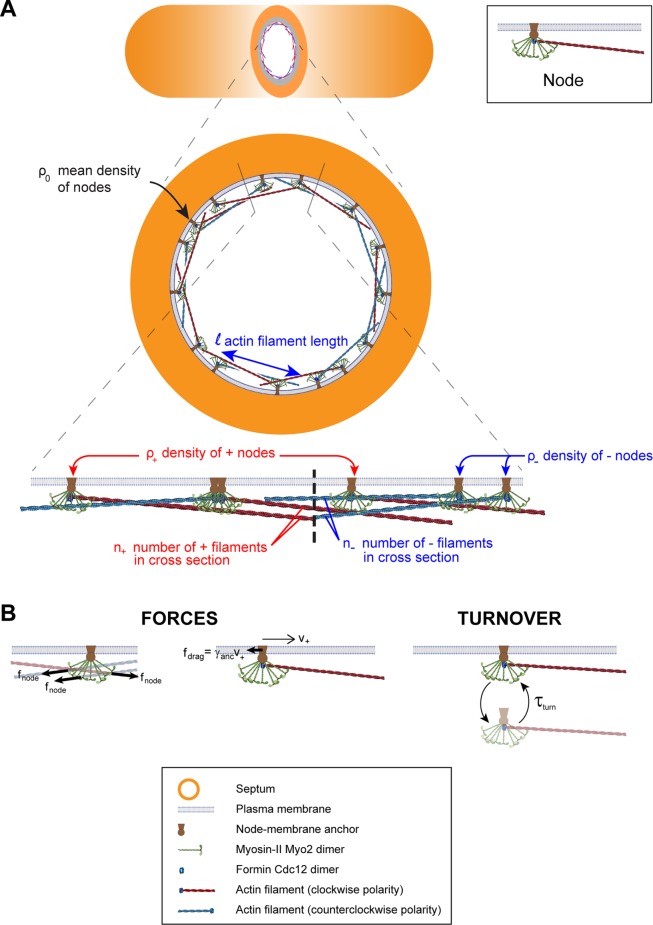
Recent experiments showed that key cytokinetic ring proteins organize into membrane-anchored complexes called nodes in the fission yeast cytokinetic ring. A mathematical model based on this organization showed that ring tension arises from myosins pulling anchored actin filaments, and component turnover and anchoring ensure structural stability.
Abstract
During cytokinesis, a contractile actomyosin ring constricts and divides the cell in two. How the ring marshals actomyosin forces to generate tension is not settled. Recently, a superresolution microscopy study of the fission yeast ring revealed that myosins and formins that nucleate actin filaments colocalize in plasma membrane-anchored complexes called nodes in the constricting ring. The nodes move bidirectionally around the ring. Here we construct and analyze a coarse-grained mathematical model of the fission yeast ring to explore essential consequences of the recently discovered ring ultrastructure. The model reproduces experimentally measured values of ring tension, explains why nodes move bidirectionally, and shows that tension is generated by myosin pulling on barbed-end-anchored actin filaments in a stochastic sliding-filament mechanism. This mechanism is not based on an ordered sarcomeric organization. We show that the ring is vulnerable to intrinsic contractile instabilities, and protection from these instabilities and organizational homeostasis require both component turnover and anchoring of components to the plasma membrane.
INTRODUCTION
During cytokinesis, the tensile actomyosin ring provides force that drives or guides division of the cell into two (Schroeder, 1972; Fujiwara and Pollard, 1976; Mabuchi and Okuno, 1977). Tension production is thought to be the primary role of the ring, and is thought to arise from forces exerted on actin filaments by nonmuscle type II myosin in the ring (Schroeder, 1975). However, the mechanism of tension production has not been settled.
A natural candidate for the tension production mechanism is a sliding-filament mechanism similar to that in striated muscle, based on the sarcomere repeat unit (Schroeder, 1975). However, for fission yeast—the subject of the present study—little evidence supports a sarcomeric-like organization in the cytokinetic ring (Kanbe et al., 1989; Bezanilla and Pollard, 2000; Kamasaki et al., 2007; Laplante et al., 2015; Courtemanche et al., 2016).
In the case of the fission yeast Schizosaccharomyces pombe, there is a real prospect of establishing realistic, detailed models of the cytokinetic ring, because many participating molecules have been identified (for a review, see Pollard and Wu, 2010) and their numbers measured during ring assembly, the maturation phase, and the actual constriction of the ring (Wu and Pollard, 2005; Courtemanche et al., 2016). Ring tensions of ∼400 pN were also recently measured in fission yeast protoplasts, and a molecularly detailed simulation constrained by the considerable body of experimental data about the fission yeast ring was able to reproduce the measured values of tension (Stachowiak et al., 2014).
Despite the abundance of information about the S. pombe ring, little was known about the organization of components in the matured, constricting ring. The organization of a number of key proteins is clearer during the process of ring assembly. The type II myosin Myo2 and the actin nucleator formin Cdc12 among other proteins are organized into plasma membrane-anchored protein complexes called nodes that assemble into a tight contractile ring from a broad band at the cell’s equator (Wu et al., 2006; Vavylonis et al., 2008).
Important new information about the detailed organization in the constricting S. pombe ring was recently provided by a study using superresolution fluorescence photoactivation localization microscopy (FPALM; Laplante et al., 2016). It was found that a membrane-anchored node-like organization of formin, myosin-II, and other proteins persists beyond assembly into the constricting ring, and the stoichiometric ratios of molecules in the nodes were measured. Because formin caps actin filament barbed ends, this suggests an organization in which actin filament barbed ends and myosin-II colocalize and are anchored to the plasma membrane.
Here we develop a coarse-grained mathematical model of the constricting fission yeast cytokinetic ring that incorporates this recently established organizational information. In the model, formin and myosin-II are anchored to the membrane in nodes as seen experimentally, and the stoichiometry of components in the ring is fixed by experiment (Wu and Pollard, 2005; Laplante et al., 2016). The model explains the origin of the observed bidirectional motion of nodes in the constricting ring, explains how tension arises from a disordered organization, and generates values of ring tension close to experimentally measured values for realistic values of the force per myosin head. We find that the actomyosin contractile ring has an intrinsic contractile instability that is controlled by turnover and by anchoring of components to the membrane. The anchoring resists lateral sliding and thereby retards the instability growth rate.
RESULTS
Mathematical model of the S. pombe cytokinetic ring: background
Our aim is to construct and analyze a coarse-grained mathematical model able to explore some essential consequences of the recently discovered ultrastructure of the fission yeast ring (Laplante et al., 2016). In that study, FPALM superresolution microscopy revealed that in the constricting ring the nonmuscle myosin-II Myo2 and the actin nucleator and polymerizer formin Cdc12 colocalize in membrane-anchored nodes, together with the IQGAP Rng2 and the F-BAR protein Cdc15. The authors argued that there are eight Myo2 dimers per node, unchanged from the number of Myo2 dimers in assembly nodes, the precursor nodes from which the ring is assembled. Previously, quantitative fluorescence microscopy had measured a mean of 1500 Myo2 dimers and 180 Cdc12 dimers in the ring at the onset of constriction (Wu and Pollard, 2005; Courtemanche et al., 2016). This suggests that, at the onset of constriction, the cytokinetic ring contains ∼190 “constriction” nodes, each containing eight Myo2 dimers and a mean of ∼1 formin Cdc12 dimer. Because Cdc12 dimers processively cap actin filament barbed ends while elongating the filament (Kovar et al., 2006), each node would contain a mean of one actin filament anchored at its barbed end.
Thus motivated, in our simplified model the ring at the onset of constriction contains 190 nodes anchored to the plasma membrane, each node containing one formin dimer from which emanates one actin filament (Figure 1A). Thus, actin filament barbed ends and Myo2 dimers are anchored to the membrane. For simplicity, we neglect fluctuations in the number of formins (and hence actin filaments) per node. Further, we assume that all actin filaments are barbed-end-capped by formin Cdc12, so there are 190 actin filaments in the ring.
FIGURE 1:
Mathematical model of the constricting fission yeast cytokinetic ring. (A) Schematic of our coarse-grained, one-dimensional mathematical model of the constricting cytokinetic ring (not to scale). The ring contains membrane-anchored cytokinetic nodes (inset), protein assemblies where eight Myo2 dimers and a formin Cdc12 dimer are attached to each other and to the node anchor that can slide along the membrane. Each node contains one actin filament of length l, which is attached to the formin Cdc12 dimer at its barbed end. Nodes are referred to as clockwise (+) or counterclockwise (-) depending on the polarity of the attached filament, shown as red and blue, respectively. The ring is attached to the plasma membrane at the leading edge of the septum. The model describes the ring using one-dimensional continuous density fields. (B) Forces and turnover rules in the model. Myosins of a given node bind and pull every actin filament that passes through the node with a pulling force fnode per filament. Actin filaments experience force fnode per node from every node that falls along their lengths (left). As nodes slide laterally along the membrane with velocity v+, they experience drag forces fdrag per node from membrane with a node anchor drag coefficient γanc (center). Nodes bind and unbind the membrane on a time scale τturn (right). We assume that nodes turnover with their attached actin filaments.
Another critical characteristic is the amount of actin in the fission yeast ring. A recent study counted 190,000 actin subunits at constriction onset, using the fluorescently tagged actin binding protein mEGFP-LifeAct (Courtemanche et al., 2016). This translates to a mean actin length of 2.7 µm per node, given an F-actin axial rise of 2.7 nm per subunit (Carlier, 1991). For simplicity, our model assumes all actin filaments have the same length, the mean value of l = 2.7 µm. As the ring thickness and width of ∼125 nm (Laplante et al., 2016) are much less than the actin filament length, to a very good approximation a node-attached filament lies parallel to the ring and can point in either a clockwise or counterclockwise direction along the ring (Figure 1A).
Turnover times of key proteins in the fission yeast ring have been reported, including Cdc12 (43 s), Myo2 (18.6 s), and myosin-II regulatory light chain Rlc1 (41 s) (Clifford et al., 2008; Yonetani et al., 2008; Sladewski et al., 2009). The precise nature of turnover in the ring is unknown, but we assume whole-node turnover in a representative time τturn = 18.6 s matching the reported Myo2 turnover time. Thus, actin turns over as whole filaments only. For simplicity, our model neglects other turnover pathways, namely, formin-mediated polymerization and cofilin-mediated disassembly (Kovar et al., 2006; Michelot et al., 2007).
Turnover is represented by stochastic association and disassociation of nodes to and from the ring, with mean dissociation time τturn = 18.6 s and a mean association rate that produces a mean of 190 nodes in the ring. We assume that formin-mediated nucleation of actin filaments is also directionally stochastic similarly to nodes during ring assembly (Vavylonis et al., 2008), so that a newly arriving node produces a filament that randomly points in either the clockwise or counterclockwise direction. We refer to nodes as clockwise or counterclockwise, depending on the polarity of the attached filament. On average, there are equal numbers of each type of node but the numbers of each fluctuate in time.
The actomyosin ring consists of the bundled actin filaments and Myo2 associated with the 190 nodes (Figure 1A), anchored to the membrane by the node-membrane anchors. Thus, an actin filament of length l belonging to a given node passes near the Myo2 dimers of all nodes within a distance l along the ring. These Myo2 molecules bind and exert pulling force on the filament and hence the node, resisted by the drag of the node anchor in the membrane. The anchor drag coefficient is γanc (Figure 1B). These forces will pull nodes clockwise or counterclockwise, depending on the node type. Consistent with this, FPALM measured clockwise and counterclockwise motions of myosin in the ring, with a mean speed 22 nm s−1 (Laplante et al., 2016). This supports our inference that on average there is one formin dimer and one actin filament per node, and that there are two classes of node.
Derivation of model equations
The above somewhat simplified representation of the fission yeast cytokinetic ring, severely constrained by experimental data, consists of 190 clockwise or counterclockwise nodes anchored to the plasma membrane. We developed a coarse-grained continuous mathematical description of this system. Our principal goals are to establish the mechanism of tension generation in the ring, to compare predictions of our model with experimentally measured values of tension (Stachowiak et al., 2014), to explore how the ring maintains structural stability, and to understand the functional significance of the two types of node motion, clockwise and counterclockwise.
We use a coarse-grained, continuous representation of the nodes. The density and velocity of clockwise nodes at time t and location x along the ring are denoted ρ+(x, t) and v+(x, t), respectively. For counterclockwise nodes the same quantities are ρ-(x, t) and v-(x, t). The total length of the fission yeast ring at constriction onset is 11.8 µm, so that 0 ≤ x ≤ 11.8 µm (Figure 1). Throughout, x represents the clockwise distance around the ring. Thus, a positive (negative) velocity or force is in the clockwise (counterclockwise) direction.
Consider first a typical clockwise node. The forces acting on a given node are 1) the reaction force  to the total force exerted by the myosins of that node that bind and pull actin filaments passing through it (note that the statistics of
to the total force exerted by the myosins of that node that bind and pull actin filaments passing through it (note that the statistics of  are the same for clockwise and counterclockwise nodes); 2) the total force
are the same for clockwise and counterclockwise nodes); 2) the total force 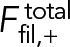 exerted by myosins of other nodes on the filament attached to that node; and 3) the drag force exerted by the membrane that resists the lateral sliding of the node anchor, characterized by anchor drag coefficient γanc (Figure 1B). The force balance reads
exerted by myosins of other nodes on the filament attached to that node; and 3) the drag force exerted by the membrane that resists the lateral sliding of the node anchor, characterized by anchor drag coefficient γanc (Figure 1B). The force balance reads
|
|
Many actin filaments of both polarities pass through this node. The net myosin force exerted by that node on these filaments,  , is proportional to the difference between the number of clockwise n+ and counterclockwise n- filaments in the cross-section of the ring at the location of the node (Figure 1B):
, is proportional to the difference between the number of clockwise n+ and counterclockwise n- filaments in the cross-section of the ring at the location of the node (Figure 1B):
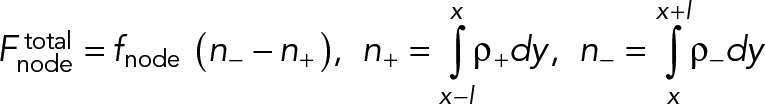 (2) (2)
|
Here, fnode is the time-averaged force exerted by the myosins of one node on one filament passing through it. Note that because each node carries one actin filament of length l, n+ (n-) equals the number of nodes within a distance l of the node in question, measured in the clockwise (counterclockwise) direction. The force acting on the node-attached actin filament is due to all nodes along its length (Figure 1B),
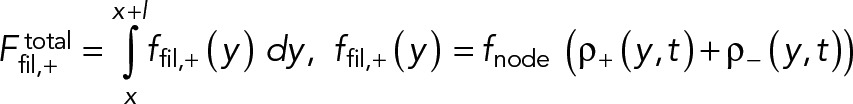 (3) (3)
|
Here, ffil, + (y) is the force per unit length experienced by a clockwise filament due to the pulling forces by myosins at y.
The evolution of the density is determined by the node velocities and turnover processes:
 (4) (4)
|
Here, ρ0 = 16.1 µm-1 is the mean number density of nodes, so that ρ0 / 2 is the mean number of clockwise nodes. The right-hand side of Eq. 4 describes the association and dissociation that maintains a mean density ρ0 over the time τturn (Figure 1B).
Equations 1– 4 describe the dynamics of clockwise (+) nodes. Similar equations are obtained for the counterclockwise (−) nodes, by replacing ρ+ (x, t), v+ (x, t) with ρ- (x, t), v- (x, t), and replacing the filament length l by −l in Eq. 3 (Materials and Methods, Eqs. M.1–M.4).
The parameters γanc and fnode are obtained as best-fit parameters by comparison of model predictions with experiment (Table 1). We solved Eqs. 1– 4 and M.1–M.4 numerically and analytically with periodic boundary conditions.
TABLE 1:
Parameters of the mathematical model of the S. pombe cytokinetic ring.
| Symbol | Meaning | Value | Legend |
|---|---|---|---|
| ρ0 | Steady-state total density of nodes at onset of constriction | 16.1 μm−1 | (A) |
| l | Length of F-actin per node at the onset of constriction | 2.7 μm | (A) |
| fmyo | Force exerted by one Myo2 head | 1.11 ± 0.43 pN | (B) |
| fnode | Force exerted by one node on one filament that passes through it | 0.41 ± 0.16 pN | (B) |
| γanc | Membrane drag coefficient of the node anchor | 810 ± 370 pN s μm−1 | (B) |
| τturn | Turnover time of nodes | 18.6 s | (C) |
| L0 | Initial length of the ring | 11.8 μm | (D) |
| frep | Repulsive force between nodes | 0.1 pN | (E) |
| brep | Range of node repulsive force | 0.1 μm | (E) |
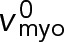 |
Load-free velocity of myosin Myo2 | 240 nm s−1 | (F) |
| - | Ring tension | 391 ± 154 pN | (G) |
| - | Myosin-II Myo2 node velocity | 22 ± 10 nm s−1 | (H) |
Errors are SDs for experimentally measured values and calculated parameters.
(A) Calculated from the experiments of Pelham and Chang (2002), Wu and Pollard (2005), Courtemanche et al. (2016), and Laplante et al. (2016).
(B) Obtained in this study. Associated error is due to uncertainty in prior experimental measurements of node velocity and ring tension.
(C) Obtained from FRAP experiments on YFP-Myo2 in constricting rings as measured in Sladewski et al. (2009).
(D) Previously measured using fluorescence microscopy on GFP-Cdc4 (Pelham and Chang, 2002).
(E) Chosen such that the final mean cluster width after aggregation was ∼150 nm (Materials and Methods).
(F) Obtained from gliding filament assays of Stark et al. (2010) as described in Supplemental Materials and Methods.
(G) Obtained from tension measurement experiments in Stachowiak et al. (2014).
(H) Obtained from FPALM myosin Myo2 node velocity measurements in Laplante et al. (2016).
Expression for fnode, the mean force exerted by a node on one actin filament
The net force exerted by a node on one filament passing through it, fnode, is a collective time-averaged force due to the individual pulling forces exerted by the 16 myosin Myo2 heads of the node. It is related to the instantaneous force fmyo that an individual myosin-II head exerts on an actin filament that it binds, as follows.
Now the mean number of filaments of each polarity passing through a given node is given by
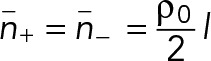 (5) (5)
|
The total force exerted by the 16 myosin heads belonging to this node, equal to 16 fmyo, is divided equally among these filaments, so that
 (6) (6)
|
Thus, the force exerted by a node on a filament passing through it depends on the number of myosin heads in the node, but also on the mean number of filaments passing through it.
The steady-state ring consists of two contra-rotating families of nodes
By inspection of Eqs. 1– 4 and M.1–M.4 it is simple to see that a steady-state solution is the homogeneous ring with equal numbers of clockwise and counterclockwise filaments and constant total node density ρ0,
|
|
Substituting Eq. 7 into Eqs. 2, 3, M.2, and M.3 gives
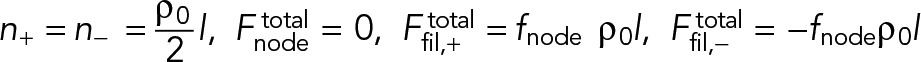 (8) (8)
|
Equation 8 states that the steady-state ring has net polarity zero; thus, the total myosin force exerted by a node  is zero, as the myosins of that node pull on as many clockwise as counterclockwise filaments passing through the node. The only unbalanced force on a node is the force on its own actin filament
is zero, as the myosins of that node pull on as many clockwise as counterclockwise filaments passing through the node. The only unbalanced force on a node is the force on its own actin filament 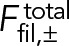 due to pulling by myosin in the ρ0l nodes that the filament of length l passes through. This force is balanced by the drag force on the node-membrane anchor. Equation 1 yields the velocities of each node family (Figure 2),
due to pulling by myosin in the ρ0l nodes that the filament of length l passes through. This force is balanced by the drag force on the node-membrane anchor. Equation 1 yields the velocities of each node family (Figure 2),
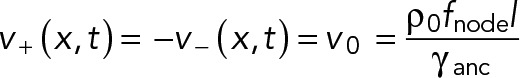 (9) (9)
|
FIGURE 2:
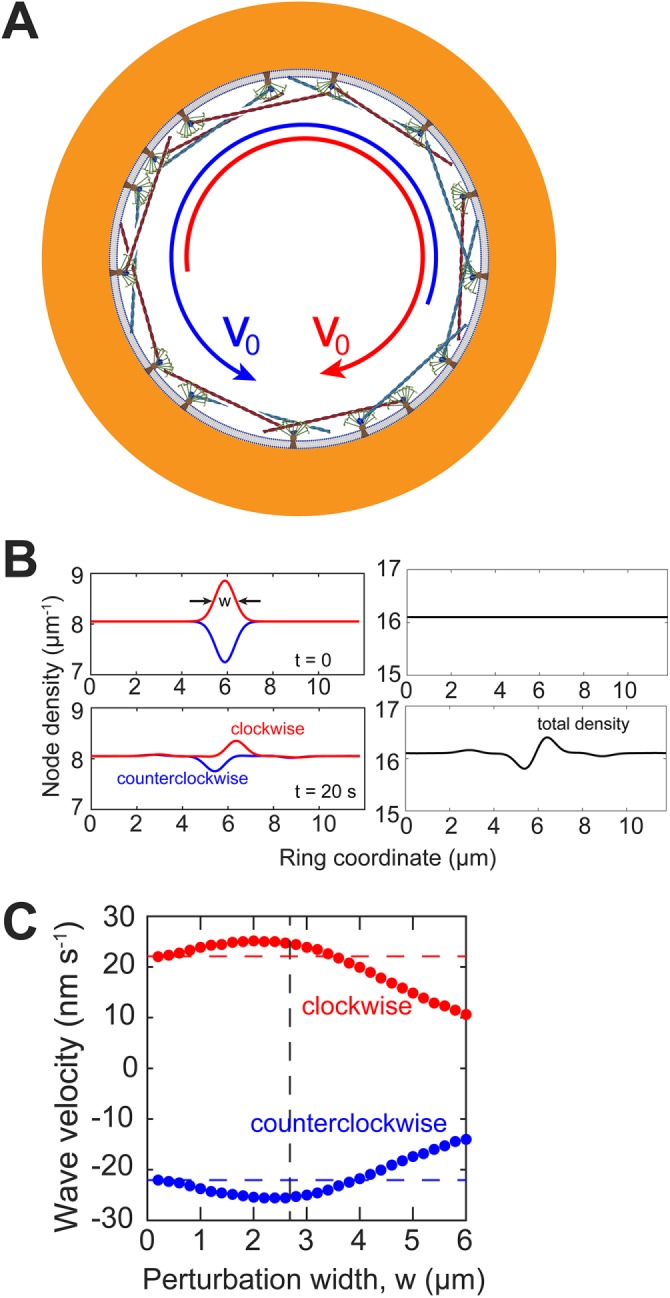
Actin and myosin in nodes contra-rotate around the steady-state cytokinetic ring. Model parameters as in Table 1. (A) Schematic of node motions in the steady-state ring. The motions of the clockwise (red filament) and counterclockwise (blue filament) nodes are indicated by curved arrows of respective colors. All nodes move at the same speed v0 = 22 nm s−1. (B) Density profiles of each type of nodes (left) and of the total density profile (right) at t = 0 and 20 s. Initial composition fluctuation had a Gaussian profile with a relative amplitude 5% and a full width half maximum w of 1 μm (black arrows). Over 20 s, perturbations traveled with a mean velocity 23 nm s−1. The amplitudes of both perturbations decreased by 63% over 20 s, approximately the turnover time τturn = 18.6 s. (C) Density wave velocities vs. perturbation full width at half maximum w. Initial condition as in B but with a varying w. Velocities are calculated by the displacement of the peak of perturbation between 10 s and 20 s. Vertical dashed line: actin filament length 2.7 µm. Horizontal dashed lines: node velocity ±v0 in the steady-state homogeneous ring. Magnitudes of cluster velocity of either type of cluster are approximately equal at every width w.
The two families move with speed v0 in opposite directions, that is, they contra-rotate. Comparing this predicted speed with the experimental node speed of 22 ± 10 nm s-1 (Laplante et al., 2016) and using the parameter values in Table 1, we obtain a membrane anchor drag coefficient γanc = 810 ± 370 pN s µm-1 (mean ± SD).
The model explains the observed bidirectional motions of nodes (Laplante et al., 2016). The origin of the contra-rotation is that + nodes are pulled clockwise because their filaments point clockwise and are pulled clockwise by myosins, while the − nodes are similarly pulled counterclockwise. This is also consistent with other confocal microscopy measurements of myosin-II motions in S. pombe (Wollrab et al., 2016).
Node composition fluctuations generate contra-propagating density waves of myosin-II and other node components
What are the experimentally measurable consequences of the contra-rotating node families? Superresolution microscopy can pick up individual node motions, but more commonly a collective fluorescence intensity distribution around the ring is measured. This intensity distribution represents the density of labeled molecules of one type, convolved with a point spread function due to optical resolution limits.
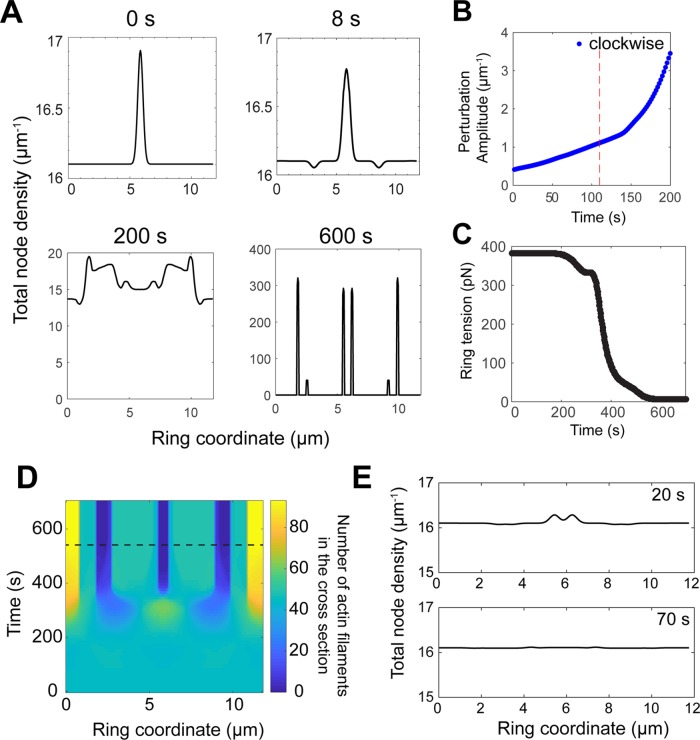
Among the most common fluorescence microscopy measurements in the cytokinetic ring is time-lapse imaging of tagged heavy or light chain myosin-II molecules. To see how the contra-rotating node families would manifest themselves in such images, consider a local node composition fluctuation producing a surfeit of + nodes (say) over - nodes. Such fluctuations are inevitable, and will constantly occur in the stochastic ring. Let us follow the fate of a simple Gaussian-shaped composition fluctuation of relative amplitude 5% and full width at half maximum of w (Figure 2B).
Numerically solving Eqs. 1– 4 and M.1–M.4, for a range of widths, 0.2 µm ≤ w ≤ 6 µm, we obtained the time course of the total density profile (that would be measured in conventional fluorescence microscopy), as well as the density profile of each type of node and the associated velocities (see Materials and Methods). The evolution of such a composition fluctuation of width 1 µm is shown in Figure 2B.
The result of such composition fluctuations is that two waves are generated in the total density field ρ(x, t), traveling in opposite directions and associated with each node type. One wave is a translating surfeit pulse, and the other a translating deficit pulse (Figure 2B). The wave speed depends on the width of the perturbation, w. Narrow initial fluctuations (w ≪ l = 2.7 μm) move at ~v0 = 22 nm s-1, whereas broader initial fluctuations generate waves that move more slowly, with a velocity that decreases with w (Figure 2, B and C). For example, a perturbation of width 6 µm generates waves moving at 10 nm s-1. The amplitude of the pulses decays over 20 s, approximately the turnover time τturn = 18.6 s (Figure 2B). These findings suggest the homogeneous ring is stable to small composition fluctuations, which we also demonstrated analytically (Supplemental Materials and Methods).
These results show that node composition fluctuations generate clockwise and counterclockwise density waves moving in opposite directions. Thus, we predict that the experimental intensity distribution of Myo2, which is associated with the nodes, exhibits intensity waves of this type, a direct reflection of the two contra-rotating node families.
Tension is generated in the cytokinetic ring by myosin pulling on barbed-end-anchored actin filaments
In this section, we calculate the steady-state ring tension predicted by the model, and we compare to experimental values measured in fission yeast protoplasts (Stachowiak et al., 2014). We start from the tension profile along the length of a clockwise filament, Tfil,+ (y), given by
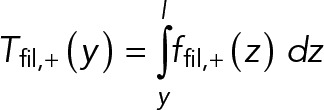 (10) (10)
|
Here, ffil,+(y) is the force per unit length experienced by the clockwise filament (Eq. 3) due to myosins at location y along the filament. Using the fact that in steady state ρ+ = ρ- = ρ0/2, Eq. 3 yields ffil,+ = fnode ρ0. Using this expression in Eq. 10 gives Tfil,+ = fnode ρ0 (l - y), showing that the tension is highest at the anchored barbed end and decreases linearly to zero at the pointed end (Figure 3A). Thus, the mean tension along the filament length is
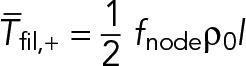 (11) (11)
|
Figure 3:
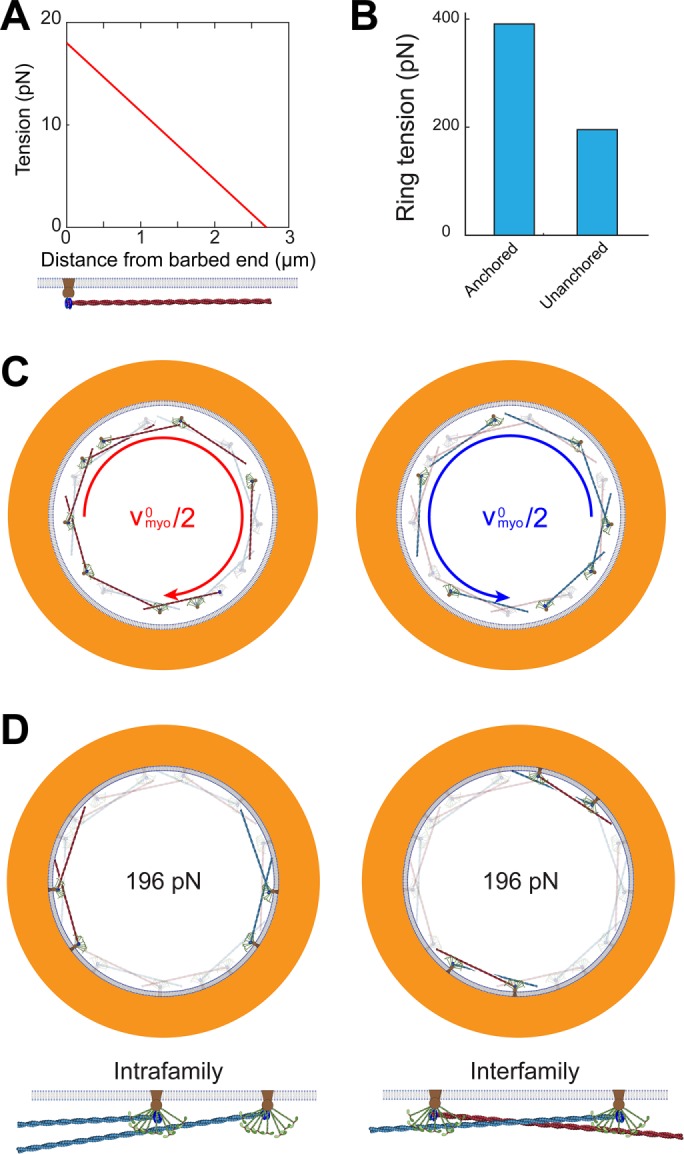
The fission yeast ring generates tension by myosin pulling on barbed-end-anchored actin filaments. Model parameters as in Table 1 unless otherwise specified. (A) Tension vs. distance from the barbed end along a filament. Tension is highest at the barbed end, and decreases linearly to zero at the pointed end. (B) Tension of rings with and without membrane anchoring. (C) Schematic of node motions in an unanchored ring, where the anchor drag coefficient γanc is zero. The motions of the clockwise (red filament) and counterclockwise (blue filament) nodes are indicated by curved arrows of respective colors. Nodes contra-rotate at the Myo2 load-free velocity 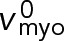 relative to each other. (D) Schematic of two types of node-node interactions in the ring: intrafamily (left) and interfamily (right). Each type of interaction contributes half of ring tension.
relative to each other. (D) Schematic of two types of node-node interactions in the ring: intrafamily (left) and interfamily (right). Each type of interaction contributes half of ring tension.
Each of the many filaments in the ring cross-section has the same mean tension in a homogeneous ring, so the net ring tension is 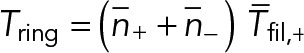 , or
, or
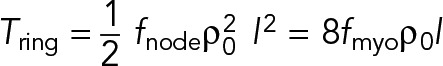 (12) (12)
|
after using Eqs. 6 and 8. This is our final expression for the tension in terms of fmyo, the force per Myo2 head.
Let us now compare this prediction with the measured ring tension in S. pombe protoplasts of 391 ± 154 pN (Stachowiak et al., 2014). Using the experimentally measured mean number of actin filaments in the cross-section of the S. pombe ring, ρ0l ≈ 44 (see Table 1), Eq. 12 then yields fnode = 0.4 ± 0.2 pN, and a myosin-II force per head of fmyo = 1.1 ± 0.4 pN (mean ± SD).This value is similar to reported stall forces ∼0.6 pN and ∼0.8 pN per head for turkey gizzard smooth muscle myosin-II and chicken pectoralis skeletal muscle myosin-II, respectively (Tyska et al., 1999). Using the above value for fnode in Eq. 11 gives an average tension per filament of 8.9 pN (Figure 3A).
The mechanism of tension generation is that myosin-II molecules bind and pull barbed-end-anchored actin filaments in nodes. The filament tension is highest at the barbed end and decreases linearly to zero at the pointed end, and is balanced by the membrane drag of the barbed-end node anchor; this lateral resistance from the anchor in the membrane is the essential feature that enables the filament to build up tension. This mechanism is supported by the fact that it leads to a myosin force per head that is very similar in magnitude to previously reported values.
Membrane anchoring of actin and myosin doubles ring tension
We have seen that membrane anchoring underlies the tension generation mechanism, suggesting that in the absence of anchoring tension would vanish. We show in this section that this is in fact not true, because in the nodes organization actin barbed ends and myosin-II are internally anchored even when removed from the membrane. However, we will see that the effect of unanchoring is to halve the tension.
To perform the “thought experiment” of unanchoring the ring, we will set the anchor drag coefficient γanc to zero. We will see below that this will cause much faster contra-rotation of the two node families; thus, we will need to incorporate a myosin force-velocity relationship into our calculations (thus far we assumed the force exerted by a Myo2 head is fixed at fmyo, a good approximation for the low velocities realized in the anchored ring).
To model the unanchored ring, we incorporate a linear force-velocity relationship for the myosin-II Myo2 in our model, with a Myo2 load-free velocity of 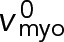 . We assume 1) the densities of clockwise and counterclockwise nodes of the unanchored ring are uniform and equal, 2) relative contra-rotation velocity between the two node families is uniform and equal to vrel, and 3) nodes of the same family move at a uniform velocity as in the anchored ring. We will self-consistently demonstrate the validity of these assumptions. Modifying Eqs. 2 and 3 to include the myosin force-velocity relationship only for the interfamily actomyosin forces leads to
. We assume 1) the densities of clockwise and counterclockwise nodes of the unanchored ring are uniform and equal, 2) relative contra-rotation velocity between the two node families is uniform and equal to vrel, and 3) nodes of the same family move at a uniform velocity as in the anchored ring. We will self-consistently demonstrate the validity of these assumptions. Modifying Eqs. 2 and 3 to include the myosin force-velocity relationship only for the interfamily actomyosin forces leads to
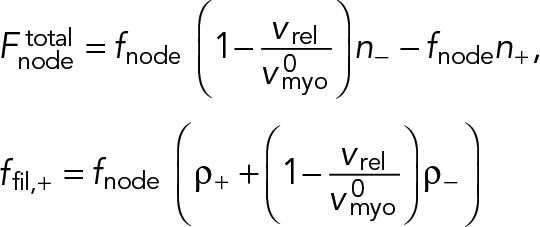 (13) (13)
|
Using the homogeneous densities and number of filaments in the cross-section from Eqs.7 and 8 into Eq. 13, we have
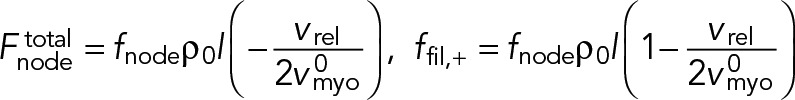 (14) (14)
|
Using these force expressions in the force balance Eq. 1 with the membrane node anchor drag γanc set to zero, we have
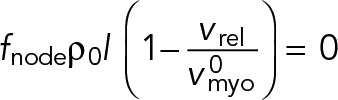 (15) (15)
|
Thus, in the unanchored ring contra-rotating nodes exert zero force on one another, and move relative to one another with the load-free velocity of myosin Myo2, 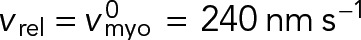 (Stark et al., 2010, and Supplemental Materials and Methods). (We note also that if formin-mediated polymerization of actin filaments is accounted for, at growth rate vpol, the relative velocity becomes
(Stark et al., 2010, and Supplemental Materials and Methods). (We note also that if formin-mediated polymerization of actin filaments is accounted for, at growth rate vpol, the relative velocity becomes 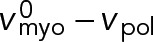 such that the relative actin-myosin sliding velocity is maintained at
such that the relative actin-myosin sliding velocity is maintained at 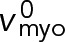 and the force is still zero.) Using Eq. 7 and a uniform node velocity in Eq. 4, we see that a homogeneous ring with uniform node velocities and a relative contra-rotation velocity of
and the force is still zero.) Using Eq. 7 and a uniform node velocity in Eq. 4, we see that a homogeneous ring with uniform node velocities and a relative contra-rotation velocity of 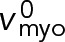 satisfies all the equations (Figure 3C). Similar equations can be written for the counterclockwise nodes (Materials and Methods, Eqs. M.5 and M.6). We now calculate the force per unit length along a filament in an unanchored ring from Eq. 13:
satisfies all the equations (Figure 3C). Similar equations can be written for the counterclockwise nodes (Materials and Methods, Eqs. M.5 and M.6). We now calculate the force per unit length along a filament in an unanchored ring from Eq. 13:
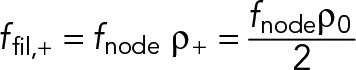 (16) (16)
|
This force per unit length is half that which a filament feels in an anchored ring (Eq. 11). In the unanchored ring, a given actin filament experiences force only from myosin from nodes whose actin filaments have the same polarity as the given filament (Eq. 15), in contrast to the anchored ring where Myo2 in nodes of both polarities exert force on a given filament (Eq. 11). Thus, the actin of one node family does not feel forces from the myosins of the other node family. Using a tension calculation procedure similar to that illustrated by Eqs. 11 and 12, we obtain  . Thus, the tension in the ring, equal to
. Thus, the tension in the ring, equal to 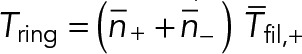 , is given by
, is given by
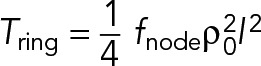 (17) (17)
|
This is our final result for the unanchored ring. We see that an unanchored ring exerts only half the tension of the anchored ring (Eqs. 12 and 17 and Figure 3, B and D). In other words, by anchoring components to the membrane, the tension is doubled because additional tension can then be generated by internode-family myosin forces.
Turnover prevents myosin aggregation that would lead to loss of tension and ring fracture
Confocal micrographs show that the density of myosin-II and other components in the ring exhibit continuous intrinsic fluctuations. In this section, we will use our model to show that such fluctuations are a constant threat to the tension and structural integrity of the ring, due to the intrinsic instability of contractile actomyosin structures. The instability arises because a fluctuation that increases density locally will tend to amplify the contractility at that location, further increasing the density and leading to a runaway instability. We will show that turnover plays a vital role in preventing this catastrophic sequence of events.
Let us use our model without turnover (turnover terms deleted from Eqs. 4 and M.4) to compute the fate, without turnover protection, of an initial 5% localized density fluctuation with full width at half maximum of 500 nm (Figure 4A). We numerically solved Eqs. 1– 4 and M.1–M.4 and obtained the time course of node density profiles (Materials and Methods). For these calculations we added to our model a small scale short-ranged node-node repulsion of magnitude frep = 0.1 pN and range brep = 0.1 µm (Materials and Methods). This measure was in order to prevent a blowup of densities to infinity as aggregation progressed.
Figure 4:
Turnover of actin and myosin in nodes prevents aggregation of nodes, loss of tension, and ring fracture. Model parameters as in Table 1, except that node turnover is switched off, unless otherwise stated. (A) Total node density vs. ring coordinate at the indicated times. In the initial condition, a Gaussian of full width at half maximum 500 nm and amplitude 5% of the mean node density ρ0 is superimposed on a homogeneous ring of node density ρ0 / 2 for both families. (B) Time evolution of the amplitude of the highest peak of clockwise node density profile, in the ring of A. Vertical dashed line: Aggregation time scale τagg defined as the time at which the amplitude of the central clockwise peak increases to a value a factor e times its initial value. (C) Mean tension of the ring in A vs. time. The ring loses 90% of its initial tension over ∼500 s. (D) Total actin filaments in the ring cross-section vs. ring coordinate and time of the ring in A. Horizontal dashed line: time of ring fracture, defined as the instant where there is <1 filament at one location in the ring. (E) Total node density vs. ring coordinate of a ring with turnover, at 20 and 70 s. Turnover prevents the aggregation of nodes in the initial condition, and the initial perturbation visibly disappears by 70 s. Initial condition as in A.
We found that the initial local density perturbation precipitated a disastrous aggregation of myosin and actin into clusters with widths much smaller than the mean actin filament length l = 2.7 µm. The detailed sequence of events is shown in Figure 4A. For smaller times, t ≤ 10 s, the perturbations grow in amplitude. Valleys appeared to the left and right of the peak, separated from the peak center by ∼l = 2.7 µm, the mean actin filament length (Figure 4A). During the next ∼200 s, the perturbation split into two distinct peaks, each associated with one node family, and several smaller peaks and valleys appeared (Figure 4A). From 200 to 600 s, peaks and valleys grew in amplitude rapidly and merged, terminating in six clusters with a mean width of ∼150 nm. This final width is set by the short-ranged node repulsion force (Table 1).
The time for aggregation during the above process was τagg = 110 s, defined as the time when the maximum density of nodes of one type increased to a value a factor e times the initial value (Figure 4B).
We also tracked the mean tension of the ring during this aggregation process (Materials and Methods), and the number of actin filaments in the cross-section from Eq. 2 (Figure 4, C and D). The mean tension dropped from 382 pN to only 6 pN after 600 s, with most of the decrease from 200 s to 400 s as peaks and valleys in density became more and more pronounced (Figure 4, A and C). The number of actin filaments in the cross-section remained approximately uniform up to ∼250 s, after which filaments rapidly aggregated. This led to ring fracture after 540 s, defined as the time when at least one location along the ring had <1 filament (Figure 4, A and D).
We then repeated this numerical calculation with the same initial localized density perturbation, but with turnover restored. The perturbation was completely smoothed within 70 s (Figure 4E). Thus, turnover prevented the aggregation and ring fracture that would have occurred after τagg = 110 s. It was essential that the turnover time τturn = 18.6 s was significantly smaller than the aggregation time for this intervention to be successful. These results show that in the absence of turnover the ring is inherently unstable to fluctuations in myosin density, which cause myosins to aggregate. Turnover intervenes and prevents such fluctuations from running their natural course, as it tends to restore the homogeneous state of the ring.
Membrane anchoring of actin and myosin stabilizes the ring
We uncovered the role of turnover in removing clumps of actin and myosin in the ring that occur due to an intrinsic aggregation instability in the ring. Presumably, the compliance of the node anchors also plays a role in such an aggregation as nodes that experience a very large drag would be reluctant to aggregate. Here, we investigated how the aggregation time varies with the membrane anchor drag coefficient.
In an earlier section, we showed that anchoring of actin and myosin to the membrane doubles ring tension. In this section, we show that anchoring serves another function, to greatly enhance stability. We will see that, were the components more weakly anchored, the intrinsic contractile instabilities described above would onset more rapidly.
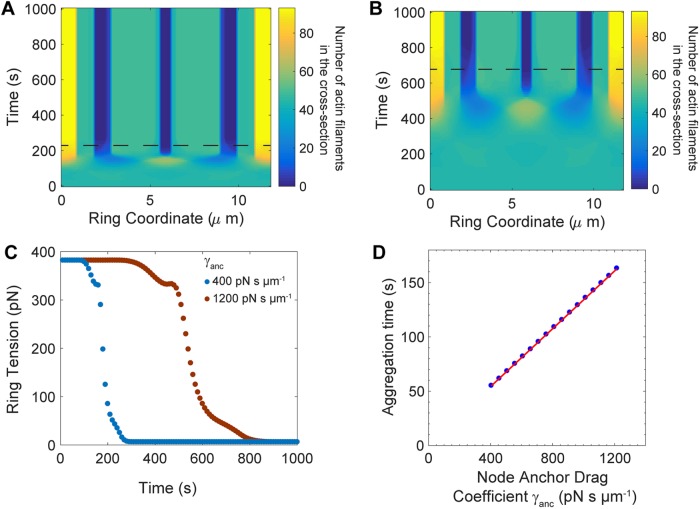
To demonstrate this we varied the anchor drag coefficient to be greater or smaller than the value γanc = 810 pN s μm−1 that we had previously obtained by comparison of predicted and experimentally measured node velocities (Table 1). We used the same initial density fluctuation as in the preceding subsection and we numerically solved Eqs. 1– 4 and M.1–M.4 with no turnover, using anchor drags of γanc = 400 and 1200 pN s μm−1. We computed the mean ring tension (Materials and Methods) and the profile of actin filaments in the cross-section from Eq. 2 (Figure 5, A–C). For both values of the drag, the mean tension decreased from 382 pN to 6 pN, the filaments aggregated and the ring fractured (Figure 5, A–C). However, for lower anchor drag these events onset sooner; for example, ring fracture occurred after 230 s (680 s) for an anchor drag of 400 (1200) pN s μm−1 (Figure 5, A and B).
Figure 5:
Weakening the lateral anchor drag leads to faster node aggregation and ring fracture in the absence of turnover. Simulation parameters as in Table 1, except node turnover is switched off by deleting turnover terms in Eqs. 4 and M.4, and the anchor drag coefficient γanc is varied as specified. (A, B) Profiles of actin filaments in the cross-section vs. time, in rings with anchor drag coefficient γanc = 400 pN s μm−1 (A) and 1200 pN s μm−1 (B). Horizontal dashed lines indicate time to ring fracture, defined as in Figure 4D. (C) Time evolution of tension in rings of A (blue) and B (red). (D) Aggregation time vs. node anchor drag coefficient γanc. Red: least-squares best-fit straight line.
We measured the aggregation time τagg for different anchor drag values in the range 400 ≤ γanc ≤ 1200 pN s μm−1. The aggregation time increased linearly with membrane drag (Figure 5D).
These results show that weakening of the lateral anchoring of the nodes makes the ring more unstable and prone to aggregation, suggesting that lateral membrane anchoring is essential to stabilize the ring.
DISCUSSION
Fission yeast is a model organism for realistic mathematical models of the cytokinetic ring
In fission yeast many contractile ring proteins have been identified and their amounts measured throughout cytokinesis (Wu and Pollard, 2005; Courtemanche et al., 2016). In addition, two recent developments have provided vital new information about the fission yeast cytokinetic ring, which opens the door to more realistic mathematical modeling. First, perhaps the most basic property of the ring, its tension, was measured in S. pombe for the first time (Stachowiak et al., 2014). The ring tension is the primary quantity for a mathematical model to generate and compare with experiment, and the absence of ring tension measurements has been a serious obstacle to modeling. Second, superresolution FPALM revealed organizational features of the ring not previously available from conventional microscopy, showing that in the constricting ring myosin and formin are organized in node-like structures anchored to the plasma membrane (Laplante et al., 2016).
Here we aimed to take advantage of this newly available information by building a realistic, coarse-grained, minimal model of the fission yeast ring whose assumptions are consistent with experimental measurements of actomyosin organization, membrane anchoring of ring components, and the amounts of key components (Wu and Pollard, 2005; Vavylonis et al., 2008; Courtemanche et al., 2016; Laplante et al., 2016). Our aim was to reveal the basic principles of tension generation and stability. Our coarse-grained approach, representing components by continuous density fields, has the advantage that it can illuminate basic mechanisms at play within the complex ring organization, which might be difficult to perceive in the context of a highly detailed molecular simulation.
Node-like organization marshals actomyosin forces to generate ring tension
Cytokinetic ring tension is thought to result from nonmuscle myosin-II binding to and exerting force on actin filaments. A difficulty is that the ring has considerable disorder, but a random actomyosin bundle exerts no tension, even when cross-linkers are present, because as much compression as tension is produced in filaments. Our model shows that ring tension is generated by myosin pulling on barbed-end-anchored actin filaments. Anchoring barbed ends to the plasma membrane is important because it limits motion both in the radial direction and in the lateral direction. Thus, barbed-end anchoring both attaches the ring to the plasma membrane, and also provides lateral resistance to myosin pulling so that filament tension can accumulate. Barbed-end anchoring is the most efficient for tension generation, as all myosins pulling a filament produce tension.
We showed that this tension production mechanism is efficient enough to generate values of tension consistent with the experimentally reported value of ∼400 pN (Figure 3B; Stachowiak et al., 2014), given the total amount of actin and myosin in the ring (Wu and Pollard, 2005; Courtemanche et al., 2016). By fitting the predicted tension to experiment, we obtained a myosin force per head for fission yeast Myo2, fmyo ∼ 1.1 pN. The force per head is actually somewhat less than the stall force, fstall, due to relative motions of the nodes and due to formin-mediated growth of the actin filaments. Assuming a linear force-velocity relation for Myo2, we estimate fmyo ∼ 0.45 fstall (see Supplemental Materials and Methods). Thus, fstall ∼ 2.4 pN, close to previously reported stall force values for myosin-II in different organisms, 0.6–2.3 pN (Kishino and Yanagida, 1988; Molloy et al., 1995; Ishijima et al., 1996; Tyska et al., 1999).
Our model neglected cross-linkers in the ring. How much do they contribute to tension? The best characterized actin filament cross-linkers in the fission yeast cytokinetic ring are α-actinin dimers, which are very dilute in the ring: given ∼250 α-actinin dimers and ∼190 nodes each with a mean of about one filament at onset of constriction (Wu and Pollard, 2005; Courtemanche et al., 2016; Li et al., 2016), there are nα ∼ 1 - 2 cross-linking connections per actin filament. Each of these has a relatively small turnover time τα ∼ 0.3 s, according to the in vitro experiments of Li et al. (2016). Using the measured α-actinin force constant kα = 0.025 pN nm−1 (Claessens et al., 2006), and taking the relative velocity of actin filaments to be twice the node velocity 2v0 ∼ 44 nm s−1, we estimate a typical force per actin filament ∼2kαv0ταnα ∼ 0.5 pN, much smaller than the ∼20 pN force that Myo2 exerts on a filament. Thus, α-actinin is likely unimportant to internal ring forces and dynamics in the constricting ring. Furthermore, given the relatively small amount of α-actinin, we suggest that myosin-II probably serves as the main cross-linker for bundling the ring.
Bidirectional motions reflect two classes of nodes in the constricting ring
From FPALM, an interesting observation about the constricting fission yeast ring is that myosin and formin move bidirectionally, with similar velocity distributions (Laplante et al., 2016). This supports the nodes hypothesis, and also indicates that few nodes remain stationary and suggests that there are two families of nodes with opposite directionality.
What defines this directionality? We used our model to show that actin filaments are responsible. A typical node, having on average one actin filament emanating from it, moves in the direction defined by the polarity of this actin filament. The actin filament produces a bias in the node motion because it points in a particular direction and is grabbed by myosin-II molecules, all pulling the filament in that same direction. By contrast, the myosin belonging to that node produces no bias in motion, because it grabs filaments in the unbiased bundle with balanced polarity. Overall, our model reproduces the phenomenon that nodes move bidirectionally in the constricting ring (Figure 2).
A previous model considered a similar but different ring organization, in which formin and myosin were assumed independently anchored rather than coanchored in nodes. The model produced a stable ring that generates tension similar to experimental values (Stachowiak et al., 2014). If we were to consider such an arrangement in the framework of the present model, the mean tension per filament, and hence the ring tension, would be unchanged, as every actin filament would interact with the same number of myosin-II heads as in the nodes organization. The formins would be predicted to execute bidirectional motion, but the anchored myosins would not move other than small fluctuations since their velocity is proportional to the local net actin polarity, which vanishes for a homogeneous ring. This latter feature is inconsistent with FPALM measurements that report bidirectional motions of both formin and myosin with similar speeds (Laplante et al., 2016).
A stochastic sliding-filament mechanism operates in the fission yeast cytokinetic ring
Long ago a sliding-filament mechanism was proposed for tension production by cytokinetic rings (Schroeder, 1975; Maupin and Pollard, 1986). How this might actually work is not obvious, however, because in muscle the mechanism is based on the sarcomere repeat unit and contraction of the unit produces thickening. In the fission yeast contractile ring neither sarcomeres nor thickening are apparent (Kanbe et al., 1989; Bezanilla and Pollard, 2000; Wu and Pollard, 2005; Kamasaki et al., 2007).
Our model shows that the mechanism at work is related to the sliding-filament system used in muscle, but is a subtly stochastic and transient version of this model in such a way as to maintain translational invariance along the ring. We find that filaments slide in opposition to one another, but do so continuously and everywhere around the ring, without being confined to fixed sarcomere-like structures. Moreover, the sliding processes are transient.
More specifically, we found that even though nodes are positioned around the ring without periodicity, pairwise interactions between nodes have characteristics of the sliding-filament mechanism. However, the interactions are transient and stochastic. A pair of nodes with opposite polarity interact like a sarcomere or contractile unit (Figure 3D, “interfamily”). As the two families of nodes contra-rotate, pairs of nodes with opposite polarity disappear as they meet and move past one other, while new pairs constantly appear as nodes first arrive within reach of one another. Turnover of nodes also contributes to the transient nature of the sliding-filament interaction.
We find, however, that the sliding-filament mechanism is not the whole story. A pair of nodes with the same polarity interacts like one half of a sarcomere, with myosins pulling on barbed-end-anchored actin filaments (Figure 3D, “intrafamily”). Unlike the transient appearing and disappearing of interfamily interacting pairs, pairs of nodes with the same polarity do not move relative to each other and maintain their relation until they leave the ring. Without sliding relative to one another, they generate tension, because a large number of such nodes are connected in a single family ring. Our model shows that each of these two suborganizations contributes to half of the ring tension.
Anchoring of components to the membrane and component turnover protect the ring from intrinsic instabilities
We found that the cytokinetic ring suffers from an instability characteristic of actively contractile organizations. A fluctuation that increases the local actin and myosin density produces higher contractile forces that draw in further actin and myosin, increasing the local contractility further and leading to runaway aggregation (Figures 4 and 5). In the absence of turnover, this instability led to loss of tension and ring fracture after a certain aggregation timescale (Figure 4). Restoring turnover, this disastrous outcome was averted provided the turnover time is less than the aggregation time. In normal cells this is indeed the case. We conclude that turnover functions to preserve organizational homeostasis and tension in the ring.
In addition, we found that node anchor drag in the membrane protects the ring from this contractile instability, because the bigger the drag the longer the aggregation and ring fracture time (Figure 5). Thus, anchoring of components in the plasma membrane and turnover work together to stabilize the ring.
Cytokinetic rings in other organisms
What can the results of the present study tell us about cytokinetic rings in organisms other than fission yeast? Although animal cells have much longer and wider contractile rings, their thicknesses are very similar to that of the fission yeast ring, ∼0.1–0.4 μm (Schroeder, 1972; Mabuchi, 1986), suggesting that anchoring of components to the plasma membrane may play a similar role in the tension mechanism as in fission yeast. Furthermore, in sea urchin embryos, contractile rings appear to assemble from clusters of myosin-II (Henson et al., 2017), reminiscent of the nodes from which the fission yeast ring is assembled (Vavylonis et al., 2008). However, light and dark bands were seen in electron microscopy images of the cleavage furrow in rat kangaroo cells (Sanger and Sanger, 1980). Fluorescence micrographs show bipolar myosin-II filaments in the contractile ring in sea urchin embryos and LLC-Pk1 cells (Beach et al., 2014; Henson et al., 2017) and lateral stacking of myosin bipolar filaments in striated patterns reminiscent of sarcomeric organization in HeLa cells (Fenix et al., 2016). Thus, cytokinetic rings in other organisms may exhibit both similarities and significant differences in organization, and a major scientific challenge will be to establish if basic mechanisms emerging in fission yeast carry over to these organisms.
MATERIALS AND METHODS
Model equations for the time evolution of counterclockwise nodes
The equations for evolution of the density and velocity of the clockwise nodes are Eqs. 1– 4. Similar equations can be written for the counterclockwise “−” nodes, using the following replacements in these equations: density ρ+(x, t) → ρ-(x, t) in Eq. 4, velocity v+(x, t) → v-(x, t) in Eqs. 1 and 4, and the filament length l → −l, in Eq. 3 as the filaments of these nodes are pointing in the opposite direction.
|
|
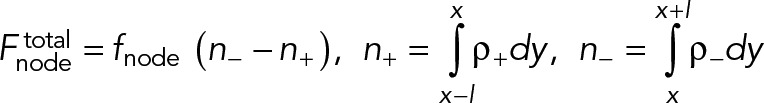 (M.2) (M.2)
|
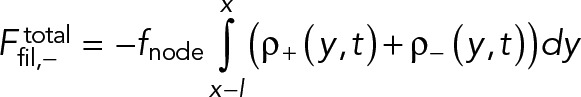 (M.3) (M.3)
|
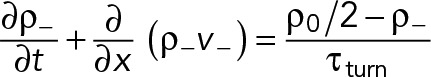 (M.4) (M.4)
|
In the presence of a force-velocity relationship for the myosin Myo2, Eqs. M.2 and M.3 get modified as
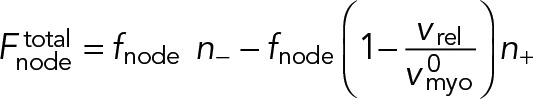 (M.5) (M.5)
|
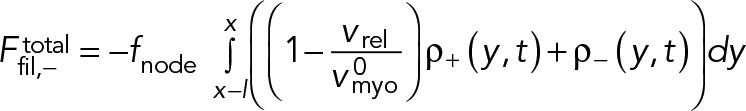 (M.6) (M.6)
|
Numerical solution of the model
We solved Eqs. 1– 4 and M.1–M.4 numerically using Runge–Kutta 4 with a spatial grid size of 50 nm and a time step of 0.2 s, except for the velocity results for Figure 2 where a finer grid of size 10 nm was used to achieve more accurate velocities. The mean velocity of the peak and the valley of Figure 2B was measured as the slope of the best-fit straight line to the peak or valley position versus time.
As the model exhibits presumably unbounded node density growth in the absence of turnover, we used a nonlinear filter to remove numerical instabilities (Engquist et al., 1989). In the absence of turnover, a node-node repulsive force was used to prevent unbounded growth. In this case, Eqs. 1 and M.1 get modified as
|
|
Here 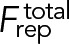 is a short-ranged repulsive force between nodes of magnitude frep and length scale brep given by
is a short-ranged repulsive force between nodes of magnitude frep and length scale brep given by
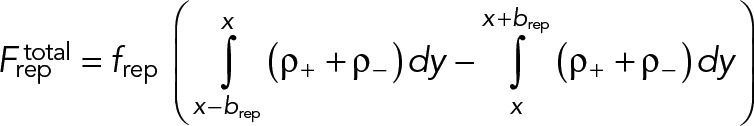 (M.8) (M.8)
|
frep and brep were chosen such that the final mean cluster width after aggregation was ∼150 nm. All numerical calculations were done in MATLAB.
Tension formula for the numerical calculations of the model
Every arrangement of nodes in the ring corresponds to a ring tension, which we calculate as follows. A clockwise filament from a + node at the point s crossing the point of interest x feels a pulling force fnode from the myosins of all the nodes lying along its length from x until its pointed end at s + l. A similar pulling force is felt by counterclockwise filaments from the nodes at x until their pointed end at s - l. The net tension is obtained by summing these contributions over all the filaments at the point x:
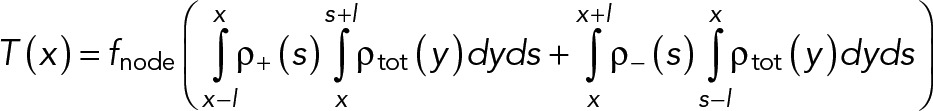 (M.9) (M.9)
|
The mean tension is then obtained by averaging T over the ring length.
Initial conditions
For the velocity measurements in Figure 2, B and C, the following initial condition was used:
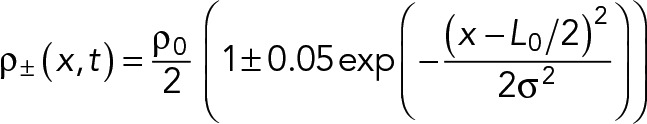 (M.10) (M.10)
|
where ρ0 = 16.1 µm-1 is the mean total density of nodes, x is the coordinate along the ring, that is, 0 ≤ x ≤ L0 where L0 = 11.8 µm is the initial length of the ring. σ is related to the full width at half maximum w as  .
.
For the results of Figures 4 and 5, a similar initial condition was used:
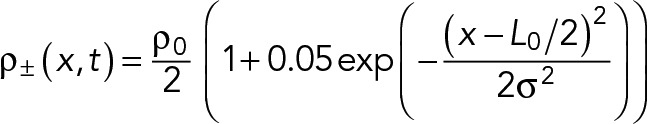 (M.11) (M.11)
|
The full width at half maximum here is w = 500 nm, which gives σ = 212 nm.
Code availability
Computer code used to obtain numerical results here can be made available on request.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant No. GM086731.
Abbreviation used:
- FPALM
fluorescence photoactivation localization microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0386) on September 27, 2017.
REFERENCES
- Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA. Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Pollard TD. Myosin-II tails confer unique functions in Schizosaccharomyces pombe: characterization of a novel myosin-II tail. Mol Biol Cell. 2000;11:79–91. doi: 10.1091/mbc.11.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF. Actin: protein structure and filament dynamics. J Biol Chem. 1991;266:1–4. [PubMed] [Google Scholar]
- Claessens MMAE, Bathe M, Frey E, Bausch AR. Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat Mater. 2006;5:748–753. doi: 10.1038/nmat1718. [DOI] [PubMed] [Google Scholar]
- Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR, Gould KL. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD, Chen Q. Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat Cell Biol. 2016;18:676–683. doi: 10.1038/ncb3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engquist B, Lotstedt P, Sjogreen B. Nonlinear filters for efficient shock computation. Math Comput. 1989;52:509–537. [Google Scholar]
- Fenix AM, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, Burnette DT. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Pollard TD. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J Cell Biol. 1976;71:848–875. doi: 10.1083/jcb.71.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Ditzler CE, Germain A, Irwin PM, Vogt ET, Yang S, Wu X, Shuster CB. The ultrastructural organization of actin and myosin II filaments in the contractile ring: new support for an old model of cytokinesis. Mol Biol Cell. 2017;28:613–623. doi: 10.1091/mbc.E16-06-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima A, Kojima H, Higuchi H, Harada Y, Funatsu T, Yanagida T. Multiple- and single-molecule analysis of the actomyosin motor by nanometer-piconewton manipulation with a microneedle: unitary steps and forces. Biophys J. 1996;70:383–400. doi: 10.1016/S0006-3495(96)79582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe T, Kobayashi I, Tanaka K. Dynamics of cytoplasmic organelles in the cell cycle of the fission yeast Schizosaccharomyces pombe: three-dimensional reconstruction from serial sections. J Cell Sci. 1989;94:647–656. doi: 10.1242/jcs.94.4.647. [DOI] [PubMed] [Google Scholar]
- Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Laplante C, Berro J, Karatekin E, Hernandez-Leyva A, Lee R, Pollard TD. Three myosins contribute uniquely to the assembly and constriction of the fission yeast cytokinetic contractile ring. Curr Biol. 2015;25:1955–1965. doi: 10.1016/j.cub.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Huang F, Tebbs IR, Bewersdorf J, Pollard TD. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc Natl Acad Sci. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Christensen JR, Homa KE, Hocky GM, Fok A, Sees JA, Voth GA, Kovar DR. The F-actin bundler α-actinin Ain1 is tailored for ring assembly and constriction during cytokinesis in fission yeast. Mol Biol Cell. 2016;27:1821–1833. doi: 10.1091/mbc.E16-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I. Biochemical aspects of cytokinesis. Int Rev Cytol. 1986;101:175–213. doi: 10.1016/s0074-7696(08)60249-1. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P, Pollard TD. Arrangement of actin filaments and myosin-like filaments in the contractile ring and of actin-like filaments in the mitotic spindle of dividing HeLa cells. J Ultrastruct Mol Struct Res. 1986;94:92–103. doi: 10.1016/0889-1605(86)90055-8. [DOI] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guérin C, Boujemaa-Paterski R, Staiger CJ, Martiel J-L, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/Cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JM, Sanger JW. Banding and polarity of actin filaments in interphase and cleaving cells. J Cell Biol. 1980;86:568–575. doi: 10.1083/jcb.86.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. The contractile ring: II. Determining its brief existence, volumetric changes, and vital role in cleaving arbacia eggs. J Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. Dynamics of the contractile ring. Soc Gen Physiol Ser. 1975;30:305–334. [PubMed] [Google Scholar]
- Sladewski TE, Previs MJ, Lord M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol Biol Cell. 2009;20:3941–3952. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MR, Laplante C, Chin HF, Guirao B, Karatekin E, Pollard TD, O’Shaughnessy B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell. 2014;29:547–561. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and Myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010;21:989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Dupuis DE, Guilford WH, Patlak JB, Waller GS, Trybus KM, Warshaw DM, Lowey S. Two heads of myosin are better than one for generating force and motion. Proc Natl Acad Sci USA. 1999;96:4402–4407. doi: 10.1073/pnas.96.8.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu J-Q, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wollrab V, Thiagarajan R, Wald A, Kruse K, Riveline D. Still and rotating myosin clusters determine cytokinetic ring constriction. Nat Commun. 2016;7:11860. doi: 10.1038/ncomms11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.