Light- and liquid-phase scanning transmission electron microscopy were used to analyze the influence of the drug trastuzumab on HER2 proteins in the plasma membranes of breast cancer cells. The drug was found to induce HER2 uptake for bulk cancer cells. In contrast, HER2 was not internalized in rare subpopulations of resting and cancer stem cells.
Abstract
The development of drug resistance in cancer poses a major clinical problem. An example is human epidermal growth factor receptor 2 (HER2) overexpressing breast cancer often treated with anti-HER2 antibody therapies, such as trastuzumab. Because drug resistance is rooted mainly in tumor cell heterogeneity, we examined the drug effect in different subpopulations of SKBR3 breast cancer cells and compared the results with those of a drug-resistant cell line, HCC1954. Correlative light microscopy and liquid-phase scanning transmission electron microscopy were used to quantitatively analyze HER2 responses upon drug binding, whereby many tens of whole cells were imaged. Trastuzumab was found to selectively cross-link and down-regulate HER2 homodimers from the plasma membranes of bulk cancer cells. In contrast, HER2 resided mainly as monomers in rare subpopulations of resting and cancer stem cells (CSCs), and these monomers were not internalized after drug binding. The HER2 distribution was hardly influenced by trastuzumab for the HCC1954 cells. These findings show that resting cells and CSCs are irresponsive to the drug and thus point toward a molecular explanation behind the origin of drug resistance. This analytical method is broadly applicable to study membrane protein interactions in the intact plasma membrane, while accounting for cell heterogeneity.
INTRODUCTION
Human epidermal growth factor receptor 2 (HER2), a member of the ErbB family of growth factor receptors, is overexpressed in the particularly aggressive HER2+ subtype of breast cancer (Yarden and Sliwkowski, 2001) diagnosed in ∼20% of breast cancer patients (Vu and Claret, 2012). The protein resides in the plasma membrane in a constitutive open conformation, ready to form active homo- or heterodimers and stimulate cellular growth signaling. As a consequence, intracellular signaling and cell growth is dysregulated in HER2-overexpressing cells (Muthuswamy et al., 1999). This is in contrast to the other HER family members that require binding of respective ligands to obtain an active conformation. State-of-the-art antibody-based therapeutics such as trastuzumab block signaling by HER2 homodimers (Ghosh et al., 2011). In early and metastatic stages of the disease, this treatment strategy significantly improves survival. However, the majority of patients develop drug resistance about a year after initially successful treatment (Vu et al., 2014) against which remedies are mostly lacking (Meacham and Morrison, 2013). Resistance development is associated with cancer cell heterogeneity enabling subpopulations of tumor cells to survive (Aguirre-Ghiso, 2007; Bedard et al., 2013). Heterogeneity of cancer cells within single tumors can arise from both genetic variations (Yates and Campbell, 2012) and epigenetic factors (Feinberg et al., 2006). The potential of tumor cells to interconvert between different cancer cell phenotypes complicates matters (Jordan et al., 2016). However, studying molecular responses upon drug binding while accounting for cell heterogeneity has suffered from the limitations of the available analytical methods (Kolch and Pitt, 2010; Burrell and Swanton, 2014; Peckys et al., 2015). For example, biochemical methods generally used to monitor the cellular responses to drugs do not examine individual cells but provide information about ensemble averages from pooled cellular material of many thousands of cells where, in addition, protein complexes are investigated devoid of their native environment. Innovations in single-cell methods, especially at the protein level, are thus needed (Chen et al., 2016).
We examined the differences in response to trastuzumab between the bulk population and rare subpopulations of the HER2-overexpressing breast cancer cell line SKBR3 and in a drug-resistant cell line, HCC1954, at the single-molecule level using liquid-phase electron microscopy (de Jonge et al., 2009; de Jonge and Ross, 2011; Peckys et al., 2015; Ross, 2015) combined with quantitative light microscopy. The influence trastuzumab exerts on HER2 complex formation was directly visualized from the positions of individual receptors while they remained in their native environment of the intact plasma membrane of whole cells. More than a thousand cells were examined via light microscopy and several tens of selected cells at high spatial resolution with electron microscopy. Significant differences in drug response were found between the bulk cancer cells and rare subpopulations.
RESULTS
Identifying subpopulations of breast cancer cells
SKBR3 breast cancer cells were first classified using light microscopy. This well-established cell line has previously served as a model system of HER2+ breast cancer in numerous in vitro studies (Hynes and Dey, 2009; Henjes et al., 2012). The cells were not sorted before analysis to preserve the undisturbed cellular diversity within the population (Fillmore and Kuperwasser, 2008), and, moreover, the cells were examined in an adhered state, avoiding altered receptor signaling due to detachment (Nagy et al., 1998). With the purpose of enhanced reproducibility of the experiments, the cells were serum starved before the experiments (Pirkmajer and Chibalin, 2011). Serum starvation leads to cell cycle arrest at the G0/G1 phase, with the consequence of an overall reduction in the proliferation rate. Because the cell cycle time under our experimental conditions was more than 48 h, our experiments were thus performed with cells in different cell cycle states but with a preference for the G0 and G1 phases. In SKBR3 cells, the expression level of membrane-bound HER2 does not significantly differ between cells cultured with and without serum starvation (Bjorkelund et al., 2011a), and so we deemed serum starvation an acceptable procedure.
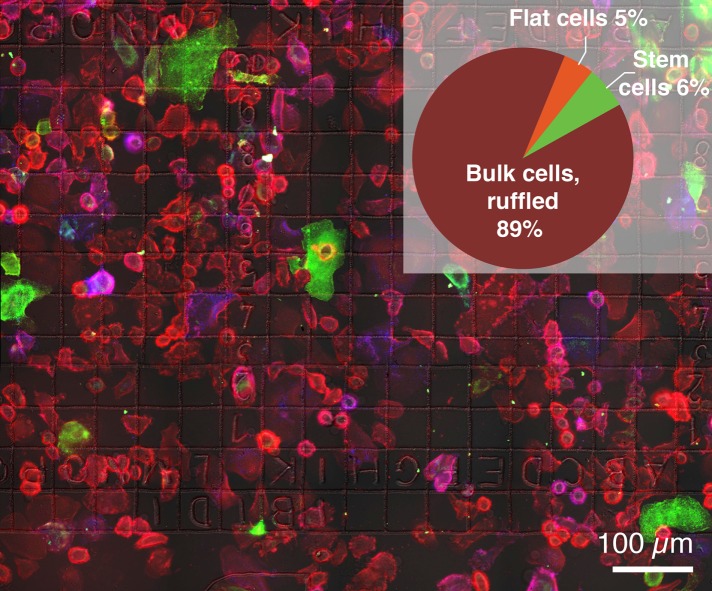
Membrane-bound HER2 in live cells was specifically labeled with an Affibody peptide (Eigenbrot et al., 2010). After fixation, quantum dot (QD) nanoparticles were coupled in a 1:1 stoichiometry to the Affibody peptides for detection with correlative fluorescence- and electron microscopy (Peckys et al., 2015). The membrane surface density of HER2 largely differed between cells, and the cells also exhibited a large degree of morphological heterogeneity (Figure 1). By far the most cells displayed a plasma membrane with membrane ruffles (e.g., cell #1 in Figure 2 and cell #1 in Figure 3, A and B). Membrane ruffles, also known as invadopodia, are highly dynamic structures involved in migration and metastasis (Weaver, 2006). In addition, some cells were identified that did not contain membrane ruffles in the peripheral plasma membrane areas (cell #2 in Figure 2). These flat cells were characterized as resting cancer cells (Sosa et al., 2014) (Supplemental Movie 1). Breast cancer stem cells (CSCs), also referred to as cancer stem-like cells (Mimeault et al., 2007; Al-Ejeh et al., 2011), were identified by the expression state of two characteristic plasma membrane proteins, namely the presence of CD44 (CD44+) combined with the absence or a low-level expression of CD24 (CD24−) (e.g., cell #2 in Figure 3). The surface marker signature CD44+/CD24– has been successfully applied in breast cancer cells for the identification of CSCs (Mimeault et al., 2007; Al-Ejeh et al., 2011), and additional proof of “stemness” for the SKBR3 cell line CD44+/CD24− is not be required (Gastl et al., 2000; Fillmore and Kuperwasser, 2008). SKBR3 cells were thus classified into three distinct phenotypic subpopulations: 1) a large fraction of ruffled bulk cells and 2) sparse flat and 3) CSCs. The ratios of these three subpopulations were determined from a total of 3367 analyzed cells (Figure 1, inset). The ruffled bulk cells formed the majority by far, while the other two subpopulations represented only 11% in total.
FIGURE 1:
Fluorescence microscopy image of adherent SKBR3 breast cancer cells. QD labels coupled to HER2 membrane proteins appear in red. Membrane-bound CD44 protein is shown in green and CD24 in blue. (Inset) Distribution into three different phenotypic subpopulations, ruffled bulk, flat, and CSCs as determined from a total of 3367 cells.
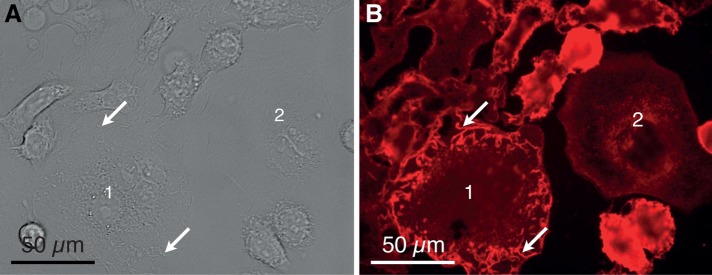
FIGURE 2:
Examples of SKBR3 breast cancer cells adhered at the bottom of a cell culture dish. (A) DIC image recorded with a 40× objective showing several adhered cells. Most cells exhibit membrane ruffles characterized as elongated membrane structures protruding from the plane of the membrane and often appearing at the cell periphery (e.g., at the arrows over cell 1). One cell does not contain membrane ruffles in its peripheral plasma membrane region and is identified as a flat cell (cell 2). (B) Fluorescence image of the same position as in A. The red fluorescence originates from the HER2-Affibody QD label.
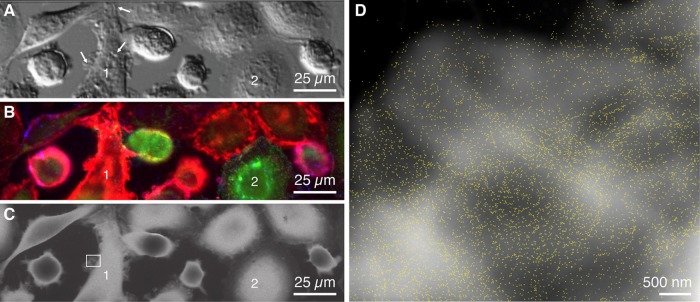
FIGURE 3:
Light- and electron microscopy images of whole SKBR3 cells in liquid. (A) DIC image of cells mostly with membrane ruffles (e.g., at the arrows) with dominating red fluorescence (e.g., cell 1) and two CSCs with green fluorescence signals (e.g., cell 2). (B) Fluorescence microscopy image showing bulk cancer cells. Red, QDs bound to HER2; green, CD44; blue, CD24. (C) Liquid-phase STEM image of the same cells. (D) STEM image recorded at the position of the rectangle in C displaying the individual HER2-QD labels. The brighter background shapes reflect membrane ruffles. The magnification was 50,000×. Label outlines are marked in yellow.
The HCC1954 cells were analyzed in a similar manner, but distinct subpopulations were not identified. Supplemental Figure S1 shows that almost all HCC1954 cells belong to the CD44+/CD24− phenotype, and are thus CSCs.
Analyzing the spatial distribution of HER2
To examine the functional state of the HER2 receptors, that is, their stoichiometric orientation in either monomers or signaling active homodimers, the spatial distribution of HER2 was studied at the single-molecule level in thin areas of the plasma membrane of selected cells. Note that heterodimers were not detected in this study, but this would be possible in principle using a multiple of labels of different size, shape, or composition. Cells classified using light microscopy were relocated in the corresponding liquid-phase scanning transmission electron microscopy (STEM) overview image (Figure 3C). The spatial distribution of HER2 was then determined based on the locations of individual HER2 proteins. Series of images from the cells at their peripheral regions were recorded at high magnification so that the dense cores of the individual nanoparticle labels bound to HER2 became visible (Figure 3D). Individual HER2 proteins were localized despite the high surface density of endogenously expressed HER2, which would hamper application of commonly used high-resolution microscopy techniques (Shivanandan et al., 2014). This image contains a total of 6902 detected QD labels. Many labels are positioned in close proximity, and many labels pairs are visible that are at such close distance that they were possibly bound to a HER2 homodimer.
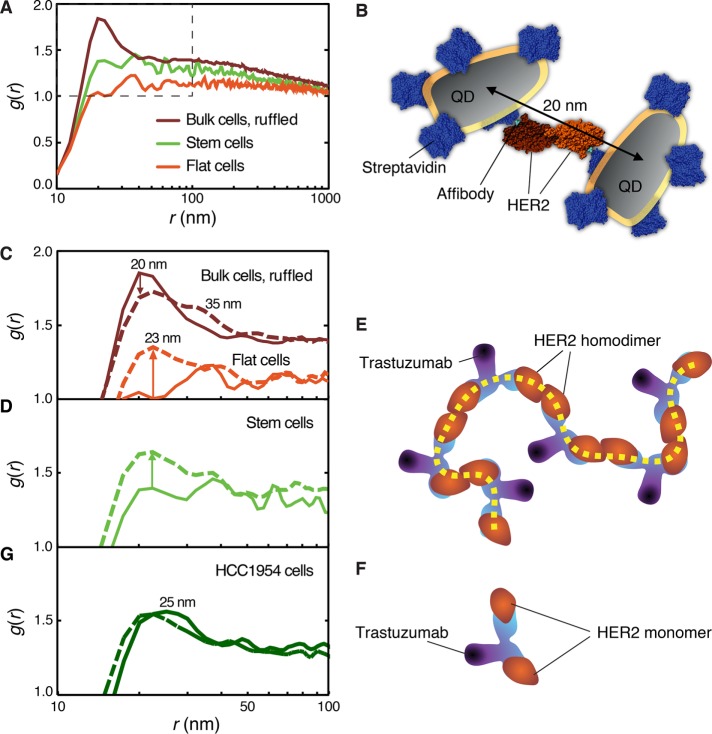
A complication in the analysis of the functional state of the receptor is that one does not know whether observed pairs of labels belong to an activated HER2 homodimer or are rather positioned in close proximity by random chance, which increases the higher the density of proteins becomes. Second, the labeling efficiency may not be 100%. Therefore the spatial distribution of the receptors was statistically analyzed using the pair correlation function g(r), with r the interlabel distance. For a random distribution g(r) = 1. Larger values indicate a higher than random probability of a certain distance to occur. The data for 86,003 labels, analyzed for 11 ruffled bulk cancer cells, show a sharp peak in g(r) at r = 20 nm (Figure 4A and Table 1). The fact that this interlabel distance was found with an above-random probability indicates an underlying molecular mechanism for the positioning of HER2, that is, dimerization (Peckys et al., 2015). In the absence of an x-ray structure for the HER2 homodimer, a schematic model was constructed using the dimensions of the monomeric HER2 protein (Figure 4B) to illustrate the origin of the measured 20 nm. The observed maximum of g(r) is thus explained by the presence of QD labels coupled to HER2 homodimers.
FIGURE 4:
Statistical analysis of the influence of trastuzumab on the HER2 positions for different phenotypic subpopulations of SKBR 3 breast cancer cells and HCC1954 drug-resistant breast cancer cells. (A) Pair correlation function g(r) for three subpopulations of cells. (B) Top view of a schematic molecular model of two QDs coupled to a HER2 homodimer via streptavidin and Affibody. Protein Data Base structures used were 3MZW and 1STP. (C) g(r) after 3-min incubation with trastuzumab compared with the control. (D) g(r) with and without 3-min trastuzumab incubation for CSCs. (E) Model of the binding of trastuzumab to HER2 homodimers leading to chain (dashed line) formation. (F) Trastuzumab binding to two HER2 monomers. (G) Analysis of HCC1954 drug-resistant cells control (solid line) and with 3-min trastuzumab (dashed line).
TABLE 1:
Breast cancer cells analyzed without drug and after trastuzumab exposure.
| Phenotype | Number of cells | Number of labels | Label surface density/μm2 | Analyzed area/μm2 |
|---|---|---|---|---|
| Ruffled bulk cells | 11 | 86,003 | 93 ± 58 | 792 |
| Flat cells | 3 | 9084 | 61 ± 27 | 139 |
| Stem-like cells | 12 | 27,785 | 47 ± 23 | 779 |
| HCC1954 | 7 | 44,879 | 84 ± 33 | 534 |
| After 3-min exposure to trastuzumab: | ||||
| Ruffled bulk cells | 19 | 112,014 | 72 ± 38 | 1713 |
| Flat cells | 5 | 21,000 | 45 ± 23 | 410 |
| Stem-like cells | 7 | 34,825 | 52 ± 23 | 721 |
| HCC1954 | 7 | 28,238 | 53 ± 27 | 530 |
SKBR3 cells were divided into three phenotype groups. One group was found for HCC1954 cells. QD labels attached to membrane-bound HER2 receptors were imaged with STEM. The cumulative number of labels reflects the amount of data used for calculating g(r) of a group. The label surface density including SD represents the average value of the surface densities of individual cells.
The peak around 20 nm is not a sharp peak at 20 nm but exists only from 15 to 25 nm, after which it flattens out. This width reflects a variation in the distances between the labels. This distance is explained by factors including 1) the flexibility of the peptides linking the QD to the HER2 protein, 2) QD size differences, and 3) different orientations of the QDs, which have an effect because the nanoparticles are elongated objects with a length of 14 nm and a width 6 nm (Peckys et al., 2015). The broad flank of the peak extending out to 1 µm indicates HER2 clustering within the membrane ruffles. It should be noted that the role of signaling active HER2 homo- and heterodimers has been a matter of lively scientific debate (Arkhipov et al., 2013; Hu et al., 2015) and that the dimerization properties of HER2 are still incompletely understood (Arkhipov et al., 2013). This lack of knowledge was presumably the consequence of a lack of analytical methods. The presence of homodimers in a cell population can be identified in principle with biochemical methods using pooled cellular material. However, this has been practically difficult for the HER2 homodimer because its extracellular domain does not homodimerize in solution (Badache and Hynes, 2004; Arkhipov et al., 2013). Direct visualizing of labeled HER2 proteins while they remained in the intact plasma membrane has shed light on this matter recently (Peckys et al., 2015). Note that it was possible to observe this difference between bulk and flat cells despite that fact that the QD labels were actually larger than the HER2 proteins themselves, and despite experimental variations between individual measurements originating from linker flexibility and differences in QD size and orientation.
In contrast to the bulk cells, flat cancer cells did not exhibit such a peak at 20 nm suggesting that HER2 homodimers were not present (Peckys et al., 2015) and instead HER2 preferentially existed in its inactive monomer conformation. A minor fraction of HER2 might have formed heterodimers with HER1 or HER3, expressed in SKBR3 cells but at two orders of magnitude lower levels (Bjorkelund et al., 2011b) than HER2; however, no ligands were present that might have activated HER1 or HER3 heterodimerization. The g(r) curve of the CSCs appears to be dominated by statistical fluctuations. No peak at 20 nm was visible above the noise level, while the broad flank reached an average value of 1.4 at around 30 nm (Figure 4A). This finding indicates that the ratio of HER2 homodimers versus monomers is similar to that of flat cells.
Visualizing the effect of trastuzumab binding on the HER2 distribution
The spatial rearrangement of HER2 initiated by trastuzumab binding to HER2 was directly examined by incubating live cells first with anti-HER2 Affibody, followed by exposure to the standard clinical dose of trastuzumab (10 ng/µl) for 3 min at 37°C. The Affibody and trastuzumab do not interfere in binding to HER2 because they bind different epitopes and functional domains (Eigenbrot et al., 2010). After trastuzumab incubation, the cells were fixed, QDs and fluorescent CSC markers were added, and 31 cells were imaged with correlative light- and electron microscopy. The observed decrease of the dimer peak at g(r = 20 nm) for the bulk cancer cells (Figure 4C) reflects a reduction of the relative number of HER2 homodimers in the plasma membrane, while the peak shift to 23 nm and the appearance of a 35-nm shoulder indicates the formation of other types of HER2 protein clusters. A model for this behavior is the binding of trastuzumab to the HER2 homodimer followed by the cross-linking of neighboring HER2 homodimers into chains (Figure 4E). This is consistent with previous work describing antibody-induced endocytosis of HER2 (Hurwitz et al., 1995; Ram et al., 2014) and stronger effects of trastuzumab on cells with higher levels of HER2 homodimers (Ghosh et al., 2011). Because the formation of cross-linked membrane proteins may lead to endocytosis (Drebin et al., 1985; Daeron, 1997), uptake of cross-linked HER2-homodimers was likely induced.
Different drug response in flat cancer cells and CSCs
Flat cells responded in a manner opposite that of bulk cells to trastuzumab incubation (Figure 4C). Whereas the HER2 homodimer peak at 20 nm decreased for the ruffled bulk cells, a new peak arose for the flat cells, but its shape was different from the typical dimer peak; it was broader and had its maximum at 23 nm. A similar effect as for the flat cells was observed for the CSCs (Figure 4D) for which a broad peak with a maximum at 23 nm appeared. The shape of the peak at r = 23 nm is different from the peak at r = 20 nm for the bulk cells reflecting HER2 homodimers and indicates a different underlying molecular structure. The cross-linking of HER2 into chains is assumed to be negligible in these cells because HER2 homodimers are mostly lacking. Instead, trastuzumab can complex only two HER2 monomers (Figure 4F) or heterodimers.
Quantification of membrane-bound HER2 in HCC1954 cells and the effect of trastuzumab
To test whether the findings from the CSC subpopulation of SKBR3 cells are specific for this cell line, we conducted experiments with the HCC1954 cell line (Table 1). This HER2-overexpressing cancer cell line is known to be trastuzumab resistant (von der Heyde et al., 2015) due to constitutive activation of the PI3K pathway, as a consequence of mutations in the PIK3CA gene. The g(r) analysis of control HCC1954 cells revealed a smaller occurrence of the 20-nm homodimer distance, and a peak value around 25 nm, with additional preferred distances up to 35 nm (Figure 4G), compared with the SKBR3 bulk cell populations. This broadening of the homodimer peak might be explained by a wider gap between the two HER2 molecules in a homodimer due to the interaction of HER2 and CD44 (Ghatak et al., 2005). In line with this hypothesis is the resemblance of the general shape of the HCC1954 g(r) curve with that of the SKBR3 CSC subpopulation under control conditions. This resemblance was also found after 3 min incubation with trastuzumab, whereby HCC1954 cells showed almost no change in the peak maximum being shifted toward 23 nm, an effect found in all g(r) subpopulation curves after 3 min drug exposure. We therefore conclude that the blunted drug responses of SKBR3 cells with a CSC phenotype are not cell line specific but relate to their stem-like phenotype, as they can also be found in the other HER2-overexpressing cell with this phenotype.
Trastuzumab-induced HER2 uptake in cancer cells
The reduction of the peak at g(r) = 20 nm for bulk cells indicates a possible cellular uptake of HER2 initiated by trastuzumab binding, while uptake does not seem to be pronounced for flat cells and CSCs. This was examined by determining the amount of membrane-bound HER2 for the three subpopulations of SKBR3 breast cancer cells, ruffled bulk, ruffled flat, and CSCs. The HER2 label was added after 1 h of trastuzumab incubation in order to measure the remaining HER2 in the plasma membrane. The total amount of HER2-bound QD fluorescence per cell was calculated by selecting the area in QD-fluorescence images within the boundary of a cell and summing all pixel intensities (Supplemental Figure S2). Because the total fluorescence signal in a cell can be assumed to be proportional to the number of QD labels, the intensity measures the relative amount of membrane-bound HER2 receptors. Figure 5 shows that the amount of HER2 remaining in the plasma membrane after 1 h of drug incubation was reduced in the bulk (ruffled) cancer cells by a factor of 2.7 of the median compared with the control. The observed cellular uptake of HER2 for bulk cells is consistent with the known association of trastuzumab binding with HER2 endocytosis (Ram et al., 2014). At least a fraction of HER2 was localized in vesicles, presumably early endosomes (Supplemental Figures S3 and S4). In addition, trastuzumab binding led to a disappearance of membrane ruffles (Supplemental Figure S5). One prominent molecular mechanism of trastuzumab in bulk cells is thus to down-regulate HER2 signaling via clearance of HER2 homodimers from the plasma membrane, fulfilled by their crosslinking into chains.
FIGURE 5:
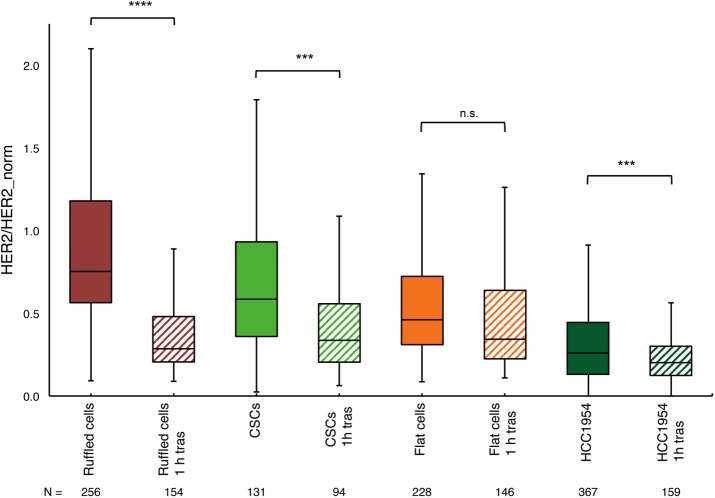
Effect of 1 h exposure to trastuzumab on the relative amount of membrane-bound HER2 per cell in three SKBR3 cell subpopulations, ruffled bulk cancer, CSCs, and flat breast cancer cells, and in HCC1954 drug-resistant breast cancer cells. The y-axis shows the measured HER2 amount per cell, normalized to the average amount of HER2 per cell in the control ruffled cancer cells (without trastuzumab). The box plot indicates the median and first and third quartiles (bottom and top of the box) and the maximum and minimum values in the data. The number of cells (N) in a group is given below the plot. One-hour exposure to trastuzumab reduced membrane-bound HER2 to very different extents in the three subpopulations of SKBR3 cells. Ruffled cells showed the strongest reduction, CSCs less reduction, and flat cells remained almost unaffected, showing only a nonsignificant reduction (two-tailed Student’s t test data from two independent experiments). The HCC1954 cell line, which is trastuzumab resistant, showed an amount of reduction similar to that for the flat cell in the SKBR3 population, and it was a significant change. The stars indicate the probability (p) values for testing the statistical significance, with *** p < 0.001 (extremely significant), **** p < 0.0001 (extremely significant), and n.s., not significant.
Flat cells responded very differently to trastuzumab incubation compared with the bulk cells, and only a small and nonsignificant uptake compared with that of the control was measured (Figure 5). The response of the CSCs was again different, although the median amount of HER2 decreased significantly by a factor of 1.7 compared with the control CSCs; this reduction indicates a much smaller uptake than was found in the ruffled bulk cells. The average amount of membrane-bound HER2 in HCC1954 cells was lower than in any of the SKBR3 subpopulations, the median being only 26%; the trastuzumab-induced effect was significant but in a similar small range as the reduction found in the flat SKBR3 cells (Figure 5), presumably because these cells contain fewer membrane ruffles, compared with SKBR3 bulk cells (Sero et al., 2015).
Trastuzumab-induced HER2 uptake was thus observed for both cell lines, and except for the SKBR3 flat cells, SKBR3 CSCs and the HCC1954 cells showed much less uptake than bulk cells.
DISCUSSION
These data show that flat cells lack the desired response to the antibody-based prescription drug trastuzumab and CSCs exhibit a reduced response, contrasting the effect observed in the ruffled bulk cells. The difference is explained at the individual receptor level by differences in HER2 dimerization status between these cellular subpopulations. For bulk cancer cells in which HER2 homodimers are present, trastuzumab binding effectively clears HER2 from the plasma membrane via cross-linking of the dimers into chains. HER2 homodimers preferentially reside in membrane ruffles (Peckys et al., 2015), and the amount of membrane ruffles, both in cultured cancer cell lines and in patient biopsies, positively correlates with the HER2 density (Chung et al., 2016). Incubation with trastuzumab led to the disappearance of membrane ruffles (see Supplemental Figure S5) and down-regulation of HER2 (Figure 5). The intracellular fate of trastuzumab-bound HER2 is a topic of scientific debate (Ram et al., 2014), and several models exist for trastuzumab-induced HER2 uptake, involving clathrin-dependent or -independent routes of endocytosis; others claim that HER2 is internalization resistant (Bertelsen and Stang, 2014).
In contrast, trastuzumab binding can cross-link only two HER2 monomers or HER2-containing heterodimers in the flat subpopulation without measurable HER2 homodimers in the plasma membrane. This is apparently insufficient for triggering significant endocytosis of HER2, and, consequently, the proteins remain in the plasma membrane. These flat cells with a resting phenotype are presumably dormant cancer cells (Guo et al., 2017), a subpopulation known to be important for acquired drug resistance and late metastasis (Sosa et al., 2014).
The third subpopulation consisting of CSCs represents a group exhibiting lower HER2 homodimer densities so that HER2 down-regulation after trastuzumab binding is reduced compared with bulk cells. The similarity in response to trastuzumab of SKBR3 CSCs and the trastuzumab-resistant HCC19654 cell line suggests a correlation between a reduced trastuzumab sensitivity and the CD44+/CD24− phentotype, consistent with findings of others that CD44 and HER2 interact (Ghatak et al., 2005; Palyi-Krekk et al., 2007; Zoller, 2011).
The aforementioned molecular model offers a new explanation for the observed poorer drug responses of in vitro CSC populations (Oliveras-Ferraros et al., 2012), the increased share of CSCs found in HER2-overexpressing cancer cells after long-term exposure to trastuzumab (Burnett et al., 2015), and the reported correlation between the frequency of the CSC phenotype and worse clinical outcome in patients with HER2 overexpression (Auvinen et al., 2013; Seo et al., 2016). Moreover, dormant residual tumor cells have been found to survive in trastuzumab-treated patients (Abravanel et al., 2015). The lack of response of the drug-resistant HCC1954 cell line to trastuzumab confirms this mechanistic model.
HER2-positive cells generally also express other members of the HER family and readily form heterodimers with HER2. Because HER2 is not cleared from the plasma membrane in the rare cell subpopulations, HER2 heterodimerization would still drive signaling once the respective ligands are present. Because certain heterodimers, for example, HER2-HER3, will trigger dysregulated cell growth (Aceto et al., 2012), the uncontrolled proliferation of the identified rare subpopulations will consequently not be prevented by trastuzumab binding. Even if a few drug-resistant cells exist, they may continue to proliferate in a dysregulated manner and so predominate in the residual tumor population after targeted drug therapy (Penault-Llorca and Radosevic-Robin, 2016). Hence the presence of small breast cancer cell subpopulations in which HER2 homodimers are absent is a possible indication for drug resistance development.
It is known that the problem of drug resistance development should be tackled by taking cancer cell heterogeneity into account (Hanahan and Weinberg, 2011). However, this has been challenging until now due to limitations of the available analytical methods (de Jonge, 2017). Biochemical methods and x-ray crystallography generally used to study the cellular responses to drugs use pooled cellular material so that information is obtained from average responses across a cell population only. Such information is insufficient for studies including cell heterogeneity, which can manifest in large differences in the composition of membrane receptor expressions between cells of the same tumor (Malinowsky et al., 2012). Sorting via flow cytometry could lead to unpredicted results because the subtypes may change after sorting (Fillmore and Kuperwasser, 2008; Jordan et al., 2016). Despite the powerful capabilities of superresolution fluorescence microscopy, these techniques exhibit insufficient spatial resolution to directly view the stoichiometry of protein complexes to determine their functional state, although clustering can be examined at the single-cell level (Sengupta et al., 2011; Shivanandan et al., 2014). Indirect optical techniques such as Förster resonance energy transfer may lead to artifacts, for example, detection of back-to-back neighbors rather than subunits in protein complexes (Piston and Kremers, 2007) and would require unnatural low membrane protein densities (Peckys et al., 2015). Note also that the cells in our study were not genetically modified as would be required for methods using proteins with genetically engineered fluorescent tags. Cryo scanning transmission electron microscopy (Wolf et al., 2014) can in principle be used to detect nanoparticle labels in thin regions of cells but the involved cryo procedures are practically incompatible with experiments requiring the analysis of many cells. Finally, methods detecting protein proximity suffer from artifacts because the measured signal depends on the density of proteins in the membrane (Leuchowius et al., 2013), and the distances between proteins are not measured as is done here.
An increasing awareness exists that single-cell methods are of key importance in cancer research (Heath et al., 2016). Drug development research should not only be based on information about the response of the average cancer cell using biochemical methods but cancer cell heterogeneity should be taken into account and in particular the responses of rare subpopulations with exceptional responses should be examined. Moreover, addressing cell heterogeneity is important for designing personalized medicine (Schmitt et al., 2016). As we show here, liquid-phase STEM of whole cancer cells combined with light microscopy for the identification of cell phenotypes fulfills this need, as it is capable of examining the drug response at the molecular level and accounts for the heterogeneity between cells. We unraveled how the target molecule HER2 responds to the prescription drug trastuzumab in the bulk population of an HER2-overexpressing breast cancer cell line. Most importantly, we discovered why rare cancer cell subpopulations remained largely unresponsive to the drug, providing a possible molecular explanation for the development of drug resistance in this type of cancer. The quantitative liquid-phase STEM method is generally applicable to study membrane protein interactions and drug responses and provides unique information for research areas in which it is important to account for cell heterogeneity and to study the proteins within the native environment of the intact plasma membrane.
MATERIALS AND METHODS
Preparation of SiN membrane microchips and cell culture dishes
Sample support microchips (Ring et al., 2011; DENSsolutions B.V., Delft, The Netherlands) of dimensions 2.00 × 2.600 × 30 mm with a central silicon nitride (SiN) membrane window of dimensions 50 × 400 μm or 150 × 400 μm and a membrane thickness of 50 nm were used for transmission electron microscopy. The preparation of silicon microchips with labeled calls silicon nitride windows (Peckys and de Jonge, 2015; Peckys et al., 2015). In addition, to those protocols, the microchips were incubated with Fibronectin-like Engineered Protein Polymer-plus (Sigma-Aldrich Chemie Gmbh, Munich, Germany) to improve cell adherence. Preparation of the glass-bottom, four-compartment cell culture dishes (Ibidi, Munich, Germany) was the same as for the microchips.
Cell culture and cell seeding on microchips or cell culture dishes
Culturing of SKBR3 cells (human breast cancer cell line overexpressing HER2) was done as previously described (Peckys et al., 2015). The SKBR3 cell line (HTB-30D) and the HCC1954 cell line were obtained from the American Type Culture Collection and authenticated by genotyping (Multiplexion, Heidelberg, Germany). Mycoplasma testing was performed in our laboratory every 3 mo on all cell lines used. The cells were passaged twice a week at 60–80% confluency, and experiments were performed with cells between passages 6 and 18. Experiments in cell culture dishes were performed for light microscopic determination of the relative shares of the three subpopulations. One-hundred-microliter volumes of cell suspension were added to each compartment of a four-compartment dish and incubated 2–4 h in a 5% CO2 atmosphere at 37°C. The cells were then incubated with cell medium without fetal bovine serum (FBS; Life Technologies, Carlsbad, CA) for 12–24 h in a 5% CO2 atmosphere at 37°C. At the time of the experiment the confluency reached 60–90%.
For correlative light microscopy and STEM, the microchips were each placed in 200 μl of FBS-supplemented cell medium in a well of a 96-well plate. SKBR3 cell suspension (20–50 μl volumes) was added to each well. After 5–10 min, 5–20 cells had adhered on the SiN window, and the microchips were transferred into new wells containing 200 μl of FBS-supplemented cell medium. The microchips were then incubated for 2–4 h in a 5% CO2 atmosphere at 37°C, placed into new wells filled with cell medium without FBS, and incubated again for 12–24 h. Also on microchips the cell confluency reached 60–90% at the time of the experiments.
Incubation with trastuzumab, labeling of HER2 and CD44/24, and fixation
Before an experiment, the biotin-conjugated anti-HER2 Affibody (HER2-AFF-B; Affibody AB, Bromma, Sweden) stock solution (20 μM) was adjusted to a final concentration of 200 nM in phosphate-buffered saline (PBS) supplemented with 1% normal goat serum (GS), 0.5% biotin-free Albumin Fraktion V (bovine serum albumin [BSA]), 0.1% gelatin from coldwater fish skin (GS-BSA-GEL-PBS). Streptavidin-QD stock solution (1 μM; Qdot 655; Life Technologies, Carlsbad, CA) was diluted 1:20 in 40 mM borate buffer (sodium tetraborate, boric acid, pH 8.3) and then diluted to a final concentration of 5 nM by adding PBS supplemented with 1% BSA (BSA-PBS). The CD44/CD24 antibody labeling solution was prepared by diluting the antibody stock solutions (Phycoerythrin-conjugated anti-CD24 antibody, clone SN3 [ab77219] and AlexaFluor 488–conjugated anti-CD44 antibody, clone MEM85 [prediluted, ab187571]; Abcam, Cambridge, UK) in BSA-PBS so that the final dilutions were 1:100 when mixed together. Trastuzumab stock solution (25 mg/ml) was diluted 1:2500 in cell culture medium without serum to yield a final concentration of 10 µg/ml.
For determination of the fractional sizes of the three cell subpopulations, cells grown in dishes were rinsed once with GS-BSA-GEL-PBS and then incubated for 10 min at room temperature (RT) in GS-BSA-GEL-PBS to block unspecific binding of HER2-AFF-B. Live cells were then incubated for 10 min at 37°C with the HER2-AFF-B labeling solution. After three rinses with PBS and once with 0.1 M cacodylate buffer supplemented with 0.1 M saccharose, pH 7.4 (CB), cells were fixed with 3% formaldehyde in CB for 10 min at RT. This was followed by a rinse with CB, three rinses with PBS, incubation in 0.1 M glycine in PBS, pH 7.4 (GLY-PBS), for 2 min, and two rinses with PBS, before incubation in Strep-QD labeling solution for 12 min at RT. This two-step labeling protocol ensured HER2 labeling in a 1:1 stoichiometry (Peckys et al., 2015) without artificial clustering. After three rinses with PBS, the cells were incubated in CD44/CD24-AB for 20–30 min at RT in the dark, followed by three rinses with PBS and one rinse with BSA-PBS. This experiment was repeated three times, with cells at different cell passages.
For experiments involving correlative light microscopy and STEM, microchips with adherently grown cells that had been serum starved were divided into two groups, one control and one drug exposure group. HER2 labeling was done as described above. Microchip samples for the drug exposure group were rinsed twice with BSA-PBS directly after the HER2-AFF-B incubation, rinsed once with cell culture medium without serum, and then incubated with trastuzumab, 10 µg/ml, in cell culture medium without FBS at 37°C for 2.5 min. Samples of the control group were rinsed three times with BSA-PBS after the HER2-AFF-B incubation and skipped the rinse in cell culture medium without serum and trastuzumab. All samples were further fixed, glycine quenched, and Strep QD- and CD44/CD24-labeled as described above. The labeled cells on the microchip samples were imaged with light microscopy. To increase the stability of the cellular material under electron beam irradiation, the cells were further fixed with glutaraldehyde (GA). Therefore the cells were rinsed once with CB and fixed for 10 min at RT with 2% GA in CB. Cells were then rinsed once with CB and three times with BSA-PBS and stored in BSA-PBS at 4°C until electron microscopy was performed, usually within the next few days.
Light microscopy
Labeled and formaldehyde-fixed cells on microchips were imaged in BSA-PBS in an inverted light microscope (DMI6000B; Leica, Germany) whereby the microchips were placed in a glass-bottom dish with the cells facing down. Four different images were recorded at each position. The first image taken from each group of cells was accomplished in bright field using direct interference contrast (DIC) to yield information about membrane borders and topography, such as membrane ruffles. Next three fluorescence images were recorded to detect CD44 and CD24 expression through fluorescein isothiocyanate, respectively phycoerythrin-filter cubes, and HER2-bound Strep-QDs through a filter cube with a 340–380 nm excitation and a >420 nm emission window.
Characterization and quantification of SKBR3 subpopulations
To determine the fraction of cells for each subpopulation (SKBR3 breast cancer ruffled bulk, flat, and CSCs), three identical labeling experiments were performed, at different days and for different cell passage numbers. DIC and fluorescence images were recorded with a 40× objective from formaldehyde-fixed cells. Images were stitched together from 25 individual frames recorded at adjacent locations on a relocation grid. All recorded cells were individually inspected using ImageJ (National Institutes of Health, Bethesda, MD). Cells of spherical shape and of exceptional small size (adherence area <600 µm2) compared with the average cell adherence area (1348 ± 944 µm2) (representing cells shortly before either division or death) and cells with a CD44+/CD24+ signature, as well as cells that were not fully captured within the image frame, were excluded from further analysis. All other cells were assigned to one of the three subpopulations.
Liquid-phase STEM
The labeled cells on the microchips were imaged in hydrated state in an environmental scanning electron microscope (Quanta 400 FEG; FEI, USA) with STEM dark field contrast (Bogner et al., 2005) as described in detail elsewhere (Peckys et al., 2013, 2015). A thin film of water was maintained over the cells at a pressure of 720–740 Pa and a temperature of 3°C around the sample. The electron beam energy was 30 keV, the spot size 1 nm, the probe current 600 pA, and the working distance 6.2 mm. Images showing HER2-bound QDs typically had a magnification of 50,000×, a pixel-dwell time of 50 μs, and an image size of 1024 × 884 pixels. The electron dose was below the limit of visible radiation damage (Hermannsdörfer et al., 2016).
Duration and reproduction of an experiment
A typical experiment included 4–10 microchip samples or 1–3 cell culture dishes. The total required experimental time amounted to 3–4 h for seeding of cells on the microchips (not counting the incubation period for initial adherence and serum starvation), the labeling, and light microscopy. STEM was the most time-consuming part of the experiment. The time to examine one microchip was typically 2 h, resulting in ∼100 images of several cells.
All experiments were reproduced at least twice from samples prepared and analyzed on different days.
Examining trastuzumab-induced HER2 uptake
SKBR3 cells were grown in two 35-mm cell culture dishes with engraved 50 μm relocation grids. CO2 independent medium without FBS was used to rinse and incubate the cells during time-lapse imaging and for drug exposure. Areas of 1–2 mm2 were selected from the grid regions, and the cells in these areas were time-lapse imaged at 37°C, with DIC contrast using a 40× objective. Thirty adjacent and slightly overlapping images covering the complete area were recorded every 2 min for 16 min. This time-lapse imaging of the dynamic nature of membrane topographies of the individual cells was later used to identify membrane ruffles. Thereafter the cells were exposed to the drug (trastuzumab, 10 µg/ml, in CO2 independent medium without FBS) for 1 h at 37°C. The cells were then washed, blocked, labeled with HER2-AFF-B, washed, fixed, and labeled with Strep-QD and CD44/CD24-AB (see above). The same areas were then relocated and imaged with DIC contrast, and with the three fluorescence channels to capture HER2-QD, CD44, and CD24 signals. For the control group the experiment was run in parallel in a second dish, with the drug incubation being replaced by incubation in CO2 independent medium without FBS. For the subsequent analysis of membrane-bound HER2 in single cells, the recorded DIC and fluorescence image series were merged into large tiled images. A description of the analysis in light microscopy images can be found in the caption of Supplemental Figure S1. In both experimental groups all identified flat and CD44+/CD24− cancer stem-like cells were included in the analysis; for the main subpopulation of ruffled bulk cells similar numbers of cells were randomly selected (using assigned numbers for every cell or random locations, i.e., lines diagonally crossing the grid). Cells of spherical shape and of a small adherence area size (<600 μm2) were excluded from the analysis. The experiment was repeated at another time with a different batch of SKBR3 cells.
Statistical analysis
An automated procedure programmed in ImageJ (National Institutes of Health) was used to detect the positions of the QD nanoparticle protein labels (Peckys et al., 2015). The label positions were statistically analyzed in software of local design in c++ via calculating the pair correlation function g(r) (Stoyan and Stoyan, 1996):
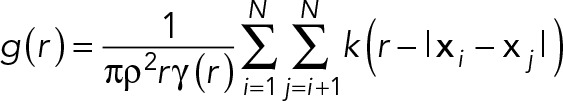 (1) (1)
|
Here r is the statistical label distance, ρ the labeling density, and N the number of particles. The covariance function (Stoyan et al., 1993) γ and the kernel (Fiksel, 1988) k are defined elsewhere (Peckys et al., 2015). The distance between the positions i and j are calculated from the modulus |xi–xj| of the position vector x. Distances smaller than 10 nm were expelled from the calculation so as to avoid counting overlapping nanoparticles. The calculation assumes the labels to be positioned approximately planar. The output of the analysis was a histogram of g(r) with a bin width in r of 2.5 nm. The bandwidth in the calculation (Peckys et al., 2015) was set equal to the bin width. The data of a group of images from cells belonging to a subpopulation were averaged. Here the average was weighted by the particle density so that cells with a lower number of labels were less strongly weighted in the average than cells with more labels.
Code availability
The codes programmed in ImageJ and in c++ are available on request from the corresponding author and may be used when this paper is cited in the results published with the code. The code is copyright of the corresponding author and may not be published, used for commercial purposes, or modified without written permission.
Supplementary Material
Acknowledgments
We thank M. Hasmann (Roche Diagnostics GmbH, Germany) for providing trastuzumab and for discussions, Stephanie Smolka for help with the experiments, and E. Arzt for his support through INM–Leibniz Institute for New Materials. The research was supported by the Leibniz Competition 2014 and by the Federal Ministry of Education and Research (BMBF; FKZ:031A429E).
Abbreviations used:
- CSC
cancer stem cell
- DIC
direct interference contrast
- EGF
epidermal growth factor
- g(r)
pair correlation function g(r)
- HER2
human EGF receptor 2
- QD
quantum dot
- STEM
scanning transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0381) on August 9, 2017.
REFERENCES
- Abravanel DL, Belka GK, Pan TC, Pant DK, Collins MA, Sterner CJ, Chodosh LA. Notch promotes recurrence of dormant tumor cells following HER2/neu-targeted therapy. J Clin Invest. 2015;125:2484–2496. doi: 10.1172/JCI74883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aceto N, Duss S, MacDonald G, Meyer DS, Roloff TC, Hynes NE, Bentires-Alj M. Co-expression of HER2 and HER3 receptor tyrosine kinases enhances invasion of breast cells via stimulation of interleukin-8 autocrine secretion. Breast Cancer Res. 2012;14:R131. doi: 10.1186/bcr3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ejeh F, Smart CE, Morrison BJ, Chenevix-Trench G, Lopez JA, Lakhani SR, Brown MP, Khanna KK. Breast cancer stem cells: treatment resistance and therapeutic opportunities. Carcinogenesis. 2011;32:650–658. doi: 10.1093/carcin/bgr028. [DOI] [PubMed] [Google Scholar]
- Arkhipov A, Shan YB, Kim ET, Dror RO, Shaw DE. Her2 activation mechanism reflects evolutionary preservation of asymmetric ectodomain dimers in the human EGFR family. eLife. 2013;2:e00708. doi: 10.7554/eLife.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvinen P, Tammi R, Kosma VM, Sironen R, Soini Y, Mannermaa A, Tumelius R, Uljas E, Tammi M. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int J Cancer. 2013;132:531–539. doi: 10.1002/ijc.27707. [DOI] [PubMed] [Google Scholar]
- Badache A, Hynes NE. A new therapeutic antibody masks ErbB2 to its partners. Cancer Cell. 2004;5:299–301. doi: 10.1016/s1535-6108(04)00088-1. [DOI] [PubMed] [Google Scholar]
- Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen V, Stang E. The mysterious ways of ErbB2/HER2 trafficking. Membranes. 2014;4:424–446. doi: 10.3390/membranes4030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkelund H, Gedda L, Andersson K. Comparing the epidermal growth factor interaction with four different cell lines: intriguing effects imply strong dependency of cellular context. PLoS One. 2011a;6:e16536. doi: 10.1371/journal.pone.0016536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkelund H, Gedda L, Barta P, Malmqvist M, Andersson K. Gefitinib induces epidermal growth factor receptor dimers which alters the interaction characteristics with I-125-EGF. PLoS One. 2011b;6:e24739. doi: 10.1371/journal.pone.0024739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner A, Thollet G, Basset D, Jouneau PH, Gauthier C. Wet STEM: a new development in environmental SEM for imaging nano-objects included in a liquid phase. Ultramicroscopy. 2005;104:290–301. doi: 10.1016/j.ultramic.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Burnett JP, Korkaya H, Ouzounova MD, Jiang H, Conley SJ, Newman BW, Sun LC, Connarn JN, Chen CS, Zhang N, et al. Trastuzumab resistance induces EMT to transform HER2(+) PTEN- to a triple negative breast cancer that requires unique treatment options. Sci Rep. 2015;5:15821. doi: 10.1038/srep15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8:1095–1111. doi: 10.1016/j.molonc.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Love JC, Navin NE, Pachter L, Stubbington MJ, Svensson V, Sweedler JV, Teichmann SA. Single-cell analysis at the threshold. Nat Biotechnol. 2016;34:1111–1118. doi: 10.1038/nbt.3721. [DOI] [PubMed] [Google Scholar]
- Chung I, Reichelt M, Shao L, Akita RW, Koeppen H, Rangell L, Schaefer G, Mellman I, Sliwkowski MX. High cell-surface density of HER2 deforms cell membranes. Nat Commun. 2016;7:12742. doi: 10.1038/ncomms12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- de Jonge N. Membrane protein stoichiometry studied in intact mammalian cells using liquid-phase electron microscopy. J Microsc. 2017 doi: 10.1111/jmi.12570. doi: 10.1111/jmi.12570. [DOI] [PubMed] [Google Scholar]
- de Jonge N, Peckys DB, Kremers GJ, Piston DW. Electron microscopy of whole cells in liquid with nanometer resolution. Proc Natl Acad Sci USA. 2009;106:2159–2164. doi: 10.1073/pnas.0809567106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge N, Ross FM. Electron microscopy of specimens in liquid. Nat Nanotechnol. 2011;6:695–704. doi: 10.1038/nnano.2011.161. [DOI] [PubMed] [Google Scholar]
- Drebin JA, Link VC, Stern DF, Weinberg RA, Greene MI. Down-modulation of an oncogene protein product and reversion of the transformed phenotype by monoclonal antibodies. Cell. 1985;41:697–706. doi: 10.1016/s0092-8674(85)80050-7. [DOI] [PubMed] [Google Scholar]
- Eigenbrot C, Ultsch M, Dubnovitsky A, Abrahmsen L, Hard T. Structural basis for high-affinity HER2 receptor binding by an engineered protein. Proc Natl Acad Sci USA. 2010;107:15039–15044. doi: 10.1073/pnas.1005025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Fiksel T. Edge-corrected density estimators for points processes. Statistics. 1988;19:67–75. [Google Scholar]
- Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastl G, Spizzo G, Obrist P, Dunser M, Mikuz G. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet. 2000;356:1981–1982. doi: 10.1016/S0140-6736(00)03312-2. [DOI] [PubMed] [Google Scholar]
- Ghatak S, Misra S, Toole BP. Hyaluronan constitutively regulates ErbB2 phosphorylation and signaling complex formation in carcinoma cells. J Biol Chem. 2005;280:8875–8883. doi: 10.1074/jbc.M410882200. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Narasanna A, Wang SE, Liu SY, Chakrabarty A, Balko JM, Gonzalez-Angulo AM, Mills GB, Penuel E, Winslow J, et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer Res. 2011;71:1871–1882. doi: 10.1158/0008-5472.CAN-10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Huang J, Moses MA. Characterization of dormant and active human cancer cells by quantitative phase imaging. Cytom Part A. 2017;91:424–432. doi: 10.1002/cyto.a.23083. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henjes F, Bender C, von der Heyde S, Braun L, Mannsperger HA, Schmidt C, Wiemann S, Hasmann M, Aulmann S, Beissbarth T, Korf U. Strong EGFR signaling in cell line models of ERBB2-amplified breast cancer attenuates response toward ERBB2-targeting drugs. Oncogenesis. 2012;1:e16. doi: 10.1038/oncsis.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermannsdörfer J, Tinnemann V, Peckys DB, de Jonge N. The effect of electron beam irradiation in environmental scanning transmission electron microscopy of whole cells in liquid. Microsc Microanal. 2016;20:656–665. doi: 10.1017/S1431927616000763. [DOI] [PubMed] [Google Scholar]
- Hu S, Sun Y, Meng Y, Wang X, Yang W, Fu W, Guo H, Qian W, Hou S, Li B, et al. Molecular architecture of the ErbB2 extracellular domain homodimer. Oncotarget. 2015;6:1695–1706. doi: 10.18632/oncotarget.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz E, Stancovski I, Sela M, Yarden Y. Suppression and promotion of tumor growth by monoclonal antibodies to ErbB-2 differentially correlate with cellular uptake. Proc Natl Acad Sci USA. 1995;92:3353–3357. doi: 10.1073/pnas.92.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Dey JH. PI3K inhibition overcomes trastuzumab resistance: blockade of ErbB2/ErbB3 is not always enough. Cancer Cell. 2009;15:353–355. doi: 10.1016/j.ccr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, Yu M, Sundaresan TK, Licausi JA, Desai R, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–106. doi: 10.1038/nature19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat Rev Cancer. 2010;10:618–629. doi: 10.1038/nrc2900. [DOI] [PubMed] [Google Scholar]
- Leuchowius KJ, Clausson CM, Grannas K, Erbilgin Y, Botling J, Zieba A, Landegren U, Soderberg O. Parallel visualization of multiple protein complexes in individual cells in tumor tissue. Mol Cell Proteomics. 2013;12:1563–1571. doi: 10.1074/mcp.O112.023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowsky K, Raychaudhuri M, Buchner T, Thulke S, Wolff C, Hofler H, Becker KF, Avril S. Common protein biomarkers assessed by reverse phase protein arrays show considerable intratumoral heterogeneity in breast cancer tissues. PloS One. 2012;7:e40285. doi: 10.1371/journal.pone.0040285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimeault M, Hauke R, Mehta PP, Batra SK. Recent advances in cancer stem/progenitor cell research: therapeutic implications for overcoming resistance to the most aggressive cancers. J Cell Mol Med. 2007;11:981–1011. doi: 10.1111/j.1582-4934.2007.00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Bene L, Balazs M, Hyun WC, Lockett SJ, Chiang NY, Waldman F, Feuerstein BG, Damjanovich S, Szollosi J. EGF-induced redistribution of erbB2 on breast tumor cells: flow and image cytometric energy transfer measurements. Cytometry. 1998;32:120–131. doi: 10.1002/(sici)1097-0320(19980601)32:2<120::aid-cyto7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Oliveras-Ferraros C, Corominas-Faja B, Cufi S, Vazquez-Martin A, Martin-Castillo B, Iglesias JM, Lopez-Bonet E, Martin AG, Menendez JA. Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin) Cell Cycle. 2012;11:4020–4032. doi: 10.4161/cc.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palyi-Krekk Z, Barok M, Isola J, Tammi M, Szollosi J, Nagy P. Hyaluronan-induced masking of ErbB2 and CD44-enhanced trastuzumab internalisation in trastuzumab resistant breast cancer. Eur J Cancer. 2007;43:2423–2433. doi: 10.1016/j.ejca.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Peckys DB, Baudoin JP, Eder M, Werner U, de Jonge N. Epidermal growth factor receptor subunit locations determined in hydrated cells with environmental scanning electron microscopy. Sci Rep. 2013:2626. doi: 10.1038/srep02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckys DB, de Jonge N. Studying the stoichiometry of epidermal growth factor receptor in intact cells using correlative microscopy. J Vis Exp. 2015 doi: 10.3791/53186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckys DB, Korf U, de Jonge N. Local variations of HER2 dimerization in breast cancer cells discovered by correlative fluorescence and liquid electron microscopy. Sci Adv. 2015;1:e1500165. doi: 10.1126/sciadv.1500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penault-Llorca F, Radosevic-Robin N. Biomarkers of residual disease after neoadjuvant therapy for breast cancer. Nat Rev Clin Oncol. 2016;13:487–503. doi: 10.1038/nrclinonc.2016.1. [DOI] [PubMed] [Google Scholar]
- Pirkmajer S, Chibalin AV. Serum starvation: caveat emptor. Am J Physiol Cell Physiol. 2011;301:C272–C279. doi: 10.1152/ajpcell.00091.2011. [DOI] [PubMed] [Google Scholar]
- Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Ram S, Kim D, Ober RJ, Ward ES. The level of HER2 expression is a predictor of antibody-HER2 trafficking behavior in cancer cells. mAbs. 2014;6:1211–1219. doi: 10.4161/mabs.29865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring EA, Peckys DB, Dukes MJ, Baudoin JP, de Jonge N. Silicon nitride windows for electron microscopy of whole cells. J Microsc. 2011;243:273–283. doi: 10.1111/j.1365-2818.2011.03501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FM. Opportunities and challenges in liquid cell electron microscopy. Science. 2015;350:aaa9886. doi: 10.1126/science.aaa9886. [DOI] [PubMed] [Google Scholar]
- Schmitt MW, Loeb LA, Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol. 2016;13:335–347. doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, Veatch SL, Lippincott-Schwartz J. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AN, Lee HJ, Kim EJ, Jang MH, Kim YJ, Kim JH, Kim SW, Ryu HS, Park IA, Im SA, et al. Expression of breast cancer stem cell markers as predictors of prognosis and response to trastuzumab in HER2-positive breast cancer. Br J Cancer. 2016;114:1109–1116. doi: 10.1038/bjc.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sero JE, Sailem HZ, Ardy RC, Almuttaqi H, Zhang TL, Bakal C. Cell shape and the microenvironment regulate nuclear translocation of NF-kappa B in breast epithelial and tumor cells. Mol Syst Biol. 2015;11:790. doi: 10.15252/msb.20145644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivanandan A, Deschout H, Scarselli M, Radenovic A. Challenges in quantitative single molecule localization microscopy. FEBS Lett. 2014;588:3595–3602. doi: 10.1016/j.febslet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyan D, Bertram U, Wendrock H. Estimation variances for estimators of product densities and pair correlation functions of planar points processes. Ann Inst Statist Math. 1993;45:211–221. [Google Scholar]
- Stoyan D, Stoyan H. Estimating pair correlation functions of planar cluster processes. Biom J. 1996;38:259–271. [Google Scholar]
- von der Heyde S, Wagner S, Czerny A, Nietert M, Ludewig F, Salinas-Riester G, Arlt D, Beissbarth T. mRNA profiling reveals determinants of trastuzumab efficiency in HER2-positive breast cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T, Claret FX. Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front Oncol. 2012;2:62. doi: 10.3389/fonc.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T, Sliwkowski MX, Claret FX. Personalized drug combinations to overcome trastuzumab resistance in HER2-positive breast cancer. BBA Rev Cancer. 2014;1846:353–365. doi: 10.1016/j.bbcan.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clin Exp Metastasis. 2006;23:97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- Wolf SG, Houben L, Elbaum M. Cryo-scanning transmission electron tomography of vitrified cells. Nat Methods. 2014;11:423–428. doi: 10.1038/nmeth.2842. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule. Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.