Abstract
The variability in Helicobacter pylori vacA and cagA genes has been related to the progression of the gastrointestinal disease; also the presence of H. pylori in the oral cavity has been associated with periodontal disease in adults, but, in children without dyspeptic symptoms, little is known about this. We evaluated the prevalence of H. pylori and the presence of vacA/cagA genotypes in the oral cavity of Mexican children without dyspeptic symptoms. The gingival status was measured, and dental plaque samples (n = 100) were taken. 38% of children were positive for H. pylori 16S rRNA gene by qPCR. A significant association between H. pylori oral infection and gingival status was observed (P < 0.001). In 34.6% (9/26) of mild gingivitis cases, s1m2 genotype was found, while s1m1 was typed in 50% (3/6) of moderate gingivitis. The cagA prevalence among H. pylori-positive children was 80.8% (21/26), 83.3% (5/6), and 16.7% (1/6) of cases of mild gingivitis, moderate gingivitis, and nongingivitis, respectively (P < 0.001). The s1m1/cagA+ combinational genotype was the most detected in children with gingivitis. Our results suggest that the prevalence of H. pylori and detection of vacA/cagA genotypes-associated gastrointestinal disease in the oral cavity could be related to the progression of gingivitis in asymptomatic children.
1. Introduction
Helicobacter pylori is a Gram-negative species which colonizes gastric mucosa in humans [1]. H. pylori infection is associated with gastrointestinal tract diseases including chronic gastritis, peptic ulcer, mucosa-associated lymphoproliferative disorders, and gastric cancer [2–4]. The global prevalence of H. pylori infection is more than 50%. This prevalence may vary significantly within and among countries, according to geography, ethnicity, age, and socioeconomic factors [5, 6]. It has been reported that H. pylori infection rate is up to 70% in developing countries, while the rates in developed countries and regions, such as Australia and Western Europe, are only 25% and 28%, respectively [7]. Socioeconomic factors also explain a significant proportion of the difference in H. pylori prevalence. In the third National Health and Nutrition Examination Survey conducted in the United States, a 25% prevalence of H. pylori infection was found in children and young adults between 6 and 19 years. In the American population, prevalence was 42%; prevalence was higher in children of a low socioeconomic status, in those whose mothers had a lower education level, and in those living in crowded conditions [8]. O'Rourke et al. evaluated the H. pylori infection in Mexican and American children living on both sides of the Rio Grande (a river that is dividing both countries); a slightly higher prevalence was observed in Mexicans compared to Americans [9]. In a similar study by Goodman et al. they found a seroprevalence of 74% and 56% of Mexican and American women, respectively [10]. The relationship between socioeconomic status and H. pylori infection has been reported too in countries such as Bolivia [11].
Epidemiologic studies have shown evidence that most infections are acquired in childhood, but the specific age of acquisition and the factors associated with its persistence are not clear [12]. In developing countries, 70 to 90% of the population become infected during childhood; in developed countries, a smaller percentage (~10%) of children become infected, and the prevalence of infection increases with age [10]. In Mexico, a seroepidemiological national survey in 1998 found a national prevalence of 66% in the general population, 20% in children younger than one year, 50% in children younger than ten years, and 70% in adults younger than 20 years. There were differences in prevalence, depending on the economic development of the regions (86.1% prevalence in southeastern Mexico, 47.1% in the northeast) [13].
H. pylori infection mainly spreads through consumption of contaminated food and drinks or is transmitted orally. Several studies have evaluated the oral cavity, and H. pylori has been detected in dental plaque and saliva [14–16]. It is known that the presence of H. pylori in the oral cavity is one of the main causes of the reappearance of gastric H. pylori infection and that treatment of oral infection significantly increases eradication of H. pylori infection in the stomach [17]. Nevertheless, the role of dental plaque as an extragastric reservoir in H. pylori transmission is controversial [18–20]. Also, the oral H. pylori infection has been related to oral cavity problems, like gingivitis, periodontitis [21, 22], and recurrent aphthous stomatitis (RAS) [23]. Nisha et al. showed a highly significant association was found between periodontal disease and colonization of H. pylori in dental plaque [24].
H. pylori has many virulence factors that allow establishment, colonization, and damage to the gastric epithelium. An important virulence factor secreted by H. pylori is VacA cytotoxin, which induces vacuolization and cell death in epithelial cells. The encoding vacA gene is present in all H. pylori strains, but it is polymorphic, comprising variable signal regions (type s1 or s2) and midregions (type m1 or m2) [3, 26]. Strains with alleles s1/m1 secrete an active toxin that is associated with the development of peptic ulcers and gastric cancer; also, this region was recently described as a determining factor in cytotoxicity and development of gastric disorders [27]. Some strains of H. pylori produce the CagA cytotoxin, as part of the cag pathogenicity island (PAI), a region of 40 kb DNA encoding about 31 genes that make up a type IV secretion system, which allows the release of oncoprotein CagA directly to the cytosol of gastric epithelial cells [28]. Upon release, CagA leads to dephosphorylation of cellular proteins, changes in morphology, increased cell proliferation, chronic inflammation, and carcinogenesis processes [29]. It is reported that individuals infected with cagA-positive and certain cagA alleles (e.g., cagA1a) have a higher risk of developing the peptic ulcer and gastric cancer [30].
Recently, some studies have evaluated the prevalence of H. pylori cagA and vacA genes in saliva and dental plaque [31–33], but its frequency has not been extensively studied in Mexico.
In this study, we evaluated the prevalence of H. pylori and the presence of vacA genotypes and cagA gene in dental plaque of Mexican children without dyspeptic symptoms.
2. Materials and Methods
2.1. Patients
From November to December 2015, one hundred Mexican children (from 3 to 16 years old) from the Back2Back nonprofit organization at Monterrey city, Nuevo León, Mexico, were selected to participate in this study. The exclusion criteria were as follows: (1) current history of antibiotic usage or during the previous two months, (2) systemic disease, (3) dyspeptic symptoms, and (4) history of gastrointestinal diseases. Before selection, the informed consent form was obtained from guardians and authorities, who declared the willingness of children to allow the use of the samples and anonymous data for research purposes, and also this study was approved by The Research Bioethics Committee in Dental Science College at Universidad Autónoma de Nuevo León (UANL).
2.2. Measurement of Gingivitis
The modified gingival index (MGI), defined by Lobene et al., is a noninvasive method (no probing) [34]. The MGI was used to assess gingival inflammation in participants, as follows: (0) normal (no inflammation), (1) mild inflammation (slight change in color, little change in texture) of any portion of the gingival unit, (2) mild inflammation of the entire gingival unit, (3) moderate inflammation (moderate glazing, redness, edema, and/or hypertrophy) of the entire gingival unit, and (4) severe inflammation (marked redness and edema/hypertrophy, spontaneous bleeding, or ulceration) of the gingival unit. The clinical examination was performed by the same observer.
2.3. Sample Collection
Dental plaque was collected with a sterile bamboo skewer by a downward scrape against the vestibular surface (supragingival) of the superior first molar. After that, the sample was suspended in 1.5 ml of trypticase soy broth (TS; Becton Dickinson, Franklin Lakes, NJ) supplemented with 30% (w/v) glycerol for the maintenance of microbial viability and then storage at −70°C, until use.
2.4. DNA Extraction
Dental plaque samples were centrifuged at 12,000 ×g for 2 min, then washed with 0.01 m Phosphate-Buffered Saline (PBS; 0.1 m, pH 7.4), and suspended in 100 μl Tris-EDTA buffer (TE; 10 mM Tris and 1 mM EDTA, at pH 7.4). For enzymatic cell lysis, 10 μl of lysozyme (10 mg/ml) and 10 μl of Proteinase K (10 mg/ml) were added and incubated at 56°C for 30 min. Total DNA was extracted using a High Pure PCR template preparation kit (Roche Diagnostics, GmbH, Mannheim, Germany) according to the manufacturer's recommendations and the DNA concentration was measured at 260 nm in a spectrophotometer (NanoDrop 8000 UV-Vis; Thermo Scientific, Wilmington, DE). The DNA samples were stored at −20°C, until use.
2.5. Detection of H. pylori and cagA Gene by qPCR
The presence of H. pylori and cagA status were assessed by qPCR using primers/probe for 16s rRNA gene and cagA gene, respectively (Table 1). The qPCR primers/probes were designed by IDT PrimerQuest tool (http://www.idtdna.com/primerquest/), DNA sequences were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/), and BLAST search (http://blast.ncbi.nlm.nih.gov/) was performed to check the specificity of the primers/probe sequences. The efficiency of primer pairs and probes was evaluated by performing serial dilutions from 0.1 to 100 ng of DNA, and the value of the slope was obtained by simple linear regression analysis (R2); the primers and probes showed ≥95% efficiency values. The qPCR reactions were performed in a 96-well plates containing 12.5 μL of 2x maxima probe qPCR master mix (Thermo Scientific, Carlsbad, California, USA), 0.3 μM primers mix forward/reverse, 0.2 μM probe, and 100 ng of the DNA sample, in free-nucleases water to a final volume of 25 μL; 100 ng of DNA from H. pylori ATCC 700824 or ATCC43504 strains and nuclease-free water were added as positive and negative control, respectively. The qPCR assay was carried out in a LightCycler 480II thermocycler (Roche, Mannheim, Germany). The thermocycler was programmed with a monocolor hydrolysis probe format (6-FAM, filter combination 465–510) as follows: one denaturalization cycle (95°C, 10 min), thirty-five amplification cycles (95°C, 10 s, 4°C/s ramp rate; 55°C, 15 s, 2°C/s ramp rate; 72°C, 15 s, 4°C/s quantification analysis ramp rate), one melting cycle (95°C, 5 s, 4°C/s ramp rate; 65°C, 1 min, 2.2°C/s ramp rate, 97°C with a 5°C continuous acquisition), and one cooling cycle (40°C, 10 s, 1.5°C/s ramp rate).
Table 1.
PCR primers and probes used for detection and genotyping of H. pylori.
| Gen | 5′-3′ sequence | Product size (bp) |
Reference |
|---|---|---|---|
| 16S rRNA | F: GATAGTCAGTCAGGTGTGAAATCC | 196 | In this study |
| R: GTTTGCTCCCCACGCTTT | |||
| Probe: AAAATCCGTAGAGATCAAGAGGA | |||
| cagA | F: GACCGACTCGATCAAATAGCA | 113 | |
| R: TTAGCTGAAAGCCCTACCTTAC | |||
| Probe: AGTGGTTTGGGTGATGTAGGGCAA | |||
|
| |||
| vacA s | vacA1F: CTGGT(C/T)TAAAGTCGCACCCTTTGTGC |
s1: 120 s2: 150 |
Koehler et al. [25] |
| vacA1R: CAATGGCTGGAATGATCACGGTTGT(A/G) | |||
| vacA2F: CAAACACACCGCAAAATCAATCGCCC | |||
| vacA m1 | m1F1: CAACAATCAAGGCACTATCAA(C/T)TA | 301 | |
| m1R1: CCGCATGCTTTAATGTCATCAG | |||
| m1F2: TGGTCCGAGGCGGG(A/C)AAGT | |||
| m1R2: TCATCAGTATTTCGCACCACAC | |||
| vacA m2 | m2F1: TTTGGAGC(C/T)CCAGGAAACATTG | 102 | |
| m2R1: C(C/T)ACACGCCCATCTTGGACAA | |||
| m2F2: ACCCTAAA(C/T)AGCAACGCAAGC | |||
| m2R2: GACAAAAAGATTCATCGTGCCTT | |||
2.6. Detection of H. pylori vacA Alleles
The vacA gene and “s” and “m” region genotyping was performed by nested PCR using a set of oligonucleotides for each gene fragment, as previously described by Koehler et al. [25]. For the signal region (s) detection, vacA1F/vacA1R and vacA2F/vacA1R oligonucleotides were used in the first and the second amplification rounds, respectively. For the middle region 1 (m1) allele detection in the first round m1F1/m1R1 primers were used; for the second round, m1F2 and m1R2 were used. For middle region 2 (m2), m2F1/m2R1 and m2F2/m2R2 primers were employed in the first and the second runs, respectively (Table 1).
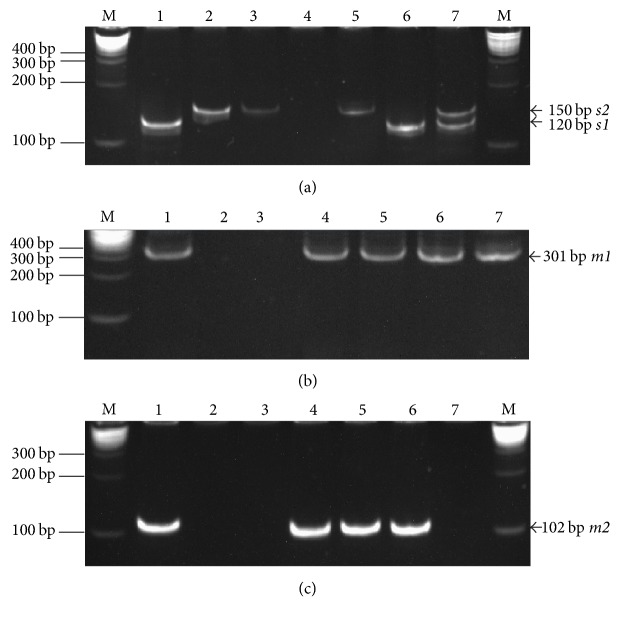
All the PCR were prepared using 1x NH4 reaction buffer, 3 mM MgCl2, 10 μM of each deoxynucleoside triphosphate, 0.2 μM of each primer, 2.5 units Taq Polymerase (Bioline USA Inc., Taunton, USA), and 200 ng of DNA up to a final volume of 25 μL. The PCR were carried out in MJ Mini Thermal Cycler (Bio-Rad Lab). The PCR program comprised 1 cycle of initial denaturation (95°C, 5 min), 35 cycles of denaturation (95°C, 1 min), annealing (58°C, 1 min) and extension (72°C, 1 min), and 1 cycle of final extension (72°C, 5 min). For the second amplification round, 1 μl DNA template from the first PCR and 0.2 μM of each primer were used. PCR was performed according to the PCR protocol described above. The PCR products were visualized on 8% (v/v) acrylamide gel (Bio-Rad Lab) in Tris-Borate-EDTA buffer (TBE; 40 mM Tris, 45 mM boric acid, 1 mM EDTA, at pH 7.4). The gel was stained with 1 μg/mL of ethidium bromide solution and transferred to ultraviolet transilluminator (Gel Doc XR+ Imager; Bio-Rad Lab). The sizes of the amplicons were estimated by comparison with a DNA size marker (Quick-Load® 2-Log DNA Ladder; New England Biolabs, MA, USA). 120 bp and 150 bp PCR products corresponded to s1 and s2 alleles, respectively, while 301 bp and 102 bp bands corresponded to m1 and m2 alleles, respectively. H. pylori ATCC43504 strain was used as positive control (Figure 1).
Figure 1.
Results of the gel electrophoresis for identification of H. pylori vacA alleles and genotypes in dental plaque samples of Mexican children. (a) Detection of s alleles. Lane M, molecular weight marker 0.1 to 10 kb (BioLabs Inc.); lane 1, positive control (DNA from H. pylori ATCC 43504 s1m1 genotype); lanes 2-3, positive control (DNA from gastric biopsy with H. pylori s2m2 genotype); lane 4, negative control (without DNA); lane 5, s2 allele positive (DNA from sample); lane 6, s1 allele positive (DNA from sample); lane 7, s1 and s2 alleles positive (DNA from sample). (b) Detection of m1 allele. Lane M, molecular weight marker 0.1 to 10 kb (BioLabs Inc.); lane 1, positive control (DNA from H. pylori ATCC 43504 s1m1 genotype); lane 2, negative control (without DNA); lane 3, m1 allele negative (DNA from sample); lanes 4–7, m1 allele positive (DNA from sample). (c) Detection of m2 alleles. Lane M, molecular weight marker 0.1 to 10 kb (BioLabs Inc.); lane 1, positive control (DNA from gastric biopsy with H. pylori vacA s2m2 genotype); lane 2, negative control (without DNA); lanes 3 and 7, m2 allele negative (DNA from sample); lanes 4–6, m2 allele positive (DNA from sample).
2.7. Statistical Analysis
The association between H. pylori oral infection and each nominal variable were determined by the Chi-square and Fisher's exact tests; one-way ANOVA tests, post hoc Tukey tests, or Student's t-tests were used for numeric variables. The P < 0.05 value was considered statistically significant. The data were analyzed using SPSS IBM statistics software v22.0.
3. Results
3.1. The History of Participants and H. pylori Detection
This study was designed to determine the frequency of H. pylori cagA and vacA genes in dental plaque of children from the northeastern Mexico. A total of 100 children without gastric clinical manifestations, 50 males and 50 females ranging in age from 3 to 16 years with an average age of 8.95 ± 3.72, were included. Also, the participants were divided according to the gingival clinical presentation, 59 (59.0%) were diagnosed as no inflammation, 35 (35.0%) were diagnosed as mild inflammation, and 6 (6.0%) were regarded as moderate inflammation (Table 2). Among 41 gingivitis cases, 62.1% (22/35) of mild inflammation cases were male, while 83% (5/6) of moderate inflammation cases were female. A significant difference (P < 0.05) was observed between the mean age of patients with moderate inflammation (4.83 ± 2.04 years old) and those who did not present inflammation (9.47 ± 3.42 years old), as shown in Table 3.
Table 2.
Prevalence of H. pylori in dental plaque from Mexican children by gender and age and the gingival clinical presentation.
| Total N = 100 | H. pylori 16S rRNA gene | P value | ||
|---|---|---|---|---|
| Positive N = 38 | Negative N = 52 | |||
| Gender n (%) | ||||
| Female | 50 (50.0) | 17 (44.7) | 33 (53.2) | 0.41a |
| Male | 50 (50.0) | 21 (55.3) | 29(46.8) | |
| Age | ||||
| Mean (±SD) | 8.95 (±3.72) | 8.37 (±4.17) | 9.31 (±3.4) | 0.2425b |
| Age groups n (%) | ||||
| 3–5 | 21 (21.0) | 12 (31.6) | 9 (14.5) | 0.084a |
| 6–8 | 33 (33.0) | 10 (26.3) | 23 (37.1) | |
| 9–11 | 14 (14.0) | 2 (5.3) | 12 (19.4) | |
| 12–14 | 26 (26.0) | 12 (31.6) | 14 (22.6) | |
| 15-16 | 6 (6.0) | 2 (5.3) | 4 (6.5) | |
| MGI n (%) | ||||
| No inflammation | 59 (59.0) | 6 (15.8) | 53 (85.5) | <0.001c |
| Mild inflammation | 35 (35.0) | 26 (68.4) | 9 (14.5) | |
| Moderate inflammation | 6 (6.0) | 6 (15.8) | 0 | |
aChi-square test. bStudent's t test. cExact Fisher test. MGI: modified gingival index.
Table 3.
Distribution of the gingival clinical presentation by gender and age.
| Modified gingival index | P value | |||
|---|---|---|---|---|
| No inflammation N = 59 |
Mild inflammation N = 35 |
Moderate inflammation N = 6 |
||
| Gender n (%) | ||||
| Female | 32 (54.2) | 13 (37.1) | 5 (83.3) | 0.078a |
| Male | 27 (45.8) | 22 (62.1) | 1 (16.7) | |
| Age | ||||
| Mean (±SD) | 9.47 (±3.42) | 8.77 (±4.02) | 4.83 (±2.04) | 0.012b |
| Age groups n (%) | ||||
| 3–5 | 8 (13.6) | 10 (28.6) | 3 (50.0) | 0.135a |
| 6–8 | 22 (37.3) | 8 (22.9) | 3 (50.0) | |
| 9–11 | 11 (18.6) | 3 (8.6) | 0 | |
| 12–14 | 14 (23.7) | 12 (34.3) | 0 | |
| 15-16 | 4 (6.8) | 2 (5.7) | 0 | |
aExact Fisher test. bOne-way ANOVA test.
The detection of H. pylori 16S rRNA gene by qPCR revealed that 38% of samples were positive. Among H. pylori-positive children, 55.3% (21/38) and 44.7% (17/38) were male and female, respectively. The mean age of H. pylori-positive individuals was 8.37 ± 4.17 years (Table 2).
3.2. Relationship between the H. pylori Oral Infection and the Gingival Clinical Presentation
A significant association between H. pylori oral detection and gingival status was observed (P < 0.001). 68.4% (26/38) of H. pylori-positive children shown mild inflammation, while 15.8% (6/38) showed no inflammation; all of the cases of moderate inflammation were positive (Table 2).
3.3. Frequency of H. pylori vacA Alleles and Association of Genotypes with the Gingival Clinical Presentation
The vacA allelic variants were determined in patients with dental plaque H. pylori-positive, as shown in Figure 1. The vacA s1 and m1 alleles were most frequent for the signal and middle region, respectively. The vacA s1m2 genotype was the most common among H. pylori-positive patients, with 28.9% (11/38), followed by s1m1 genotypes with 26.3% (10/38) (Table 4). The coinfection of s1m1 with s2m1 was observed in 13.2% (5/38) of H. pylori-positive samples. In 5.3% (2/38) of patients, the s1 allele was detected, but the m region was undetectable (s1m0); the s2 allele was identified in 1 patient, but the m region could not be identified (s2m0).
Table 4.
Distribution of vacA genotypes, cagA status, and combinational vacA/cagA genotypes in dental plaque from H. pylori-positive children with gingival clinical presentation.
| Total N = 38 |
H. pylori-positive | |||
|---|---|---|---|---|
| No inflammation N = 6 |
Mild inflammation N = 26 |
Moderate inflammation N = 6 |
||
| a vacA alleles n (%) | ||||
| s1 | 31 (81.6) | 3 (50.0) | 22 (84.6) | 6 (100.0) |
| s2 | 10 (26.3) | 3 (50.0) | 6 (23.1) | 1 (16.7) |
| m1 | 24 (63.2) | 4 (66.7) | 15 (57.7) | 5 (83.3) |
| m2 | 16 (42.1) | 1 (16.7) | 13 (50.0) | 2 (33.3) |
| m0 | 3 (7.9) | 1 (16.7) | 2 (7.7) | 0 |
| vacA genotypes | ||||
| s1m1 | 10 (26.3) | 1 (16.7) | 6 (23.1) | 3 (50.0) |
| s1m2 | 11 (28.9) | 1 (16.7) | 9 (34.6) | 1 (16.7) |
| s2m1 | 6 (15.8) | 3 (50.0) | 3 (11.5) | 0 |
| s1m0 | 2 (5.3) | 1 (16.7) | 1 (3.8) | 0 |
| s2m0 | 1 (2.6) | 0 | 1 (3.8) | 0 |
| bs1m1/s1m2 | 5 (13.2) | 0 | 4 (15.4) | 1 (16.7) |
| bs1m1/s2m1 | 3 (7.9) | 0 | 2 (7.7) | 1 (16.7) |
| cagA status | ||||
| Positive | 27 (71.1) | 1 (16.7) | 21 (80.8) | 5 (83.3) |
| Negative | 11 (28.9) | 5 (83.3) | 5 (19.2) | 1 (16.7) |
| vacA/cagA genotypes | ||||
| s1m1/cagA+ | 9 (23.7) | 1 (16.7) | 5 (19.2) | 3 (50.0) |
| s1m1/cagA− | 1 (2.6) | 0 | 1 (3.8) | 0 |
| s1m2/cagA+ | 5 (13.2) | 0 | 5 (19.2) | 0 |
| s1m2/cagA− | 5 (13.2) | 1 (16.7) | 3 (11.5) | 1 (16.7) |
| s2m1/cagA+ | 4 (10.5) | 0 | 4 (15.4) | 0 |
| s2m1/cagA− | 3 (7.9) | 3 (50.0) | 0 | 0 |
| s1m1/s1m2/cagA+ | 4 (10.5) | 0 | 3 (11.5) | 1 (16.7) |
| s1m1/s2m1/cagA+ | 3 (7.9) | 0 | 2 (7.7) | 1 (16.7) |
| s1m1/s2m1/cagA− | 1 (2.6) | 0 | 1 (3.8) | 0 |
| cUndetermined | 3 (7.9) | 1 (16.7) | 2 (7.7) | 0 |
aFrequency and percentages based on H. pylori-positive patients. bCoexistence of genotypes; cvacA genotypes with m0 alleles; m0: nontypeable for middle region.
The prevalence of vacA genotypes and alleles varied with gingival status. The s2m1 genotype was detected in 50% (3/6) of H. pylori-positive children without gingivitis. In 34.6% (9/26) of mild inflammation cases, s1m2 genotype was found, while s1m1 allele combination was typed in 50% (3/6) of moderate inflammation (Table 4). The coinfection of s1m1/s1m2 was observed only in patients with gingivitis (4 and 1 for mild and moderate inflammation, resp.).
3.4. Association between the Presence of H. pylori cagA Gene and Gingival Clinical Presentation
We found that the prevalence of cagA gene was 27% (27/100). In 71.1% (27/38) of H. pylori-positive samples, cagA gene was detected (Table 4). The cagA gene status has shown a significant relationship with gingival status (P < 0.001). Also, the cagA prevalence among H. pylori-positive children was 80.8% (21/26), 83.3% (5/6), and 16.7% (1/6) of cases of mild gingivitis, moderate gingivitis, and nongingivitis, respectively.
The combinational s1m1/cagA+ genotype was detected in 23.7% (9/38) of H. pylori-positive children. The same genotypic combination of vacA/cagA was the most frequent in children with gingivitis (5/26 and 3/6 of cases with mild and moderate gingivitis, resp.), while s2m1/cagA− combinational genotype was most prevalent in children without gingivitis (Table 4).
4. Discussion
It has been demonstrated that H. pylori infection is a risk factor for the development of gastric pathologies, including gastric cancer [4]. Several studies of H. pylori prevalence have shown the strong association with sociodemographic and socioeconomic factors where the developing countries are usually the most affected [8, 10–13]. The gastric infection is mainly acquired during the first years of life, where common symptoms are not present. Several studies have concluded that the high frequency of H. pylori could be due to its multiple mechanisms of transmission, where the oral cavity plays a determinant role as a source of transmission to other hosts and a focus for the continuous gastric reinfection after eradication therapy [14]. However, some researchers do not agree with this approach.
In this study, the detection of H. pylori and cagA gene was performed by qPCR. The use of TaqMan qPCR probes would allow accurate identification of H. pylori and would prevent the detection in the oral cavity of other microorganisms that are close phylogenetically to H. pylori, as Campylobacter species [35]. Our results show that 38% of dental plaque samples from children were H. pylori-positive. All participants were apparently healthy and did not report any gastric disease at the time of the evaluation (Table 2). Our results agree with those found by Duque et al. and those reported by Mendoza et al., both studies in Mexican school children, but these reports used urea breath test (UBT) for the diagnosis of infection [8, 36]. The agreement would be attributed to the fact that both diagnosis methods (UBT and PCR) have an estimated sensitivity of 75% to 100%. However, PCR has a higher specificity than UBT due to the presence of other urease-producing microorganisms in the stomach [37]. In 2014, Valdez-Gonzalez et al. evaluated the presence of H. pylori in dental plaques of 40 Mexican children using qPCR, and 35% of the samples were positive [16]. Our results are different from those reported by Castro-Muñoz et al.; they studied 162 asymptomatic Mexican children, and 13% of oral swab sample (from inside cheeks) were positive by PCR [15]. This difference may be due to the site from which samples were collected because the detection rates in dental plaques were more than those in other locations of oral cavity [14]. Zheng and Zhou evaluated the detection rate of H. pylori in subgingival and supragingival plaque samples from adults with/without periodontitis, and they observed a higher detection rate in subgingival plaques than that in supragingival plaques (P < 0.05) [22]. In our study, supragingival plaque samples were taken because it is a noninvasive sampling method (not probing) for children. It is possible that the microaerobic conditions and microbial associations in dental plaque could favor the persistence of H. pylori in the oral cavity, constituting an ideal site for sampling in H. pylori oral detection studies.
The presence of H. pylori in oral cavity would provide a foundation for a role in H. pylori transmission. Miyabayashi et al. investigated the effect of oral H. pylori on the stomach. The investigators observed that their patients with oral H. pylori were found to be at a significantly higher risk for gastric reinfection after having received adequate treatment. The authors also determined that drugs used for the eradication of the gastric H. pylori did not affect the oral H. pylori [38]. Most published studies have reported the oral cavity (dental plaque and saliva) as a source for subsequent gastric infections or reinfections in the same patient and as a reservoir for oral-oral transmission [19, 39]. However, some authors suggest that H. pylori is a microorganism transitory in the human oral cavity [40]. The mechanism by which H. pylori reaches the oral cavity is unknown. It is possible that the occasional reflux from the gastric reservoir allows colonization of the oral cavity. It is also possible that the reverse is true.
Our results indicate a significant frequency of H. pylori in dental plaque of asymptomatic children where the dental plaque would act as a reservoir for oral-oral dispersal to the population. However, our results cannot confirm the bacterial viability of oral H. pylori nor the ability to transmit to other individuals. This subject requires considerably more investigation due to the fact that the involvement of dental plaque in the transmission of H. pylori can be confirmed and that new measures can be tailored toward the prevention of oral spread.
It has been demonstrated that exposure to an H. pylori-positive family is a risk factor for persistence of infection in children under five years. Cervantes et al. followed a large cohort of children throughout the first years of life and concluded that when siblings are close in age, the older sibling may be an important source of H. pylori transmission for younger siblings [41]. In our study, we observed that 31.6% of H. pylori-positive subjects corresponded to the groups of age from 3 to 5 years (12/38) and 12 to 14 years (12/38). The children come from a care center for orphans where the coexistence between the older children and the younger ones would have favored the transmission among the children. However, this possibility could not be confirmed in this study. On the other hand, other studies have reported the familial transmission between parents and children [42, 43]. After many years of research, very little is known about the details of the modes of transmission of H. pylori and its propagation pathways. The primary modes of transmission are thought to be fecal-oral and oral-oral, but some indirect evidence has also been published for transmission via drinking water and other environmental sources [44].
Dental plaque harbors at least 400 different bacterial species and forms a biofilm in which organisms are intimately associated with each other and embedded in an exopolymeric matrix (salivary polymers and microbial extracellular products). This biofilm adheres to supragingival and subgingival tooth surfaces [19]. The accumulation of microorganisms and increased concentration of bacterial metabolites in gingiva induces an inflammatory process, called gingivitis. Gingivitis is one of the first clinical manifestations of the periodontal disease [45]. H. pylori oral infection has been related to oral pathologies, including gingivitis/periodontal disease, aphthous stomatitis, and oral cancer [5]. In this study, a significant relationship between the presence of H. pylori in dental plaque and the gingival clinical presentation was found (Table 2). In 1999, in Great Britain, Riggio and Lennon demonstrated the presence of H. pylori in 11/29 (38%) subgingival plaques of patients with chronic periodontitis. They suggested that, in this patient group at least, subgingival plaque may be a reservoir for H. pylori infection [46]. In 2012, Agarwal and Jithendra found that 60% of the samples were H. pylori-positive by PCR in periodontitis group compared to 15% in without-periodontitis group [47]. In Mexico, there are few reports about this association; De la Garza-Ramos et al. found a relationship between the detection of H. pylori in dental plaque and the depth of the periodontal pockets in adults [48].
It is known that certain genotypes of H. pylori are related to the severity of gastric pathology. Rudi et al. found that H. pylori strains of the vacA s1 allele and the cagA gene are more likely to result in peptic ulcer disease [49]. In Mexico, González-Valencia et al. found that vacA s1 and cagA+ strains were more frequent in adults than in children [50]. Another study, conducted by Martínez-Carrillo et al., confirmed that vacA s1m1 genotypes are the most common in patients with gastric ulcer and chronic gastritis [51]. However, Garza-Gonzalez et al. found that the most frequent genotype was s2m2 in H. pylori isolates from the northeastern region of Mexico, but the s1m1 genotype was associated with cagA-positive strains (P < 0.05) [52]. Our results show that the H. pylori strains with the vacA s1m1 and s1m2 genotypes are predominant in dental plaque from Mexican children without dyspeptic symptoms and agree with most of the ones found in Mexican patients with severe gastric pathology [52, 53].
In another study, the most frequent genotype in the oral cavity and gastric mucosa was vacA s1bm1 [54]. Román-Román et al. found that 47/196 (24%) patients were coinfected (saliva and gastric biopsy). H. pylori vacA s1m1 or s1m2 genotypes (either or both) were detected in 41.5% of the patients with chronic gastritis. The s1m1/s1m2 genotypes, alone or together, were found simultaneously in saliva and gastric biopsy from the same patient [32]. These results and others [33] support our findings; however, as far as we know this is the first study of genotyping vacA in oral samples from Mexican children without dyspepsia symptoms. Otherwise, we found that s1m1/s1m2 genotypes alone or together were mostly detected in participants with mild or moderate gingivitis (Table 4).
People infected with H. pylori who have a functional cag-PAI have increased mucosal concentrations of IL8, neutrophilic infiltration into the gastric mucosa, and increased risk of developing the gastric ulcer and cancer [29, 55, 56]; however other researchers did not agree to this [31]. Román-Román et al. conducted a study in a population from Southern Mexico, and they observed an association between H. pylori and the s1m1 genotype with gastric cancer, but cagA was not associated with the diagnosis [57]. In our study, the cagA status was related to the gingival clinical presentation in H. pylori-positive children (P < 0.001). The s1m1/cagA+ or s1m2/cagA+ combinational genotypes alone or together were mostly detected in dental plaque of children with mild or moderate gingivitis. It is possible that these genotypes play a role in the development of periodontal disease in asymptomatic people with poor oral hygiene measures.
5. Conclusions
Our results demonstrate that H. pylori oral infection is frequent among asymptomatic Mexican children and that, in one individual, more than one H. pylori strain may exist in dental plaque. The prevalence of H. pylori and detection of some vacA/cagA genotypes-associated gastrointestinal disease in the oral cavity could be related to the progression of gingivitis in children without dyspeptic symptoms.
Acknowledgments
This study was partly supported by grants from Programa de Apoyo a la Investigación Científica y Tecnológica from Universidad Autónoma de Nuevo León (PAICYT-UANL) to Myriam Angélica De la Garza-Ramos and by Consejo Nacional de Ciencia y Tecnología (CONACYT-México) to Alejandra Mendoza-Cantú. The authors also thank Unidad de Odontología Integral y Especialidades of Centro de Investigación y Desarrollo en Ciencias de la Salud (UOIE-CIDICS) for supporting the developing of this study.
Ethical Approval
All procedures were conducted in accordance with the Declaration of Helsinki (1964) and with the ethical standard of the Institutional Research Bioethics Committee in Dental Science College at Universidad Autónoma de Nuevo León (UANL).
Consent
The informed consent form was obtained from guardians and authorities, who declared the willingness of children to participate in this study.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
References
- 1.Marshall B. J., Warren J. R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. The Lancet. 1984;323(8390):1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Marshall B. J., McGechie D. B., Rogers P. A., Glancy R. J. Pyloric Campylobacter infection and gastroduodenal disease. Medical Journal of Australia. 1985;142(8):439–444. doi: 10.5694/j.1326-5377.1985.tb113444.x. [DOI] [PubMed] [Google Scholar]
- 3.Wroblewski L. E., Peek R. M., Jr., Wilson K. T. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clinical Microbiology Reviews. 2010;23(4):713–739. doi: 10.1128/cmr.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.IARC. (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans).Schistosomes, Liver Flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. 1994;61 [PMC free article] [PubMed] [Google Scholar]
- 5.Adler I., Muino A., Aguas S., et al. Helicobacter pylori and oral pathology: relationship with the gastric infection. World Journal of Gastroenterology. 2014;20(29):9922–9935. doi: 10.3748/wjg.v20.i29.9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nabwera H. M., Logan R. P. H. Epidemiology of Helicobacter pylori: transmission, translocation and extragastric reservoirs. Journal of Physiology and Pharmacology. 1999;50(5):711–722. [PubMed] [Google Scholar]
- 7.Rothenbacher D., Brenner H. Burden of Helicobacter pylori and H. pylori-related diseases in developed countries: recent developments and future implications. Microbes and Infection. 2003;5(8):693–703. doi: 10.1016/S1286-4579(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 8.Duque X., Vilchis J., Mera R., et al. Natural history of Helicobacter pylori infection in Mexican schoolchildren: incidence and spontaneous clearance. Journal of Pediatric Gastroenterology and Nutrition. 2012;55(2):209–216. doi: 10.1097/MPG.0b013e318248877f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Rourke K., Goodman K. J., Grazioplene M., Redlinger T., Day R. S. Determinants of geographic variation in Helicobacter pylori infection among children on the US-Mexico border. American Journal of Epidemiology. 2003;158(8):816–824. doi: 10.1093/aje/kwg219. [DOI] [PubMed] [Google Scholar]
- 10.Goodman K. J., O'Rourke K., Day R. S., et al. Helicobacter pylori infection in pregnant women from a U.S.-Mexico border population. Journal of Immigrant and Minority Health. 2003;5(3):99–107. doi: 10.1023/A:1023935701082. [DOI] [PubMed] [Google Scholar]
- 11.Glynn M. K., Friedman C. R., Gold B. D., et al. Seroincidence of Helicobacter pylori infection in a cohort of rural Bolivian children: acquisition and analysis of possible risk factors. Clinical Infectious Diseases. 2002;35(9):1059–1065. doi: 10.1086/342910. [DOI] [PubMed] [Google Scholar]
- 12.Torres J., Pérez-Pérez G., Goodman K. J., et al. A comprehensive review of the natural history of Helicobacter pylori infection in children. Archives of Medical Research. 2000;31(5):431–469. doi: 10.1016/s0188-4409(00)00099-0. [DOI] [PubMed] [Google Scholar]
- 13.Torres J., Leal-Herrera Y., Perez-Perez G., et al. A community-based seroepidemiologic study of Helicobacter pylori infection in Mexico. The Journal of Infectious Diseases. 1998;178(4):1089–1094. doi: 10.1086/515663. [DOI] [PubMed] [Google Scholar]
- 14.Anand P. S., Kamath K. P., Anil S. Role of dental plaque, saliva and periodontal disease in Helicobacter pylori infection. World Journal of Gastroenterology. 2014;20(19):5639–5653. doi: 10.3748/wjg.v20.i19.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro-Muñoz L. J., González-Díaz C. A., Muñoz-Escobar A., et al. Prevalence of Helicobacter pylori from the oral cavity of Mexican asymptomatic children under 5 years of age through PCR. Archives of Oral Biolog. 2017;73:55–59. doi: 10.1016/j.archoralbio.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Valdez-Gonzalez J. A., Mares-Moreno P. C., Kowolik M. J., Vargas-Villlarreal J., Gonzalez-Salazar F., De la Garza-Ramos M. A. Detection of Helicobacter pylori in dental plaque of mexican children by real-time PCR. Health. 2014;6(4):231–235. doi: 10.4236/health.2014.64034. [DOI] [Google Scholar]
- 17.Wang X. M., Yee K. C., Hazeki-Taylor N., et al. Oral Helicobacter pylori, its relationship to successful eradication of gastric H. pylori and saliva culture confirmation. Journal of Physiology and Pharmacology. 2014;65(4):559–566. [PubMed] [Google Scholar]
- 18.Perez-Perez G. I., Rothenbacher D., Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2004;9(1):1–6. doi: 10.1111/j.1083-4389.2004.00248.x. [DOI] [PubMed] [Google Scholar]
- 19.Al Sayed A., Anand P. S., Kamath K. P., Patil S., Preethanath R. S., Anil S. Oral cavity as an extragastric reservoir of Helicobacter pylori. ISRN Gastroenterology. 2014;2014:16. doi: 10.1155/2014/261369.261369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Salazar F., Gerardo-Aviles J., Castro G. J., et al. Dental plaque of children as probable Helicobacter pylori reservoir. African Journal of Microbiology Research. 2014;8(35):3220–3229. doi: 10.5897/AJMR2013.6465. [DOI] [Google Scholar]
- 21.Dye B. A., Kruszon-Moran D., McQuillan G. The relationship between periodontal disease attributes and Helicobacter pylori infection among adults in the United States. American Journal of Public Health. 2002;92(11):1809–1815. doi: 10.2105/AJPH.92.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng P., Zhou W. Relation between periodontitis and Helicobacter pylori infection. International Journal of Clinical and Experimental Medicine. 2015;8(9):16741–16744. [PMC free article] [PubMed] [Google Scholar]
- 23.Gülseren D., Karaduman A., Kutsal D., Nohutcu R. M. The relationship between recurrent aphthous stomatitis, and periodontal disease and Helicobacter pylori infection. Clinical Oral Investigations. 2016;20(8):2055–2060. doi: 10.1007/s00784-015-1704-0. [DOI] [PubMed] [Google Scholar]
- 24.Nisha K. J., Nandakumar K., Shenoy K. T., Janam P. Periodontal disease and Helicobacter pylori infection: a community-based study using serology and rapid urease test. Journal of Investigative and Clinical Dentistry. 2016;7(1):37–45. doi: 10.1111/jicd.12122. [DOI] [PubMed] [Google Scholar]
- 25.Koehler C. I., Mues M. B., Dienes H. P., Kriegsmann J., Schirmacher P., Odenthal M. Helicobacter pylori genotyping in gastric adenocarcinoma and MALT lymphoma by multiplex PCR analyses of paraffin wax embedded tissues. Molecular Pathology. 2003;56(1):36–42. doi: 10.1136/mp.56.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argent R. H., Thomas R. J., Letley D. P., Rittig M. G., Hardie K. R., Atherton J. C. Functional association between the Helicobacter pylori virulence factors VacA and CagA. Journal of Medical Microbiology. 2008;57(2):145–150. doi: 10.1099/jmm.0.47465-0. [DOI] [PubMed] [Google Scholar]
- 27.Jafari F., Shokrzadeh L., Dabiri H., et al. vacA genotypes of Helicobacter pylori in relation to cagA status and clinical outcomes in Iranian populations. Japanese Journal of Infectious Diseases. 2008;61(4):290–293. [PMC free article] [PubMed] [Google Scholar]
- 28.Chiarini A., Calà C., Bonura C., et al. Prevalence of virulence-associated genotypes of Helicobacter pylori and correlation with severity of gastric pathology in patients from western Sicily, Italy. European Journal of Clinical Microbiology & Infectious Diseases. 2009;28(5):437–446. doi: 10.1007/s10096-008-0644-x. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira R. M., Pinto-Ribeiro I., Wen X., et al. Helicobacter pylori cagA promoter region sequences influence CagA expression and interleukin 8 secretion. The Journal of Infectious Diseases. 2016;213(4):669–673. doi: 10.1093/infdis/jiv467. [DOI] [PubMed] [Google Scholar]
- 30.Momtaz H., Souod N., Dabiri H. Comparison of the virulence factors of Helicobacter pylori isolated in stomach and saliva in Iran. The American Journal of the Medical Sciences. 2010;340(5):345–349. doi: 10.1097/maj.0b013e3181d94fbc. [DOI] [PubMed] [Google Scholar]
- 31.Momtaz H., Souod N., Dabiri H., Sarshar M. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World Journal of Gastroenterology. 2012;18(17):2105–2111. doi: 10.3748/wjg.v18.i17.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Román-Román A., Giono-Cerezo S., Camorlinga-Ponce M., Martínez-Carrillo D. N., Loaiza-Loeza S., Fernández-Tilapa G. vacA genotypes of Helicobacter pylori in the oral cavity and stomach of patients with chronic gastritis and gastric ulcer. Enfermedades Infecciosas y Microbiología Clínica. 2013;31(3):130–135. doi: 10.1016/j.eimc.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Tilapa G., Axinecuilteco-Hilera J., Giono-Cerezo S., Martínez-Carrillo D.-N., Illades-Aguiar B., Román-Román A. vacA genotypes in oral cavity and Helicobacter pylori seropositivity among adults without dyspepsia. Medicina Oral Patología Oral y Cirugía Bucal. 2011;16(2):175–180. doi: 10.4317/medoral.16.e175. [DOI] [PubMed] [Google Scholar]
- 34.Lobene R. R., Weatherford T., Ross N. M., Lamm R. A., Menaker L. A modified gingival index for use in clinical trials. Clinical Preventive Dentistry. 1986;8(1):3–6. [PubMed] [Google Scholar]
- 35.Silva D. G., Tinoco E. M. B., Rocha G. A., et al. Helicobacter pylori transiently in the mouth may participate in the transmission of infection. Memórias do Instituto Oswaldo Cruz. 2010;105(5):657–660. doi: 10.1590/S0074-02762010000500009. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza E., Camorlinga-Ponce M., Perez-Perez G., et al. Present and past Helicobacter pylori infection in Mexican school children. Helicobacter. 2014;19(1):55–64. doi: 10.1111/hel.12098. [DOI] [PubMed] [Google Scholar]
- 37.Patel S. K., Pratap C. B., Jain A. K., Gulati A. K., Nath G. Diagnosis of Helicobacter pylori: what should be the gold standard? World Journal of Gastroenterology. 2014;20(36):12847–12859. doi: 10.3748/wjg.v20.i36.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyabayashi H., Furihata K., Shimizu T., Ueno I., Akamatsu T. Influence of oral Helicobocter pylori on the success of eradication therapy against gastric Helicobacter pylori. Helicobacter. 2000;5(1):30–37. doi: 10.1046/j.1523-5378.2000.00004.x. [DOI] [PubMed] [Google Scholar]
- 39.Payão S. L., Rasmussen L. T. Helicobacter pylori and its reservoirs: a correlation with the gastric infection. World Journal of Gastrointestinal Pharmacology and Therapeutics. 2016;7(1):126–132. doi: 10.4292/wjgpt.v7.i1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Ahmad A., Kürschner A., Weckesser S., et al. Is Helicobacter pylori resident or transient in the human oral cavity? Journal of Medical Microbiology. 2012;61(8):1146–1152. doi: 10.1099/jmm.0.043893-0. [DOI] [PubMed] [Google Scholar]
- 41.Cervantes D. T., Fischbach L. A., Goodman K. J., Phillips C. V., Chen S., Broussard C. S. Exposure to Helicobacter pylori-positive siblings and persistence of Helicobacter pylori infection in early childhood. Journal of Pediatric Gastroenterology and Nutrition. 2010;50(5):481–485. doi: 10.1097/MPG.0b013e3181bab2ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kivi M., Tindberg Y. Helicobacter pylori occurrence and transmission: a family affair? Infectious Diseases. 2006;38(6-7):407–417. doi: 10.1080/00365540600585131. [DOI] [PubMed] [Google Scholar]
- 43.Yokota S.-I., Konno M., Fujiwara S.-I., et al. Intrafamilial, preferentially mother-to-child and intraspousal, Helicobacter pylori infection in Japan determined by mutilocus sequence typing and random amplified polymorphic DNA fingerprinting. Helicobacter. 2015;20(5):334–342. doi: 10.1111/hel.12217. [DOI] [PubMed] [Google Scholar]
- 44.Bellack N. R., Koehoorn M. W., MacNab Y. C., Morshed M. G. A conceptual model of water's role as a reservoir in Helicobacter pylori transmission: a review of the evidence. Epidemiology and Infection. 2006;134(3):439–449. doi: 10.1017/S0950268806006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pari A., Ilango P., Subbareddy V., Katamreddy V., Parthasarthy H. Gingival diseases in childhood—a review. Journal of Clinical and Diagnostic Research. 2014;8(10):ZE01–ZE04. doi: 10.7860/JCDR/2014/9004.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riggio M. P., Lennon A. Identification by PCR of Helicobacter pylori in subgingival plaque of adult periodontitis patients. Journal of Medical Microbiology. 1999;48(3):317–322. doi: 10.1099/00222615-48-3-317. [DOI] [PubMed] [Google Scholar]
- 47.Agarwal S., Jithendra K. D. Presence of Helicobacter pylori in subgingival plaque of periodontitis patients with and without dyspepsia, detected by polymerase chain reaction and culture. Journal of Indian Society of Periodontology. 2012;16(3):398–403. doi: 10.4103/0972-124X.100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De la Garza-Ramos M. A., Valdez-Gonzalez J. A., Elizondo-Perez R. A., Pereyra Alférez B., Caffesse R. G., González-Salazar F. Prevalence of Helicobacer pylori in saliva and dental plaque related to periodontal disease and gastritis. African Journal of Microbiology Research. 2013;7(21):2505–2509. [Google Scholar]
- 49.Rudi J., Rudy A., Maiwald M., Kuck D., Sieg A., Stremmel W. Direct determination of Helicobacter pylori vacA genotypes and cagA gene in gastric biopsies and relationship to gastrointestinal diseases. The American Journal of Gastroenterology. 1999;94(6):1525–1531. doi: 10.1111/j.1572-0241.1999.1138_a.x. [DOI] [PubMed] [Google Scholar]
- 50.González-Valencia G., Atherton J. C., Muñoz O., Dehesa M., Madrazo-De la Garza A., Torres J. Helicobacter pylori vacA and cagA genotypes in Mexican adults and children. The Journal of Infectious Diseases. 2000;182(5):1450–1454. doi: 10.1086/315864. [DOI] [PubMed] [Google Scholar]
- 51.Martínez-Carrillo D. N., Garza-González E., Betancourt-Linares R., et al. Association of IL1B -511C/-31T haplotype and Helicobacter pylori vacA genotypes with gastric ulcer and chronic gastritis. BMC Gastroenterology. 2010;10, article 126 doi: 10.1186/1471-230x-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garza-Gonzalez E., Bosques-Padilla F. J., Tijerina-Menchaca R., Perez-Perez G. I. Characterisation of Helicobacter pylori isolates from the north-eastern region of Mexico. Clinical Microbiology and Infection. 2004;10(1):41–45. doi: 10.1111/j.1469-0691.2004.00731.x. [DOI] [PubMed] [Google Scholar]
- 53.Morales-Espinosa R., Castillo-Rojas G., Gonzalez-Valencia G., et al. Colonization of Mexican patients by multiple Helicobacter pylori strains with different vacA and cagA genotypes. Journal of Clinical Microbiology. 1999;37(9):3001–3004. doi: 10.1128/jcm.37.9.3001-3004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Assumpção M. B., Martins L. C., Barbosa H. P. M., et al. Helicobacter pylori in dental plaque and stomach of patients from Northern Brazil. World Journal of Gastroenterology. 2010;16(24):3033–3039. doi: 10.3748/wjg.v16.i24.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres J., Pérez-Pérez G. I., Leal-Herrera Y., Muñoz O. Infection with cagA+ Helicobacter pylori strains as a possible predictor of risk in the development of gastric adenocarcinoma in Mexico. International Journal of Cancer. 1998;78(3):298–300. doi: 10.1002/(SICI)1097-0215(19981029)78:3<298::AID-IJC6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 56.Bosques-Padilla F. J., Tijerina-Menchaca R., Pérez-Pérez G. I., Flores-Gutiérrez J. P., Garza-González E. Comparison of Helicobacter pylori prevalence in symptomatic patients in northeastern Mexico with the rest of the country: its association with gastrointestinal disease. Archives of Medical Research. 2003;34(1):60–63. doi: 10.1016/S0188-4409(02)00459-9. [DOI] [PubMed] [Google Scholar]
- 57.Román-Román A., Martínez-Carrillo D. N., Atrisco-Morales J., et al. Helicobacter pylori vacA s1m1 genotype but not cagA or babA2 increase the risk of ulcer and gastric cancer in patients from Southern Mexico. Gut Pathogens. 2017;9(1):18. doi: 10.1186/s13099-017-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]