Tissue profiles and MSC transcriptomics separately indicate that LBR varies inversely with lamin-A,C. Such anti-correlations are recapitulated in MSC adipogenesis and osteogenesis as well as with matrix elasticity, knockdowns, overexpression, and myosin-II inhibition, which together suggest the competitive binding of lamin-A,C and LBR for lamin-B.
Abstract
Matrix stiffness that is sensed by a cell or measured by a purely physical probe reflects the intrinsic elasticity of the matrix and also how thick or thin the matrix is. Here, mesenchymal stem cells (MSCs) and their nuclei spread in response to thickness-corrected matrix microelasticity, with increases in nuclear tension and nuclear stiffness resulting from increases in myosin-II and lamin-A,C. Linearity between the widely varying projected area of a cell and its nucleus across many matrices, timescales, and myosin-II activity levels indicates a constant ratio of nucleus-to-cell volume, despite MSCs’ lineage plasticity. Nuclear envelope fluctuations are suppressed on the stiffest matrices, and fluctuation spectra reveal a high nuclear tension that matches trends from traction force microscopy and from increased lamin-A,C. Transcriptomes of many diverse tissues and MSCs further show that lamin-A,C’s increase with tissue or matrix stiffness anti-correlates with lamin-B receptor (LBR), which contributes to lipid/sterol biosynthesis. Adipogenesis (a soft lineage) indeed increases LBR:lamin-A,C protein stoichiometry in MSCs versus osteogenesis (stiff). The two factors compete for lamin-B in response to matrix elasticity, knockdown, myosin-II inhibition, and even constricted migration that disrupts and segregates lamins in situ. Matrix stiffness-driven contractility thus tenses the nucleus to favor lamin-A,C accumulation and suppress soft tissue phenotypes.
INTRODUCTION
As a cell adheres and pulls on its surroundings (Nicolas et al., 2004; Discher et al., 2005; Sawada et al., 2006; Levental et al., 2009; Gardel et al., 2010; Kassianidou et al., 2017), the shape of the nucleus changes in a manner similar to the shape of the cell—as quantified long ago (Weiss and Garber, 1952). The stiffness of a cultured cell’s microenvironment is now understood to drive the spreading and flattening of most types of cells (Pelham and Wang, 1997) and their nuclei (Swift et al., 2013). Such studies have typically used gel matrices that are thick relative to the size of a cell. However, thin matrices present a means to maintain constant gel chemistry and surface density of adhesion ligand as a function of matrix thickness, h (Engler et al., 2006; Hadden et al., 2017), with early studies showing that soft gels on top of rigid substrates can drive spreading when minimal polymer is used (Ben-Ze’ev et al., 1980). Thin layers of extracellular matrix are also evident in some tissues, including the collagenous “osteoid” film on top of calcified bone, which promotes osteogenesis (Sodek and McKee, 2000). Although studies of the interplay between nucleus and cytoskeleton in cells cultured on matrices of controlled elasticity and/or thickness remain limited (Buxboim et al., 2014; Alam et al., 2015), plating of cells on surfaces with different mechanical properties has been well studied in terms of the effects on the acto-myosin cytoskeleton (Beningo et al., 2004; Engler et al., 2004b; Prager-Khoutorsky et al., 2011; Talele et al., 2015; Meacci et al., 2016; Kassianidou et al., 2017). Tractions generated by the cytoskeleton propagate into any flexible matrix, and the resistance sensed by a cell can include an underlying hidden rigidity—like the proverbial “princess and the pea.” However, many quantitative aspects of mechanical depth-sensing remain obscure, particularly nuclear phenotypes that include the nuclear-to-cell volume ratio (Kume et al., 2017). Such features can be complicated for stem cells including the mesenchymal stem cells (MSCs) studied here because phenotype-defining changes in gene expression occur in differentiation.
The nuclear envelope is composed of the nuclear lamina proteins plus many more factors that attach the lamina to the nuclear membrane or link it to the cytoskeleton (Goldman et al., 2002; Gruenbaum et al., 2005). Akin to keratins of nails or hair, lamins are nuclear intermediate filament proteins that assemble into ~300–600 nm multimers (Turgay et al., 2017) in juxtaposed meshworks of A-type versus B-type lamins (Shimi et al., 2008). Across tissues, lamin-A,C varies widely but increases systematically with tissue stiffness E, and the in vitro levels of lamin-A,C have also been found to be matrix-mechanosensitive in MSCs and other cell types (Swift et al., 2013). The roundworm Caenorhabditis elegans likewise shows less lamin in neuronal tissue (soft) compared to muscle tissue (stiff) (Zuela et al., 2016). Interphase phosphorylation of lamin-A,C (Kochin et al., 2014) favors its turnover in cells on soft matrix (Buxboim et al., 2014). In comparison, B-type lamins differ minimally across solid tissues and do not exhibit much interphase phosphorylation. Lamin knockout mice generate all embryonic lineages (Kim et al., 2013), but lamin-A and -C nulls die within weeks of birth with defects only in stiff tissues, including contractile heart and skeletal muscle (Sullivan et al., 1999; Kubben et al., 2011). Key relationships between lamins, other nuclear envelope proteins, and the acto-myosin cytoskeleton are thus emerging, but there is still a need to understand the effective stiffness and/or thickness of matrix in relation to cytosketelal tension, nuclear stress, and tissue-relevant differentiation. Here we estimate the nuclear-to-cell volume ratio and changes in nuclear envelope tension as MSCs spread on gels of varied stiffness and thickness, and then we place the in vitro findings in a tissue context that takes advantage of MSC lineage plasticity. Our findings help identify lamin-B receptor (LBR), which contributes to lipid/sterol synthesis (Subramanian et al., 2012), as an additional matrix-mechanosensor in the nucleus that is downstream from nuclear tension and lamin-A,C.
RESULTS
Cells sense rigidity that is buried in soft matrices
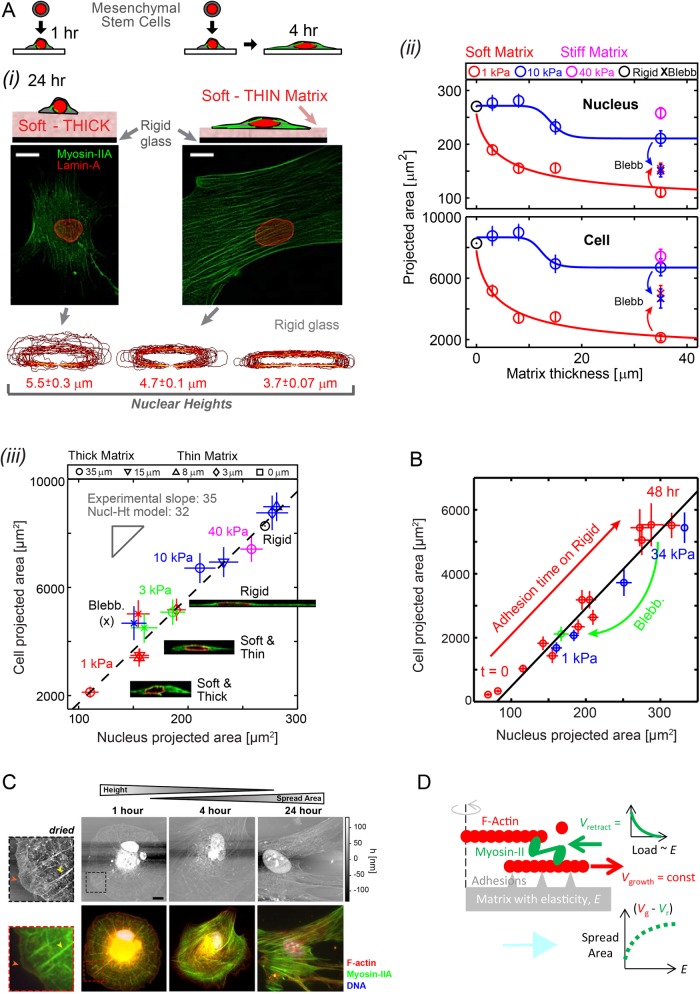
Sparse cultures of MSCs on soft-and-thin gels (1 kPa, 3–15 µm; Buxboim et al., 2010b) that are collagen-functionalized show greater cell spreading and more prominent stress fibers of myosin-II than cells cultured on soft-and-thick matrices (>35 µm) made in parallel (Figure 1Ai). Stiff matrices (e.g., 40 kPa) promote both spreading (Pelham and Wang, 1997; Engler et al., 2004a) and stress fibers (Yeung et al., 2005); MSCs clearly feel through thin gels to the rigid glass beneath. Even with matrices of intermediate stiffness (10 kPa), differences are evident between thin and thick matrices of the same stiffness E (Figure 1Aii). Nuclei exhibit the same trends in spreading as cells, consistent with past observations of morphologies on diverse matrices (Weiss and Garber, 1952). Simple hyperbolic models (Zemel et al., 2010) typically fit cell spreading as well as nuclear spreading (Supplemental Figure S1) in the coupled processes of protrusion, matrix binding, and myosin-II–driven contraction resisted (or not) by matrix. Such spreading profiles reveal tactile length scales (ht) for both cells and nuclei that systematically decrease from ∼25 µm for soft matrices to ∼0 µm for very stiff matrices.
FIGURE 1:
Cells feel many microns into soft microenvironments. (A) (i) Mesenchymal stem cells (MSCs) and their nuclei are spread more on gels that are soft-and-thin (1 kPa; 2–3 μm) compared to soft-and-thick (∼35 μm) at 24–36 h in culture (scale bars = 10 μm). Bottom, x-z contours of lamina from confocal stacks of immunostained lamin-A,C. Nuclear height (average ± SEM; n > 25) is maximal on thick and soft gels but nuclei become increasingly flattened on thin-and-soft gels (p = 0.006) and rigid glass (p < 0.001). (ii) Mean projected areas of nuclei and cells vs. matrix thickness. Hill function exponents are α = 0.8 and 15 for 1 and 10 kPa gels, respectively. Tactile length scales are defined as the thickness below which cells or nuclei spread more than a measurable 10% relative to cells on thick gels of the same E: ht ≈ 25 μm for 1 kPa, ht ≈ 15 μm for 10 kPa, and ht ≈ 0 μm for 40 kPa, making the latter indistinguishable from collagen-coated glass (i.e., “rigid”). Blebbistatin (Blebb) inhibits myosin-II and eliminates spreading differences on different matrices. (iii) Linearity of cell vs. nucleus projected area is maintained across matrices of different elasticities and thicknesses and is also satisfied by Blebb-treated myosin-inhibited cells. Inset images of x-z cross sections show spread cell height is constrained by nuclear height. (B) Cell vs. nuclear spreading kinetics on rigid glass (red) tracks the “steady-state” projected area of cells on diverse gels (blue) or with myosin-II inhibition by Blebb (green). (C) The dynamics of cell adhesion and spreading were interrogated by AFM (top) and immunostaining (bottom) to show organization of protein of interests with z-axis topography. MSCs were cultured in a standard cell-culture plastic plate and fixed at times ranging between 1 h and 1 d. To facilitate AFM imaging, cells were lipid-membrane stripped and air dried before immunostaining. With adhesion time, cells increasingly spread, and nuclei stretch and flatten down against the substrate and prominent stress fibers developed, consistent with increased generation of contractile forces. At 1 h, cells maintain a spherical morphology and stress fibers are radially distributed (yellow arrowheads, zoom-in images) with a wide lamellipodium structure (red arrowheads) but after 4 h of adhesion symmetry is broken and within 24 h cells obtain a typical mesenchymal-like morphology typical of stiff matrices. Unlike the immunofluorescence (IF) images, AFM micrographs are in scale (scale bar: 10 µm). z-Axis heights measured by AFM is an underestimate, due to fixing, membrane-stripping, and dehydration of the cells. (D) A simple Hill-type model of cell spreading in which myosin-II rectifies F-actin polymerization depending on the resistance provided by matrix elasticity.
Collecting all morphology results for all matrices together shows that a linear relationship between projected areas of cells and nuclei (Figure 1Aiii) is maintained, even when sensitivity to matrix elasticity is inhibited by relaxing cells with the myosin-II inhibitor, blebbistatin (blebb). An invariant ratio of cell-to-nuclear projected areas is a key biophysical parameter that is independent of microenvironment even for high passage MSCs that spread more, express distinct markers of differentiation such as α-smooth muscle actin (Dingal et al., 2015; Talele et al., 2015), and eventually become senescent (Supplemental Figure S1). As a function of adhesion time on rigid substrates, cell and nuclear spreading achieve within ~1 h the same proportionality as the steady-state morphologies on gels of different stiffness (Figure 1B and Supplemental Figure S2). Rigid substrates allow for nanoresolution imaging by atomic force microscopy (AFM) of dried samples that show the expected round cell morphology that breaks symmetry with spreading (Yam et al., 2007; Prager-Khoutorsky et al., 2011; Wolfenson et al., 2014). Consistent with such past studies, the accompanying fluorescence images for actin and myosin-IIA confirms the leading edge polymerization of the dense actin meshwork with myosin-II further back that assembles into both radial and circumferential stress fibers (Figure 1C). A muscle-inspired, Hill-type model of cell spreading is thus suggested: the steady state in retrograde actin flow is tuned by myosin-II minifilaments pulling F-actin back quickly (on soft matrix) or slowly (on stiff matrix), such that the load or resistance is provided by matrix elasticity (Figure 1D).
A systematic flattening of the spread nucleus on stiffer matrices is evident in confocal z-stacks of hydrated samples (Figure 1Ai, bottom). For all substrates the bottom of the nucleus is nearly in contact with the underlying substrate (Figure 1Aiii, inset images), and the top of the nucleus is near the maximum height of the cell. These observations suggest that the height of the nucleus can limit the spreading of the cell. This idea is mathematically formulated in a nuclear height model of spreading that yields an invariant ratio of cell-to-nuclear volume (see Materials and Methods: Nucl-Ht Model). Cell-to-nuclear volume ratio is a long-recognized characteristic of a given cell type (Kume et al., 2017) and is, for example, small with embryonic stem cells. However, the MSC results here further indicate that this key biophysical characteristic is independent of both time and matrix elasticity as well as senescence and the associated differentiation, although other lineages need to be studied further. Factors that control nuclear height or volume, including both transport factors (Kume et al., 2017) and osmolarity (Irianto et al., 2013), will thus regulate spreading of both the cell and its nucleus. Based on the matrix-desensitizing effects of blebbistatin (Figure 1), such factors certainly also include the stress applied by the cytoskeleton on the nucleus.
Cells sense an apparent microelasticity that accounts for geometry
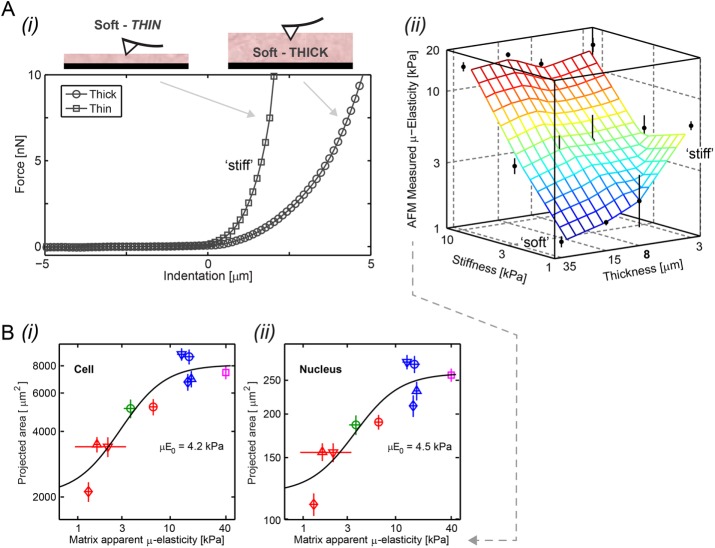
A soft-and-thin gel is clearly equivalent to a stiff-and-thick gel of high stiffness in terms of cell and nuclear morphology. Physical probing of a soft-and-thin gel with the tip of an atomic force microscope indeed yields a high value for the “apparent” microelasticity (µ-elasticity; Figure 2A). Remarkably, all of the data for cell and nuclear projected areas versus the apparent µ-elasticity measured by AFM fitted a “universal” Hill-type curve with half-max (µE0) between soft and stiff of ∼4 kPa (Figure 2B).
FIGURE 2:
The apparent microelasticity sensed by a cell on thin or thick gels is similar in trend to that measured by an inanimate probe. (A)(i) AFM force-indentation representative curves show a sharp deflection of the cantilever on gels that are soft (1 kPa) but thin. Gel contact points are at the origin. (ii) Apparent microelasticity (μ-elasticity) of polyacrylamide gels, as measured by AFM nanoindentation, is plotted as a function of the elastic modulus (bulk stiffness calibrated by desktop rheometer) and thickness. While AFM nanoindentation of thick gels shows agreement with gels’ elastic moduli, μ-elasticity increases with decreasing gel height, exhibiting effective stiffening at the micron scale most profoundly for soft gels. (B) As the spreading of cells (i) and the projected area of nuclei (ii) are distinctively shown to increase with gels’ stiffness and thinness (Figure 1Aii and Supplemental Figure S1B), we plot them here as a function of the apparent matrix μ-elasticity. Cell and nucleus data (average ± SEM, n > 25 cells) are collectively fit to 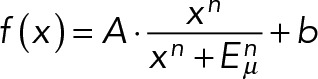 , with an exponent of cooperativity n = 0.5. The transition between soft and stiff matrices is set by Eµ, amounting to 4–4.5 kPa, thus discriminating between compliant tissues such as brain, marrow, and fat and stiffer tissues such as muscle, cartilage, and bone.
, with an exponent of cooperativity n = 0.5. The transition between soft and stiff matrices is set by Eµ, amounting to 4–4.5 kPa, thus discriminating between compliant tissues such as brain, marrow, and fat and stiffer tissues such as muscle, cartilage, and bone.
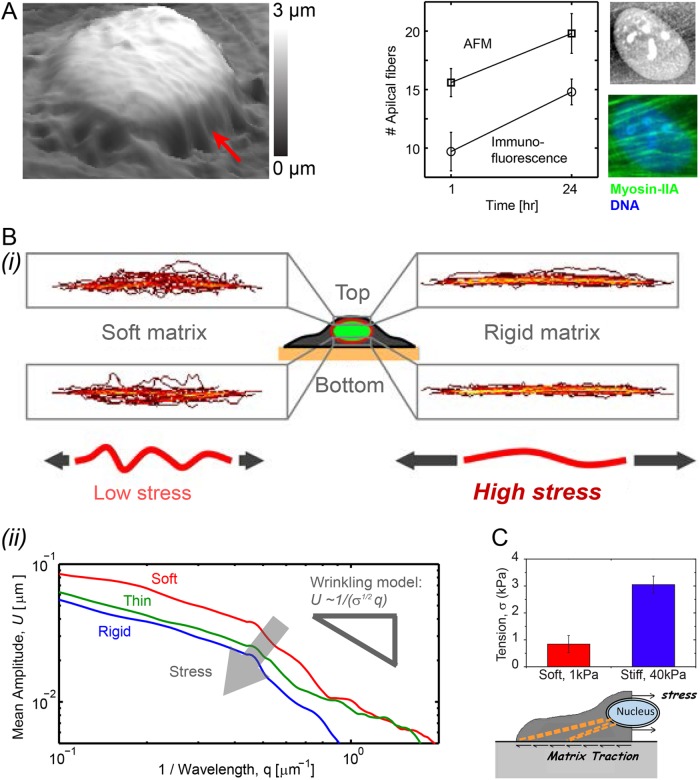
Compared to soft-and-thick matrices, MSCs on soft-and-thin or stiff matrices produce high contractile forces, with a progressive increase in apical stress fibers above the nucleus, that stretch and squash the otherwise wrinkled nucleus against the matrix. Nuclear height measurements of ∼3 µm by AFM (Figure 3A) are consistent with confocal imaging (Figure 1A). Fourier spectra of quenched nuclear fluctuations or roughness, top-and-bottom averaged per cell and smoothed across many nuclei shows that the mean amplitude of the wrinkles (U) decays with wave number q (Figure 3B). The scaling (∼1/q) is reminiscent of the tension-suppressed undulations on lipid membranes (Sackmann, 1994). According to such a model, the approximate twofold suppression of wrinkle amplitude from soft matrix to rigid substrate corresponds to an approximate fourfold increase in nuclear tension or stress, σ. Traction force microscopy applied to MSCs shows that the cell tensions in the midplane of the cell near the nucleus likewise increase approximately fourfold from similarly soft to stiff substrates (Figure 3C; Engler et al., 2006). Nuclei of cells on soft-and-thin substrates are thus under intermediate tension. Sustaining such nuclear tension is the expected role for the main structural proteins of the nucleus, the lamins, but evidence of this was sought from expression profiles of cells on various soft matrices and in diverse tissues.
FIGURE 3:
Nuclear roughness is suppressed by matrix stiffness, yielding an estimate of increased nuclear tension. (A) Nanoscopic imaging by atomic force microscopy (AFM) of a membrane-stripped but hydrated MSC shows apical stress fibers that flatten the nucleus against the matrix (red arrow). Nucleus-draped fibers quantified by AFM and by immunofluorescence of myosin-IIA increase in number with adhesion time. (Bi) Lamin-A,C immunostained MSCs imaged by confocal fluorescence microscopy have apical and basal contours of the nuclear envelope that exhibit larger wrinkles on soft gels (0.3 kPa) than on rigid substrates (average ± SEM, n > 25 cells). (ii) The amplitude of nuclear wrinkles is quantified by Fourier-transformed spectra U(q) vs. spatial wave number q. Averaged across apical and basal profiles of all nuclei and smoothed, U(q) decreases as ∼1/q with a prefactor related to nuclear stress σ based on wrinkled membrane theory (i.e., 1/σ1/2). (C) Traction force microscopy (Engler et al., 2006) indicates an increase in nuclear stress σ in cells on soft gels vs. stiff gels (average ± SEM, n > 10 cells) that is similar to that estimated from wrinkled membrane theory.
Mechanosensitive nuclear envelope: four genes in vitro and in vivo
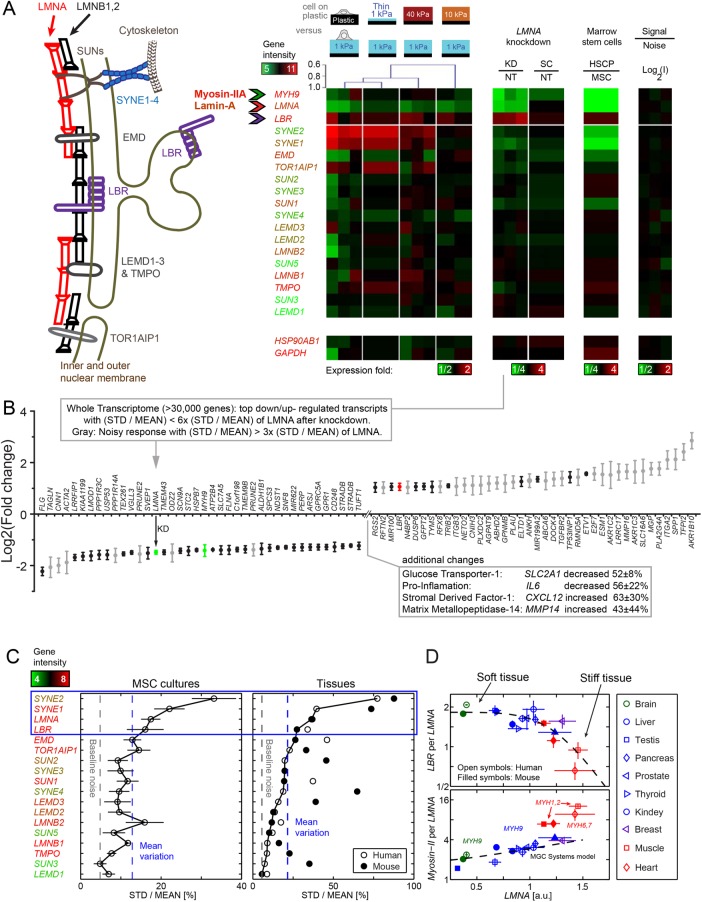
Gene expression profiles of MSCs differ significantly after just 24 h on matrices of varied elasticity and thickness as well as standard plastic flasks, particularly for some of the most widely studied nuclear envelope structural components (Figure 4A). Key components include the three lamin isoforms (LMNA, LMNB1, LMNB2), four nesprins (SYNE1 to SYNE4), and 11 more components involved in linking the lamina to the nesprins or the nuclear envelope to chromatin, such as the lamin-B receptor, LBR. Dendrogram clustering of the heat map comparing all conditions to soft-and-thick matrix shows the expression profiles for stiff gels and soft-and-thin gels are similar to plastic (Pearson correlation 0.9), consistent with these latter substrates feeling rigid to cells. Importantly, LMNA anti-correlates with LBR (Pearson: p = −0.3) in this matrix mechanosensing by the nucleus.
FIGURE 4:
Transcript profiles reveal mechano-responsive nucleo-structural genes. (A) Nuclear envelope schematic and variations in transcript levels. Consistent with matrix-directed morphologies of nuclei, heatmaps of MSCs cultured (for 36 h) on soft-and-thin gels correlate best with cultures on rigid plastic: Dendrogram shows a Pearson correlation p = 0.9. Absolute gene expression intensities averaged across matrix conditions are color-coded by gene symbols (e.g., MYH9 is high, LMNA is intermediate, SYNE2 is very low). Second heatmap: Knockdown of lamin-A produces a low contractility MSC phenotype with down-regulation of MYH9 relative to nontreated (NT) or scrambled siRNA (SC). Third heatmap: Hematopoietic stem cells and progenitors (HSCPs) likewise exhibit a low contractility phenotype with low MYH9 levels correlating with LMNA. In all heatmaps, LBR is anti-correlated. Fourth heatmap: Technical noise across triplicate hybridizations on three microarrays is <4% on average and no greater than 7% STD of mean intensity. Bottommost “housekeeping” genes validate intensity (n = 3 unless indicated). (B) Whole-genome transcriptome changes as indicated after LMNA knockdown. (C) Four mechano-malleable nuclear envelope genes (LMNA, SYNE1, SYNE2, and LBR) exhibit maximal transcriptional variation (STD normalized by mean) across matrix elasticity and thickness conditions in MSC cultures (left) and also in mouse and human tissues (right) of various stiffnesses (brain, liver, kidney, skeletal muscle, heart). For the latter, the species with the least variation provides the highest confidence for comparison. As indicated by the text color of the gene names, the top four nuclear envelope components are expressed at moderate to high levels, whereas similar genes such as the B-type lamins show little variation. (D) Consistent with the anti-correlation in MSC cultures, LBR transcript levels decrease with LMNA for the stiffest mouse and human tissues. Consistent with the noted correlation in MSC cultures, the most abundant myosin-II (e.g., MYH9) is positively correlated with LMNA. LBR/LMNA fits a Hill function for inhibition with exponent n = 5.5. Myosin-II isoform switching and muscle specialization are evident in skeletal (MYH1,2) and cardiac (MYH6,7) muscle, which are more than twofold above the trend for nonmuscle MYH9 that fits a Mechanobiological Gene Circuit Systems model (Buxboim et al., 2014). Consistent with tissue stiffness trends, brain (ectodermal tissue: green) is softer than liver (endodermal: blue), which is softer than muscle (mesoderm: red), and LMNA levels increase (n = 3 per tissue).
Anti-correlations of LMNA and LBR have been reported in other contexts (Solovei et al., 2013), and so to begin assessing whether such relationships are generic, we partially knocked down LMNA with small interfering RNA (siRNA) versus scrambled siRNA (Figure 4A, second heatmap). Not only did the same four-gene set change the most after direct LMNA knockdown relative to scrambled (and nontreated [NT]), but LMNA and LBR remained anti-correlated in the MSCs. At the whole-genome level, changes in transcripts after LMNA knockdown show that LBR is one of the most up-regulated of all transcripts and also exhibits a tight variation (STD/mean) rather than “noisy” variation after the perturbation (Figure 4B). Low noise likely indicates a more deterministic than stochastic propagation of a perturbation; most down-regulated transcripts are noisy in comparison to LBR.
With lamin-A,C knockdown, the acto-myosin cytoskeleton seems to disassemble based on deep down-regulation of actin-binding factors including transgelin (TAGLN), calponin-1 (CNN1), smooth muscle actin (ACTA2), and other less well-known actin binders (LRRFIP1, LMOD1, etc.). Importantly, nonmuscle myosin-IIA (MYH9) decreases with knockdown and is clearly part of the repression of the serum response factor (SRF) pathway that regulates the actin cytoskeleton (Buxboim et al., 2014). Among the most up-regulated of genes, AKR1B10 is an aldo-keto reductase that controls cellular retinoic acid levels (Ruiz et al., 2009), consistent perhaps with regulation of LMNA by retinoic acid (Solovei et al., 2013); ENPP1 is an anti-aging enzymatic factor (Watanabe et al., 2017), consistent with well-known roles of LMNA mutations in aging; and SPP1 encodes for osteopontin that anchors osteoclasts to bone for degradation (Reinholt et al., 1990), suggesting a switch away from osteogenesis of these MSCs. Up-regulation of MMP16 as well as β3-integrin (ITGB3) suggest a shift toward an enhanced responsiveness to soft matrix with more randomly oriented traction forces (Balcioglu et al., 2015). The profile perturbation is also broad, affecting other metabolic factors (e.g., GLUT1), inflammation (e.g., IL6), and adhesive recruitment (e.g., CXCL12) of CXCR4+ cells such as monocytes that can become osteoclasts. Such changes in expression after lamin-A,C knockdown—sometimes very noisy—hint at induction of MSCs toward lineage(s) that lack actin cytoskeleton (i.e., a soft lineage) and that possess altered metabolism and inflammatory properties. Fat is one tissue with such properties, and it is notable that human adult marrow (unlike mouse) is filled with adipocytes derived in part from MSCs and that tend to adversely affect bone mass and strength (Veldhuis-Vlug and Rosen, 2017).
Narrowing the focus to nuclear envelope genes, LMNA, LBR, SYNE1, and SYNE2 proved to be the most mechanosensitive to matrix thickness and stiffness in showing the largest variations above the mean in gene expression (Figure 4C, left plot). The nesprins link the nucleus and cytoskeleton to transmit stress to the nuclear envelope (Starr and Fridolfsson, 2010) and understandably vary greatly between tissues under different stresses. However, the LBR variations motivated study in broader contexts.
Hematopoietic stem cells and progenitors (HSCPs) are bone marrow derived as are MSCs, but HSCPs do not adhere and spread on plastic, which is how MSCs are classically isolated (Supplemental Figure S3). Marrow is of course very soft, and the human HSCPs indeed show low LMNA levels compared to MSCs (Figure 4A, third heatmap) consistent with low levels of lamin-A,C protein in HSCPs and a soft nucleus (Pajerowski et al., 2007). Nuclear envelope genes that show the largest %-difference in HSCPs relative to MSCs are again LMNA, SYNE1, SYNE2, and LBR, with a clear anti-correlation between LBR and LMNA. In neutrophils that derive from HSCPs, lamin-A,C is strongly down-regulated and LBR increases, with LBR’s sterol reductase domain being important to lipid droplet formation (Subramanian et al., 2012). Given that HSCPs are not strongly adherent cells, they might not need much myosin-II to mechanosense matrix (Engler et al., 2004a; Raab et al., 2012). Transcript levels of MYH9 are indeed low in HSCPs, and MYH9 generally correlates with LMNA across conditions. In contrast, the constitutively expressed heat shock stress gene HSP90 isoform, HSP90AB1, remains nearly constant across all conditions.
Human and mouse tissue profiles show that LMNA, LBR, and SYNE1,2 are the most mechanosensitive nuclear envelope genes in five tissues that range from soft (brain) to stiff (heart) (Figure 4C, right plot). In these plots of the coefficient of variation across the same five tissues (soft to stiff: brain < liver < kidney < skeletal muscle ∼ heart), we take as the most reliable measure for a given gene the species with the smallest variation (man or mouse). Although all tissues are now understood to harbor MSCs as perivascular cells with a highly plastic phenotype (Crisan et al., 2008), it is surprising that a single stem cell type in culture, the MSC, shows the same four components with the same trend in variation, that is, SYNE2 > SYNE1 > LMNA > LBR. For the broad range of human and mouse tissues, systematic relationships were then sought between LMNA and LBR as well as myosin-II isoforms (MYH#) that vary widely across tissues. Transcript ratios for (LBR per LMNA) and (most abundant MYH# per LMNA) plotted versus LMNA provide a measure of stoichiometry changes relevant to directly competitive as well as indirectly synergistic association networks, and LMNA is of course expected to increase with tissue stiffness (Figure 4D). The softest tissue profiled is brain (from ectoderm), which shows the lowest relative levels of LMNA and MYH9 but also the highest relative level of LBR. For the other nonmuscle tissues (mostly endodermal) we find MYH9 has again the highest gene expression among myosin-II isoforms, which is consistent with myosin-IIA being most abundant in MSCs at the transcript level here and at the protein level in both MSCs (Raab et al., 2012) and related cell lines (Ma et al., 2010). For muscle tissues (mesoderm), the most abundant isoforms are MYH1,2 in skeletal muscle and MYH6,7 in heart, and the expression levels of these isoforms in highly contractile tissue lie understandably above the MYH9 curve, consistent with tissue-specific regulation of distinct genes. Nonetheless, these muscle tissues are the stiffest tissues profiled here, and have the highest LMNA levels and the lowest LBR. All of these trends in transcriptomes suggest mechanosensing involves an anti-correlated expression of lamin-A,C and LBR, which should be scrutinized at protein levels, starting with a tissue-relevant context of differentiation.
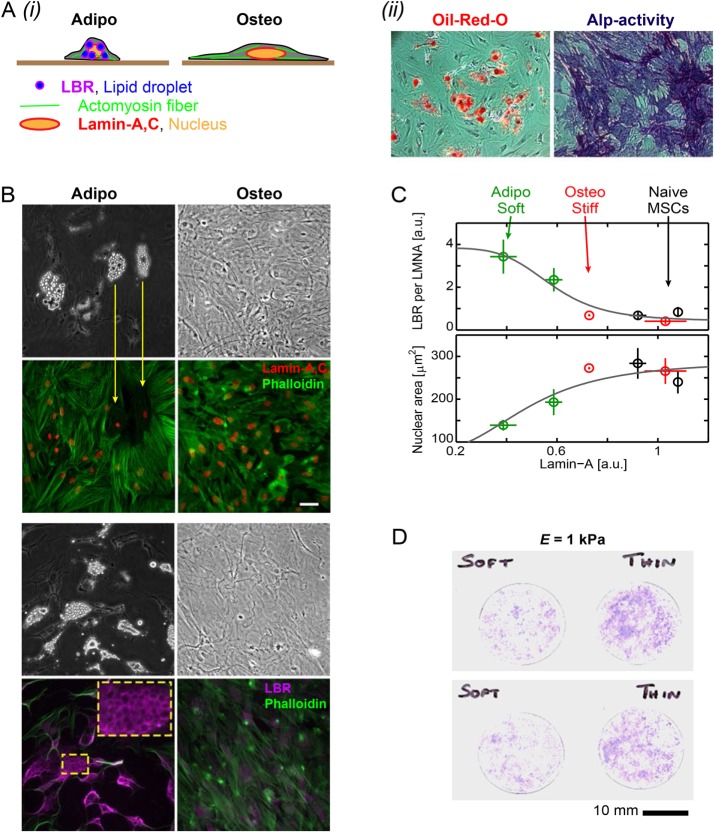
Adipogenesis (soft) favors LBR, while osteogenesis (stiff) favors lamin-A,C
Tissue-relevant functions of MSCs were confirmed in conventional cultures by inducing adipogenesis and osteogenesis as typical soft and stiff tissue lineages, respectively (Figure 5Ai,ii), and then we focused on the cytoskeleton and nucleus. In adipogenesis, lipid droplets completely replace the actin cytoskeleton, and LBR is highly abundant in localizing to the periphery of lipid droplets (Figure 5B). This seems appropriate given LBR’s role in lipid synthesis (Subramanian et al., 2012). Lamin-A,C is lowest in adipogenesis, whereas LBR is highest (Figure 5C, top), consistent with transcriptomics showing of soft lineages and repression of acto-myosin (Figure 4).
FIGURE 5:
Adipogenesis of MSCs and LBR, osteogenesis and lamin-A,C, and enhanced osteogenesis with thin gels. (A)(i) As part of the canonical MSC differentiation capacity, osteogenically induced cells are highly spread with mature stress fibers and flattened nucleus and adipogenically induced cells exhibit cytoplasmic lipid droplets that are associated with LBR. (ii) Adipogenesis and osteogenesis were induced by respectively supplemented media within 2 wk in culture. Oil-red-O positive lipid vacuoles and alkaline phosphatase (ALP) activity, as assayed by blue RR-salt staining, confirm adipogenic and osteogenic differentiation capacity (bottom). (B) Bright-field (BF) and immunofluorescence (IF) images of lamin-A,C and LBR with F-actin staining (phalloidin). Adipogenically differentiated cells clearly show lipid droplets in association with LBR staining (yellow framed zoomed-in). (C) Consistent with “relaxed” vs. “contractile” cellular phenotypes, lamin-A,C is low and LBR is high and nucleus projected area is small in adipogenically induced cells (average ± SEM, n > 25 cells). (D) Enhanced osteogenesis, as shown by increased ALP activity staining, is observed in cells that were cultured on soft gels that are also thin after 1 wk in basal media and 5 d of induction.
Nuclear area is greatest in osteogenesis (Figure 5C, bottom), consistent with matrix stiffness effects (Figure 1A). Indeed, soft-and-thin gels increase osteogenesis of MSCs relative to soft-and-thick gels with the same E (1 kPa; Figure 5D). This finding is in line with osteoid being a thin layer of matrix on rigid bone (Sodek and McKee, 2000; Buxboim et al., 2010a). Such differentiation takes many days, whereas cell and nuclear morphology changes occur within hours (Figure 1B and Supplemental Figure S2). This considerable difference in time scales raised questions about the interactions of lamin-A,C, LBR, and myosin-IIA in the absence of differentiation.
Matrix stiffness coordinately increases myosin-IIA and lamin-A,C
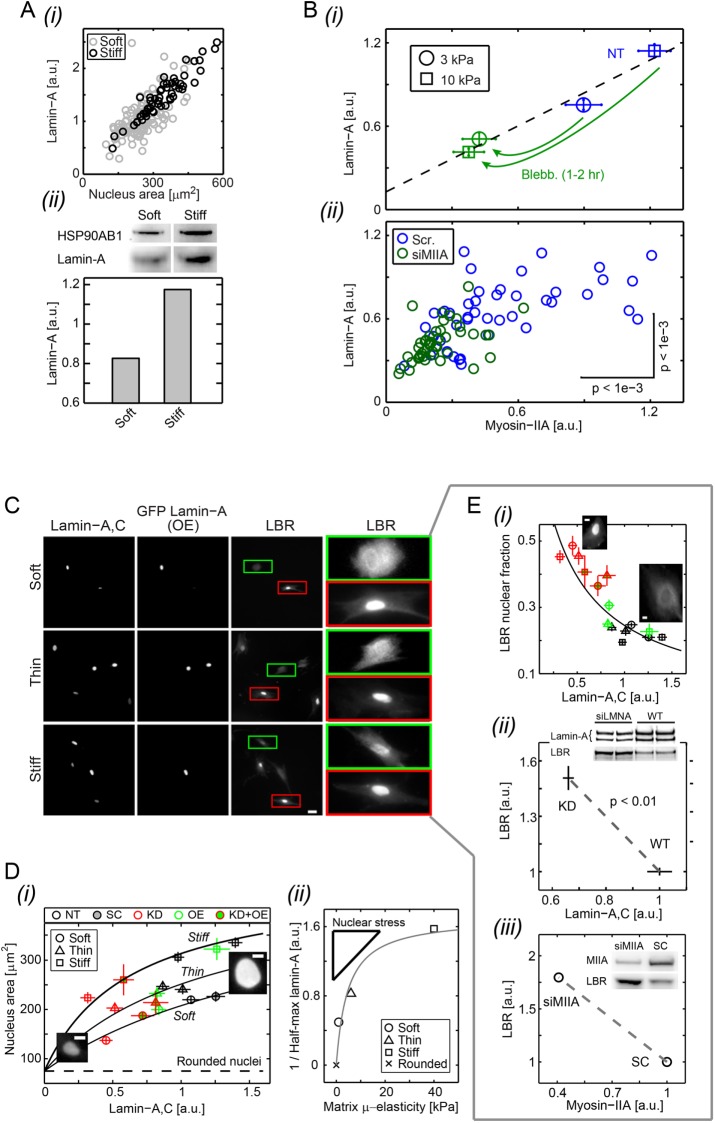
Consistent with our recent studies (Swift et al., 2013) and those of others (Hadden et al., 2017), lamin-A,C nuclear intensities are lower in cells on soft matrices (Figure 6Ai), as confirmed by immunoblotting after a few hours of adhesion (Figure 6Aii). The increase of lamin-A,C with matrix stiffness is also greatest with low passage MSCs (Supplemental Figure S1), and a positive correlation in single cell analyses between mechanosensitive lamin-A,C levels and the projected area of nuclei was particularly evident with fresh MSCs that were stressed in isolation by fluid shear (Supplemental Figure S3). Importantly, cells on stiff matrix also express more myosin-IIA (Figure 6Bi), consistent with more stress fibers (Figure 1). Decreased spreading of MSCs on stiff substrates with both blebbistatin or knockdown of myosin-IIA (siMIIA) is accompanied by decreases in both myosin-IIA and lamin-A,C protein (Figure 6Bii) and transcript (Supplemental Figure S4). LBR shows the opposite response. Likewise, increases in cell and nuclear spreading with adhesion time (Supplemental Figure S2) show increased myosin-IIA and lamin-A,C unless inhibited by blebbistatin (Supplemental Figure S5). Inhibition of myosin-II indeed disassembles stress fibers within hours (Engler et al., 2006; Raab et al., 2012). Myosin-IIA and lamin-A,C thus respond in a highly coordinated fashion to matrix mechanics, myosin inhibition, and adhesion time.
FIGURE 6:
Expression profiles of lamin-A,C, myosin-IIA, and LBR are consistent with cell mechanosensitivity. (A) Lamin-A,C levels increase with matrix µ-elasticity (for low passage MSCs, P2), as shown by single cell immunofluorescence at 36 h (i) and also by immunoblot normalized to HSP90AB1 housekeeping levels (ii) after 5 h of cell adhesion. (B) Myosin-IIA levels also increase with matrix µ-elasticity, unless inhibited by Blebb (i) or by knockdown (ii: siMIIA), which suppress lamin-A,C on soft and stiff gels. (C) MSCs transduced with GFP-lamin-A were knocked down for lamin-A,C and cultured on soft-and-thick (0.3 kPa), soft-and-thin (2–3 μm), or stiff (40 kPa) gels for 36 h, and then fixed and immunostained. High lamin-A,C–expressing cells show diffuse cytoplasmic LBR (green frames; image contrast is readjusted to show cytoplasmic pool); low lamin-A,C–expressing cells show nucleus-localized LBR (red frames). (D) Overexpression and knockdown allow lamin-A,C levels to be controlled independent of matrix. (i) Nucleus projected area increases with the lamin-A,C level on a given matrix but is greatest on stiff matrices at any given lamin-A,C level. Hyperbolic fits of form 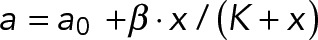 intersect at x = 0, which corresponds to the projected area of rounded nuclei in cells in suspension. (ii) As A0 and β are the same across matrix fits and K is the lamin-A,C level for half-maximum nuclear stretching, 1/K relates to nuclear stress σ and increases with matrix μ-elasticity. (Ei) LBR nuclear fraction is anti-correlated with lamin-A,C across the various matrix conditions (legend in panel Di), and fits a competitive binding model. Overall protein levels of LBR are enhanced by knockdown of either lamin-A,C (i, siLMNA) or myosin-IIA (iii, siMIIA). Cell extract volumes were calibrated using Bradford total protein assay (i) or based on the immunoblotting intensities of HSP90AB1 housekeeping protein (ii). Average ± SEM, n > 25 cells. Scale bars = 10 µm.
intersect at x = 0, which corresponds to the projected area of rounded nuclei in cells in suspension. (ii) As A0 and β are the same across matrix fits and K is the lamin-A,C level for half-maximum nuclear stretching, 1/K relates to nuclear stress σ and increases with matrix μ-elasticity. (Ei) LBR nuclear fraction is anti-correlated with lamin-A,C across the various matrix conditions (legend in panel Di), and fits a competitive binding model. Overall protein levels of LBR are enhanced by knockdown of either lamin-A,C (i, siLMNA) or myosin-IIA (iii, siMIIA). Cell extract volumes were calibrated using Bradford total protein assay (i) or based on the immunoblotting intensities of HSP90AB1 housekeeping protein (ii). Average ± SEM, n > 25 cells. Scale bars = 10 µm.
The finding that myosin-II contractility regulates nuclear morphology and ultimately impacts lamin-A,C expression is perhaps reasonable, but the reciprocal relationship also seemed likely because lamin-A,C interacts with nuclear actin and other actin-binding proteins (Holaska et al., 2004; Zastrow et al., 2004) that might regulate the transcription factor SRF, which controls MYH9 transcription (Olson and Nordheim 2010). Partial knockdown (KD) of lamin-A,C in MSCs shows, by quantitative immunofluorescence, that myosin-IIA levels are similar in wild-type (WT) and KD cells after just 1 h of adhesion on rigid coverslips starting with suspended cells (Supplemental Figure S5). However, 2 h after adhesion begins, both myosin-IIA and (of course) lamin-A,C remain low in the siLMNA treated cells, whereas myosin-IIA levels have increased in WT cells together with lamin-A,C (consistent with cell and nuclear spreading; Figure 1 and Supplemental Figure S2). At 36 h, a similar difference in both proteins was evident in lysates by both standard immunoblotting and by mass spectrometry quantitation of a dozen or more peptides from each protein (Supplemental Figure S5).
Overexpression of transduced GFP-LMNA (OE) sustained high levels of lamin-A (Supplemental Figure S4), and with increasing lamin-A in A549 lung-epithelial cells the stiffness of the nucleus increased based on micropipette aspiration (Supplemental Figure S4). The effective nuclear elasticity scales with the square root of the increase in the amount of lamin-A (Supplemental Figure S4), which is weaker than expected of typical cross-linked polymers and perhaps consistent with recent images of the lamina meshwork (Turgay et al., 2017), although nuclear viscosity increases more strongly with lamin-A,C levels (Swift et al., 2013). Cells cultured on soft-thick, soft-thin, or stiff matrices and then immunostained for lamin-A,C (Figure 6C) showed that for the same lamin-A,C level (set by OE or siLMNA), which establishes nuclear stiffness, the spreading of the nucleus is greater on stiff and thin versus soft matrices (Figure 6Di). For a given amount of lamin-A,C, the nucleus thus deforms and strains more on a stiff or thin matrix where myosin-II–generated stresses are higher compared to a soft-and-thick matrix.
The various observations suggest a simple model for the stresses that drive changes in nuclear spreading that should relate to the increased stress that suppresses nuclear roughness in cells on stiff matrices (Figure 3). At low lamin-A,C, the projected nuclear area increases linearly with lamin-A,C level, but then nuclear area saturates. Such a hyperbolic form (with or without a cooperativity exponent),
is typical of increases in cell area or nuclear area (a) from low tension states of cells on soft matrix to high tension states of cells on stiff matrix (e.g., x = E in Figure 2B). Importantly, with lamin-A,C levels controlled here (= x), the half-max characteristic (K) is seen to vary with E. K is relatively low for cells on stiff matrix where cell stress is high, and K is relatively high for cells on soft matrix where cell stress is low. Soft-and-thin substrates exhibit an intermediate K. Assuming that stress in the cell approximates the stress on the nucleus and relates inversely to K (i.e., σ ∼ K−1), then nuclear stress or tension increases about fourfold from soft to stiff matrix (Figure 6Dii). This result is quantitatively consistent with estimates from nuclear roughness and with traction force–based measurements (Engler et al., 2006) of increasing cell tensions in MSCs on soft versus stiff matrices (Figure 3). Moreover, in the limit of no contribution of lamin-A,C to nucleus deformability (lamin-A,C → 0) all three hyperbolic fits intersect at the nuclear area of cells rounded in suspension (Supplemental Figure S2) where matrix-directed, cytoskeleton-imposed nuclear stress vanishes (σ = 0 as K → ∞).
Increased lamin-A,C in stressed nuclei outcompetes LBR
The mechanosensitivity of lamin-A,C to matrix stiffness was assessed from a distinct perspective by taking advantage of our finding that LBR is anti-correlated with LMNA in transcriptomics of both MSCs (Figure 3A) and human and mouse tissues (Figure 3C). By immunofluorescence, cells with high lamin-A,C also showed abundant cytoplasmic LBR and nuclear staining (green frames in Figure 6C), whereas high intensity nuclear localization of LBR was observed exclusively in low-expressing lamin-A,C nuclei (red frames). Combining all of the imaging results together for all levels of expression in cells on soft, soft-and-thin, and stiff matrices, nuclear LBR decreased as expected with lamin-A,C and collapsed onto a single hyperbolic curve consistent with competitive inhibition (Figure 6Ei). Such a fit suggests that lamin-A,C can outcompete LBR for binding to lamin-B (Ye and Worman, 1994). Knockdown of lamin-A,C indeed increases overall LBR by immunoblotting (Figure 6Eii). Consistent with an indirect coupling to contractility through lamin-A,C, LBR is also up-regulated by knockdown of myosin-IIA (Figure 6Eiii). Knockdown of myosin-IIA as well as LMNA in mesenchymal-like A549 cells (Ye and Worman, 1994) likewise shows the respective decrease or increase in stoichiometry of LBR per LMNA (Supplemental Figure S6). The LBR results thus reinforce the working model of a link between matrix stiffness, contractility, and lamin-A,C.
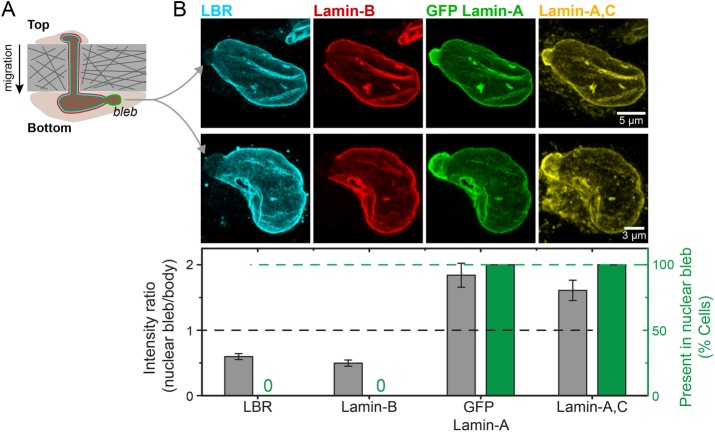
An alternative approach to visualizing the complementary localization of LBR and lamin-A,C in stressed nuclei was taken with the A549 cells after constricted migration. Nuclear blebs enriched in lamin-A and depleted in lamin-B have been seen at one or both ends of the elongated nuclei postmigration (Harada et al., 2014). Immunostaining reveals that LBR is always depleted from the nuclear bleb (Figure 7). One simple interpretation of such images is that LBR binds lamin-B (as discussed further below), and that abundant lamin-A,C somehow obstructs and thereby outcompetes the lamin-B interaction with LBR.
FIGURE 7:
LBR depletes from the lamin-A,C–enriched nuclear blebs and distributes similar to lamin-B. (A) Migration of A549 cells through 3 µm Transwell pores leads to nuclear blebs formation. (B) These nuclear blebs are always (green bars) enriched in lamin-A,C and deficient in both lamin-B and LBR (gray bars), as shown by the representative images (average ± SEM, n > 10 cells).
DISCUSSION
Microenvironments influence nuclear phenotypes
Although a constant cell-to-nuclear volume ratio is a long-recognized characteristic of a given cell type (Kume et al., 2017), it is not obvious that it should apply to a stem cell such as an MSC under diverse adhesion and spreading conditions (Figure 1) that greatly influence differentiation (Engler et al., 2006; Swift et al., 2013). Differentiation of hematopoietic stem cells, for example, yields lineages that have zero nucleus-to-cell volume ratio (red blood cells), small nucleus-to-cell volume ratio (neutrophils), and a larger nucleus-to-cell volume ratio (T-cells). The correlations for MSCs are nonetheless clear in suggesting a constant cell-to-nuclear volume ratio. Matrix elasticity might therefore be an early inductive cue for lineage specification, so that definitively differentiated cells such as those filled with lipid droplets in adipogenesis or else undergoing osteogenesis (Figure 5) should have nucleus-to-cell volume ratios carefully quantified for comparison.
Nuclear flattening and nuclear smoothing with spreading on stiff versus soft matrices are clear (Figures 1 and 4), and the latter suggests about a fourfold increase in tension on the nucleus, which is in agreement with both traction force estimations and changes in lamin-A,C (Figure 6). However, it is unclear whether such tensions (or other mechanisms) can drive decreases in nuclear volume in cells on stiff matrices, given that the nucleus-to-cell volume ratio remains constant. Nuclear volume changes can certainly occur in parallel with cell volume changes when the external osmolarity of a cell medium is altered (Irianto et al., 2013). The latter study approximated both cells and nuclei as ellipsoids and fitted the data to a van der Waals–type model that applies widely to nonideal fluids with excluded volume: water can certainly be extracted from the nucleus, but the nuclear envelope is impermeable to DNA, and so DNA concentration increases when water is extracted (Irianto et al., 2016). The ellipsoidal approximation of nuclear shape was therefore assessed further here in order to describe DNA concentration changes that might result from the decrease in nuclear spreading upon knockdown of lamin-A,C (Figure 6). Voxel approaches have resolution limits given the large nuclear roughness (Figures 1 and 3), but one simple complementary analysis uses the intensity of Hoechst-labeled DNA (Supplemetal Figure S7). Data fits show that nuclear height H multiplied by DNA concentration [DNA] increases inversely with projected nuclear area A after knockdown, that is, H [DNA] ~ 1/A. Nuclei become taller and A becomes smaller for soft matrix (Figure 1A), and so if [DNA] remains constant, then H = 1/A, which is roughly consistent with observations as well as an ellipsoidal geometry. Furthermore, if DNA concentration does somehow change as a function of nuclear spreading as [DNA] ∼ Ax, then nuclear height must vary as H ∼ Ax−1. For example, if [DNA] condenses with cell and nuclear spreading as [DNA] ∼ A−0.5, then nuclear height varies as H ∼ 1/A1.5, but this is not consistent with initial measurements that show weak increases in average nuclear height upon rounding (Figure 1A). DNA concentration and nuclear volume are thus unlikely to change much as nuclei round up. The approach needs to be applied more widely in efforts to assess whether nuclear volume and [DNA] remain constant (or not) as nuclear tension differences and other changes occur for soft versus stiff matrices.
Apparent microelasticity of matrix regulates relaxed versus contractile cell states
A biophysical picture is emerging in which stiff matrices including soft-and-thin matrices strongly favor a contractile phenotype owing to an increase in the apparent µ-elasticity sensed by a cell (Figure 8A). Expression levels of nonmuscle myosins in cells on stiff matrices increase with assembled filaments that are stabilized by high tension (Raab et al., 2012). Stiff matrices thereby enhance cell-generated traction forces (Engler et al., 2006; Solon et al., 2007) that exert a tension on the nucleus that is evident as the nucleus is thus pulled and flattened against the matrix. Lamin-A,C responds by increasing on stiff or thin matrices, mirroring tissue profiles (Swift et al., 2013), and conferring nuclear rigidity. Additional factors in the cytoplasm and extracellular matrix might also contribute to nuclear mechanical properties and likely to mechanosensing. Such factors range from nesprins that transfer tension to the nucleus (Starr and Fridolfsson, 2010) to at least one matrix metalloprotease, MMP14 (Gutiérrez-Fernández et al., 2015), that when knocked out in mice leads to increases in collagen fibers (in heart), nesprins, and lamin-A,C levels, consistent with a stiff, fibrotic phenotype. Interestingly, lamin-A,C knockdown in human MSCs increases MMP14 transcripts (Figure 4B), but the effect is very noisy compared to LBR, suggesting a more direct regulation of the latter.
FIGURE 8:
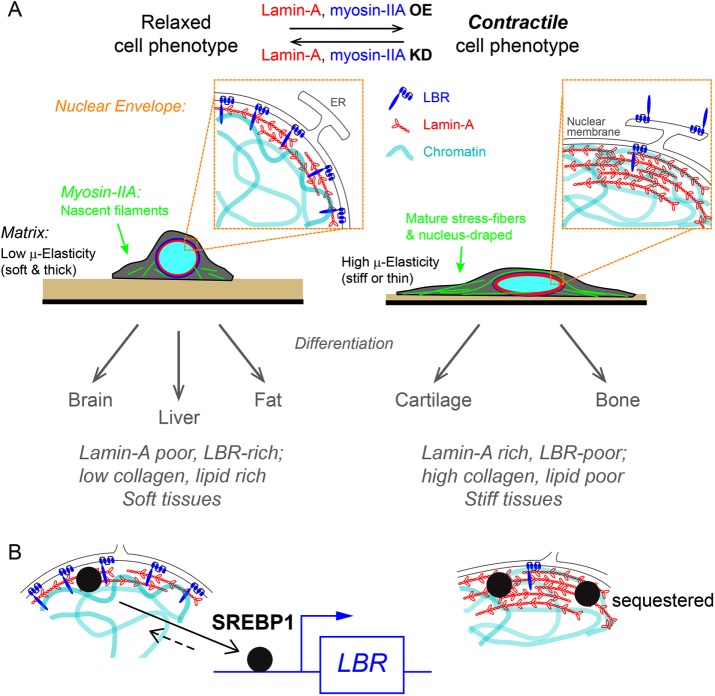
Matrix apparent microelasticity determines physical, molecular, and functional cell contractile phenotypes. Cells on stiff matrices or soft ones that are sufficiently thin and are attached to rigid surfaces generate elevated contractile forces that enhance lamin-A,C expression at the nuclear envelope. Acting as a mechano-sensor, lamin-A,C directs myosin-IIA expression and suppression of LBR. Reduction in lamin-A,C, for example in soft environments, enables LBR translocation at the nuclear envelope. Together, cells obtain a contractile phenotype in accordance with matrix mechanics to direct differentiation programs of soft or stiff tissue lineages.
Coordinated changes in the cytoskeleton and nuclear lamina contributes to matrix-directed cellular differentiation toward soft versus stiff tissue lineages (Engler et al., 2006; Gilbert et al., 2010), and some aspects of mechanism are becoming clearer. As a nuclear mechano-sensor, lamin-A,C orchestrates matrix-directed remodeling of both the nuclear envelope and cytoskeletal reorganization. Down-regulation of lamin-A,C results in suppression of myosin-IIA levels, which in turn relaxes cell contractility to feedback on lamin-A,C. Additionally, LBR levels are elevated with translocation of cytoplasmic pools toward the nuclear envelope. Tissue profiles of LBR expression thus reflect a correlation with tissue softness that anti-correlates with lamin-A,C levels.
Fat is indeed soft, and so it is intriguing that LBR has a carboxy-terminal sterol reductase domain for lipid biosynthesis, consistent with increasing LBR expression levels observed during adipogenic differentiation (Subramanian et al., 2012). Like fat, the brain is very soft and also makes its own cholesterol, amounting to 30% of whole-body levels (Bjorkhem and Meaney, 2004). High LBR and low lamin-A,C in the brain (Figure 3C) is consistent with the complementarity of these two nuclear envelope factors that is evident in adpiogenesis (Figure 5C). In contrast, stiff tissues express high lamin-A,C, high myosin-IIA, and low LBR that reflect the more contractile phenotype of cells in stiffer tissues such as bone (Figures 1A, 4, 5, and 7). Knockdown of lamin-A,C in MSCs on rigid plastic not only suggests an induction away from an osteoblastic phenotype toward a more adipogenic favored phenotype, but the transcriptome changes suggest a phenotype that promotes bone degradation by osteoclasts (Supplemental Figure S8A) consistent with the noted up-regulation of osteopontin (SPP1 in Figure 4B).
LBR is downstream from lamin-A,C mechanosensing
Lamin-A,C and LBR were found here in three different analyses of tissues or MSCs on different matrices to fit a competitive binding model in terms of either lamin-A,C protein or transcript (Figures 4 – 7). Anti-correlations between these two nuclear envelope components have been noted before (Olins et al., 2001; Zwerger et al., 2008) and most recently found to be almost interchangeable in tethering heterochromatin to the nuclear envelope (Solovei et al., 2013); lack of both proteins generally leads to centralized heterochromatin. In early (E8–E17) embryonic lineages where observations could be made in these studies, LBR is detectable, but in tissues that are subject to early mechanical stress LBR decreases as lamin-A,C increases. The observations includes stiff tissue cells such as those of adult cartilage and bone, and myotubes, cardiomyocytes, and smooth muscle as well as endothelial cells subject to flow stresses, but not the earliest muscle stem cells (satellite cells), or the earliest intestine or skin cells, which are all softer tissues. However, the reasons for these expression patterns had not yet been explained. The initial systematics is perhaps clear from the results here: when and where mechanical stress is high in a tissue, lamin-A,C is favored, which inhibits LBR at the protein and transcript level.
LBR interactions with lamin-B are well established and although the site of interaction remains unclear (Worman et al., 1988; Georgatos et al., 1989; Ye and Worman 1994), the findings here lead us to hypothesize that LBR competes with lamin-A,C for binding lamin-B. Heterotypic interactions between lamin-A,C and B-type lamins have also long been reported (Fisher et al., 1986; Georgatos et al., 1988) and are facilitated by the unstructured N-termini that precede the coiled-coil, dimerizing rod domains. Phosphorylation of lamin-A,C in both the N- and C-termini drive disassembly in mitosis and in interphase mechanosensing of soft microenvironments (Buxboim et al., 2014), which will thereby enable binding of LBR to B-type lamins.
LBR transcription could occur through the transcription factor SREBP1 (sterol regulatory element-binding protein 1), which chromatin-IP has shown to bind the LBR promoter (Supplemental Figure S8B; Rosenbloom et al., 2013). SREBP1 also associates with lamin-A,C and is visually sequestered in high-density aggregates of lamin-A,C (Lloyd et al., 2002; Yang et al., 2013). SREBP1 might thus be sequestered by lamin-A,C at the nuclear envelope and be released to bind promoters only when stiffness, stress, and therefore lamin-A,C are low, thereby promoting expression of LBR (Figure 8B). Understanding such a mechanosensitive process thus requires continued efforts at quantifying cytoskeleton-generated nuclear tension.
MATERIALS AND METHODS
Isolation of fresh MSCs from bone marrow
Bone marrow aspirates were obtained from posterior iliac crest of human donors (University of Pennsylvania School of Medicine) under the procedures and regulations defined by the Helsinki agreement. Mononucleated cells (MNCs) were obtained using a Ficoll density gradient (Ficoll-Paque PLUS; GE Healthcare) and depleted from CD34-positive cells by a Micro-bead kit (Direct CD34 Progenitor Cell Isolation Kit; Miltenyi Biotec) and screened by automated cell separation (AutoMacs; Miltenyi Biotec) according to manufacturer’s protocols. MNCs were resuspended in 10% FBS (Sigma-Aldrich) and 1% penicillin/streptomycin antibiotics (P/S; GE Healthcare) supplemented low-glucose basal medium (DMEM; Life Technologies). Typically, MNCs were seeded at 10,000–100,000 cells/cm2 in standard tissue-culture plastic flasks and incubated at 37°C and 5% CO2 humidified conditions. Cells were thoroughly rinsed in phosphate-buffered saline (PBS) ×3 after 24 h to remove nonadherent cells. Fibroblastic colony forming unit (CFU) stromal cells appeared within 3–4 d and cells expansion included medium exchange every 4 d. Cells confluence was maintained <80% by passaging cells and reseeding at >50% confluence. Expression of stromal stem cells markers (CD105, CD166, CD44, CD90) and the lack of hematopoietic markers (CD45-RA, CD34) was verified by flow cytometry (Supplemental Figure S3C and unpublished data). Differentiation capacity toward fat and bone was verified using adipogenic and osteogenic induction media (R&D: following manufacturer’s protocols; Figure 4). Fresh human CD34-positive cells were obtained by cell sorting from the mononucleated fraction of donor bone marrow cells. Cells were cultured for 7 d with stem cells factor (SCF) and thrombopoietin (TPO) for 4 d.
Glass coverslips silanization and polyacrylamide gel preparation
The preparation of polyacrylamide (PA) gels with controlled elasticity, thickness, and covalent attachment to glass coverslips is described in detail in a published methods paper (Buxboim et al., 2010b). In brief, glass coverslips (thickness #1.5; Fisher Scientific) were placed in boiling ethanol for 10 min, rinsed in distilled water (DW), and immersed in RCA (DW, hydrogen peroxidase [30%; Fisher Scientific], ammonium hydroxide [30%; Fisher Scientific] at 3:1:1 [vol/vol/vol]) at 80°C for 10 min and rinsed in DW. To remove water traces, glasses were rinsed in ethanol and then in chloroform and silanized in 0.1% allyltrichlorosilane (ATCS; Aldrich) in chloroform (Fisher Scientific) for 30 min. Silanized glasses were then rinsed in chloroform → ethanol → DW and dried under vacuum. Successful silanization can be evaluated by assessing hydrophobicity, for example, by water droplet contact angle. PA gel precursors were prepared by mixing acrylamide (AA, 40%; Sigma) and N,N′-methylenebisacrylamide (bis-AA, 1.5% [wt/vol] in DW; Sigma) in PBS (Sigma). Nominal gel elasticity was specified by varying acrylamide and cross-linker concentrations as calibrated by desktop rheometer: 0.3 kPa, 7.5% AA, 4.7% bis-AA; 1 kPa, 9% AA, 5.6% bis-AA; 3 kPa, 11.2% AA, 7% bis-AA; 10 kPa, 15% AA, 9.3% bis-AA; 40 kPa, 2.5% AA, 20% bis-AA. Gel thickness was specified by varying the volume of gel precursor and by using 1-μm-diameter monodispersed silica microsphere spacers (Thermo Scientific) as calibrated by confocal imaging (Buxboim et al., 2010b). Gelation was initiated by adding 0.1% (vol/vol) tetramethylethylenediamine (TEMED; Sigma) and 0.1% (wt/vol) ammonium persulfate (Sigma) to gel precursor just before placing it at the center of the silanized coverslips and covering with RCA-treated glasses. Thin gels were cured with weights placed on top of the cover glasses, pressing them against the beads spacers. Gels were allowed to polymerize while covalently binding the silanized glasses for 30–60 min. Nonsilanized glasses were gently removed after immersing in PBS for 1–2 h.
Gels collagen coating and sterilization
Gels were immersed in 10 mg/ml sulfo-SANPAH (Fisher Scientific) in 50 mM, pH 8.5, HEPES and reacted under 365 nm i-line exposure for 10 min. Collagen was mixed in 0.1 M acetic acid (Fisher Scientific) at equal volume and in 50 mM, pH 8.5, HEPES to reach 0.2 mg/ml final concentration. Gels were immersed with collagen while agitated overnight at 37°C. Prior to seeding cells, gels were UV-sterilized (cell-culture hood UV light source) for 3 h. During all preparation steps, gels were maintained in a hydrated state.
Nuclear height model of spreading for an invariant cell-to-nuclear volume ratio
The projected area of the cell is seen to scale linearly with the projected area of the nucleus regardless of matrix elasticity or adhesion time (e.g., Figure 1Aiii). Designating the projected area of the nucleus in state n as pn and that of the cell as Pn, the proportionality constant is the slope s between the projected areas of cells and nuclei in any two states:
|
|
In this nuclear height model (Nucl-Ht Model), nuclear height limits spreading of the cell and its nucleus. We designate cell height as An, and nuclear height as an, so that An ≈ an is consistent with observations. For simplicity, we approximate the nucleus by an ellipsoid of volume vn = (4π/3) an bn cn and that of the cell by half of an ellipsoid Vn = (2π/3) An Bn Cn. Projected areas are pn = π bn cn = (3/4) vn/an and Pn = π Bn Cn = (3/2) Vn/An. Hence,
|
|
Although the choice of state 1 is arbitrary, we obtain the most accurate measurements of projected areas in the maximally spread state, so that (P1 / p1) = (Pmax / pmax). Importantly, we observe that sexpt ≈ (Pmax / pmax) within 10%, which implies that the ratio of terms in square brackets is near unity, thus reducing Eq. 2 to
|
|
The Nucl-Ht Model thus reveals an invariant ratio of cell-to-nuclear volume. Such a ratio is often considered phenotypic of cells and is characteristically small for cells such as blood stem cells but generally much larger for various mesenchymal cells. Indeed, with chondrocytes embedded in agarose, osmotically driven changes in cell and nuclear volume (Irianto et al., 2013) also indicate proportional changes within <3%, even with perturbations in excess of 100 mOsm (∼300 kPa hydrostatic stress) from iso-osmotic conditions (∼300 mOsm). The Nucl-Ht Model with the deduced invariance of cell-to-nuclear volume implies that factors that control nuclear height, such as nuclear stiffness conferred by lamins or stress applied by the cytoskeleton, will generally regulate spreading of both the cell and its nucleus.
Transcriptional profiling by titrated DNA microarrays
Total RNA was extracted from cells using Trizol and purified by RNeasy (Qiagen) with on-column DNase digestion according to manufacturer’s protocol. Adhesive cells were gently scraped in Trizol. Total RNA was amplified and converted to cDNA using WT-Ovation Pico kit (NuGen). Fragmented and biotin-labeled ST-cDNA was generated using WT-Ovation Exon Module (NuGen). Titrated X/3, 2X/3, and 2X hybridization cocktails (42 μl each) were prepared with 20, 40, and 120 ng/μl ST-cDNA, respectively. Eukaryotic hybridization controls (GeneChip) were included at proportional concentrations. Each sample was interrogated by a series of hybridization, rinse, and scan cycles in which X/3, 2X/3, and 2X titrated hybridization cocktails were applied sequential to individual human gene 1.0 ST DNA microarrays (Affymetrix). Two additional rinse-scan cycles were included in which only hybridization buffer was applied to the arrays (42 μl). Hybridization, washing, and array scanning were performed according to the manufacturer’s instructions. Transcriptional profiling of mouse and human tissues was obtained from Affymetrix (publically available data). Original and public gene expression data sets were RMA-summarized.
Immunostaining
Cells were fixed with 3.7% formaldehyde (Sigma-Aldrich) in PBS for 10 min at room temperature (RT) followed by PBS washing 2× for 5 min. Blocking and primary antibody staining were performed in 1% bovine serum albumin in PBS. Primary antibody concentrations ranged between 1/300 and 1/500, depending on the stock concentration, and all primary antibodies were incubated at RT for 2 h or overnight at 4°C. All donkey secondary antibodies (Alexa Fluor dyes 488, 564, and 647) were stained for 1–2 h at RT at 1:500 dilution in PBS and TRITC-phalloidin (Sigma-Aldrich) was used at a concentration of 100 ng/ml. Imaging for quantitative immunofluorescence of lamin-A,C, myosin-IIA, and LBR was performed using an inverted microscope (IX-71; Olympus) with either 20× (Olympus; NA 0.75) or 40× (NA 0.60) objectives, and a cooled charge-coupled device (CCD) camera (Cascade; Photometrics) and image acquisition was performed with Image Pro software (Media Cybernetics). Fixed cells were immunostained using the following antibodies and reagents: myosin-IIA (mouse), monoclonal HSP90AB1, polyclonal SYNE1 (Abcam); polyclonal myosin-IIA, monoclonal lamin-A,C (mouse), polyclonal lamin-A,C (goat), polyclonal lamin-B1, polyclonal cleaved lamin-A,C (Santa Cruz); polyclonal LBR (Novus Biologicals); phalloidin (Sigma). Fixed cells were mounted in mounting medium (Axell). To prevent volume distortions, samples prepared for confocal z-stacks quantification were not mounted.
Quantitative immunofluorescence
Lamp intensity and field of view homogeneity were calibrated relative to a fluorescent plastic standard pixel by pixel per each experiment. Image analysis was performed by a Matlab-based (Mathworks) custom designated application that included background subtraction, cell and nucleus objects registration and intensity integration, morphological and statistical analyses.
Immunoblotting
Cells were trypsinized, pelleted, and stored at −20°C until analysis. Pellets were thawed and resuspended in 1× LDS lysis buffer supplemented with 1% protease and 1% phosphatase inhibitors and sonicated on ice (3 × 15 × 1 s pulses, intermediate power setting). After resting for 30 min on ice, samples were denatured at 80°C with 0.5% β-mercaptoethanol (vol/vol) for 10 min. Samples were loaded onto bis-Tris 4–12% gradient gels for electrophoresis (100 V × 10 min; 160 V × 55 min) and then blotted (iBlot; Life Technologies; settings P3, 7 min) onto blotting membrane. Band intensities were quantified using Fiji/ImageJ, relative to local background levels flanking the specific bands.
Mass spectrometry
SDS–PAGE gels (NuPAGE 4–12% bis-Tris; Invitrogen) were run at 100 V for 10 min and 160 V for 25 min. Gel sections were washed (50% 0.2 M ammonium bicarbonate [AB] solution, 50% acetonitrile [ACN], 30 min at 37°C), dried by lyophilization, incubated with a reducing agent (20 mM tris(2-carboxyethyl)phosphine [TCEP] in 25 mM AB solution at pH 8.0, 15 min at 37°C), and alkylated (40 mM iodoacetamide [IAM] in 25 mM AB solution at pH 8.0, 30 min at 37°C). The gel sections were dried by lyophilization before in-gel trypsinization (20 μg/ml sequencing grade modified trypsin in buffer as described in the manufacturer’s protocol [Promega], 18 h at 37°C with gentle shaking). The resulting solutions of tryptic peptides were acidified by addition of 50% digest dilution buffer (60 mM AM solution with 3% methanoic acid).
Peptide separations (5 μl injection volume) were performed on 15-cm PicoFrit column (75 μm inner diameter; New Objective) packed with Magic 5 μm C18 reversed-phase resin (Michrom Bioresources) using a nanoflow high-pressure liquid chromatography system (Eksigent Technologies), which was coupled online to a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ion source. Chromatography was performed with solvent A (Milli-Q water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Peptides were eluted at 200 nl/min for 3–28% B over 42 min, 28–50% B over 26 min, 50–80% B over 5 min, 80% B for 4.5 min before returning to 3% B over 0.5 min. To minimize sample carryover, a fast blank gradient was run between each sample. The LTQ-Orbitrap XL was operated in the data-dependent mode to automatically switch between full scan MS (m/z = 350–2000) in the Orbitrap analyzer (with resolution of 60,000 at m/z 400) and the fragmentation of the six most intense ions by collision-induced dissociation in the ion trap mass analyzer.
Raw mass spectroscopy data was processed using Elucidator (version 3.3; Rosetta Biosoftware). The software was set up to align peaks in data from samples derived from corresponding molecular weight regions of the 1D gels. Peptide and protein annotations were made using SEQUEST (version 28; Thermo Fisher Scientific) with full tryptic digestion and up to two missed cleavage sites. Peptide masses were selected between 800 and 4500 amu with peptide mass tolerance of 1.1 amu and fragment ion mass tolerance of 1.0 amu. Peptides were searched against a database compiled from UniRef100 (November 2010) mouse, plus contaminants and a reverse decoy database. A deltaCn of 0.01 and mass error limit of 20 ppm was used, resulting in a false-positive rate of ∼10%. In these experiments, only proteins detected with three or more peptides were considered. The peptide database was modified to search for alkylated cysteine residues (monoisotopic mass change; Δ = +57.021 Da) and oxidized methionine (Δ = +15.995 Da). In proteomic profiling experiments, we also considered the acetylation of lysine (Δ = +42.011 Da), methylation of lysine and arginine (Δ = +14.016 Da), and phosphorylation of serine, tyrosine, threonine, histidine, and aspartate (Δ = +79.966 Da). Ion currents of modified peptides were summed with their parent peptide. Peptides derived from trypsin or keratin were considered to be contaminants and were not used in subsequent calculations. When evaluating total ion current, only signals from annotated peptides were summed. The PRF algorithm was coded for Mathematica (version 8; Wolfram Research) and was used for all MS protein quantitation (Shin et al., 2011).
siRNA and short hairpin RNA knockdown of MYH9, LMNA, LBR, and inhibitors
All siRNAs used in this study were purchased from Dharmacon. Cells were passaged > 24 h before transfection and incubated with a complex of siRNA (30 nM; siLMNA: 5′-GGUGGUGACGAUCUGGGCU-3′; siMYH9: 5′-GGCCAAACCUGCCGAAUAAAUU-3′ with complement sequence 5′-UUUAUUCGGCAGGUUUGGCCUU-3′ or scrambled-siRNA siGENOME nontargeting siRNA #1 [Thermo Fisher Scientific]) or shLBR (TRCN0000060460; Sigma) and 1 μg/ml Lipofectamine 2000 according to the manufacturer’s instructions for 24 h (in low-glucose DMEM with 10% FBS). Racemic blebbistatin (EMD) was used at 30 μM. For shLMNA treatment, A549 cells were infected with lentiviral supernatants targeting lamin-A:C (TRCN000061833; Sigma) at a multiplicity of infection (MOI) of 10 in the presence of 80 µg/ml polybrene (Sigma), and cultured for 24 h. The cells were then selected by 2 µg/ml puromycin (Sigma) for 30 d.
Transfection and transductions of lamin-A
A construct expressing GFP-lamin-A under the EF1α promoter (Izumi et al., 2000) was packed into a lentiviral delivery system. Cells were transduced at MOI 50 and evaluated for survival and proliferation. The fraction of GFP-lamin-A–positive cells before experiment was ∼50%. Colonies of GFP-lamin-A–expressing MSCs were isolated using a cloning cylinder (Bel-Art Products, Pequannock, NJ), trypsinized and further expanded for experiments. Lamin-A constructs were transfected via electroporation following manufacturer’s protocols (MSCs kit, Nucleofector; Lonza). Lamin-A transfections of A549 cells and all myosin-IIA constructs in MSCs transfections used Lipofectamine LTX (Invitrogen) with Plus reagent using 0.5 g DNA per well in a six-well plate. Transfection levels were similar across all constructs (within 20–30%) based on GFP densitometry of Western blots.
Transwell migration
For migration through transwells (Corning), A549 cells were seeded at 300,000 cells/cm2 onto the top side of the filter membrane and left to migrate in normal culture condition for 24 h.
Confocal microscopy and deconvolution of fixed samples
Laser scanning confocal fluorescence microcopy was carried out using the following systems: Leica Microsystems TCS SP8, 63× / NA 1.4 oil immersion objective (Figures 1Ai,iii and 2Ei). Nuclear orthogonal view was generated from 0.21 μm z-stack and constructed using Fiji (Schindelin et al., 2012). To evaluate the corresponding nuclear roughness spectra, iterative self-organizing data algorithm thresholding (Ridler and Calvard, 1978) was applied and nuclei were horizontally aligned per fitted ellipses. Apical and dorsal nuclear perimeter contours were obtained while discarding 25% edge regions, and roughness Fourier spectra were evaluated and smoothed using MatLab. Epi-fluorescence deconvolution was applied using the Deltavision Deconvolution IX70 Olympus system (Figure 1A).
Rheology
The nominal elasticity of the gels was calibrated using a strain-controlled rheometer (TA instruments; RFS-II). A flat titanium plate, 25 mm diameter, was used with 0.5 ml sample volume and 0.95 mm gap. Platform surface temperature was set to 25°C, with 0.1% strain and 1 rad/s rate.
Apparent μ-elasticity of thin gels
A detailed description of the AFM force-indentation measurements was published in a methods paper (Buxboim et al., 2010b). In brief, AFM (MFP-3D; Asylum Research) force-indentation curves were obtained using pyramidal tip cantilevers (ksp ∼ 24 pN/nm, TR400PB; Olympus). Force-indentation data was exported to MatLab for computing the apparent μ-elasticity. The Young’s modulus E was calculated by fitting (MatLab) the classical parabolic Hertz model (Hertz, 1882) assuming a Poisson ratio of υ = 0.5 as we estimated for PA gels (Buxboim et al., 2010b) and a pyramidal indenter (Domke and Radmacher 1998; Rehfeldt et al., 2007):
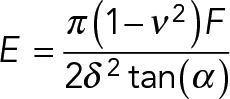 |
Supplementary Material
Acknowledgments
We thank Yun-Ruei C. Kao and Takamasa Harada for considerable technical assistance. The research was supported by National Institutes of Health/National Cancer Institute PSOC Award U54 CA193417, National Heart Lung and Blood Institute Awards R01 HL124106 and R21 HL128187, the US–Israel Binational Science Foundation, and a National Science Foundation Materials Science and Engineering Center grant to the University of Pennsylvania. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other granting agencies.
Abbreviations used:
- AFM
atomic force microscopy
- blebb
blebbistatin
- HSCP
hematopoietic stem cells and progenitor
- KD
knockdown
- LBR
lamin-B receptor
- MGC
mechanobiological gene circuit
- MSC
mesenchymal stem cells
- OE
overexpression
- RT
room temperature
- SRF
serum response factor
- STD
standard deviation
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0393) on September 20, 2017.
REFERENCES
- Alam SG, Lovett D, Kim DI, Roux KJ, Dickinson RB, Lele TP. The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J Cell Sci. 2015;128:1901–1911. doi: 10.1242/jcs.161703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcioglu HE, van Hoorn H, Donato DM, Schmidt T, Danen EH. The integrin expression profile modulates orientation and dynamics of force transmission at cell matrix adhesions. J Cell Sci. 2015;128:1316–1326. doi: 10.1242/jcs.156950. [DOI] [PubMed] [Google Scholar]
- Ben-Ze’ev A, Farmer SR, Penman S. Protein synthesis requires cell-surface contact while nuclear events respond to cell shape in anchorage-dependent fibroblasts. Cell. 1980;21:365–372. doi: 10.1016/0092-8674(80)90473-0. [DOI] [PubMed] [Google Scholar]
- Beningo KA, Dembo M, Wang Y-L. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci USA. 2004;101:18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- Buxboim A, Ivanovska IL, Discher DE. Matrix elasticity, cytoskeletal forces and physics of the nucleus: how deeply do cells “feel” outside and in? J Cell Sci. 2010a;123:297–308. doi: 10.1242/jcs.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Rajagopal K, Brown AEX, Discher DE. How deeply cells feel: methods for thin gels. J Phys Condens Matter. 2010b;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PC, Athirasala A, Kao YR, Cho S, Harada T, Shin JW, Discher DE. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr Biol. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Dingal PC, Bradshaw AM, Cho S, Raab M, Buxboim A, Swift J, Discher DE. Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nat Mater. 2015;14:951–960. doi: 10.1038/nmat4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Domke J, Radmacher M. Measuring the elastic properties of thin polymer films with the atomic force microscope. Langmuir. 1998;14:3320–3325. [Google Scholar]
- Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004a;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Griffin MA, Sen S, Bonnetnann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004b;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamin-A and lamin-C reveals primary and secondary structural homology to intermediate filament proteins. Proc Natl Acad Sci USA. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatos SD, Maroulakou I, Blobel G. Lamin-A, lamin-B, and lamin-B receptor analogs in yeast. J Cell Biol. 1989;108:2069–2082. doi: 10.1083/jcb.108.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatos SD, Stournaras C, Blobel G. Heterotypic and homotypic associations between the nuclear lamins: site-specificity and control by phosphorylation. Proc Natl Acad Sci USA. 1988;85:4325–4329. doi: 10.1073/pnas.85.12.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Fernández A, Soria-Valles C, Osorio FG, Gutiérrez-Abril J, Garabaya C, Aguirre A, Fueyo A, Fernández-García MS, Puente XS, López-Otín C. Loss of MT1-MMP causes cell senescence and nuclear defects which can be reversed by retinoic acid. EMBO J. 2015;34:1875–1888. doi: 10.15252/embj.201490594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden WJ, Young JL, Holle AW, McFetridge ML, Kim DY, Wijesinghe P, Taylor-Weiner H, Wen JH, Lee AR, Bieback K, et al. Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc Natl Acad Sci USA. 2017;114:5647–5652. doi: 10.1073/pnas.1618239114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz H. Über die Berührung fester elastischer Körper. J Reine Angew Math. 1882;92:156–171. [Google Scholar]
- Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:1354–1362. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanovska IL, Liu AJ, Greenberg RA, Discher DE. Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol Biol Cell. 2016;27:4011–4020. doi: 10.1091/mbc.E16-06-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Swift J, Martins RP, McPhail GD, Knight MM, Discher DE, Lee DA. Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophys J. 2013;104:759–769. doi: 10.1016/j.bpj.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Vaughan OA, Hutchison CJ, Gilbert DM. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol Biol Cell. 2000;11:4323–4337. doi: 10.1091/mbc.11.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E, Brand CA, Schwarz US, Kumar S. Geometry and network connectivity govern the mechanics of stress fibers. Proc Natl Acad Sci USA. 2017;114:2622–2627. doi: 10.1073/pnas.1606649114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res. 2013;23:1420–1423. doi: 10.1038/cr.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochin V, Shimi T, Torvaldson E, Adam SA, Goldman A, Pack CG, Melo-Cardenas J, Imanishi SY, Goldman RD, Eriksson JE. Interphase phosphorylation of lamin A. J Cell Sci. 2014;15:2683–2696. doi: 10.1242/jcs.141820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N, Voncken JW, Konings G, van Weeghel M, van den Hoogenhof MMG, Gijbels M, van Erk A, Schoonderwoerd K, van den Bosch B, Dahlmans V, et al. Post-natal myogenic and adipogenic developmental defects and metabolic impairment upon loss of A-type lamins. Nucleus-Austin. 2011;2:195–207. doi: 10.4161/nucl.2.3.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Neumann FR, Jones AW, Snijders AP, Nurse P. A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control. PLos Genet. 2017;13:e1006767. doi: 10.1371/journal.pgen.1006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu HM, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum Mol Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- Ma XF, Jana SS, Conti MA, Kawamoto S, Claycomb WC, Adelstein RS. Ablation of nonmuscle myosin II-B and II-C reveals a role for nonmuscle myosin II in cardiac myocyte karyokinesis. Mol Biol Cell. 2010;21:3952–3962. doi: 10.1091/mbc.E10-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacci G, Wolfenson H, Liu S, Stachowiak MR, Iskratsch T, Mathur A, Ghassemi S, Gauthier N, Tabdanov E, Lohner J, et al. α-Actinin links ECM rigidity sensing contractile units with periodic cell edge retractions. Mol Biol Cell. 2016;27:3471–3479. doi: 10.1091/mbc.E16-02-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Geiger B, Safran SA. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proc Natl Acad Sci USA. 2004;101:12520–12525. doi: 10.1073/pnas.0403539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Raab M, Swift J, Dingal PCDP, Shah P, Shin JW, Discher DE. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt F, Engler AJ, Eckhardt A, Ahmed F, Discher DE. Cell responses to the mechanochemical microenvironment—implications for regenerative medicine and drug delivery. Adv Drug Deliv Rev. 2007;59:1329–1339. doi: 10.1016/j.addr.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholt FP, Hultenby K, Oldberg A, Heinegård D. Osteopontin—a possible anchor of osteoclasts to bone. Proc Natl Acad Sci USA. 1990;87:4473–4475. doi: 10.1073/pnas.87.12.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridler TW, Calvard S. Picture thresholding using an iterative selection method. IEEE Trans Syst Man Cybern. 1978;8:630–632. [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang RH, Heitner SG, et al. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz FX, Gallego O, Ardèvol A, Moro A, Domínguez M, Alvarez S, Alvarez R, de Lera AR, Rovira C, Fita I, et al. Aldo-keto reductases from the AKR1B subfamily: retinoid specificity and control of cellular retinoic acid levels. Chem Biol Interact. 2009;178:171–177. doi: 10.1016/j.cbi.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Sackmann E. Membrane bending energy concept of vesicle-shape and cell-shape and shape-transitions. FEBS Lett. 1994;346:3–16. doi: 10.1016/0014-5793(94)00484-6. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JW, Swift J, Spinler KR, Discher DE. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 2011;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek J, McKee MD. Molecular and cellular biology of alveolar bone. Periodontol 2000. 2000;24:99–126. doi: 10.1034/j.1600-0757.2000.2240106.x. [DOI] [PubMed] [Google Scholar]
- Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian G, Chaudhury P, Malu K, Fowler S, Manmode R, Gotur D, Zwerger M, Ryan D, Roberti R, Gaines P. Lamin B receptor regulates the growth and maturation of myeloid progenitors via its sterol reductase domain: implications for cholesterol biosynthesis in regulating myelopoiesis. J Immunol. 2012;188:85–102. doi: 10.4049/jimmunol.1003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talele NP, Fradette J, Davies JE, Kapus A, Hinz B. Expression of α-smooth muscle actin determines the fate of mesenchymal stromal cells. Stem Cell Rep. 2015;4:1016–1030. doi: 10.1016/j.stemcr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O. The molecular architecture of lamins in somatic cells. Nature. 2017;543:261–264. doi: 10.1038/nature21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis-Vlug AG, Rosen CJ. Mechanisms of marrow adiposity and its implications for skeletal health. Metabolism. 2017;67:106–114. doi: 10.1016/j.metabol.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Fujita N, Sato Y, Kobayashi T, Morita M, Oike T, Miyamoto K, Kuro OM, Michigami T, Fukumoto S, et al. Enpp1 is an anti-aging factor that regulates Klotho under phosphate overload conditions. Sci Rep. 2017;7:7786. doi: 10.1038/s41598-017-07341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P, Garber B. Shape and movement of mesenchyme cells as functions of the physical structure of the medium—contributions to a quantitative morphology. Proc Natl Acad Sci USA. 1952;38:264–280. doi: 10.1073/pnas.38.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenson H, Iskratsch T, Sheetz MP. Early events in cell spreading as a model for quantitative analysis of biomechanical events. Biophys J. 2014;107:2508–2514. doi: 10.1016/j.bpj.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Yuan J, Blobel G, Georgatos SD. A lamin B-receptor in the nuclear-envelope. Proc Natl Acad Sci USA. 1988;85:8531–8534. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam PT, Wilson CA, Ji L, Hebert B, Barnhart EL, Dye NA, Wiseman PW, Danuser G, Theriot JA. Actin–myosin network reorganization breaks symmetry at the cell rear to spontaneously initiate polarized cell motility. J Cell Biol. 2007;178:1207–1221. doi: 10.1083/jcb.200706012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Munck M, Swaminathan K, Kapinos LE, Noegel AA, Neumann S. Mutations in LMNA modulate the lamin A–Nesprin-2 interaction and cause LINC complex alterations. PLoS One. 2013;8:e71850. doi: 10.1371/journal.pone.0071850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Primary structure-Analysis and lamin-B and DNA-binding of human LBR, an integral protein of the nuclear-envelope inner membrane. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming WY, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- Zemel A, Rehfeldt F, Brown AEX, Discher DE, Safran SA. Optimal matrix rigidity for stress-fibre polarization in stem cells. Nat Phys. 2010;6:468–473. doi: 10.1038/nphys1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuela N, Zwerger M, Levin T, Medalia O, Gruenbaum Y. Impaired mechanical response of an EDMD mutation leads to motility phenotypes that are repaired by loss of prenylation. J Cell Sci. 2016;129:1781–1791. doi: 10.1242/jcs.184309. [DOI] [PubMed] [Google Scholar]
- Zwerger M, Herrmann H, Gaines P, Olins AL, Olins DE. Granulocytic nuclear differentiation of lamin B receptor-deficient mouse EPRO cells. Exp Hematol. 2008;36:977–987. doi: 10.1016/j.exphem.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.