Abstract
Age-related macular degeneration (AMD) is the main reason of blindness in developed countries. Aging is the main AMD risk factor. Oxidative stress, inflammation and some genetic factors play a role in AMD pathogenesis. AMD is associated with the degradation of retinal pigment epithelium (RPE) cells, photoreceptors, and choriocapillaris. Lost RPE cells in the central retina can be replaced by their peripheral counterparts. However, if they are senescent, degenerated regions in the macula cannot be regenerated. Oxidative stress, a main factor of AMD pathogenesis, can induce DNA damage response (DDR), autophagy, and cell senescence. Moreover, cell senescence is involved in the pathogenesis of many age-related diseases. Cell senescence is the state of permanent cellular division arrest and concerns only mitotic cells. RPE cells, although quiescent in the retina, can proliferate in vitro. They can also undergo oxidative stress-induced senescence. Therefore, cellular senescence can be considered as an important molecular pathway of AMD pathology, resulting in an inability of the macula to regenerate after degeneration of RPE cells caused by a factor inducing DDR and autophagy. It is too early to speculate about the role of the mutual interplay between cell senescence, autophagy, and DDR, but this subject is worth further studies.
1. Introduction
Age-related macular degeneration affects the macula, a specific structure in the central retina, leading to worsening of visual acuity. It is the major cause of blindness in the elderly in developed countries. Its global pooled prevalence is estimated to be more than 8%. It is an emerging problem, as it is estimated that the number of people affected by AMD in 2020 will be about 200 million, increasing to almost 300 million in 2040 [1, 2]. Medical cost of care about AMD patients is high reaching over 2 billion dollars in the USA and Australia and about a hundred million euros in some European countries [3]. Therefore, AMD is an emerging element of the global issue of vision loss and medical care.
AMD is a complex disease in which both genetic and environmental factors play a role, but the exact mechanism of its pathogenesis is unknown. The disease occurs in two forms: dry and wet. Molecular studies addressing AMD are impeded by inaccessibility of the live retina tissue from AMD patients. No effective treatment for the more common, dry form of AMD has been established yet.
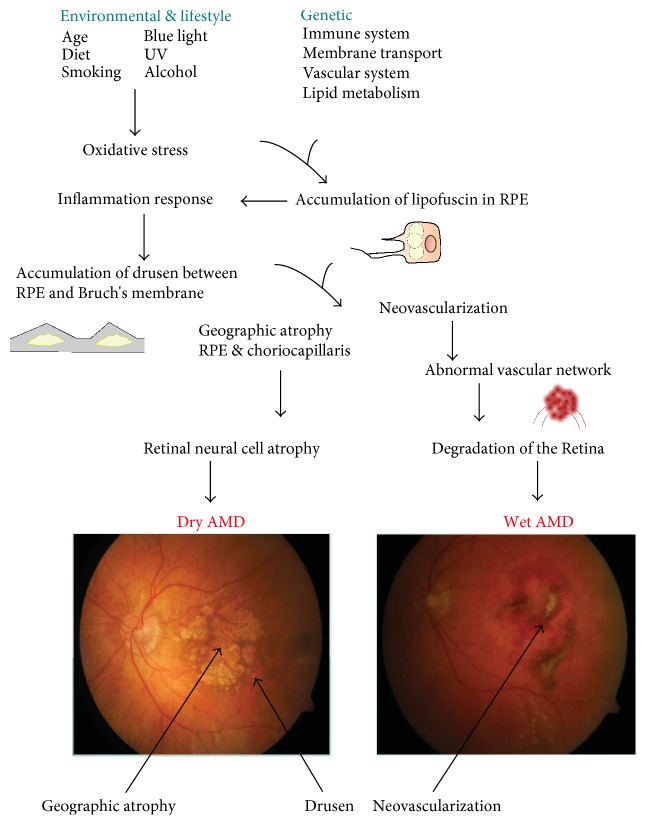
AMD affects mainly elderly people, and it is a major reason for blindness among individuals over 65 years in developed countries [4]. Aging is the most important risk factor for AMD. Although the exact mechanism of AMD pathogenesis is not known, oxidative stress, protein aggregation, and inflammation as well as some genetic factors play a central role in AMD development (Figure 1) [5]. Early dry AMD is hardly detectable and usually asymptomatic. Its advanced form, called geographic atrophy (GA), is associated with a massive loss of photoreceptors that evokes central visual loss [6]. A clinical hallmark of wet AMD is the presence of neovascular vessels sprouting from the choriocapillaris into the retina.
Figure 1.
The exact mechanism of AMD pathogenesis is not known, but several factors can be implicated with a distinct role of aging. Besides aging, various oxidative stress-related environmental and lifestyle influences can be involved. The complement gene mutations play a major role in AMD. Oxidative stress and presumably other factors lead to accumulation of heterogenous lysosomal lipofuscin in retinal pigment epithelium (RPE), which induces a proinflammatory response. This, in turn, can lead to accumulation of extracellular drusen. Lipofuscin contains proangiogenic factors, such as A2E, that may develop choroidal neovascularization typical for wet AMD.
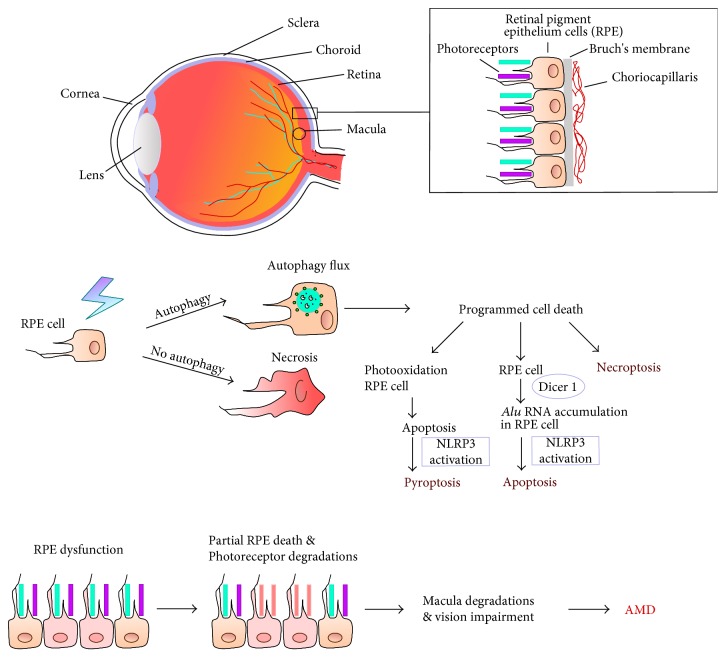
Progression of AMD ultimately leads to RPE and photoreceptors death via several mechanisms, including apoptosis, pyroptosis, necroptosis, and necrosis [7]. Autophagy may be involved in the regulation of the cell death mode in AMD [8] (Figure 2).
Figure 2.
Cell death and autophagy in AMD progression. AMD affects the macula, a part in the central retina, and is associated with degradation of retinal pigment epithelium (RPE) cells, photoreceptors, and choriocapillaris. Autophagy can be decisive in switching between programmed and nonprogrammed cell death mode. Apoptosis of RPE cells can be linked to blue light exposure (photooxidation), oxidative stress, accumulation of Alu transposons due to impaired functioning of the DICER1 endonuclease, and the activation of the NLRP3 inflammasome. Pyroptosis can also result from photooxidation and activation of the NLRP3 inflammasome. Oxidative stress and other factors can induce necroptosis, a programmed version of necrosis.
2. Cell Senescence and Aging in AMD
Kozlowski proposed that cellular senescence of RPE cells played a role in the etiology of AMD [9]. It seems that many studies on the role of cell senescence in organismal aging and age-related pathologies support this idea.
Senescence of human fibroblasts, described for the first time by Hayflick and Moorehead [10, 11] as a cell division limit in culture, affects not only fibroblasts but also other proliferating somatic human cells, such as keratinocytes and melanocytes [12], lymphocytes [13], epithelial [14] and endothelial [15] cells, vascular smooth muscle [16, 17], mesothelial cells [18], mesenchymal stem cells [19], and even cancer cells [20, 21].
Many studies suggest involvement or even a causative role of cell senescence in aging and age-related diseases [22–25]. Indeed, using different set of markers, senescent cells were found in vivo in human, baboon, and mouse skin, human and rodent vascular endothelium, smooth and skeletal muscles, fat tissue and liver [26], skeletal muscle of rodents and primates [27], and human T cells [13]. There is emerging experimental evidence of the accumulation of senescent cells at sites of pathology, such as type 2 diabetes, atherosclerosis, hypertension, chronic pulmonary disease, cataracts, and glaucoma [28]. Senescent cells were also found in RPE of primates [29].
It was postulated that the exposure of cells to recurrent or chronic nonlethal stress might contribute to an increase in the accumulation of stress-induced senescent cells, thereby accelerating tissue aging [30]. Although we believe that senescent cells accumulate with age partially due to their resistance to apoptosis [31], one cannot exclude that at least some of them are cleared by the immune system, as recently reported [32] or that in certain circumstances they can die. Eradication of senescent cells by forcing them to undergo apoptosis is a subject of genetic manipulation [33, 34] or pharmacological interventions by using senolytic agents and can prolong health span [35]. On the other hand, age-dependent apoptosis of muscle cells (sarcopenia) is an undesirable hallmark of the process of organismal senescence, which can be more common than expected [36].
From a mechanistic point of view, a growing body of evidence proves that persistent DNA damage, especially double-strand breaks (DSBs) and DNA damage response (DDR), are closely associated with cell senescence [37]. Number of DSB sensor, γ-H2AX foci, a marker of DSBs, increased in both mouse and human senescent primary cells in tissue culture [38] and in the skin of old primates [39]. Senescence-associated galactosidase- (SA-β-gal-) positive cells and γ-H2AX-positive cells colocalize in old mice [40], and the number of γ-H2AX foci in lymphocytes in humans increases with age [41, 42]. Fibroblasts from individuals suffering from progeria (Hutchinson-Gilford syndrome) persistently displayed many markers of increased basal DDR [43]. Recently, it has been shown that a controlled induction of DSBs in mouse liver induces features of tissue aging [44].
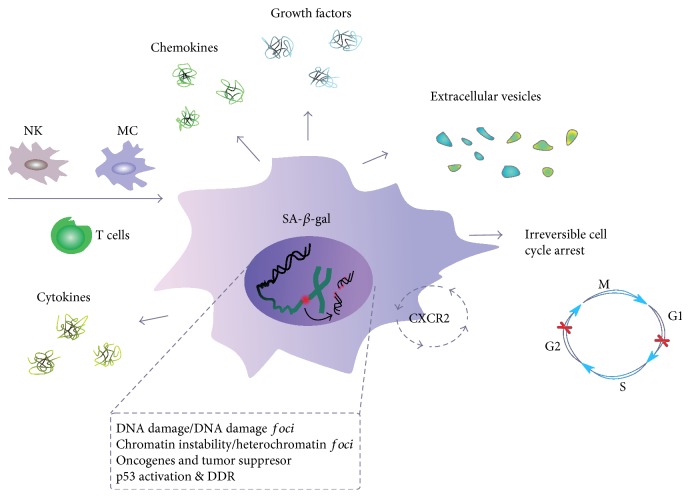
All senescent cells display common features, such as arrest in the G1 or G2 phase of the cell cycle, increased cell size, granularity, and increased activity of SA-β-gal (Figure 3) [24]. Senescent cells stay alive and are metabolically active and secrete a lot of factors [45] that can be classified into inflammatory chemokines and cytokines, matrix-remodeling proteases, and growth factors [46]. SASP can not only influence tissue surveillance in a way which can promote tissue repair, prevent fibrosis, and signal to the innate immune system to clear the senescent cells but also induce cancer development and other age-related diseases [47]. It also promotes a low-grade inflammation, which can drive organismal aging (inflammaging) [48].
Figure 3.
Senescent cells and senescence-associated secretory phenotype (SASP). A cell undergoing senescence is permanently arrested in the G1 or G2 phase of the cell cycle and has changed morphology. It is featured by an increased activity of senescence-associated-β-galactosidase (SA-β-gal) and can be targeted by the immunological system, with natural killer (NK) cells, macrophages (MS), and T-lymphocytes involved. Released various soluble agents, including cytokines, chemokines, growth factors, and extracellular vesicles, are main determinants of SASP. A senescent cell is characterized by an elevated level of DNA damage and chromosomes aberrations, which are also signs of genomic and chromosomal instability, typical for cancer cells. Chemokine signaling through the CXCR2 protein increases senescence.
There is a growing body of evidence linking DNA damage with inflammation and disease, particularly age-dependent diseases [49]. This is sort of a vicious cycle as DNA damage-dependent senescence can lead to secretion of molecules, which can reinforce senescence [50] and can induce DNA damage and DNA damage-dependent bystander senescence [51].
Initiation and maintenance of the SASP requires the DDR proteins ATM, NBS1, and CHK2, but not p53 and pRb. NF-κB signaling is another pathway involved in generating SASP that can be linked with DDR [52]. Recently, it has been shown that the GATA4 protein is directly involved in SASP. GATA4 is normally degraded, but it is stabilized in cells undergoing senescence. GATA4 is activated by DNA damage response regulators, ATM and ATR, but not by p53 or p16. This transcription factor activates NF-κB to initiate SASP and facilitate senescence [53]. However, NF-κB can be also activated by p38MAPK independently of DDR [54]. Also, mTOR can be involved in SASP as its inhibition by rapamycin substantially reduces the level of secreted cytokines [55, 56].
Although the causative role of reactive oxygen species (ROS) in aging is disputable, the paradigm assuming that oxidative stress and ROS produced by mitochondria play an important role in cell senescence has been supported [57, 58]. Hydrogen peroxide was the first factor used to show oxidative stress-induced senescence [59]. We also used this compound to show a canonical signaling pathway involved in cell senescence [60]. Oxidative stress, which can induce cellular damage, has been closely connected with the pathogenesis of AMD as the retina is particularly susceptible to the stress because of its high consumption of oxygen, high proportion of polyunsaturated fatty acids, and exposure to visible light [61]. Therefore, retinal cells can be prone to stress-induced senescence. The retina is built of three layers of neural cells and one layer of RPE cells, which are quiescent.
Although cell senescence per definition denotes a permanent growth arrest of proliferation competent cells, recently, Jurk and others have shown that some features of cell senescence, including DDR, also apply to postmitotic neurons in vivo [62]. However, we showed that the SA-β-gal phenotype in neurons could not be attributed uniquely to cell senescence either in vitro or in vivo [63].
Although epithelial cells stay quiescent in the retina, they are proliferation-prone and vulnerable to oxidative stress-induced senescence. Indeed, in several studies using proliferating human RPE-derived ARPE-19 cells, which proliferate in vitro, the cell senescence process was documented upon oxidative stress. In several studies, senescence was induced by hydroxyl peroxide in nontoxic concentrations [64–67]. In other studies, tert-butyl hydroperoxide [68] or cigarette smoke [69] was applied. Arend et al. observed a significant increase in cell viability and reduced SA-β-gal activity, ROS amount, and DNA damage foci in ARPE-19 cells induced to senescence with H2O2 and pretreated with idebenone, which is a derivative of coenzyme Q10, but with a tenfold higher antioxidant capacity than its parental compound [64]. Similarly, fullerenol, a ROS scavenger and antioxidant, protected ARPE-19 cells from H2O2-induced senescence. Interestingly, fullerenol activated SIRT1, which belongs to the family of “proteins of youth”—sirtuins [70].
The use of ARPE-19 cells in senescence studies have some limitations. ARPE-19 population can contain a substantial, if not the major, fraction of cells which are able to double their population to over 270 times, so they can be considered as immortal [23]. Unlike cells with limited number of divisions, immortal cells do not undergo replicative senescence. However, it was shown that they are prone to stress-induced senescence [71, 72].
3. Autophagy and DDR-Dependent or DDR-Independent Players in AMD Pathogenesis
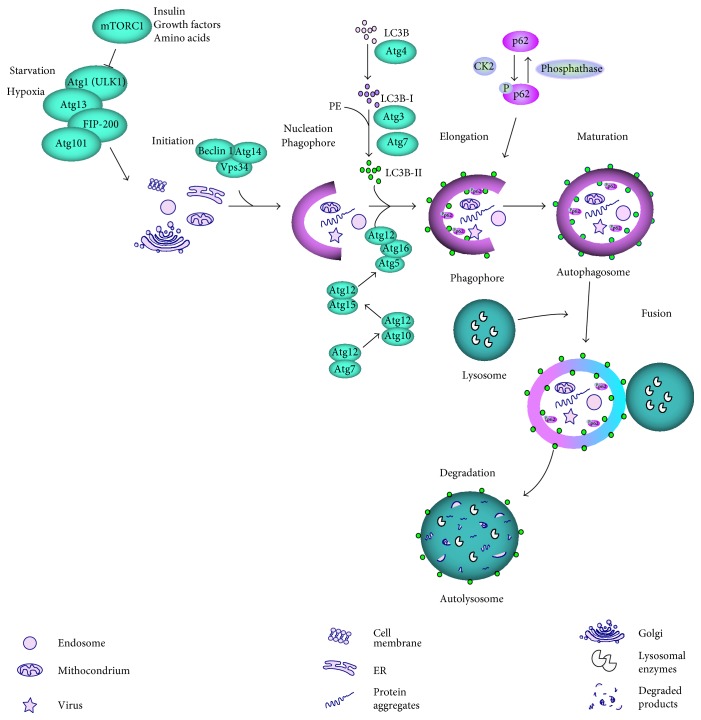
Autophagy controls cellular homeostasis by degrading in lysosomes damaged, nonfunctional or no longer needed cellular components, including organelles. Autophagic degradation provides energy, and lysosomal machinery can deliver amino acids and other degradation products back to the cytoplasm, where they can be reused as building blocks in cellular metabolism (“recycling”) [73]. This process can be carried out through at least three distinguished pathways: macroautophagy (further referred to as autophagy), chaperone-mediated autophagy (CMA), and microautophagy. Many proteins are involved in autophagy, including autophagy-related proteins (ATGs), mammalian target of rapamycin (mTOR), the serine/threonine kinase (ULK1), FIP-200, p62 (SQSTM1), and microtubule-associated protein light chain 3 (LC3) [74]. The hallmark of autophagy and its critical stage is the formation of a double-membraned vesicle enclosing materials for degradation (cargo), called the autophagosome (Figure 4). It then fuses with the lysosome forming autolysosome, in which the cargo is degraded [75].
Figure 4.
Autophagy dependent on mTOR. In normal nutrient conditions, the mTOR complex 1 (mTORC1) inhibits the ULK1 complex, consisting of ULK1, Atg13, Atg101, and FIP200, which can activate autophagy in stress conditions, including starvation and hypoxia or when the inhibitory effect of mTORC1 is abolished by growth factors, insulin, amino acids, or other agents. The material to be degraded (cargo) is then enclosed by a nucleating phagophore, which requires a translocation of ULK1 to endoplasmic reticulum (ER). ER membrane is used to form the phagophore, but other sources are also possible. The phagophore membrane is elongated, which leads to the formation of autophagosome, a vesicle with the enclosed cargo. This process is assisted by LC3 lipidated by phosphatidylethanolamine (PE) and many individual proteins, including Beclin 1, Vps34, and autophagy-related proteins (Atgs). The p62 protein functions as a selective autophagy receptor for degradation of ubiqutinated substrates, but it is itself a specific substrate for autophagy after its phosphorylation and can be selectively incorporated into the autophagosome and degraded. Fusion of autophagosome with lysosome creates autolysosome in which the cargo is degraded by lysosomal enzymes. Autophagy can be also activated by mTOR-independent pathways.
Impaired autophagy was observed in serious human disorders, such as cancer and neurodegenerative diseases including AMD [76]. In general, autophagy plays an important role in the functioning of RPE cells [77–79]. Drusen are a clinical hallmark of AMD and an important element of its pathogenesis [5, 80, 81]. They are yellowish deposits between RPE cells and Bruch's membrane. Consequently, impaired autophagy can lead to drusen accumulation contributing to AMD development [82]. However, the relationship between autophagy and aging is not fully known and the activity of this process changes during lifetime. In addition, detailed autophagic pathways involved in the development of AMD have not been identified as different mechanisms of autophagy can function in normal and pathological retinas [83].
There is an emerging body of experimental evidence on the involvement of autophagy in AMD pathogenesis. These experiments are performed mainly on animal model of AMD and retinas obtained postmortem from AMD donors [78]. Autophagy is closely associated with cellular response to oxidative stress with the involvement of the p62/Keap1/Nrf2 pathway [84, 85]. Moreover, autophagy can be considered as an element of DDR, in which DNA repair is a major component, being a major constituent of cellular antioxidant defense [86–88]. We and others showed that AMD could be associated with disturbed DNA repair [49, 89–91].
Oxidative stress, a main factor in AMD pathogenesis, results in various DNA lesions, and 8-oxo-7,8-dihydroguanine (8-oxoG) is a hallmark of oxidative DNA damage and a major mutagenic intermediate of oxidative stress [92]. In most cases, the repair of 8-oxoG is initiated by the hOGG1 glycosylase via the base excision repair (BER) pathway. If 8-oxoG escapes this process and replicative DNA polymerase misinserts adenine instead of cytosine opposite to 8-oxoG, an alternative pathway of BER can be activated with the hMYH (MUTYH) glycosylase, which removes that adenine. Our observations revealed that genetic variability in the hOGG1 and hMYH genes may be associated with AMD occurrence and progression [93]. It was reported that the level of 8-oxoG was higher in patients with exudative AMD than in control individuals. This led to the conclusion that DNA damage may underline the role of oxidative stress in AMD pathology.
Our earlier data indicated that lymphocytes isolated from AMD patients displayed a higher level of endogenous DNA damage than lymphocytes from control individuals [89]. Also, oxidative DNA damage was higher in AMD patients than in controls and cells from the patients were more sensitive to hydrogen peroxide and UV radiation, which allowed us to speculate that the combination of impaired DNA repair and elevated sensitivity to UV radiation can be important for AMD pathogenesis.
Not only nuclear DNA (nDNA) but also its mitochondrial counterpart (mtDNA) was reported to have elevated extent of damage in AMD (Figure 5) [90, 94–96]. Moreover, in those studies, performed on macular and peripheral RPE cells obtained from AMD patients and rodents, an increase in heteroplasmic mutations and a decrease in the efficacy of mtDNA repair were observed. In fact, there are reports showing that mtDNA in cultured RPE cells is more prone to DNA damage, at least induced by certain agents, than nDNA [91, 97–101]. Although the efficacy of DNA repair declines with age for both kinds of DNA, mtDNA from the retina was shown to have more potentially detrimental changes, than mtDNA from blood, for both normal and AMD samples [102]. These changes are potentially detrimental, if not repaired, as mtDNA contains almost exclusively coding sequences. The repair of mtDNA is generally considered as poorer than in the nucleus. All DNA repair proteins are encoded in nDNA, so its damage can affect protection from damage in mtDNA. Therefore, DNA damage can be associated with AMD occurrence and progression, suggesting that DDR can play a role in the pathogenesis of this disease.
Figure 5.

Nuclear and mitochondrial DNA (nDNA and mtDNA) can be damaged by AMD risk factors, which can also affect proteins, including DNA repair proteins. Nonrepaired or misrepaired DNA can contribute to retinal cell death occurring in AMD [89].
4. Relationship between Senescence, Autophagy, and DNA Damage Response in RPE Cells
Many reports link oxidative stress with autophagy showing that this association is regulated in a highly coordinated pathway [103]. It is clearly illustrated by the activation of Nrf2, a transcription factor crucial for cellular antioxidant defense, by p62, a key regulator of autophagy [104]. Likely, the most direct association between oxidative stress and autophagy is expressed by mitophagy, when mitochondria with highly damaged DNA are degraded [105]. This is a specific feature of DDR in mitochondria, as in the nucleus heavily damaged DNA can induce a programmed death or in certain circumstances can be tolerated, which usually results in mutations [106]. Although ROS can induce autophagy in starvation conditions, it is not known which species are responsible for this effect [107] and both superoxide radical (O2•−) and hydrogen peroxide were considered to trigger autophagy in starvation [108–111]. In general, ROS are inducers of autophagy [112, 113]. Moreover, some data suggest that mitochondria are the main source of ROS needed for the induction of autophagy [109, 110, 114].
Several DDR pathways prevent or cope with DNA damage. However, if DNA is highly damaged, cells remain quiescent or undergo programmed cell death. Persistent, unrepaired DNA damage is typical for cell senescence. Autophagy acts as both a prosurvival mechanism and a kind of cell death, making a critical contribution to cell fate after DNA damage. Some reports suggest that autophagy delays apoptosis induced by DNA damage, providing energy required for DNA repair [1]. In general, autophagy participates in DDR by elimination of toxic aggregates, which can be a source of ROS and in this way indirectly decrease DNA damage [115, 116].
Several DDR proteins are involved in the regulation of autophagy. PolyADP-ribose polymerase 1 (PARP1), which is essential for DNA single-strand break repair, catalyzes polyribosylation of nuclear proteins converting NAD+ into polymers of polyADP-ribose. This leads to NAD+ utilization and ATP depletion, resulting in an energetic imbalance, which activates autophagy via the AMPK pathway, to recycle metabolic precursors for ATP and provide energy needed for DDR [117]. ATM, a crucial protein for DDR signaling, is another protein linking DDR to autophagy. It can activate TSC2, a tumor suppressor, to inhibit mTORC1 and induce autophagy [118]. The p53 protein, a key DDR regulator, and members of its family were shown to affect expression of several genes encoding autophagic proteins [119, 120].
It is generally accepted that autophagy declines with aging of model organism, resulting in the accumulation of cellular debris and turning the cell brown. Decreased autophagy is often associated with accelerated aging, whereas stimulated autophagy can exert a potent antiaging effect [121]. It is associated with reduced expression of proteins important for autophagy induction, including ATGs, Sirtuin 1, Beclin 1, ULK1, and LC3 [122].
DNA damage, with the key proteins ATM and ATR is a causative signal to cellular senescence. DNA damage-dependent senescence was shown in human and murine fibroblasts upon different stimuli [37]. The role of mitochondrial homeostasis and ROS generation in the process of aging has been subject of debate. Free-radical and mitochondrial theories of aging speculate that cumulative damage to mitochondria and mitochondrial DNA induced by ROS is one of the causes of aging. Oxidative damage affects replication and transcription of mtDNA and results in a decline in mitochondrial functions, which in turn leads to enhanced ROS production and further damage to mtDNA [58]. However, we showed that a decreased ROS level did not protect cells against senescence [21, 123]. Correia-Melo et al. observed that senescent cells had an increased mitochondrial mass driven by mitochondrial biogenesis, which resulted in increased cellular oxygen consumption. They have also uncovered a novel senescence regulatory pathway, in which the activation of the ATM, AKT, and mTOR phosphorylation cascades downstream of DNA damage triggered PGC-1α- (peroxisome proliferator-activated receptor-gamma coactivator 1 alpha-) dependent mitochondrial biogenesis [57]. However, other recent studies have highlighted mTOR as a SASP regulator by alternative mechanisms emphasizing mTOR rather as an antisenescence target [55]. Therefore, the role of mitochondrial biogenesis, ROS, and mTOR in cell senescence and SASP is still an open question.
The interplay between cell senescence and autophagy is yet unclear. Young et al. have shown that autophagy is activated upon an induction of cell senescence and contributes to the establishment of senescence [124], but there are contradictory data showing that inhibition of autophagy can favor cell senescence and that autophagy is necessary for senescence [21, 123]. We showed an impaired autophagy in RPE upon chronic oxidative stress, but senescence was not induced in that study [125]. Senescence of RPE cells may be associated with alterations in PGC-1α function. In neurons as well as in RPE cells, PGC-1α was shown to regulate lysosomal activity by TFEB protein, which might be important for improvement of autophagy flux and removal of cell damage [126, 127]. It was also demonstrated in that work that PGC-1α deficient mice developed some abnormalities in RPE, which were associated with their accelerated senescence. Next, PGC-1α alpha silencing in ARPE-19 cells aggravated H2O2-induced senescence. These cells displayed a significantly higher SA-β-gal activity than control cells. Senescence of RPE cells has been associated with an altered mTOR signaling [128, 129].
GATA4 is a member of GATA transcription factors, and Kang and coworkers identified this protein as a key regulator of cellular senescence [53, 130]. GATA4 is also important for DDR and is regulated by autophagy, so it can be at the crossroad of these three cellular phenomena: senescence, autophagy, and DDR (Figure 6). It is also important that GATA4 is involved in the mechanisms inducing SASP phenotype, which can promote chronic inflammation associated with most age-related diseases, including AMD [5, 53, 131]. GATA4 is normally degraded by p62-mediated autophagy and it was shown to be stabilized in cells undergoing senescence, possibly due to a decreased association with p62 [53]. As GATA4 activation depends on the key DDR signaling proteins, ATM and ATR and it is accumulated in some aging tissues, it is a good candidate to orchestrate interplay between senescence, autophagy, and DDR. Kang and coworkers postulated that the GATA4-mediated relationship between autophagy and senescence is different for different modes of autophagy [53]. GATA4 can be a positive regulator of senescence and selective autophagy, but a signal inducing senescence can stimulate GATA4 to avoid selective autophagy.
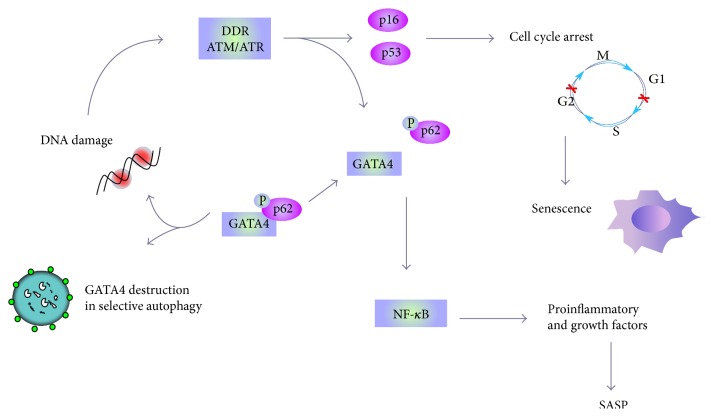
Figure 6.
GATA4 can be involved in autophagy, senescence, and DNA damage response (DDR). The level of GATA4 is normally regulated by p62-dependent selective autophagy, but DNA damage and resulting DDR can release GATA4 from p62 control by its ATM-induced phosphorylation. If DNA damage cannot be repaired, DDR effectors induce permanent and irreversible cell cycle arrest, which is a hallmark of senescence with senescence-associated phenotype (SASP). GATA4 released from autophagic degradation can transactivate several genes that activate NF-κB, resulting in the release of growth factors, chemokines, cytokines, and other molecules typical for SASP.
In searching for a mechanism by which GATA4 regulates senescence, Kang et al. observed that it upregulated (mostly) and downregulated genes important for senescence [53]. Therefore, GATA4 could be involved in the stimulation of a considerable fraction of genes, including the SASP genes, whose expression determines the senescence phenotype. GATA4 was shown to act upstream of NF-κB during senescence induction and depletion of RelA (p65), a component of NF-κB [53]. This effect was associated with the repression of almost all genes involved in SASP, except IL1A. Subsequent experiments showed that GATA4 regulated senescence response independently of the p53 and p16INK4a/Rb pathways.
When key DDR regulators ATM and ATR were inhibited, the GATA4 pathway was inhibited during senescence, suggesting that it is a new independent branch of DDR. The role of DDR in inhibition of autophagy-mediated degradation of GATA4 is not known yet. GATA4 was observed to accumulate in organs of 22-month-old mice as compared to their 6-month-old counterparts, which correlates with the accumulation of senescent cells in aging organism [26, 53, 132–134].
In summary, GATA4 can be involved in DDR and this involvement is independent of p53 and p16INK4a/Rb pathways. GATA4 is closely associated with senescence and SASP and is controlled by selective autophagy, but can also stimulate general autophagy [53]. Therefore, it is justified to consider a role of GATA4 in coordinating senescence, autophagy, and DDR. In addition, as GATA4 associates with inflammation, studies on its role in AMD pathogenesis are justified.
5. Senescence-Based Pathogenesis of AMD with the Contribution of Autophagy and DDR
RPE cells in the central retina are quiescent due to spatial constraints and contact with neuroretina, and when damaged, they can be replaced by their proliferating counterparts at RPE periphery in an endogenous compensatory mechanism [135]. This endogenous regenerative mechanism is activated in pathological conditions, which can increase with age [136]. Oxidative stress can induce senescence in RPE cells if they are prone to and result in inability of peripheral RPE cells to rescue their central RPE counterparts, which can lead to a massive loss of RPE cells observed in clinically detected AMD. If most of macular peripheral RPE cells are affected by senescence, this mechanism can fail leading to AMD. Senescent RPE will be the source of pathology and have a detrimental impact on surrounding tissue through SASP.
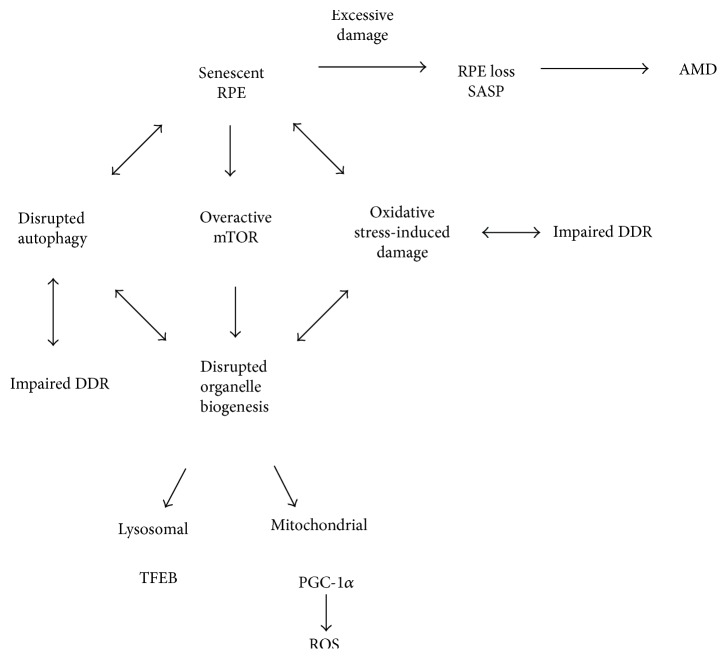
We hypothesize that under some circumstances, RPE senescence may contribute to or/and precede irreversible pathological events in the retina specific for AMD, such as RPE loss and inflammation. Senescent RPE cells may be excessively damaged, dysfunctional, and capable of overexpression of SASP. Figure 7 illustrates this novel concept of RPE senescence as a critical contributor to AMD induction and progression.
Figure 7.
Senescence as a critical factor in AMD pathogenesis. In certain stress conditions, which can be induced by environmental or/and lifestyle factors in aging retina, a major fraction of RPE cells become senescent and are no longer able to regenerate damaged RPE cells, which leads to AMD. The senescence of RPE cells can result from an interplay between aging, autophagy, and DDR in stress conditions. This interplay is a kind of vicious cycle as impaired DNA damage response (DDR) can lead to an increased damage to biomolecules by ROS. Damage to biomolecules induces the degradation of organelles via mTOR-dependent autophagy. This may lead to aggravation of oxidative stress and cellular damage as well as continue to impair autophagy and antioxidant defense by altered TFEB (transcription factor E-box binding) and PGC-1α signaling and increased ROS generation.
We believe that senescence associates with autophagy and DDR. As mentioned, cell senescence can be causative for aging and age-dependent diseases. All these three effects, senescence, autophagy, and DDR, can be provoked by oxidative stress, which is a major factor in AMD pathogenesis. Moreover, aging is the main risk factor of pathogenesis of AMD and can be related to oxidative stress [46]. Inflammation associates with oxidative stress, aging (inflammaging), and AMD [5, 137, 138]. Therefore, it is logical and justified to hypothesize that senescence can play a role in AMD and this process can be influenced or regulated by autophagy and DDR. Consequently, GATA4, as an identified factor to be involved in cell senescence, autophagy, DDR, and inflammation, seems to be a natural candidate to play a major role in the proposed mechanism of AMD pathogenesis. However, this is only a hypothesis, which should be verified, but we tried to show some arguments that this subject is worth further study and development.
6. Conclusions and Perspectives
Molecular studies on AMD pathogenesis in humans are limited. Therefore, choosing an optimal experimental model for these studies is essential. ARPE-19 cell line is commonly used in molecular research on AMD, even though it is a heterogeneous cell population, including dividing and nondividing cells. However, as AMD is an age-related disease, the process of cell senescence should be included in in vitro models. As oxidative stress is a main AMD pathogenesis factor, cellular antioxidant defense is important in the disease prevention. That is why one can use RPE cells from mice knockout of genes essential for antioxidant defense. To include aging in that study, animals at different ages can be used. As the main AMD genetic risk factor are mutations in the gene encoding complement factor H, one can use RPE cells derived from induced pluripotent stem cells (iPSCs) obtained from AMD patients having such mutations.
It seems that the relationship between DDR and autophagy in mitochondria can be especially important for AMD pathogenesis due to many reasons [139]. First, mitochondrial DDR is different from DDR in the nucleus [140]. Second, autophagic mtDNA degradation can be considered as a DDR pathway dealing with heavily damaged molecules of mtDNA [141]. Third, mitochondrial mutagenesis was reported to play a role in AMD pathogenesis [89–91]. Furthermore, the role of mTOR, a crucial autophagy protein, in lysosomal and mitochondrial biogenesis has been recently appreciated [57, 142]. Therefore, studies of DDR and autophagy in mitochondria in the context of AMD are warranted.
To summarize, cellular senescence and SASP can be related to age-related chronic diseases [137, 143]; chronic inflammation (inflammaging) is also involved in age-related chronic diseases [137]; autophagy and senescence seem to be closely related [144]; and lastly the GATA4 protein can be involved in DDR, senescence, and autophagy as well as in inflammation and aging [53, 130].
Work to determine the relationship between aging (senescence), autophagy, and DDR and relate it to AMD can bring information important for AMD clinic and basic molecular biology as there are many essential unanswered questions and problems concerning mutual relationships between aging, autophagy, and cellular reaction to DNA damage.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Wong W. L., Su X., Li X., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. The Lancet Global Health. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Jonas J. B. Global prevalence of age-related macular degeneration. The Lancet Global Health. 2014;2(2):e65–e66. doi: 10.1016/S2214-109X(13)70163-3. [DOI] [PubMed] [Google Scholar]
- 3.Gordois A., Cutler H., Pezzullo L., et al. An estimation of the worldwide economic and health burden of visual impairment. Global Public Health. 2012;7(5):465–481. doi: 10.1080/17441692.2011.634815. [DOI] [PubMed] [Google Scholar]
- 4.Reibaldi M., Longo A., Pulvirenti A., et al. Geo-epidemiology of age-related macular degeneration: new clues into the pathogenesis. American Journal of Ophthalmology. 2016;161:78.e1–93.e2. doi: 10.1016/j.ajo.2015.09.031. [DOI] [PubMed] [Google Scholar]
- 5.Kauppinen A., Paterno J. J., Blasiak J., Salminen A., Kaarniranta K. Inflammation and its role in age-related macular degeneration. Cellular and Molecular Life Sciences. 2016;73(9):1765–1786. doi: 10.1007/s00018-016-2147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein R., Klein B. E., Knudtson M. D., Meuer S. M., Swift M., Gangnon R. E. Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 2007;114(2):253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 7.Kaarniranta K., Tokarz P., Koskela A., Paterno J., Blasiak J. Autophagy regulates death of retinal pigment epithelium cells in age-related macular degeneration. Cell Biology and Toxicology. 2017;33(2):113–128. doi: 10.1007/s10565-016-9371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klettner A., Kauppinen A., Blasiak J., Roider J., Salminen A., Kaarniranta K. Cellular and molecular mechanisms of age-related macular degeneration: from impaired autophagy to neovascularization. The International Journal of Biochemistry & Cell Biology. 2013;45(7):1457–1467. doi: 10.1016/j.biocel.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Kozlowski M. R. RPE cell senescence: a key contributor to age-related macular degeneration. Medical Hypotheses. 2012;78(4):505–510. doi: 10.1016/j.mehy.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Hayflick L., Moorhead P. S. The serial cultivation of human diploid cell strains. Experimental Cell Research. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 11.Moorhead P. S., Nicholls W. W., Perkins F. T., Hayflick L. Standards of karyology for human diploid cells. Journal of Biological Standardization. 1974;2(2):95–101. doi: 10.1016/0092-1157(74)90023-7. [DOI] [PubMed] [Google Scholar]
- 12.Bandyopadhyay D., Timchenko N., Suwa T., Hornsby P. J., Campisi J., Medrano E. E. The human melanocyte: a model system to study the complexity of cellular aging and transformation in non-fibroblastic cells. Experimental Gerontology. 2001;36(8):1265–1275. doi: 10.1016/s0531-5565(01)00098-5. [DOI] [PubMed] [Google Scholar]
- 13.Brzezinska A., Magalska A., Szybinska A., Sikora E. Proliferation and apoptosis of human CD8+CD28+ and CD8+CD28− lymphocytes during aging. Experimental Gerontology. 2004;39(4):539–544. doi: 10.1016/j.exger.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Romanov S. R., Kozakiewicz B. K., Holst C. R., Stampfer M. R., Haupt L. M., Tlsty T. D. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409(6820):633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 15.Erusalimsky J. D., Kurz D. J. Endothelial cell senescence. Handbook of Experimental Pharmacology. 2006;176, Part 2:213–248. doi: 10.1007/3-540-36028-X_7. [DOI] [PubMed] [Google Scholar]
- 16.Bielak-Zmijewska A., Wnuk M., Przybylska D., et al. A comparison of replicative senescence and doxorubicin-induced premature senescence of vascular smooth muscle cells isolated from human aorta. Biogerontology. 2014;15(1):47–64. doi: 10.1007/s10522-013-9477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorenne I., Kavurma M., Scott S., Bennett M. Vascular smooth muscle cell senescence in atherosclerosis. Cardiovascular Research. 2006;72(1):9–17. doi: 10.1016/j.cardiores.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Ksiazek K., Piwocka K., Brzezinska A., et al. Early loss of proliferative potential of human peritoneal mesothelial cells in culture: the role of p16INK4a-mediated premature senescence. Journal of Applied Physiology. 2006;100(3):988–995. doi: 10.1152/japplphysiol.01086.2005. [DOI] [PubMed] [Google Scholar]
- 19.Shibata K. R., Aoyama T., Shima Y., et al. Expression of the p16INK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem Cells. 2007;25(9):2371–2382. doi: 10.1634/stemcells.2007-0225. [DOI] [PubMed] [Google Scholar]
- 20.Roninson I. B., Broude E. V., Chang B. D. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resistance Updates. 2001;4(5):303–313. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- 21.Sikora E., Mosieniak G., Sliwinska M. A. Morphological and functional characteristic of senescent cancer cells. Current Drug Targets. 2016;17(4):377–387. doi: 10.2174/1389450116666151019094724. [DOI] [PubMed] [Google Scholar]
- 22.Childs B. G., Durik M., Baker D. J., van Deursen J. M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nature Medicine. 2015;21(12):1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikora E. Rejuvenation of senescent cells-the road to postponing human aging and age-related disease? Experimental Gerontology. 2013;48(7):661–666. doi: 10.1016/j.exger.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Sikora E., Arendt T., Bennett M., Narita M. Impact of cellular senescence signature on ageing research. Ageing Research Reviews. 2011;10(1):146–152. doi: 10.1016/j.arr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Sikora E., Bielak-Zmijewska A., Mosieniak G. Cellular senescence in ageing, age-related disease and longevity. Current Vascular Pharmacology. 2014;12(5):698–706. doi: 10.2174/1570161111666131219094045. [DOI] [PubMed] [Google Scholar]
- 26.Jeyapalan J. C., Sedivy J. M. Cellular senescence and organismal aging. Mechanisms of Ageing and Development. 2008;129(7-8):467–474. doi: 10.1016/j.mad.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreiling J. A., Tamamori-Adachi M., Sexton A. N., et al. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 2011;10(2):292–304. doi: 10.1111/j.1474-9726.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naylor R. M., Baker D. J., van Deursen J. M. Senescent cells: a novel therapeutic target for aging and age-related diseases. Clinical Pharmacology and Therapeutics. 2013;93(1):105–116. doi: 10.1038/clpt.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishima K., Handa J. T., Aotaki-Keen A., Lutty G. A., Morse L. S., Hjelmeland L. M. Senescence-associated beta-galactosidase histochemistry for the primate eye. Investigative Ophthalmology & Visual Science. 1999;40(7):1590–1593. [PubMed] [Google Scholar]
- 30.Toussaint O., Dumont P., Dierick J. F., et al. Stress-induced premature senescence. Essence of life, evolution, stress, and aging. Annals of the New York Academy of Sciences. 2000;908:85–98. doi: 10.1111/j.1749-6632.2000.tb06638.x. [DOI] [PubMed] [Google Scholar]
- 31.Campisi J. Cancer and ageing: rival demons? Nature Reviews Cancer. 2003;3(5):339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- 32.Sagiv A., Krizhanovsky V. Immunosurveillance of senescent cells: the bright side of the senescence program. Biogerontology. 2013;14(6):617–628. doi: 10.1007/s10522-013-9473-0. [DOI] [PubMed] [Google Scholar]
- 33.Baker D. J., Childs B. G., Durik M., et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker D. J., Wijshake T., Tchkonia T., et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovadya Y., Krizhanovsky V. Senescent cells: SASPected drivers of age-related pathologies. Biogerontology. 2014;15(6):627–642. doi: 10.1007/s10522-014-9529-9. [DOI] [PubMed] [Google Scholar]
- 36.Marzetti E., Privitera G., Simili V., et al. Multiple pathways to the same end: mechanisms of myonuclear apoptosis in sarcopenia of aging. Scientific World Journal. 2010;10:340–349. doi: 10.1100/tsw.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nature Reviews Cancer. 2008;8(7):512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura A. J., Chiang Y. J., Hathcock K. S., et al. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics & Chromatin. 2008;1(1):p. 6. doi: 10.1186/1756-8935-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herbig U., Ferreira M., Condel L., Carey D., Sedivy J. M. Cellular senescence in aging primates. Science. 2006;311(5765):p. 1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 40.Wang C., Jurk D., Maddick M., Nelson G., Martin-Ruiz C., von Zglinicki T. DNA damage response and cellular senescence in tissues of aging mice. Aging Cell. 2009;8(3):311–323. doi: 10.1111/j.1474-9726.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- 41.Sedelnikova O. A., Horikawa I., Redon C., et al. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7(1):89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- 42.Sedelnikova O. A., Horikawa I., Zimonjic D. B., Popescu N. C., Bonner W. M., Barrett J. C. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nature Cell Biology. 2004;6(2):168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 43.Burtner C. R., Kennedy B. K. Progeria syndromes and ageing: what is the connection? Nature Reviews Molecular Cell Biology. 2012;11(8):567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- 44.White R. R., Milholland B., de Bruin A., et al. Controlled induction of DNA double-strand breaks in the mouse liver induces features of tissue ageing. Nature Communications. 2015;6:p. 6790. doi: 10.1038/ncomms7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freund A., Orjalo A. V., Desprez P. Y., Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends in Molecular Medicine. 2010;16(5):238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Deursen J. M. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoare M., Narita M. Transmitting senescence to the cell neighbourhood. Nature Cell Biology. 2013;15(8):887–889. doi: 10.1038/ncb2811. [DOI] [PubMed] [Google Scholar]
- 48.Franceschi C., Bonafe M., Valensin S., et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Annals of the New York Academy of Sciences. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 49.Ioannidou A., Goulielmaki E., Garinis G. A. DNA damage: from chronic inflammation to age-related deterioration. Frontiers in Genetics. 2016;7:p. 187. doi: 10.3389/fgene.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuilman T., Michaloglou C., Vredeveld L. C., et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 51.Hubackova S., Krejcikova K., Bartek J., Hodny Z. IL1- and TGFβ-Nox4 signaling, oxidative stress and DNA damage response are shared features of replicative, oncogene-induced, and drug-induced paracrine ‘bystander senescence’. Aging. 2012;4(12):932–951. doi: 10.18632/aging.100520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salminen A., Kauppinen A., Kaarniranta K. Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP) Cellular Signalling. 2012;24(4):835–845. doi: 10.1016/j.cellsig.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Kang C., Xu Q., Martin T. D., et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349(6255, article aaa5612) doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freund A., Patil C. K., Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. The EMBO Journal. 2011;30(8):1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herranz N., Gallage S., Mellone M., et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nature Cell Biology. 2015;17(9):1205–1217. doi: 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laberge R. M., Sun Y., Orjalo A. V., et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nature Cell Biology. 2015;17(8):1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Correia-Melo C., Marques F. D., Anderson R., et al. Mitochondria are required for pro-ageing features of the senescent phenotype. The EMBO Journal. 2016;35(7):724–742. doi: 10.15252/embj.201592862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Correia-Melo C., Passos J. F. Mitochondria: are they causal players in cellular senescence? Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2015;1847(11):1373–1379. doi: 10.1016/j.bbabio.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 59.Chen Q., Ames B. N. Senescence-like growth arrest induced by hydrogen peroxide in human diploid fibroblast F65 cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(10):4130–4134. doi: 10.1073/pnas.91.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Przybylska D., Janiszewska D., Gozdzik A., et al. NOX4 downregulation leads to senescence of human vascular smooth muscle cells. Oncotarget. 2016;7(41):66429–66443. doi: 10.18632/oncotarget.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beatty S., Koh H., Phil M., Henson D., Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Survey of Ophthalmology. 2000;45(2):115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 62.Jurk D., Wang C., Miwa S., et al. Postmitotic neurons develop a p21-dependent senescence-like phenotype driven by a DNA damage response. Aging Cell. 2012;11(6):996–1004. doi: 10.1111/j.1474-9726.2012.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piechota M., Sunderland P., Wysocka A., et al. Is senescence-associated β-galactosidase a marker of neuronal senescence? Oncotarget. 2016;7(49):81099–81109. doi: 10.18632/oncotarget.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arend N., Wertheimer C., Laubichler P., Wolf A., Kampik A., Kernt M. Idebenone prevents oxidative stress, cell death and senescence of retinal pigment epithelium cells by stabilizing BAX/Bcl-2 ratio. Ophthalmologica. 2015;234(2):73–82. doi: 10.1159/000381726. [DOI] [PubMed] [Google Scholar]
- 65.Aryan N., Betts-Obregon B. S., Perry G., Tsin A. T. Oxidative stress induces senescence in cultured RPE cells. The Open Neurology Journal. 2016;10:83–87. doi: 10.2174/1874205X01610010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Supanji, Shimomachi M., Hasan M. Z., Kawaichi M., Oka C. HtrA1 is induced by oxidative stress and enhances cell senescence through p38 MAPK pathway. Experimental Eye Research. 2013;112:79–92. doi: 10.1016/j.exer.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 67.Zhuge C. C., Xu J. Y., Zhang J., et al. Fullerenol protects retinal pigment epithelial cells from oxidative stress-induced premature senescence via activating SIRT1. Investigative Ophthalmology & Visual Science. 2014;55(7):4628–4638. doi: 10.1167/iovs.13-13732. [DOI] [PubMed] [Google Scholar]
- 68.Glotin A. L., Debacq-Chainiaux F., Brossas J. Y., et al. Prematurely senescent ARPE-19 cells display features of age-related macular degeneration. Free Radical Biology & Medicine. 2008;44(7):1348–1361. doi: 10.1016/j.freeradbiomed.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 69.Marazita M. C., Dugour A., Marquioni-Ramella M. D., Figueroa J. M., Suburo A. M. Oxidative stress-induced premature senescence dysregulates VEGF and CFH expression in retinal pigment epithelial cells: implications for age-related macular degeneration. Redox Biology. 2016;7:78–87. doi: 10.1016/j.redox.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grabowska W., Sikora E., Bielak-Zmijewska A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology. 2017;18(4):447–476. doi: 10.1007/s10522-017-9685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alster O., Bielak-Zmijewska A., Mosieniak G., et al. The role of nibrin in doxorubicin-induced apoptosis and cell senescence in Nijmegen breakage syndrome patients lymphocytes. PLoS One. 2014;9(8, article e104964) doi: 10.1371/journal.pone.0104964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pospelova T. V., Demidenko Z. N., Bukreeva E. I., Pospelov V. A., Gudkov A. V., Blagosklonny M. V. Pseudo-DNA damage response in senescent cells. Cell Cycle. 2009;8(24):4112–4118. doi: 10.4161/cc.8.24.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 74.Gallagher L. E., Williamson L. E., Chan E. Y. Advances in autophagy regulatory mechanisms. Cell. 2016;5(2) doi: 10.3390/cells5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li L. C., Wang D. L., Wu Y. Z., et al. Gastric tumor-initiating CD44+ cells and epithelial-mesenchymal transition are inhibited by γ-secretase inhibitor DAPT. Oncology Letters. 2015;10(5):3293–3299. doi: 10.3892/ol.2015.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang Y. C., Hsieh M. C., Wu H. J., Wu W. C., Kao Y. H. Methylglyoxal, a reactive glucose metabolite, enhances autophagy flux and suppresses proliferation of human retinal pigment epithelial ARPE-19 cells. Toxicology In Vitro. 2015;29(7):1358–1368. doi: 10.1016/j.tiv.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 78.Mitter S. K., Song C., Qi X., et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. 2014;10(11):1989–2005. doi: 10.4161/auto.36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Y., Zhao K. K., Tong Y., et al. Exogenous NAD+ decreases oxidative stress and protects H2O2-treated RPE cells against necrotic death through the up-regulation of autophagy. Scientific Reports. 2016;6, article 26322 doi: 10.1038/srep26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Algvere P. V., Kvanta A., Seregard S. Drusen maculopathy: a risk factor for visual deterioration. Acta Ophthalmologica. 2016;94(5):427–433. doi: 10.1111/aos.13011. [DOI] [PubMed] [Google Scholar]
- 81.Handa J. T., Cano M., Wang L., Datta S., Liu T. Lipids, oxidized lipids, oxidation-specific epitopes, and age-related macular degeneration. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2017;1862(4):430–440. doi: 10.1016/j.bbalip.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaarniranta K., Sinha D., Blasiak J., et al. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy. 2013;9(7):973–984. doi: 10.4161/auto.24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rodriguez-Muela N., Koga H., Garcia-Ledo L., et al. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell. 2013;12(3):478–488. doi: 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johansson I., Monsen V. T., Pettersen K., et al. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy. 2015;11(9):1636–1651. doi: 10.1080/15548627.2015.1061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L., Cano M., Handa J. T. p62 provides dual cytoprotection against oxidative stress in the retinal pigment epithelium. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843(7):1248–1258. doi: 10.1016/j.bbamcr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eliopoulos A. G., Havaki S., Gorgoulis V. G. DNA damage response and autophagy: a meaningful partnership. Frontiers in Genetics. 2016;7:p. 204. doi: 10.3389/fgene.2016.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hewitt G., Korolchuk V. I. Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends in Cell Biology. 2017;27(5):340–351. doi: 10.1016/j.tcb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 88.Sample A., He Y. Y. Autophagy in UV damage response. Photochemistry and Photobiology. 2016;93(4):943–955. doi: 10.1111/php.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blasiak J., Glowacki S., Kauppinen A., Kaarniranta K. Mitochondrial and nuclear DNA damage and repair in age-related macular degeneration. International Journal of Molecular Sciences. 2013;14(2):2996–3010. doi: 10.3390/ijms14022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jarrett S. G., Lin H., Godley B. F., Boulton M. E. Mitochondrial DNA damage and its potential role in retinal degeneration. Progress in Retinal and Eye Research. 2008;27(6):596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 91.Jarrett S. G., Rohrer B., Perron N. R., Beeson C., Boulton M. E. Assessment of mitochondrial damage in retinal cells and tissues using quantitative polymerase chain reaction for mitochondrial DNA damage and extracellular flux assay for mitochondrial respiration activity. Methods in Molecular Biology. 2013;935:227–243. doi: 10.1007/978-1-62703-080-9_16. [DOI] [PubMed] [Google Scholar]
- 92.Testa U., Labbaye C., Castelli G., Pelosi E. Oxidative stress and hypoxia in normal and leukemic stem cells. Experimental Hematology. 2016;44(7):540–560. doi: 10.1016/j.exphem.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 93.Synowiec E., Blasiak J., Zaras M., Szaflik J., Szaflik J. P. Association between polymorphisms of the DNA base excision repair genes MUTYH and hOGG1 and age-related macular degeneration. Experimental Eye Research. 2012;98:58–66. doi: 10.1016/j.exer.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 94.Liang F. Q., Godley B. F. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Experimental Eye Research. 2003;76(4):397–403. doi: 10.1016/s0014-4835(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 95.Lin H., Xu H., Liang F. Q., et al. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Investigative Ophthalmology & Visual Science. 2011;52(6):3521–3529. doi: 10.1167/iovs.10-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang A. L., Lukas T. J., Yuan M., Neufeld A. H. Increased mitochondrial DNA damage and down-regulation of DNA repair enzymes in aged rodent retinal pigment epithelium and choroid. Molecular Vision. 2008;14:644–651. [PMC free article] [PubMed] [Google Scholar]
- 97.Ballinger S. W., Van Houten B., Jin G. F., Conklin C. A., Godley B. F. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Experimental Eye Research. 1999;68(6):765–772. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- 98.Ferrington D. A., Kapphahn R. J., Leary M. M., et al. Increased retinal mtDNA damage in the CFH variant associated with age-related macular degeneration. Experimental Eye Research. 2016;145:269–277. doi: 10.1016/j.exer.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gramajo A. L., Zacharias L. C., Neekhra A., et al. Mitochondrial DNA damage induced by 7-ketocholesterol in human retinal pigment epithelial cells in vitro. Investigative Ophthalmology & Visual Science. 2010;51(2):1164–1170. doi: 10.1167/iovs.09-3443. [DOI] [PubMed] [Google Scholar]
- 100.Jarrett S. G., Boulton M. E. Poly(ADP-ribose) polymerase offers protection against oxidative and alkylation damage to the nuclear and mitochondrial genomes of the retinal pigment epithelium. Ophthalmic Research. 2007;39(4):213–223. doi: 10.1159/000104683. [DOI] [PubMed] [Google Scholar]
- 101.Jarrett S. G., Boulton M. E. Consequences of oxidative stress in age-related macular degeneration. Molecular Aspects of Medicine. 2012;33(4):399–417. doi: 10.1016/j.mam.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kenney M. C., Atilano S. R., Boyer D., et al. Characterization of retinal and blood mitochondrial DNA from age-related macular degeneration patients. Investigative Ophthalmology & Visual Science. 2010;51(8):4289–4297. doi: 10.1167/iovs.09-4778. [DOI] [PubMed] [Google Scholar]
- 103.Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death and Differentiation. 2015;22(3):377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Komatsu M., Kurokawa H., Waguri S., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biology. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 105.Kurihara Y., Kanki T., Aoki Y., et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. The Journal of Biological Chemistry. 2012;287(5):3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma S., Helchowski C. M., Canman C. E. The roles of DNA polymerase zeta and the Y family DNA polymerases in promoting or preventing genome instability. Mutation Research. 2013;743-744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Filomeni G., Desideri E., Cardaci S., Rotilio G., Ciriolo M. R. Under the ROS...thiol network is the principal suspect for autophagy commitment. Autophagy. 2010;6(7):999–1005. doi: 10.4161/auto.6.7.12754. [DOI] [PubMed] [Google Scholar]
- 108.Chen Y., Azad M. B., Gibson S. B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death and Differentiation. 2009;16(7):1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 109.Scherz-Shouval R., Shvets E., Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3(4):371–373. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 110.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. The EMBO Journal. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang C., Yang L., Wang X. B., et al. Calyxin Y induces hydrogen peroxide-dependent autophagy and apoptosis via JNK activation in human non-small cell lung cancer NCI-H460 cells. Cancer Letters. 2013;340(1):51–62. doi: 10.1016/j.canlet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 112.Azad M. B., Chen Y., Gibson S. B. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxidants & Redox Signaling. 2009;11(4):777–790. doi: 10.1089/ARS.2008.2270. [DOI] [PubMed] [Google Scholar]
- 113.Levonen A. L., Hill B. G., Kansanen E., Zhang J., Darley-Usmar V. M. Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics. Free Radical Biology & Medicine. 2014;71:196–207. doi: 10.1016/j.freeradbiomed.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scherz-Shouval R., Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends in Cell Biology. 2007;17(9):422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 115.Mathew R., Karp C. M., Beaudoin B., et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mathew R., Kongara S., Beaudoin B., et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes & Development. 2007;21(11):1367–1381. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Munoz-Gamez J. A., Rodriguez-Vargas J. M., Quiles-Perez R., et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy. 2009;5(1):61–74. doi: 10.4161/auto.5.1.7272. [DOI] [PubMed] [Google Scholar]
- 118.Alexander A., Cai S. L., Kim J., et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kenzelmann Broz D., Spano Mello S., Bieging K. T., et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes & Development. 2013;27(9):1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maiuri M. C., Galluzzi L., Morselli E., Kepp O., Malik S. A., Kroemer G. Autophagy regulation by p53. Current Opinion in Cell Biology. 2010;22(2):181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 121.Madeo F., Tavernarakis N., Kroemer G. Can autophagy promote longevity? Nature Cell Biology. 2010;12(9):842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 122.Rubinsztein D. C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 123.Mosieniak G., Sliwinska M. A., Alster O., et al. Polyploidy formation in doxorubicin-treated cancer cells can favor escape from senescence. Neoplasia. 2015;17(12):882–893. doi: 10.1016/j.neo.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Young A. R., Narita M., Ferreira M., et al. Autophagy mediates the mitotic senescence transition. Genes & Development. 2009;23(7):798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tokarz P., Kaarniranta K., Blasiak J. Inhibition of DNA methyltransferase or histone deacetylase protects retinal pigment epithelial cells from DNA damage induced by oxidative stress by the stimulation of antioxidant enzymes. European Journal of Pharmacology. 2016;776:167–175. doi: 10.1016/j.ejphar.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 126.Roggia M. F., Ueta T. αvβ5 integrin/FAK/PGC-1α pathway confers protective effects on retinal pigment epithelium. PLoS One. 2015;10(8, article e0134870) doi: 10.1371/journal.pone.0134870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsunemi T., Ashe T. D., Morrison B. E., et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Science Translational Medicine. 2012;4(142, article 142ra97) doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen Y., Wang J., Cai J., Sternberg P. Altered mTOR signaling in senescent retinal pigment epithelium. Investigative Ophthalmology & Visual Science. 2010;51(10):5314–5319. doi: 10.1167/iovs.10-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu B., Xu P., Zhao Z., Cai J., Sternberg P., Chen Y. Subcellular distribution and activity of mechanistic target of rapamycin in aged retinal pigment epithelium. Investigative Ophthalmology & Visual Science. 2014;55(12):8638–8650. doi: 10.1167/iovs.14-14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kang C., Elledge S. J. How autophagy both activates and inhibits cellular senescence. Autophagy. 2016;12(5):898–899. doi: 10.1080/15548627.2015.1121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Coppe J. P., Desprez P. Y., Krtolica A., Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual Review of Pathology. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Campisi J., d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature Reviews Molecular Cell Biology. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 133.Jeyapalan J. C., Ferreira M., Sedivy J. M., Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mechanisms of Ageing and Development. 2007;128(1):36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Al-Hussaini H., Kam J. H., Vugler A., Semo M., Jeffery G. Mature retinal pigment epithelium cells are retained in the cell cycle and proliferate in vivo. Molecular Vision. 2008;14:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 136.Xia H., Krebs M. P., Kaushal S., Scott E. W. Enhanced retinal pigment epithelium regeneration after injury in MRL/MpJ mice. Experimental Eye Research. 2011;93(6):862–872. doi: 10.1016/j.exer.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Franceschi C., Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2014;69(Supplement 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 138.Khansari N., Shakiba Y., Mahmoudi M. Chronic inflammation and oxidative stress as a major cause of age-related diseases and cancer. Recent Patents on Inflammation & Allergy Drug Discovery. 2009;3(1):73–80. doi: 10.2174/187221309787158371. [DOI] [PubMed] [Google Scholar]
- 139.Hyttinen J. M., Blasiak J., Niittykoski M., et al. DNA damage response and autophagy in the degeneration of retinal pigment epithelial cells-implications for age-related macular degeneration (AMD) Ageing Research Reviews. 2017;36:64–77. doi: 10.1016/j.arr.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 140.Van Houten B., Hunter S. E., Meyer J. N. Mitochondrial DNA damage induced autophagy, cell death, and disease. Frontiers in Bioscience. 2016;21:42–54. doi: 10.2741/4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Labbe K., Murley A., Nunnari J. Determinants and functions of mitochondrial behavior. Annual Review of Cell and Developmental Biology. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 142.Roczniak-Ferguson A., Petit C. S., Froehlich F., et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science Signaling. 2012;5(228, article ra42) doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhu Y., Armstrong J. L., Tchkonia T., Kirkland J. L. Cellular senescence and the senescent secretory phenotype in age-related chronic diseases. Current Opinion in Clinical Nutrition and Metabolic Care. 2014;17(4):324–328. doi: 10.1097/MCO.0000000000000065. [DOI] [PubMed] [Google Scholar]
- 144.Gewirtz D. A. Autophagy and senescence: a partnership in search of definition. Autophagy. 2013;9(5):808–812. doi: 10.4161/auto.23922. [DOI] [PMC free article] [PubMed] [Google Scholar]