and the other to report tension above a designed threshold on a ligand: 1) tension gauge tethers (TGTs), for which the idea is to tether ligands to a substrate with a double-stranded DNA that has a defined tension tolerance for rupture (Wang and Ha, 2013; Luca et al., 2017) (Figure 3H). When force generated by the cell and applied through the receptor–ligand bonds exceeds the tolerance force, the TGT irreversibly ruptures and reduces the availability of the tethered ligands, thereby inhibiting force-dependent cellular functions such as spreading, migration, or signaling. By observing whether such functions are inhibited in cells seeded on ligands tagged to TGTs with a range of tolerance forces (from 12 to 56 pN), one can define the force required by such functions. 2) Molecular tension-based fluorescence microscopy probes, in which a DNA hairpin is used that unfolds under a threshold force (ranging from 4 to 16 pN) but refolds upon force removal. The two feet of the hairpin are conjugated with a fluorophore–quencher pair that prevents/allows fluorescent signal when the hairpin is folded/unfolded, thereby reporting whether the force exceeds the unfolding threshold (Blakely et al., 2014; Zhang et al., 2014; Liu et al., 2016) (Figure 3I). Because unfolding is reversible, this design should not affect cell functions, which allows molecular tension-based fluorescence microscopy probes to visualize the spatiotemporal distributions of tensions on the receptor–ligand bonds under more physiological conditions. The sensitivity of these DNA-based force probes can be enhanced by immobilizing DNA hairpins on gold nanoparticles, because the fluorophore will be dual-quenched by both the molecular quencher and gold nanoparticle to yield ∼100-fold higher signal-to-noise ratio (Liu et al., 2016). In addition, the immobility of the probes can be overcome by anchoring on supported lipid bilayers (Figure 3J) (Ma et al., 2016), allowing the probes to provide diffusible ligands for mechanosensing by cells. Taking advantage of the fluorescence reporting method, DNA-based force probes also afford the possibilities of dual/multicolor imaging of multiple receptor–ligand species to study their interplay in complex systems.
The third class of microscopic force probes is similar to DNA-based force probes, only with the DNA strands replaced by elastic polypeptides. It can report the dynamics and distribution of forces not only extracellularly (Morimatsu et al., 2013) (Figure 3K) but also intracellularly when the polypeptides and the reporter (e.g., fluorescence resonance energy transfer [FRET] pair or an organic fluorophore) are genetically encoded into an intracellular protein (Grashoff et al., 2010) (Figure 3L). A well-known example of polypeptide-based force probes is the tension sensor module (TSMod), which consists of an elastic domain flanked by two fluorophores whose fluorescence intensity is most sensitive to force in the regime of 1–6 pN. TSMod was originally encoded into vinculin to measure force applied on intracellular vinculin molecules during their recruitment to developing focal adhesions (Grashoff et al., 2010). It has also been inserted along the cytoplasmic tails of the integrin αL and β2 subunits to show that migrating cells exert force through the β but not the α subunit (Figure 3L) (Nordenfelt et al., 2016). Incorporation of these intracellular tension probes into various key players in the cell will allow researchers to monitor tensions inside the cell in response to specific mechanical perturbations, which is especially advantageous for studying mechanotransduction.
The various tension-sensing probes have provided researchers with a set of tools to measure force requirement of cellular behaviors and observe force loading and relaxation on a cell. Although the current force ranges of these probes cannot provide a “one-size-fits-all” solution to all receptor types, the iteration of new probe designs continues to push boundaries on the capabilities of these probes. Currently, single-molecule imaging of the probes has not been attained due to limitations in signal detection. Nevertheless, single molecule–level sensitivity and receptor force monitoring using current probe designs may become a reality in the near future as better imaging methods continue to mature.
PLATELET MECHANOSENSING VIA SINGLE GPIb
Next, we will use three systems to illustrate the concepts of receptor-mediated cell mechanosensing outlined earlier. The first example to be looked at in this section is platelet mechanosensing via GPIb receptor. Platelets play a central role in hemostasis and thrombosis. Being anuclear cells with simplified signaling machinery and rapid responses to highly variable mechanical environments, platelets represent a natural model for studying mechanosensing. The subunit of GPIb (GPIbα) contains an N-terminal LRRD that interacts with the VWF-A1, a highly glycosylated long stalk region (macroglycopeptide region, MP) (Fox et al., 1988), a juxtamembrane MSD (Zhang et al., 2015), a single-span TMD, and a short cytoplasmic tail (CT) (Figure 4A). GPIbα is covalently linked to GPIbβ through disulfides, and together they associate tightly with GPIX to form the GPIb–IX complex (referred as GPIbβ from here below) (Luo et al., 2007b; McEwan et al., 2011) and interact with cytoplasmic adaptor proteins such as 14-3-3ζ (Figure 1B). The plasma protein VWF has a limited binding potential for platelet GPIb in circulation due to autoinhibitory mechanisms that mask the GPIbα binding site on VWF-A1 by the adjacent D′D3 domain (Ulrichts et al., 2006), the N-terminal flanking region of the A1 domain (residues 1238–1260) (Auton et al., 2012; Ju et al., 2013; Deng et al., 2017), and the A2 domain (Martin et al., 2007; Aponte-Santamaria et al., 2015). Once VWF is immobilized onto subendothelial collagen and subject to flow, hemodynamic drag forces stretch the macromolecule to adopt an elongated conformation, which facilitates the engagement of A1 to GPIbα (Barg et al., 2007; Schneider et al., 2007). Thus, VWF is mechanosensitive and responds to force by unfolding and exposing its A1 domain (VWF activation). Upon binding of GPIbα to immobilized VWF in the presence of shear, intraplatelet Ca2+ is rapidly triggered to activate integrin αIIbβ3. Thus, GPIb-mediated mechanosensing is the first step of the platelet adhesion and signaling cascade for which GPIb has long been indicated to be the initiating mechanoreceptor (Mazzucato et al., 2002; Nesbitt et al., 2002), and VWF-A1 is the mechanopresenter in this hemostatic process.
VWF–GPIbα binding kinetics
Platelet adhesion to the vascular surface usually occurs in a mechanically stressful environment of blood circulation. Upon vascular injury, VWF in plasma is immobilized onto the subendothelium. It becomes a ligand for mechanopresentation, because it presents the binding motif to GPIbα, the mechanoreceptor, on embarking platelets under shear flow. The rapid VWF–GPIbα association enables the capture of fast-flowing platelets, and rapid bond dissociation allows platelets to translocate (Doggett et al., 2002, 2003; Kumar et al., 2003; Yago et al., 2008; Coburn et al., 2011). In blood flow, physical transport—convection and diffusion—drives blood cells to collide with the vascular surface, bringing interacting receptors and ligands into close proximity (Yago et al., 2007). The three distinct steps of the transport mechanism have been demonstrated to regulate the VWF–GPIbα association: tethering of platelet to the vascular surface, Brownian motion of the platelet, and rotational diffusion of the interacting molecules (Ju et al., 2015c).
Moreover, flow shear results in force on VWF–GPIbα bonds to modulate their dissociation kinetics, eliciting catch bond to prolong bond lifetime at forces <22 pN (Yago et al., 2008; Ju et al., 2013). At forces >22 pN, VWF–GPIbα interaction displays the ordinary slip bond behavior. Such force-dependent kinetics governs the counterintuitive flow enhancement of platelet adhesion on VWF (Savage et al., 1996; Zhu et al., 2008a), which is crucial to the recruitment of platelets to sites of injury in the arterioles. On the other hand, VWF-A1 mutations exhibiting the phenotype of type 2B VWD form slip-only bonds with GPIbα and eliminate flow-enhanced platelet adhesion (Yago et al., 2008; Ju et al., 2013). Furthermore, force-dependent activation of VWF is also affected by its interaction with collagen upon immobilization at sites of vascular injury. DFS and two-dimensional affinity measurements suggest relative contributions of distinct VWF domains, such that the initial VWF capture is mediated by collagen interaction with the A3 domain while the subsequent VWF activation is mediated by interaction with the A1 domain (Ju et al., 2015a). Given that VWF represents an excellent example of a mechanopresenter (Figure 1B), the relative contributions of its distinct domains (and their synergy) to mechanopresentation remain as an interesting topic for future studies.
Taken together, the flow/force-enhanced platelet adhesion mediated by VWF–GPIbα involves at least three mechanisms that directly affect the binding kinetics: 1) relief of the VWF autoinhibitory mechanism involving interdomain associations within the A1A2A3 tridomain and between A1 and D′D3 to expose the GPIbα binding site; 2) enhancement of VWF–GPIbα association by transport; 3) enhancement of A1 interaction with GPIbα by the catch bond. As discussed in the section that demonstrates Platelet mechanosensing via single GPIb, the force-dependent kinetic properties of VWF–GPIbα interaction have a significant impact on platelet mechanosensing via GPIbα, not only in their effect on the duration of the force application, but also in both mechanotransmission and mechanotransduction.
Force-induced GPIbα domain unfolding mechanics
Two types of mechanical events have been observed when the GPIbα molecule is pulled by force, namely, unfolding of the LRRD and MSD (Ju et al., 2015b; Zhang et al., 2015). The former was identified using combined molecular dynamics (MD) simulations and BFP experiments (Ju et al., 2015b, 2016). The latter was identified by optical tweezers experiments and mutagenesis studies (Zhang et al., 2015). Pulling GPIbα on the platelet surface, both types of unfolding events are possible, but the two can be distinguished by their different unfolding lengths and by their differential susceptibilities to varying force waveforms. Unfolding of LRRD and MSD generates lengths of around 36 and 20 nm, respectively. Ramped force can unfold both LRRD and MSD (Figure 4, C and D), whereas clamped force can only unfold MSD (Figure 4, E and F). Unfolding of the juxtamembrane MSD following pulling on the GPIbα headpiece indicates the propagation of force along the GPIbα MP (macroglycopeptide) stalk, which defines its role of mechanotransmission.
Coupling between unbinding kinetics and the unfolding mechanics
Interestingly, the occurrence frequency of MSD unfolding was found to depend on the level of clamped force in the same manner as does the VWF–GPIbα bond lifetime. This is true for both cases of the WT VWF that forms a catch-slip bond with GPIbα and the type 2B VWD mutant VWF that forms a slip-only bond (Ju et al., 2016), despite the fact that MSD unfolding rate is accelerated by force, that is, behaving as a slip bond regardless of whether GPIbα is pulled by WT or the mutant VWF. These data suggest a coupling between the unbinding kinetics and the unfolding mechanics. Indeed, BFP experiments showed that LRRD unfolding prolonged VWF–GPIbα bond lifetime (Ju et al., 2015b, 2016). A simple interpretation for this is a force-induced fit mechanism wherein LRRD unfolding exposes cryptic binding site(s) for VWF-A1, as suggested by MD simulations (Ju et al., 2015b).
In contrast to the membrane-distal LRRD, the juxtamembrane MSD is separated from the VWF binding site by the >30-nm MP region. The highly glycosylated MP region is thought to be poorly structured and not known to propagate conformational changes allosterically (Ju et al., 2016). However, it can transmit tensile force from the LRRD to MSD. Thus, the simplest explanation for the coupling between VWF unbinding kinetics and MSD unfolding kinetics is that unfolding of MSD requires sustained force and cannot occur after force removal by VWF unbinding. Indeed, a mathematical model based on this idea predicts the MSD unfolding frequencies very well (Ju et al., 2016).
Importantly, the coupling between ligand binding kinetics and the domain unfolding mechanics generates cooperativity between LRRD and MSD unfolding, such that LRRD unfolding greatly increases the probability of MSD unfolding. This cooperativity occurs only when GPIbα is pulled by WT VWF but not by type 2B VWD mutant VWF, and reaches maximum at 25 pN, where the WT VWF–GPIbα bond lifetime is the longest (Ju et al., 2016).
MSD unfolding and GPIb mechanotransduction
Recent work has demonstrated that unfolding of MSD results in platelet intracellular signaling (Deng et al., 2016; Ju et al., 2016). The R. Li and X. Zhang groups have combined protein engineering and structural analysis with force ramp DFS via optical tweezers to identify the MSD (Zhang et al., 2015) and its Trigger sequence (Deng et al., 2016). Binding of VWF under physiological shear induced MSD unfolding and intracellular signaling in the platelet. Furthermore, mutations that unfolded the MSD and the Trigger sequence therein induced calcium fluxes, filopodia formation, and P-selectin expression in the absence of ligand binding (Deng et al., 2016).
At the level of single molecular interactions, the role of MSD unfolding in GPIb-mediated mechanosensing has been demonstrated using a fluorescence BFP (fBFP), which enables real-time single-molecule, single-cell correlative analyses of ligand binding kinetics, receptor domain unfolding, and intraplatelet calcium imaging (Ju et al., 2016). Single-platelet calcium imaging has revealed two types of intracellular Ca2+ fluxes induced in platelets interrogated by a VWF-coated probe that forms infrequent, sequential, and intermittent single bonds with GPIbα (Figure 4, G and H): 1) α type, which features an initial latent phase followed by a high spike with a quick decay (Figure 4G); and 2) β type, which features fluctuating signals around the baseline or gradually increasing signals to an intermediate level followed by a gradual decay to baseline (Figure 4H) (Ju et al., 2016). In-depth time-lapse correlative analyses have revealed that MSD unfolding is required to trigger α-type Ca2+. Furthermore, the GPIb example meets the criterion for receptor-mediated cell mechanosensing, that differential information embedded in the force waveform received by the receptor is transduced into distinct biochemical signals. Indeed, it was found that it is the durable force at an optimal level, not the transient force, that triggers maximum calcium. This provides a mechanistic explanation for the bleeding phenotype of VWD patients. The WT VWF–GPIbα catch bond enables sustained binding under high forces to unfold MSD and trigger robust Ca2+ for platelet activation. The conversion of the VWF–GPIbα catch bond to a slip bond in type 2B VWD patients (Yago et al., 2008; Ju et al., 2013) cannot support sustained binding under high forces, and thus fails to trigger adequate mechanosensing (Ju et al., 2016). Moreover, an interfering peptide that disrupts the interaction of the cytoplasmic adaptor protein 14-3-3ζ to GPIbα (Dai et al., 2005) abolishes the α-type Ca2+, implicating the role of 14-3-3ζ as a signal-transducing protein for GPIb-mediated mechanosensing (Ju et al., 2016).
These findings concerning GPIb-mediated mechanosensing can be summarized as follows:
Mechanopresentation (Figure 1B, step 1): The ligand VWF undergoes local and global conformational changes to enhance the GPIbα binding capacity to its A1 domain.
Mechanoreception (Figure 1B, step 2): The LRRD binds the VWF-A1 and receives the force signal. Force induces LRRD unfolding, which in turn prolongs the duration of force application.
Mechanotransmission (Figure 1B, step 3): Force propagates from the LRRD through the MP stalk (Figure 4A) to induce unfolding of the MSD in the juxtamembrane region. The interplay between the VWF–GPIbα bond lifetime and time required to unfold the MSD drives them to follow the same force dependency. The coupling between ligand binding and receptor unfolding results in the cooperative unfolding of LRRD and MSD without the need for allosteric changes in the MP.
Mechanotransduction (Figure 1B, step 4): To convert mechanical cues to biochemical signals requires exposure of the Trigger sequence within the MSD and association of 14-3-3ζ to GPIb without inhibition by the interfering peptide.
Structurally, GPIbα may represent a class of mechanoreceptors, containing a distal ligand binding domain and a long, repeated sequence and/or a heavily glycosylated region to connect to the cell membrane. Notch may be another example in this class of receptors. Like GPIbα, Notch1 forms a catch-slip bond with ligand Jagged1 to prolong interactions in the range of forces required for Notch activation (Luca et al., 2017). Pulling generated by endocytosis induces unfolding of the juxtamembrane negative regulatory region, resulting in shedding of its extracellular domain (Stephenson and Avis, 2012) and initiating Notch biochemical signaling inside the cell (Luca et al., 2015). Although GPIbα bears little resemblance to Notch in sequence, structure, or function, their mechanosensing models are remarkably similar in three key aspects. First, both receptors form catch-slip bonds with their respective ligands to prolong the duration of mechanoreception. Second, both receptors contain a polypeptide sequence for long-distance mechanotransmission. Third, force induces unfolding of a juxtamembrane MSD, converting the mechanical cue into a conformational change. Therefore, it seems reasonable to propose that GPIbα may represent a fundamental and evolutionarily conserved signaling mechanism used by cells to sense their mechanical environment.
T-CELL MECHANOSENSING VIA TCR
Engagement of TCR by pMHC on antigen-presenting cells (APCs) to trigger intracellular signaling is central to T-cell development and function. The TCR complex includes the αβTCR heterodimer noncovalently associated with CD3εγ and CD3εδ heterodimers and a CD3ζζ homodimer (Figure 1C). The αβTCR binds pMHC ligand via the membrane distal end, but its very short CT contains no signaling motif. Conversely, the CTs of the CD3 chains contain a total of 10 immunoreceptor tyrosine-based activation motifs (ITAMs). TCR signaling has been extensively analyzed experimentally and modeled based on a number of hypothetical mechanisms. Yet it is still not clear how TCR–pMHC interaction gives the sensitivity and specificity for T-cells to recognize rare pathogenic antigens and discriminate them from abundant self-peptides. Nor is it clear how the TCR is triggered, that is, how information embedded in the pMHC is sensed by the TCR at the ligand binding site, transmitted across the membrane, and transduced into signaling events in terms of phosphorylation of CD3 ITAMs (van der Merwe and Dushek, 2011; Zhu et al., 2013a; Malissen and Bongrand, 2015). Unlike GPIb, whose functions in hemostasis include providing adhesion force, hence giving force an ample opportunity to carry mechanical signals, TCR-mediated mechanosensing is a relatively new idea. In the next section, we compare features of TCR-mediated mechanosensing with those described earlier for GPIb-mediated mechanosensing. We suggest that viewing the TCR signaling process through the lens of the four-step model for receptor-mediated cell mechanosensing helps delineate key properties of T-cell antigen recognition.
TCR-mediated mechanosensing
Increasing evidence has demonstrated that the TCR can mediate T-cell mechanosensing. T-cells can sense the rigidity of anti-CD3 antibody–coated substrate at least in part by TCR signaling (Judokusumo et al., 2012; O’Connor et al., 2012). AFM measurements show that T-cells develop adhesion forces with pMHC-presenting APCs in a manner that correlates with the peptide’s potency to induce T-cell calcium and interleukin-2 secretion (Lim et al., 2011). Measurements by traction force microscopy (Hui et al., 2015) and micro/nanoposts (Figure 3G) (Bashour et al., 2014a, b) revealed that cytoskeletal forces are generated and exerted by T-cells during activation through TCR (Basu and Huse, 2016). As more direct evidence, several groups have observed intracellular calcium induction by exerting force through the TCR or CD3 chains, both tangential (Kim et al., 2009; Li et al., 2010; Feng et al., 2017) and normal (Li et al., 2010; Liu et al., 2014a; Pryshchep et al., 2014; Feng et al., 2017) to the cell surface. More recently, oscillatory forces applied to TCR–pMHC bonds was shown to rescue F-actin–dependent T-cell signaling, decoupling the actin network from TCR proximal signaling (Hu and Butte, 2016). All these studies highlighted the importance of force in TCR signal triggering.
Although exerting external force to the TCR could induce T-cell signaling, it was not clear how the forces applied were related to endogenous forces experienced by the TCR. The rigidity-responsive mechanosensing and force-generation processes were clearly mediated by the TCR, but they occurred on the order of minutes when T-cells usually form an organized immunological synapse of adhesion and signaling molecules that convolute the independent mechanosensing of, and force exerted on, the TCR. These issues were addressed by two recent studies using the DNA hairpin force probe (Figure 3J), demonstrating that T-cells could exert 12-19 pN tension on pMHC via TCR and/or CD8 bonding (Liu et al., 2016; Ma et al., 2016). This finding also supports the proposal that, even at early time points, the TCR–pMHC bond is likely subject to mechanical forces (Malissen and Bongrand, 2015).
Force-modulated TCR–pMHC kinetics
Although most kinetic analyses of TCR–pMHC interactions were conducted when (or assumed that) no force was exerted on these bonds, several recent studies demonstrated that externally applied forces alter TCR–pMHC dissociation kinetics (Robert et al., 2012; Liu et al., 2014a; Das et al., 2015; Hong et al., 2015). Similar to VWF–GPIbα interactions, agonist pMHCs form catch-slip bonds with TCR, and like the type 2B VWD mutant VWF, weak agonist/antagonist pMHCs form slip-only bonds with TCR. This peptide-dependent formation of catch and slip bonds enables force to amplify antigen discrimination (Liu et al., 2014a; Das et al., 2015; Hong et al., 2015). Differential prolongation of bond lifetimes for rare agonists over abundant self-peptides can enhance context discrimination, a well-known property of T-cell antigen recognition, to distinguish between appropriate immune response and autoimmunity, respectively. The idea follows the TCR kinetic proofreading model, which postulates that completion of a series of reaction steps must occur while the TCR is bound to pMHC (McKeithan, 1995; Rabinowitz et al., 1996) or upon rebinding (Dushek et al., 2009; Jansson, 2011) to achieve T-cell activation.
Force-induced mechanical events and TCR mechanotransmission
Like GPIbα that can be unfolded by tension applied via engaged VWF, pulling a TCR–pMHC bond can induce structural changes in the molecular complex, yielding an ∼10-nm extension (Figure 5, left). Interestingly, similar to GPIbα LRRD unfolding that prolongs GPIbα–VWF lifetime, this structural change impacts the TCR–pMHC dissociation such that catch bonds are only formed after the extension, suggesting a correlation between the binding kinetics and the structure of the TCR–pMHC complex. This idea has also been supported by other work (Das et al., 2016; Schamel et al., 2017). It is believed that the well-structured FG loop that is present in the Cβ chains of both TCR and pre-TCR contributes to such coupling, because its elimination diminished or abolished the catch bonds for TCR and pre-TCR, respectively, while stabilizing the FG loop greatly enhances those catch bonds (Das et al., 2016). Moreover, the correlation between manipulating hydrophobic hot spots and influencing force-driven kinetic readouts such as double catch bonds highlighted the importance of studying TCR triggering under force (Das et al., 2016).
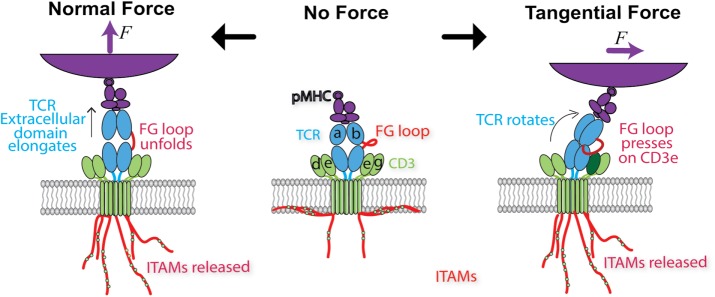
FIGURE 5:
Models of how force may trigger TCR signaling. Middle, schematic of the ligated, unloaded, and untriggered TCR. Soluble pMHC binds to the TCR V domains, while the CTs of the TCR-associated CD3 chains remain buried in the lower leaflet of the cell membrane, preventing ITAM phosphorylation. Left, a force normal to the cell membrane pulls on the TCR, extending the length of the complex by ∼10 nm. While the structural region responsible for such conformational change has not been identified, here the FG loop connecting the Cβ and Vβ domains is assumed to unfold to result in an extended conformer and in catch-bond formation. Force propagated across the TCR-CD3 connection is assumed to release the CD3 CTs for phosphorylation of the ITAMs. Right, when a force tangential to the cell surface is applied to the ligand binding site of the TCR that also experiences a lateral reaction force from its membrane anchor, a torque is generated to rotate the complex, which is assumed to allow the FG loop to press down on the CD3ε ectodomain to expose the cytoplasmic ITAMs in a piston-like manner.
The extension in the TCR–pMHC complex correlates with the biological activity of the peptide, suggesting a role of such a structural change in TCR-mediated mechanosensing (Das et al., 2015, 2016). Because the location where this structural change occurs is far from cytoplasm signaling motifs, its role in mechanosensing is likely that of mechanotransmission. The mechanoreception and mechanotransmission is naturally coupled, because the longer the application time, the more likely the force would induce structural changes in the TCR–pMHC complex. This coupling also allows the antigen discrimination property of TCR that produces distinctive force-dependent bond lifetimes for different antigens (Liu et al., 2014a; Das et al., 2015; Hong et al., 2015) to be passed on from the mechanoreception step to the mechanotransmission step.
Models of TCR mechanotransduction and the effect of force direction
Both force and the change in the TCR–pMHC structure that induces extension propagate along the binding axis, until reaching downstream of the TCR C domains, where a conformational change is thought to be responsible for dislodging the CD3ε and ζ CTs from the inner leaflet of the plasma membrane, to make ITAMs accessible for phosphorylation, resulting in mechanotransduction (Figure 5) (DeFord-Watts et al., 2011; Bettini et al., 2014; Dobbins et al., 2016; Li et al., 2017b). This view is supported by a recent finding that three different conformational states of CD3ε cytoplasmic tails exist with various degrees of association with the plasma membrane (Guo et al., 2017). The authors suggested that changes in these conformations could be triggered by mechanical force generated by TCR–pMHC binding. However, how the extension in the TCR–pMHC complex is related to conformational changes in the TCR-CD3 complex has not been defined. This is partially due to the fact that the structural data of the membrane-embedded TCR-CD3 complex are still not available.
The concept of making ITAMs more available for phosphorylation provides a basis for two models of TCR-mediated mechanotransduction. First, a structurally detailed mechanosensor model (Wang and Reinherz, 2012) proposes that lateral force applied via the pMHC generates a torque to rotate the αβTCR. With the TCRβ TMD acting as a fulcrum, the FG loop pushes on the CD3εγ to cause a piston-like movement downward through the membrane, exposing the cytoplasmic ITAMs for phosphorylation (Figure 5, right). The second model, which also highlights the importance of the TMD (Lee et al., 2015), postulates a steady-state divarication of the CD3ζζ juxtamembrane regions, with the N-terminal CD3ζζ TMD acting as the pivot point. Upon pMHC engagement, a conformational change occurs, forcing the CD3ζζ CTs toward each other from the divarication, providing a potential means of transferring mechanical energy into biochemical energy by exposing ITAMs for phosphorylation by kinases (Lee et al., 2015). Unlike the GPIb-mediated mechanosensing model, in which only the magnitude of force is considered, the above TCR-triggering models consider the direction of force as a key parameter (Figure 5). Also, unlike the GPIb-mediated mechanosensing model, in which the mechanoreceptor can only support linear tension, the TCR-triggering models require that the mechanoreceptor also supports rotational torque.
The mechanical changes in the TCR-CD3 complex may depend on the direction of force applied on the TCR–pMHC bond for several reasons. From a structural perspective, the TCR Cα and Cβ domains vary in size and form different angles with their respective V domains, creating a cavity below the β chain (Kim et al., 2012). Taken together with the CD3 heterodimers that organize around this asymmetry (Birnbaum et al., 2014), force applied to the αβTCR may be productive only in specific directions. Furthermore, the FG loop, which connects the Vβ and Cβ domains, adds to the structural asymmetry and appears to play a functional role, as described earlier. From the ligand perspective, different peptide or antibody docking topologies of TCR have differential capacities to trigger T-cell signaling (Kim et al., 2009; Adams et al., 2011). When the force direction does not coincide with the line from the point of force application (which is determined by the ligand docking topology) to the membrane anchor point of the TCR-CD3 complex (which is determined by its asymmetric structure), a moment will be generated. This moment depends on both the magnitude and direction of force and on the lever arm that could be influenced by the length of the TCR–pMHC complex (which could be extended, as discussed earlier) (Kim et al., 2012). It has therefore been suggested that tangential force, derived from lateral motions, may provide the specific directionality to trigger the TCR (Kim et al., 2009).
However, it should be noted that, in reality, applying directional force through a micrometer-sized force probe to the cell does not guarantee the same directionality when the force is applied on the nanometer-sized molecule. Applying force tangential to the interface between a pMHC-coated bead and the T-cell (Kim et al., 2009; Feng et al., 2017) may cause the bead to pivot at the bead–cell anchor point to pull on the TCR–pMHC bond instead of shearing it. Conversely, pulling a bead away from the T-cell (Liu et al., 2014a) could also induce shear if alignment is not perfect. Recently, both normal and tangential forces applied to T-cells through as few as a single antigenic MHC interaction were shown to induce calcium release, although tangential forces tended to improve activation efficiency (Feng et al., 2017). Additionally, the surface roughness of the T-cell, due to dynamic microvilli, adds another layer of complexity to affect the force direction (Hivroz and Saitakis, 2016). Anisotropy may play an important role in TCR triggering, but further studies are required to address the influence of force direction.
Maintenance and amplification of TCR triggering
A sufficiently long TCR–pMHC bond lifetime would sustain the propagation of force waveform and allow conformational changes to reach the tail end of the TCR-CD3 complex. Yet the time to expose CD3 ITAMs is transitively finite. Is the window of opportunity for TCR signaling limited to the single TCR–pMHC bond lifetime? On the contrary, Liu and colleagues determined that intracellular calcium flux can be triggered if a T-cell accumulates durable TCR–pMHC bonds with a cumulative lifetime surpassing a threshold of 10 s within a 60-s window in the first 3 min of contact. Cells not meeting this threshold failed to trigger calcium, even if single bonds with relatively long lifetimes were experienced (Liu et al., 2014a). This work unraveled a connection between mechanoreception and mechanotransduction in the process of TCR-mediated mechanosensing. Agonist pMHCs that form catch bonds with the TCR meet this requirement and hence can trigger T-cell activation, whereas antagonist pMHCs that form slip bonds with the TCR are defective in activating T-cells, due, at least in part, to the inability to support a durable force at sufficient levels. This may allow the T-cell to distinguish different ligands and react to the correct one only (Liu et al., 2014a). Further, upon including CD8 to form the trimolecular complex, sustained calcium signaling can even be achieved by force-ramping the complex to bond rupture (Pryshchep et al., 2014). These studies de-emphasize the need of single-bond durability in robust mechanosensing and implicate a memory mechanism involving sequential accumulation of a number of bonds, allowing reconciliation between seemingly conflicting requirements of disengagement and signal maintenance within a serial triggering and kinetic proofreading framework.
The subsequent progression of the membrane proximal TCR signaling process is regulated by the spatiotemporal balance between kinases and phosphatases such as CD45. Highlighting this balance concept, the kinetic segregation model of TCR signaling postulates that phosphatases with large ectodomains are excluded by the narrow cell junction brought together by TCR–pMHC binding, tilting the balance to phosphorylation of ITAMs to trigger signaling (Davis and van der Merwe, 2006). While details of this model require further elaboration, the concept of force on the TCR–pMHC bond due to the large CD45 ectodomains is at the core of this model and must be considered in future revisions. Exactly what biochemical activity directly follows the above mechanotransduction remains elusive. Nonetheless, “come and stay” (Stepanek et al., 2014), “warm-up” (Malissen and Bongrand, 2015), and “catch-and-release” (Katz et al., 2017) models have been proposed to address the timing of the biochemical initiation. Specifically, they each describe how signaling molecules can nucleate amplification of T-cell signaling from the CD3 ITAM mechanotransduction.
From available data, a combination of the cumulative bond lifetime model (Liu et al., 2014a) and models for TCR mechanosensing (van der Merwe and Dushek, 2011; Wang and Reinherz, 2012; Bettini et al., 2014; Das et al., 2015; Liu et al., 2016) is summarized below:
Mechanopresentation (Figure 1C, step 1): pMHC as the mechanopresenter binds to the TCR, and a force is applied to this bond externally by relative cell–cell motion or spontaneous cell membrane motility, or internally by basal actomyosin transport of TCR clusters. Although the source of force can be internal to the T-cell, from the perspective of the TCR, force is modulated and presented by the pMHC for the TCR to sense.
Mechanoreception (Figure 1C, step 2): The TCR forms a specific bond with the pMHC to support the force, thereby acting as the mechanoreceptor. Force amplifies the information embedded in the pMHC, eliciting catch bonds to strengthen engagement with agonists but slip bonds to weaken engagement with weak agonists/antagonists. In this way, the mechanically modulated information is received in a form readily transmittable across the TCR-CD3 complex.
Mechanotransmission (Figure 1C, step 3): The catch bond may be further amplified by the FG loop, which may undergo structural changes to stabilize the extended state of the stressed TCR–pMHC complex (Das et al., 2015). The observation that peptide potency correlates with mechanical extension of the TCR–pMHC complex suggests that information embedded in the pMHC has been translated into a mechanical variable from the binding site to the site of structural transition under force (Das et al., 2016). This mechanical change further propagates to the CD3 chains, resulting in the release of CD3 CTs from the membrane.
Mechanotransduction (Figure 1C, step 4): Exposure of the CD3 ITAMs permits phosphorylation, which initiates further signal propagation. This is regulated by the balance of kinases and phosphatases and occurs in a cumulative manner.
Recently, emerging cancer immunotherapies such as the TCR-based chimeric antigen receptor approach have demonstrated high response rates but lack broad applicability (Lim and June, 2017). Genetically modifying TCRs of autologous, tumor-infiltrating T-cells (Lim and June, 2017) to target a vast array of antigens on solid tumors also appears to be a promising strategy to combat cancer (Maus and June, 2016). By further vetting the four-step process of TCR mechanosensing, one could more rationally engineer TCRs and chimeric antigen receptors to detect and respond exclusively to tumor antigens and neoantigens. Receptors could be designed to amplify different responses between cancerous and healthy cells with minute differences in the antigen structure and abundance, through distinctive force-regulated binding and mechanosensing (Hinrichs and Restifo, 2013). To take such a reverse-engineering approach requires further understanding of TCR-mediated mechanosensing. For example, specific conformational changes under force within the TCR V domains to elicit catch bonds have yet to be identified. Additionally, because conformational changes were found through the FG loop only when the TCR–pMHC complex was under force, it will be instructive to monitor the CD3 CTs under force to study the force transmission beyond the cell membrane. Elucidating these processes will inform how the T-cell can detect and respond to appropriate antigens with such exquisite sensitivity and specificity, key knowledge needed to improve immunotherapies.
INTEGRIN-MEDIATED CELL ADHESION AND MECHANOSENSING
Integrins mediate many cellular processes, including adhesion, spreading, migration, proliferation, and differentiation (Roca-Cusachs et al., 2009; Elosegui-Artola et al., 2014; Qiu et al., 2014). As physical linkages for the mechanochemical coupling between the ECM and the intracellular structures (Li et al., 2016b), integrins have long been known for their role in mediating force transmission from the ECM across the cell membrane (Volk et al., 1990; Welch et al., 1990). The discovery that integrins are a mechanoreceptor was made more than two decades ago in an elegant experiment using MTC, showing that twisting ligand-bearing beads bound to β1 integrins caused endothelial cells to stiffen (Wang et al., 1993). The integrin family consists of 18 α and 8 β subunits that combine to form 24 αβ heterodimeric membrane receptors. Each integrin subunit contains an N-terminal ectodomain consisting of a large head with the ligand binding site and a long leg, a single-pass TMD, and a short C-terminal CT (Figure 6A). The heads and the upper legs of the αβ heterodimer form the headpiece that connects with the α and β lower legs at the knees (Figure 6A).
FIGURE 6:
Integrin structure and signaling models and illustrative BFP signals that detect integrin bending and unbending. (A) Integrin structure. Domains of the integrin and the ranges of integrin head, upper leg, headpiece, and tailpiece (or lower leg) are annotated. αI domain that exists only in αI-bearing integrins is distinguished by a lighter color and dashed lines. (B) A model of integrin inside-out signaling mediated by RAP1 and RIAM. (C, D) Illustrative BFP force vs. time traces depicting respective integrin unbending (C) and bending (D) conformational change events, denoted by red arrows. The conformations of the integrin are inserted accordingly, with red and blue indicating the extended and bent conformers of the ectodomain, respectively. Widths of the shaded regions reflect the thermal fluctuation amplitude of each trace in the clamping phase. (E) Postulated model of integrin-mediated mechanosensing without prior talin engagement. External force pulls on the i) bent integrin headpiece and allosterically induces ii) ectodomain extension, iii) hybrid domain swing-out, and αβ-subunit separation below the headpiece due to an endogenous lateral force pulling on the βCT. The αβ separation at the CT presumably recruits intracellular molecules and initiates downstream signaling. The ligand, integrin domains, and talin are colored based on their respective roles in the four-step mechanosensing model (cf. Figure 1). Proposed regulatory loops 1 (between mechanoreception and mechanotransmission) and 2 (between mechanotransmission and mechanotransduction) are denoted.
In a resting state, integrin adopts a bent conformation with a closed headpiece (Figure 6, B and Ei). Upon activation, integrins undergo a series of conformational changes: ectodomain extension, αβ separation below the headpiece (including the ectodomain lower leg, TMD, and CT), and hybrid domain swing-out and headpiece opening (Takagi et al., 2002, 2003; Xiao et al., 2004; Nishida et al., 2006; Xie et al., 2010) (Figure 6, B and E, middle, right). The coordinated transitions between these conformations are essential for the cell to regulate integrins’ adhesive and signaling functions dynamically (Takagi et al., 2002; Nishida et al., 2006; Campbell and Humphries, 2011; Springer and Dustin, 2012; Cheng et al., 2013; Zhu et al., 2013b; Nordenfelt et al., 2016; Su et al., 2016). Such mechanical changes are accompanied by biochemical changes, making integrins a key component of a cell’s mechanosensing machinery. Especially, the separated integrin βCT can associate with talin, kindlin, and several other adaptors to transduce mechanical/biochemical signals (Arnaout et al., 2005; Luo and Springer, 2006). In inside-out signaling, cytoplasmic signals are generated from other cell receptors and relayed by an integrin from the βCT to the headpiece for high-affinity ligand binding. Moreover, it is generally assumed that integrin conformational changes are allosteric, hence also operating reversely to achieve outside-in signaling, in which extracellular ligand binding acts as a signal to propagate from the integrin headpiece to the βCT to induce intracellular signaling. Although outside-in signaling of integrins has long been recognized, the mechanical aspects of this process often are not sufficiently emphasized. We suggest that integrin outside-in signaling in many cases is a mechanosensing process and hence can be analyzed by our four-step model.
Mechanopresentation by the ECM
Besides blood flow, (sub)endothelium, and adjacent adherent cells, the ECM represents a major mechanical environment sensed by integrins for many cell types. The mechanical properties of the ECM, for example, stiffness, can directly affect cell functions (Engler et al., 2006; Fu et al., 2010). Interestingly, in addition to passively receiving mechanical cues from the ECM, cells can also actively modify the ECM as a response of mechanosensing. For example, certain tissues exhibit a “stress-relaxation” property, such that stretching decreases its stiffness (Chaudhuri et al., 2016); conversely, some biological gels and fibrin networks display a “strain-stiffening” property, such that force-induced deformation increases their stiffness (Storm et al., 2005; Jansen et al., 2013). Such properties add a dynamic confounding factor into the ECM mechanopresentation in the integrin-mediated cell mechanosensing (Jansen et al., 2017). In-depth study of cell mechanosensing in this context requires techniques that can adjust the system stiffness in real-time. An example of this is the feedback loop added to an AFM to tune the cantilever stiffness “felt” by the cell in real-time (Lam et al., 2011).
Force-modulated integrin binding kinetics and mechanoreception
The integrin binding kinetics is modulated by force in many biological processes, for example, in leukocyte inflammatory response and in platelet thrombus formation, wherein hemodynamic forces acting on rolling or adherent cells are balanced by adhesive forces on integrin–ligand bonds. Like GPIb and TCR, several integrins have been shown to form catch-slip bonds with their ligands, allowing force to prolong bond lifetimes (Chen and Zhu, 2013). These include α5β1–fibronectin (Kong et al., 2009), αLβ2–ICAM-1 (intercellular adhesion molecule 1) (Chen et al., 2010), α4β1–VCAM-1 (vascular adhesion molecule 1) (Choi et al., 2014), αMβ2–ICAM-1 (Rosetti et al., 2015), and αVβ3–fibronectin (Chen et al., 2016; Elosegui-Artola et al., 2016) interactions. Structural analyses (Kallen et al., 1999; Luo et al., 2007a) and molecular dynamics simulations (Jin et al., 2004; Xiang et al., 2011) on αI-bearing integrins suggest that pulling on the metal ion–dependent adhesion site (MIDAS) of the αI domain causes its anchored α7 helix to shift downward like a piston relative to the upward-moving αI domain, thereby allosterically inducing the αI domain to bind ligand more stably (Chen et al., 2010). Here, the anchor of the αI domain is not at the point of its C-terminal insertion to the β-propeller domain but is provided by the binding of an intrinsic ligand at the end of the α7 helix to the βI domain MIDAS, which also forms an interdomain catch bond (Chen et al., 2010). Using this “dual-catch” mechanism, the suppression of αMβ2–ICAM-1 catch bonds by an αMβ2 variant R77H (Rosetti et al., 2015), which is associated with systemic lupus erythematosus (SLE) (Nath et al., 2008; Han et al., 2009), can be well explained. The difficulty here is that R77H occurs in the β-propeller domain away from the αI domain MIDAS, which cannot directly alter the αI ligand binding epitope. However, mechanical analysis suggests that R77H loosens the connection of αI to the β-propeller domain, thereby shifting a greater portion of the ligand-transmitted force to the αI–βI interdomain bond. Consequently, before the total force on the external bond (between the ligand and the αI domain MIDAS) reaches a sufficient level to pull the αI domain upward, the component force on the internal bond (between the αI domain α7 helix and the βI domain MIDAS) already exceeds the optimal level. The resulting short-lived slippery internal bond cannot effectively pull down the α7 helix to induce conformational changes in the αI domain MIDAS, thereby suppressing its ability to form a catch bond with the external ligand. This example underscores the pathophysiological relevance of integrin catch bonds. Conversely, cellular signaling can change a receptor’s ligand binding behavior under force. For example, the α4β1–VCAM-1 catch bond is suppressed by inside-out signaling upon binding of sema3E to plexinD1 (Choi et al., 2014). This in turn regulates the signaling outcomes of mechanosensing.
Force is inevitably involved in many cases of integrin signaling, owing to the immobilization of the ligand and cytoskeletal linkage of the integrin. Many studies have demonstrated that integrin signal transduction exhibits force sensitivity, recognizing integrins’ role as a mechanoreceptor (Lefort and Ley, 2012; Case and Waterman, 2015). Mutations that reduce integrin binding, for example, those in the α1/α1′-helix of the αIIbβ3 ligand binding βI domain and adjacent to the MIDAS (ADMIDAS) region, cause defects in outside-in signaling in transfected cells (Raborn et al., 2011; Zhang et al., 2013a). Conversely, T188I, a heterozygous ligand binding–increasing mutation in the β1 integrin headpiece, promotes cell spreading (Evans et al., 2003). Furthermore, like TCR and GPIb, the catch-bond property of an integrin mediates its mechanoreception (Janostiak et al., 2014; Qiu et al., 2014; Sun et al., 2016). For example, the endothelial surface molecule Thy-1 (CD90) forms a slip bond with integrin α5β1 or syndecan-4 when only one receptor is presented. Thy-1 forms two additive slip bonds at low forces with the two receptors when both are present. However, higher forces induce cooperative binding of both receptors to Thy-1, forming a trimolecular catch bond (Fiore et al., 2014). This “dynamic catch” correlates with inhibition of intracellular tension– and substrate rigidity–dependent FAK activity, indicating its relevance to mechanosensing and, with alterations in tumor cell metastasis, suggesting its pathophysiological relevance (Fiore et al., 2014).
Force-induced integrin conformational changes and integrin mechanotransmission
Unlike GPIb and TCR, whose extended or unfolded conformations are induced by force, integrins can exist in three distinct global conformations in the absence of external force: bent with a closed headpiece (Figure 6Ei), extended with a closed headpiece (Figure 6Eii), and extended with an open headpiece (Figure 6Eiii) (Springer and Dustin, 2012). The transitions between these stable conformations may occur spontaneously, giving rise to the coexistence of multiple conformers in equilibrium (Xiao et al., 2004; Zhu et al., 2008b; Ye et al., 2010). From a mechanical standpoint, the multidomain quaternary structure of integrins resembles a protein machine, with structural parts for force transmission and moving parts for conformational motion connected by ratchets, ropes, and hinges. It is therefore intuitive that the integrin conformation would modulate the transmission of force from the ligand binding site across the main ectodomain down to the cytoplasmic anchor or vice versa. An example is the SLE-associated αMβ2 variant discussed earlier, where the single-residue replacement R77H is suggested to tilt the partition of forces between the N- and C-termini of the αI domain that respectively transmit to the β-propeller and βI domains (Rosetti et al., 2015).
Conversely, force can directly induce integrin conformational changes. MD simulations have observed that force can activate the headpiece in silico (Puklin-Faucher and Vogel, 2009), swing out the hybrid domain (Zhu et al., 2008b), separate the αβ tailpieces (Zhu et al., 2008b), and extend the ectodomain (Chen et al., 2011). Real-time observations of force-regulated dynamic bending (Figure 6C) and unbending (Figure 6D) of single integrin αLβ2 (Chen et al., 2012) and αVβ3 (Chen et al., 2016) using the BFP show that force-induced integrin unbending tilts the conformational equilibrium to favor the extended over the bent conformation. Specifically, tensile force increases the rate (how soon it occurs) and speed (how fast it proceeds), and therefore the frequency, of unbending, and at the same time, decreases the corresponding parameters of bending in a magnitude-dependent manner (Chen et al., 2012, 2016). Thus, integrins are mechanosensitive via a hinge-motion mechanism (Figure 2C) to allow force to facilitate their extension (unbending). The extension in turn facilitates swing-out of the hybrid domain by freeing it from close contact with the tailpiece (Zhu et al., 2008b). Thus, along with force, conformational changes can also propagate in the integrin in a relayed manner. The force-modulated propagation of conformational changes transmits mechanical signals along the molecule, which is an important part of the integrin mechanotransmission step.
The cytoplasmic linkage between integrins and actin is thought to be critical to stabilizing integrins in the extended-open conformation (Zhu et al., 2008b). Recent structural analyses and MD simulations suggest a “cytoskeletal force model,” in which lateral pulling on the βCT via talin/kindlin bound to actin retrograde flow could pull the α and β CTs and tailpieces apart to stabilize the extended-open conformation for high-affinity ligand binding. Using integrin βCT-TSMod fusion (Figure 3L) constructs in cell-spreading experiments (Nordenfelt et al., 2016), it has been found that an activated integrin requires the balance between internal forces exerted by retrograde actin flow and external forces applied through engaged ligands by flow-induced shear or substrate stretch. By using the DNA-based force probes, the external force on integrin has been estimated to range from 4–13 pN at the initial spreading (Zhang et al., 2014) and ∼40 pN at a stable adhesion (Wang and Ha, 2013). With the TSMod method, the internal force transmitted across the integrin has been estimated as 2.5 pN in focal adhesion (Grashoff et al., 2010).
Mechanical coupling of integrin mechanoreception and mechanotransmission
Integrin conformation and affinity are closely coupled: a bent integrin has a low affinity for ligand. Upon the activation, the hybrid domain swings out to open the headpiece for high-affinity ligand binding. For αI-absence integrins, this requires the headpiece to transition through six intermediate conformations, starting from a closed to a fully open βI (Zhu et al., 2013b). The opening of the headpiece is allosterically associated with the ectodomain extension (Liu et al., 2015b). Conversely, locking the integrin in the bent or extended-closed conformation prevents the emergence of the high-affinity state (Li et al., 2017a). Promoting ectodomain extension, TMD and CT separation and hybrid-domain swing-out by point mutation or functional activating agents greatly enhances integrin reactivity (Vanhoorelbeke et al., 2009; Rosetti et al., 2015; Su et al., 2016). Although supported by many studies, not all aspects of this model have been validated, leaving room for alternative models. For example, a recent study revealed a bent but high-affinity conformation in the β2 integrins, implying that ectodomain extension is not necessarily required for the integrin activation (Fan et al., 2016).
Similar to the force-induced GPIb binding reinforcement by LRRD unfolding (Figure 4B) (Ju et al., 2015b), force-induced extension of αLβ2 immediately prolongs its binding lifetime (Chen et al., 2012). In addition, cyclic forces applied to integrin α5β1–fibronectin and αLβ2–ICAM-1 bonds substantially reinforce their lifetimes (Kong et al., 2013), suggesting a highly localized force-induced dynamic affinity–conformation interplay within the molecule. Considering the coupling between integrin conformation and affinity, integrin mechanoreception (Figure 1D, step 2) and mechanotransmission (Figure 1D, step 3) may be able to provide feedback to each other in a back-and-forth “regulatory loop” (Figure 6Eii, regulatory loop 1), providing force a role in the integrin affinity maturation process.
Signal transmission across the membrane and downstream mechanotransduction
Downstream of integrins and the linked cytoskeletal proteins, mechanotransduction involves the recruitment and phosphorylation of kinases. For example, FAK is phosphorylated by integrin signaling (Harburger and Calderwood, 2009) and regulates Rho-family GTPase activation, which mediates cell migration (Mitra et al., 2005). As another example, following integrin signaling, the Src-family protein tyrosine kinases contribute to the formation of integrin-mediated firm adhesion (Ginsberg et al., 2005; Giannone and Sheetz, 2006; Jansen et al., 2017). Previously, it was believed that induction of these signals requires integrins to cluster, and mechanotransduction is via rearrangement of a cytoskeletal structure like actin (de)polymerization and accumulation of cytoskeletal proteins (Miyamoto et al., 1995; Yamada and Miyamoto, 1995). Yet more recent work has suggested that single integrins may be capable of transducing mechanical signals as well (Harburger and Calderwood, 2009; Lefort et al., 2009; Hu and Luo, 2015). Below we provide three models of single integrin-mediated mechanotransduction that rely on different components of the molecular network connected to integrin. These models are not mutually exclusive; rather, they may respectively modulate different stages/scenarios of cell functioning.
The αβ-subunit separation below the headpiece is the most likely mechanical event that allows a single integrin to transmit signal across the membrane (Kim et al., 2003). As mentioned earlier, during mechanotransmission, the extension of integrin is stabilized by a tensile force pulling on the headpiece. Using molecular dynamic simulations on an integrin ectodomain structure, Zhu and colleagues found that a combination of a tensile force normal to the cell membrane and a lateral force on the lower β leg applied on an extended integrin swings out the hybrid domain and separates the lower legs (Zhu et al., 2008b). Integrin βCT is engaged to a number of adaptor proteins, including talin, kindlin, filamin, and migfilin, among others (Figure 6B), which provide constraints to its lateral diffusion (Zhang et al., 2004; Roca-Cusachs et al., 2012). Therefore, the above evidence from Zhu et al. (2008b) suggests that pulling on the headpiece of an extended integrin that is not well aligned with its cytoplasmic anchor may result in a lateral component force on the β tail causing it to detach from the α tail. The αβ separation in the CT may in turn unmask binding/catalytic sites within the cytoplasmic domains (e.g., enable talin association), resulting in initiation of biochemical signaling and the fulfillment of mechanotransduction (Jani and Schock, 2009) (Figure 6E).
It is widely accepted that talin binding to integrin βCT represents a final common step in integrin activation (Shattil et al., 2010). For this reason, talin can play a major role in the mechanotransduction step beneath the membrane. Talin associates with integrin βCT through its head domain and with actin through its rod domain, making the sequences in between susceptible to conformational change upon force pulling (Critchley, 2009). In integrin outside-in signaling, force transmitted through the βCT unfolds the attached talin (Yao et al., 2016), which in turn recruits vinculin and facilitates downstream signaling (Yao et al., 2014). A recent study has provided support for a talin-dependent mechanotransduction mechanism in relation to cell sensing of substrate stiffness. On a soft surface, integrin–ligand engagement is weak and fails to unfold talin. On a stiff surface, by contrast, integrin binding is reinforced by a catch bond, allowing talin to unfold before ligand dissociation (Elosegui-Artola et al., 2016; Jansen et al., 2017). Because the αβ separation below the headpiece would affect the stability of the extended-open conformation of the integrin (Zhu et al., 2008b), a second “regulatory loop” between mechanotransduction (Figure 1D, step 3) and mechanotransmission (Figure 1D, step 4) may exist (Figure 6Eiii, regulatory loop 2). The two regulatory loops described here may even work cooperatively to allow back-and-forth communications among mechanoreception, mechanotransmission, and mechanotransduction (Figure 6E).
As force propagates through intracellular structures, several other proteins besides talin that are indirectly linked to integrins may also change conformation and/or function. Examples include vinculin unfolding that exposes the buried motifs (Huveneers and Danen, 2009), p130Cas extension (Sawada et al., 2006), Src activation (Wang et al., 2005), myosin translocation on actin (Figure 2E) (Cross, 2006), and deformation of the nuclear membrane (Wang et al., 2009). These events respectively facilitate focal adhesion maturation (vinculin, p130Cas, and Src), cell deformation (myosin translocation), and directed gene expression and nuclear transport (nuclear membrane deformation), and thus represent additional choices of intracellular mechanotransduction.
Among the three mechanosensing-mediating receptors discussed in this review, integrins represent the most sophisticated system, due to its feature of bidirectional signal transduction, multiple conformational states, and a combination of conformational changes associated with its activation. While multiple mechanisms of integrin-mediated mechanosensing coexist, we can nevertheless summarize the integrin mechanosensing process into the following four steps:
Mechanopresentation (Figure 1D, step 1): Integrin binds to the ligands in its headpiece and external force is applied to this bond by relative cell–matrix motion or spontaneous cell membrane movement.
Mechanoreception (Figure 1D, step 2): The ligand binding site of the integrin headpiece receives the ligand together with the force signal. Catch bonds are formed to reinforce the binding state under a tensile force.
Mechanotransmission: Force propagates along the integrin, unbends integrin through a “hinge” mechanism (Figure 6Eii), and swings out the hybrid domain. In certain scenarios, force directly propagates onto talin (Figure 1D, step 3) or other cytoplasmic molecules, and the integrin unbending and hybrid domain swing-out may not be necessary.
Mechanotransduction: Propagation of the conformational changes leads to the αβ separation at the lower leg, TMD, and finally CT to expose cytoplasmic sequences to recruit signaling molecules (Figure 6Eiii). Force propagated through the βCT unfolds MSDs in talin rod and exposes multiple cryptic vinculin binding sites. The recruited vinculin then recruits more F-actin and strengthens the talin–actin linkage (Figure 1D, step 4). In addition, mechanotransduction may also be accomplished via conformational changes of several other cytoplasmic proteins with varied functions.
OTHER CELL MECHANOSENSING MODELS
Besides the aforementioned examples, other molecular systems have been shown to be used by cells to sense their mechanical environment. These systems do not necessarily follow the proposed four-step model. However, certain steps are shared, despite the variations among different biological systems and distinctive functions to be served. Especially, the step of mechanotransduction has to be included in any mechanosensing mechanism, because it is the step of converting mechanical signals to biochemical signals. Here, we will briefly discuss other cell mechanosensing models that include some, but not all, of the four steps of receptor-mediated cell mechanosensing.
In the sound reception of an auditory system, the mechanical signal adopts the form of a soundwave, which is a pure physical signal instead of a mechanical–biological combination. Thus, the mechanopresentation step is not required and is replaced by sound generation. The mechanoreception step, on the other hand, is required, but exists in another form—instead of force-bearing binding to a ligand, the soundwave exerts mechanical load on the hair bundles to cause vibration and deflection. Following that is the mechanotransmission step, wherein the force borne in the deflecting hair bundles propagates across connecting molecules to reach the anchorage of the cell surface ion channels. Force then controls the opening and closing of the mechanically gated ion channels to convert to the first chemical signal, resulting in mechanotransduction with the “clamp movement” mechanism (Figure 2B) (Gillespie and Walker, 2001; Gillespie and Muller, 2009). Adopting a similar mechanism, mechanosensing has also been observed in bristle-based haptic systems of animals, for example, flies (Gillespie and Walker, 2001). Primary cilia that extend from most mammalian cell types have been proposed to perform cell mechanosensing of pure physical signals. Although the underlying mechanotransduction mechanism of this process is still under debate (Delling et al., 2016), its mechaotransmission should also be achieved by the force-induced deflection of the primary cilium (Hoey et al., 2012).
In comparison, the force-sensitive ion channels of somatosensory systems adopt another form of mechanosensing. It also lacks mechanopresentation, with a mechanical force being physically exerted onto the skin cell membrane to trigger the mechanoreception. But in contrast to the auditory system, the mechanotransmission here is the propagation of force on the cell that causes it to deform and change shape. The deformation leads to cell membrane stretching, and opens up ion channels that adopt the “deformation” mechanosensitive mechanism (Figure 2A) to achieve mechanotransduction. This mechanosensing mechanism was suggested long ago (Orr et al., 2006) and has recently been demonstrated on mouse epidermal Merkel cells (Maksimovic et al., 2014). The same mechanism is shared by muscle mechanosensing, in which stretching of the membrane of the vascular smooth muscle cells opens cation channels. In a similar manner, red blood cells release ATP in response to mechanical deformation, with the only difference being that the mechanotransducer here is the cytoskeletal spectrin–actin network, which is rearranged under force, instead of ion channels (Wan et al., 2008).
Yet in some other systems, the cell can use surface structures to directly receive mechanical signals without binding to a ligand, hence having a mechanoreceptor but no mechanopresenter. For example, glycocalyx and G proteins on the endothelial cell surface have been found to directly transduce shear stress for mechanosensing without binding to any ligand (Chachisvilis et al., 2006; Makino et al., 2006; Orr et al., 2006). Corresponding mechanisms have been proposed (Fu and Tarbell, 2013).
Although examples in this review only discuss monovalent molecular interactions, multivalent interactions do exist in many receptor–ligand systems. For example, P- and E-selectins (but not L-selectin) are dimeric and can form dimeric bonds with the dimeric P-selectin GP ligand 1 (Ramachandran et al., 2001; Zhang et al., 2013b); fibrinogen and VWF are both capable of binding multiple copies of their corresponding receptors on the same or even different platelets (Rooney et al., 1998; Zhou et al., 2012). The association and dissociation of these complexed bonds follow multistep kinetic mechanisms that involve intermediate steps with additional rate parameters. Also, the force burden is, likely unevenly, dispersed onto multiple bonds, making the force dependency of the bond lifetime more complicated (Friddle et al., 2012). Nonetheless, it is likely that a multimeric ligand has a higher chance to engage cell receptors for more frequent mechanopresentation. The overall bond lifetime is also likely prolonged, because dissociated members of the receptor–ligand cluster are still kept within close proximity by other members, remaining associated so that they can quickly re-engage, resulting in more effective mechanoreception. Although receptor cross-linking by multimeric ligand is known to facilitate cell signaling in many situations, multivalency may or may not guarantee stronger mechanosensing. While clustering creates a higher local concentration of signaling molecules to strengthen signaling, the formation of more bonds must be accompanied by the decrease of force dispersed over each of the bonds, which may weaken the signaling of each receptor. Should mechanosensing adopt a threshold mechanism that requires certain amplitude of force on single receptors, excessive force dispersion could even eliminate signaling. In addition, despite not having a dimeric/multimeric structure, some proteins contain dual or multiple sites that can bind independently or cooperatively. For example, fibronectin contains a synergy site in the III9 domain that can strengthen the binding of the III10 domain RGD sequence to integrin α5β1 (Friedland et al., 2009; Kong et al., 2009). Thy-1 can simultaneously bind integrin α5β1 and syndecan-4 in a force-induced cooperative manner (Fiore et al., 2014). Accordingly, the mechanoreception is reinforced in both cases. Conversely, cellular signaling can directly affect the multimericity of a molecular system. For example, when pMHC binds TCR to induce signaling, the co-receptor CD8 is up-regulated to bind the pMHC pre-engaged with the TCR, which converts the TCR–pMHC bimolecular bond to a TCR–pMHC–CD8 trimolecular bond (Jiang et al., 2011; Casas et al., 2014; Liu et al., 2014b).
CONCLUSIONS
Research in the past few decades focused on developing biochemical and molecular biological approaches, which allowed scientists to identify and characterize molecular players of mechanosensing pathways. Recently, new biophysical nanotools have moved the mechanobiology field into a new era, when it has become possible to address a fundamental question: how are mechanical cues presented, received, transmitted, and transduced by cell surface and intracellular proteins? Furthermore, the time has come to elucidate the design and working principles of the mechanosensing apparatus. As summarized in this review, the use of these nanotools allows the investigation of receptor-mediated cell mechanosensing in many biologically significant contexts, including how platelets balance their functions between hemostasis and thrombosis, how T-cells recognize pathogens, how stem cells differentiate into distinct cell types, how cancer cells metastasize, and how neutrophils migrate to fight infections. More importantly, the combined kinetic analysis of molecular interactions and mechanical analysis of the molecular conformational changes reveal the working mechanisms of each mechanosensing receptor and its MSDs—GPIbα unfolds the LRRD and MSD, TCR–pMHC increases its length, and integrins bend and unbend between the upper and lower legs. In the future, full characterization of these and other modular structures of mechanosensing machineries will provide synthetic biology design principles for a generic mechanosensing machine. The similarities and differences between the GPIb and TCR systems imply that, in addition to the general principles, each receptor may have its own details to accommodate its specific function. Future studies will relate the mechanosensing mechanisms of distinct receptors to their physiological context and allow customization based on the general principles and re-engineering of the details.
Acknowledgments
This study was completed during C.Z.’s sabbatical at the Heart Research Institute in Australia. We thank Shaun P. Jackson and his lab for helpful discussion and support for the sabbatical and writing of the paper. L.J. is supported by the postdoctoral fellowship (101285) from the National Heart Foundation of Australia, This work was supported by National Institutes of Health grants AI044902, AI124680, HL132019, and CA214354 (C.Z.); Army Research Office grant DODW911NF-16-1-0257 (C.Z.); Royal College of Pathologists of Australasia Kanematsu Research Award (L.J.); a Diabetes Australia research grant G179720 (L.J.); a Sydney Medical School 2016 early-career researcher kickstart grant (L.J.); and National Science Foundation Graduate Research Fellowship DGE-1650044 (M.R.).
Abbreviations used:
- ADAMTS-13
a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13
- AFM
atomic force microscopy
- APC
antigen-presenting cell
- BCR
B-cell receptor
- BFP
biomembrane force probe
- CT
cytoplasmic tail
- DFS
dynamic force spectroscopy
- ECM
extracellular matrix
- FAK
focal adhesion kinase
- fBFP
fluorescence BFP
- FRET
fluorescence resonance energy transfer
- GP
glycoprotein
- ICAM-1
intercellular adhesion molecule 1
- ITAM
immunoreceptor tyrosine-based activation motif
- LRRD
leucine-rich repeat domain
- MD
molecular dynamics
- MIDAS
metal ion–dependent adhesion site
- MP
macroglycopeptide
- MSD
mechanosensitive domain
- MTC
magnetic twisting cytometry
- pMHC
peptide–major histocompatibility complex
- SLE
systemic lupus erythematosus
- TCR
T-cell receptor
- TGT
tension gauge tethers
- TMD
transmembrane domain
- TSMod
tension sensor module
- VCAM-1
vascular adhesion molecule 1
- VWD
von Willebrand disease
- VWF
von Willebrand factor
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-04-0228) on September 27, 2017.
REFERENCES
- Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, Chen W, Levin AM, Connolly JM, Zhu C, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen Eguiluz RC, Kaylan KB, Underhill GH, Leckband DE. Substrate stiffness and VE-cadherin mechano-transduction coordinate to regulate endothelial monolayer integrity. Biomaterials. 2017;140:45–57. doi: 10.1016/j.biomaterials.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte-Santamaria C, Huck V, Posch S, Bronowska AK, Grassle S, Brehm MA, Obser T, Schneppenheim R, Hinterdorfer P, Schneider SW, et al. Force-sensitive autoinhibition of the von Willebrand factor is mediated by interdomain interactions. Biophys J. 2015;108:2312–2321. doi: 10.1016/j.bpj.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- Auton M, Sowa KE, Behymer M, Cruz MA. N-terminal flanking region of A1 domain in von Willebrand factor stabilizes structure of A1A2A3 complex and modulates platelet activation under shear stress. J Biol Chem. 2012;287:14579–14585. doi: 10.1074/jbc.M112.348573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axmann M, Huppa JB, Davis MM, Schutz GJ. Determination of interaction kinetics between the T cell receptor and peptide-loaded MHC class II via single-molecule diffusion measurements. Biophys J. 2012;103:L17–L19. doi: 10.1016/j.bpj.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barg A, Ossig R, Goerge T, Schneider MF, Schillers H, Oberleithner H, Schneider SW. Soluble plasma-derived von Willebrand factor assembles to a haemostatically active filamentous network. Thromb Haemost. 2007;97:514–526. [PubMed] [Google Scholar]
- Barry AK, Tabdili H, Muhamed I, Wu J, Shashikanth N, Gomez GA, Yap AS, Gottardi CJ, de Rooij J, Wang N, Leckband DE. α-Catenin cytomechanics—role in cadherin-dependent adhesion and mechanotransduction. J Cell Sci. 2014;127:1779–1791. doi: 10.1242/jcs.139014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashour KT, Gondarenko A, Chen H, Shen K, Liu X, Huse M, Hone JC, Kam LC. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci USA. 2014a;111:2241–2246. doi: 10.1073/pnas.1315606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashour KT, Tsai J, Shen K, Lee JH, Sun E, Milone MC, Dustin ML, Kam LC. Cross talk between CD3 and CD28 is spatially modulated by protein lateral mobility. Mol Cell Biol. 2014b;34:955–964. doi: 10.1128/MCB.00842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Huse M. Mechanical communication at the immunological synapse. Trends Cell Biol. 2016;27:241–254. doi: 10.1016/j.tcb.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Whitlock BM, Husson J, Le Floc’h A, Jin W, Oyler-Yaniv A, Dotiwala F, Giannone G, Hivroz C, Biais N, et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 2016;165:100–110. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini ML, Guy C, Dash P, Vignali KM, Hamm DE, Dobbins J, Gagnon E, Thomas PG, Wucherpfennig KW, Vignali DAA. Membrane association of the CD3 epsilon signaling domain is required for optimal T cell development and function. J Immunol. 2014;193:258–267. doi: 10.4049/jimmunol.1400322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum ME, Berry R, Hsiao YS, Chen Z, Shingu-Vazquez MA, Yu X, Waghray D, Fischer S, McCluskey J, Rossjohn J, et al. Molecular architecture of the αβ T cell receptor-CD3 complex. Proc Natl Acad Sci USA. 2014;111:17576–17581. doi: 10.1073/pnas.1420936111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, Chen CS. A DNA-based molecular probe for optically reporting cellular traction forces. Nat Methods. 2014;11:1229–1232. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3:a004994. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas J, Brzostek J, Zarnitsyna VI, Hong JS, Wei Q, Hoerter JA, Fu G, Ampudia J, Zamoyska R, Zhu C, Gascoigne NR. Ligand-engaged TCR is triggered by Lck not associated with CD8 coreceptor. Nat Commun. 2014;5:5624. doi: 10.1038/ncomms6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachisvilis M, Zhang YL, Frangos JA. G protein-coupled receptors sense fluid shear stress in endothelial cells. Proc Natl Acad Sci USA. 2006;103:15463–15468. doi: 10.1073/pnas.0607224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, Mooney DJ. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lou J, Hsin J, Schulten K, Harvey SC, Zhu C. Molecular dynamics simulations of forced unbending of integrin α(v)β(3) PLoS Comput Biol. 2011;7:e1001086. doi: 10.1371/journal.pcbi.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. 2010;285:35967–35978. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhu C. Mechanical regulation of T-cell functions. Immunol Rev. 2013;256:160–176. doi: 10.1111/imr.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lee H, Tong H, Schwartz M, Zhu C. Force regulated conformational change of integrin αVβ3. Matrix Biol. 2016;60–61:70–85. doi: 10.1016/j.matbio.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu B, Ju L, Hong J, Ji Q, Chen W, Zhu C. Fluorescence biomembrane force probe: concurrent quantitation of receptor-ligand kinetics and binding-induced intracellular signaling on a single cell. J Vis Exp. 2015:e52975. doi: 10.3791/52975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Li J, Negri A, Coller BS. Swing-out of the β3 hybrid domain is required for αIIbβ3 priming and normal cytoskeletal reorganization, but not adhesion to immobilized fibrinogen. PLoS ONE. 2013;8:e81609. doi: 10.1371/journal.pone.0081609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YI, Duke-Cohan JS, Chen W, Liu B, Rossy J, Tabarin T, Ju L, Gui J, Gaus K, Zhu C, Reinherz EL. Dynamic control of β1 integrin adhesion by the plexinD1-sema3E axis. Proc Natl Acad Sci USA. 2014;111:379–384. doi: 10.1073/pnas.1314209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn LA, Damaraju VS, Dozic S, Eskin SG, Cruz MA, McIntire LV. GPIbα-vWF rolling under shear stress shows differences between type 2B and 2M von Willebrand disease. Biophys J. 2011;100:304–312. doi: 10.1016/j.bpj.2010.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;4:165–178. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley DR ( Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- Cross RA ( Myosin’s mechanical ratchet. Proc Natl Acad Sci USA. 2006;103:8911–8912. doi: 10.1073/pnas.0603213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RA, McAinsh A. Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Bodnar R, Berndt MC, Du X. A critical role for 14–3-3ζ protein in regulating the VWF binding function of platelet glycoprotein Ib-IX and its therapeutic implications. Blood. 2005;106:1975–1981. doi: 10.1182/blood-2005-01-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL, Lang MJ. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA. 2015;112:1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Mallis RJ, Duke-Cohan JS, Hussey RE, Tetteh PW, Hilton M, Wagner G, Lang MJ, Reinherz EL. Pre-T cell receptors (pre-TCRs) leverage Vβ complementarity determining regions (CDRs) and hydrophobic patch in mechanosensing thymic self-ligands. J Biol Chem. 2016;291:25292–25305. doi: 10.1074/jbc.M116.752865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol. 2006;7:803–809. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- DeFord-Watts LM, Dougall DS, Belkaya S, Johnson BA, Eitson JL, Roybal KT, Barylko B, Albanesi JP, Wulfing C, van Oers NSC. The CD3 zeta subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR-CD3 complex at the immunological synapse. J Immunol. 2011;186:6839–6847. doi: 10.4049/jimmunol.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, Indzhykulian AA, Liu X, Li Y, Xie T, Corey DP, Clapham DE. Primary cilia are not calcium-responsive mechanosensors. Nature. 2016;531:656–660. doi: 10.1038/nature17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Deng W, Wang Y, Druzak SA, Healey JF, Syed AK, Lollar P, Li R. A discontinuous autoinhibitory module masks the A1 domain of von Willebrand factor. J Thromb Haemost. 2017;15:1867–1877. doi: 10.1111/jth.13775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Xu Y, Chen W, Paul DS, Syed AK, Dragovich MA, Liang X, Zakas P, Berndt MC, Di Paola J, et al. Platelet clearance via shear-induced unfolding of a membrane mechanoreceptor. Nat Commun. 2016;7:12863. doi: 10.1038/ncomms12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbins J, Gagnon E, Godec J, Pyrdol J, Vignali DA, Sharpe AH, Wucherpfennig KW. Binding of the cytoplasmic domain of CD28 to the plasma membrane inhibits Lck recruitment and signaling. Sci Signal. 2016;9:ra75. doi: 10.1126/scisignal.aaf0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett TA, Girdhar G, Lawshe A, Miller JL, Laurenzi IJ, Diamond SL, Diacovo TG. Alterations in the intrinsic properties of the GPIbα-VWF tether bond define the kinetics of the platelet-type von Willebrand disease mutation, Gly233Val. Blood. 2003;102:152–160. doi: 10.1182/blood-2003-01-0072. [DOI] [PubMed] [Google Scholar]
- Doggett TA, Girdhar G, Lawshe A, Schmidtke DW, Laurenzi IJ, Diamond SL, Diacovo TG. Selectin-like kinetics and biomechanics promote rapid platelet adhesion in flow: the GPIb(α)-vWF tether bond. Biophys J. 2002;83:194–205. doi: 10.1016/S0006-3495(02)75161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D, Lipfert J, Moolman MC, Dekker NH. Studying genomic processes at the single-molecule level: introducing the tools and applications. Nat Rev Genet. 2013;14:9–22. doi: 10.1038/nrg3316. [DOI] [PubMed] [Google Scholar]
- Dushek O, Das R, Coombs D. A role for rebinding in rapid and reliable T Cell responses to antigen. PLoS Comput Biol. 2009;5:e1000578. doi: 10.1371/journal.pcbi.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev. 2008;221:77–89. doi: 10.1111/j.1600-065X.2008.00589.x. [DOI] [PubMed] [Google Scholar]
- Elosegui-Artola A, Bazellieres E, Allen MD, Andreu I, Oria R, Sunyer R, Gomm JJ, Marshall JF, Jones JL, Trepat X, Roca-Cusachs P. Rigidity sensing and adaptation through regulation of integrin types. Nat Mater. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X, Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RD, Perkins VC, Henry A, Stephens PE, Robinson MK, Watt FM. A tumor-associated β1 integrin mutation that abrogates epithelial differentiation control. J Cell Biol. 2003;160:589–596. doi: 10.1083/jcb.200209016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, McArdle S, Marki A, Mikulski Z, Gutierrez E, Engelhardt B, Deutsch U, Ginsberg M, Groisman A, Ley K. Neutrophil recruitment limited by high-affinity bent β2 integrin binding ligand in cis. Nat Commun. 2016;7:12658. doi: 10.1038/ncomms12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghhi S, Munday AD, Tooley WW, Rajsekar S, Fura AM, Kulman JD, Lopez JA, Sniadecki NJ. Glycoprotein Ib-IX-V complex transmits cytoskeletal forces that enhance platelet adhesion. Biophys J. 2016;111:601–608. doi: 10.1016/j.bpj.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Brazin KN, Kobayashi E, Mallis RJ, Reinherz EL, Lang MJ. Mechanosensing drives acuity of αβ T-cell recognition. Proc Natl Acad Sci USA. 2017;114:E8204–E8213. doi: 10.1073/pnas.1703559114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore VF, Ju L, Chen Y, Zhu C, Barker TH. Dynamic catch of a Thy-1-α5β1+syndecan-4 trimolecular complex. Nat Commun. 2014;5:4886. doi: 10.1038/ncomms5886. [DOI] [PubMed] [Google Scholar]
- Fox JE, Aggerbeck LP, Berndt MC. Structure of the glycoprotein Ib.IX complex from platelet membranes. J Biol Chem. 1988;263:4882–4890. [PubMed] [Google Scholar]
- Friddle RW, Noy A, De Yoreo JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci USA. 2012;109:13573–13578. doi: 10.1073/pnas.1202946109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls α5β1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- Fu BM, Tarbell JM. Mechano-sensing and transduction by endothelial surface glycocalyx: composition, structure, function. Wiley Interdiscip Rev Syst Biol Med. 2013;5:381–390. doi: 10.1002/wsbm.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Jiang Y, Yang D, Scheiflinger F, Wong WP, Springer TA. Flow-induced elongation of von Willebrand factor precedes tension-dependent activation. Nat Commun. 2017;8:324. doi: 10.1038/s41467-017-00230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JR, Garcia AJ. Cellular mechanotransduction: sensing rigidity. Nat Mater. 2014;13:539–540. doi: 10.1038/nmat3996. [DOI] [PubMed] [Google Scholar]
- Gebhardt JC, Clemen AE, Jaud J, Rief M. Myosin-V is a mechanical ratchet. Proc Natl Acad Sci USA. 2006;103:8680–8685. doi: 10.1073/pnas.0510191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Sheetz MP. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol. 2006;16:213–223. doi: 10.1016/j.tcb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, Muller U. Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell. 2009;139:33–44. doi: 10.1016/j.cell.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PG, Walker RG. Molecular basis of mechanosensory transduction. Nature. 2001;413:194–202. doi: 10.1038/35093011. [DOI] [PubMed] [Google Scholar]
- Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou RB, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Yan C, Li H, Huang W, Shi X, Huang M, Wang Y, Pan W, Cai M, Li L, et al. Lipid-dependent conformational dynamics underlie the functional versatility of T-cell receptor. Cell Res. 2017;27:505–525. doi: 10.1038/cr.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining AW, Lieberthal TJ, Del Rio Hernandez A. Talin: a mechanosensitive molecule in health and disease. FASEB J. 2016;30:2073–2085. doi: 10.1096/fj.201500080R. [DOI] [PubMed] [Google Scholar]
- Halvorsen K, Wong WP. Massively parallel single-molecule manipulation using centrifugal force. Biophys J. 2010;98:L53–L55. doi: 10.1016/j.bpj.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer JA, III, Burkhardt JK. Controversy and consensus regarding myosin II function at the immunological synapse. Curr Opin Immunol. 2013;25:300–306. doi: 10.1016/j.coi.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SZ, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, Thomas K, Kaufman KM, Ojwang J, Rojas-Villarraga A, et al. Evaluation of imputation-based association in and around the integrin-α-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Hum Mol Genet. 2009;18:1171–1180. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122:159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs CS, Restifo NP. Reassessing target antigens for adoptive T-cell therapy. Nat Biotechnol. 2013;31:999–1008. doi: 10.1038/nbt.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivroz C, Saitakis M. Biophysical aspects of T lymphocyte activation at the immune synapse. Front Immunol. 2016;7:46. doi: 10.3389/fimmu.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey DA, Chen JC, Jacobs CR. The primary cilium as a novel extracellular sensor in bone. Front Endocrinol. 2012;3:75. doi: 10.3389/fendo.2012.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle AW, Engler AJ. More than a feeling: discovering, understanding, and influencing mechanosensing pathways. Curr Opin Biotechnol. 2011;22:648–654. doi: 10.1016/j.copbio.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Murugesan S, Betzig E, Hammer JA. Contractile actomyosin arcs promote the activation of primary mouse T cells in a ligand-dependent manner. PLoS ONE. 2017;12:e0183174. doi: 10.1371/journal.pone.0183174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Persaud SP, Horvath S, Allen PM, Evavold BD, Zhu C. Force-regulated in situ TCR-peptide-bound MHC class II kinetics determine functions of CD4+ T cells. J Immunol. 2015;195:3557–3564. doi: 10.4049/jimmunol.1501407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KH, Butte MJ. T cell activation requires force generation. J Cell Biol. 2016;213:535–542. doi: 10.1083/jcb.201511053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Luo BH. Integrin αIIbβ3 transmembrane domain separation mediates bi-directional signaling across the plasma membrane. PLoS ONE. 2015;10:e0116208. doi: 10.1371/journal.pone.0116208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol Biol Cell. 2015;26:685–695. doi: 10.1091/mbc.E14-03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, Danen EHJ. Adhesion signaling—crosstalk between integrins, Src and Rho. J Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat Immunol. 2009;10:531–539. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape-the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- Jani K, Schock F. Molecular mechanisms of mechanosensing in muscle development. Dev Dyn. 2009;238:1526–1534. doi: 10.1002/dvdy.21972. [DOI] [PubMed] [Google Scholar]
- Janostiak R, Pataki AC, Brabek J, Rosel D. Mechanosensors in integrin signaling: the emerging role of p130Cas. Eur J Cell Biol. 2014;93:445–454. doi: 10.1016/j.ejcb.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Jansen KA, Atherton P, Ballestrem C. Mechanotransduction at the cell-matrix interface. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.07.027. (in press) [DOI] [PubMed] [Google Scholar]
- Jansen KA, Bacabac RG, Piechocka IK, Koenderink GH. Cells actively stiffen fibrin networks by generating contractile stress. Biophys J. 2013;105:2240–2251. doi: 10.1016/j.bpj.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson A. Kinetic proofreading and the search for nonself-peptides. Self/nonself. 2011;2:1–3. doi: 10.4161/self.2.1.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, Zhu C. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity. 2011;34:13–23. doi: 10.1016/j.immuni.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Andricioaei I, Springer TA. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure. 2004;12:2137–2147. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Jin W, Black CT, Kam LC, Huse M. Probing synaptic biomechanics using micropillar arrays. Methods Mol Biol. 2017;1584:333–346. doi: 10.1007/978-1-4939-6881-7_19. [DOI] [PubMed] [Google Scholar]
- Ju L, Chen Y, Li K, Yuan Z, Liu B, Jackson SP, Zhu C, et al. Dual biomembrane force probe enables single-cell mechanical analysis of signal crosstalk between multiple molecular species. Sci Rep 7. 2017:14185. doi: 10.1038/s41598-017-13793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Chen Y, Xue L, Du X, Zhu C. Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. eLife. 2016;5:e15447. doi: 10.7554/eLife.15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Chen Y, Zhou F, Lu H, Cruz MA, Zhu C. Von Willebrand factor-A1 domain binds platelet glycoprotein Ibα in multiple states with distinctive force-dependent dissociation kinetics. Thromb Res. 2015a;136:606–612. doi: 10.1016/j.thromres.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Dong J-F, Cruz MA, Zhu C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ib. J Biol Chem. 2013;288:32289–32301. doi: 10.1074/jbc.M113.504001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Lou J, Chen Y, Li Z, Zhu C. Force-induced unfolding of leucine-rich repeats of glycoprotein Ibα strengthens ligand interaction. Biophys J. 2015b;109:1781–1784. doi: 10.1016/j.bpj.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Qian J, Zhu C. Transport regulation of two-dimensional receptor-ligand association. Biophys J. 2015c;108:1773–1784. doi: 10.1016/j.bpj.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. 2012;102:L5–L7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen J, Welzenbach K, Ramage P, Geyl D, Kriwacki R, Legge G, Cottens S, Weitz-Schmidt G, Hommel U. Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain. J Mol Biol. 1999;292:1–9. doi: 10.1006/jmbi.1999.3047. [DOI] [PubMed] [Google Scholar]
- Kasza KE, Vader D, Koster S, Wang N, Weitz DA. Magnetic twisting cytometry. Cold Spring Harb Protoc. 2011;2011:pdb prot5599. doi: 10.1101/pdb.prot5599. [DOI] [PubMed] [Google Scholar]
- Katz ZB, Novotna L, Blount A, Lillemeier BF. A cycle of Zap70 kinase activation and release from the TCR amplifies and disperses antigenic stimuli. Nat Immunol. 2017;18:86–95. doi: 10.1038/ni.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- Kim ST, Shin Y, Brazin K, Mallis RJ, Sun ZY, Wagner G, Lang MJ, Reinherz EL. TCR mechanobiology: torques and tunable structures linked to early T cell signaling. Front Immunol. 2012;3:76. doi: 10.3389/fimmu.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL. The αβ T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TJ, Zheng S, Sun J, Muhamed I, Wu J, Lei L, Kong X, Leckband DE, Wang Y. Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr Biol. 2015;25:218–224. doi: 10.1016/j.cub.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Li Z, Parks WM, Dumbauld DW, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell. 2013;49:1060–1068. doi: 10.1016/j.molcel.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Dong JF, Thaggard JA, Cruz MA, Lopez JA, McIntire LV. Kinetics of GPIbα-vWF-A1 tether bond under flow: effect of GPIbα mutations on the association and dissociation rates. Biophys J. 2003;85:4099–4109. doi: 10.1016/S0006-3495(03)74822-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JC ( Mechanotransduction at focal adhesions: integrating cytoskeletal mechanics in migrating cells. J Cell Mol Med. 2013;17:704–712. doi: 10.1111/jcmm.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam WA, Chaudhuri O, Crow A, Webster KD, Li TD, Kita A, Huang J, Fletcher DA. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nat Mater. 2011;10:61–66. doi: 10.1038/nmat2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Glassman CR, Deshpande NR, Badgandi HB, Parrish HL, Uttamapinant C, Stawski PS, Ting AY, Kuhns MS. A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3ζζ. Immunity. 2015;43:227–239. doi: 10.1016/j.immuni.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort CT, Hyun YM, Schultz JB, Law FY, Waugh RE, Knauf PA, Kim M. Outside-in signal transmission by conformational changes in integrin Mac-1. J Immunol. 2009;183:6460–6468. doi: 10.4049/jimmunol.0900983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort CT, Ley K. Neutrophil arrest by LFA-1 activation. Front Immunol. 2012;3:157. doi: 10.3389/fimmu.2012.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, Rodriguez V, Interlandi G, Klevit R, Vogel V, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Su Y, Xia W, Qin Y, Humphries MJ, Vestweber D, Cabanas C, Lu C, Springer TA. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 2017a;36:629–645. doi: 10.15252/embj.201695803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Guo X, Shi X, Li C, Wu W, Yan C, Wang H, Li H, Xu C. Ionic CD3-Lck interaction regulates the initiation of T-cell receptor signaling. Proc Natl Acad Sci USA. 2017b;114:E5891–E5899. doi: 10.1073/pnas.1701990114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Chen BM, Wu PC, Cheng TL, Kao LS, Tao MH, Lieber A, Roffler SR. Cutting edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol. 2010;184:5959–5963. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- Li Z, Kong F, Zhu C. A model for cyclic mechanical reinforcement. Sci Rep. 2016a;6:35954. doi: 10.1038/srep35954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lee H, Zhu C. Molecular mechanisms of mechanotransduction in integrin-mediated cell-matrix adhesion. Exp Cell Res. 2016b;349:85–94. doi: 10.1016/j.yexcr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim TS, Mortellaro A, Lim CT, Hammerling GJ, Ricciardi-Castagnoli P. Mechanical interactions between dendritic cells and T cells correlate with T cell responsiveness. J Immunol. 2011;187:258–265. doi: 10.4049/jimmunol.1100267. [DOI] [PubMed] [Google Scholar]
- Lim WA, June CH. The principles of engineering immune cells to treat cancer. Cell. 2017;168:724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014a;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen W, Zhu C. Molecular force spectroscopy on cells. Annu Rev Phys Chem. 2015a;66:427–451. doi: 10.1146/annurev-physchem-040214-121742. [DOI] [PubMed] [Google Scholar]
- Liu B, Zhong S, Malecek K, Johnson LA, Rosenberg SA, Zhu C, Krogsgaard M. 2D TCR-pMHC-CD8 kinetics determines T-cell responses in a self-antigen-specific TCR system. Eur J Immunol. 2014b;44:239–250. doi: 10.1002/eji.201343774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Z, Thinn AM, Ma YQ, Zhu J. The dual structural roles of the membrane distal region of the α-integrin cytoplasmic tail during integrin inside-out activation. J Cell Sci. 2015b;128:1718–1731. doi: 10.1242/jcs.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA. 2016;113:5610–5615. doi: 10.1073/pnas.1600163113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–853. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, Haltiwanger RS, Zhu C, Ha T, Garcia KC. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355:1320–1324. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007a;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Springer TA. Integrin structures and conformational signaling. Curr Opin Cell Biol. 2006;18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SZ, Mo X, Afshar-Kharghan V, Srinivasan S, Lopez JA, Li R. Glycoprotein Ibα forms disulfide bonds with 2 glycoprotein Ibβ subunits in the resting platelet. Blood. 2007b;109:603–609. doi: 10.1182/blood-2006-05-024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma VP, Liu Y, Blanchfield L, Su H, Evavold BD, Salaita K. Ratiometric tension probes for mapping receptor forces and clustering at intermembrane junctions. Nano Lett. 2016;16:4552–4559. doi: 10.1021/acs.nanolett.6b01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Prossnitz ER, Bunemann M, Wang JM, Yao W, Schmid-Schonbein GW. G protein-coupled receptors serve as mechanosensors for fluid shear stress in neutrophils. Am J Physiol Cell Physiol. 2006;290:C1633–1639. doi: 10.1152/ajpcell.00576.2005. [DOI] [PubMed] [Google Scholar]
- Maksimovic S, Nakatani M, Baba Y, Nelson AM, Marshall KL, Wellnitz SA, Firozi P, Woo SH, Ranade S, Patapoutian A, Lumpkin EA. Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature. 2014;509:617–621. doi: 10.1038/nature13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen B, Bongrand P. Early T cell activation: integrating biochemical, structural, and biophysical cues. Annu Rev Immunol. 2015;33:539–561. doi: 10.1146/annurev-immunol-032414-112158. [DOI] [PubMed] [Google Scholar]
- Mallis RJ, Bai K, Arthanari H, Hussey RE, Handley M, Li Z, Chingozha L, Duke-Cohan JS, Lu H, Wang JH, et al. Pre-TCR ligand binding impacts thymocyte development before αβTCR expression. Proc Natl Acad Sci USA. 2015;112:8373–8378. doi: 10.1073/pnas.1504971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Martin C, Morales LD, Cruz MA. Purified A2 domain of von Willebrand factor binds to the active conformation of von Willebrand factor and blocks the interaction with platelet glycoprotein Ibα. J Thromb Haemost. 2007;5:1363–1370. doi: 10.1111/j.1538-7836.2007.02536.x. [DOI] [PubMed] [Google Scholar]
- Maus MV, June CH. Making better chimeric antigen receptors for adoptive T-cell therapy. Clin Cancer Res. 2016;22:1875–1884. doi: 10.1158/1078-0432.CCR-15-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucato M, Pradella P, Cozzi MR, De Marco L, Ruggeri ZM. Sequential cytoplasmic calcium signals in a 2-stage platelet activation process induced by the glycoprotein Ibα mechanoreceptor. Blood. 2002;100:2793–2800. doi: 10.1182/blood-2002-02-0514. [DOI] [PubMed] [Google Scholar]
- McEwan PA, Yang WJ, Carr KH, Mo X, Zheng XF, Li RH, Emsley J. Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the crystal structures of GPIb β ectodomain and a GPIb β/GPIX chimera. Blood. 2011;118:958–958. doi: 10.1182/blood-2011-05-356253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan TW. Kinetic proofreading in T-cell receptor signal-transduction. Proc Natl Acad Sci USA. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- Mendolicchio GL, Ruggeri ZM. New perspectives on von Willebrand factor functions in hemostasis and thrombosis. Semin Hematol. 2005;42:5–14. doi: 10.1053/j.seminhematol.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Akiyama SK, Yamada KM. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science. 1995;267:883–885. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- Moore SW, Sheetz MP. Biophysics of substrate interaction: influence on neural motility, differentiation, and repair. Dev Neurobiol. 2011;71:1090–1101. doi: 10.1002/dneu.20947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- Muhamed I, Wu J, Sehgal P, Kong X, Tajik A, Wang N, Leckband DE. E-cadherin-mediated force transduction signals regulate global cell mechanics. J Cell Sci. 2016;129:1843–1854. doi: 10.1242/jcs.185447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan S, Hong J, Yi J, Li D, Beach JR, Shao L, Meinhardt J, Madison G, Wu X, Betzig E, Hammer JA. Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse. J Cell Biol. 2016;215:383–399. doi: 10.1083/jcb.201603080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers DR, Qiu Y, Fay ME, Tennenbaum M, Chester D, Cuadrado J, Sakurai Y, Baek J, Tran R, Ciciliano JC, et al. Single-platelet nanomechanics measured by high-throughput cytometry. Nat Mater. 2016;16:230–235. doi: 10.1038/nmat4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi-Matsui M, Sekiya M, Nakamoto RK, Futai M. The mechanism of rotating proton pumping ATPases. Biochim Biophys Acta. 2010;1797:1343–1352. doi: 10.1016/j.bbabio.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, Chen W, Zhu C, McEver RP, Kimberly RP, et al. A nonsynonymous functional variant in integrin-α(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- Natkanski E, Lee WY, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science. 2013;340:1587–1590. doi: 10.1126/science.1237572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt WS, Kulkarni S, Giuliano S, Goncalves I, Dopheide SM, Yap CL, Harper IS, Salem HH, Jackson SP. Distinct glycoprotein Ib/V/IX and integrin α IIbβ 3-dependent calcium signals cooperatively regulate platelet adhesion under flow. J Biol Chem. 2002;277:2965–2972. doi: 10.1074/jbc.M110070200. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- Nordenfelt P, Elliott HL, Springer TA. Coordinated integrin activation by actin-dependent force during T-cell migration. Nat Commun. 2016;7:13119. doi: 10.1038/ncomms13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, Milone MC. Substrate rigidity regulates human T cell activation and proliferation. J Immunol. 2012;189:1330–1339. doi: 10.4049/jimmunol.1102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell. 2006;10:11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Paluch EK, Aspalter IM, Sixt M. Focal adhesion-independent cell migration. Annu Rev Cell Dev Biol. 2016;32:469–490. doi: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc Natl Acad Sci USA. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov SV, Sabass B, Schwarz US, Waterman CM. High-resolution traction force microscopy. Methods Cell Biol. 2014;123:367–394. doi: 10.1016/B978-0-12-420138-5.00020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD, Zhu C. Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J Immunol. 2014;193:68–76. doi: 10.4049/jimmunol.1303436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E, Vogel V. Integrin activation dynamics between the RGD-binding site and the headpiece hinge. J Biol Chem. 2009;284:36557–36568. doi: 10.1074/jbc.M109.041194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Brown AC, Myers DR, Sakurai Y, Mannino RG, Tran R, Ahn B, Hardy ET, Kee MF, Kumar S, et al. Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc Natl Acad Sci USA. 2014;111:14430–14435. doi: 10.1073/pnas.1322917111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Beeson C, Lyons DS, Davis MM, McConnell HM. Kinetic discrimination in T-cell activation. Proc Natl Acad Sci USA. 1996;93:1401–1405. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raborn J, Wang W, Luo BH. Regulation of integrin αIIbβ3 ligand binding and signaling by the metal ion binding sites in the βI domain. Biochemistry. 2011;50:2084–2091. doi: 10.1021/bi2000092. [DOI] [PubMed] [Google Scholar]
- Rakshit S, Sivasankar S. Biomechanics of cell adhesion: how force regulates the lifetime of adhesive bonds at the single molecule level. Phys Chem Chem Phys. 2014;16:2211–2223. doi: 10.1039/c3cp53963f. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Yago T, Epperson TK, Kobzdej MM, Nollert MU, Cummings RD, Zhu C, McEver RP. Dimerization of a selectin and its ligand stabilizes cell rolling and enhances tether strength in shear flow. Proc Natl Acad Sci USA. 2001;98:10166–10171. doi: 10.1073/pnas.171248098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert P, Aleksic M, Dushek O, Cerundolo V, Bongrand P, van der Merwe PA. Kinetics and mechanics of two-dimensional interactions between T cell receptors and different activating ligands. Biophys J. 2012;102:248–257. doi: 10.1016/j.bpj.2011.11.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Conte V, Trepat X. Quantifying forces in cell biology. Nat Cell Biol. 2017;19:742–751. doi: 10.1038/ncb3564. [DOI] [PubMed] [Google Scholar]
- Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of α(5)β(1) integrins determines adhesion strength whereas α(v)β(3) and talin enable mechanotransduction. Proc Natl Acad Sci USA. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Iskratsch T, Sheetz MP. Finding the weakest link: exploring integrin-mediated mechanical molecular pathways. J Cell Sci. 2012;125:3025–3038. doi: 10.1242/jcs.095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romet-Lemonne G, Jegou A. Mechanotransduction down to individual actin filaments. Eur J Cell Biol. 2013;92:333–338. doi: 10.1016/j.ejcb.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Rondelez Y, Tresset G, Nakashima T, Kato-Yamada Y, Fujita H, Takeuchi S, Noji H. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature. 2005;433:773–777. doi: 10.1038/nature03277. [DOI] [PubMed] [Google Scholar]
- Rooney MM, Farrell DH, van Hemel BM, de Groot PG, Lord ST. The contribution of the three hypothesized integrin-binding sites in fibrinogen to platelet-mediated clot retraction. Blood. 1998;92:2374–2381. [PubMed] [Google Scholar]
- Rosetti F, Chen Y, Sen M, Thayer E, Azcutia V, Herter JM, Luscinskas FW, Cullere X, Zhu C, Mayadas TN. A lupus-associated Mac-1 variant has defects in integrin allostery and interaction with ligands under force. Cell Rep. 2015;10:1655–1664. doi: 10.1016/j.celrep.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage B, Saldivar E, Ruggeri ZM. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84:289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Hordijk PL. Cell-stiffness-induced mechanosignaling—a key driver of leukocyte transendothelial migration. J Cell Sci. 2015;128:2221–2230. doi: 10.1242/jcs.163055. [DOI] [PubMed] [Google Scholar]
- Schamel WW, Alarcon B, Hofer T, Minguet S. The allostery model of TCR regulation. J Immunol. 2017;198:47–52. doi: 10.4049/jimmunol.1601661. [DOI] [PubMed] [Google Scholar]
- Schneider SW, Nuschele S, Wixforth A, Gorzelanny C, Alexander-Katz A, Netz RR, Schneider MF. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci USA. 2007;104:7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Curr Opin Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong J, Wang N, Wang Y. Mechanotransduction at focal adhesions: from physiology to cancer development. J Cell Mol Med. 2013;17:597–604. doi: 10.1111/jcmm.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy SP, Grosberg A, Parker KK. The contribution of cellular mechanotransduction to cardiomyocyte form and function. Biomech Model Mechanobiol. 2012;11:1227–1239. doi: 10.1007/s10237-012-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims TN, Soos TJ, Xenias HS, Dubin-Thaler B, Hofman JM, Waite JC, Cameron TO, Thomas VK, Varma R, Wiggins CH, et al. Opposing effects of PKCθ and WASp on symmetry breaking and relocation of the immunological synapse. Cell. 2007;129:773–785. doi: 10.1016/j.cell.2007.03.037. [DOI] [PubMed] [Google Scholar]
- Sing CE, Alexander-Katz A. Elongational flow induces the unfolding of von Willebrand factor at physiological flow rates. Biophys J. 2010;98:L35–L37. doi: 10.1016/j.bpj.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24:107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, Munster S, Aifantis KE, Fabry B. Three-dimensional force microscopy of cells in biopolymer networks. Nat Methods. 2016;13:171–176. doi: 10.1038/nmeth.3685. [DOI] [PubMed] [Google Scholar]
- Stepanek O, Prabhakar AS, Osswald C, King CG, Bulek A, Naeher D, Beaufils-Hugot M, Abanto ML, Galati V, Hausmann B, et al. Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell. 2014;159:333–345. doi: 10.1016/j.cell.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc Natl Acad Sci USA. 2012;109:E2757–E2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- Su Y, Xia W, Li J, Walz T, Humphries MJ, Vestweber D, Cabanas C, Lu C, Springer TA. Relating conformation to function in integrin α5β1. Proc Natl Acad Sci USA. 2016;113:E3872–E3881. doi: 10.1073/pnas.1605074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev SI, Sigurdson WJ, Kung C, Sachs F. Energetic and spatial parameters for gating of the bacterial large conductance mechanosensitive channel, MscL. J Gen Physiol. 1999;113:525–540. doi: 10.1085/jgp.113.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Guo SS, Fassler R. Integrin-mediated mechanotransduction. J Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater. 2016;15:1287–1296. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–511. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyjanova J, Hannen E, Bar-Kochba E, Darling EM, Henann DL, Franck C. 3D viscoelastic traction force microscopy. Soft Matter. 2014;10:8095–8106. doi: 10.1039/c4sm01271b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X, Fredberg JJ. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 2011;21:638–646. doi: 10.1016/j.tcb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrichts H, Udvardy M, Lenting PJ, Pareyn I, Vandeputte N, Vanhoorelbeke K, Deckmyn H. Shielding of the A1 domain by the D′D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J Biol Chem. 2006;281:4699–4707. doi: 10.1074/jbc.M513314200. [DOI] [PubMed] [Google Scholar]
- Upadhyaya A ( Mechanosensing in the immune response. Semin Cell Dev Biol. 2017;71:137–145. doi: 10.1016/j.semcdb.2017.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahidkhah K, Diamond SL, Bagchi P. Platelet dynamics in three-dimensional simulation of whole blood. Biophys J. 2014;106:2529–2540. doi: 10.1016/j.bpj.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- Vanhoorelbeke K, De Meyer SF Pareyn I, Melchior C, Plancon S, Margue C, Pradier O, Fondu P, Kieffer N, Springer TA, Deckmyn H. The novel S527F mutation in the integrin β3 chain induces a high affinity αIIbβ3 receptor by hindering adoption of the bent conformation. J Biol Chem. 2009;284:14914–14920. doi: 10.1074/jbc.M809167200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu Rev Biophys Biomol Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Volk T, Fessler LI, Fessler JH. A role for integrin in the formation of sarcomeric cytoarchitecture. Cell. 1990;63:525–536. doi: 10.1016/0092-8674(90)90449-o. [DOI] [PubMed] [Google Scholar]
- Wan J, Ristenpart WD, Stone HA. Dynamics of shear-induced ATP release from red blood cells. Proc Natl Acad Sci USA. 2008;105:16432–16437. doi: 10.1073/pnas.0805779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Chen X, Chen H, Ji Q, Chen Y, Wang J, Cao Y, Wang F, Lou J, Tang Z, Liu W. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. eLife. 2015;4:e06925. doi: 10.7554/eLife.06925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Reinherz EL. The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev. 2012;250:102–119. doi: 10.1111/j.1600-065X.2012.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- Welch MP, Odland GF, Clark RA. Temporal relationships of F-actin bundle formation, collagen and fibronectin matrix assembly, and fibronectin receptor expression to wound contraction. J Cell Biol. 1990;110:133–145. doi: 10.1083/jcb.110.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Wu T, Lin J, Cruz MA, Dong JF, Zhu C. Force-induced cleavage of single VWFA1A2A3 tridomains by ADAMTS-13. Blood. 2010;115:370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Lee CY, Li T, Chen W, Lou J, Zhu C. Structural basis and kinetics of force-induced conformational changes of an αA domain-containing integrin. PLoS ONE. 2011;6:e27946. doi: 10.1371/journal.pone.0027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhu J, Chen X, Mi L, Nishida N, Springer TA. Structure of an integrin with an αI domain, complement receptor type 4. EMBO J. 2010;29:666–679. doi: 10.1038/emboj.2009.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yago T, Lou J, Wu T, Yang J, Miner JJ, Coburn L, Lopez JA, Cruz MA, Dong JF, McIntire LV, et al. Platelet glycoprotein Ibα forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 2008;118:3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yago T, Zarnitsyna VI, Klopocki AG, McEver RP, Zhu C. Transport governs flow-enhanced cell tethering through L-selectin at threshold shear. Biophys J. 2007;92:330–342. doi: 10.1529/biophysj.106.090969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Miyamoto S. Integrin transmembrane signaling and cytoskeletal control. Curr Opin Cell Biol. 1995;7:681–689. doi: 10.1016/0955-0674(95)80110-3. [DOI] [PubMed] [Google Scholar]
- Yang D, Ward A, Halvorsen K, Wong WP. Multiplexed single-molecule force spectroscopy using a centrifuge. Nat Commun. 2016;7:11026. doi: 10.1038/ncomms11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Klapholz B, Hu X, Toseland CP, Guo Y, Cong P, Sheetz MP, Yan J. The mechanical response of talin. Nat Commun. 2016;7:11966. doi: 10.1038/ncomms11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Jiang X, Haydar N, Zhang C, Shan H, Zhu J. Modulation of integrin activation and signaling by α1/α1’-helix unbending at the junction. J Cell Sci. 2013a;126:5735–5747. doi: 10.1242/jcs.137828. [DOI] [PubMed] [Google Scholar]
- Zhang HQ, Berg JS, Li ZL, Wang YL, Lang P, Sousa AD, Bhaskar A, Cheney RE, Stromblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- Zhang W, Deng W, Zhou L, Xu Y, Yang W, Liang X, Wang Y, Kulman JD, Zhang XF, Li R. Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood. 2015;125:562–569. doi: 10.1182/blood-2014-07-589507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiang N, Zarnitsyna VI, Klopocki AG, McEver RP, Zhu C. P-selectin glycoprotein ligand-1 forms dimeric interactions with E-selectin but monomeric interactions with L-selectin on cell surfaces. PLoS ONE. 2013b;8:e57202. doi: 10.1371/journal.pone.0057202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wei F, Poh YC, Jia Q, Chen J, Chen J, Luo J, Yao W, Zhou W, Huang W, et al. Interfacing 3D magnetic twisting cytometry with confocal fluorescence microscopy to image force responses in living cells. Nat Protoc. 2017;12:1437–1450. doi: 10.1038/nprot.2017.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA. Sequence and structure relationships within von Willebrand factor. Blood. 2012;120:449–458. doi: 10.1182/blood-2012-01-405134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Jiang N, Huang J, Zarnitsyna VI, Evavold BD. Insights from in situ analysis of TCR-pMHC recognition: response of an interaction network. Immunol Rev. 2013a;251:49–64. doi: 10.1111/imr.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Yago T, Lou J, Zarnitsyna VI, McEver RP. Mechanisms for flow-enhanced cell adhesion. Ann Biomed Eng. 2008a;36:604–621. doi: 10.1007/s10439-008-9464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008b;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu J, Springer TA. Complete integrin headpiece opening in eight steps. J Cell Biol. 2013b;201:1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]