KIF16B is a motor that binds early endosomes and controls the recycling of receptors. It has a PX domain that binds PI(3)P, which has been proposed to mechanically link motor to cargo. We determined the strength of this bond and its lifetime under load, and found it to be a suitable mechanical linkage for the KIF16B motor under working conditions.
Abstract
KIF16B is a highly processive kinesin-3 family member that participates in the trafficking and tubulation of early endosomes along microtubules. KIF16B attaches to lipid cargoes via a PX motif at its C-terminus, which has nanomolar affinity for bilayers containing phosphatidylinositol-3-phosphate (PI[3]P). As the PX domain has been proposed to be a primary mechanical anchor for the KIF16B-cargo attachment, we measured the adhesion forces and detachment kinetics of the PX domain as it interacts with membranes containing 2% PI(3)P and 98% phosphatidylcholine. Using optical tweezers, we found that the adhesion strength of a single PX domain ranged between 19 and 54 pN at loading rates between 80 and 1500 pN/s. These forces are substantially larger than the interaction of the adhesion of a pleckstrin homology domain with phosphatidylinositol 4,5-bisphosphate. This increased adhesion is the result of the membrane insertion of hydrophobic residues adjacent to the PI(3)P binding site, in addition to electrostatic interactions with PI(3)P. Attachment lifetimes under load decrease monotonically with force, indicating slip-bond behavior. However, the lifetime of membrane attachment under load appears to be well matched to the duration of processive motility of the KIF16B motor, indicating the PX domain is a suitable mechanical anchor for intracellular transport.
INTRODUCTION
Intracellular trafficking of vesicular organelles is powered by cytoskeletal motors recruited through their attachment to signaling and scaffolding proteins and/or lipids. These signaling and scaffolding molecules define not only the identity of the organelle but also their motile properties through motor linkages (Akhmanova and Hammer, 2010). Different membrane compartments have distinct phosphoinositide contents that change as the compartments mature or transform, which has been shown to affect motor recruitment (Di Paolo and De Camilli, 2006; Picas et al., 2016). In some cases, this motor recruitment is due to the direct interaction between specific phosphoinositide headgroups and protein subdomains within the motor tails (Hoepfner et al., 2005; Hokanson and Ostap, 2006; Spudich et al., 2007; McKenna and Ostap, 2009).
The direct interactions between motors and membranes via physiological attachments can suffice for in vitro transport of synthetic liposomes along immobilized cytoskeletal tracks or the movement of cytoskeletal filaments by motors attached to fluid bilayers under very low forces (Zot et al., 1992; Klopfenstein et al., 2002; Hoepfner et al., 2005; Pyrpassopoulos et al., 2012 , 2016), as can motors bound to membranes via engineered attachments (e.g., Pyrpassopoulos et al. [2012], Nelson et al. [2014], Grover et al. [2016]). Despite this demonstration of motility, little is known about the mechanical and kinetic properties of physiological motor-membrane interactions under working conditions. To be a suitable anchor, the duration of membrane attachments under mechanical force should be well matched to the kinetics and force generation of the cytoskeletal motor.
We previously explored the attachment lifetime of the pleckstrin-homology (PH) domain containing tail-region of vertebrate Myo1c to membrane bilayers containing 2% phosphatidylinositol-4,5-bisphosphate (PI[4,5]P2) under tensile forces (Pyrpassopoulos et al., 2010). The binding of Myo1c-PH domain to PI(4,5)P2, which is crucial for defining the motor’s subcellular localization (Hokanson et al., 2006), is due to electrostatic interactions between positively charged residues in the PH domain and the anionic headgroup of PI(4,5)P2 (Hokanson et al., 2006; McKenna and Ostap, 2009). We found the membrane attachment lifetime to be highly force sensitive. The durations of attachments loaded to 2.5 pN in a direction normal to the plane of the membrane is substantially shorter than the actin-attachment lifetime of actively cycling Myo1c (Manceva et al., 2007; Pyrpassopoulos et al., 2010). Although we found that ensembles of PI(4,5)P2-bound Myo1c molecules can generate and sustain forces (Pyrpassopoulos et al., 2016), the duration of attachment to these model membranes would be far too short for a low number of motors to processively carry a cargo under load.
KIF16B, a highly processive kinesin 3 family member that binds to early endosomes and controls the recycling of receptors, has a C-terminal PX domain with high specificity for phosphatidylinositol-3-phosphate (PI[3]P) (Hoepfner et al., 2005; Blatner et al., 2007; Soppina et al., 2014). Like the Myo1c-PH domain described above, there is a substantial electrostatic component to the binding of the PX domain to the phosphoinositide head group. However, kinetic and high-resolution structural studies of KIF16B-PX domain revealed that membrane binding is greatly enhanced by the penetration of hydrophobic residues that are adjacent to the PI(3)P binding site into the lipid membrane (Blatner et al., 2007), which may impart a mechanical advantage to the attachment.
To better understand the mechanical properties of the KIF16B-PX-membrane interaction, and to determine whether this linkage is an appropriate anchor for a processive kinesin, we measured membrane adhesion forces and the lifetime of membrane attachment under tensile forces. Our results indicate that binding of the PX domain to PI(3)P is a suitable anchor for processive motility, as determined by adhesion strength and lifetime, made possible by both the electrostatic interaction with PI(3)P and the insertion of hydrophobic residues into the membrane bilayer.
RESULTS
Mechanical strength of KIF16B-PX cargo binding domain with supported lipid membranes containing 2% PI(3)P
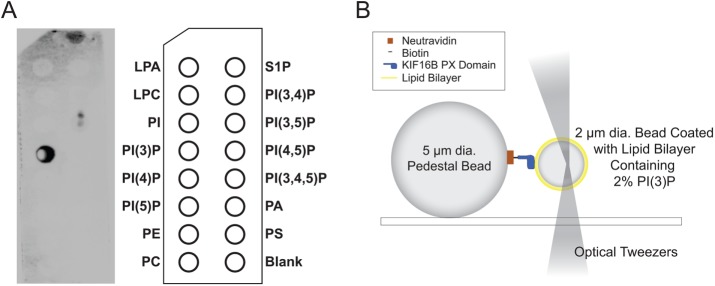
We measured the forces required to detach a single, monomeric KIF16B PX-domain construct from beads coated with a lipid bilayer composed of 2% PI(3)P and 98% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). KIF16B-PX was site-specifically biotinylated (Figure 1A) and attached to spherical pedestals that were coated with neutravidin at surface densities that resulted in single molecule interactions (see Materials and Methods; Figure 1B). Interactions between pedestal-bound KIF16B-PX and lipid-coated beads were detected as rupture peaks (Figure 2A) after the lipid-coated beads were compressed against immobilized pedestals and then retracted using the optical trap.
FIGURE 1:
(A) PIP-Strip showing the binding of biotinylated KIF16B-PX to PI(3)P as detected by chemiluminescence imaging of HRP-streptavidin. (B) Diagram of experimental setup used to measure the interaction of pedestal attached KIF16B-PX with membrane-coated beads held in an optical trap. The trap position was oscillated, resulting in the compression of the trapped beads against KIF16B-PX–coated pedestals, followed by retraction. Adhesions forces displace the bead from the trap center during retraction. Formation and subsequent rupture of bonds appeared as negative peaks in the data traces (Figure 2A). Beads and protein molecules are not drawn to scale. Adapted from Pyrpassopoulos et al. (2013).
FIGURE 2:
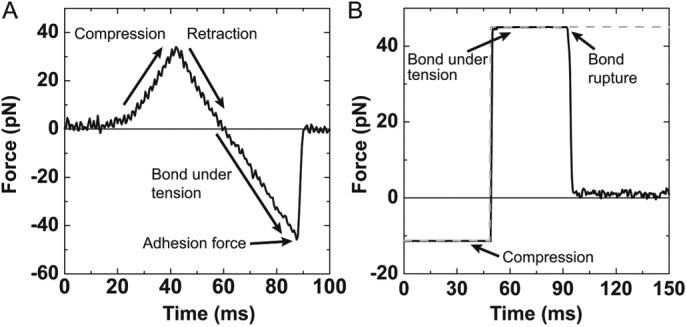
(A) Example of an experimental ramp-force rupture event between 2% PI(3)P lipid-coated bead and KIF16B-PX at a loading rate of 1500 pN/s. The three regions are the compression of the optically trapped bead on the pedestal, retraction of the bead in the opposite direction until the compressive force reaches zero, and the bond under tension as the optical trap is moved until the bond ruptures. The reported adhesion force is the point where the bond ruptures. (B) The attachment duration of an experimental single adhesive bond between a 2% PI(3)P lipid-coated bead and KIF16B-PX under 45 pN of load. A square pulse (dashed gray trace) is the signal to drive compression and retraction of the membrane-coated bead. Adhesive forces (solid black trace) are recorded during attachments. A constant force is maintained via a feedback loop until the bond ruptures.
The forces required to rupture adhesion between KIF16B-PX and membranes containing 2% PI(3)P were determined at loading rates of 80 and 1500 pN/s (Figure 3A). Contributions of nonspecific interactions to the experimental distributions were removed by subtracting distributions of forces with either KIf16B-PX or PI(3)P absent. With either KIf16B-PX or PI(3)P absent, the magnitude and the frequency of the nonspecific adhesion forces were similar and all control data were consolidated to one distribution (Supplemental Figure 1).
FIGURE 3:
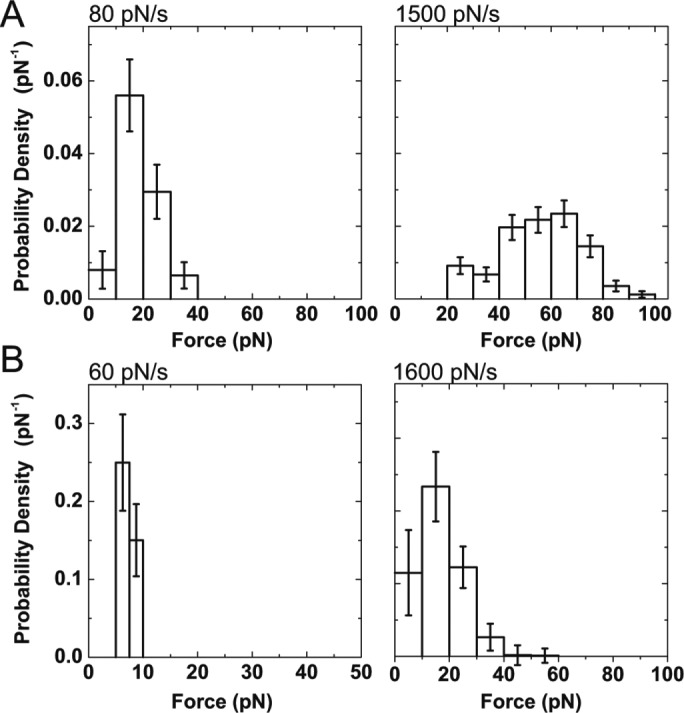
Probability density distributions of forces calculated from ramp-force experiments (Figure 2A) for the dissociation of KIF16B-PX from (A) supported lipid membranes containing 2% PI(3)P at loading rates of 80 ± 20 and 1500 ± 120 pN/s, and (B) B-PI(3)P in the absence of lipid membrane at loading rates of 60 ± 20 and 1600 ± 270 pN/s. Bin widths are 10 pN, except for B (left), which are 2.5 pN. Errors are SDs calculated from bootstrapped data sets.
The average forces at the two loading rates of 80 and 1500 pN/s were 19 and 56 pN, respectively (Table 1). Interestingly, the force for the higher loading rate is substantially larger than the force required to extract a biotinylated DOPC molecule from a lipid bilayer using a similar experimental geometry and loading rate (Pyrpassopoulos et al., 2010; Table 1), suggesting that there is an additional attachment between KIF16B-PX and the membrane besides the one with the headgroup of a single PI(3)P lipid molecule.
TABLE 1:
Adhesion forces from ramp measurements.
| Loading rate (pN/s) | Average force (pN) |
|---|---|
| KIF16B-PX and 2% PI(3)P | |
| 80 ± 20 | 19 ± 7.2 |
| 1500 ± 120 | 56 ± 16 |
| KIF16B-PX and B-PI(3)P | |
| 60 ± 20 | 7.2 ± 1.3 |
| 1600 ± 270 | 16 ± 8.5 |
| Extraction of biotinylated lipid DOPC (Pyrpassopoulos et al., 2010) | |
| 1100 ± 130 | 27 ± 15 |
Errors are SDs.
Structural studies of the KIF16B-PX domain suggest that in addition to the electrostatic interactions between the positively charged residues in the PX domain and the negatively charged headgroup of PI(3)P, the membrane binding of KIF16B-PX is also mediated by the insertion of hydrophobic residues in the PX domain into lipid membranes (Blatner et al., 2007). To determine the contribution of the electrostatic interactions to adhesion in the absence of the hydrophobic insertion, we measured the force required to detach KIF16B-PX from B-PI(3)P, a soluble analogue of PI(3)P that is biotinylated on its acyl chain allowing attachment to streptavidin-coated polystyrene beads in the absence of a lipid bilayer. The frequency distributions of adhesion forces between B-PI(3)P beads and pedestals decorated with KIF16B-PX were determined for loading rates of 60 and 1600 pN/s (Figure 3B). Average forces were 7 and 16 pN, with forces increasing with loading rate. These forces are significantly lower than those observed for adhesion to PI(3)P in lipid bilayers, pointing to the important contribution of the hydrophobic residues that insert into the lipid bilayer (Table 1).
Lifetime of KIF16B-PX attachment to lipid membranes containing 2% PI(3)P under constant force
We used a force feedback system to measure the lifetime of adhesion at constant separating force between KIF16B-PX and 2% PI(3)P in a DOPC bilayer or B-PI(3)P in the absence of a bilayer (Figure 2B). Attachment lifetimes were determined for a range of force values that fell within the distribution of ramp force experiments (11–45 pN; Figure 4). The contributions of nonspecific interactions were removed from the frequency distributions of the adhesion lifetime at each force using similar control experiments as in the ramp of force measurements. The data were normalized with respect to the duration of the bin width and the total number of events to obtain survival probability densities. The final survival probability density distributions were well fit to a single exponential decay function (Eq. 4; Figure 4), resulting in adhesion lifetimes calculated at each force (Figure 4).
FIGURE 4:
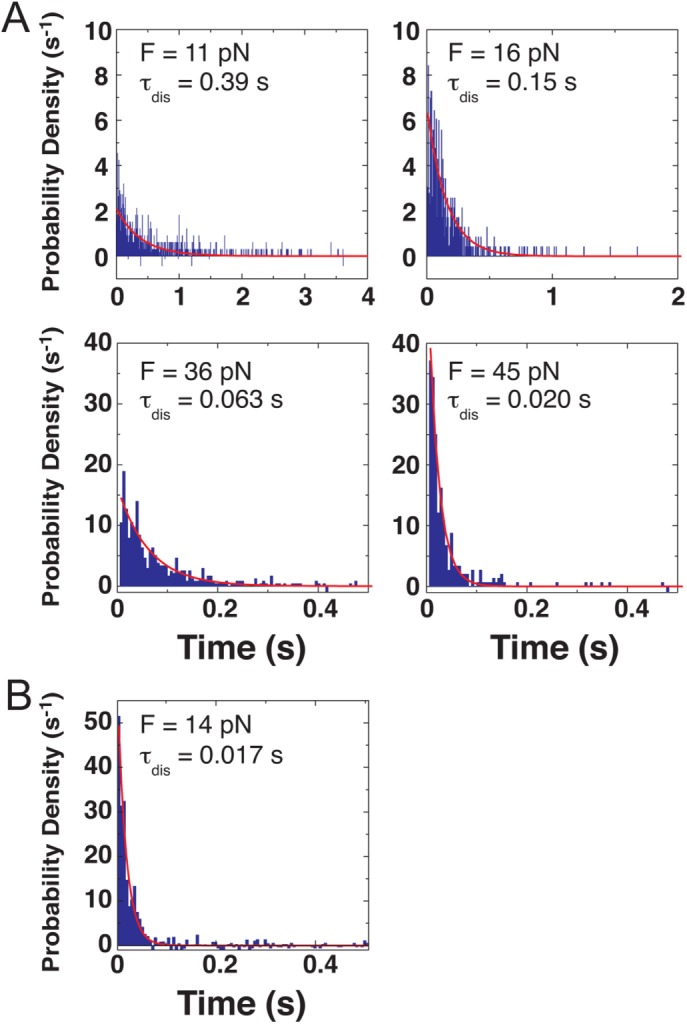
Probability density distributions showing the duration of attachments between KIF16B-PX and (A) 2%PI(3)P in lipid membranes and (B) B-PI(3)P in the absence of lipid membrane under constant tensile forces determined from force-feedback experiments (Figure 2B). Single exponential fits to the data are shown in red.
The lifetime of the bonds between KIF16B-PX and PI(3)P decreased monotonically from 390 ms at 11 pN to 20 ms at 45 pN (Figure 4), indicating slip-bond-like behavior. The lifetime of the interaction of KIF16B-PX with B-PI(3)P at 14 pN (17 ms) is more than 20-fold shorter than with PI(3)P in the presence of membrane, again pointing to the key role of the hydrophobic insertion in membrane adhesion. The very short lifetime under load prevented acquisition at higher forces.
Force dependence of the lifetime of KIF16B-PX attachment to lipid membranes containing 2% PI(3)P
Since the adhesion lifetime under constant separating forces can be described by a single exponential decay (Figure 4), the formalism proposed by Dudko et al. (2008) can be applied to extract the force dependence of the adhesion lifetimes from the probability density distribution of adhesion forces obtained at differed loading rates (Figure 5),
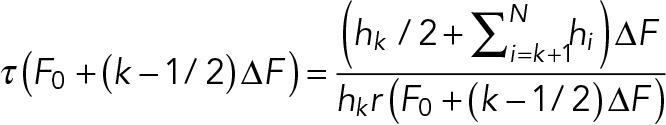 (1) (1)
|
where hi is the probability density (pN–1) for the ith bin, N is the number of bins, r is the loading rate, ΔF the width of the bin (pN), and F0 is the first force value in the histogram of the probability density distribution. Strikingly, the attachment lifetime as a function of force calculated from ramp force distributions follows a similar curve as lifetimes measured directly from the constant force experiments.
FIGURE 5:
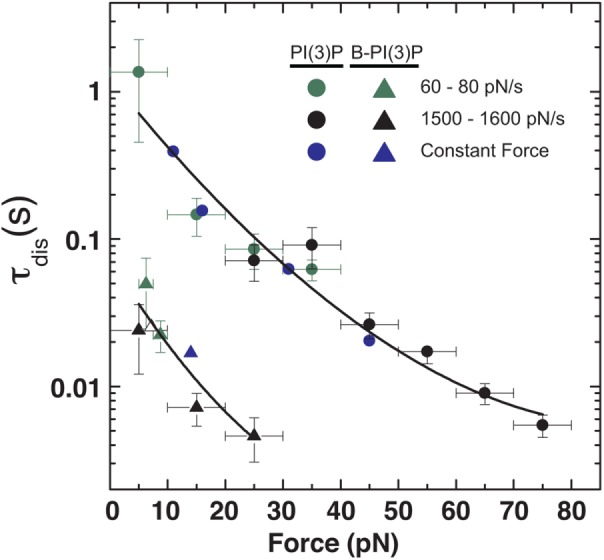
Lifetime of adhesion between KIF16B-PX and (circles) PI(3)P and (triangles) B-PI(3)P as a function of force. Lifetimes of adhesion calculated from (green) 60–80 pN/s and (black) 1500–1600 pN/s ramp-force experiments are shown overlaid with those obtained from (blue) constant-force experiments. Solid lines are least-square fits to Eq. 2, where ν = 1/2. Parameters from the fit are provided in Table 2.
The force dependence of the adhesion lifetime τ can be well fitted by the formula derived from Kramer’s theory for a simple one-dimensional free-energy profile (Dudko et al., 2006, 2008),
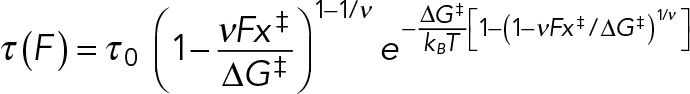 (2) (2)
|
where τ0 is the lifetime in the absence of force, ΔG‡ is the free energy of activation in the absence of force, x‡ is the distance to the transition state, kB is Boltzmann’s constant, T is the absolute temperature in K, and ν is a scaling factor that takes values 1/2 (harmonic well with a cusplike barrier), 2/3 (linear-cubic well), and 1 (Bell’s model). The resulting fits for ν = 1/2 and 2/3 are almost identical (Table 2), and we therefore show the fitting curve for ν = 1/2 (Figure 5). For ν = 1 the model fails to describe the force dependence of the lifetime and in particular the curvature in the logarithmic plot for higher forces (fit not shown).
TABLE 2:
Fitting parameters for force dependence of adhesion lifetime as determined using Eq. 2.
| 2% PI(3)P | B-PI(3)P (no lipid membrane) | |||
|---|---|---|---|---|
| Fitting parameter | ν = 1/2 | ν = 2/3 | ν = 1/2 | ν = 2/3 |
| τ0 (s) | 1.2 ± 0.43 | 1.1 ± 0.34 | 0.073 ± 0.062 | 0.073 ± 0.056 |
| x‡ (nm) | 0.50 ± 0.085 | 0.44 ± 0.062 | 0.66 ± 0.66 | 0.65 ± 0.49 |
| ΔG‡ (kBT) | 7.2 ± 0.65 | 6.5 ± 0.65 | 5.3 ± 5.4 | 4.3 ± 2.6 |
Errors are SEs of the fit.
DISCUSSION
Adhesion of KIF16B-PX to PI(3)P-containing membranes
Our results show that in addition to the interaction of KIF16B-PX with the headgroup of PI(3)P, the lipid membrane itself is essential for the mechanical strength of bond. Indeed, the average adhesion force due to the electrostatic interaction between KIF16B-PX and B-PI(3)P in the absence of lipid membrane is less than half of the adhesion force between the KIF16B-PX and 2% PI(3)P in a DOPC membrane (Table 1). The additional contribution to the mechanical strength of the adhesion is likely due to the hydrophobic insertion of PX residues L1248 and F1249 into the lipid membrane (Blatner et al., 2007). The adhesion force between the PX and PI(3)P is significantly higher than the force required to extract a single phospholipid from the membrane at a similar loading rate, which is made possible by this network of interactions rather than the specific PI(3)P recognition alone.
Interestingly, the average adhesion forces of KIF16B-PX domain to PI(3)P in the absence of lipid membrane are very similar in magnitude to those between the pleckstrin homology (PH) domain-containing tail of Myo1c (an actin-based, membrane-binding molecular motor) and 2% PI(4,5)P2 at corresponding loading rates (Pyrpassopoulos et al., 2010). The PH domains of myosin-I isoforms are not thought to contain residues that penetrate the membrane but rather interact with lipid membranes exclusively via electrostatic interactions (Hokanson et al., 2006; McKenna and Ostap, 2009; Feeser et al., 2010; Patino-Lopez et al., 2010). The above comparison may indicate that the average adhesion forces due to phosphoinositide recognition, regardless of the headgroup specificity, may be of similar mechanical strengths, which are in the low pN range for physiological loading rates.
The adhesion forces between KIF16B-PX and lipid membranes of 100% DOPC were similar to the nonspecific adhesion forces measured for membranes containing 2% PI(3)P when KIF16B-PX was not present in the experimental chamber. Therefore, in our mechanical assay, KIF16B-PX cannot appreciably adhere to DOPC lipid membranes in the absence of PI(3)P. This result agrees with in vitro motility assays that reconstitute KIF16B microtubule-based transport of liposomes that contain 2% PI(3)P and 98% DOPC, but not 100% DOPC (Hoepfner et al., 2005). This result is also consistent with the electrostatic potential calculations, binding measurements, and monolayer penetration assays that suggest PI(3)P is required to enhance the penetration of hydrophobic residues L1248 and F1249 into the membrane (Blatner et al., 2007).
In addition to the magnitude of the PX-membrane adhesion force, the kinetic lifetime of membrane attachment under load is of critical biological importance to a processive cytoskeletal motor anchored to a biological membrane. Our data for the force dependence of the adhesion lifetime between KIF16B-PX and PI(3)P in the presence or absence of lipid membrane (Figure 5) can be well described by the simple model of a single potential well with an energy barrier as described by Eq. 2 (Dudko et al., 2008). The fitting parameters that describe the binding kinetics in the absence of force are insensitive to the shape of the energy barrier (values of the scaling factor ν) and show a 30% increase of the activation energy due to the insertion of hydrophobic residues in to the membrane (Table 2). Comparison between the fitting curves in the presence and absence of lipid membrane shows that for forces of 0–10 pN, the adhesion lifetime between KIF16B-PX domain and PI(3)P in the presence of lipid membrane is 16- to 20-fold higher than without it. This corroborates the enhanced stability provided by additional interactions between KIF16B-PX and the lipid membrane besides the specific recognition of PI(3)P.
Comparison of membrane adhesion to KIF16B motor activity
From the fitting results (Table 2), the lifetime of KIF16B-PX on 2% PI(3)P lipid membranes at zero force is 1.2 s. Assuming that for a dimeric KIF16B motor the adhesion lifetime to PI(3)P membranes doubles due to the attachment of two PX domains, a single processive KIF16B motor could transport an early endosome for a distance of 2.4 um, since its velocity and run length in the absence of resisting loads are 1 μm/s and 10 µm, respectively (Soppina et al., 2014). Indeed, data acquired at a fourfold higher KIF16B-PX concentration than used to achieve single-molecule conditions resulted in two- to ninefold increases in adhesion for 5- to 35-pN loads, respectively (Supplemental Figure 2). If we assume Kif16B has a stall force of ∼4 pN, as for other kinesins (Schroeder et al., 2012), the dissociation time of monomeric KIF16B-PX is 0.8 s, and we would predict a dimeric PX to have an attachment lifetime 1.6 s (Table 2 and Eq. 2). Although the force sensitivity of KIF16B motor is not known, previous studies indicate that the dissociation of kinesin 3 from stable microtubules is highly force sensitive in comparison to other kinesin families (Arpag et al., 2014; Norris et al., 2014). Therefore, recruitment of more than a single KIF16B motor may be required for longer transport distances under resisting load. We note the adhesion lifetime of KIF16B-PX on early endosome may increase by the presence of additional anionic phospholipids such as phosphatidylserine (PS) (Kobayashi et al., 1998) and/or higher local concentrations of PI(3)P modulated by Rab5 (Christoforidis et al., 1999; Gillooly et al., 2000). Overall our results support the view that the direct interaction alone between PI(3)P-containing membranes and KIF16B is sufficient for the function of KIF16B as an early endosome transporter in the cell.
MATERIALS AND METHODS
Proteins and reagents
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) and the 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′-phosphate) form of PI(3)P were purchased from Avanti Polar Lipids (Alabaster, AL). Biotin phosphatidylinositol-3-phosphate (B-PI[3]P), a water-soluble analogue of PI(3)P labeled with biotin at the sn-1 position, was purchased from Echelon Biosciences (Salt Lake City, UT). Silica beads with diameters of 2 μm (9.98% solid) and 5 μm (9.92% solid) were purchased from Polysciences (Warrington, PA). Streptavidin-coated polystyrene beads with diameter of 2.11 μm (0.5% wt/vol) were purchased from Spherotech (Lake Forest, IL). Lyophilized neutravidin and casein were purchased from Sigma-Aldrich (St. Louis, MO).
A construct that contains the PX domain of KIF16B (residues 1179–1317) and N-terminal His6 tag and avitag (Schatz, 1993) sequences was expressed in Escherichia coli. Cells were harvested, lysed in lysis buffer (25 mM Tris, pH 7.5, 20 mM imidazole, 300 mM NaCl, 0.5 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid), 0.5% Igepal, 1 mM-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.01 mg/ml aprotinin, and 0.01 mg/ml leupeptin) by tip sonication for 6 × 10 s with pauses of 15 s in between. Lysed cells were centrifuged, and the supernatant was applied onto a Cobalt resin (Clontech Laboratories, Mountain View, CA) at a flow rate of 1 ml/min. PX domain was eluted with 125 mM imidazole and dialyzed overnight in 10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM EGTA, 1 mM DTT (dithiothreitol). The lysine in the avitag sequence was specifically biotinylated using 20 mg/ml BirA (Avidity, Denver, CO) at 30°C for 1 h. Biotinylated PX domain was dialyzed overnight against HNa100 (10 mM HEPES, pH 7, 100 mM NaCl, 1 mM EGTA, and 1 mM DTT), aliquoted, and stored at –80°C.
PIP-strip assay
PIP-strips spotted with 15 different lipids at 100 pmol per spot were purchased from Echelon Biosciences (Salt Lake City, UT) and were used to confirm the successful biotinylation of KIF16B-PX domain as well as its specificity for PI(3)P (Figure 1A). The assay was performed at room temperature in the sequence of the following steps: a) membrane was blocked with 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) at room temperature for 1 h, b) incubated with 7.9 μg/ml biotinylated KIF16B-PX in 3% BSA PBS for 1 h, c) washed three times for 5 min with 0.1% Tween PBS, d) incubated with diluted 1:4666 HRP-streptavidin (Thermo Fisher) in 3% BSA PBS for 1 h, e) washed three times for 5 min with 0.1% Tween PBS, f) incubated with 8 ml of chemiluminescent substrate solution for HRP (Thermo Fischer) for 10 min, and then imaged using G:BOX (Syngene, Frederick, MD).
Preparation of small unilamellar vesicles
Small unilamellar vesicles (SUVs) were prepared as follows: 98% DOPC was mixed with 2 mol % of PI(3)P in a 50-ml round-bottom flask. The solution was thermally equilibrated in a water bath at ∼35°C for 5 min and dried rapidly (∼1 min) in a rotary evaporator. The lipid film was kept under hard vacuum for at least 30 min. Multilamellar vesicles were formed by vortexing the flask for 2 min at maximum speed after adding 2 ml of HNa100 (0.5 mM total lipid concentration). The lipid mixture was subjected to four freeze-thawing cycles using an isopropanol/dry ice mix and a water bath at 37°C. SUVs were formed by extruding the lipid solution at least 11 times through polycarbonate membranes with pores of 30 nm in diameter using a mini extruder (Avanti Polar Lipids).
Preparation of lipid-coated beads
Lipid-coated beads were prepared as described previously (Pyrpassopoulos et al., 2010, 2013). Briefly, 2.0-μm-diameter silica beads (40 μl) were washed as follows: 1 ml methanol, 1 ml 1N KOH (water bath sonicated for 15 min), 7 × 1 ml DI H2O. All washes were done at room temperature (RT), 735 × g for 2 min, using a benchtop centrifuge. After the last wash the final volume was 40 μl and beads were mixed with ∼600 μl of SUVs containing either 2% PI(3)P and 98% DOPC or 100% DOPC. The mixture was vortexed at maximum speed for 1 min and incubated at RT overnight. Finally, beads were washed three times with 1 ml of HNa100 using a benchtop centrifuge (735 × g, 2 min, RT) at a final volume of ∼250 μl in buffer HNa100. Lipid-coated beads were kept at RT and used within a day.
Preparation of beads coated with biotinylated PI(3)P
Streptavidin-coated polystyrene beads (8 μl), 2.11 μm in diameter, were mixed with 1 μl of 10 mg/ml casein and 0.48 μM biotinylated PI(3)P (1 μl) and kept on ice at least 1 h before experiments. Freshly prepared beads were used within a day.
Ramp-force measurements
Nitrocellulose-coated chambers containing 5-μm-diameter silica pedestals were prepared as described previously (Pyrpassopoulos et al., 2010, 2013). Chamber volumes were ∼20 μl. All proteins and reagents were prepared in HNa100. Solutions were added to the chamber in the following sequence: 1) 0.01 mg/ml neutravidin for 5 min, 2) 1 mg/ml casein as a nitrocellulose blocking agent for 5 min, 3) 0.1–1 nM biotinylated KIF16B-PX domain, 1 mg/ml casein for 5 min, and 4) three chamber volumes 1 mg/ml casein.
Lipid-coated beads or beads with B-PI(3)P were diluted 1:10 in HNa100 that contained 1 mg/ml casein, and 4–6 μl was injected to one side of the chamber. The chamber was sealed with silicon vacuum grease (Dow Corning, Midland, MI). Chambers were used within 60 min of preparation. For control experiments, either a) step 3 of the chamber preparation was skipped or b) beads coated with only 100% DOPC or without B-PI(3)P were introduced into the chamber at the final step.
Optical trap instrumentation was as described but with a water instead of oil objective (Takagi et al., 2006; Pyrpassopoulos et al., 2010). The trap stiffness and the force calibration coefficient were determined from the power spectrum of the thermal motion of a trapped bead. Custom LabVIEW software routines (National Instruments, Austin, TX) were used for data collection and data analysis.
Adhesion forces were determined by performing ramp-force measurements as described (Pyrpassopoulos et al., 2010, 2013). Briefly, lipid-coated beads were compressed against immobilized pedestals as the trap position was oscillated in a triangular waveform. Formation of bonds between 2% PI(3)P lipid-coated beads and KIF16B-PX and their subsequent rupture on retraction appeared as negative peaks in the traces, as can be seen in the data trace in Figure 2A. Data were digitized with a 2-kHz sampling rate, amplified, and filtered at 1 kHz. The nominal rate at which load was exerted on KIF16B-PX domain attachments (loading rate) was set by controlling the frequency and amplitude of the triangular oscillation and the trap stiffness. The trap stiffness in ramp force experiments was 0.1–0.2 pN/nm. The oscillation amplitude was ∼0.44–1.5 μm with frequencies between 0.1 and 2 Hz. For each loading rate, data were collected using 3–10 different beads against at least four different pedestals per bead for no more than 2 min at each pedestal. The number of total oscillation cycles for each loading rate ranged from 217 to 5295. Experimental loading rates (60–1600 pN/s) were selected to fulfill two criteria: 1) to match the effective loading rate of single kinesin motors walking against a stationary optical trap (Visscher et al., 1999; Carter and Cross, 2005) and 2) to obtain a wide range of forces to allow the calculation of the force dependence of adhesion lifetime (Evans and Ritchie, 1997; Dudko et al., 2006, 2008).
Ramp-force data collection using lipid-coated beads
Ramp-force control experiments that omitted either KIF16B-PX from the pedestals or PI(3)P from the lipid-coated beads were performed to assess the frequency and magnitude of nonspecific interactions. Attachments between the oscillating lipid-coated beads and the spherical pedestals were rare and resulted in adhesion forces of less than 20 pN, which are substantially lower than when both KIF16B-PX and PI(3)P were present (Supplemental Figure 1). Single molecule conditions were achieved by titrating the KIF16B-PX concentration to ensure that specific interactions were no more than 10% above the background of nonspecific interactions. Adhesion forces were determined for loading rates of 80 and 1500 pN/s.
Experiments performed at a loading rate of 80 pN/s required acquisition at low oscillation frequencies (0.1 Hz), which was time-consuming. Therefore, titrations to achieve single-molecule conditions were performed at 1500 pN/s. Once a region of the pedestal with a specific interaction was identified (>20 pN), the loading rate was decreased to 80 pN/s. Notably, stage displacements as small as ∼30 nm along the contact surface between the pedestal and the lipid-coated bead were sufficient to move away from the specific interactions, which is indicative of the low surface density of KIF16B-PX on the pedestals. Confirmation of single-molecule conditions is supported by the finding that the force dependence of the KIF16B-PX–PI(3)P lifetime calculated from the two loading rates follow the same curve (see below). A data set acquired at a slower pulling rate (20 pN/s) and with fourfold higher KIF16B-PX concentration than used to achieve single-molecule conditions resulted in increased adhesion times and deviation from this curve (Supplemental Figure 2).
Ramp-force data collection using streptavidin-coated polystyrene beads:
Ramp-force control experiments were performed to assess the nonspecific interaction between pedestal beads and the polystyrene beads used as a vehicle for B-PI(3)P. Omitting B-PI(3)P from polystyrene beads or KIF16B-PX from the pedestals revealed more nonspecific interactions than with lipid beads (Supplemental Figure 1). To achieve single molecule densities for the reported experiments, the concentration of KIF16B-PX was titrated such that specific interactions between B-PI(3)P polystyrene beads and KIF16B-PX-decorated pedestals was no more than 10% above the background of nonspecific interactions. Adhesion forces were determined for loading rates of 60 and 1600 pN/s.
Ramp-force data analysis
Compliance in the lipid membrane attached to the bead resulted in variability of the loading rate. To ensure accurate reporting, the loading rates in all data sets were determined for each cycle by the slope of linear fits of the segment between the point of maximum compressive force and the point where the force becomes zero on retraction (Figure 2A). Only rupture events that had experimental loading rates within 30% of the defined rates were selected for analysis. The reported loading rates and their errors (Figure 3 and Table 1) correspond to the average slope and the SD for all the interactions in each data set.
We calculated the distribution for the frequency of interactions per oscillation cycle for the experimental and control data over the range of forces observed (Supplemental Figure 1) by measuring the number of rupture events falling within a given force bin and dividing by the total number of contact cycles. The units (frequency/contact cycle) allow us to calculate and confirm that the specific interaction events are sparse and do not contribute more than 10% above the background of nonspecific interactions. Errors in the frequency distributions of interactions per oscillation cycle were calculated by the bootstrap method (1000 calculated data sets) (Press et al., 2002). Assuming that the probability of specific and nonspecific interactions occurring simultaneously is negligible, the frequency distributions of the nonspecific interactions were subtracted from the corresponding frequency distributions that contained the specific interactions in each case. After subtraction of the nonspecific interactions, the resulting distributions were renormalized with respect to the total number of events instead of contact cycles and the size of the force bin to give the probability density (pN–1), such that
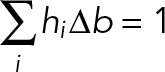 (3) (3)
|
where hi is the probability density (pN–1), Δb the size of the bin (pN), and the product hi Δb is the probability of the bond rupturing at a force value that falls within the ith force bin (Figure 3). The probability density distributions were then used to calculate from Eq. 1 the force dependence of the adhesion lifetime between KIF16B-PX and PI(3)P or B-PI(3)P (Figure 5).
Constant force measurements
Attachment durations under constant tensile force between pedestal-immobilized KIF16B-PX domain and PI(3)P on the lipid-coated beads or B-PI(3)P on streptavidin-coated polystyrene beads were measured using a force-clamp (Pyrpassopoulos et al., 2010, 2013). Briefly, lipid-coated beads were compressed against immobilized pedestals as the trap position was oscillated in a square waveform (Figure 2B). The desired compressive and separating force were set by the amplitudes of the square pulse, with the negative one corresponding to compression of the trapped bead against the pedestal and the positive one to retraction from the pedestal as can be seen in the data trace in Figure 2B. Data points were recorded every 0.5 ms for forces in the range 11–45 pN. For each value of pulling force, data were collected using three to eight different beads against at least four different pedestals per bead and no more than 2 min at each pedestal. The number of total oscillation cycles for each force was between 1330 and 3782. The laser trap stiffness was 0.1–0.2 pN/nm. The KIF16B-PX concentration was titrated to achieve single-molecule conditions as above. Data for nonspecific interactions were collected in the absence of KIF16B-PX or PI(3)P or B-PI(3)P in the experimental chamber. Similarly as in the ramp of force data analysis, the frequency distributions of attachment’s durations per contact cycle for specific and nonspecific interactions were calculated separately. After subtraction of the nonspecific interactions, the distributions of attachments’ duration for every force were renormalized with respect to the total number of events instead of contact cycles and the time width of the bin to give the probability density (s–1) so that Eq. 3 is satisfied.
In this case, hi is the probability density (s−1), Δb the size of the bin (s), and the product hi Δb the probability that the duration of the bond under constant force falls within the ith time bin (Figure 4). Dissociation rates were determined by fitting the probability density distributions to the exponential probability density distribution:
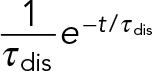 (4) (4)
|
Supplementary Material
Acknowledgments
The KIF16B full-length construct was a generous gift of Kristen Verhey (University of Michigan). We thank Tianming Lin for his technical assistance in developing the KIF16B-PX constructs. This work was supported by National Institutes of Health grant GM087253 to E.M.O. and H.S.
Abbreviations used:
- B-PI(3)P
biotin phosphatidylinositol-3-phosphate
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- KIF16B-PX
biotinylated PX domain construct from KIF16B
- PH
pleckstrin homology
- PI(3)P
phosphatidylinositol-3-phosphate PI(3)P
- PI(4,5)P2
phosphatidylinositol-4,5-bisphosphate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-05-0324) on September 20, 2017.
REFERENCES
- Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpag G, Shastry S, Hancock WO, Tuzel E. Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys J. 2014;107:1896–1904. doi: 10.1016/j.bpj.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatner NR, Wilson MI, Lei C, Hong W, Murray D, Williams RL, Cho W. The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 2007;26:3709–3719. doi: 10.1038/sj.emboj.7601800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci USA. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeser EA, Ignacio CM, Krendel M, Ostap EM. Myo1e binds anionic phospholipids with high affinity. Biochemistry. 2010;49:9353–9360. doi: 10.1021/bi1012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R, Fischer J, Schwarz FW, Walter WJ, Schwille P, Diez S. Transport efficiency of membrane-anchored kinesin-1 motors depends on motor density and diffusivity. Proc Natl Acad Sci USA. 2016;113:E7185–E7193. doi: 10.1073/pnas.1611398113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative Pleckstrin homology domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Ostap EM. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci USA. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Holleran EA, Vale RD. Kinesin motors and microtubule-based organelle transport in Dictyostelium discoideum. J Muscle Res Cell Motil. 2002;23:631–638. doi: 10.1023/a:1024403006680. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Manceva S, Lin T, Pham H, Lewis JH, Goldman YE, Ostap EM. Calcium regulation of calmodulin binding to and dissociation from the myo1c regulatory domain. Biochemistry. 2007;46:11718–11726. doi: 10.1021/bi700894h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna JM, Ostap EM. Kinetics of the interaction of myo1c with phosphoinositides. J Biol Chem. 2009;284:28650–28659. doi: 10.1074/jbc.M109.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SR, Trybus KM, Warshaw DM. Motor coupling through lipid membranes enhances transport velocities for ensembles of myosin Va. Proc Natl Acad Sci USA. 2014;111:E3986–E3995. doi: 10.1073/pnas.1406535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Soppina V, Dizaji AS, Schimert KI, Sept D, Cai D, Sivaramakrishnan S, Verhey KJ. A method for multiprotein assembly in cells reveals independent action of kinesins in complex. J Cell Biol. 2014;207:393–406. doi: 10.1083/jcb.201407086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino-Lopez G, Aravind L, Dong X, Kruhlak MJ, Ostap EM, Shaw S. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH) J Biol Chem. 2010;285:8675–8686. doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picas L, Gaits-Iacovoni F, Goud B. The emerging role of phosphoinositide clustering in intracellular trafficking and signal transduction. F1000Res. 2016;5 doi: 10.12688/f1000research.7537.1. doi: 10.12688/f1000research.7537.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Revipes in C++ New York: Cambridge University Press; 2002. [Google Scholar]
- Pyrpassopoulos S, Arpag G, Feeser EA, Shuman H, Tuzel E, Ostap EM. Force generation by membrane-associated Myosin-I. Sci Rep. 2016;6:25524. doi: 10.1038/srep25524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpassopoulos S, Feeser EA, Mazerik JN, Tyska MJ, Ostap EM. Membrane-bound myo1c powers asymmetric motility of actin filaments. Curr Biol. 2012;22:1688–1692. doi: 10.1016/j.cub.2012.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpassopoulos S, Shuman H, Ostap EM. Single-molecule adhesion forces and attachment lifetimes of myosin-I phosphoinositide interactions. Biophys J. 2010;99:3916–3922. doi: 10.1016/j.bpj.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpassopoulos S, Shuman H, Ostap EM. Method for measuring single-molecule adhesion forces and attachment lifetimes of protein-membrane interactions. Methods Mol Biol. 2013;1046:389–403. doi: 10.1007/978-1-62703-538-5_24. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Nat Biotechnol. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, 3rd, Hendricks AG, Ikeda K, Shuman H, Rodionov V, Ikebe M, Goldman YE, Holzbaur EL. Force-dependent detachment of kinesin-2 biases track switching at cytoskeletal filament intersections. Biophys J. 2012;103:48–58. doi: 10.1016/j.bpj.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, Verhey KJ. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich G, Chibalina MV, Au JS, Arden SD, Buss F, Kendrick-Jones J. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- Zot HG, Doberstein SK, Pollard TD. Myosin-I moves actin filaments on a phospholipid substrate: implications for membrane targeting. J Cell Biol. 1992;116:367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.